Abstract
Phosphoinositides (PIs) are lipid signaling molecules that operate by recruiting proteins to cellular membranes via PI recognition domains. The dominant PI of the plasma membrane is phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2]. One of only two PI(4,5)P2 recognition domains characterized in detail is the tubby domain. It is essential for targeting proteins into cilia involving reversible membrane association. However, the PI(4,5)P2 binding properties of tubby domains have remained enigmatic. Here, we used coarse-grained molecular dynamics simulations to explore PI(4,5)P2 binding by the prototypic tubby domain. The comparatively low PI(4,5)P2 affinity of the previously described canonical binding site is underpinned in a cooperative manner by a previously unknown, adjacent second binding site. Mutations in the previously unknown site impaired PI(4,5)P2-dependent plasma membrane localization in living cells and PI(4,5)P2 interaction in silico, emphasizing its importance for PI(4,5)P2 affinity. The two-ligand binding mode may serve to sharpen the membrane association-dissociation cycle of tubby-like proteins that underlies delivery of ciliary cargo.
A previously unknown membrane binding mechanism for decoding of spatiotemporal phospholipid signals in cells is identified.
INTRODUCTION
Phosphoinositides (PIs) have multifaceted signaling functions, acting as a cellular membrane identity code (1, 2) and providing instructive molecular signals in various pathways and membrane dynamics. Prominent examples are the generation of phosphatidylinositol 3,4,5-trisphosphate (PIP3) downstream of growth factor receptors and the depletion of phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] by phospholipase C–β (PLCβ) downstream of Gq-coupled receptors. In general, the impact of PIs on cellular processes is mediated by the binding of proteins to the membrane via PI recognition domains including pleckstrin homology (PH), phox (PX), Fab1-YOTB-Vac1-EEA1 (FYVE), and epsin N-terminal homology (ENTH) domains among others [reviewed in (1, 3–5)].
Beyond their eminent role in cell biology, ligand-specific PI-binding domains have emerged as indispensable biosensors for their cognate PI lipid in living cells (4). Encoded genetically to yield fusions with fluorescent (or luminescent) modules such as green fluorescent protein (GFP), they are used widely to interrogate PI cell biology in model systems across a range of biological complexity from isolated membrane fragments (6) to intact living animals (7).
PI(4,5)P2 is the most abundant PI of the plasma membrane (PM) and, besides being a precursor of the canonical second messengers PIP3, inositol 1,4,5-trisphosphate (IP3), and diacylglycerol, has multiple roles as bona fide messenger (3). Nevertheless, only few specific PI(4,5)P2 binding domains have been identified (4), and fewer have been characterized in depth (8–10). The first lipid-binding domain used as a biosensor is the PI(4,5)P2-specific PH domain of phospholipase C–δ1 (PLCδ1-PH) (11, 12) that has been used in countless studies (4). The only alternative PI(4,5)P2 sensor used fairly frequently is the C-terminal domain of the tubby protein (“tubby domain,” hereafter abbreviated tubbyCT), which is conserved in the family of tubby-like proteins (TULPs) (13). Despite being structurally unrelated, the binding mode of PLCδ1-PH domain and tubbyCT is highly similar as shown by crystal structures (13, 14). Both domains feature a positively charged binding grove that accommodates the inositol head group by electrostatic coordination of the phosphate groups. In the crystal structures, both domains tightly bind soluble head group analogs (IP3 or glycero-IP3). Physiologically, PLCδ1-PH also has a high affinity for the second messenger IP3, and therefore, its cellular localization is effectively defined by competition between PI(4,5)P2 in the PM and cytosolic IP3, such that an increase in IP3 [e.g., during G protein–coupled receptor (GPCR) signaling] is sufficient to dislodge PLCδ1-PH from the membrane without depletion of PI(4,5)P2 (15). In contrast, membrane binding of tubbyCT is unaffected by IP3 (9, 16), which seems unexpected given the similar head group interactions.
The differential IP3 recognition must affect fundamentally on the cell biology of the PI(4,5)P2 recognition domains. While PLCδ1-PH function is intrinsically sensitive to IP3-producing Gq/PLCβ pathways (11, 17), the IP3-insensitive tubbyCT determines function in a manner selectively dependent on local PI(4,5)P2 concentration and dynamics. Tubby and related TULPs mediate trafficking of cargo proteins including GPCRs from PI(4,5)P2-rich PM domains into primary cilia that are sparse in PI(4,5)P2. This allows dissociation of tubby and thus release of the cargo GPCR at its ciliary destination (18, 19). Various ciliopathies have been linked to dysfunction of TULPs, underscoring the central importance of dynamic PI(4,5)P2 binding of the tubby domain for ciliary function. Mutational disruption of the PI(4,5)P2-binding C-terminal domain of tubby in a mouse model leads to severe obesity and retinal degeneration, which is thought to result from defective ciliary function in the hypothalamus (pivotal for regulation of energy balance) and the retina, respectively (20–22). In humans, a mutation within the tubby domain is associated with early onset obesity and retinal degeneration (23). Recently, mutations in human TULP3, a tubby homolog that shares the conserved tubby domain, were found to cause multisystemic ciliopathy associated with liver fibrosis, fibrocystic kidney disease, and cardiomyopathy (24).
In addition to ciliary localization, we recently found preferential and dynamic accumulation of tubby in PI(4,5)P2 microdomains formed by endoplasmic reticulum (ER)–PM junctions, a property not shared by PLCδ1-PH (25).
These differential membrane binding properties and their important cell biological implications prompted us to explore binding of tubbyCT to PI(4,5)P2 using coarse-grained (CG) molecular dynamics (MD) simulations with the Martini 3 force field (26, 27). CG MD simulations are a suitable and well-established tool to study protein-lipid interactions (28) and specifically protein-PI interactions (29–34). Our simulations of membrane binding showed that tubbyCT’s PI(4,5)P2 binding affinity is lower than that of PLCδ1-PH, confirming previous experimental data (8, 35). Unexpectedly, the MD data revealed a second PI(4,5)P2 binding site within tubbyCT’s membrane-oriented surface, comprising a cluster of positively charged residues. Mutations within this second binding site strongly reduced PI(4,5)P2 binding both in silico and experimentally in living cells, demonstrating that it contributes essentially to the PI(4,5)P2-dependent membrane association of tubby. The positively charged cluster is conserved throughout TULP family, indicating that simultaneous PI(4,5)P2 binding by two binding sites is a conserved feature of TULPs. The second binding site constrains high affinity binding to the PI(4,5)P2-containing membrane surface and may explain the lack of sensitivity to soluble IP3. Moreover, cooperative PI(4,5)P2 binding can explain differential association to membrane microdomains that differ in PI(4,5)P2 content. This property may facilitate delivery of cargo into primary cilia by TULPs, by ensuring strong binding to the PI(4,5)P2-rich PM but brisk dissociation at lowered PI(4,5)P2 level of the ciliary membrane.
RESULTS
TubbyCT requires interaction with multiple PI(4,5)P2 lipids for stable membrane binding
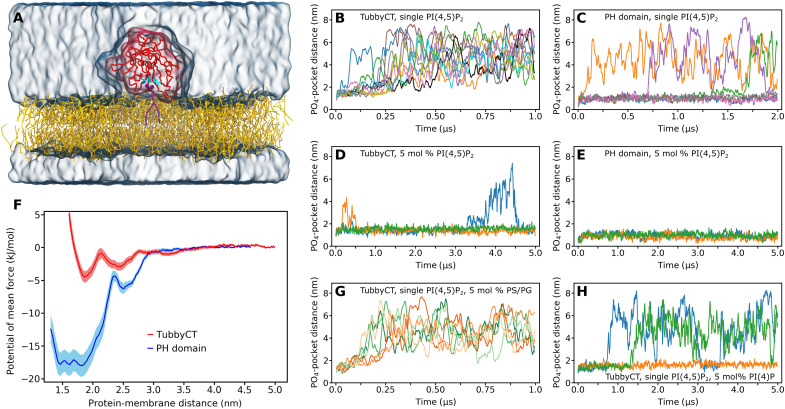
To gain insights into the PI(4,5)P2 binding behavior of tubbyCT, we first performed CG MD simulations of tubbyCT bound to a single PI(4,5)P2 embedded in a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer using the open-beta version of the Martini 3 force field (Fig. 1A). Figure 1B shows the time evolution of the distance between the phosphate (PO4) plane of the binding leaflet and the previously characterized binding pocket (13) for 10 simulations of 1 μs each. It can be clearly seen that tubbyCT unbinds from the membrane and hence from the PI(4,5)P2 lipid in all cases within 250 ns. In addition, no stable rebinding event to the membrane is observed within the simulation time.
Fig. 1. Binding a single PI(4,5)P2 lipid does not target tubbyCT stably to a model membrane.
