Abstract
TET (ten-eleven translocation) enzymes catalyze the oxidation of 5-methylcytosine bases in DNA, thus driving active and passive DNA demethylation. Here, we report that the catalytic domain of mammalian TET enzymes favor CGs embedded within basic helix-loop-helix and basic leucine zipper domain transcription factor–binding sites, with up to 250-fold preference in vitro. Crystal structures and molecular dynamics calculations show that sequence preference is caused by intrasubstrate interactions and CG flanking sequence indirectly affecting enzyme conformation. TET sequence preferences are physiologically relevant as they explain the rates of DNA demethylation in TET-rescue experiments in culture and in vivo within the zygote and germ line. Most and least favorable TET motifs represent DNA sites that are bound by methylation-sensitive immediate-early transcription factors and octamer-binding transcription factor 4 (OCT4), respectively, illuminating TET function in transcriptional responses and pluripotency support.
TET enzyme catalytic domain has intrinsic sequence preference, demethylating bHLH and bZIP transcription factor–binding sites.
INTRODUCTION
DNA methylation in the form of 5-methylcytosine (5mC) is an epigenetic modification essential for mammalian development and cellular differentiation (1). TET enzymes catalyze the oxidation of 5mC bases in DNA to hydroxymethylcytosine (5hmC), formylcytosine (5fC), or carboxylcytosine (5caC) bases (2, 3). Ten-eleven translocation (TET) oxidation initiates active, replication-uncoupled demethylation, and although this can occur by other means, TETs are also responsible for passive, replication-coupled DNA demethylation. Active DNA demethylation is primed by the oxidized 5mC derivatives, 5fC and 5caC, which resemble damaged nucleobases (4). These are recognized and excised by the DNA repair machinery, particularly base excision repair, ultimately leading to the replacement of methylated 2′-deoxynucleotides by their unmodified congeners (3). Passive DNA demethylation is facilitated by the most abundant 5mC oxidation product, 5hmC, which prevents remethylation of the daughter strand at the replisome by the maintenance methyltransferase (5). In driving demethylation of cytosine residues, TET proteins enhance the reprogramming of cultured cells to a pluripotent state (6–8) and allow the germ line to achieve full developmental potency in vivo (9). TETs also play a role as tumor suppressors, judging from their frequent loss in acute myelogenic leukemia and other malignancies (10), further emphasizing their importance for modulating epigenetic regulation.
Although central to understanding of TET function, the mechanism by which TET proteins are targeted to specific DNA sequences is not clear. CXXC (cys-x-x-cys)- or IDAX (inhibitor of disheveled and axin)–mediated recruitment of TET proteins targets nonmethylated CGs, or more generally, regions of low cytosine methylation (11). While this may suggest that TETs can function as epigenomic repair enzymes, it does not explain their strong demethylation capacity elsewhere. Posttranslational modification is known to alter the binding of TET to DNA (12), and formation of TET complexes with transcription factors and other DNA binding proteins (6, 13, 14) is thought to alter TET targeting and support pluripotency. Histone marks and epigenomic chromatin states are good predictors for sites of TET activity (15); however, this alone does not provide insight into how TET proteins select sites for catalysis.
Here, we show that the TET catalytic domain, previously considered solely a catalytic engine, significantly contributes to DNA target selection with a pronounced, up to 250-fold, preference for some CG sequence contexts over others. Moreover, both the most and least favorable motifs constitute methylation-sensitive transcription factor–binding sites and contribute to a new understanding of TET enzyme function.
RESULTS
Specificity of the TET catalytic domain in vitro
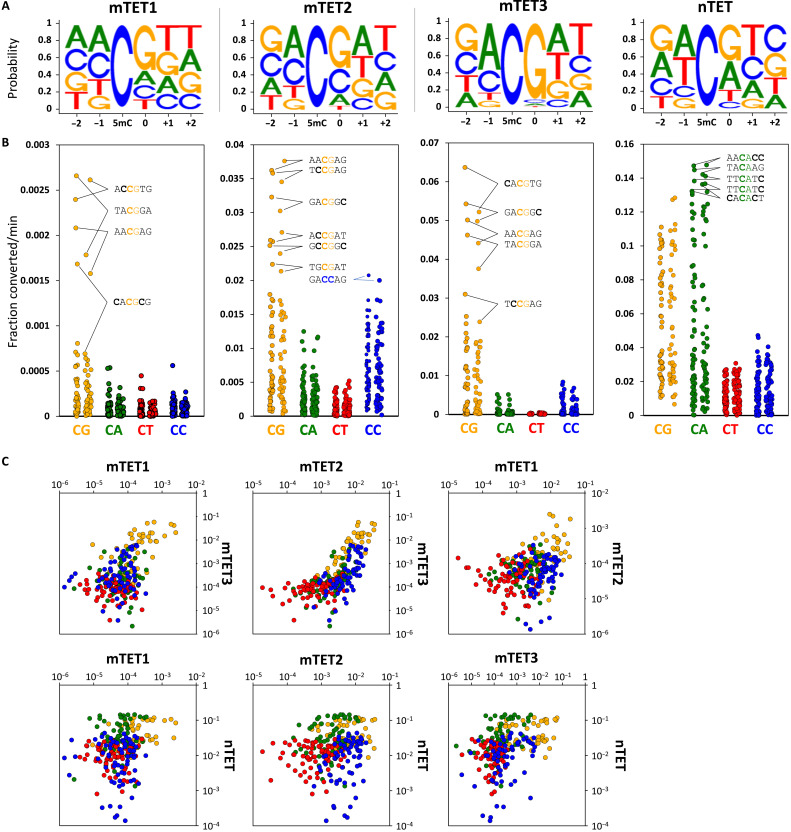
Throughout this work, we used isolated catalytic domains of TETs (for precise constructs, see fig. S1A). We initially found TET sequence preference using in vitro assays where the recombinant catalytic domain of mouse TET3 (mTET3) was incubated with libraries of DNA substrates where all cytosine positions were methylated (Fig. 1A and fig. S1B). To read out the mTET3-oxidized products, we used a bisulfite assay that exploits the resistance of 5mC and 5hmC, but not their higher-oxidation products 5fC and 5caC, to bisulfite-driven deamination. Therefore, 5mC bases oxidized by TET to 5fC/5caC in the substrate DNA could be identified and quantified using next-generation sequencing as the enzymatic reaction progressed (Fig. 1, B and D, and fig. S1, B and C). As expected, mTET3 oxidized non-CG sequences to 5fC/5caC at a much slower rate than CG sites (Figs. 1D and 2, A and B), with decreasing activity toward CG>>CC>CA>CT. Among the CG-containing sequences, we uncovered a 250-fold dynamic range between the most rapidly and the most slowly oxidized sites. The top-ranked oxidized sequence was the CACGTG hexamer (in vitro oxidation velocity 0.058 ± 0.008 fraction converted per minute) (Fig. 1B). This sequence represents the canonical E-box motif, a well-known recognition site for many basic helix-loop-helix (bHLH) and basic leucine zipper domain (bZIP) transcription factors, the most iconic of which is the c-MYC oncogene. Many of the other sequences rapidly oxidized by mTET3 had an adenine upstream (−1 position) and a thymine downstream (+1 position) of the CG, with 7 of the top 13 (53.8%) fastest demethylating sites having this motif. At the other end of the spectrum, the most slowly oxidized sequence was GGCGGG (0.00023 ± 0.00009 fraction converted per minute) (Fig. 1B).
Fig. 1. In vitro flanking sequence preference of TET enzymes.
(A) Outline of the in vitro demethylation kinetics setup. 5mC-modified double-stranded DNA (dsDNA) substrates were incubated with recombinant TET catalytic domains for various lengths of time, and products were purified, ligated to Illumina adapters, bisulfite-converted, and sequenced. NGS, next-generation sequencing. (B) Comparison of mTET3 reaction kinetics on CACGTG (fastest), CCCGCA (middle), and GGCGGG (slowest) 5mCG-containing substrate identified in the screen. Fastest non-5mCG–containing substrate (TTCCCG) is shown in blue. 5mC bases are bolded. (C) Comparison of in vitro reaction kinetics of mTET3 oxidation of synthetic CA5mCGTG (fastest) and GG5mCGGG (slowest) substrates measured using LC-MS. For (C) and (D), error bars denote SD from two biological replicates. (D) TET activity profile on fully 5mC-modified mouse Esrrb (mEsrrb) promoter fragment. The seven Cs on the 5′ end of the DNA substrates are devoid of modification, as these were part of the unmethylated primers used for substrate generation (colored with light gray). Sequence on top represents the analyzed 5mC flanked by three bases upstream (−3 to −1) and four bases downstream (0 to +3). Please note that in these substrates, all the Cs are 5mC. The rows represent the time points of reaction progression, the columns represent each potential 5mC site that could be modified. The color of the boxes denotes the methylation level of the site, according to the legend at the bottom of the panel. For convenience, CG sites are marked with gray boxes on top of the substrate panels.
Fig. 2. In vitro specificity profiles of TET enzymes.
(A) Sequence logos of the TET enzymes based on the in vitro reaction kinetics. (B) Dot plot of TET catalytic activities observed for 5mCs embedded in CG and non-CG contexts in all four tested in vitro substrates (mEsrrb, CG-rich, Tcl1, and Nanog). Each dot represents a 5mC site in either CG, CA, CT, or CC context also differing in the flanking sequences beyond the central dinucleotide. The top activity hexamers are labeled with 5mC bases bolded. (C) Pairwise comparison of activity profiles of mTET1, mTET2, mTET3, and nTET. Orange dots represent CG; green, CA; red, CT; and blue, CC sites. X and y axes represent fraction of 5mC converted per minute.
Oxidation of the least favorable CG sequences was even slower (23-fold slower) than for the most favorable non-CG substrates (GCCCTT; 0.0055 ± 0.00086 fraction converted per minute), suggesting that the flanking sequences strongly affect mTET3 activity.
