Abstract
PARP inhibitors (PARPi) have emerged as promising cancer therapeutics capable of targeting specific DNA repair pathways, but their mechanism of action with respect to PARP1-DNA retention remains unclear. Here, we developed single-molecule assays to directly monitor the retention of PARP1 on DNA lesions in real time. Our study reveals a two-step mechanism by which PARPi modulate the retention of PARP1 on DNA lesions, consisting of a primary step of catalytic inhibition via binding competition with NAD+ followed by an allosteric modulation of bound PARPi. While clinically relevant PARPi exhibit distinct allosteric modulation activities that can either increase retention of PARP1 on DNA or induce its release, their retention potencies are predominantly determined by their ability to outcompete NAD+ binding. These findings provide a mechanistic basis for improved PARPi selection according to their characteristic activities and enable further development of more potent inhibitors.
PARP1:DNA retention is governed by catalytic inhibition and allosteric modulation.
INTRODUCTION
PARP1 is a key DNA repair protein involved in the repair of various DNA lesions, including base damage and single-strand and double-strand breaks (1–4). The activation and DNA repair functions of PARP1 are mediated via its multidomain architecture (Fig. 1A) containing N-terminal DNA binding domains, an automodification domain bearing BRCT (BReast cancer gene 1 C-Terminal) and automodification sites, a WGR [Trp(W)-Gly(G)-Arg(R)] domain bridging the Zn1 and catalytic domains, and a C-terminal catalytic domain, which are thought to form a “beads-on-a-string”–like structure in the absence of DNA lesions (5). Upon encountering a DNA lesion, PARP1 binds to the lesion to undergo conformational changes, providing an allosteric structure-function framework for its activity and allowing its substrate nicotinamide adenine dinucleotide (NAD+) to access the catalytic pocket (6, 7). This initiates PARP1 polymerase activity of poly(ADP)ribosylation (PARylation) by hydrolyzing NAD+ to add adenosine diphosphate (ADP)–ribose moieties onto itself and its target proteins, nucleic acids, and metabolites (4). Extensive auto-PARylation of PARP1 triggers its release from DNA due to the high negative charge of PAR chains (8). The central roles of PARP1 in DNA repair present a promising venue for targeting of cancers with deficiencies in DNA damage response and repair pathways (9–11). This concept was further realized with the identification of small molecule inhibitors of PARP1 activity, which mimic its substrate NAD+ and compete for binding onto the catalytic pocket (11–13). Several PARP inhibitors (PARPi) have been clinically approved to treat breast, ovarian, prostate, and pancreatic cancers (14–19), yet their mechanism of action is still not fully understood and has been the subject of much debate.
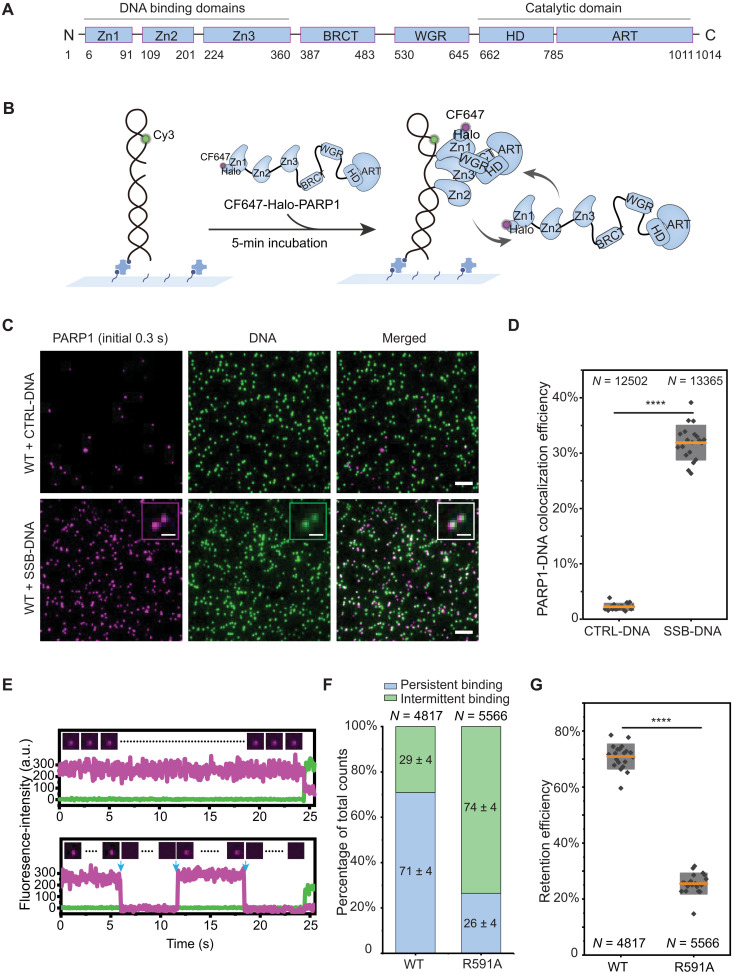
Fig. 1. Single-molecule kinetics of PARP1 retention on DNA lesions.
(A) PARP1 protein domains. (B) Schematic of smCL assays. Biotinylated DNA substrates labeled with Cy3 (shown in green) were immobilized onto the surface of a microfluidic chamber via biotin-neutravidin linkages. PARP1 (HaloTag-PARP1, 2 nM) labeled with CF647 shown in magenta was injected into the chamber, and data were acquired following a 5-min incubation. (C) Representative images of DNA or PARP1 molecules, showing nearly no PARP1 colocalization with CTRL-DNA (top) and PARP1 colocalization with SSB-DNA (bottom). Insets show magnified images of PARP1:DNA colocalization. Scale bars, 5 μm (original images) and 1 μm (insets). (D) Quantification of colocalization efficiency of PARP1 with CTRL-DNA or SSB-DNA. N is the total number of DNA molecules measured. Orange lines and gray boxes indicate means ± SD. Individual data points represent colocalization efficiency analyzed using data from a single imaging area. (E) Representative single-molecule trajectories of PARP1:SSB-DNA colocalization as a function of time, showing persistent binding (top) and intermittent binding (bottom) of PARP1 to SSB-DNA. Insets show representative frames of PARP1 from the trajectories, and blue arrows denote dissociation and binding events. a.u., arbitrary unit. (F) Quantification of the percentage of persistent (blue) or intermittent (green) binding events of WT-PARP1 or R591A on SSB-DNA. N is the total number of PARP1:DNA trajectories, and the mean ± SD of each population is shown on the column. (G) Quantification of retention efficiency of PARP1 or R591A on SSB-DNA. N is the total number of PARP1:DNA trajectories. The P value of two-sample t test is as indicated. ****P ≤ 0.0001.