(A) CG system setup of tubbyCT (red) with one PI(4,5)P2 lipid (violet) in the binding pocket known from the crystal structure (cyan residues). The PI(4,5)P2 is embedded in a POPC bilayer (yellow); water and ions are shown as transparent surface. (B and C) Distance between the tubbyCT/PLCδ1-PH domain binding pocket and the phosphate layer (PO4 beads) of the binding leaflet containing a single PI(4,5)P2. Ten unbiased simulations of 1 μs [tubbyCT (B)] and seven unbiased simulations of 2 μs [PLCδ1-PH (C)] are shown, respectively. (D and E) Distance between the tubbyCT/PLCδ1-PH domain binding pocket and the phosphate layer (PO4 beads) of the binding leaflet containing 5 mol % of PI(4,5)P2. Three unbiased simulations of 5 μs each are shown. (F) Potential of mean force (PMF) for the PI(4,5)P2 binding of tubbyCT (red) and PLCδ1-PH domain (blue). (G) Control simulations of tubbyCT bound to one PI(4,5)P2 lipid embedded in a POPC membrane containing 5 mol % of POPS (PS) (green; 3 × 1 μs) and POPG (PG) (orange; 3 × 1 μs) lipids, respectively. (H) Control simulations of tubbyCT bound to one PI(4,5)P2 lipid embedded in a POPC membrane containing 5 mol % of PI(4)P (3 × 5 μs).
As a reference for PI(4,5)P2 binding proteins, we also performed simulations of the PLCδ1-PH domain. Figure 1C shows the time evolution of the PO4-binding pocket distance for seven simulations of 2 μs each. The PLCδ1-PH domain binds much more stably to the PI(4,5)P2 lipid. Despite the doubled simulation time, unbinding was only observed in three cases, and one rebinding event occurred.
To clarify whether an increased PI(4,5)P2 concentration affects the binding stability of both proteins, we increased the PI(4,5)P2 concentration in the bilayer to 5 mole percent (mol %) and performed three simulations of 5 μs each. The observed time evolution of the PO4-binding pocket distance is shown in Fig. 1D (tubbyCT) and Fig. 1E (PLCδ1-PH domain), respectively. Both PI(4,5)P2 sensors bind more stably to the membrane containing 5 mol % of PI(4,5)P2. However, the increase in binding stability is more pronounced in the case of tubbyCT, where a single PI(4,5)P2 was not sufficient to target the protein for more than 250 ns to the bilayer surface. Moreover, observed unbinding events of tubbyCT from the membrane with high PI(4,5)P2 concentration are only transient and followed by rebinding.
We next asked whether the increased membrane binding is specific for higher PI(4,5)P2 concentrations or whether any negatively charged lipid could support the binding of tubbyCT to a single PI(4,5)P2. This is particularly relevant because the inner leaflet of the PM contains several negatively charged lipid species (36, 37). We decided to test three phospholipids abundant in the eukaryotic PM, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG), as well as the mono-phosphorylated PI phosphatidylinositol 4-phosphate [PI(4)P]. The PI(4,5)P2 bound to tubbyCT was embedded in a POPC bilayer containing 5 mol % of POPS, POPG, or PI(4)P. Figure 1G shows the PO4-binding pocket distance for three simulations of 1 μs with POPS (green) and POPG (orange), respectively. In each case, no additional stabilization of tubbyCT membrane binding was observed. PI(4)P clearly increased membrane binding (Fig. 1H); however, the stabilization was less pronounced than with the same concentration of PI(4,5)P2. In addition, we tested a 5 mol % fraction of POPG with doubly negatively charged head group, which showed minor additional stabilization compared to regular POPG and less stabilization than PI(4)P (fig. S1).
To quantitatively compare the PI(4,5)P2 affinity of the two proteins, we calculated the potential of mean force (PMF) for the protein-PI(4,5)P2 binding. The resulting free energy profiles (Fig. 1F) confirm that tubbyCT binds much weaker to PI(4,5)P2. The PMF minima differ by more than a factor of 3 (tubbyCT, −5.0 kJ/mol; PLCδ1-PH domain, −18.0 kJ/mol). The total binding free energy ΔGbind can be calculated by integrating the PMF profile over the region of the bound state while taking into account the constraints in the membrane plane (38). This yields a ΔGbind = −2.4 kJ/mol for tubbyCT and ΔGbind = −15.8 kJ/mol for the PLCδ1-PH domain. Notably, the differential binding energies qualitatively agree with experimental data where PI(4,5)P2 affinities were estimated in living cells from the domain’s membrane unbinding in response to gradual depletion of PI(4,5)P2 with a voltage-sensitive phosphatase (VSP) (8, 35).
Identification of a previously unknown PI(4,5)P2 binding site of tubbyCT
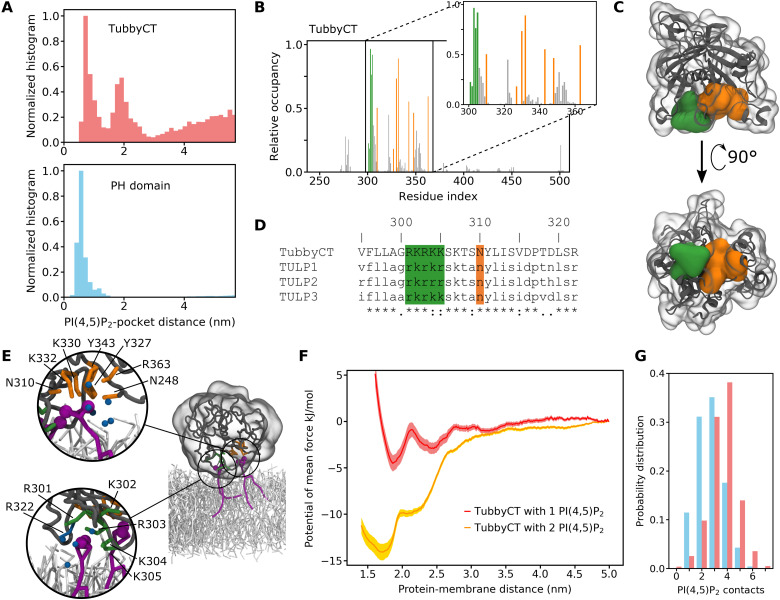
The CG MD simulations allow a microscopic investigation of the membrane binding behavior of tubbyCT and PLCδ1-PH domain. Figure 2A depicts a normalized histogram of the distance between the PI(4,5)P2 head group and the crystal structure binding pocket for the simulations with a single PI(4,5)P2 embedded in a POPC bilayer. Note that this distance is different from the distances depicted in Fig. 1 (B to E, G, and H) where the distance between the PO4 plane of the binding leaflet and the binding pocket was analyzed. The distance to the head group shown here provides more detailed microscopic information about the protein-PI(4,5)P2 contacts.
Fig. 2. Identification of the second PI(4,5)P2 binding hotspot of tubbyCT.
(A) Normalized distribution for the distance between the PI(4,5)P2 head group and the crystal structure binding pocket of tubbyCT (top) and PLCδ1-PH domain (bottom). The simulations were performed with one PI(4,5)P2 in the bilayer. (B) Relative PI(4,5)P2 occupancy of the tubbyCT residues calculated with a distance cutoff of 0.5 nm. Orange bars represent residues of the crystal structure binding pocket; green bars highlight residues of the binding hotspot identified here; gray bars represent all other residues. (C) Crystal structure of tubbyCT including the modeled loops. The surface of the crystal structure binding pocket is shown in orange; the surface of the previously unknown binding hotspot is shown in green. (D) Sequence alignment of tubbyCT with other proteins of the TULP family; color code according to (B). (E) Representative snapshot showing the binding mode of two PI(4,5)P2 lipids in the binding pockets of tubbyCT. The magnified details show the binding mode in the crystal structure binding site (top) and the previously unknown second binding site (bottom). Additional particles close to the coordinated PI(4,5)P2 lipid are colored in dark cyan [six water beads (top); four water beads and R322 (bottom)]. Protein colors are the same as in (D); PI(4,5)P2 is colored purple, and POPC is colored light gray. (F) PMF for the binding of tubbyCT to a membrane with one PI(4,5)P2 lipid (red) and two PI(4,5)P2 lipids (orange), respectively. (G) Probability distribution of the simultaneous PI(4,5)P2-protein contacts for tubbyCT (red) and PLCδ1-PH domain (blue) binding to a membrane with 5 mol % of PI(4,5)P2.
The difference in height of the distributions at long distance is due to the different PI(4,5)P2 binding affinity of tubbyCT (Fig. 2A, top) and PLCδ1-PH domain (Fig. 2A, bottom). Notably, the histogram for tubbyCT clearly shows two distinct maxima at which a stabilizing interaction exists, while in the case of the PLCδ1-PH domain, only one maximum appears. The two maxima in the case of tubbyCT coincide with the two minima of the PMF (red line in Fig. 1F). Despite both reaction coordinates not being identical, they are closely related, and at the second minimum of the PMF, tubbyCT mostly interacts with PI(4,5)P2 by means of residues not being part of the canonical binding site.
The second maximum indicated an additional PI(4,5)P2 binding site on the tubbyCT surface. To evaluate this more closely, we calculated the average PI(4,5)P2 occupancy of each residue of tubbyCT for the simulations in which tubbyCT interacted with a membrane containing 5 mol % of PI(4,5)P2 (Fig. 2B). The orange bars highlight the residues of the binding pocket previously identified in the crystal structure (13). Besides those residues, there are five consecutive positively charged residues (amino acids 301 to 305, colored in green) of which, in particular, residues 303 to 305 exhibit a notably high PI(4,5)P2 occupancy. Accordingly, these residues constitute the previously unknown second binding site identified in the PI(4,5)P2 head group canonical binding site distance histogram (Fig. 2A). The sequence alignment of TULPs depicted in Fig. 2D reveals that these five positively charged residues are conserved within the TULP family. The newly identified binding site is located in proximity to the crystal structure binding pocket, and both PI(4,5)P2 binding sites are oriented in parallel so that simultaneous PI(4,5)P2 binding is possible (see Fig. 2, C and E). Thus, the binding affinities of both binding sites act together and target tubbyCT to the membrane.