To independently corroborate these findings using an alternative in vitro assay, we compared the conversion of 5mCG in the E-box sequence (CA5mCGTG) by mTET3 to the most slowly oxidizing motif identified in the screen (GG5mCGGG) using quantitative liquid chromatography–coupled mass spectrometry (LC-MS) (Fig. 1C). This analysis confirmed rapid transition of the central methylated cytosine within the E-box sequence to 5hmC (4.5%), 5fC (38.6%), and 5caC (51.8%) during 5 min of the reaction, whereas in the same time, the GGCGGG sequence had the central 5mC only oxidized to 5hmC (44%), 5fC (15.6%), and 5caC (1.5%).
Fully 5mC-modified double-stranded DNA (dsDNA) is not a natural substrate for TET enzymes. The presence of a large number of 5mC bases in DNA can influence its structure (16) and therefore potentially also its interaction with TET enzymes. We therefore generated M.SssI-methylated substrates containing 5mC only at CG dinucleotides and observed a very similar sequence preference as with the fully modified substrate (r2 = 0.74, P = 3.45 × 10−15) (fig. S1, B and C). The similarity of the results obtained with the fully methylated products and those only methylated at CG sites (fig. S1C) indicates that the high prevalence of 5mC in DNA does not prevent or significantly alter TET activity.
To check whether this sequence preference is shared between all three mouse TET paralogs, we assayed the specificity of mouse TET1 and TET2 (mTET1 and mTET2) catalytic domains. For mTET1 and mTET2, we observed a similar sequence preference (MCGW, i.e., A or C in the −1 position and T or A in the +1 position) (Figs. 1D and 2, A to C, and fig. S1B) despite lower overall activity of mTET1 in the assays. Unexpectedly, in contrast to mTET3, mTET2 showed reduced selectivity for 5mCG and also acted on 5mCC sites with ~50% efficiency.
In addition to mammalian TETs, we also examined the activity of the recombinant Naegleria gruberi TET (nTET) enzyme from the amoeba Naegleria gruberi. nTET is distantly related to mammalian enzymes, whereby it shares a similar overall structure yet is missing the Cys-rich region and binds DNA somewhat differently. nTET was shown to efficiently convert 5mC all the way to 5caC in vitro (17). We also observed a very robust catalytic activity of nTET on all the provided substrates and found that when compared to mammalian enzymes, nTET was able to oxidize 5mC in a broader sequence context, preferentially oxidizing CA and CG sites over CC and CT sites (Figs. 1D and 2). This indicates that the sequence specificity profile obtained for mTET1, mTET2, and mTET3 is characteristic for mammalian enzymes.
To analyze sequence preferences quantitatively, we built predictive models for the demethylation rates, assuming an independent site model (i.e., overall preferences are products of individual site preferences). According to this model, logarithms of catalytic rates should be amenable to linear regression. This was indeed the case, with good correlation coefficients, indicating that preferences in the flanking regions of the central CG dinucleotide were not strongly interdependent (fig. S2).
Overall, our in vitro results show that activity of mammalian TETs, and, to a lesser extent, also nTET, depends on sequence context flanking the target 5mC. The bases outside the central CG have a strong influence on the rate of catalysis. The mTET1, mTET2, and mTET3 sequence preferences are similar (MCGW), suggesting that the TET sequence preferences may be shared among paralogs (Fig. 2 and fig. S1C).
Structural basis of TET sequence preference
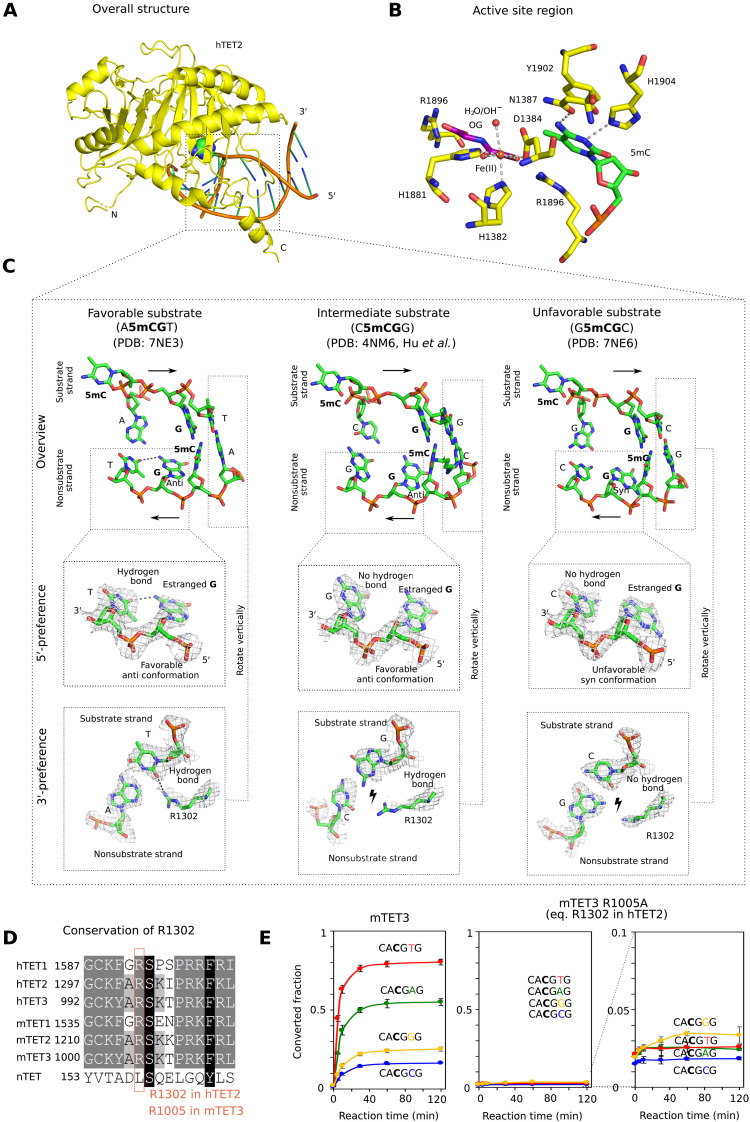
To understand the structural basis for TET sequence preferences and their conservation among TET paralogs, we aimed to crystallize vertebrate TET protein complexes with the most and least favorable substrates. Among the vertebrate TET paralogs, only human TET2 catalytic domain (hTET2) could be crystallized in our hands. We used the truncated version of the protein (residues 1129 to 1936) with a 15-residue glycine-serine (GS) linker replacing the internal disordered region (residues 1481 to 1843) similar to that used in the original report on the TET2 structure (18). We grew crystals in the previously published C222(1) crystal form, with oligoduplexes containing the most and least favorable sequences, CA5mCGTG and GG5mCGCC, respectively. For crystallization, we used the enzyme with the native Fe2+ (favorable substrate) or Mn2+ (unfavorable substrate) in the active site and replaced the cosubstrate 2-oxoglutarate with oxalylglycine, which does not support the reaction.
Overall, the structures with the two substrates are very similar to each other and resemble the previously published hTET2:DNA costructure [Protein Data Bank (PDB) accession: 4NM6 (19), <0.4-Å Cα root mean square deviation between any pair of structures] (figs. S3 and S4, and Fig. 3, A and B). Recognition of both the 5′- and 3′-flanking sequences is indirect, not because of direct discriminating amino acid contacts.
Fig. 3. Structural basis for TET sequence specificity.
(A) Structure of the core region of hTET2 (residues 1129 to 1936, with a 15-residue GS linker replacing disordered residues 1481 to 1843) with the most favorable substrate. Protein is shown in yellow in ribbon representation and DNA in schematic representation (brown backbone and green/blue nucleobases). The substrate 5mC base is highlighted in all-atom representation. The structure with the least favorable substrate is indistinguishable at this level of detail, except at the very N terminus, which is very uncertain because of high B factors (fig. S3). (B) Active site region with key hTET2 residues (yellow), Fe2+ (brown), the cosubstrate analog N-oxalylglycine (purple), and the substrate 5-methyl-2′-deoxycytidine monophosphate (green). At the level of resolution of the crystal structures, the active site regions are indistinguishable for the complexes with the most and least favorable substrates. (C) Conformation of the central four 2′-deoxynucleotides of substrate and nonsubstrate strands. In the magnified regions, composite omit densities contoured at 1σ are shown. (D) Conservation of the arginine residue (R1302 in hTET2) responsible for 3′-substrate preferences in the TET paralogs. (E) Confirmation of the relevance of the conserved arginine (R1302 in hTET2 and R1005 in mTET3) for the 3′-substrate preference. A synthetic substrate containing four 5mCG sites embedded in CA5mCGNG context differing only in the base pair immediately downstream of the methylated CG was subjected to oxidation by mTET3 or mTET3 R1005A, followed by quantification of conversion of 5mC to 5fC and 5caC by bisulfite sequencing.
The TET sequence preference on the 5′ side of a 5mCG (in the substrate strand) is due to interactions in the nonsubstrate strand. For the favorable substrate with an A upstream of the 5mCG, the estranged guanine in the nonsubstrate strand (that was originally base paired with the substrate 5mC) has its glycosidic bond to the 2′-deoxyribose in favorable anti-orientation. As a result, it can donate a hydrogen bond from its exocyclic amino group to the T in −1 position in the nonsubstrate strand (Fig. 3C, left). For the intermediate substrate, which has a C upstream of the 5mCG in the substrate strand, the estranged guanine still adopts the favorable anti-conformation, but the hydrogen bond is not formed (Fig. 3C, middle). For the least favorable substrate with a G upstream of the 5mCG in the substrate strand, the complementary C in the bottom strand is in steric conflict with the estranged guanine in anti-orientation and therefore drives this nucleobase into the unfavorable syn conformation (Fig. 3C, right). A similar steric conflict is also expected for T in the substrate and hence A in the nonsubstrate strand. Therefore, the intrastrand interactions in the nonsubstrate strand favor A, followed by C (together abbreviated as M for a base with an exocyclic amino group) in the substrate strand upstream of the 5mCG.