The mechanisms by which PARPi affect PARP1:DNA interaction and DNA repair have been the focus of several key studies that measured different aspects of PARPi both biochemically and in cells to provide confounding data and opposing interpretations (12, 20–24). Initial studies demonstrated that PARPi treatment led to an enrichment of chromatin-bound PARP1 upon induction of DNA damage in cells and impeded PARP1 dissociation from DNA in vitro, suggesting that their toxicity is due to PARP1 trapping at DNA lesions (20, 21). Because the magnitude of trapping and the cellular toxicity of different PARPi did not correlate with their potency for inhibiting PARP1 catalytic activity, it was proposed that their effect is mediated by two linked mechanisms (13, 20). In the first mechanism, inhibition of PARP1 catalytic activity prevents auto-PARylation, thereby hindering its dissociation from DNA. In the second mechanism, the docking configuration of the inhibitor at the catalytic core induces allosteric modulation that increases PARP1 stability on DNA. It was proposed that the level of PARP1 trapping via allosteric modulation could be the underlying cause of the differing efficacies of PARPi in cells rather than simply differences in catalytic inhibition potencies (13, 20, 21). However, subsequent biochemical and cellular studies of PARPi reported that trapping is solely caused by inhibition of catalytic activity and not due to allosteric modulation (22). Consistent with this study, biochemical analysis of PARP1 dissociation from DNA upon the addition of competitor DNA showed that PARPi did not affect its rate of dissociation (23). Moreover, cellular fluorescence recovery after photobleaching measurements found that the turnover rates of fluorescent PARP1 at DNA lesions were largely unchanged in the presence of clinical PARPi (24). Recently, comprehensive structural and biochemical analyses of the organization of PARP1 domains induced by different PARPi demonstrated that some allosteric configurations enhance PARP1 retention while others can facilitate release (12), proposing a model for PARPi potency that includes their allosteric proretention and prorelease activities. Despite the important insights provided by these studies, the exact contribution of catalytic activity inhibition and allosteric modulation to the potency of different clinically relevant PARPi (talazoparib, olaparib, veliparib, niraparib, and rucaparib) remains unknown.
Here, we addressed these unknowns using single-molecule assays to directly visualize the real-time kinetics of PARP1 retention on DNA lesions. Our approach enabled us to resolve the specific retention of PARP1 and relevant PARP1 mutants on DNA that is induced by different PARPi in the presence or absence of NAD+. This provided a means to detect and quantify PARP1 kinetics even at short time scales that correspond to its rapid release from DNA upon automodification, thereby allowing us to distinguish the contributions of catalytic inhibition from that of allosteric modulation. Our results indicate that under conditions that support PARP1 catalytic activity, its retention on DNA is predominantly determined by the ability of PARPi to compete with NAD+ and inhibit PARP1 enzymatic activity. Moreover, our data provides unique insights into the specific involvement of key PARP1 functional and structural mutations in driving allosteric modulation induced by different PARPi. On the basis of these findings, we propose a refined model linking the activity of clinically relevant PARPi and the retention of PARP1 on DNA lesions.
RESULTS
To measure the binding and stability of PARP1 onto a DNA lesion, we designed and optimized a specific single-molecule assay that can faithfully capture such dynamics. This assay is illustrated in Fig. 1B and uses single-molecule colocalization (smCL) to monitor the binding and dissociation of individual fluorescent PARP1 proteins (CF647-Halo-PARP1) on fluorescently labeled DNA substrates (internally Cy3-labeling DNA, end-biotinylated). DNA substrates were first tethered to the surface of a flow chamber followed by injection of fluorescent PARP1 molecules, and their association with surface-bound DNA was monitored after a 5-min incubation using a multicolor total internal reflection fluorescence (TIRF) microscopy system (fig. S1, A and B, and Materials and Methods). To model a single-strand break (SSB) lesion, we designed a double-stranded DNA (dsDNA) substrate containing a single-base gap (SSB-DNA), while a continuous dsDNA substrate (CTRL-DNA) was used to mimic undamaged DNA (fig. S1C and Materials and Methods). Quantification of the percentage of DNA molecules with bound PARP1 revealed clear colocalization of PARP1 with SSB-DNA (~32%) but not with CTRL-DNA (Fig. 1, C and D).
To characterize the single-molecule kinetics of PARP1 retention at equilibrium, we analyzed the trajectories of individual PARP1 molecules that colocalized with SSB-DNA. Because the single-turnover assays showed that the half-life of SSB-DNA associated PARP1 is longer than 6 min (fig. S4), the trajectories of the single molecules at equilibrium were classified into two categories on the basis of their distinct behaviors: persistent-binding, wherein PARP1 is bound to DNA throughout the trajectory, and intermittent-binding, wherein PARP1 dissociates, binds, or rebinds to DNA (Fig. 1E). To assess how these behaviors correspond to the interaction of PARP1 with DNA and overall binding stability, we quantified the percentage of trajectories in each category for wild type (WT)–PARP1 and for a WGR domain mutant, R591A (6). We found that the majority (~71%) of WT molecules displayed persistent binding, while a reversed trend was observed for R591A, which is in line with the reduced foci formation in cells shown for a PARP1 patient mutant, R591C (Fig. 1F and fig. S5, A and B) (25). Accordingly, the percentage of persistent-binding molecules provides a direct quantification for PARP1 retention efficiency (Fig. 1G and fig. S5C).
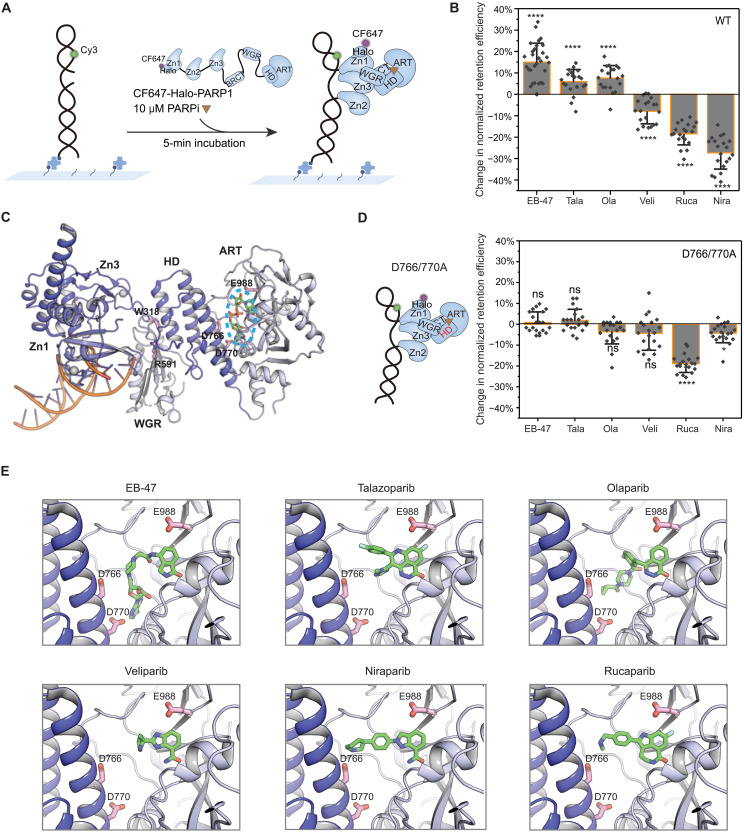
A recent study of the allosteric modulation of PARP1 has categorized PARPi into three types on the basis of how their induced allostery affects retention: proretention (type I), modest proretention or no effect (type II), and prorelease (type III) (12). Accordingly, type I PARPi [EB-47 and benzamide adenine dinucleotide (BAD)] yielded strong allosteric retention, whereas clinically relevant PARPi were classified as type II (talazoparib and olaparib) and type III (rucaparib, niraparib, and veliparib). Inspired by these observations, to determine the specific contributions of different PARPi in inducing allosteric proretention or prorelease activities at the single-molecule level, we measured changes in PARP1:SSB-DNA retention in the absence of NAD+ and in the presence of a high concentration (10 μM) of PARPi, ensuring that all DNA-bound PARP1 were bound with PARPi (Fig. 2, A and B, and fig. S6). We observed a noticeable increase (~15% or more) in PARP1 retention for EB-47 and BAD and a modest increase for talazoparib and olaparib, whereas veliparib, rucaparib, and niraparib induced a decrease in retention that was most substantial for niraparib (~25%). These results demonstrated that PARP1 allostery for BAD, EB-47, talazoparib, and olaparib enhance retention, whereas veliparib, rucaparib, and niraparib facilitate release, which are consistent with the reported dissociation rate constants measured by surface plasmon resonance (12). To verify that the observed trends are due to PARPi-induced allosteric modulation, we analyzed their effects on the PARP1 helical domain (HD) mutant D766/770A (Fig. 2, C and D) shown to abolish the reverse allosteric activation and proretention induced by EB-47 (12). While the retention efficiency of D766/770A was similar to WT (fig. S7B), the retention/release trends were abolished for all PARPi with the exception of rucaparib, which stimulated considerable prorelease activity (Fig. 2D and fig. S7C), indicating that its allostery likely involves other PARP1 residues (26). The structural models of PARPi bound onto PARP1 indicated that the loss of allosteric modulation of the D766/770A mutant is likely due to the removal of a steric conflict with the inhibitor (Fig. 2E).