The PMF for tubbyCT binding to two PI(4,5)P2 lipids (Fig. 2F) shows a strong increase in binding affinity compared to a single PI(4,5)P2. In addition, the minimum is shifted closer to the membrane, to a distance of 1.7 nm [one PI(4,5)P2, 1.9 nm] indicative for reinforced binding. Two exemplary snapshots of tubbyCT binding to one and two PI(4,5)P2 lipids, respectively, are depicted in fig. S2. The snapshots are taken from the simulation at the respective minimum. Integration of the PMF yields a binding free energy of ΔGbind = −12.5 kJ/mol compared to the previously obtained of ΔGbind = −2.4 kJ/mol. The fivefold binding free energy indicates cooperativity between the two binding sites. For noncooperative binding, a maximum of a doubling of ΔGbind would be expected.
As a consequence of the two binding sites oriented toward the membrane, we observed a strong enrichment of PI(4,5)P2 close to tubbyCT. Figure 2G depicts the probability distribution of the number of simultaneous protein-PI(4,5)P2 contacts with a distance of ≤0.5 nm for tubbyCT (red) and the PLCδ1-PH domain (blue) when binding to a membrane with 5 mol % of PI(4,5)P2. Multiple contacts to the same lipid are only counted once. It can be clearly seen that in the case of tubbyCT, the distribution exhibits a higher number of protein-PI(4,5)P2 contacts. The average number of PI(4,5)P2 molecules in direct contact with tubbyCT shows a PI(4,5)P2 enrichment of 44% (distance, ≤0.5 nm; Table 1). It decreases to 19% for a distance of ≤0.9 nm, which is still about four times higher than the PI(4,5)P2 concentration in the membrane (5%). Thus, the lipid fingerprint—i.e., the lipid composition of the annular lipid shell—of tubbyCT is highly enriched of PI(4,5)P2, while POPC is depleted. Similarly, the lipid fingerprint of PLCδ1-PH is enriched with PI(4,5)P2; however, PLCδ1-PH is in contact with fewer PI(4,5)P2 lipids. Another consequence of the membrane binding of tubbyCT is a reduced flexibility of three consecutive loops between residues 300 and 340 (see Supplementary Text and fig. S5), which includes the previously unknown second binding site.
Table 1. Number of lipids in contact with tubbyCT and the PLCδ1-PH domain depending on the cutoff distance.
Abs., absolute; rel., relative.
| TubbyCT | PLCδ1-PH domain | |||||||
| PI(4,5)P2 | POPC | PI(4,5)P2 | POPC | |||||
| Cutoff (nm) | Abs. | Rel. | Abs. | Rel. | Abs. | Rel. | Abs. | Rel. |
| 0.5 | 3.4 ± 0.2 | 0.44 | 4.4 ± 0.4 | 0.56 | 2.7 ± 0.2 | 0.34 | 5.4 ± 0.2 | 0.66 |
| 0.7 | 3.6 ± 0.2 | 0.31 | 8.0 ± 0.7 | 0.69 | 2.9 ± 0.2 | 0.25 | 8.4 ± 0.2 | 0.75 |
| 0.9 | 3.9 ± 0.2 | 0.19 | 16.4 ± 1.0 | 0.81 | 3.1 ± 0.3 | 0.18 | 14.6 ± 1.0 | 0.82 |
| Membrane composition | 18 | 0.05 | 334 | 0.95 | 18 | 0.05 | 334 | 0.95 |
Validation of the secondary binding site with mutants
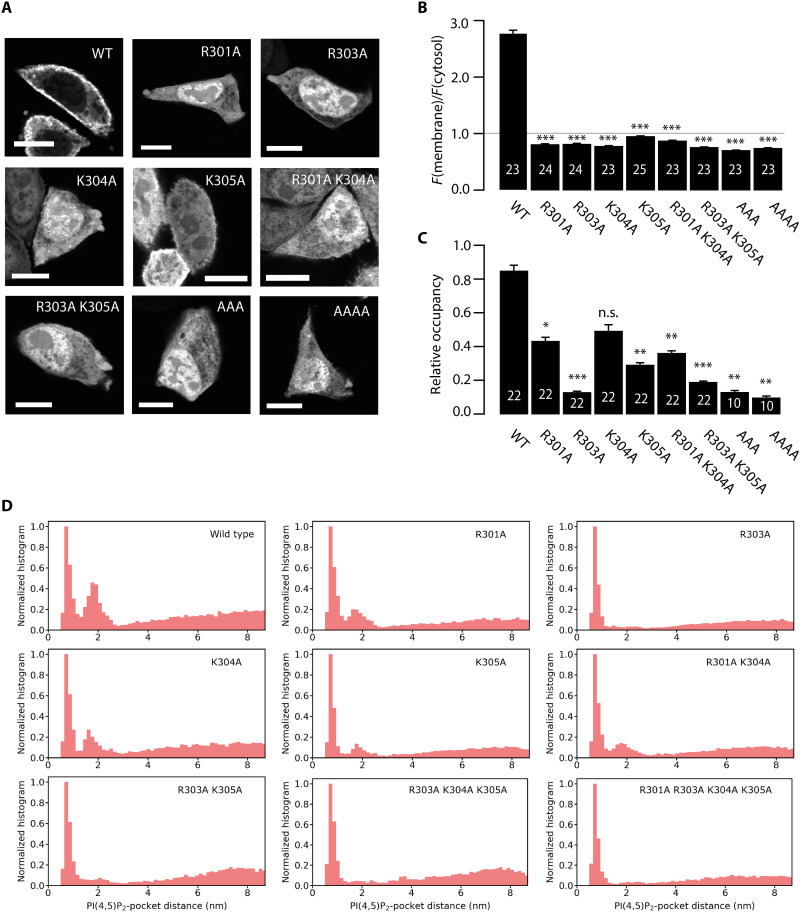
To evaluate the role of the newly identified second binding site in membrane association of tubbyCT in living cells, we mutated the positively charged amino acids 301 to 305 that constitute the previously unknown binding site to alanine, which lacks electrostatic attraction to the anionic PI(4,5)P2. GFP-fused tubbyCT and the various mutants were expressed in Chinese hamster ovary (CHO) cells and examined for their membrane localization by confocal microscopy. As shown in Fig. 3 (A and B), the single point mutants R301A, R303A, and K304A lost the strong membrane association of the wild-type (WT) domain and predominantly localized to the cytoplasm and nucleus. TubbyCT K305A showed a similar phenotype, although this mutant retained distinct but reduced membrane localization. We did not analyze position R302 by mutation as it binds D499 by hydrogen bonding and thus might be important for the tubbyCT secondary structure. Combined neutralization of several of the site’s positive charges even further reduced PM localization (Fig. 3, A and B).
Fig. 3. Mutational analysis of the second binding site.
(A) Representative confocal images of tubbyCT WT and mutants expressed in CHO cells show the different degrees of membrane localization of the constructs. Scale bars, 5 μm. (B) Membrane-to-cytosol fluorescence ratios obtained from images as shown in (A). Localization of the membrane was defined as the local fluorescence maximum of a coexpressed RFP-fused membrane marker. White numbers show the number of analyzed cells, and asterisks indicate significant differences compared to the WT control (***P < 0.0001). (C) Ratio of occupancy of the second binding site relative to the canonical site obtained from occupancy distributions as shown in (D). White numbers show the number of analyzed replicas, and asterisks indicate significant differences compared to the WT control (*P < 0.05, **P < 0.01, and ***P < 0.0001). n.s., not significant. (D) Normalized distribution for the distance between the PI(4,5)P2 head group and the canonical binding pocket of tubbyCT mutants. Simulations were performed with one PI(4,5)P2 in the bilayer.
In a complimentary manner, we investigated PI(4,5)P2 binding of the mutants by CG MD simulations. We simulated 10 replicas of 1 μs simulation time for each tubbyCT mutant where the protein was initially bound to a single PI(4,5)P2 lipid embedded in a POPC bilayer, and for the WT, single, and double mutants, we simulated additional 12 replicas of 0.5-μs simulation time. As a measure for PI(4,5)P2 binding behavior, we calculated the distance between the PI(4,5)P2 head group and the canonical binding site over the duration of each simulation. For tubbyCT WT, the histogram of the resulting distances shows two maxima corresponding to the two binding sites (c.f., Fig. 2A). The occupancy of the second maximum was reduced by the various mutations (Fig. 3D). To estimate the relative binding strength of the second binding site in comparison to the canonical one, we integrated both maxima and calculated their relative occupancy, i.e., the ratio of the integrals of both binding site maxima. While for tubbyCT WT, the relative occupancy is >0.8, all mutants show a reduced relative occupancy with the triple and quadruple mutants exhibiting the lowest relative occupancy close to zero (Fig. 3C).