The TET sequence preference on the 3′ side of the 5mCG (in the substrate strand) is due to interactions with a conserved arginine residue (R1302 in hTET2; Fig. 3D). For the favorable A or T (often abbreviated as W for a base taking part in a weak base pair), the arginine adopts an “in” conformation that enables the hydrogen bond formation with a universal acceptor site in the minor groove of the DNA. By contrast, for the unfavorable C or G, the arginine is pushed into an “out” conformation by the presence of the two-amino group of the guanine in the central minor groove, which abolishes the favorable interaction (Fig. 3C). We confirmed the relevance of this arginine residue (R1302 in hTET2 and R1005 in mTET3) by a comparison of the ability of wild-type mTET3 and the R1005A variant to oxidize 5mC to 5fC and 5caC, as assayed by bisulfite conversion. We designed a synthetic dsDNA substrate containing four 5mCGs embedded in CA5mCGNG sequence context whereby different sites varied only in the base pair immediately downstream of the 5mCG (in the substrate strand). As expected, the wild-type enzyme oxidized substrates with T or A in this position faster than substrates with G or C (Fig. 3E, left). The mTET3 R1005A variant was severely compromised in its ability to oxidize any of the four substrates, presumably because of lost favorable electrostatic interactions between the arginine residue and the DNA phosphodiester backbone and showed no preference for A or T in this position (G was now best, but differences were close to bisulfite conversion noise) (Fig. 3E, middle and right).
Together, the biochemical and structural data suggest that TET sequence specificity is best described as MCGW. This conclusion was independently confirmed by molecular dynamics (MD) simulations, which generated very similar results to the crystallographic analysis, except for the transition to the disfavored syn conformation of the glycosidic bond for the unfavorable substrate that was not seen in the simulations (fig. S5).
TET sequence specificity in culture and in vivo
To test the contribution of this inherent flanking sequence specificity of TETs on the genomic demethylation pattern, we expressed the mouse TET3 catalytic domain transgene (mTET3) in mouse embryonic stem cells (mESCs) using an inducible piggyBAC construct (Fig. 4A and fig. S6A). To ensure that there were no confounding interactions with endogenous TET proteins, we used the mESC line background with a triple genetic knockout for TET1 to TET3 (TET-TKO) (20). As 5fC and 5caC are very rare and unstable in vivo, we used whole-genome bisulfite sequencing (WGBS) as a readout of cytosine demethylation (i.e., loss of 5mC and 5hmC) in 6-hour intervals over 72 hours of doxycycline (dox) induction (Fig. 4A). We found that global CG methylation was reduced by 12.8 percentage points (pp) at 30 hours after dox treatment and then slowly regained 3.0 pp by 72 hours (red line, Fig. 4B). In contrast, global methylation did not change in control TET-TKO cells over the same period (fig. S6B).
Fig. 4. TET3 catalytic domain selectivity in vitro and in cultured mESCs is proportional.
(A) The TET3 catalytic domain (mTET3) was overexpressed in a TET triple-knockout (TET-TKO) mESC line using an inducible piggyBAC transposon system. Dox and control (no-dox) samples were collected, in triplicate, every 6 hours over a 72-hour period. (B) Absolute methylation levels in non-CGI contexts (red) and the difference in methylation of individual motifs (gray lines) following dox-induced mTET3 expression. (C) Demethylation velocities of CG-containing hexamer motifs from (B) (calculated from linear phase, 6 to 18 hours), compared to log2 demethylation velocities in vitro (Fig. 2B). (D) Absolute methylation levels in non-CGI contexts (red) and the difference in methylation of individual motifs (gray lines) following demethylation by the small molecule decitabine. (E) Demethylation velocities of CG-containing hexamer motifs from (E) (calculated from linear phase, 0 to 12 hours), compared to log2 demethylation velocity in vitro (Fig. 2B). (F) 5hmC levels at all 256 hexamers (gray line), 0 to 12 hours after dox treatment. Global 5hmC is represented in red. (G) Demethylation velocities of CG-containing hexamer motifs from (B), compared to 5hmC accumulation rate (0 to 12 hours).
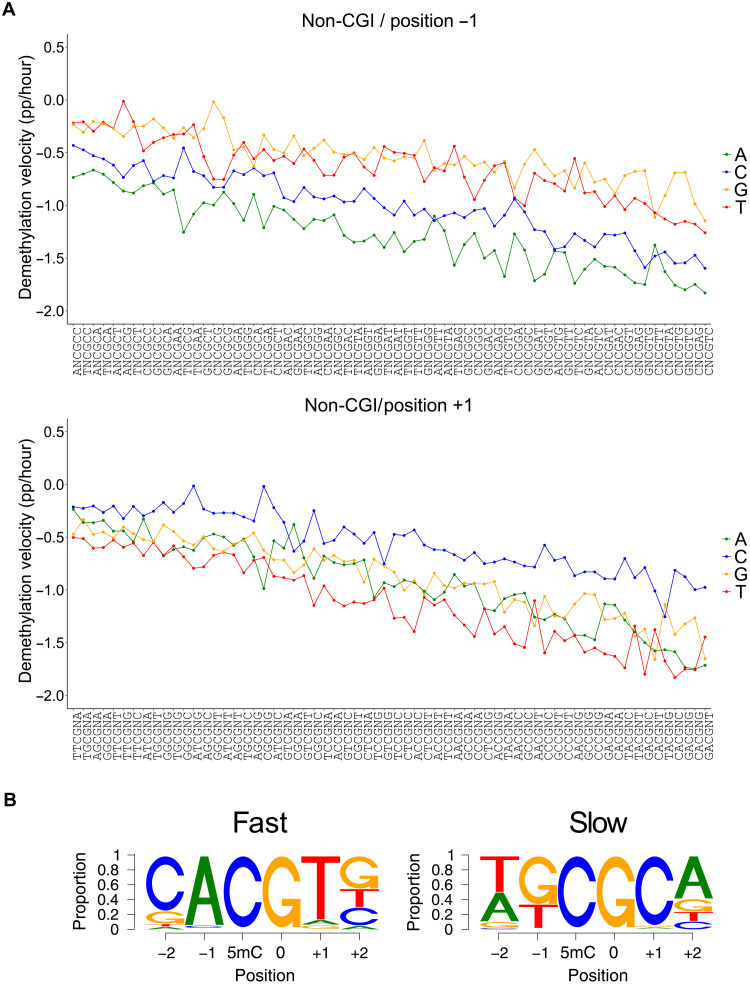
To determine whether mTET3 displayed any sequence specificity within this system, we binned each mapped CG dinucleotide according to the 2 base pairs (bp) flanking it in each direction. Inspection of individual CG-containing hexamer sites revealed a large variation in the demethylation velocity between motifs. We predicted that at least some of this rate variation was due to low starting methylation of some motifs (49 motifs had <65% of starting 5mC; fig. S6C, left). These lowly methylated motifs often contained CGs that were in addition to the central CG (red dots), indicating that they were likely restricted to CpG island regions (CGI), which are well known for significantly reduced methylation levels. When we considered only motifs located in non-CGI regions, starting methylation for all 256 motifs was much more consistent at 75.8 to 90.2% (fig. S6C, right) and was thus used for further analysis. Moreover, for motifs containing CG in addition to central CG, demethylation velocities were shown to be higher within non-CGI contexts (fig. S6D).
Most CG-containing hexamer motifs showed linear demethylation 6 to 18 hours after dox treatment, allowing calculation of maximum demethylation velocity for each motif and comparison to the 38 CG-containing motifs from the in vitro experiment (Fig. 4C). Despite a much larger range of demethylation velocities observed in the in vitro experiment, a significant correlation was observed between the demethylation velocities in the cell culture experiment and log2-transformed values from the synthetic in vitro data (r2 = 0.61, P = 6.5 × 10−9). This indicates that the unique selectivity of TETs observed in in vitro biochemical reaction also exists in a cellular context.
To be more confident that the observed variation in mTET3-targeting was not a technical artifact, we performed two further experiments. The first was to examine motif demethylation dynamics in wild-type V6.5 mESCs following treatment with the demethylating small molecule decitabine (Fig. 4D). Because of the fact that decitabine drives demethylation by inhibition of DNA methyltransferase 1 (DNMT1) (21, 22) (and thus operates in a TET-independent manner), we hypothesized that similar sequence preference should not be observed. Overall, there was minimal variation in motif-demethylation rate following decitabine treatment (Fig. 4D), but most importantly, when compared to the in vitro demethylation, no significant correlation in demethylation velocity was uncovered (Fig. 4E) (r2 = 0.016, P = 0.45). In a second validation experiment, we quantified 5hmC levels from the 0- to 12-hour time points (Fig. 4F) using APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide)–coupled epigenetic sequencing (ACE-seq) (23). As for the bisulfite-based experiment, we found that 5hmC accumulation varied greatly between the 256 CG-containing hexamers (Fig. 4F). Further, we saw that 5hmC was an excellent predictor of demethylation rate determined by WGBS (r2 = 0.57, P = 6 × 10−49; Fig. 4G). We conclude that, perhaps not unexpectedly, the motifs that rapidly gain 5hmC are also those which become rapidly demethylated following mTET3 overexpression and that our measures of TET activity are robust despite using independent bisulfite- and enzymatic-based sequencing techniques.
To further characterize mTET3 catalytic selectivity in mESC cells, we investigated which CG sites were demethylated the fastest and slowest according to the nucleotides at each flanking position when all other bases were kept the same (Fig. 5A); a measure we termed intramotif positional preference (IMPP). In doing so, we found that for 95.3% of motifs (i.e., 61 of 64), those with adenine immediately upstream of CG (i.e., −1 position) were demethylated faster than any other motif with base in that position (Fig. 5A, top). In the three remaining cases, the preferred base at −1 was C—a result perfectly matching expectations from our in vitro data, as well as the structural analysis and modeling. Moreover, when the +1 position was examined, for 79.6% of motifs (51 of 64), those with T demethylated faster than another base, with A being the next most common (Fig. 5A, bottom). Favored nucleotides at the −2 and + 2 position were less obvious; C- and G-containing motifs were the most rapid demethylating in 54.6% (35 of 64) and 40.6% (25 of 64) of instances, respectively. The most preferred nucleotides at each position recapitulated the CACGTG E-box motif (Fig. 5B), as initially uncovered in the in vitro experiments.
Fig. 5. Intramotif positional preference of mTET3-induced demethylation in cultured mESCs.
