Abstract
Vibrio cholerae strains of the classical biotype express the genes encoding cholera toxin (CT) and toxin-coregulated pilus (TCP) under a variety of environmental conditions in vitro, whereas El Tor biotype strains express these genes only under specialized culture conditions. We show here that a single base-pair difference at positions −65 and −66 of the classical and El Tor tcpPH promoters, respectively, is responsible for the differential regulation of virulence gene expression in these two disease-causing biotypes. Analysis of tcpP-lacZ fusions in both V. cholerae and Escherichia coli indicated that transcriptional activation of the El Tor tcpPH promoter by the LysR regulator AphB was significantly reduced relative to that of the classical promoter. Reciprocal exchange of the tcpPH promoter between the two biotypes in V. cholerae showed that the ability to activate the transcription of tcpPH is not dependent on the biotype of the strain per se but on the tcpPH promoter itself. Classical and El Tor tcpP-lacZ promoter chimeras in E. coli localized the region responsible for the differential activation of tcpPH by AphB to within 75 bp of the transcriptional start site. Individual base-pair changes within this region showed that the presence of either an A or a G at position −65 or −66 conferred the classical or El Tor, respectively, pattern of tcpPH activation by AphB. Reciprocal exchange of these base pairs between biotypes in V. cholerae switched the biotype-specific pattern of expression of tcpPH as well as the production of CT and TCP in response to environmental stimuli.
Two serotypes of Vibrio cholerae, O1 and O139, are capable of causing the life-threatening epidemic disease cholera. These strains have acquired a large pathogenicity island, TCP-ACF (20) or VPI, for Vibrio pathogenicity island (18), which has recently been reported to be the genome of a filamentous phage, VPIφ (19). Carried on this element are a number of genes required for the expression and production of the primary colonization factor of V. cholerae, the toxin-coregulated pilus (TCP) (35). Disease-causing strains of V. cholerae have also acquired the genome of a second filamentous phage, CTXφ (37). Within this element are the genes encoding the subunits of cholera toxin (CT), which is responsible for the profuse diarrhea associated with the disease. Strains capable of elaborating TCP apparently acquired CTXφ by virtue of the fact that TCP serves as the receptor for the phage (37).
The expression of the genes encoding TCP and CT is positively activated by the AraC regulator ToxT (4, 9, 15). ToxT is itself encoded within the TCP-ACF pathogenicity element, and its expression is dependent upon two transcriptional activator pairs, ToxRS and TcpPH, as well as by stimuli from the environment (7, 13). ToxR and TcpP are homologous transmembrane DNA binding proteins which cooperate to activate toxT transcription (13, 26). The abilities of ToxR and TcpP to activate transcription are enhanced by the accessory transmembrane proteins ToxS and TcpH, respectively (7, 13, 24). The toxR and toxS genes, which are expressed as an operon, are not carried on either the TCP-ACF or the CTX element and have other important regulatory roles in V. cholerae (6, 25). The tcpPH operon is located within the TCP-ACF element, immediately upstream of the gene encoding the major pilin subunit, tcpA (28). TcpP and TcpH have no additional known roles in V. cholerae other than their involvement in toxT transcription. The toxT gene is located within the tcpA operon, and its expression is influenced by ToxRS and TcpPH at a promoter located immediately upstream of the gene (14) as well as by a promoter upstream of tcpA which appears to function in an autoregulatory capacity (1, 38).
The expression of the tcpPH operon is itself under the control of two regulatory proteins, AphA and AphB, which function synergistically to activate transcription (21, 33). AphB is a member of the LysR family of transcriptional regulators, and AphA presently has no known homologs. Neither aphA nor aphB is carried on the TCP-ACF or CTX elements and, like toxR and toxS, these genes presumably have other regulatory roles in V. cholerae.
The expression of tcp, ctx, toxT, and tcpPH is influenced by environmental stimuli, such as pH, temperature, and osmolarity (3, 9, 25, 35). The mechanisms involved in this environmental regulation are different between the two disease-causing biotypes of V. cholerae O1, classical and El Tor. Classical biotype strains typically exhibit maximal expression of these virulence genes in vitro in Luria-Bertani (LB) medium (pH 6.5) at 30°C, whereas El Tor biotype strains require a bicarbonate-containing medium (AKI medium) at 37°C for high-level expression (16). This differential regulation of virulence genes between the two biotypes may account for why infections with classical strains are generally more severe than those with El Tor biotype strains (17).
The overproduction of AphB in El Tor biotype strains has been shown to increase the expression of tcpPH and tcpA to close to classical levels in LB medium (pH 6.5) at 30°C (21). Since the expression of aphB is similar in both biotypes (21), these results suggested that some aspect of AphB function was involved in the differential regulation of virulence genes between the biotypes. We show here that the molecular basis for the biotype specificity of virulence gene regulation is a single base-pair difference 65 (classical) or 66 (El Tor) bp upstream from the tcpPH transcriptional start site, which is critical for activation by AphB.
MATERIALS AND METHODS
Bacterial strains and growth.
The V. cholerae and Escherichia coli strains and plasmids analyzed in this study are listed in Table 1. Strains were maintained at −70°C in LB medium (23) containing 30% (vol/vol) glycerol. Antibiotics were used at the following concentrations in LB medium: ampicillin, 100 μg/ml; kanamycin, 45 μg/ml; polymyxin B, 50 IU/ml; and streptomycin, 1 mg/ml. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was used in LB agar at 40 μg/ml.
TABLE 1.