Fig. 2. Distinct modes of allosteric modulation induced by different PARPi.
(A) Schematic of smCL assays of PARP1:DNA measured in the presence of PARPi. (B) Quantification of change in retention efficiency of WT-PARP1 in the presence of different PARPi. Data for 10 μM EB-47 (N = 5072), Tala (talazoparib; N = 6161), Ola (olaparib; N = 4657), Veli (veliparib; N = 5303), Rucap (rucaparib; N = 4542), or Nira (niraparib; N = 5565) was plotted relative to PARP1 only control (DMSO). (C) A model of crystal structure of PARP1 essential domains (Protein Data Bank code 4DQY) binding with nonhydrolyzable NAD+ analog indicated in the blue dashed ellipse at catalytic pocket. All mutated amino acids [W318 in Zn3, R591 in WGR, D766/770 in HD, and E988 in ADP-ribosyl-transferase (ART) domain] in this study are shown in the model. (D) Left: Schematic of HD domain (highlighted in red) mutant D766/770A. Right: Quantification of change in retention efficiency of D766/770A mutant in the presence of 10 μM EB-47 (N = 4882), Tala (N = 4785), Ola (N = 4682), Veli (N = 4985), Rucap (N = 4598), or Nira (N = 4814). (E) Zoomed-in panels of PARPi binding to the catalytic pocket. In (B) and (D), color bars and error bars indicate mean and SD, respectively. N is the total number of PARP1:DNA trajectories. Individual data points represent the measured retention efficiency analyzed using data from one imaging area. The P value of two-sample t test is as indicated. ns, nonsignificant difference; *P ≤ 0.05 and ****P ≤ 0.0001.
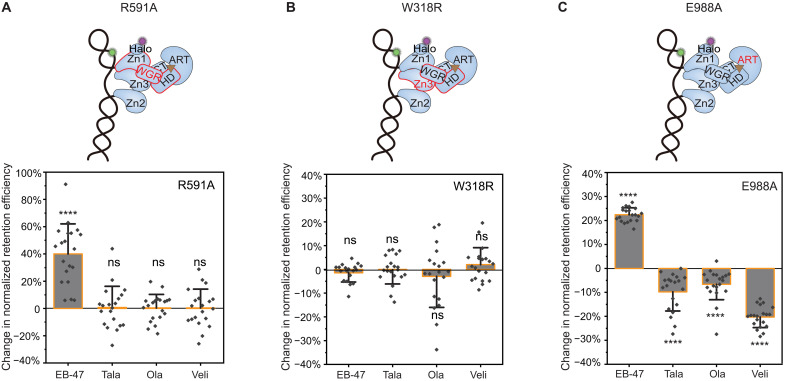
To further analyze the specific allosteric modulation of different PARPi, we tested two PARP1 mutants (R591A and W318R) shown to disrupt communication interfaces between the DNA binding domains and the catalytic domain. R591A is a mutation in the WGR domain that disrupts allostery through the Zn1-WGR-HD interface; W318R is a mutation in the Zn3 domain that disrupts allostery through the WGR-Zn3-HD interface (Fig. 2C) (6). Analysis of the changes in retention efficiency for these mutants revealed that EB-47 induced an increase in retention of R591A, whereas no changes were observed for W318R and other PARPi (Fig. 3, A and B, and fig. S8, A and B). These results indicate that EB-47-induced allostery can overcome the R591A-induced deficiency at the Zn1-WGR-HD interface, while that of talazoparib, olaparib, and veliparib are unable to overcome the deficiency at either the Zn1-WGR-HD or the WGR-Zn3-HD interface. We note that these single-molecule assays are particularly advantageous in detecting such changes since the overall retention efficiency of these mutants was considerably lower than the WT (Fig. 1G and fig. S8D, for R591A and W318R, respectively). To examine whether the coordination of PARPi at the catalytic pocket also contribute to their induced allostery, we tested a catalytic mutant of PARP1, E988A (Fig. 3C), having substantially reduced catalytic activity and PAR-polymer elongation (fig. S9) but little effect on NAD+ binding (27). Notably, while the retention efficiency of E988A was similar to WT (fig. S10B), we found that talazoparib and olaparib induced a decrease in E988A retention opposite to the trend observed with the WT (Fig. 2B), whereas EB-47 and veliparib maintained the trends that were observed with the WT (Fig. 3C and fig. S10C). These changes indicate that altered coordination of the NAD+ moiety of PARPi at the catalytic core can modify their induced allostery depending on their size, enabling a switch from proretention to prorelease.
Fig. 3. Mechanisms of allosteric modulation induced by different PARPi.
(A) Top: Schematic of the WGR domain mutation R591A disrupting PARP1 allostery through the Zn1-WGR-HD interface highlighted in red. Bottom: Quantification of change in retention efficiency of R591A mutant in the presence of 10 μM EB-47 (N = 5112), Tala (N = 5817), Ola (N = 5629), and Veli (N = 4067). (B) Top: Schematic of the Zn3 domain mutation W318R disrupting PARP1 allostery through the WGR-Zn3-HD interface highlighted in red. Bottom: Quantification of change in retention efficiency of W318R mutant in the presence of 10 μM EB-47 (N = 4470), Tala (N = 4100), Ola (N = 4198), and Veli (N = 3598). (C) Top: Schematic of the ART domain (highlighted in red) mutant E988A. Bottom: Quantification of change in retention efficiency of E988A mutant in the presence of 10 μM EB-47 (N = 4114), Tala (N = 4971), Ola (N = 4799), or Veli (N = 4822). Color bars and error bars indicate mean and SD, respectively. N is the total number of PARP1:DNA trajectories. Individual data points represent the measured retention efficiency analyzed using data from one imaging area. The P value of two-sample t test is as indicated. ****P ≤ 0.0001.