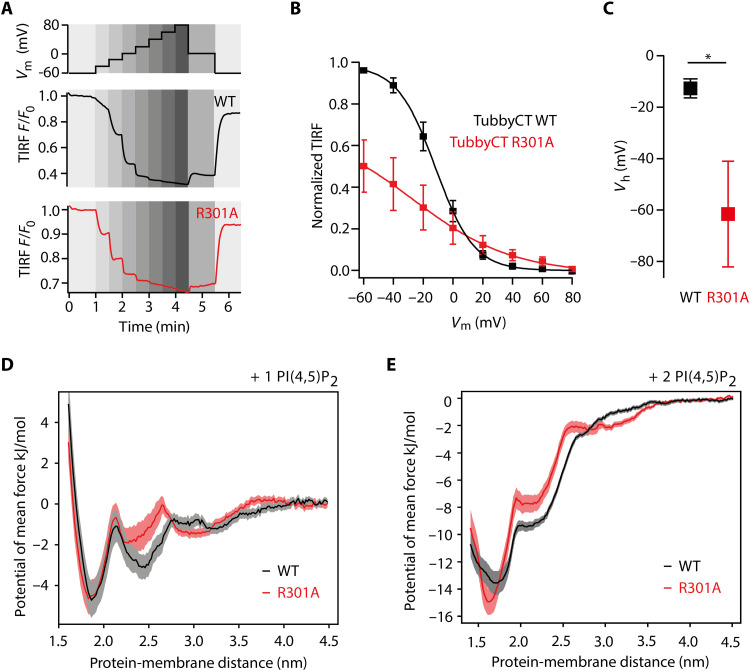
Next, we chose tubbyCT R301A as a representative mutant for a more detailed experimental characterization of the PI(4,5)P2 binding affinity in living cells. To this end, we used the VSP (from Ciona intestinalis; Ci-VSP) for gradual and stepwise change of the PM level of PI(4,5)P2. VSPs are 5-phosphatases that dephosphorylate PI(4,5)P2 to PI(4)P with a gradual dependency of their enzymatic activity on the membrane potential, where depolarization increases the activity. As the PI(4,5)P2 concentration depends on the counteracting activities of the VSP and intrinsic PI(4)P 5-kinase, a stepwise activation of Ci-VSP (Fig. 4A, top) allows for titration of PI(4,5)P2 levels [cf., (8, 35, 39)]. To this end, we coexpressed the respective GFP-tubbyCT mutant or WT together with red fluorescent protein (RFP)–tagged Ci-VSP in CHO cells. RFP-positive cells were whole-cell voltage-clamped, while the membrane association of GFP-tubbyCT constructs was monitored by total internal reflection fluorescence (TIRF) microscopy. As shown in Fig. 4A, incremental depolarization from −60 to +80 mV induced progressive dissociation of tubbyCT WT and R301A mutant from the PM, as reported by a decreasing TIRF signal, consistent with the membrane binding being PI(4,5)P2 dependent. TubbyCT R301A translocation occurred at less depolarized membrane potential compared to tubbyCT WT (Fig. 4, A and B). For quantitative assessment, we fitted TIRF signal amplitudes with a Boltzmann function that describes the voltage dependency of VSP enzymatic activity (35). As shown in Fig. 4 (B and C), half-maximum translocation of the R301A mutant occurred at much more negative potentials (Vh = −61.5 ± 20.6 mV) compared to the WT construct (Vh = −12.7 ± 3.7 mV). Thus, much less activation of the phosphatase was required for unbinding of the mutant domain, which is equivalent to the dissociation at a higher PI(4,5)P2 concentration, indicating a reduced PI(4,5)P2 affinity. Notably, the lower overall reduction in normalized signal amplitude observed with the mutant is explained by the lower basal membrane association (Fig. 3A), consistent with the reduced affinity. Moreover, Fig. 4B also shows that the dependence of the domain’s membrane association on voltage was much steeper for the WT domain with both PI(4,5)P2 binding sites intact (slope factor, 11.9 ± 1.1 mV) compared to the R301A mutant with an impaired second binding site (30.2 ± 2.7 mV; P = 0.000145). This voltage sensitivity translates into a steeper PI(4,5)P2 concentration dependence of WT tubbyCT, as expected for cooperative binding.
Fig. 4. Contribution of the second binding site to PI(4,5)P2 affinity of tubbyCT.
(A) Changes in membrane association of GFP-tubbyCT in response to activation of Ci-VSP as recorded by TIRF microscopy. CHO cells cotransfected with GFP-tubbyCT and RFP-Ci-VSP were whole-cell voltage-clamped and depolarized gradually (staircase voltage protocol) (top) while measuring membrane-localized fluorescence by TIRF microscopy. Bottom shows representative TIRF recordings for WT and R301A mutant, normalized to resting signal at −60 mV. (B) Fluorescence-voltage curves obtained from experiments as in (A) were fitted by a Boltzmann function and normalized to maximal fitted fluorescence change. Shown are averaged data from n = 9 and 7 cells for WT (black) and R301A (red), respectively. (C) Mean voltage required for half-maximal dissociation from the membrane from curves shown in (B). Asterisk indicates statistical significance (*P < 0.05). (D) PMF for the binding of tubbyCT WT (black) and tubbyCT R301A (red) to a single PI(4,5)P2 lipid. (E) PMF for the binding of tubbyCT WT (black) and tubbyCT R301A (red) to two PI(4,5)P2 lipids.
Last, we quantified the impact of the R301A mutation on the PI(4,5)P2 binding free energy, by calculating the PMF of the R301A mutant and WT tubbyCT. Figure 4D shows that the potential depth at the second binding site (protein-membrane distance of 2.4 nm) is reduced, while the canonical binding site (at 1.8 nm) is unaffected. The total binding free energy ΔGbind calculated from the PMF profile yields a ΔGbind = −2.4 kJ/mol for tubbyCT WT and a ΔGbind = −2.0 kJ/mol for the R301A mutant. Despite the moderate difference, it confirms the reduced affinity measured in living CHO cells. In the case of two PI(4,5)P2 lipids binding to tubbyCT R301A, the strongest impact on the PMF is between 1.9 and 2.6 nm, where the R301A shows weaker binding than the WT (Fig. 4E). However, the minimum of the PMF at a shorter protein-membrane distance becomes deeper by about −1 kJ/mol and narrower. Furthermore, the PMFs of the R301 mutant indicate that the pronounced increase in binding affinity due to a second PI(4,5)P2 is still present. In addition, we calculated the PMFs for the binding to a single PI(4,5)P2 for the two mutants R303A and K305A (fig. S3). In accordance with the R301A mutant, the binding strength in the second binding site is reduced by the mutations with a more pronounced effect for the R303A mutant compared to R301A and R305A. This trend agrees with the experimental membrane association (Fig. 3B).
Together, experimental and computational analyses agree in showing that the PI(4,5)P2 affinity of tubbyCT critically depends on the previously unknown second binding site, such that PM association at physiological PI(4,5)P2 concentration requires lipid interaction at both binding sites.
Dual binding mode directs tubbyCT to ER-PM junctions
The dual, cooperative binding mode provides steeper concentration dependence, which can facilitate the sensing of subtle PI(4,5)P2 concentration differences. Thus, it may promote the sharp-cut dissociation of tubby from the ciliary membrane following delivery of cargo from the PM into this PI(4,5)P2-poor compartment (18, 19). Vice versa, this binding mode could also account for the accumulation of tubbyCT at PM domains with relatively high PI(4,5)P2 content. We recently found that tubbyCT preferentially localizes to E-Syt3–rich ER-PM junctions (25). Accumulation of tubbyCT at ER-PM junctions was dependent on PI(4,5)P2 synthesis, suggesting that the junctional PM compartment features a higher PI(4,5)P2 level than the bulk PM (25).
We therefore investigated the impact of the second PI(4,5)P2 binding site on the localization of tubbyCT to these contact sites. GFP-tagged tubbyCT mutants were expressed in CHO cells, and PM-associated tubbyCT was examined by TIRF microscopy. Association of tubbyCT with ER-PM junctions was assessed by quantification of its clustering (Fig. 5, A and B). As expected, tubbyCT WT displayed a prominent localization to ER-PM junctions at approximately 12% of the PM area. All analyzed mutations reduced the recruitment to ER-PM junctions, with K305A showing the smallest effect, consistent with the strongest residual PI(4,5)P2 binding (Fig. 4, A and B) of this mutant. Mutations R301A, R303A, K304A, and the double mutations almost completely abolished preferential junctional localization of tubbyCT (Fig. 5, A and B).
Fig. 5. Dual binding mode directs tubbyCT to ER-PM junctions.
(A) Representative TIRF images of tubbyCT WT and mutants expressed in CHO cells show the different degrees of membrane clustering of the constructs. Scale bars, 5 μm. (B) Quantification of clustered area of tubbyCT WT and mutants calculated from images as in (A). White numbers show the number of analyzed cells, and asterisks indicate significant differences compared to the WT control (*P < 0.05 and ***P < 0.0001).
DISCUSSION
Here, we identify a previously unknown second PI(4,5)P2-binding site in the C-terminal domain of the tubby protein (tubby domain). It consists of a conserved cluster of positively charged amino acids and is located next to the classical or canonical binding site at the essentially planar protein-lipid interface of the tubby domain. PI(4,5)P2 binding by the canonical binding site as previously shown by crystallography and mutational analysis (13) was fully reproduced by our CG MD simulations. However, occupancy of the second binding was nearly as high (ratio, 0.8) as the occupancy of that primary site and indeed proved essential for PI(4,5)P2 binding and hence membrane association of the domain at physiological PI(4,5)P2 levels.