(A) Demethylation kinetics of the four nucleotides in a given CG containing hexamer position (shown are −1 and + 1 positions flanking the central CG). (B) Proportion of nucleotides that are demethylated the fastest (left) and slowest (right) at each motif position when all other nucleotides in that motif are kept constant.
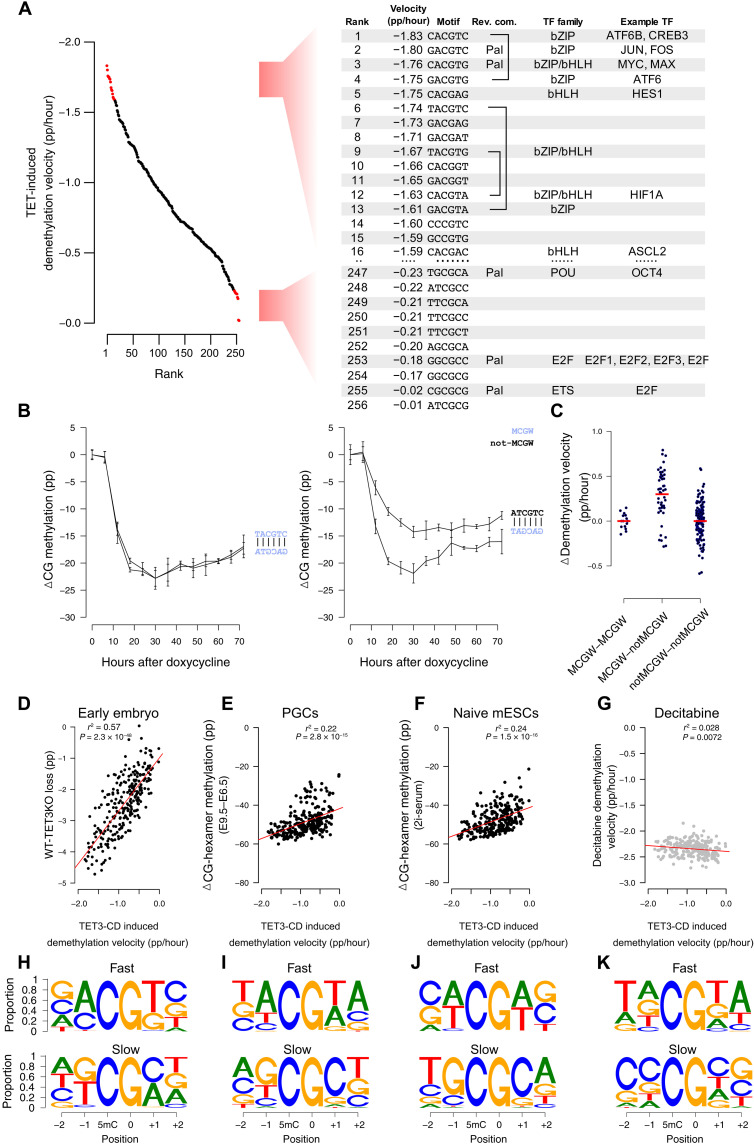
When demethylation rates of each of the 256 CG-containing hexamers were considered individually, the CACGTG E-box sequence was the third most preferentially targeted motif (−1.76% per hour) (Fig. 6A). As mentioned, E-box is notable for binding c-MYC, an iconic “immediate-early” bHLH and bZIP domain–containing protein that is among the first to be transcribed in response to a wide variety of cellular stimuli. Significantly, the two motifs that demethylated faster (CACGTC and GACGTC) also constitute binding sites for bZIP-domain, methylation-sensitive immediate-early transcription factors, i.e., CREB (adenosine 3′,5′-monophosphate response element–binding protein) and JUN/FOS, respectively. A further five motifs in the top 15 favorable recognition sites are bound by bZIP or bHLH domain–containing transcription factors, many of which display methylation-sensitive binding (24) (Fig. 6A). In contrast, the least preferred bases at each position most commonly featured G and C at the −1 and +1 position, with the TGCGCA OCT4-binding motif as the least preferred (Fig. 6A). OCT4 and other members of the POU (pit-oct-unc) transcription factor family are known to bind TGCGCA specifically when methylated (24), indicating that resistance to TET demethylation may be a prerequisite for OCT4 binding.
Fig. 6. TET selectivity in cultured cells coincides with methylation-sensitive transcription factors, is strand dependent, and correlates with global demethylation in vivo.
(A) Many fast demethylating CG-containing hexamer motifs following mTET3 expression (ranks 1 to 16) bind bZIP and bHLH methylation-sensitive transcription factors. Slow demethylating sites (ranks 247 to 256) bind methylation-sensitive E2F and POU family transcription factors. (B) Many complementary motifs have similar demethylation kinetics (e.g., TACGTC; left); however, some are significantly discordant (e.g., ATCGTC; right). (C) CG-containing hexamers with MCGW in one strand demethylate faster than complementary ones without MCGW (MCGW-notMCGW). In contrast, motifs where MCGW is present on both strands (MCGW-MCGW) or not (notMCGW-notMCGW) show equal demethylation rates. (D to G) CG-containing hexamer demethylation velocities following mTET3 overexpression (x axis, mTET3 induced demethylation velocity) and the demethylation found in mouse (y axis) (D) postfertilization embryos following TET3-knockout (WT-TET3KO), (E) primordial germ cells (E9.5 to E6.5), and (F) naive mESCs. Only a weak negative correlation exists with (G) decitabine-treated cells. (H to K) IMPP for the demethylating scenarios listed in (D) to (G). Shown is the proportion of nucleotides that are demethylated the fastest (top) and slowest (bottom) at each motif position when all other nucleotides in that motif are kept constant.
Many rapidly demethylating nonpalindromic sites showed demethylation velocities on each strand that were similar. For example, of the 10 fastest demethylating CG-containing hexamers, 3 had reverse-complement motifs also in the top 20 (Fig. 6A), implying that both strands of a CG-containing hexamer may be targeted with similar efficiency. Despite this, we uncovered notable exceptions, where certain CG-containing hexamer sequences were much preferred over their antisense counterparts (Fig. 6B). We noticed that preferred motifs in a discordant antisense pair were often MCGW, whereas the nonfavored strand was non-MCGW (Fig. 6C). In addition to confirming our biochemical and structural predictions, this result is significant because it shows that demethylation rates on each strand are clearly separable in situations of discordant binding preferences. Hence, we conclude that they are likely demethylated by independent binding events.
Mammalian DNA is subject to two significant waves of demethylation during normal development. The first erasure event occurs in the zygote immediately following fertilization (25), while the second occurs during primordial germ cell specification and proliferation (25). These genome-wide erasure events can also be modeled, at least to some extent, using naive mESC culture conditions (26, 27); however, in all cases, passive demethylation is thought to be the driving demethylating force (28–33). Nevertheless, we found significant correlations between all global demethylation experiments tested (26, 30, 34) and our results (Fig. 6, D to F), particularly when lowly methylated CGI-rich motifs were removed. Moreover, when we analyzed the IMPP in these datasets, we found that all three datasets recapitulated preference for A at −1 and A or T at +1 positions (Fig. 6, H to J, top). The clearest signal of TET activity was uncovered in the early embryo data when we examined the absolute difference in methylation between TET3-KO mice and wild-type mice (Fig. 6D). This is significant because using TET3-KO data allowed us to separate passive and active and/or active-passive contributions to demethylation. In addition, the oocyte-specific “TET3o” variant that predominates in the early embryo resembles the TET3 overexpression construct we used, perhaps explaining in part the particularly high correlation seen for this comparison. In addition, other examples of high correlation included examined WGBS data from three independent human embryonic stem cells (hESCs) whereby the de novo methyltransferases were removed (15), allowing unopposed TET activity (r2 > 0.43, P < 7 × 10−33, n = 3; fig. S7A). Likewise, the CG-containing hexamer demethylation rates from our cell culture experiment were significantly correlated with the change in methylation caused by TET1 to TET3 rescue in TET and DNMT3 “pentuple-knockout” cells (15) (fig. S7B) and average-predicted “TET activity” on 800,000 CG sites in mESCs from a previous study (r2 = 0.29, P = 8.9 × 10−21; fig. S7C) (35).
To exclude the possibility that some other factor was driving this relationship in CG-containing hexamer demethylation rate (e.g., chromatin structure), we assessed TET-independent global demethylation driven by decitabine treatment and found no correlation (Fig. 6G). Moreover, IMPP for those sites losing methylation the fastest following decitabine treatment did not feature any selectivity at −1 or + 1 positions (where we find TET favored A and T in the strict sense, or more loosely M and W), but instead, we uncovered a previously described DNMT1 target motif preference for −2 and + 2 positions, TNCGNW (36) (Fig. 6K).
DISCUSSION
Together, our data demonstrates that the TET catalytic domains have a previously unknown intrinsic sequence specificity, as also recently shown in vitro for TET1 and TET2 (37). We provide a structural understanding for the shared sequence preference of the TET paralogs and demonstrate that it can be detected in a wide range of methylation erasure scenarios, both in vivo and in culture. We show that the intrinsic sequence preference of the TET catalytic domains significantly contributes to the establishment of DNA methylation patterns and TET function in the cell, in addition to other identified targeting mechanisms, in particular CXXC domain targeting and chromatin factors. Highly favored motifs constitute targets of immediate-early transcription factors, which are the first to respond to a range of stimuli such as mitogens or infection and proceed to initiate expression of downstream effector genes by binding to DNA in a methylation-sensitive manner. Thus, it makes biological sense that their binding sites should be efficiently targeted for DNA methylation erasure. Furthermore, while TET apparently acts to remove methylation from DNA to allow binding of immediate-early effectors, it may also help preserving DNA methylation at OCT4 sites (by avoiding them) to equally stimulate OCT4 binding and maintenance of developmental potency (fig. S8). Together, our data support a model where, on multiple levels, the kinetics of TET-mediated demethylation is inextricably tied to the biological function of the TET enzymes.