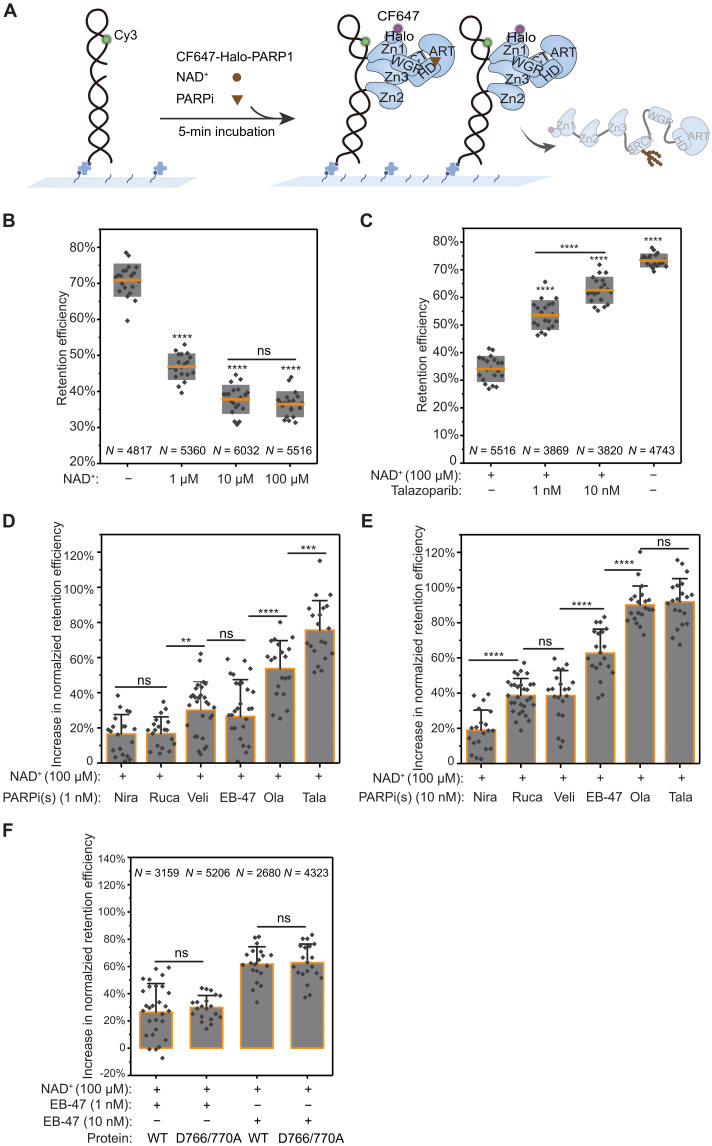
Next, we sought to determine how PARPi affect PARP1 retention on SSB-DNA in the presence of NAD+ (Fig. 4A). To examine these under conditions supporting automodification, we first obtained a baseline for PARP1 retention efficiency in the presence of NAD+ at various concentrations (Fig. 4B). These measurements showed a noticeable decrease in retention efficiency with increase in NAD+ concentrations, demonstrating the rapid release of PARP1 from DNA due to auto-PARylation (Fig. 4B and fig. S11), which was further confirmed by single-molecule fluorescence resonance energy transfer (FRET) assays (fig. S12). Because PARPi inhibit PARP1 activity by competing with NAD+ binding at its catalytic site, we next analyzed the concentration dependence of this competition by measuring PARP1:SSB-DNA retention in the presence of 100 μM NAD+, which is at the range of intracellular NAD+ concentration (28, 29), at varying concentrations of the potent inhibitor talazoparib (Fig. 4C). This analysis showed that the respective levels of PARP1 retention follow the concentration ratio of PARPi to NAD+, confirming that retention is strongly dependent on competitive inhibition. To further evaluate whether the potencies of the different clinically relevant PARPi and their competition with NAD+ correspond to PARP1 retention efficiency, we measured their effects on PARP1 retention in the presence of 100 μM NAD+. We quantified the percent increase in retention relative to NAD+ only (normalized), for two different concentrations of PARPi (1 and 10 nM; Fig. 4, D and E, and fig. S13, A to C). These measurements showed that at a concentration of 1 nM PARPi and 100 μM NAD+, talazoparib elicited the highest increase in retention efficiency (~76%), followed by olaparib (~53%), whereas veliparib and EB-47 showed similar increase (~26–30%), and rucaparib and niraparib resulted in a modest increase (~17%; Fig. 4D and fig. S13, A and C). Higher PARPi concentration (10 nM) led to further increase in retention efficiency, where both talazoparib and olaparib yielded the highest retention (~ 90–92%), while EB-47 also exhibited a noticeable increase (~63%), but veliparib and rucaparib showed modest increase in retention (~39%), and no notable change was observed for niraparib (Fig. 4E and fig. S13, B and C). These trends are consistent with the dissociation constant values of different PARPi measured by ensemble methods (26, 30).
Fig. 4. PARP1 retention on DNA is governed by PARPi binding competition with NAD+.
(A) Schematic of smCL assays of PARP1:DNA measured in the presence of NAD+ and PARPi. (B) Quantification of PARP1 retention efficiency in the absence or presence of 1, 10, or 100 μM NAD+. Orange lines and gray boxes indicate means ± SD. (C) Quantification of PARP1 retention efficiency in the presence of 100 μM NAD+ alone, in the presence of 1 or 10 nM talazoparib together with 100 μM NAD+, and in the absence of both NAD+ and talazoparib. (D) Quantification of the increase in PARP1 retention efficiency induced by the different PARPi (1 nM) added together with 100 μM NAD+ compared to 100 μM NAD+ alone. Nira (N = 5110), Rucap (N = 4809), Veli (N = 3323), EB-47 (N = 3159), Ola (N = 3553), and Tala (N = 3869). Colored bars and error bars indicate means and SD, respectively. Data were plotted as an increase from 100 μM NAD+ control. (E) Quantification of the increase in PARP1 retention efficiency shows changes in retention at higher PARPi concentration (10 nM) added together with 100 μM NAD+ compared to 100 μM NAD+ alone. Nira (N = 5320), Rucap (N = 5427), Veli (N = 3209), EB-47 (N = 2680), Ola (N = 3604), and Tala (N = 3820). (F) Quantitative comparison of the increase in retention efficiency of WT-PARP1 and D766/770A induced by 1 nM or 10 nM EB-47 added together with 100 μM NAD+. N is the total number of PARP1:DNA trajectories. Individual data points represent the measured retention efficiency analyzed using data from one imaging area. The P value of two-sample t test is as indicated. **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001.
To further differentiate the specific contribution of PARPi catalytic inhibition from that of allosteric modulation to PARP1 retention, we used the D766/770A mutant, which completely abolishes allosteric proretention effects of the different PARPi except rucaparib (Fig. 2D) while retaining its NAD+ catalytic activity. Analysis of the changes in normalized retention efficiency of WT-PARP1 and D766/770A mutant in the presence of 1 nM or 10 nM EB-47 together with 100 μM NAD+ (Fig. 4F) showed a similar increase in retention for D766/770A as the WT despite loss of allosteric proretention activity in the mutant. In addition, retention of D766/770A in the presence of NAD+ was higher for talazoparib and olaparib than for EB-47 and similar to the increase in the WT (fig. S15), consistent with the direct contribution of catalytic inhibition to overall retention efficiency. These results demonstrate that PARP1 retention in the presence of NAD+ is strongly governed by inhibition of enzymatic activity of clinically relevant PARPi via competition with NAD+.
DISCUSSION
In this study, we address the specific effect of clinically relevant PARPi on PARP1 trapping on DNA lesions by direct measurement of PARP1:DNA retention at the single-molecule level, establishing a mechanism for their activities with distinct contributions of inhibition and allosteric modulation. The mechanism governing PARP1 retention consists of two steps where PARPi first inhibit enzymatic activity by competing with NAD+ at its binding site, and once bound to the catalytic pocket, they induce allosteric modulation that change PARP1 retention on DNA (Fig. 5). Our results indicate that the retention potency of clinically relevant PARPi is mainly determined by the first step, whereas different PARPi induce distinct modes of allosteric modulation that can be uniquely adjusted by PARP1 mutations. We demonstrate that EB-47, talazoparib, and olaparib induce proretention allostery but with distinctive dependence on domain interfaces whereby EB-47 can still support SSB-DNA association of PARP1 harboring mutations that weakens Zn1-WGR-HD interaction. Accordingly, EB-47 and its derivatives can potentially be used to treat patients with certain PARP1 mutations, such as R591C found in patients with ovarian cancer that displayed resistance to olaparib (25). Furthermore, we demonstrate that a mutation in PARP1 catalytic core affects the coordination of the different inhibitors, resulting in changes in the allostery induced by the different PARPi that ranges from switching from proretention to prorelease. Similar modification of allostery was achieved using chemical changes to a given PARPi (12); here, we show that such an effect can also be produced by a mutation that affects the coordination of the inhibitor within the PARP1 catalytic core. Combined, we provide a mechanistic basis for improved PARPi selection according to their characteristic activities and patient mutations that can facilitate further development of more potent and selective therapeutics.