Bacterial strains and expression plasmids
| Strain or plasmid | Relevant genotype | Reference or source |
|---|---|---|
| Strains | ||
| V. cholerae | ||
| KSK618 | CG842 tcpP-lacZ (classical Ogawa Smr) | 33 |
| KSK805 | KSK618 ΔaphA1 ΔaphB1 | 21 |
| KSK725 | KSK262 tcpP-lacZ (El Tor Inaba Smr) | 21 |
| GK184 | KSK725 ΔaphA1 ΔaphB1 | This work |
| GK250 | KSK725 ΔtcpPH promoter | This work |
| GK300 | KSK618 ΔtcpPH promoter | This work |
| GK318 | GK300 with classical tcpPH promoter (C/C control) | This work |
| GK319 | GK300 with El Tor tcpPH promoter (C/E hybrid) | This work |
| GK321 | GK250 with classical tcpPH promoter (E/C hybrid) | This work |
| GK323 | GK250 with El Tor tcpPH promoter (E/E control) | This work |
| GK404 | GK300 with classical tcpPH promoter A→G change at position −65 | This work |
| GK436 | GK250 with El Tor tcpPH promoter G→A change at position −66 | This work |
| KSK218 | CG842 ctx-lacZ Smr Cmr | 32 |
| KSK1019 | KSK218 with TAA codon in tcpP coding region | This work |
| O395 | Classical Ogawa Smr | Laboratory collection |
| KSK1093 | O395 tcpPH promoter A→G change at position −65 | This work |
| C6706 str2 | El Tor Inaba Smr | Laboratory collection |
| KSK1117 | C6706 str2 tcpPH promoter G→A change at position −66 | This work |
| E. coli | ||
| MC1061 | Δ(ara-leu)7697 Δ(lac)X74 | Laboratory collection |
| KSK782 | MC1061 λKSPL1 (tcpP-lacZ wild-type classical) | 21 |
| KSK864 | MC1061 λKSPL2 (tcpP-lacZ wild-type El Tor) | This work |
| GK334 | MC1061 λGK91 (tcpP-lacZ C4/C3 control) | This work |
| GK335 | MC1061 λGK92 (tcpP-lacZ C4/E3 chimera) | This work |
| GK336 | MC1061 λGK93 (tcpP-lacZ E4/C3 chimera) | This work |
| GK337 | MC1061 λGK94 (tcpP-lacZ E4/E3 control) | This work |
| GK370 | MC1061 λGK101 (tcpP-lacZ classical A→G change at position −65) | This work |
| GK372 | MC1061 λGK102 (tcpP-lacZ classical G→T change at position −7) | This work |
| GK407 | MC1061 λGK109 (tcpP-lacZ El Tor G→A −66) | This work |
| GK409 | MC1061 λGK111 (tcpP-lacZ classical C→A change at position −13) | This work |
| GK411 | MC1061 λGK112 (tcpP-lacZ classical A addition at position −19) | This work |
| GK441 | MC1061 λGK117 (tcpP-lacZ class A→C change at position −65) | This work |
| GK442 | MC1061 λGK118 (tcpP-lacZ El Tor G→C change at position −66) | This work |
| GK443 | MC1061 λGK119 (tcpP-lacZ classical A→T change at position −65) | This work |
| GK444 | MC1061 λGK120 (tcpP-lacZ El Tor G→T change at position −66) | This work |
| Expression plasmids | ||
| pKAS107 | pMMB66EH aphA (classical) Apr | 33 |
| pKAS117 | pMMB66EH aphB (classical) Apr | 21 |
| pKAS121 | pBAD-TOPO aphB (classical) Kanr | This work |
| pKAS126 | pBAD-TOPO aphA (classical) Kanr | This work |
Identification of the tcpPH transcriptional start site.
El Tor strain GK184 was generated by introducing the ΔaphA1 deletion from pGKK35 into GK138 (KSK725 ΔaphB1) (21). Total RNA was isolated from GK184 and KSK805 carrying both pKAS107 and pKAS121 after growth for 4 h in LB medium (pH 6.5) at 30°C in the presence of 0.2% arabinose and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). RNA was purified on an RNeasy column (Qiagen) and subjected to 5′ rapid amplification of cDNA ends (RACE) (Gibco BRL). Briefly, first-strand cDNA synthesis was carried out with 1 μg of RNA, the tcpP-specific primer TP-Bam (33), and reverse transcriptase. The cDNA was purified on a PCR purification column (Qiagen), and a poly(dC) tail was added to the 3′ end by use of terminal deoxynucleotidyl transferase. PCR of the cDNA was carried out initially using a 5′ RACE-abridged anchor primer (5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG) and the tcpP nested primer TP-Kpn (5′-GATCGGGTACCCCGGCTAATTCATGTTGATACC). PCR was carried out subsequently with a 5′ RACE-abridged universal amplification primer (5′-GGCCACGCGTCGACTAGTAC) and a second tcpP nested primer, TP-Kpn2 (5′-GATCGGGTACCTCCACCAAATCACAGGTAGC). The resulting DNA products were sequenced using the ABI PRISM Dye System (Perkin-Elmer).
Introduction of a termination codon into the TcpP coding region.
The termination codon (TAA) was introduced into the TcpP coding region 24 bp downstream of the first potential ATG by converting a C to a T and simultaneously creating a restriction site for AseI (see Fig. 2). Two overlapping PCR products containing the desired change (underlined) were amplified from classical biotype DNA using primer pair I-Eco2 (5′-GATCGGAATTCTCTAGAGTACCAATATCTGTAAAC) and P-Ase2 (5′-GATCGCTCTTCGAATAAATCACGCGGACATACC) and primer pair P-Ase1 (5′-GATCGCTCTTCGATTAATTTCCCGATAACCTTTGG) and TP-Kpn. Sites for the restriction enzyme EarI, which cuts outside of its recognition sequence, were incorporated into the P-Ase primers so as to create a seamless junction with no unnatural base pairs (29) upon ligation of the two PCR products into pKAS46 (31). The resulting plasmid, pKAS148, was sequenced, and the mutation was introduced into V. cholerae by allelic exchange.
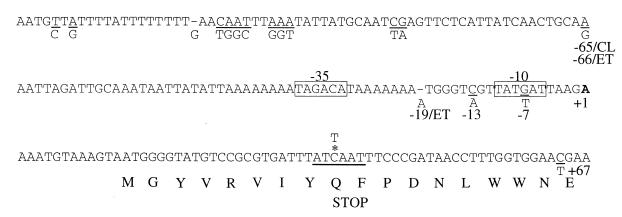
FIG. 2.
Nucleotide sequence of the tcpPH promoter. The El Tor residues which differ from the classical residues are shown below the corresponding (underlined) residue. The start of AphB-dependent transcription is indicated by the bold A at +1. Where certain positions differ between the two biotypes relative to the start of transcription, the designations CL for classical and ET for El Tor are used. Boxes delineate putative −35 and −10 promoter consensus sequences. The asterisk denotes the C which was changed to a T to create the stop codon; the thick underlining shows the resulting AseI restriction site.
Construction of V. cholerae tcpPH promoter replacements.
The classical and El Tor tcpPH promoter deletion plasmids, pGKK79 and pGKK63, respectively, each contain two DNA fragments of approximately 500 bp which flank the tcpPH promoter region (see Fig. 3A) in a derivative of pKAS46 (31), pKAS125, lacking the unique ScaI site in the bla gene. The upstream fragment, amplified with primer pair I-Eco (5′-GATCGGAATTCATAGTGAGAACGTGTTGCCC) and I-ScaNot (5′-GATCGGCGGCCGCTTATCACGAAGTACTCCGTG), extends from the ScaI site 34 bp from the initiation codon of the divergently transcribed tcpI gene into the tcpI coding region. The downstream fragment, amplified with primer pair TP-Not1 (5′-GATCGGCGGCCGCTTTCCCGATAACCTTTGGTGG) and TP-Kpn, lies within the tcpP coding sequence (from +40 relative to the start site of tcpPH transcription). Introduction of these plasmids into the classical and El Tor tcpP-lacZ fusion strains KSK618 and KSK725, respectively, generated the ΔtcpPH promoter strains GK300 and GK250, respectively. The tcpPH promoter replacement plasmids were constructed by reamplifying the downstream tcpP fragments using a primer containing an EarI site, TP-EarI (5′-GATCGCTCTTCGATCAATTTCCCGATAACCTTTG), and TP-Kpn. Each of these fragments was ligated with the appropriate promoter region amplified from either classical or El Tor strains using primer pair TP-Ear2 (5′-GATCGCTCTTCGGATAAATCACGCGGACATACC) and I-Sca (5′-TTATCACGGAGTACTTCGTG), into the plasmids carrying the upstream tcpI fragments. Allelic exchange was carried out with the resulting plasmids, pGKK83, pGKK84, pGKK85, and pGKK86, in the classical and El Tor ΔtcpPH promoter strains GK300 and GK250. The tcpPH promoter regions of the resulting strains were sequenced.