PI(4,5)P2 binding affinity of the tubby domain
There has been considerable confusion concerning the PI(4,5)P2 binding properties of tubbyCT. In some cell types, tubbyCT resists dissociation from the PM during strong activation of PLCβ-mediated depletion of PI(4,5)P2, despite confirmation of PI(4,5)P2 loss by independent readouts (8, 9, 16). This has been taken to indicate a higher PI(4,5)P2 affinity of tubbyCT compared to PLCδ1-PH. However, titration of PI(4,5)P2 with a voltage-activated phosphatase (Ci-VSP) (40) provided evidence that the affinity of tubbyCT for PI(4,5)P2 is actually lower compared to PLCδ1-PH (8, 35). Our CG MD simulations show a low binding affinity of tubbyCT to a single PI(4,5)P2 lipid compared to the PLCδ1-PH domain. Only a second PI(4,5)P2 lipid bound to the previously unknown binding site results in a binding strength comparable to the PLCδ1-PH domain with one PI(4,5)P2 lipid. This is in accordance with the Ci-VSP titration, where tubby unbinding occurred at a higher PI(4,5)P2 concentration.
Selective ligand binding of the tubby domain
Polybasic motifs similar to the second binding site mediate membrane association of many cytosolic proteins by electrostatic interactions with PI(4,5)P2 and PIP3 (41, 42) or PI(4)P (43). Here, we observed substantial specificity of the previously unknown binding site for PI(4,5)P2. Thus, PI(4)P had a much weaker, although detectable capacity to bind tubbyCT to the membrane, and the anionic POPS and POPG were ineffective. This specificity may help to ensure targeting to the PM rather than to other negatively charged membrane compartments.
The high selectivity for distinct PI species characteristic for some PH domains is achieved by stereospecificity of the interaction between the binding site and the anionic head group of the ligand, and differences in binding selectivity arise from well-defined variations in the amino acid sequence of the binding site (14, 44–46). However, the steric interaction with the anionic head group of the lipid also goes along with high-affinity binding of the isolated head group, e.g., the soluble second messenger IP3 in the case of the PI(4,5)P2-binding PH domain of PLCδ1 (14). This second cellular high affinity ligand is a major confounding problem in the use of this domain as a reliable PI(4,5)P2 biosensor (4, 15). In contrast, by restricting high affinity binding to the membrane surface, the combinatorial PI(4,5)P2 binding by the two sterically parallel binding sites likely contributes to the fact that tubbyCT has no detectable IP3 affinity in its cellular environment (9, 16).
PI(4,5)P2 binding mode of the tubby domain
How does the dual binding mode of tubbyCT compare to well-characterized PI-binding domains? Most of the PH domains bind their PI ligand by a single binding pocket (14). However, several studies on different PH domains identified more than one lipid binding site (30, 33, 47, 48). The functional consequences of multiple lipid binding sites include (i) nonspecific binding of anionic lipids, (ii) alternative ligand binding conformations, and (iii) cooperative binding.
Let us first focus on nonspecific binding of anionic lipids. For instance, binding of general receptor for phosphoinositides (GRP)1-PH domain to its specific ligand, PIP3, in the membrane is substantially enhanced by the presence of bulk anionic lipids such as PS and PI (49). However, this interaction is thought to be mediated by weak electrostatic interaction with the canonical binding site that precedes the high affinity binding of the specific substrate in a “search mode,” rather than by a secondary interaction site (49, 50). Similar observations have been made for a number of yeast PH domains (48).
Second, an alternative occupation of a second binding site (ii) can allow for an orientational fine-tuning depending on the concentration of the specific lipid. Such an orientational fine-tuning was reported for the GRP1-PH domain, for which differently oriented bound states depending on the PIP3 concentration were obtained in CG MD simulations (33).
Last, more similar to the two–binding site mode identified here for tubbyCT, some PH domain proteins [GRP1 and ADP-Ribosylation factor Nucleotide-binding site Opener (ARNO)] feature a polybasic motif outside of the PH domain, which enhances affinity for negatively charged membranes in a cooperative manner (51, 52). Similarly, the PH domain of ASter-Associated Protein (ASAP)1 features two binding sites that confer cooperative PI(4,5)P2 binding in liposome binding assays (47). Nevertheless, one of these sites is nonspecific, with a substantial affinity to PS. Some other classes of PI recognition domains, such as the FYVE and PX domains, also use a dual binding mode where, in addition to head group recognition, insertion of a hydrophobic moiety into the membrane mediates membrane association and increases affinity for the PI ligand in the membrane environment [reviewed in (5)].
Distinct from these well-studied domains, the secondary binding site of tubby is highly specific for PI(4,5)P2, indicated by lack of membrane stabilization by negatively charged lipids of the PM such as PS and only minor efficacy of the related PI(4)P. Moreover, because the two binding sites are located on the same face of the protein, orientational fine-tuning does not play a role either. The close proximity of both sites enables simultaneous occupancy and strongly enhances the binding affinity in a cooperative manner. A possible explanation for the strong cooperativity could be the fact that the positive residues of the second binding site are already immobilized by the binding of the first PI(4,5)P2 lipid. This potentially provides an entropic driving force for the favorable binding energy of the second PI(4,5)P2 lipid. Despite their charges, PI(4,5)P2 lipids are often being observed to form small clusters in membranes and around proteins (32, 33, 37, 42, 53). Overall, among PI recognition domains that feature a cooperative binding mode, the second binding site has the most decisive impact in tubbyCT in enhancing both specificity and concentration sensitivity.
Functional implications of the binding mode
PI(4,5)P2 is considered the major signaling PI of the PM, and a plethora of proteins are known as PI(4,5)P2 interactors. However, beyond the PH domain of PLCδ1, detailed characterization of binding properties is largely lacking, although the binding mode critically determines when, where, and in response to which signaling events PI(4,5)P2 effector proteins are active in the complex cell.
The simultaneous binding of two PI(4,5)P2 molecules by tubbyCT not only increases the overall affinity but also enables a steeper concentration dependence of membrane binding compared to binding by a single binding pocket as in the classical PH domains. Such a steep concentration dependence may be particularly relevant for differential membrane association under conditions of spatial or temporal PI(4,5)P2 inhomogeneities. For example, tubby domain–containing proteins may robustly bind at basal PM levels of PI(4,5)P2 while readily dissociating from the membrane at moderately decreased levels. Vice versa, cooperative binding may strongly favor binding to PI(4,5)P2-enriched membrane domains, e.g., ER-PM junctions as opposed to the bulk membrane.
Given the conservation of the previously unknown binding site motif in mammalian TULPs (Fig. 2D), we consider this idea in the context of known common functions of TULPs. Although the cell biology of TULPs is only beginning to be illuminated, a congruent theme is the trafficking of proteins into cilia (54). The following mechanistic model emerged (18): Tubby domains can bind to cargo proteins designated for ciliary delivery by interacting with a ciliary localization signal of these proteins, and this interaction requires membrane association of the tubby domain through PI(4,5)P2 binding (18). The TULP also binds to the intraflagellar transport complex A, which shuttles the entire complex into the cilium (19). Once localized in the cilium, the cargo would be released from the TULP, because the ciliary membrane is poor in PI(4,5)P2 (18). The discrimination between different PI(4,5)P2 concentrations underlying this cyclic process may be facilitated by steep cooperative PI(4,5)P2 binding.
In summary, our combined computational and experimental study revealed a previously unknown second PI(4,5)P2 binding site of the tubby domain. This binding site is pivotal for the membrane targeting of the tubby domain contributing to a large extent to the binding affinity via cooperativity. Moreover, our data emphasize the key role of the second binding site to target specific regions of the PM such as ER-PM junctions. In addition, our findings contribute to better understand the functioning of TULPs in ciliary trafficking.
MATERIALS AND METHODS
MD simulations
System setup and simulation details
All simulations were performed using the CG force field Martini 3 [open-beta version (27)] and the program package Gromacs (version 2018.1) (55). We used the tubbyCT crystal structure [Protein Data Bank (PDB) code: 1I7E] (13) and modeled the missing loops with the Iterative Threading ASSEmbly Refinement (I-TASSER) server (56). Two of the residues involved in the previously unknown PI(4,5)P2 binding site—K304 and K305—are part of a missing loop consisting of six residues, and their initial structure is based on the model. Figure S4 depicts the crystal structure together with the I-TASSER model and a more recent model by AlphaFold (57, 58) for comparison including a magnified view of residues 301 to 305. To generate the CG model, the missing loops were ligated to the crystal structure. This step was necessary to maintain the side chain orientations around the PI(4,5)P2 binding pocket of tubbyCT identified in the crystal structure. On the basis of the Martini protein model without any elastic network (59), side chain corrections (60) were added. To maintain the secondary and tertiary protein structure, a Gō-like model was used according to the procedure in (61). On the basis of contacts defined by an overlap and contacts of structural units-criterion evaluated only for the residues resolved in the crystal structure, Lennard-Jones interactions were added up to a cutoff distance of 1.1 nm (62). The dissociation energy of the Lennard-Jones potential was set to ε = 12.0 kJ/mol (63). In total, 541 Gō-like bonds were added for tubbyCT. All intermolecular interactions are solely described by the nonbonded terms of the Martini force field.