MATERIALS AND METHODS
Cloning, expression, and purification of TET enzymes
Construction of the pET28a vectors containing the catalytic domains of mouse TET1, TET2, and TET3 has been described previously (8). Full-length Naegleria gruberi TET (nTET) was cloned from a synthetic gene [Integrated DNA Technologies (IDT)] into pET28a bacterial expression vector in fusion with the N-terminal His-tag. The pET28a vectors encoding each TET catalytic domain were transformed in Escherichia coli BL21 (DE3) CodonPlus RIL (Novagen) and grown on kanamycin and chloramphenicol selection plates overnight. For protein expression, single colonies were inoculated in 100 ml of LB media supplemented with appropriate antibiotics and grown overnight at 37°C in a shaking incubator. For each protein expression, 4 liters of LB media was inoculated with the overnight culture, and the cells were grown at 37°C until optical density at 600 nm (OD600) of 0.6 was reached, recombinant protein expression was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and culture was grown at 20°C for ~14 to 15 hours. The cells were harvested by centrifugation [Lynx 6000 (Thermo Fisher Scientific), Fiberlite F9-6 1000 LEX, at 4200 rpm for 15 min), washed with 1× Sodium chloride-Tris-EDTA (STE) buffer [10 mM tris-HCl (pH 8.0), 100 mM NaCl, and 1 mM EDTA], and stored at −20°C until further use. For purification, cells were resuspended in lysis buffer [50 mM Hepes (pH 6.8), 35 mM imidazole, 1 mM α-ketoglutarate, 1 mM dithiothreitol (DTT), 500 mM NaCl, and 10% glycerol, supplemented with protease inhibitor cocktail (Roche)] and disrupted using a Bandelin Sonoplus ultrasonic homogenizer. The cell lysates were cleared by centrifugation [Lynx 600 (Thermo Fisher Scientific), Fiberlite F21-8x50y] at 38,300g for 75 min at 4°C, and the supernatant was loaded onto an affinity column containing 2 ml of Ni–nitrilotriacetic acid (NTA) agarose beads (Genaxxon, Germany). The beads were washed thoroughly with wash buffer [50 mM Hepes (pH 6.8), 35 mM imidazole, 1 mM α-ketoglutarate, 1 mM DTT, 500 mM NaCl, and 10% glycerol] and eluted with elution buffer [50 mM Hepes (pH 6.8), 300 mM imidazole, 1 mM DTT, 500 mM NaCl, and 10% glycerol]. The concentrated fractions were pooled, dialyzed against dialysis buffer [50 mM Hepes (pH 6.8), 1 mM α-ketoglutarate, 1 mM DTT, 300 mM NaCl, and 10% glycerol], subsequently aliquoted, flash-frozen in liquid nitrogen, and stored at −80°C until further use.
Preparation of TET in vitro substrates
The substrates for in vitro reactions—human CG-rich (249 bp), promotor fragments of mouse Esrrb (249 bp), mouse Nanog (260 bp), and mouse Tcl1 (251 bp)—were amplified from genomic DNA, subcloned into pCR2.1 vector using the TOPO TA Cloning Kit (Invitrogen), and sequence-confirmed (substrate sequences are listed in table S2). Fully 5mC-modified substrates with methylated cytosine in both CG and non-CG contexts were amplified from subcloned template plasmids using primers listed in table S2. Fully 5mC-modified substrates with methylated cytosine in both CG and non-CG contexts were amplified by polymerase chain reaction (PCR) from subcloned template plasmids (primers listed in table S2) using in-house–made Taq polymerase and deoxynucleotide triphosphate (dNTP) mixture containing 2′-deoxy-5-methylcytidine 5′-triphosphate (5mdCTP) (NEB, N0356S) instead of 2′-deoxycytidine 5′-triphosphate. To generate substrates only methylated at CG sites, unmethylated templates were amplified by PCR using in-house–made TaqPol with standard dNTP mix, and the purified PCR products were further methylated with M.SssI methyltransferase (NEB, M0226S) following the manufacturer’s protocol. The amplified substrates were purified using #4.1 Bio-On-Magnetic-Beads (BOMB) protocol (38) and verified with gel electrophoresis.
For LC-MS, a 26-bp hemimethylated substrate was prepared by annealing complementary oligonucleotides (IDT, listed in table S2). Briefly, 20 μM of each oligo was resuspended in annealing buffer [10 mM Hepes (pH 7.4) and 50 mM NaCl]. The oligos were heated at 85°C for 5 min and were cooled down gradually to room temperature over several hours. The annealed oligos were stored at −20°C until further use.
TET activity assays
To analyze the activity of mammalian TETs, a 0.15 μM DNA substrate was incubated with 2 μM enzymes in a reaction mixture containing 50 mM Hepes (pH 6.8), 100 μM Fe2+, 1 mM α-ketoglutarate, 1 mM ascorbic acid, and 150 mM NaCl at 37°C for up to 120 min. The reaction was stopped at specified time points by adding an aliquot of the reaction mixture to 1% SDS and proteinase K (NEB) for an hour at 50°C. Proteinase K was inactivated, and the DNA was purified using a DNA purification kit (Macherey-Nagel) following the manufacturer’s protocol. The purified DNA was ligated with methylated TruSeq LT Illumina adapters and subjected to bisulfite conversion using the EZ-DNA Methylation-Lightning Kit (Zymo Research, D5030) according to the manufacturer’s protocol. The bisulfite-converted DNA was amplified by PCR (primer details in table S2), and successful amplification was verified using gel electrophoresis. The amplified products were quantified using NEBNext kit (E7630S, NEB) and sequenced on an Illumina MiSeq 2 × 300-bp platform (Illumina).
For the nTET activity assay, the reaction was carried out as outlined above with slight modifications. Briefly, 0.5 μM DNA substrate was treated with 2 μM enzyme in a reaction mixture containing 50 mM bis-tris (Sigma-Aldrich, 14879-100G-F; pH 6.0), 75 μM Fe2+, 1 mM α-ketoglutarate (Sigma-Aldrich, 75890-25G), 1 mM ascorbic acid, and 100 mM NaCl for specified time at 34°C. After enzymatic treatment, the DNA was processed using same procedure as described above for the mammalian enzymes.
Liquid chromatography–mass spectrometry
To quantify the oxidized products of TET enzyme using LC-MS, 0.5 μM hemimethylated 26-bp dsDNA substrates (oligonucleotide sequences listed in table S2) were treated with mTET3 CD for 1, 5, 10, 20, and 40 min. The reactions were stopped at specified time points by adding 10 mM EDTA and flash-frozen in liquid nitrogen. Subsequently, the enzyme was inactivated at 95°C for 5 min, followed by proteinase K (New England Biolabs, P8107S) treatment for an hour at 50°C. The DNA was recovered by ethanol precipitation and analyzed by LC-MS.
For this, the purified DNA oligonucleotides were digested into nucleosides using the following protocol: 160 ng of each DNA, 0.6 U of nuclease P1 from Penicillium citrinum (Sigma-Aldrich), 0.2 U of snake venom phosphodiesterase from Crotalus adamanteus (Worthington), 200 ng of pentostatin (Sigma-Aldrich), and 500 ng of tetrahydrouridine (Merck-Millipore) were incubated at 37°C in 5 mM ammonium acetate (pH 5.3; Sigma-Aldrich) for 2 hours. Afterward, 1/10 volume of 10× fast alkaline phosphatase buffer (50 mM NH4OAc; pH 9.0) and 1 U of fast alkaline phosphatase (Fermentas) were added, followed by incubation at 37°C for 1 hour. The nucleosides were then spiked with labeled internal standard (D3-dm5C and D2-hdm5C) and subjected to analysis. For each time point, 400 fmol of digested DNA oligo and 100 fmol of each internal standard were injected and analyzed via LC-MS [Agilent 1260 series and Agilent 6460 Triple Quadrupole mass spectrometer equipped with an electrospray ion source (ESI)]. The solvents consisted of 5 mM ammonium acetate buffer (pH 5.3; solvent A) and LC-MS grade acetonitrile (solvent B; Honeywell). The elution started with 100% solvent A with a flow rate of 0.35 ml/min, followed by a linear gradient to 20% solvent B at 10 min and back to 100% solvent A in 2 min. Initial conditions were regenerated with 100% solvent A for 5 min. The column used was a Synergi Fusion (4 μM particle size, 80-Å pore size, 250 mm by 2.0 mm; Phenomenex). The ultraviolet signal at 254 nm was recorded via a diode array detector to monitor the main nucleosides. ESI parameters were as follows: gas temperature of 350°C, gas flow of 8 liters/min, nebulizer pressure of 344 kPa (50 psi), sheath gas temperature of 350°C, sheath gas flow of 12 liters/min, and capillary voltage of 3000 V. The MS was operated in the positive ion mode using Agilent MassHunter software in the dynamic multiple reaction monitoring mode. For quantification, a combination of external and internal calibration was applied as described previously (39).
Bioinformatic analysis of sequencing from in vitro demethylation
The quality control of the next-generation sequences in FASTQ format was done using FastQC (Babraham Bioinformatics). The adapters from the sequences were trimmed and quality-controlled using TrimGalore (Babraham Bioinformatics). Files were then demultiplexed using a custom script and processed further using BiQ Analyzer HT (Max-Plank-Institut Informatik), where all Cs in CpN contexts were analyzed and exported to an Excel readable file. The average oxidation at each C was calculated, and the overall oxidation for each time point is obtained by subtracting the oxidation at respective time point from the control. Linear regression of the initial reaction points was used to estimate the demethylation velocity for each site.
Linear regression model to predict reaction rates
For quantitative analysis of the sequence dependence of demethylation rates, we assumed uncorrelated sequence preferences. Under this assumption, the logarithms of reaction rates should be amenable to linear regression (in the R language) with DNA base identities as categorical variables. Two bases upstream of the C and two bases downstream of the C were included in all models. For the in vitro data, the position of the G was also kept variable, but for the in cellulo data, the G was fixed (as rates were only available in CG context). For the in vitro data, reaction rates were available from two independent experiments. As the reaction rate logarithm for a given sequence, we used the average of the rate logarithms. Regression was done with weighting, using the inverse absolute difference of the reaction rate logarithms (augmented by 0.1 to avoid overemphasis on data points with accidentally good agreement). This way, sequences with good reaction rate agreement in the two experimental repeats contribute more to the regression analysis than sequences with larger discrepancies. For the in cellulo data, only a single set of reaction rates was available from the upstream analysis, and hence, weighting was not used. To exclude overfitting, data were split into “training” and “testing” sets. Except for the TET1 in vitro data, correlation coefficients were very similar when the full dataset was used for training and testing and when training and testing data were nonoverlapping.