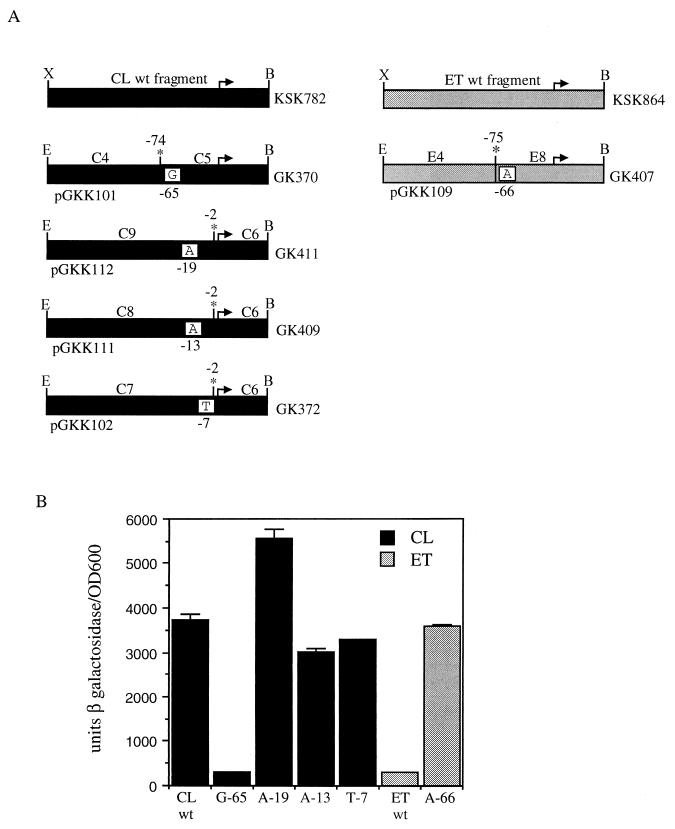
FIG. 3.
Reciprocal exchange of the classical and El Tor tcpPH promoter regions in V. cholerae. (A) Promoter deletion and replacement plasmids and strains. The divergently transcribed coding regions of tcpI and tcpP are indicated by the large black arrows at the top, and the intergenic region is shown by the solid line. The small arrow denotes the start of the tcpPH message. Striped black-white and gray-black boxes represent the regions of tcpI and tcpP from classical and El Tor, respectively, present in the plasmids (indicated at the left), and dotted lines represent intervening DNA which was deleted. Solid black and gray boxes represent classical and El Tor promoter regions, respectively. The strains generated by allelic exchange from the plasmids are indicated to the right. CL, classical; ET, El Tor; C/C, classical with classical promoter; C/E, classical with El Tor promoter; E/E, El Tor with El Tor promoter; E/C, El Tor with classical promoter. E, EcoRI; S, ScaI; N, NotI; K, KpnI. The asterisk shows the position of the seamless junction created using EarI. (B) Expression of tcpP-lacZ. Cultures were grown in LB medium (pH 6.5) at 30°C (left) and in AKI medium at 37°C (right). Black bars, strains with classical (CL) promoter regions: KSK618 (wild type [WT]); GK318 (C/C control); GK321 (E/C hybrid). Gray bars, strains with El Tor (ET) promoter regions: KSK725 (wild type); GK323 (E/E control); GK319 (C/E hybrid). Error bars show standard deviations.
Construction of E. coli tcpP-lacZ fusion strains.
The E. coli strain carrying the El Tor tcpP promoter-lacZ fusion, KSK864, was constructed in a manner similar to that described previously for the classical tcpP promoter-lacZ fusion strain KSK782 (21). Briefly, a 1.3-kb fragment encompassing a region from tcpI to tcpP was amplified from El Tor DNA using primer pair I-Xba (33) and TP-BamE (21). This fragment was ligated into the lacZ operon fusion vector pRS415 to generate pGKK41. The resulting fusion was recombined onto λRS45, generating λKSPL2, and then integrated into the chromosome of E. coli MC1061. To construct the classical and El Tor tcpP promoter-lacZ chimera constructs, the 1.3-kb fragments present in the wild-type fusions were PCR amplified from either biotype as two 650-bp fragments joined at positions −74 and −75, respectively, relative to the start of tcpPH transcription using the restriction enzyme EarI (see Fig 4A). The classical C4 and C3 fragments were amplified with primer pair EarC4 (5′-GATCGCTCTTCGTGATAATGAGAACTCGATTG) and I-Eco2 and with primer pair EarC3 (5′-GATCGCTCTTCGTCAACTGCAAAATTAGATTG) and TP-Bam, respectively. The El Tor E4 and E3 fragments were amplified with primer pair EarE4 (5′-GATCGCTCTTCGTGATAATGAGAACTTAATTGC) and I-Eco2 and with primer pair EarE3 (5′-GATCGCTCTTCGTCAACTGCAGAATTAGATTG) and TP-BamE, respectively. The resulting fragments were ligated into pRS415, generating pGKK91, pGKK92, pGKK93, and pGKK94, and introduced into E. coli MC1061. The tcpPH promoter regions of the resulting strains were sequenced.
FIG. 4.
AphB-dependent expression of classical and El Tor biotype tcpP-lacZ promoter chimeras in E. coli. (A) PCR-generated V. cholerae DNA fragments cloned in pRS415. Black and gray boxes represent classical (CL) and El Tor (ET) DNAs, respectively. Arrows denote the start and direction of transcription. The positions where the two fragments were joined after EarI digestion are shown by the asterisk. E. coli lysogens generated from the various constructs are indicated to the right. wt, wild type; X, XbaI; B, BamHI; E, EcoRI. (B) Expression of tcpP-lacZ. Strains containing pKAS117 (AphB) were grown in LB medium (pH 7.0) at 37°C and induced with 1 mM IPTG. Black bars, strains with the classical region from positions −74 to +1: KSK782 (wild type [wt]); GK334 (C4/C3 control); GK336 (E4/C3 chimera). Gray bars, strains with the El Tor region from positions −75 to +1: KSK864 (wild type); GK337 (E4/E3 control); GK335 (C4/E3 chimera). Error bars show standard deviations.
Introduction of single base-pair changes into E. coli tcpP-lacZ fusions.