Different membrane compositions were used to study the PI(4,5)P2 binding of tubbyCT: (i) a simple POPC membrane containing one additional PI(4,5)P2 lipid, which was embedded in the pocket identified in the crystal structure in the starting structure. (ii) In a second setup, the membrane consisted of POPC and PI(4,5)P2 lipids in a molar ratio of 95:5. (iii) As control setup, POPC membranes with 5 mol % of different negatively charged lipids [POPG, POPS, and PI(4)P] were used to test the impact of other negatively charged lipids. In addition, in cases (ii) and (iii), one PI(4,5)P2 lipid was initially embedded in the crystal structure binding pocket of tubbyCT. In all simulations, the recently refined bonded parameters of the PI(4,5)P2 lipids were used (32). The membrane patches had a size of 15 nm by 15 nm containing approximately 700 lipids in total. They were generated using the program insane.py (64). Last, the system was neutralized and solvated in a 0.15 M NaCl solution. The rectangular box with an initial size of 15 nm by 15 nm by 14 nm contained ~17,300 water beads corresponding to ~69,200 water molecules.
The simulation parameters were chosen in accordance to the new reaction-field settings given in (65). After the equilibration, setup (i) was simulated for 1 μs (10 replicas), setup (ii) for 5 μs (3 replicas), and setup (iii) for 1 μs in the case of POPG and POPS and for 5 μs in the case of PI(4)P (three replicas for each lipid type).
For comparison, we also simulated the PLCδ1-PH domain (PDB code: 1MAI) (14), a well-characterized stable PI(4,5)P2 binder (11, 12). We modeled the missing termini with the I-TASSER server (56) and ligated them to the crystal structure. The Gō-like model was set up similar to the tubbyCT resulting in 234 added Lennard-Jones interactions. Simulations were performed using the setups (i) and (ii) as described earlier for tubbyCT.
CG models for tubbyCT and PLCδ1-PH domain were based on the amino acid sequence used experimentally. Thus, tubbyCT had a size of 263 residues (amino acids 243 to 505), and PLCδ1-PH domain had a size of 170 residues (amino acids 1 to 170). The size difference and the different number of resolved residues in the crystal structures (tubbyCT, 237; PLCδ1-PH, 119) explain the difference in the number of Lennard-Jones interactions in both Gō-like models, because the contact map was calculated for the crystal structure without taking into account the modeled parts. The missing residues of the PLCδ1-PH domain are solely located at the termini (C terminus, 40; N terminus, 11), which are both on the opposite side of the PI(4,5)P2 binding site.
Analysis
To identify the unbinding of the proteins from the membrane, the distance between the plane of the phosphate groups of the lipids (PO4 beads) and the crystal binding pocket was calculated using the gmx distance tool. The phosphate plane is formed by the PO4− groups, which are directly linked to the glycerol moiety. The phosphate groups of the inositide head groups are not considered here. Note that this indirectly indicates the binding of PI(4,5)P2 to the peripheral membrane proteins tubbyCT and PLCδ1-PH domain, which unbind from the membrane if not binding PI(4,5)P2. This reaction coordinate—the distance between PO4 plane and binding pocket—has the advantage that an exchange of individual PI(4,5)P2 lipids in contact with the protein in systems with multiple PI(4,5)P2 lipids does not alter the calculated distance as it would be the case for an individual PI(4,5)P2-binding pocket distance.
To obtain more detailed information about the position of the PI(4,5)P2 lipid with respect to the protein surface, the distance between the PI(4,5)P2 head group and the crystal structure binding pocket was also calculated for the systems containing only one PI(4,5)P2 lipid [setup (i)]. Again, the gmx distance tool was used.
To calculate the protein-PI(4,5)P2 contacts, the number of PI(4,5)P2 lipids with a distance ≤0.5/0.7/0.9 nm between the head group and the protein was analyzed (using gmx select), while the protein was considered being bound to the membrane. The number of contacts to the individual residues was also calculated using a distance cutoff of 0.5 nm.
To estimate the relative binding strength of the previously unknown second binding site in comparison to the canonical binding site, we first integrated the population of the respective binding site in the histogram of the PI(4,5)P2-binding pocket distance and then calculated their ratio. The boarders for the integration were 0.00 to 1.40 nm for the canonical and 1.40 to 2.45 nm for the previously unknown binding site, respectively. Means and SEs were calculated; for statistical analysis, a two-tailed Dunnett’s test was performed (*P < 0.05, **P < 0.01, and ***P < 0.0001) using IGOR Pro software. Ten replicas of 1-μs-long simulations were performed for the WT and all mutants binding to a single PI(4,5)P2. Additional 12 replicas of 0.5-μs-long simulations were performed for the WT, single, and double mutants. Because the unbinding from a single PI(4,5)P2 happens quickly (Fig. 1B) and only the relative population of the two binding sites in the bound state was analyzed, 0.5 μs of simulation was performed for the additional replicas.
To calculate the root mean square fluctuations (RMSFs) of the backbone of membrane-bound and free tubbyCT, we used 10 windows of 500 ns each of membrane-bound tubbyCT [POPC membrane with 5 mol % of PI(4,5)P2] and free tubbyCT. We fitted the protein backbone within each of the 500-ns windows before calculating the backbone RMSF using gmx rmsf. Last, we calculated the mean value of the 10 windows, and its SE was depicted in fig. S5.
PMF calculations
To estimate the binding free energy of WT tubbyCT, tubbyCT R301A, R303A, R305A, and PLCδ1-PH domain, we calculated the PMF using umbrella sampling. We followed the procedure described in (30). Briefly, we generated conformations along the reaction coordinate by pulling the protein center of mass away from the PI(4,5)P2 head group perpendicular to the membrane plane [z direction; force constant, 1000 kJ/(mol nm2); pulling rate, 0.001 nm/ps], while the PI(4,5)P2 lipid was restrained in space using a harmonic potential [force constant, 1000 kJ/(mol nm2)], and the PI(4,5)P2 head group and the protein center of mass were kept at the same point in the plane of the membrane [x/y plane; force constant, 100 kJ/(mol nm2)]. For the umbrella sampling simulations, the harmonic constraint of the PI(4,5)P2 lipid in z direction was released, and the distance between the PI(4,5)P2 head group and the protein center of mass was constrained with a harmonic potential [force constant, 1000 kJ/(mol nm2)]. This distance was sampled between 1.6 and 5 nm for tubbyCT and 1.2 and 4.6 nm for the PLCδ1-PH domain, respectively, using an interval of 0.1 nm. The resulting 35 windows were sampled for 2 μs up to 4 nm and 1 μs for larger distances in the case of tubbyCT and 1 μs in the case of the PH domain. The sampling for tubbyCT was increased to better distinguish the WT tubbyCT from its R301A mutant. To calculate the PMFs, the gmx wham tool was used, and the initial 200 ns of each umbrella window was discarded; error estimation was done using a bootstrap analysis with 100 bootstraps (66). The convergence behavior of the PMFs is depicted in figs. S6 to S12.
Experimental assessment of membrane binding in living cells
Cell culture
CHO dhFr− cells (CRL-9096, American Type Culture Collection) were cultured in MEM (minimum essential medium) Alpha medium (Gibco, Thermo Fisher Scientific, Waltham, USA) supplemented with 10% fetal bovine serum, 1% penicillin, and 1% streptomycin. Cells were seeded on glass bottom dishes and kept at 37°C and 5% CO2. Two days after seeding, cells were transfected using JetPEI DNA transfection reagent (Polyplus-transfection, Illkirch-Graffenstaden, France) according to the manufacturer’s instructions. Experiments were performed 24 hours after transfection. For each experiment, at least three individual transfections were performed (biological replicates at the transfection level). The number of cells analyzed is given in the respective figure.
Molecular biology
Expression constructs used for transfection were as follows: mouse tubbyCT (amino acids 243 to 505) in pEGFP-C1 (NM_021885.4) and Ci-VSP and Ci-VSP-C363S in pRFP-C1 (AB183035.1). Mutagenesis of tubbyCT was done using a QuikChange II XL Site-Directed mutagenesis kit (Stratagene, Agilent Technologies, Waldbronn, Germany). Used mutagenesis primers are listed in the Supplementary Materials (table S3).
Confocal microscopy
Experiments were performed on an upright LSM 710 Axio Examiner Z1 microscope equipped with a 100×/1.30 oil ultraviolet objective (Carl Zeiss, Oberkochen, Germany). For determination of membrane localization of GFP-tubbyCT constructs, CHO cells were cotransfected with an RFP-tagged membrane marker (catalytically inactive Ci-VSP C363S or Lyn11). Cells were fixed with 4% paraformaldehyde before imaging. To quantify membrane localization of tubbyCT, line profiles across the cells were derived, and ratios of membrane-localized GFP-tubbyCT fluorescence intensities (averaged from the two intersections with the PM, defined by the membrane marker fluorescence peaks) and cytosolic fluorescence were calculated.
TIRF microscopy
TIRF imaging was done on a Dmi8 upright microscope (Leica, Wetzlar, Germany) equipped with an Infinity TIRF module (Leica), a HC PL APO 100×/1.47 oil objective (Leica), and a widefield laser (Leica). GFP fluorescence was excited at 488 nm and imaged through a GFP-T (505 to 555 nm) emission filter (Leica). Images were acquired every 6 s with an ORCA-Flash4.0 C13440-20C camera (Hamamatsu photonics, Hamamatsu, Japan) controlled by LAS X software (Leica). During imaging, cells were perfused with an extracellular solution [5.8 mM KCl, 144 mM NaCl, 0.9 mM MgCl2, 1.3 mM CaCl2, 0.7 mM NaH2PO4, 5.6 mM d-glucose, and 10 mM Hepes (pH 7.4)]. Data were analyzed by ImageJ software, and tubbyCT clusters were detected by an algorithm described previously (25).