Expression of human TET2 for structural studies
For structural studies, we used a fragment of human TET2 (1129 to 1936) with residues 1481 to 1843 replaced by a 15-residue GS linker. The protein was expressed with an N-terminal His6-Gly3-His6-SUMO-tag. This was done in E. coli BL21 (DE3) CodonPlus RIL cells (Novagen) under T7 promoter control from a pET28a-derived plasmid, which was maintained under kanamycin and chloramphenicol selection. A preculture at OD = 0.8 1/cm was induced with 0.1 mM IPTG and grown overnight at 16°C (shaking at 130 rpm). Cells were harvested by centrifugation (4000g at 4°C for 30 min), and bacterial pellets were frozen and stored at −20°C until use.
Protein purification of human TET2 for structural studies
The hTET2 bacterial pellets from 2 liters of culture were resuspended in 50 ml of sonication buffer [50 mM Hepes (pH 8.0), 500 mM NaCl, 10 mM imidazole, 20% glycerol, 5 mM β-mercaptoethanol, and 2 mM phenylmethylsulfonyl fluoride] and sonicated on ice (15 s of pulse, 45 s of rest, 5.5 power, and total sonication time of 6 min). The lysate was cleared by ultracentrifugation (4°C, 40 min, and 40,000g) and applied two times on a column containing 5 ml of Ni-NTA resin (Qiagen) equilibrated with the Sonication buffer. The column was washed sequentially with 100 ml of Wash1 buffer [20 mM Hepes (pH 8.0), 500 mM NaCl, 10 mM imidazole, and 5 mM β-mercaptoethanol], 500 ml of Wash buffer [20 mM Hepes (pH 8.0), 1750 mM NaCl, 1 mM imidazole, and 5 mM β-mercaptoethanol], DnaK buffer to remove DnaK chaperone [20 mM Hepes (pH 8.0), 50 mM NaCl, 2 mM MgCl2, 2 mM adenosine 5′-triphosphate, and 5 mM β-mercaptoethanol], and Ulp1 buffer [20 mM Hepes (pH 8.0), 150 mM NaCl, 15 mM imidazole, and 5 mM β-mercaptoethanol). The protein was cleaved from the tag on-column (170 μg of Ulp1 in Ulp1 buffer for protein from 10 liters of bacterial culture, 15 hours at 6°C). Eluted protein was diluted in dilution buffer [20 mM Hepes (pH 8.0) and 5 mM β-mercaptoethanol] and applied to 5 ml of heparin column (GE Healthcare) equilibrated in Hep1 buffer [20 mM Hepes (pH 8.0), 40 mM NaCl, and 5 mM β-mercaptoethanol] and eluted with a gradient between this buffer and Hep2 buffer [20 mM Hepes (pH 8.0), 2000 mM NaCl, and 5 mM β-mercaptoethanol]. hTET2 fractions were then concentrated to 1 ml and subjected to gel filtration on a Superdex 200 (GE Healthcare) column equilibrated in gel filtration (GF) buffer [10 mM Hepes (pH 7.4), 100 mM NaCl, and 1 mM DTT].
Crystallization and structure determination
For crystallization, we used equimolar mixtures of hTET2 [from a stock (70 mg/ml) in GF buffer] and 12-mer dsDNA (from IDT, in water). Top strand sequences of most and least optimal substrates were 5′-ACACA5mCGTGTGT 3′ and 5′-ACAGG5mCGCCTGT-3′, respectively. Complementary strands, also with DNA methylation, were annealed into top strands by heating to 95°C for 10 min and subsequent slow cooling. For crystallization, 1.5 μl of a 0.5 mM solution of hTET2-dsDNA complex were mixed with 1.5 μl of reservoir buffer [100 mM MES (pH 6.3) and polyethylene glycol (PEG) 2000 monomethyl ether] and supplemented with 2 mM of the cosubstrate analog N-oxalylglycine and 1 mM Fe2+ (favorable substrate) or Mn2+ (unfavorable substrate). Crystals were grown in hanging drops at 4°C by equilibration of the crystallization mix against reservoir buffer. For the complex of hTET2 with the least optimal substrate, seeding was required. A crystal of insufficient quality for diffraction experiments was crushed, crystal seeds were suspended in 50 μl of reservoir buffer, and this stock was then used for seeding (seed solution was 10% of the crystallization drop volume). Diffraction data up to 2.0-Å resolution for the complexes with most and least optimal substrates were collected at beamlines of the Deutsches Elektronen-Synchrotron (P11, DESY, Hamburg) and Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung m. b. H. (BESSY, Berlin), respectively, at 100 K. Both datasets were processed with XDSapp (40). Structures were solved by the Phaser program (41), using the previous PDB TET2 model (PDB accession: 4NM6) as template, and refined using Phenix software (42) (table S1).
MD simulations
All MD and energy minimization were performed using the Amber16 simulation package (43). Using the published TET2:DNA complex structure (PDB accession: 4NM6) as a reference, the hexameric recognition sequence AC5mCGGT was systematically varied at positions 2 and 5 (the positions flanking the CG) to generate all 16 possible sequence variants using the xleap module of Amber16. The resulting complex structures were conformationally relaxed using energy minimization (5000 conjugated gradient steps) and short MD simulations at 290 K for 1 ns followed by another energy minimization (5000 steps). The parm14SB (for the protein) and the parmBsc1 force field (for the DNA) were used in combination with a generalized Born implicit solvent as implemented in Amber16 (igb = 5 option). During each minimization and MD phase, the backbone atoms of the DNA and of the protein were restrained to the positions found in the 4NM6 reference structure but allowing full mobility of the side chains.
ESC lines and culturing conditions
mESC culture was performed using standard media conditions in the presence of serum and leukemia inhibitory factor (LIF) [Dulbecco’s modified Eagle’s medium, glucose (4500 mg/liter), 4 mM l-glutamine, sodium pyruvate (110 mg/liter), 15% fetal bovine serum, penicillin (1 U/ml), streptomycin (1 mg/ml), 0.1 mM nonessential amino acids, 50 mM β-mercaptoethanol, and LIF (1000 U/ml)]. All cells were grown “feeder-free” on gelatin-coated plates and were maintained at 37°C in a humidified 5% CO2-containing atmosphere.
To create an ESC line with inducible TET expression, we first created a PiggyBAC construct that contained the mouse TET3 catalytic domain (mTET3) under the control of a tetracycline responsive promoter (pB-tetO2-mTET3cd-mCherry). The correct identity of this construct was confirmed using shotgun Illumina sequencing on the MiSeq platform. The assembled construct sequence can be found on the NCBI sequence archive under accession (MW139646).
A construct mix containing 4 μg of pB-tetO2-mTET3cd-mCherry, 4 μg of pB-CAG-rtTA-Puro [containing the reverse tetracycline-controlled transactivator (rtTA) gene] and 4 μg of the pCAG-pBASE vector [containing a PiggyBac transposase allowing “cut and paste” insertion of the construct into the host genome (44)] was prepared in 125 ml of Opti-MEM Reduced Serum Media (Thermo Fisher Scientific, catalog no. 31985062). Transfection Reagent was prepared separately by adding 89 μl of Opti-MEM to 36 μl of FuGENE HD (Promega, E2311) and then incubated for 5 min. Once the construct mix and FuGENE solutions were incubated separately for 5 min, they were added together and incubated for an additional 20 min before being added dropwise to TET triple-knockout ESCs, thus following a previously established protocol (20). Cell cultures were then incubated at 37°C for 2 to 6 hours in 250 μl of media, allowing transfection to take place. Selection for positive transfectants was performed over 7 days by adding puromycin to the media (2 μg/ml).
Once a stable mTET3-CD complemented TET-TKO line was produced, media supplemented with ascorbate (50 μg/ml; Sigma-Aldrich, A7631) was added to cells plated at low density in 12-well plates overnight. The following morning, mTET3-CD expression was initiated by the addition of 1 μM dox. Control and dox-treated cells were harvested in triplicate every 6 hours over the next 72 hours.
For the decitabine treatment experiment, wild-type V6.5 hybrid ESCs (i.e., C57BL/6 X 129/sv cross; a gift from B. Oback) were seeded at low density and treated with 0.215 μM decitabine (5-aza-2′-deoxycytidine) over a period of 48 hours. This level of decitabine had been determined empirically in a prior experiment as being the highest concentration that did not cause overt cell death. In both experiments, cells were harvested by media removal and addition of a 4 M guanidinium isothiocyanate-based lysis buffer. Cell lysates were then stored at −80°C ahead of total nucleic acid purification using the BOMB system (38). Briefly, cell lysate was combined with TE-diluted Sera-Mag Magnetic SpeedBeads (GE Healthcare, GEHE45152105050250) and isopropanol in a volumetric ratio of 2:3:4 (beads:lysate:isopropanol). Beads were captured with a neodymium magnet, washed once with isopropanol and twice with 70% ethanol, and resuspended in Milli-Q water.
Postbisulfite adapter tagging library preparation and sequencing
Bisulfite-converted libraries were prepared using a modified postbisulfite adapter tagging (PBAT) method (45). Purified DNA was subjected to bisulfite conversion using the EZ-96 DNA methylation DirectTM MagPrep Lit (Zymo Research, D5044), according to the manufacturer’s user guide, with reagent volumes scaled down to 25% of the recommended volume, with the exception of the final elution step that remained at 25 μl. To synthesize the first strand, we used converted DNA and 5′-biotinylated adapter primers containing seven random nucleotides at its 3′ end (BioP5N7, biotin, ACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNNN). The first-strand product was purified using streptavidin-coated magnetic beads (Thermo Fisher Scientific, 11205D) and alkaline denaturation. Second-strand DNA was synthesized using the immobilized first-strand DNA and another adapter primer also containing seven random nucleotides at its 3′ end (P7N7, GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNNN). Unique molecular barcodes and sequences essential for binding to Illumina flow-cells were added to the second-strand DNA by PCR using 1 × HiFi HotStart Uracil+ Mix (KAPA, KK2801) and 10 μM indexed TruSeq-type oligos, amplified by 15 cycles of PCR and size-selected by PEG-diluted SPRI (solid phase reversible immobilization) beads. Library integrity was determined by agarose gel electrophoresis and sequenced on an Illumina HiSeq using single-end 100-bp chemistry.