Single base pairs within the classical or El Tor tcpPH promoters were converted to those of the other biotype by incorporating the changes into primers (underlined in sequences shown below) containing a restriction site for EarI and performing a seamless cloning strategy similar to that described above (see Fig. 5A). Conversion of classical A at position −65 to G was carried out by amplification with primer pair EarC5 (5′-GATCGCTCTTCGTCAACTGCAGAATTAGATTGC) and TP-Bam. The resulting C5 fragment was ligated with the classical C4 fragment from above at position −74 to generate pGKK101. The classical C9 fragment containing the addition of A at position −19 was generated with primer pair EarC9 (5′-GATCGCTCTTCGTCTTAATCATAACGACCCATTTTTTTTATG) and I-Eco2. The classical C8 fragment containing the C-to-A change at position −13 was generated with EarC8 (5′-GATCGCTCTTCGTCTTAATCATAACTACCCATTT) and I-Eco2. The classical C7 fragment containing the G-to-T change at position −7 was generated with EarC7 (5′-GATCGCTCTTCGTCTTAATAATAACGACCCATT) and I-Eco2. Each of these fragments was ligated with a classical C6 fragment generated with primer pair EarC6 (5′-GATCGCTCTTCGAGAAAATGTAAAGTAATGGGG) and TP-Bam at position −2 to obtain plasmids pGKK112, pGKK111, and pGKK102, respectively.
FIG. 5.
AphB-dependent expression in E. coli tcpP-lacZ fusions containing single base-pair alterations. (A) See the legend to Fig. 4A. Single base-pair alterations are indicated in white boxes with the positions from the start of transcription shown below. (B) Expression of tcpP-lacZ. Strains containing pKAS117 (AphB) were grown as described in the legend to Fig. 4B. Black bars, strains with classical (CL) DNA: KSK782 (wild type [wt]); GK370 (A→G change at position −65); GK411 (addition of A at position −19); GK409 (C→A change at position −13); GK372 (G→T change at position −7). Gray bars, strains with El Tor (ET) DNA: KSK864 (wild type); GK407, (G→A change at position −66). Error bars show standard deviations.
Conversion of El Tor G at position −66 to A was carried out by amplification with primer pair EarC3 and TP-BamE. The resulting E8 fragment was ligated with the El Tor E4 fragment from above at position −75 to generate pGKK109. To introduce either T or C into positions −65 and −66 of the classical and El Tor promoters, respectively, classical C10 and C11 fragments, respectively, were generated with primer TP-Bam and primer EarC10 (5′-GATCGCTCTTCGTCAACTGCATAATTAGATTGC) or EarC11 (5′-GATCGCTCTTCGTCAACTGCACAATTAGATTGC). These fragments were each ligated with the classical C4 fragment from above at position −74 to generate pGKK119 and pGKK117, respectively. Similarly, El Tor E10 and E11 fragments were amplified with primer TP-BamE and EarC10 or EarC11. These fragments were each ligated with the El Tor E4 fragment from above at position −75 to generate pGKK120 and pGKK118, respectively. The tcpPH promoter regions of the resulting strains were sequenced.
Introduction of single base-pair changes into V. cholerae tcpPH.
The fragment of DNA containing the classical A→G change at position −65 was isolated from pGKK101 and inserted into pKAS32 (31), and that containing the El Tor G→A change at position −66 was isolated from pGKK109 and inserted into pKAS46 (31). The resulting plasmids, pGKK110 and pGKK121, respectively, were used for allelic exchange into classical and El Tor strains. The tcpPH promoter regions of the resulting strains were sequenced.
Expression plasmids.
A Kmr fragment was inserted into the aphB and aphA expression plasmids pKAS118 and pKAS119 (21) to generate pKAS121 and pKAS126, respectively (Table 1).
β-Galactosidase assays.
β-Galactosidase assays (23) with V. cholerae tcpP-lacZ fusions were carried out after growth for 4 h in either LB medium (pH 6.5) at 30°C with rotation or in AKI medium at 37°C without rotation. Overnight cultures of E. coli tcpP-lacZ fusions in LB medium (pH 7.0) at 37°C were diluted 1:100 into fresh medium, induced after 2 to 3 h with arabinose and/or IPTG, and assayed after 4 h.
CT assays.
GM1 ganglioside enzyme-linked immunosorbent CT assays (12) were carried out after overnight growth in LB medium (pH 6.5) at 30°C.
Immunoblot analysis.
Cell extracts prepared from the overnight cultures used for CT assays were subjected to sodium dodecyl sulfate–12.5% polyacrylamide gel electrophoresis, transferred to nitrocellulose, probed with anti-TcpA antibody (34), and visualized using the enhanced chemiluminescence detection system (Amersham).
RESULTS
Activation of the El Tor tcpPH promoter by AphB is significantly reduced relative to that of the classical promoter.
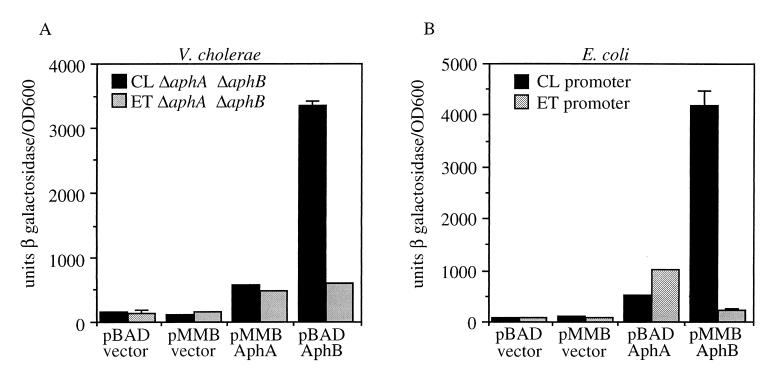
AphA and AphB are both required for transcriptional activation of the tcpPH promoter in the classical and El Tor biotypes of V. cholerae (21, 33). Strains of the El Tor biotype show reduced expression of the tcpPH operon relative to classical biotype strains in vitro (21, 27). The introduction of a plasmid expressing aphB into El Tor strains increased the expression from this operon to a level similar to that in classical strains (21). The finding that the expression of aphB was similar in both biotypes (21) suggested that some aspect of AphB function might be responsible for the reduced expression of tcpPH in El Tor biotype strains. To address whether AphA and AphB function differently at the classical and El Tor tcpPH promoters to activate transcription, an El Tor tcpP-lacZ ΔaphA ΔaphB double mutant, GK184, was constructed and compared with a classical tcpP-lacZ ΔaphA ΔaphB double mutant, KSK805, in the presence of plasmids expressing either AphA or AphB (Fig. 1A). Although AphA expressed from plasmid pKAS107 activated both biotype promoters more or less equally, approximately 3- to 5-fold, AphB from pKAS121 activated the classical promoter more than 20-fold, whereas it activated the El Tor promoter only 4-fold. These results indicate that the transcriptional activation of tcpPH by AphB is significantly reduced in El Tor biotype strains relative to classical strains.
FIG. 1.
Differential activation of classical and El Tor biotype tcpP-lacZ fusions by AphB. (A) V. cholerae cultures were grown in LB medium (pH 6.5) at 30°C. Those with pMMB66EH or pKAS107 (AphA) contained 1 mM IPTG, and those with pBAD or pKAS121 (AphB) contained 0.2% arabinose. Black bars, KSK805 (classical [CL]) ΔaphA1 ΔaphB1; gray bars, GK184 (El Tor [ET]) ΔaphA1 ΔaphB1. (B) E. coli cultures were grown in LB medium (pH 7.0) at 37°C. Those with pBAD or pKAS126 (AphA) were induced with 0.2% arabinose, and those with pMMB66EH or pKAS117 (AphB) were induced with 1 mM IPTG. Black bars, KSK782 (classical promoter); gray bars, KSK864 (El Tor promoter). Error bars show standard deviations.