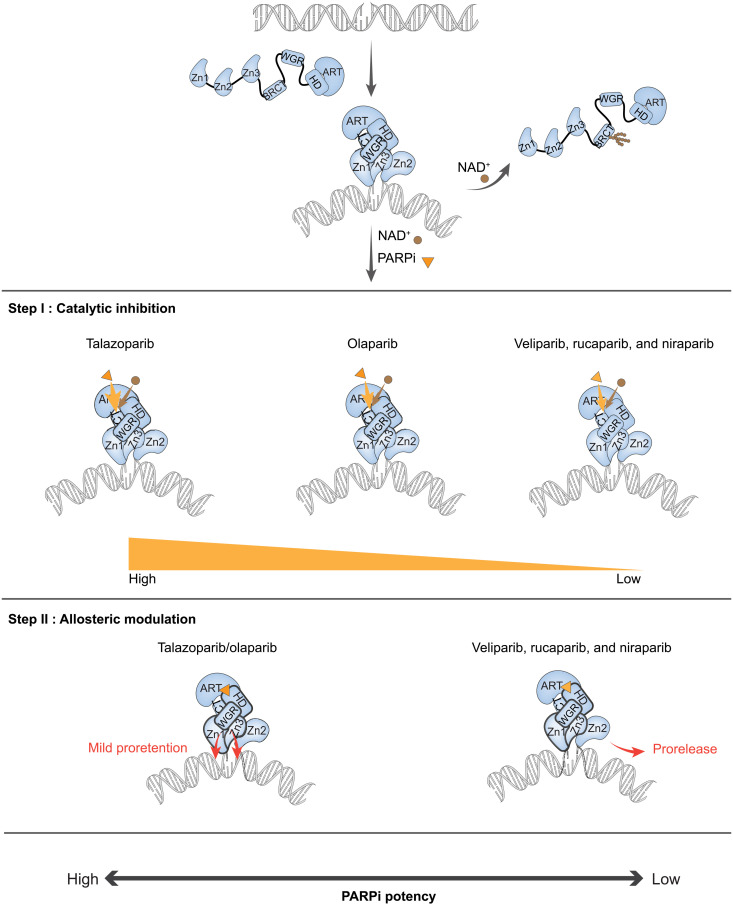
Fig. 5. A two-step model for clinical PARPi mediated PARP1 retention on DNA lesions.
NAD+ is depicted as a brown dot, and PARPi is depicted as an orange triangle. (i) PARP1 recognizes and binds to the DNA lesion through its N-terminal DNA binding domains, which activates its C-terminal domain polymerase activity to add ADP-ribose to its automodification domain. Extensive PARylation of PARP1 leads to release from DNA due to the high negative charge of poly-ADP ribose. (ii) Step I catalytic inhibition: PARPi retain PARP1 at the lesion by competing with NAD+ for the catalytic pocket binding site. (iii) Step II allosteric modulation: Once PARPi are bound at the catalytic pocket of PARP1, they further affect PARP1 retention on DNA by inducing allosteric modulation. PARPi mediate allostery through the Zn1-WGR-HD and WGR-Zn3-HD interfaces. Talazoparib and olaparib show mild proretention activity, while veliparib, rucaparib, and niraparib support prorelease.
Last, we note that many other factors and interactions likely contribute to the association and release kinetics of PARP1 from DNA lesions in cells. For example, the association of PARP1 with damaged chromatin has been shown to be stabilized by γH2A.X (31), whereas the adenosine triphosphatase p97 can facilitate the removal of PARP1 from chromatin in a ubiquitin-dependent manner (32). Another important player is histone PARylation factor 1 (HPF1), which affects PARP1 and PARP2 catalytic activity by controlling the balance between the initiation and elongation of ADP ribosylation to switch the modification from aspartate or glutamate to serine residues (33–35). Moreover, a recent in vitro study showed that HPF1 contributes to PARPi-mediated PARP1 retention on DNA lesions (36). Future studies of the effect of PARPi on PARP1 and PARP2 retention in the presence of nucleosomes and related factors are needed to further understand the roles of PARP1 and PARPi mechanism of action in vivo.
MATERIALS AND METHODS
Gene cloning and mutagenesis
The DNA fragments of N-terminal Halo-tagged WT human PARP1 (Halo-PARP1) or its mutants (Halo-PARP1 W318R, R591A, D766/770A, and E988A) were integrated between Nde I and Hind III enzyme sites of pET-28a vector using an infusion cloning kit (638948, Takara). The DNA fragment of Halo-PARP1 containing a Tobacco Etch Virus (TEV) linker was amplified from pBABE_Halo-PARP1_puro vector. DNA fragments of PARP1 mutants were obtained by overlap polymerase chain reaction and verified by sequencing by Genewiz. Expression plasmids are listed in table S1.
Protein purification and labeling
All proteins were purified from Escherichia coli as described previously (37). PARP1 without Halo-tag was obtained by cutting the Halo-tag using TEV enzyme (P8112S, New England Biolabs). Purified WT Halo-tagged PARP1 or its mutants were labeled with CF647-conjugated Halo-tag ligands at a 1:3.8 molar ratio for half an hour at room temperature in the dark. Excess dye-conjugated ligands were removed using a desalting column (52-1308-00, GE Healthcare). The labeling efficiency of Halo-tagged PARP1 was determined via absorbance measurements using NanoDrop and calculated according to the established protocols described previously (38) using the following equations and parameters: Cd = Aλmax/εd, F = Ad(280)/Ad, Cp = [A280 − (AλmaxF)]/εp, and D/P = Cd/Cp. The extinction coefficient of the free CF647 at 650 nm is εd = 240,000. F (A280/650) of CF647 is 0.03. A647 and A280 were measured using NanoDrop. The extinction coefficient of the protein εp was obtained according to the protein parameters (https://web.expasy.org/protparam/). The labeling efficiency was found to be ~78%. CF647-conjugated Halo-tag ligands were obtained by conjugating CF647 dyes with Halo-tag Amine(O2) ligands (P671A, Promega) using a labeling kit (#92359, Biotium). We used SDS–polyacrylamide gel electrophoresis (SDS-PAGE) for PARylation assays and agarose gels for gel shift assays (fig. S2 and S3) to verify that the catalytic activities of Halo-PARP1 were not affected by the Halo-tag or conjugation to CF647 (via HaloTag ligand) and also to compare the activity with that of Halo-PARP1 mutant (R591A).
SDS-PAGE PARylation assay
The auto-PARylation of PARP1, Halo-PARP1, and CF647-Halo-PARP1 were detected by SDS-PAGE PARylation assay as previously described (37). For unlabeled proteins, the experiments were performed by incubating 1 μM 18–base pair (bp) DNA duplex, 3.5 μM (6 μg) PARP1, and 2.6 μM (6 μg) Halo-PARP1 in the presence or absence of 5 mM NAD+ (B9007S, New England Biolabs) in reaction buffer [40 mM tris (pH7.5), 50 mM NaCl, 5 mM MgCl2, and 0.1 mM TCEP (tris(2-carboxyethyl)phosphine)] for 10 min at room temperature. For CF647-Halo-PARP1, 0.33 μM 18-bp DNA duplex and 0.33 μM CF647-Halo-PARP1 were incubated in the presence or absence of 1.6 mM NAD+ in imaging buffer [20 mM Hepes (pH8.0), 50 mM NaCl, 5 mM MgCl2, and 25% glucose] for 10 min at room temperature in the dark. The reaction was stopped by adding 4× Laemmli sample buffer (1610747, Bio-Rad), and samples were run on a 4 to 20% precast polyacrylamide gel (456-1094, Bio-Rad) and stained using Coomassie Blue. Images were captured by iBright imaging systems.