APOBEC-coupled epigenetic sequencing
For ACE-seq, 10 ng of purified DNA was enzymatically fragmented to ~300 bp with dsDNA Fragmentase (#M0348 NEB), according to the manufacturer instructions, with the reaction scaled down 50% and incubation time of 37°C for 32 min. Fragmented DNA was concentrated using PEG-diluted SPRI beads. In a total volume of 5 μl, 5hmC was glucosylated with 5 U of T4 phage β-glucosyltransferase in NEB buffer 4 supplemented with 2 mM uridine 5′-diphosphate–glucose at 37°C for 1 hour. Glucosylated DNA was denatured in the presence of 20% dimethyl sulfoxide at 95°C for 5 min then swiftly transferred to a precooled PCR tube rack on dry ice for snap-cooling. The single-stranded glucosylated DNA was then deaminated in 50 mM bis-tris (pH 6.0), 0.1% Triton X-100 (APOBEC Reaction Buffer, EM-seq component E7134AA, NEB), and 20 μg of bovine serum albumin (NEB) using 0.2 μg of APOBEC3A (EM-seq component E7133AA, NEB), in a total volume of 10 μl. DNA was deaminated by APOPBEC3A at 4°C for 10 min, then ramping from 4° to 50°C over 2 hours followed by 50°C for 10 min as previously reported (46). First-strand synthesis, second-strand synthesis for adapter tagging, and PCR were completed as described above for PBAT. Library integrity was determined by agarose gel electrophoresis, libraries were pooled and size-selected by gel extraction (MinElute Gel Extraction Kit, Qiagen) and sequenced over three Illumina iSeq 100 runs using paired-end 150-bp chemistry.
Bioinformatic analysis of the TET3-CD overexpression experiment
The quality of the raw FASTQ files was evaluated using FastQC software [www.bioinformatics.babraham.ac.uk/projects/fastqc/] (v0.11.9). Raw reads were trimmed using Trim Galore! [www.bioinformatics.babraham.ac.uk/projects/trim_galore/] (v0.6.4), in a two-step process. First, adapters were removed, 10 bp was hard-trimmed from the 5′ end of all reads, and then low-quality base calls (Phred score < 20) were removed. Read mapping and base calling were performed using Bismark (v0.22.3) with the option --pbat specified (47). Mus musculus genome assembly GRCm38 (mm10) was used as reference. Bismark output files were deduplicated, and methylation calls were obtained. The nonconversion rate during the bisulfite treatment was evaluated by calculating the proportion of non-CG methylation; by this measure, all libraries had a bisulfite conversion efficiency of at least 97.5% (data file S1).
Methylation in the CG context for each hexamer was calculated using an in-house Python script. Briefly, methylation calls were tracked to the reference genome, validated as CG context, and the −2, −1, +1, and + 2 nucleotides were examined to determine the identity of the CG-containing hexamer (NNCGNN). Then, methylation status for each call was extracted and added to a count table containing each hexamer. Last, hexamer methylation was calculated as the proportion of total methylated cytosines over total cytosines. To examine methylation in CGI regions, previously published CGI coordinates (13) were converted from mm9 to GRCm38 (mm10) using the liftOver tool [https://genome.ucsc.edu/cgi-bin/hgLiftOver] (48). Then, methylation calls were classified as CGI or non-CGI and used as input for the methylation by hexamer script. Although some of CG-containing hexamers are more common than others in the mouse genome (meaning that they were more likely to have overlapping reads associated with them), the average number of methylation calls for each motif was 6332. Theoretical asymptotic estimators predict technical error associated with this level of sequencing at ±1.03 pp. (95% confidence) (49).
The demethylation velocity for each hexamer, following TET3-CD overexpression and decitabine treatment, was calculated using the linear phase of demethylation (6 to 18 hours and 0 to 32 hours after treatment, respectively). Further in-house R-scripts were developed to assess the characteristics of demethylation velocity for a range of motif parameters and groups (e.g., Intra-Motif Positional Preference and reverse complement motifs), all of which can be found on our GitHub code repository (https://github.com/TimHore-Otago/TET_specificity). Last, these demethylation velocities were compared to those calculated from previously published BS-seq datasets (data file S2) (15, 26, 30, 34, 35), using the same workflow as outlined above.
Acknowledgments
We thank the High Throughput Sequencing unit of the Genomics & Proteomics Core Facility, German Cancer Research Center (DKFZ), Cardiff University Genome Research Hub, and the Otago Genomics Facility for providing excellent next-generation sequencing services. We are grateful to K. Skowronek (IIMCB), R. Pluta (IIMCB), and D. Bond for the technical advice and assistance in the project. We want to thank Y. Xu (Fudan University) for sharing the hTET2 expression construct used for crystallization. Diffraction data for the hTET2:DNA complexes have been collected on BL14.1 at the BESSY II electron storage ring (Helmholtz-Zentrum Berlin) and the DESY P11 beamline (PETRA III, Hamburg). We want to acknowledge M. Weiss (Helmholtz-Zentrum Berlin) and A. Burkhardt (DESY) for their assistance during the diffraction data collection.
Funding: This work was supported by University of Otago (to T.A.H.), Deutsche Forschungsgemeinschaft SPP1784 - JU2773/2-1 (to T.P.J.), Deutsche Forschungsgemeinschaft SPP1784 - HE3397/13-2 (to M.H.), Foundation for Polish Science/European Union POIR.04.04.00-00-5D81/17-00 (to M.B.), Polish National Science Centre 2018/30/Q/NZ2/00669 (to M.B.), Polish National Agency for Academic Exchange PPI/APM/2018/1/00034 (to M.B.), and Deutsche Forschungsgemeinschaft ZA153/28-1 (to M.Z.).
Author contributions: Conceptualization: T.P.J., T.A.H., and M.B. Methodology: M.Rav., D.R., O.O.-R., C.I.D., X.N., C.R.G., A.H.W., M.W., M.Raż., I.M.Ma., K.M., U.S., O.K., K.Z., I.M.Mo., D.W., F.K., T.P.J., T.A.H., M.B., A.K., and M.H. Investigation: M.Rav., D.R., O.O.-R., C.I.D., X.N., C.R.G., A.K., K.M., O.K., U.S., K.Z., T.P.J., T.A.H., and M.B. Visualization: M.Rav., D.R., O.O.-R., C.I.D., C.R.G., U.S., R.Z.J., F.K., T.P.J., T.A.H., M.B., M.H., and A.K. Funding acquisition: M.Z., T.P.J., T.A.H., and M.B. Project administration: T.P.J., T.A.H., and M.B. Supervision: T.P.J., T.A.H., M.B., C.P., M.Z., R.Z.J., and M.H. Writing (original draft): T.P.J., T.A.H., and M.B. Writing (review and editing): All authors.
Competing interests: T.A.H. is a director and shareholder of a small biotech/agricultural consultancy, TOTOVISION/TOTOGEN Ltd. T.P.J. is a director and shareholder of a small biotech company, Magnacell Ltd. M.H. is a consultant for Moderna Inc. The other authors declare that they have no competing interests.