To rule out possible biotype-specific effects on the activity of AphB or its production from pKAS121, a similar experiment was carried out with E. coli with classical (KSK782) and El Tor (KSK864) tcpP-lacZ fusions crossed onto λ lysogens (Fig. 1B). AphA from plasmid pKAS126 activated both fusions more or less similarly, between 6- and 12-fold, whereas AphB from pKAS117 activated the classical fusion 40-fold and the El Tor fusion only 2-fold. These results further indicate that the ability of AphB to activate the transcription of the El Tor tcpPH promoter is reduced compared to that of the classical promoter.
Determination of the AphB-dependent tcpPH transcriptional start site.
The start of AphB-dependent tcpPH transcription was determined using 5′ RACE (10). In this procedure, RNA was isolated from cultures of V. cholerae KSK805 (classical) and GK184 (El Tor) expressing both aphA and aphB from plasmids. Synthesis of cDNA was carried out using an antisense primer within the tcpP gene, and a homopolymeric tail was added to the resulting 3′ end. The cDNA was then amplified by PCR using tail-specific primers and nested tcpP primers. A prominent PCR product of approximately 500 bp was observed from both the classical and the El Tor strains; this size corresponded to the size of a product expected for a start site upstream of tcpP (data not shown). Direct sequencing of the DNA products revealed an identical start site for both the classical and the El Tor strains: an A residue located 13 bp upstream of the first potential tcpP translational initiation codon (Fig. 2). Examination of the 5′ upstream region revealed the presence of putative −35 (TAGACA) and −10 (TAT[G or T]AT) sequences (boxed in Fig. 2) separated by 16 bp in the classical promoter and 17 bp in the El Tor promoter. For both regions, five out of six positions matched the E. coli consensus sequence.
A potential ATG codon for TcpP translation initiation is located 13 bp downstream from the start site identified above. To rule out the possibility that a second ATG codon, located 85 bp downstream from the first, is actually used for TcpP initiation, a termination codon was introduced 24 bp downstream of the first ATG codon by converting a C to a T. This change converts a CAA codon to TAA and simultaneously creates a restriction site for AseI (Fig. 2). Since the introduction of this termination codon abolished tcpPH-mediated activation of virulence gene expression, as indicated by significantly reduced β-galactosidase production from a V. cholerae strain carrying a ctx-lacZ fusion, strain KSK1019 (data not shown), these results indicate that the second ATG codon is not used for TcpP initiation. It remains possible, however, that a GTG codon located 15 bp downstream of the first ATG codon is used for TcpP initiation. Neither of these potential start codons appears to have a consensus ribosome binding site.
Exchange of the tcpPH promoter between classical and El Tor biotypes.
The observation that AphB strongly activates the classical but not the El Tor tcpPH promoter in E. coli suggests that the biotype specificity of tcpPH expression in V. cholerae is the result of differences in the promoters rather than in the activities of AphB. A comparison of the nucleotide sequence of the region between the upstream divergently transcribed tcpI gene and tcpP in classical and El Tor biotypes reveals close to 50 base-pair differences. To determine if these differences influence biotype-specific expression of tcpPH in V. cholerae, the entire region between the tcpI and tcpP genes (from a ScaI site 34 bp downstream of the tcpI initiation codon into the tcpP gene at position +40) was exchanged between the two biotypes (Fig. 3A). To accomplish this, it was first necessary to delete this entire region from the chromosomes of both tcpP-lacZ strains by allelic exchange. The region from the other biotype was then introduced into each deletion strain. The various promoter deletion and replacement plasmids used for the strain constructions are shown in Fig. 3A. Each region that was deleted was also replaced with the region from the same biotype strain to ensure that wild-type expression specific for that biotype was restored.
The results of the tcpPH promoter exchange experiment are shown in Fig. 3B. In LB medium (pH 6.5) at 30°C, conditions optimal for tcpPH expression in the classical biotype, the wild-type classical fusion, KSK618, produced approximately 3,000 U of β-galactosidase, and the wild-type El Tor fusion, KSK725, produced about 400. Each of the control replacement strains, GK318 (classical with classical promoter) and GK323 (El Tor with El Tor promoter) for El Tor, showed levels of expression comparable to those of the corresponding wild-type fusion strains. Insertion of the El Tor tcpPH promoter into the classical biotype, strain GK319 (classical with El Tor promoter), decreased the expression of the fusion to a level close to that observed with the wild-type El Tor strain. Conversely, insertion of the classical tcpPH promoter into the El Tor biotype, strain GK321 (El Tor with classical promoter), increased expression to close to the wild-type classical strain level. A similar promoter-specific result was observed after growth of the strains under AKI medium conditions, which are known to induce tcpPH expression in El Tor strains (Fig. 3B). Under these conditions, however, the strains containing the El Tor promoters still displayed reduced tcpPH expression relative to those with the classical promoters. These results indicate that sequence differences in the respective promoter regions are important for the biotype specificity of tcpPH expression in V. cholerae.
Analysis of biotype tcpP-lacZ promoter chimeras in E. coli.
To discern which regions within the classical and El Tor tcpPH promoters are responsible for biotype-specific activation by AphB, classical and El Tor tcpP-lacZ promoter chimeras were constructed and analyzed in E. coli. To accomplish this, the 1.3-kb fragments present in the wild-type E. coli tcpP-lacZ fusions were PCR amplified from the classical and El Tor biotypes as two approximately 650-bp fragments using overlapping primers at positions −74 and −75, respectively, relative to the start site of tcpPH transcription. As shown in Fig. 2, these positions separated a promoter-proximal region containing four nucleotide differences from a distal region containing all of the remaining nucleotide differences. The two fragments from the different biotypes were then seamlessly joined together in pRS415 (Fig. 4A). Fragments from the same biotype were also joined to ensure that wild-type expression was restored. The resulting fusions were then introduced into the E. coli chromosome and analyzed in the presence of AphB. Figure 4B shows that the region downstream of position −74 or −75 determines the biotype specificity of tcpPH expression. When the upstream classical fragment was joined with the downstream El Tor fragment, resulting in strain GK335 (C4 fragment + E3 fragment), AphB-dependent expression was decreased to a level virtually identical to that seen with the wild-type El Tor fusion. Conversely, when the upstream El Tor fragment was joined with the downstream classical fragment, resulting in strain GK336 (E4 fragment + C3 fragment), expression was increased to a level similar to that seen with the wild-type classical fusion. These results suggest that either one or several of the four base-pair differences between classical and El Tor strains in the region from positions −74 or −75 to +1 of the tcpPH promoter (Fig. 2) are responsible for biotype-specific activation by AphB.
Introduction of single-base-pair changes into E. coli tcpP-lacZ fusions.