Combined TIRF microscopy and voltage-clamp experiments
TIRF imaging was performed as described above, and images were taken every 3 s. Simultaneously, cells were whole-cell patch-clamped for control of Ci-VSP activity as described previously (8, 35). Briefly, voltage clamp recordings were done with an EPC 10 amplifier controlled by PATCHMASTER software (HEKA Elektronik, Lambrecht, Germany). Patch pipettes were pulled from borosilicate glass (Sutter Instrument Company, Novato, CA, USA) and had an open pipette resistance of 2 to 3 megohms after backfilling with intracellular solution containing 135 mM KCl, 2.41 mM CaCl2, 3.5 mM MgCl2, 5 mM Hepes, 5 mM EGTA, 2.5 mM Na2ATP, and 0.1 mM Na3GTP (pH 7.3) (with KOH), 290 to 295 mosmol/kg. Series resistance (Rs) typically was below 6 megohms. Cells were held at −60 mV and depolarized in a stairstep command. During these experiments, the experimental chamber was continuously fed with an extracellular solution [5.8 mM KCl, 144 mM NaCl, 0.9 mM MgCl2, 1.3 mM CaCl2, 0.7 mM NaH2PO4, 5.6 mM d-glucose, and 10 mM Hepes (pH 7.4)].
Analysis and statistical evaluation
Mean values and SEs were calculated on the basis of the number of cells examined (biological replicates at the cellular level) as given in Figs. 3 to 5. For the imaging experiments (confocal and TIRF microscopy), cells from at least three individual transfections were analyzed for each condition. For statistical analysis, a two-tailed Dunnett’s test was performed (*P < 0.05 and ***P < 0.0001) using IGOR Pro software (WaveMetrics, Lake Oswego, OR, USA). In patch-clamp experiments, one or two cells were recorded per independent transfection. For statistical comparison, Student’s t tests were applied with an asterisk indicating statistical significance at P < 0.05. All data are displayed as means ± SEM.
Acknowledgments
We like to thank L. Shapiro and Y. Okamura for providing expression plasmids. S.T. thanks the Center for Information Technology of the University of Groningen for providing access to the Peregrine high-performance computing cluster and the Center for Scientific Computing of the Goethe University Frankfurt for providing access to the Goethe-HLR high-performance computing cluster.
Funding: This work was supported by Deutsche Forschungsgemeinschaft (DFG) Research Training Group 2213 “Membrane Plasticity in Tissue Development and Remodeling” (to D.O.), European Commission ERC Advanced Grant COMP-MICR-CROW-MEM (grant agreement 669723) (to S.J.M.), Hessian Ministry of Science and Arts LOEWE program (Center for Multiscale Modeling in Life Sciences, CMMS) (to S.T.), Alfons und Gertrud Kassel foundation (to S.T.), and Dr. Rolf M. Schwiete foundation (to S.T.).
Author contributions: Conceptualization: V.T., S.T., and D.O. Investigation: V.T., L.S., S.T., and W.Z. Visualization: V.T. and S.T. Discussion: V.T., S.T., D.O., S.J.M., L.S., and W.Z. Writing—original draft: S.T., D.O., and V.T. Writing—review and editing: S.T., D.O., V.T., and S.J.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The I-TASSER model of tubbyCT, starting configurations for the simulations, and Gromacs files (mdp, itp, and index files) are publicly available at https://doi.org/10.5281/zenodo.6587156. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All expression plasmids described are available from the authors.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S12
Tables S1 to S3
REFERENCES AND NOTES
- 1.Dickson E. J., Hille B., Understanding phosphoinositides: Rare, dynamic, and essential membrane phospholipids. Biochem. J. 476, 1–23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Paolo G., De Camilli P., Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Balla T., Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammond G. R. V., Balla T., Polyphosphoinositide binding domains: Key to inositol lipid biology. Biochim. Biophys. Acta 1851, 746–758 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemmon M. A., Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Milosevic I., Sørensen J. B., Lang T., Krauss M., Nagy G., Haucke V., Jahn R., Neher E., Plasmalemmal phosphatidylinositol-4,5-bisphosphate level regulates the releasable vesicle pool size in chromaffin cells. J. Neurosci. 25, 2557–2565 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardie R. C., Liu C. H., Randall A. S., Sengupta S., In vivo tracking of phosphoinositides in Drosophila photoreceptors. J. Cell Sci. 128, 4328–4340 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leitner M. G., Thallmair V., Wilke B. U., Neubert V., Kronimus Y., Halaszovich C. R., Oliver D., The N-terminal homology (ENTH) domain of Epsin 1 is a sensitive reporter of physiological PI(4,5)P2 dynamics. Biochim. Biophys. Acta 1864, 433–442 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Szentpetery Z., Balla A., Kim Y. J., Lemmon M. A., Balla T., Live cell imaging with protein domains capable of recognizing phosphatidylinositol 4,5-bisphosphate; a comparative study. BMC Cell Biol. 10, 67 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoon Y., Lee P. J., Kurilova S., Cho W., In situ quantitative imaging of cellular lipids using molecular sensors. Nat. Chem. 3, 868–874 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stauffer T. P., Ahn S., Meyer T., Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 8, 343–346 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Várnai P., Balla T., Visualization of phosphoinositides that bind pleckstrin homology domains: Calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 143, 501–510 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santagata S., Boggon T. J., Baird C. L., Gomez C. A., Zhao J., Shan W. S., Myszka D. G., Shapiro L., G-protein signaling through tubby proteins. Science 292, 2041–2050 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Ferguson K. M., Lemmon M. A., Schlessinger J., Sigler P. B., Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell 83, 1037–1046 (1995). [DOI] [PubMed] [Google Scholar]
- 15.Hirose K., Kadowaki S., Tanabe M., Takeshima H., Iino M., Spatiotemporal dynamics of inositol 1,4,5-trisphosphate that underlies complex Ca2+ mobilization patterns. Science 284, 1527–1530 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Quinn K. V., Behe P., Tinker A., Monitoring changes in membrane phosphatidylinositol 4,5-bisphosphate in living cells using a domain from the transcription factor tubby. J. Physiol. 586, 2855–2871 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varnai P., Thyagarajan B., Rohacs T., Balla T., Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J. Cell Biol. 175, 377–382 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Badgandi H. B., Hwang S. H., Shimada I. S., Loriot E., Mukhopadhyay S., Tubby family proteins are adapters for ciliary trafficking of integral membrane proteins. J. Cell Biol. 216, 743–760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukhopadhyay S., Wen X., Chih B., Nelson C. D., Lane W. S., Scales S. J., Jackson P. K., TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 24, 2180–2193 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan X. M., Yu H., Van der Ploeg L. H., Evidence of altered hypothalamic pro-opiomelanocortin/ neuropeptide Y mRNA expression in tubby mice. Brain Res. Mol. Brain Res. 59, 273–279 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Kleyn P. W., Fan W., Kovats S. G., Lee J. J., Pulido J. C., Wu Y., Berkemeier L. R., Misumi D. J., Holmgren L., Charlat O., Woolf E. A., Tayber O., Brody T., Shu P., Hawkins F., Kennedy B., Baldini L., Ebeling C., Alperin G. D., Deeds J., Lakey N. D., Culpepper J., Chen H., Glücksmann-Kuis M. A., Carlson G. A., Duyk G. M., Moore K. J., Identification and characterization of the mouse obesity gene tubby: A member of a novel gene family. Cell 85, 281–290 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Mukhopadhyay S., Jackson P. K., Cilia, tubby mice, and obesity. Cilia 2, 1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borman A. D., Pearce L. R., Mackay D. S., Nagel-Wolfrum K., Davidson A. E., Henderson R., Garg S., Waseem N. H., Webster A. R., Plagnol V., Wolfrum U., Farooqi I. S., Moore A. T., A homozygous mutation in the TUB gene associated with retinal dystrophy and obesity. Hum. Mutat. 35, 289–293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devane J., Ott E., Olinger E. G., Epting D., Decker E., Friedrich A., Bachmann N., Renschler G., Eisenberger T., Briem-Richter A., Grabhorn E. F., Powell L., Wilson I. J., Rice S. J., Miles C. G., Wood K.; Genomics England Research Consortium, Trivedi P., Hirschfield G., Pietrobattista A., Wohler E., Mezina A., Sobreira N., Agolini E., Maggiore G., Dahmer-Heath M., Yilmaz A., Boerries M., Metzger P., Schell C., Grünewald I., Konrad M., König J., Schlevogt B., Sayer J. A., Bergmann C., Progressive liver, kidney, and heart degeneration in children and adults affected by TULP3 mutations. Am. J. Hum. Genet. 109, 928–943 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.V. Thallmair, L. Schultz, S. Evers, C. Goecke, S. Thallmair, M. G. Leitner, D. Oliver, Local PI(4,5)P2 pool dynamics detected by the coincidence biosensor tubbyCT maintain the integrity of ER-PM junctions during PLC signaling. bioRxiv 2020.09.25.313403 [Preprint]. 25 September 2020. 10.1101/2020.09.25.313403. [DOI]
- 26.Souza P. C. T., Alessandri R., Barnoud J., Thallmair S., Faustino I., Grünewald F., Patmanidis I., Abdizadeh H., Bruininks B. M. H., Wassenaar T. A., Kroon P. C., Melcr J., Nieto V., Corradi V., Khan H. M., Domański J., Javanainen M., Martinez-Seara H., Reuter N., Best R. B., Vattulainen I., Monticelli L., Periole X., Tieleman D. P., de Vries A. H., Marrink S. J., Martini 3: A general purpose force field for coarse-grained molecular dynamics. Nat. Methods 18, 382–388 (2021). [DOI] [PubMed] [Google Scholar]
- 27.P. C. T. Souza, S. J. Marrink, Martini 3 - Open Beta-Release (2020); http://cgmartini.nl.