Gel shift assay
The binding and PARylation activities of CF647-Halo-PARP1 were detected by agarose gel shift assays as previously described (6). The experiments were performed by incubating 0.33 μM 18-bp DNA duplex and 0.33 μM CF647-Halo-PARP1 in the presence or absence of 1.6 mM NAD+ in imaging buffer [20 mM Hepes (pH8.0), 50 mM NaCl, 5 mM MgCl2, and 25% glucose] for 30 min at room temperature in dark. Samples were run on a 1% agarose gel containing 0.01% SYBR Gold dye (S11494, Thermo Fisher Scientific) for 40 min at 100 V at 4°C in Tris/Borate/EDTA (TBE) (B52, Thermo Fisher Scientific) in the dark. Images were captured by iBright imaging systems.
DNA substrate preparation
DNA oligos were ordered from IDT. DNA substrates were obtained by annealing two oligos or one oligo containing complementary sequence at 1 μM working concentration in T50 buffer [10 mM tris-Cl 8.0 and 50 mM NaCl] for 7 min at 95°C, followed by slowly cooling down. A continuous dsDNA substrate [control DNA (CTRL-DNA)] was obtained by annealing an oligo-named hairpin DNA. The DNA substrate containing a single-base gap (SSB-DNA) used in smCL assays was annealed by SSB-DNA-smCL top strand and SSB-DNA-smCL bottom strand at a 1:1.25 molar ratio. The FRET DNA substrate containing a single-base gap (SSB-DNA) used in fig. S12 was annealed by SSB-DNA-smFRET top strand and SSB-DNA-smFRET bottom strand at a 1:1.25 molar ratio. Oligos are listed in table S2.
Flow chamber preparation
All experiments were performed in microfluidic chambers made by sandwiching functionalized coverslips and microscope slides with double-sided tapes (fig. S1, A and B) as previously described (39). Briefly, functionalized coverslips and microscope slides were obtained by cleaning with 4 M KOH (06005, Sigma-Aldrich), followed by incubation in salinization solution consisting of 93% methanol (LC168104, LabChem), 5% acetic acid (BP1185, Thermo Fisher Scientific), and 2% 3-(2-aminoethylamino) propyltrimethoxysilane (catalog no. A0774, TCI), and followed by coating with Methoxy poly(ethylene glycol) succinimidyl valerate (MPEG-SVA-5000-5g, Laysan) with 2.5% biotin-PEG-SVA (biotin-PEG-SVA-5000, Laysan) dissolved in freshly prepared NaHCO3 (31437, Sigma-Aldrich). Slides were placed in 50-ml Falcon tubes and stored at −80°C.
TIRF microscope setup
A custom-built objective-based TIRF microscope was used for single-molecule imaging as previously described (39). A 532-nm laser (Ultra-Laser, MGL-FN-532-1000) was used for exciting Cy3, and a 639-nm laser (Ultra-Laser, MGL-FN-532-800) was used for exciting CF647 at ~10 mW. Emissions of Cy3 and CF647 or Cy5 were split by a beam splitter (Semrock, Di03-635-t1), filtered by a 582 ± 32 filter (Semrock, FF01-582/64) and 680 ± 21 filter (Semrock, FF01-680/42), respectively, and collected by an electron-multiplying charge–coupled device (Andor, iXon 897).
smCL assay
First, 10 μl of neutravidin (1 mg/ml; dissolved in water; A2666, Thermo Fisher Scientific) were injected into the microfluidic chamber, followed by 10-min incubation. Then, excess neutravidin were washed-off, followed by 50 μl of bovine serum albumin (30 mg/ml dissolved in water; A7030, Sigma-Aldrich) injection and 30-min incubation to block the surface of the chamber. Biotinylated Cy3-labeled DNA molecules were diluted to 20 to 50 pM using imaging buffer and tethered onto the surface of the chamber via biotin-neutravidin linkages, followed by a wash to remove unbound DNA. Fluorescent PARP1 proteins (0.5 to 2 nM) were injected into the chamber using PARP1 imaging buffer containing 20 mM Hepes (pH 8.0), 100 mM NaCl, 5 mM MgCl2, 25% glucose, and trolox (238813, Sigma-Aldrich) adding to 1% Gloxy. Gloxy was made of 80% 20 mM Hepes, 20% catalase (v/v; C30, Sigma-Aldrich), and 10 mg of glucose oxidase (G2133, Sigma-Aldrich). Movies were acquired after 5 min of incubation. For all the smCL experiments in this study, at least 18 movies of different imaging areas obtained from a minimum of two independent experiments were analyzed for each condition. Each movie contains 850 frames in total with 30-ms exposure time, with frames 1 through 810 corresponding to protein signals excited by a 639-nm laser, and the remaining 40 ± 10 frames were DNA signals excited by a 532-nm laser. For the competition assays of PARPi and NAD+, 1 or 10 nM PARPi with 100 μM NAD+ was mixed together with fluorescent PARP1 proteins in imaging buffer and then injected into a chamber. All PARPi were dissolved in dimethyl sulfoxide (DMSO) at a 5 mM concentration, aliquoted, and stored at −80°C. Talazoparib (S7048), olaparib (S1060), veliparib (S1004), niraparib (S2741), and rucaparib (S4948) were purchased from Selleck Biochem. EB-47 (E8405) and BAD were purchased from Sigma-Aldrich and Viva Biotech Ltd. (Shanghai), respectively. For the experiments in the presence of 10 μM PARPi, DMSO was added into the reactions of PARP1 only reaction as control. For all PARP1 retention measurements as a function of PARPi, we also measured the effect of the NAD+ analog, BAD, at comparable concentrations to PARPi. As expected, addition of BAD (shown in fig. S14) did not result in any increase in retention, as inhibition of PARP1 can only be achieved when BAD is at equimolar with or higher concentrations than NAD+ (40).
Single-molecule dissociation assay
Biotinylated Cy3-labeled DNA molecules were diluted to 20 to 50 pM using imaging buffer and tethered on the surface of the chamber, followed by washing off the excessive ones. Fluorescent PARP1 proteins were injected into the chamber. Excess proteins were washed off after 5 min of incubation. After buffer or buffer containing 100 μM NAD+ was injected into the chamber, three consecutive short movies (each less than 5 s) of different imaging areas were acquired immediately and correspond to time zero (0 min). Each movie contained 150 frames in total with 30-ms exposure time, with frames 1 through 110 corresponding to protein signals excited by a 639-nm laser, and the remaining 40 ± 10 frames were DNA signals excited by a 532-nm laser. Three movies were acquired every 3 min over a span of 12 min.
Single-molecule FRET assay
Biotinylated Cy3 and Cy5 dual-labeled DNA molecules were diluted to 20 to 50 pM using imaging buffer and tethered on the surface of the chamber followed by washing off the excess. Unconjugated Halo-PARP1 protein was injected into the chamber using PARP1 imaging buffer, followed by a 5-min incubation. DNA molecules were excited by a 532-nm laser. Ten movies of different imaging areas obtained from two independent experiments were analyzed for each condition. Each movie contains 1000 frames in total with 30-ms exposure time.