To elucidate which of the four base-pair differences in the region from positions −74 or −75 to +1 of the tcpPH promoter might be important for biotype-specific expression, the four El Tor residues, G at −66, A at −19, A at −13, and T at −7, were separately introduced in place of the corresponding classical residues (or lack thereof) in an E. coli tcpP-lacZ fusion, and the resulting strains were analyzed in the presence of AphB. The constructs containing these single base-pair changes (Fig. 5A) were made by a procedure similar to that used to create the chimeras shown in Fig. 4. Replacement of the classical A at position −65 with G, strain GK370, had a significant effect on AphB-dependent expression, reducing activation from 40- to 4-fold (Fig. 5B). This reduced level of expression was essentially identical to that observed with the wild-type El Tor fusion strain KSK864. The addition of an A at position −19, strain GK411, actually increased tcpPH expression somewhat. This result might be expected since the alteration increased the spacing between the −35 and −10 regions from 16 bp to the more optimal distance of 17 bp. Replacement of the classical C at position −13 with A and the G at position −7 with T had no significant effect on AphB-dependent transcriptional activation; both strains, GK372 and GK409, respectively, still showed approximately 40-fold activation by AphB. Thus, of the four El Tor base-pair replacements made in the classical fusion, only the G at position −65 significantly reduced AphB-dependent transcription. When the G at position −66 of the El Tor tcpP-lacZ fusion was converted to A, AphB-dependent transcription in the resulting strain, GK426, was activated 40-fold, a level similar to that observed with the classical fusion strain KSK782 (Fig. 5B). These results indicate that the presence of either an A or a G at position −65 or −66 of the classical or El Tor tcpPH promoter, respectively, determines biotype-specific expression by influencing the ability of AphB to activate transcription.
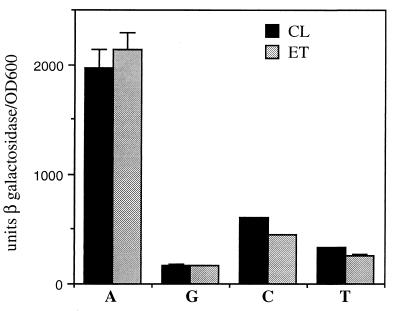
Since the presence of an A or a G at position −65 or −66 of the tcpPH promoter dramatically influenced the ability of AphB to activate transcription, it was of interest to determine the effect of replacing the base pair at this position with either C or T. Whereas the presence of A or G at position −65 or −66 permitted 40- or 4-fold activation of tcpPH transcription by AphB, respectively, replacement of this base pair with either C or T resulted in approximately 10- or 5-fold activation, respectively (Fig. 6). As was observed with A or G, the presence of either C or T produced nearly equivalent effects on AphB-dependent transcription from both biotype promoters. These results indicate that an adenine residue at position −65 or −66 of the tcpPH promoter is critical for optimal AphB-dependent activation of transcription.
FIG. 6.
AphB-dependent expression in E. coli tcpP-lacZ fusions containing A, G, C, or T at position −65 or −66. Strains containing pKAS117 (AphB) were grown as described in the legend to Fig. 4B. Black bars, classical (CL) fusions: KSK782 (wild type, A at position −65); GK370 (G at −65); GK441 (C at −65); GK443 (T at −65). Gray bars, El Tor (ET) fusions: GK407 (A at position −66); KSK864, (wild type, G at −66); GK442 (C at −66); GK444 (T at −66). Error bars show standard deviations.
Influence of A or G at position −65 or −66 on virulence gene expression in V. cholerae.
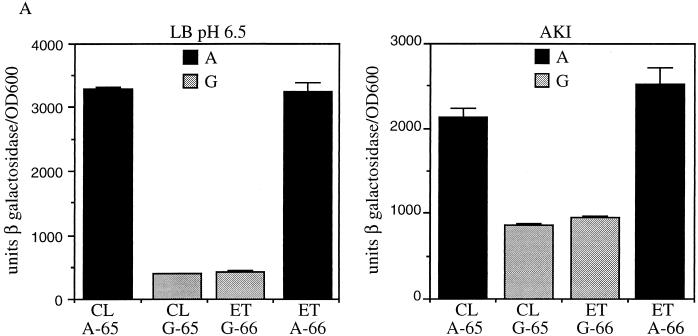
To determine the effect in V. cholerae of reciprocal exchange between biotypes of the A or G residue at position −65 or −66 of the tcpPH promoter on tcpPH expression, fragments of DNA containing the single base-pair changes from above were cloned into allelic exchange vectors and introduced into the chromosomes of the ΔtcpPH promoter tcpP-lacZ fusion strains GK300 and GK250. Classical strain GK404, which has a classical tcpPH promoter except for a G at position −65, showed a significantly reduced level of β-galactosidase production similar to that of the wild-type El Tor fusion strain KSK725 in either LB medium (pH 6.5) at 30°C or AKI medium (Fig. 7A). In contrast, β-galactosidase production in El Tor strain GK436, which has an El Tor tcpPH promoter except for an A at position −66, mirrored that observed with the wild-type classical fusion strain KSK618. These results support those obtained with E. coli indicating that the biotype-specific pattern of tcpPH expression is determined by the presence of an A or a G at position −65 or −66 of the tcpPH promoter.
FIG. 7.
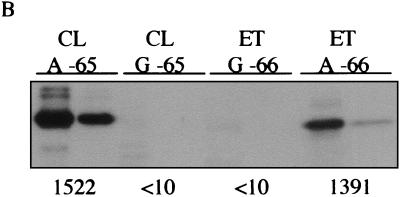
Reciprocal exchange of A and G at positions −65 and −66 of the classical and El Tor tcpPH promoters in V. cholerae. (A) Expression of tcpP-lacZ. Cultures were grown in LB medium (pH 6.5) at 30°C (left) and in AKI medium at 37°C (right). Black bars, strains with an A at position −65 or −66: KSK618 (wild type, classical [CL], A at position −65); GK436 (El Tor [ET], A at −66). Gray bars, strains with a G at position −65 or −66: KSK725 (wild type, El Tor, G at position −66); GK404 (classical, G at −65). Error bars show standard deviations. (B) Production of TcpA and CT. Cultures were grown overnight in LB medium (pH 6.5) at 30°C. For each strain, 16 μg (left) and 3 μg (right) of total protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, electroblotted to nitrocellulose paper, and probed with anti-TCP antiserum. From left to right: O395 (classical, A at position −65); KSK1093 (classical, G at −65); C6706 str2 (El Tor, G at −66); KSK1117 (El Tor, A at −66). The amount of CT in each culture supernatant is shown at the bottom (in nanograms per milliliter per OD600 unit).
To determine the influence of these base-pair changes on the production of TCP and CT in the two biotypes, the G at position −65 in the classical tcpPH promoter was introduced into classical strain O395, and the A at position −66 in the El Tor tcpPH promoter was introduced into El Tor strain C6706 str2. As shown in Fig. 7B, the presence of a G at position −65 in the classical tcpPH promoter, strain KSK1093, reduced the production of TCP in LB medium (pH 6.5) at 30°C to a level undetectable by Western blotting and decreased toxin production from 1,500 ng/ml per unit of optical density at 600 nm (OD600) to less than 10 ng/ml. Conversely, an A at position −66 in the El Tor tcpPH promoter, strain KSK1117, increased TCP production to a detectable level in LB medium (pH 6.5) at 30°C and increased toxin production from less than 10 ng/ml/OD600 unit to almost 1,400 ng/ml. These results indicate that it is the ability of AphB to activate tcpPH expression which largely determines the biotype-specific pattern of virulence gene expression that is observed in V. cholerae.