- 28.Corradi V., Sejdiu B. I., Mesa-Galloso H., Abdizadeh H., Noskov S. Y., Marrink S. J., Tieleman D. P., Emerging diversity in lipid-protein interactions. Chem. Rev. 119, 5775–5848 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corradi V., Mendez-Villuendas E., Ingólfsson H. I., Gu R. X., Siuda I., Melo M. N., Moussatova A., DeGagné L. J., Sejdiu B. I., Singh G., Wassenaar T. A., Delgado Magnero K., Marrink S. J., Tieleman D. P., Lipid-protein interactions are unique fingerprints for membrane proteins. ACS Cent. Sci. 4, 709–717 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naughton F. B., Kalli A. C., Sansom M. S. P., Association of peripheral membrane proteins with membranes: Free energy of binding of GRP1 PH domain with phosphatidylinositol phosphate-containing model bilayers. J. Phys. Chem. Lett. 7, 1219–1224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naughton F. B., Kalli A. C., Sansom M. S. P., Modes of interaction of pleckstrin homology domains with membranes: Toward a computational biochemistry of membrane recognition. J. Mol. Biol. 430, 372–388 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Sun F., Schroer C. F. E., Palacios C. R., Xu L., Luo S. Z., Marrink S. J., Molecular mechanism for bidirectional regulation of CD44 for lipid raft affiliation by palmitoylations and PIP2. PLoS Comput. Biol. 16, e1007777 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto E., Domański J., Naughton F. B., Best R. B., Kalli A. C., Stansfeld P. J., Sansom M. S. P., Multiple lipid binding sites determine the affinity of PH domains for phosphoinositide-containing membranes. Sci. Adv. 6, eaay5736 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto E., Kalli A. C., Yasuoka K., Sansom M. S. P., Interactions of pleckstrin homology domains with membranes: Adding back the bilayer via high-throughput molecular dynamics. Structure 24, 1421–1431 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halaszovich C. R., Schreiber D. N., Oliver D., Ci-VSP is a depolarization-activated phosphatidylinositol-4,5-bisphosphate and phosphatidylinositol-3,4,5-trisphosphate 5′-phosphatase. J. Biol. Chem. 284, 2106–2113 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Lorent J. H., Levental K. R., Ganesan L., Rivera-Longsworth G., Sezgin E., Doktorova M., Lyman E., Levental I., Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape. Nat. Chem. Biol. 16, 644–652 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ingólfsson H. I., Melo M. N., van Eerden F. J., Arnarez C., Lopez C. A., Wassenaar T. A., Periole X., de Vries A. H., Tieleman D. P., Marrink S. J., Lipid organization of the plasma membrane. J. Am. Chem. Soc. 136, 14554–14559 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Doudou S., Burton N. A., Henchman R. H., Standard free energy of binding from a one-dimensional potential of mean force. J. Chem. Theory Comput. 5, 909–918 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Costa H. A., Leitner M. G., Sos M. L., Mavrantoni A., Rychkova A., Johnson J. R., Newton B. W., Yee M. C., de la Vega F. M., Ford J. M., Krogan N. J., Shokat K. M., Oliver D., Halaszovich C. R., Bustamante C. D., Discovery and functional characterization of a neomorphic PTEN mutation. Proc. Natl. Acad. Sci. U.S.A. 112, 13976–13981 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murata Y., Iwasaki H., Sasaki M., Inaba K., Okamura Y., Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature 435, 1239–1243 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Heo W. D., Inoue T., Park W. S., Kim M. L., Park B. O., Wandless T. J., Meyer T., PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science 314, 1458–1461 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun F., Schroer C. F. E., Xu L., Yin H., Marrink S. J., Luo S. Z., Molecular dynamics of the association of L-selectin and FERM regulated by PIP2. Biophys. J. 114, 1858–1868 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hammond G. R. V., Fischer M. J., Anderson K. E., Holdich J., Koteci A., Balla T., Irvine R. F., PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science 337, 727–730 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park W. S., Heo W. D., Whalen J. H., O’Rourke N. A., Bryan H. M., Meyer T., Teruel M. N., Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol. Cell 30, 381–392 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lietzke S. E., Bose S., Cronin T., Klarlund J., Chawla A., Czech M. P., Lambright D. G., Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol. Cell 6, 385–394 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Ferguson K. M., Kavran J. M., Sankaran V. G., Fournier E., Isakoff S. J., Skolnik E. Y., Lemmon M. A., Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell 6, 373–384 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Jian X., Tang W. K., Zhai P., Roy N. S., Luo R., Gruschus J. M., Yohe M. E., Chen P. W., Li Y., Byrd R. A., Xia D., Randazzo P. A., Molecular basis for cooperative binding of anionic phospholipids to the PH domain of the arf GAP ASAP1. Structure 23, 1977–1988 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vonkova I., Saliba A. E., Deghou S., Anand K., Ceschia S., Doerks T., Galih A., Kugler K. G., Maeda K., Rybin V., van Noort V., Ellenberg J., Bork P., Gavin A. C., Lipid cooperativity as a general membrane-recruitment principle for PH domains. Cell Rep. 12, 1519–1530 (2015). [DOI] [PubMed] [Google Scholar]
- 49.He J., Haney R. M., Vora M., Verkhusha V. V., Stahelin R. V., Kutateladze T. G., Molecular mechanism of membrane targeting by the GRP1 PH domain. J. Lipid Res. 49, 1807–1815 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corbin J. A., Dirkx R. A., Falke J. J., GRP1 pleckstrin homology domain: Activation parameters and novel search mechanism for rare target lipid. Biochemistry 43, 16161–16173 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santy L. C., Frank S. R., Hatfield J. C., Casanova J. E., Regulation of ARNO nucleotide exchange by a PH domain electrostatic switch. Curr. Biol. 9, 1173–1176 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Nagel W., Schilcher P., Zeitlmann L., Kolanus W., The PH domain and the polybasic c domain of cytohesin-1 cooperate specifically in plasma membrane association and cellular function. Mol. Biol. Cell 9, 1981–1994 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van den Bogaart G., Meyenberg K., Risselada H. J., Amin H., Willig K. I., Hubrich B. E., Dier M., Hell S. W., Grubmüller H., Diederichsen U., Jahn R., Membrane protein sequestering by ionic protein–lipid interactions. Nature 479, 552–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukhopadhyay S., Jackson P. K., The tubby family proteins. Genome Biol. 12, 225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., Lindahl E., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 (2015). [Google Scholar]
- 56.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y., The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A., Žídek A., Green T., Tunyasuvunakool K., Petersen S., Jumper J., Clancy E., Green R., Vora A., Lutfi M., Figurnov M., Cowie A., Hobbs N., Kohli P., Kleywegt G., Birney E., Hassabis D., Velankar S., AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., Hassabis D., Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Jong D. H., Singh G., Bennett W. F. D., Arnarez C., Wassenaar T. A., Schäfer L. V., Periole X., Tieleman D. P., Marrink S. J., Improved parameters for the martini coarse-grained protein force field. J. Chem. Theory Comput. 9, 687–697 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Herzog F. A., Braun L., Schoen I., Vogel V., Improved side chain dynamics in MARTINI simulations of protein-lipid interfaces. J. Chem. Theory Comput. 12, 2446–2458 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Poma A. B., Cieplak M., Theodorakis P. E., Combining the MARTINI and structure-based coarse-grained approaches for the molecular dynamics studies of conformational transitions in proteins. J. Chem. Theory Comput. 13, 1366–1374 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Thallmair S., Vainikka P. A., Marrink S. J., Lipid fingerprints and cofactor dynamics of light-harvesting complex II in different membranes. Biophys. J. 116, 1446–1455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Souza P. C. T., Thallmair S., Marrink S. J., Mera-Adasme R., An allosteric pathway in copper, zinc superoxide dismutase unravels the molecular mechanism of the G93A amyotrophic lateral sclerosis-linked mutation. J. Phys. Chem. Lett. 10, 7740–7744 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wassenaar T. A., Ingolfsson H. I., Bockmann R. A., Tieleman D. P., Marrink S. J., Computational lipidomics with insane: A versatile tool for generating custom membranes for molecular simulations. J. Chem. Theory Comput. 11, 2144–2155 (2015). [DOI] [PubMed] [Google Scholar]
- 65.de Jong D. H., Baoukina S., Ingólfsson H. I., Marrink S. J., Martini straight: Boosting performance using a shorter cutoff and GPUs. Comput. Phys. Commun. 199, 1–7 (2016). [Google Scholar]
- 66.Hub J. S., de Groot B. L., van der Spoel D., g_wham—A free weighted histogram analysis implementation including robust error and autocorrelation estimates. J. Chem. Theory Comput. 6, 3713–3720 (2010). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S12
Tables S1 to S3