Single-molecule data extraction
Before the smCL/FRET experiments, the donor and acceptor channels were aligned by registering the fluorescent beads, presenting in both channels (T7279, Thermo Fisher Scientific), via polynomial fit as previously described (39). For the smCL/FRET experiments, the localizations of each single-molecule point spread function (PSF) in each of the donor and acceptor channel were first identified by their maximum projection. A colocalization/FRET pair was then identified if a localization pair from the two channels is no more than two pixels apart from each other. To read the PSF trajectory of each single molecule, a 7 pixels × 7 pixels × 1000 frame stack around each of the single-molecule localization was cropped for analyses. At each time point (frame), the median of the edge pixels of the 7 pixels × 7 pixels was read as the background, and the PSF intensity was read as the mean value of the central 3 pixels × 3 pixels subtracted by the background.
Single-molecule FRET data analysis
FRET efficiency (EFRET) was estimated as previously described (39) on the basis of the ratio of the acceptor intensity to the total intensity of donor and acceptor. In fig. S12, 300 FRET trajectories from duplicate experiments in each condition were submitted for hidden Markov model (HMM) analysis (41). Note that instead of arbitrarily assigning a known number state for HMM analysis, one to four states were attempted for each trajectory and the best-fitted number of states was determined via maximum evidence (42). The presence of the FRET states was histogrammed from the HMM-idealized FRET trajectories and normalized by the number of molecules.
smCL data analysis and quantification of retention
PARP1-DNA colocalization efficiencies were defined as the ratio of PARP1-DNA colocalized pairs to the total number of DNA molecules in the first 10 frames. The trajectories of smCL at equilibrium were classified into two categories, persistent binding events and intermittent binding events, on the basis of whether the intensity of CF647 signals are above zero during all the analyzed frames or not.
Quantification of retention efficiency (raw retention)
Retention efficiency was defined as the ratio of persistent-bound molecules to the total number of colocalized PARP1-DNA pairs. These provide the raw retention efficiency as the percentage of persistently bound molecules for each of the PARP1 mutants and directly show how these values compare to the WT. These measurements consistently showed that the levels of E988A and D766/770A are similar to that of the WT, whereas W318R shows reduced retention, while R591A shows substantial reduction in retention as compared to the WT (figs. S7B, S8D, and S10B and Fig. 1G).
Quantification of normalized retention (relative retention)
To accurately show the specific changes in retention of PARP1 and PARP1 mutants for the different PARPi, we plotted these values as percent change in retention from the mutant’s “baseline” retention value that was measured in the presence of DMSO or NAD+. These plots provide a corrected normalized scale that captures changes in retention efficiency relative to PARP1 and PARP1 mutant’s baseline retention, showing the exact effect of PARPi on retention that is specific for each mutant. Specifically, in the experiments, in the absence of NAD+, the retention efficiency in the presence of different PARPi was normalized by the mean of its corresponding retention efficiency of DMSO control (figs. S6; S7C; S8, A and B; and S10C). In the experiments, in the presence of NAD+, the retention efficiency in the presence of different PARPi and NAD+ was normalized by the mean of its corresponding retention efficiency of NAD+ only control.
Except for the analyses in figs. S5C and S11A, the cutoff of the trajectories in all other analyses at equilibrium was 24 s (800 frames × 30 ms). The results in figs. S5C and S11A showed that similar trends were obtained, while the cutoffs were less than 24 s. At least 2500 colocalized PARP1-DNA pairs in at least 18 movies from experiments performed in duplicate or triplet were analyzed for each condition. Two-sample t test was performed to compare the retention efficiency of two populations in different experimental conditions.
Acknowledgments
We thank S. Zha, T. Huang, and members of the Rothenberg laboratory for helpful comments and suggestions. G. Rona (New York University Grossman School of Medicine) provided the pBABE_Halo-PARP1_puro vector.
Funding: This work was supported by NIH grants: R35 GM134947, AI153040, and CA247773 (to E.R.), and the V Foundation BRCA Research collaborative grant (to E.R.) and Pfizer.
Author contributions: H.X. and E.R. designed the experiments, H.X., Y.Y., and E.R. wrote the paper. L.Z., X.D., and T.L.V. participated in discussions of experiment designs during the project and provided suggestions. J.M.P provided the structural models and suggestions and edited the paper. H.X. performed the single-molecule experiments. A.B. and H.X. purified the proteins. H.X. and A.E. generated the plasmids. C.F. designed and provided the SSB-DNA-smFRET substrate. H.X. and E.R. analyzed the data. Y.Y., E.R., and A.B. wrote the scripts for single-molecule data analysis.
Competing interests: E.R. received research funding from Pfizer. E.R. received honoraria payment from Ionis Pharmaceuticals. L.Z., X.D., and T.L.V. are employees of Pfizer. J.M.P. has served as a scientific consultant for XinThera Inc. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Tables S1 and S2
Figs. S1 to S15
REFERENCES AND NOTES
- 1.Ray Chaudhuri A., Nussenzweig A., The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 18, 610–621 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin-Hernandez K., Rodriguez-Vargas J. M., Schreiber V., Dantzer F., Expanding functions of ADP-ribosylation in the maintenance of genome integrity. Semin. Cell Dev. Biol. 63, 92–101 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Krishnakumar R., Kraus W. L., The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol. Cell 39, 8–24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lüscher B., Ahel I., Altmeyer M., Ashworth A., Bai P., Chang P., Cohen M., Corda D., Dantzer F., Daugherty M. D., Dawson T. M., Dawson V. L., Deindl S., Fehr A. R., Feijs K. L. H., Filippov D. V., Gagne J. P., Grimaldi G., Guettler S., Hoch N. C., Hottiger M. O., Korn P., Kraus W. L., Ladurner A., Lehtio L., Leung A. K. L., Lord C. J., Mangerich A., Matic I., Matthews J., Moldovan G. L., Moss J., Natoli G., Nielsen M. L., Niepel M., Nolte F., Pascal J., Paschal B. M., Pawlowski K., Poirier G. G., Smith S., Timin zky G., Wang Z. Q., Yelamos J., Yu X., Zaja R., Ziegler M., ADP-ribosyltransferases, an update on function and nomenclature. FEBS J. 10.1111/febs.16142 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langelier M. F., Pascal J. M., PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Curr. Opin. Struct. Biol. 23, 134–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langelier M. F., Planck J. L., Roy R., Pascal J. M., Structural basis for DNA damage–dependent poly(ADP-ribosyl)ation by human PARP-1. Science 336, 728–732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langelier M. F., Eisemann T., Riccio A. A., Pascal J. M., PARP family enzymes: Regulation and catalysis of the poly(ADP-ribose) posttranslational modification. Curr. Opin. Struct. Biol. 53, 187–198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satoh M. S., Lindahl T., Role of poly(ADP-ribose) formation in DNA repair. Nature 356, 356–358 (1992). [DOI] [PubMed] [Google Scholar]
- 9.Kim J. H., Kim B., Cai L., Choi H. J., Ohgi K. A., Tran C., Chen C., Chung C. H., Huber O., Rose D. W., Sawyers C. L., Rosenfeld M. G., Baek S. H., Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Bryant H. E., Schultz N., Thomas H. D., Parker K. M., Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Rouleau M., Patel A., Hendzel M. J., Kaufmann S. H., Poirier G. G., PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer 10, 293–301 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zandarashvili L., Langelier M. F., Velagapudi U. K., Hancock M. A., Steffen J. D., Billur R., Hannan Z. M., Wicks A. J., Krastev D. B., Pettitt S. J., Lord C. J., Talele T. T., Pascal J. M., Black B. E., Structural basis for allosteric PARP-1 retention on DNA breaks. Science 368, eaax6367 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pommier Y., O’Connor M. J., de Bono J., Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci. Transl. Med. 8, 362ps17 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Fong P. C., Boss D. S., Yap T. A., Tutt A., Wu P., Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 361, 123–134 (2009). [DOI] [PubMed] [Google Scholar]