DISCUSSION
The expression of the genes encoding TCP and CT is regulated by a transcriptional cascade involving multiple activator proteins. The AraC regulator ToxT directly activates the transcription of the tcpA and ctx operons (4, 9, 15). The expression of toxT is in turn regulated by two pairs of transmembrane transcriptional activator proteins, ToxRS and TcpPH (7, 13, 24, 26). The expression of the tcpPH operon is activated by the LysR regulator AphB in cooperation with a second protein, AphA (21, 33). Each of these activator proteins appears to play a similar role in both disease-causing biotypes of V. cholerae. However, the expression of tcpA, ctx, toxT, and tcpPH is significantly lower in the El Tor biotype than in the classical biotype under most in vitro culture conditions (8, 27). The overproduction of AphB from a plasmid in the El Tor biotype has recently been observed to dramatically increase tcpPH and tcpA expression to close to classical levels in vitro (21). This result, together with the finding that the expression of aphB is similar in both biotypes (21), raised the possibility that the biotype-specific expression of tcpPH might be related to the ability of AphB to activate transcription at each of these promoters.
To address this issue, experiments carried out with both V. cholerae and E. coli showed that when expressed from a plasmid, AphB alone activated the classical tcpPH promoter 20- to 40-fold, whereas it activated the El Tor promoter only 2- to 4-fold. That AphB showed reduced transcriptional activation of the El Tor tcpPH promoter relative to the classical promoter in E. coli suggested that the molecular basis for this difference might be at the level of the promoter rather than a biotype-specific influence over the activity of the protein. Indeed, replacement of the entire region between the tcpI and tcpP genes in V. cholerae with that of the other biotype showed that it is the tcpPH promoter itself which determines the biotype-specific pattern of expression. Biotype promoter chimera constructs in E. coli localized the region of the tcpPH promoter responsible for this difference to within 75 bp of the start of transcription. Of the four base-pair differences between the two biotypes in this region from positions −75 to +1, only conversion of the A at position −65 in the classical promoter to the El Tor G dramatically reduced AphB-dependent expression of tcpPH to the level typically observed with El Tor. The presence of a C or a T residue in either biotype promoter at this position only marginally increased AphB-dependent expression over that observed with a G residue. These results indicate that an A at position −65 or −66 in the tcpPH promoter is optimal for transcriptional activation by AphB.
DNA footprinting studies have shown that many LysR transcriptional regulators bind to promoter regions via a 15-bp dyad sequence with a common structure and location near the −65 region of the promoter (30). Two well-characterized LysR regulators are MetR from Salmonella enterica serovar Typhimurium (2, 22, 36) and TrpI from Pseudomonas (5, 11). MetR positively regulates several genes which encode enzymes in the methionine biosynthetic pathway. The protein binds to a region upstream of the metE transcriptional start site that contains the interrupted dyad TGAA-N5-TTCA from −67 to −55 (36). A similar sequence is also found in the MetR binding regions of the metH and metA promoters (2, 22). TrpI positively regulates the trpBA genes encoding tryptophan synthetase. The protein similarly binds to a region containing an interrupted dyad between −71 and −57 relative to the trpBA transcriptional start site: GTgAG-N5-CTgAC (5, 11). Examination of the DNA sequence around position −65 in the classical tcpPH promoter revealed the presence of an interrupted dyad from −69 to −53 (TGCAA-N7-TTGCA) that is a potential site for the interaction of AphB with DNA, based on similarities with other LysR-regulated promoters; the A at −65 is in the left arm of the dyad (indicated by the underline). It is possible that the conversion to G at this position reduces the binding of AphB to DNA. Substitution mutations in the conserved dyad arms of the consensus sequence upstream of the metH gene have been found to significantly reduce MetR binding and transcriptional activation (2). Experiments are currently in progress to determine whether this position is the actual binding site for AphB in the tcpPH promoter.
In V. cholerae as well as in E. coli, the biotype-specific pattern of tcpPH expression is determined by the presence of an A or a G at position −65 or −66 in the respective promoters. Substitution of the A at position −65 of the classical tcpPH promoter with a G decreased the expression of tcpPH in LB medium (pH 6.5) at 30°C to a level typically observed with El Tor. In addition, the production of TCP and CT was reduced under these conditions to levels comparable to those observed with El Tor. This classical A→G (position −65) mutant also failed to autoagglutinate. These results suggest that the lower levels of TcpP and TcpH in this mutant prevent the normally high-level expression of toxT that occurs with classical strains under these conditions.
Conversely, substitution of the G at position −66 of the El Tor tcpPH promoter with an A in V. cholerae increased the expression of tcpPH to classical levels in LB medium (pH 6.5) at 30°C. The observation that the production of both TCP and CT was increased by the G-to-A substitution suggests that the expression of toxT has been increased as a consequence of the higher levels of TcpP and TcpH present in the cells. However, the level of TcpA in the El Tor G→A (position −66) mutant, although dramatically increased from the level in the wild-type strain, still appeared to be approximately fivefold lower than that observed in the classical strain. In addition, the mutant did not strongly autoagglutinate in LB medium (pH 6.5) at 30°C like the classical strain did. This result suggests that additional differences with regard to the expression of tcpA may still exist between the two biotype strains. Consistent with this hypothesis, β-galactosidase production in an El Tor tcpA-lacZ fusion strain containing a G- to-A substitution at position −66 of the tcpPH promoter is approximately twofold lower than that in a wild-type classical tcpA-lacZ fusion strain (data not shown). In contrast, a wild-type El Tor tcpA-lacZ fusion strain (with a G at position −66 of the tcpPH promoter) shows 150-fold lower β-galactosidase production than the classical tcpA-lacZ fusion strain. Thus, although the G→A (position −66) substitution in the tcpPH promoter dramatically increased the expression of tcpA-lacZ in the El Tor strain, this increased level of expression was still lower than that observed with the classical strain. There are a number of nucleotide differences between the classical and El Tor tcpA promoters which could potentially influence transcription.
The evolution of toxigenic V. cholerae from environmental strains incapable of causing disease involves the acquisition of the TCP-ACF element (18, 19, 20) and the CTXφ phage (37). It is interesting that ToxR, ToxS, AphA, and AphB are not encoded within either of these elements, yet they play essential roles in activating the expression of virulence genes that are present on them. How the virulence gene promoters on these elements evolved to come under the control of these activator proteins can only be speculated. In addition, it is not obvious why the expression of tcpPH in the classical and El Tor biotype strains evolved to become differentially activated by AphB. One possibility is that the reduced virulence of the strain containing a G at position −66 of the tcpPH promoter enabled the organism to persist within the host to a greater degree and thus be more efficiently disseminated into the environment.