Data and materials availability: All biological materials are available on request (T.A.H., tim.hore@otago.ac.nz; M.B., mbochtler@iimcb.gov.pl; T.P.J., jurkowskit@cardiff.ac.uk). The assembled construct sequence for pB-tetO2-mTET3cd-mCherry can be found on the NCBI sequence archive under accession (MW139646). In-house developed analysis scripts and processed data are available on GitHub. (https://github.com/TimHore-Otago/TET_specificity). Raw high-throughput BS-seq data on TET3-overexpressing cells are available on GEO under accession GSE159205; ACE-seq on TET3-overexpressing cells are available on GEO under accession number GSE199083. Data and code are available at https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/UTMEWY.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 and S2
Other Supplementary Material for this manuscript includes the following:
Data files S1 and S2
REFERENCES AND NOTES
- 1.Goll M. G., Bestor T. H., Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., Agarwal S., Iyer L. M., Liu D. R., Aravind L., Rao A., Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He Y.-F., Li B.-Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., Sun Y., Li X., Dai Q., Song C.-X., Zhang K., He C., Xu G.-L., Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochtler M., Kolano A., Xu G.-L., DNA demethylation pathways: Additional players and regulators. Bioessays 39, 1–13 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Hashimoto H., Liu Y., Upadhyay A. K., Chang Y., Howerton S. B., Vertino P. M., Zhang X., Cheng X., Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 40, 4841–4849 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costa Y., Ding J., Theunissen T. W., Faiola F., Hore T. A., Shliaha P. V., Fidalgo M., Saunders A., Lawrence M., Dietmann S., Das S., Levasseur D. N., Li Z., Xu M., Reik W., Silva J. C. R., Wang J., NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 495, 370–374 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doege C. A., Inoue K., Yamashita T., Rhee D. B., Travis S., Fujita R., Guarnieri P., Bhagat G., Vanti W. B., Shih A., Levine R. L., Nik S., Chen E. I., Abeliovich A., Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 488, 652–655 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hore T. A., von Meyenn F., Ravichandran M., Bachman M., Ficz G., Oxley D., Santos F., Balasubramanian S., Jurkowski T. P., Reik W., Retinol and ascorbate drive erasure of epigenetic memory and enhance reprogramming to naïve pluripotency by complementary mechanisms. Proc. Natl. Acad. Sci. U.S.A. 113, 12202–12207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu T.-P., Guo F., Yang H., Wu H.-P., Xu G.-F., Liu W., Xie Z.-G., Shi L., He X., Jin S., Iqbal K., Shi Y. G., Deng Z., Szabó P. E., Pfeifer G. P., Li J., Xu G.-L., The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Abdel-Wahab O., Mullally A., Hedvat C., Garcia-Manero G., Patel J., Wadleigh M., Malinge S., Yao J., Kilpivaara O., Bhat R., Huberman K., Thomas S., Dolgalev I., Heguy A., Paietta E., Le Beau M. M., Beran M., Tallman M. S., Ebert B. L., Kantarjian H. M., Stone R. M., Gilliland D. G., Crispino J. D., Levine R. L., Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood 114, 144–147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ko M., An J., Bandukwala H. S., Chavez L., Äijö T., Pastor W. A., Segal M. F., Li H., Koh K. P., Lähdesmäki H., Hogan P. G., Aravind L., Rao A., Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature 497, 122–126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakagawa T., Lv L., Nakagawa M., Yu Y., Yu C., D’Alessio A. C., Nakayama K., Fan H.-Y., Chen X., Xiong Y., CRL4(VprBP) E3 ligase promotes monoubiquitylation and chromatin binding of TET dioxygenases. Mol. Cell 57, 247–260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams K., Christensen J., Pedersen M. T., Johansen J. V., Cloos P. A. C., Rappsilber J., Helin K., TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravichandran M., Jurkowska R. Z., Jurkowski T. P., Target specificity of mammalian DNA methylation and demethylation machinery. Org. Biomol. Chem. 16, 1419–1435 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Charlton J., Jung E. J., Mattei A. L., Bailly N., Liao J., Martin E. J., Giesselmann P., Brändl B., Stamenova E. K., Müller F.-J., Kiskinis E., Gnirke A., Smith Z. D., Meissner A., TETs compete with DNMT3 activity in pluripotent cells at thousands of methylated somatic enhancers. Nat. Genet. 52, 819–827 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez A., Castellazzi C. L., Battistini F., Collinet K., Flores O., Deniz O., Ruiz M. L., Torrents D., Eritja R., Soler-López M., Orozco M., Impact of methylation on the physical properties of DNA. Biophys. J. 102, 2140–2148 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hashimoto H., Pais J. E., Zhang X., Saleh L., Fu Z.-Q., Dai N., Corrêa I. R., Zheng Y., Cheng X., Structure of a naegleria TET-like dioxygenase in complex with 5-methylcytosine DNA. Nature 506, 391–395 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu L., Li Z., Cheng J., Rao Q., Gong W., Liu M., Shi Y. G., Zhu J., Wang P., Xu Y., Crystal structure of TET2-DNA complex: Insight into TET-mediated 5mC oxidation. Cell 155, 1545–1555 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Hu L., Lu J., Cheng J., Rao Q., Li Z., Hou H., Lou Z., Zhang L., Li W., Gong W., Liu M., Sun C., Yin X., Li J., Tan X., Wang P., Wang Y., Fang D., Cui Q., Yang P., He C., Jiang H., Luo C., Xu Y., Structural insight into substrate preference for TET-mediated oxidation. Nature 527, 118–122 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Hu X., Zhang L., Mao S.-Q., Li Z., Chen J., Zhang R.-R., Wu H.-P., Gao J., Guo F., Liu W., Xu G.-F., Dai H.-Q., Shi Y. G., Li X., Hu B., Tang F., Pei D., Xu G.-L., Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell 14, 512–522 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Jones P. A., Taylor S. M., Cellular differentiation, cytidine analogs and DNA methylation. Cell 20, 85–93 (1980). [DOI] [PubMed] [Google Scholar]
- 22.Mayyas I. M., Weeks R. J., Day R. C., Magrath H. E., O’Connor K. M., Kardailsky O., Hore T. A., Hampton M. B., Morison I. M., Hairpin-bisulfite sequencing of cells exposed to decitabine documents the process of DNA demethylation. Epigenetics 16, 1251–1259 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schutsky E. K., DeNizio J. E., Hu P., Liu M. Y., Nabel C. S., Fabyanic E. B., Hwang Y., Bushman F. D., Wu H., Kohli R. M., Nondestructive, base-resolution sequencing of 5-hydroxymethylcytosine using a DNA deaminase. Nat. Biotechnol. 36, 1083–1090 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin Y., Morgunova E., Jolma A., Kaasinen E., Sahu B., Khund-Sayeed S., Das P. K., Kivioja T., Dave K., Zhong F., Nitta K. R., Taipale M., Popov A., Ginno P. A., Domcke S., Yan J., Schübeler D., Vinson C., Taipale J., Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356, eaaj2239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reik W., Dean W., Walter J., Epigenetic reprogramming in mammalian development. Science 293, 1089–1093 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Ficz G., Hore T. A., Santos F., Lee H. J., Dean W., Arand J., Krueger F., Oxley D., Paul Y.-L., Walter J., Cook S. J., Andrews S., Branco M. R., Reik W., FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell 13, 351–359 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leitch H. G., McEwen K. R., Turp A., Encheva V., Carroll T., Grabole N., Mansfield W., Nashun B., Knezovich J. G., Smith A., Surani M. A., Hajkova P., Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 20, 311–316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen L., Song C.-X., He C., Zhang Y., Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu. Rev. Biochem. 83, 585–614 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo F., Li X., Liang D., Li T., Zhu P., Guo H., Wu X., Wen L., Gu T.-P., Hu B., Walsh C. P., Li J., Tang F., Xu G.-L., Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 15, 447–459 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Peat J. R., Dean W., Clark S. J., Krueger F., Smallwood S. A., Ficz G., Kim J. K., Marioni J. C., Hore T. A., Reik W., Genome-wide bisulfite sequencing in zygotes identifies demethylation targets and maps the contribution of TET3 oxidation. Cell Rep. 9, 1990–2000 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amouroux R., Nashun B., Shirane K., Nakagawa S., Hill P. W., D’Souza Z., Nakayama M., Matsuda M., Turp A., Ndjetehe E., Encheva V., Kudo N. R., Koseki H., Sasaki H., Hajkova P., De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat. Cell Biol. 18, 225–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Meyenn F., Iurlaro M., Habibi E., Liu N. Q., Salehzadeh-Yazdi A., Santos F., Petrini E., Milagre I., Yu M., Xie Z., Kroeze L. I., Nesterova T. B., Jansen J. H., Xie H., He C., Reik W., Stunnenberg H. G., Impairment of DNA methylation maintenance is the main cause of global demethylation in naive embryonic stem cells. Mol. Cell 62, 848–861 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hackett J. A., Sengupta R., Zylicz J. J., Murakami K., Lee C., Down T. A., Surani M. A., Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339, 448–452 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seisenberger S., Peat J. R., Hore T. A., Santos F., Dean W., Reik W., Reprogramming DNA methylation in the mammalian life cycle: Building and breaking epigenetic barriers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20110330 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ginno P. A., Gaidatzis D., Feldmann A., Hoerner L., Imanci D., Burger L., Zilbermann F., Peters A. H. F. M., Edenhofer F., Smallwood S. A., Krebs A. R., Schübeler D., A genome-scale map of DNA methylation turnover identifies site-specific dependencies of DNMT and TET activity. Nat. Commun. 11, 2680 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adam S., Anteneh H., Hornisch M., Wagner V., Lu J., Radde N. E., Bashtrykov P., Song J., Jeltsch A., DNA sequence-dependent activity and base flipping mechanisms of DNMT1 regulate genome-wide DNA methylation. Nat. Commun. 11, 3723 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adam S., Bräcker J., Klingel V., Osteresch B., Radde N. E., Brockmeyer J., Bashtrykov P., Jeltsch A., Flanking sequences influence the activity of TET1 and TET2 methylcytosine dioxygenases and affect genomic 5hmC patterns. Commun. Biol. 5, 92 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oberacker P., Stepper P., Bond D. M., Höhn S., Focken J., Meyer V., Schelle L., Sugrue V. J., Jeunen G.-J., Moser T., Hore S. R., von Meyenn F., Hipp K., Hore T. A., Jurkowski T. P., Bio-on-magnetic-beads (BOMB): Open platform for high-throughput nucleic acid extraction and manipulation. PLoS Biol. 17, e3000107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kellner S., Neumann J., Rosenkranz D., Lebedeva S., Ketting R. F., Zischler H., Schneider D., Helm M., Profiling of RNA modifications by multiplexed stable isotope labelling. Chem. Commun. (Camb.) 50, 3516–3518 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Krug M., Weiss M. S., Heinemann U., Mueller U., XDSAPP: A graphical user interface for the convenient processing of diffraction data using XDS. J. Appl. Cryst. 45, 568–572 (2012). [Google Scholar]
- 41.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Echols N., Headd J. J., Hung L.-W., Jain S., Kapral G. J., Kunstleve R. W. G., McCoy A. J., Moriarty N. W., Oeffner R. D., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Case D. A., Cheatham T. E., Darden T., Gohlke H., Luo R., Merz K. M., Onufriev A., Simmerling C., Wang B., Woods R. J., The amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X., Cui J., Yan Z., Zhang H., Chen X., Wang N., Shah P., Deng F., Zhao C., Geng N., Li M., Denduluri S. K., Haydon R. C., Luu H. H., Reid R. R., He T.-C., Sustained high level transgene expression in mammalian cells mediated by the optimized piggyBac transposon system. Genes Dis. 2, 96–105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peat J. R., Ortega-Recalde O., Kardailsky O., Hore T. A., The elephant shark methylome reveals conservation of epigenetic regulation across jawed vertebrates. F1000Res 6, 526 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang T., Luo M., Berrios K. N., Schutsky E. K., Wu H., Kohli R. M., Bisulfite-free sequencing of 5-hydroxymethylcytosine with APOBEC-coupled epigenetic sequencing (ACE-Seq). Methods Mol. Biol. 2198, 349–367 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Krueger F., Andrews S. R., Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Illingworth R. S., Gruenewald-Schneider U., Webb S., Kerr A. R. W., James K. D., Turner D. J., Smith C., Harrison D. J., Andrews R., Bird A. P., Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLOS Genet. 6, e1001134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ortega-Recalde O., Peat J. R., Bond D. M., Hore T. A., Estimating global and erasure using low-coverage whole-genome bisulfite (WGBS). Methods Mol. Biol. 2272, 29–44 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 and S2
Data files S1 and S2