- 15.de Bono J., Mateo J., Fizazi K., Saad F., Shore N., Sandhu S., Chi K. N., Sartor O., Agarwal N., Olmos D., Thiery-Vuillemin A., Twardowski P., Mehra N., Goessl C., Kang J., Burgents J., Wu W., Kohlmann A., Adelman C. A., Hussain M., Olaparib for metastatic castration-resistant prostate cancer. N. Engl. J. Med. 382, 2091–2102 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Moore K., Colombo N., Scambia G., Kim B. G., Oaknin A., Friedlander M., Lisyanskaya A., Floquet A., Leary A., Sonke G. S., Gourley C., Banerjee S., Oza A., Gonzalez-Martin A., Aghajanian C., Bradley W., Mathews C., Liu J., Lowe E. S., Bloomfield R., DiSilvestro P., Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 379, 2495–2505 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Litton J. K., Rugo H. S., Ettl J., Hurvitz S. A., Goncalves A., Lee K. H., Fehrenbacher L., Yerushalmi R., Mina L. A., Martin M., Roche H., Im Y. H., Quek R. G. W., Markova D., Tudor I. C., Hannah A. L., Eiermann W., Blum J. L., Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N. Engl. J. Med. 379, 753–763 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robson M., Im S. A., Senkus E., Xu B., Domchek S. M., Masuda N., Delaloge S., Li W., Tung N., Armstrong A., Wu W., Goessl C., Runswick S., Conte P., Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N. Engl. J. Med. 377, 523–533 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Golan T., Hammel P., Reni M., Van Cutsem E., Macarulla T., Hall M. J., Park J. O., Hochhauser D., Arnold D., Oh D. Y., Reinacher-Schick A., Tortora G., Algul H., O’Reilly E. M., McGuinness D., Cui K. Y., Schlienger K., Locker G. Y., Kindler H. L., Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med. 381, 317–327 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murai J., Huang S. Y., Das B. B., Renaud A., Zhang Y., Doroshow J. H., Ji J., Takeda S., Pommier Y., Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 72, 5588–5599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murai J., Huang S. Y., Renaud A., Zhang Y., Ji J., Takeda S., Morris J., Teicher B., Doroshow J. H., Pommier Y., Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol. Cancer Ther. 13, 433–443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopkins T. A., Shi Y., Rodriguez L. E., Solomon L. R., Donawho C. K., DiGiammarino E. L., Panchal S. C., Wilsbacher J. L., Gao W., Olson A. M., Stolarik D. F., Osterling D. J., Johnson E. F., Maag D., Mechanistic dissection of PARP1 trapping and the impact on in vivo tolerability and efficacy of PARP inhibitors. Mol. Cancer Res. 13, 1465–1477 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Rudolph J., Mahadevan J., Dyer P., Luger K., Poly(ADP-ribose) polymerase 1 searches DNA via a “monkey bar” mechanism. eLife 7, e37818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shao Z., Lee B. J., Rouleau-Turcotte E., Langelier M. F., Lin X., Estes V. M., Pascal J. M., Zha S., Clinical PARP inhibitors do not abrogate PARP1 exchange at DNA damage sites in vivo. Nucleic Acids Res. 48, 9694–9709 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettitt S. J., Krastev D. B., Brandsma I., Drean A., Song F., Aleksandrov R., Harrell M. I., Menon M., Brough R., Campbell J., Frankum J., Ranes M., Pemberton H. N., Rafiq R., Fenwick K., Swain A., Guettler S., Lee J. M., Swisher E. M., Stoynov S., Yusa K., Ashworth A., Lord C. J., Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat. Commun. 9, 1849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogden T. E. H., Yang J. C., Schimpl M., Easton L. E., Underwood E., Rawlins P. B., McCauley M. M., Langelier M. F., Pascal J. M., Embrey K. J., Neuhaus D., Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition. Nucleic Acids Res. 49, 2266–2288 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marsischky G. T., Wilson B. A., Collier R. J., Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation. Evidence for active site similarities to the ADP-ribosylating toxins. J. Biol. Chem. 270, 3247–3254 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Xie N., Zhang L., Gao W., Huang C., Huber P. E., Zhou X., Li C., Shen G., Zou B., NAD+ metabolism: Pathophysiologic mechanisms and therapeutic potential. Signal Transduct. Target. Ther. 5, 227 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cambronne X. A., Stewart M. L., Kim D., Jones-Brunette A. M., Morgan R. K., Farrens D. L., Cohen M. S., Goodman R. H., Biosensor reveals multiple sources for mitochondrial NAD+. Science 352, 1474–1477 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryan K., Bolanos B., Smith M., Palde P. B., Cuenca P. D., VanArsdale T. L., Niessen S., Zhang L., Behenna D., Ornelas M. A., Tran K. T., Kaiser S., Lum L., Stewart A., Gajiwala K. S., Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1. J. Biol. Chem. 296, 100251 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma D., De Falco L., Padavattan S., Rao C., Geifman-Shochat S., Liu C. F., Davey C. A., PARP1 exhibits enhanced association and catalytic efficiency with gammaH2A.X-nucleosome. Nat. Commun. 10, 5751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krastev D. B., Li S., Sun Y., Wicks A. J., Hoslett G., Weekes D., Badder L. M., Knight E. G., Marlow R., Pardo M. C., Yu L., Talele T. T., Bartek J., Choudhary J. S., Pommier Y., Pettitt S. J., Tutt A. N. J., Ramadan K., Lord C. J., The ubiquitin-dependent ATPase p97 removes cytotoxic trapped PARP1 from chromatin. Nat. Cell Biol. 24, 62–73 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun F. H., Zhao P., Zhang N., Kong L. L., Wong C. C. L., Yun C. H., HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones. Nat. Commun. 12, 1028 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suskiewicz M. J., Zobel F., Ogden T. E. H., Fontana P., Ariza A., Yang J. C., Zhu K., Bracken L., Hawthorne W. J., Ahel D., Neuhaus D., Ahel I., HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation. Nature 579, 598–602 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langelier M. F., Billur R., Sverzhinsky A., Black B. E., Pascal J. M., HPF1 dynamically controls the PARP1/2 balance between initiating and elongating ADP-ribose modifications. Nat. Commun. 12, 6675 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rudolph J., Roberts G., Luger K., Histone parylation factor 1 contributes to the inhibition of PARP1 by cancer drugs. Nat. Commun. 12, 736 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langelier M. F., Planck J. L., Servent K. M., Pascal J. M., Purification of human PARP-1 and PARP-1 domains from Escherichia coli for structural and biochemical analysis. Methods Mol. Biol. 780, 209–226 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Brinkley M., A brief survey of methods for preparing protein conjugates with dyes, haptens, and cross-linking reagents. Bioconjug. Chem. 3, 2–13 (1992). [DOI] [PubMed] [Google Scholar]
- 39.Yin Y., Lee W. T. C., Gupta D., Xue H., Tonzi P., Borowiec J. A., Huang T. T., Modesti M., Rothenberg E., A basal-level activity of ATR links replication fork surveillance and stress response. Mol. Cell 81, 4243–4257.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langelier M. F., Zandarashvili L., Aguiar P. M., Black B. E., Pascal J. M., NAD+ analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains. Nat. Commun. 9, 844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKinney S. A., Joo C., Ha T., Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 91, 1941–1951 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bronson J. E., Fei J., Hofman J. M., Gonzalez R. L. Jr., Wiggins C. H., Learning rates and states from biophysical time series: A Bayesian approach to model selection and single-molecule FRET data. Biophys. J. 97, 3196–3205 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2
Figs. S1 to S15