The work presented here establishes a basis for the differential regulation of virulence gene expression between the two disease-causing biotypes of V. cholerae, classical and El Tor. The insights gained from this work will aid in the further analysis of how AphA and AphB interact with the tcpPH promoter to influence virulence gene expression.
ACKNOWLEDGMENTS
We thank Ronald Taylor for helpful suggestions and for critical reading of the manuscript.
This work was supported by Public Health Service grant AI-41558 to K.S.
REFERENCES
- 1.Brown R C, Taylor R K. Organization of tcp, acf, and toxT genes within a ToxT-dependent operon. Mol Microbiol. 1995;16:425–439. doi: 10.1111/j.1365-2958.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- 2.Byerly K A, Urbanowski M L, Stauffer G V. The MetR binding site in the Salmonella typhimurium metH gene: DNA sequence constraints on activation. J Bacteriol. 1991;173:3547–3553. doi: 10.1128/jb.173.11.3547-3553.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll P A, Tashima K T, Rogers M B, DiRita V J, Calderwood S B. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol Microbiol. 1997;25:1099–1111. doi: 10.1046/j.1365-2958.1997.5371901.x. [DOI] [PubMed] [Google Scholar]
- 4.Champion G A, Neely M N, Brennan M A, DiRita V J. A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxT mutant strains. Mol Microbiol. 1997;23:323–331. doi: 10.1046/j.1365-2958.1997.2191585.x. [DOI] [PubMed] [Google Scholar]
- 5.Chang M, Crawford I P. In vitro determination of the effect of indoleglycerol phosphate on the interaction of purified TrpI protein with its DNA-binding sites. J Bacteriol. 1991;173:1590–1597. doi: 10.1128/jb.173.5.1590-1597.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawford J A, Kaper J B, DiRita V J. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol Microbiol. 1998;29:235–246. doi: 10.1046/j.1365-2958.1998.00925.x. [DOI] [PubMed] [Google Scholar]
- 7.DiRita V J, Mekalanos J J. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell. 1991;64:29–37. doi: 10.1016/0092-8674(91)90206-e. [DOI] [PubMed] [Google Scholar]
- 8.DiRita V J, Neely M, Taylor R K, Bruss P M. Differential expression of the ToxR regulon in classical and El Tor biotypes of Vibrio cholerae is due to biotype-specific control over toxT expression. Proc Natl Acad Sci USA. 1996;93:7991–7995. doi: 10.1073/pnas.93.15.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiRita V J, Parsot C, Jander G, Mekalanos J J. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frohman M A, Dush M K, Martin G R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao J, Gussin G N. Activation of the trpBA promoter of Pseudomonas aeruginosa by the TrpI protein in vitro. J Bacteriol. 1991;173:3763–3769. doi: 10.1128/jb.173.12.3763-3769.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardel C L, Mekalanos J J. Regulation of cholera toxin by temperature, pH and osmolarity. Methods Enzymol. 1994;235:517–526. doi: 10.1016/0076-6879(94)35167-8. [DOI] [PubMed] [Google Scholar]
- 13.Häse C C, Mekalanos J J. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins D E, DiRita V J. Transcriptional control of toxT, a regulatory gene in the ToxR regulon of Vibrio cholerae. Mol Microbiol. 1994;14:17–29. doi: 10.1111/j.1365-2958.1994.tb01263.x. [DOI] [PubMed] [Google Scholar]
- 15.Higgins D E, Nazareno E, DiRita V J. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. J Bacteriol. 1992;174:6974–6980. doi: 10.1128/jb.174.21.6974-6980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwanaga M, Yamamoto K, Higa N, Ichinose Y, Nakasone N, Tanabe M. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol Immunol. 1986;30:1075–1083. doi: 10.1111/j.1348-0421.1986.tb03037.x. [DOI] [PubMed] [Google Scholar]
- 17.Kaper J B, Morris J G, Jr, Levine M M. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karaolis D K R, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karaolis D K R, Somara S, Maneval D R, Jr, Johnson J A, Kaper J B. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature. 1999;399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 20.Kovach M E, Shaffer M D, Peterson K M. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology. 1996;142:2165–2174. doi: 10.1099/13500872-142-8-2165. [DOI] [PubMed] [Google Scholar]
- 21.Kovacikova G, Skorupski K. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J Bacteriol. 1999;181:4250–4256. doi: 10.1128/jb.181.14.4250-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mares R, Urbanowski M L, Stauffer G V. Regulation of the Salmonella typhimurium metA gene by the MetR protein and homocysteine. J Bacteriol. 1992;174:390–397. doi: 10.1128/jb.174.2.390-397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 24.Miller V L, DiRita V J, Mekalanos J J. Identification of toxS, a regulatory gene whose product enhances ToxR-mediated activation of the cholera toxin promoter. J Bacteriol. 1989;171:1288–1293. doi: 10.1128/jb.171.3.1288-1293.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 27.Murley Y M, Carroll P A, Skorupski K, Taylor R K, Calderwood S B. Differential transcription of the tcpPH operon confers biotype-specific control of the Vibrio cholerae ToxR virulence regulon. Infect Immun. 1999;67:5117–5123. doi: 10.1128/iai.67.10.5117-5123.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogierman M A, Voss E, Meaney C, Faast R, Attridge S R, Manning P A. Comparison of the promoter proximal regions of the toxin-co-regulated tcp gene cluster in classical and El Tor strains of Vibrio cholerae O1. Gene. 1996;170:9–16. doi: 10.1016/0378-1119(95)00744-x. [DOI] [PubMed] [Google Scholar]
- 29.Padgett K A, Sorge J A. Creating seamless junctions independent of restriction sites in PCR cloning. Gene. 1996;168:31–35. doi: 10.1016/0378-1119(95)00731-8. [DOI] [PubMed] [Google Scholar]
- 30.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 31.Skorupski K, Taylor R K. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 32.Skorupski K, Taylor R K. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc Natl Acad Sci USA. 1997;94:265–270. doi: 10.1073/pnas.94.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skorupski K, Taylor R K. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol Microbiol. 1999;31:763–771. doi: 10.1046/j.1365-2958.1999.01215.x. [DOI] [PubMed] [Google Scholar]
- 34.Sun D, Seyer J M, Kovari I, Sumrada R A, Taylor R K. Localization of protective epitopes within the pilin subunit of the Vibrio cholerae toxin-coregulated pilus. Infect Immun. 1991;59:114–118. doi: 10.1128/iai.59.1.114-118.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urbanowski M L, Stauffer G V. Genetic and biochemical analysis of the MetR activator-binding site in the metE metR control region of Salmonella typhimurium. J Bacteriol. 1989;171:5620–5629. doi: 10.1128/jb.171.10.5620-5629.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 38.Yu R R, DiRita V J. Analysis of an autoregulatory loop controlling ToxT, cholera toxin, and toxin-coregulated pilus production in Vibrio cholerae. J Bacteriol. 1999;181:2584–2592. doi: 10.1128/jb.181.8.2584-2592.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]