Abstract
Gamete fusion is a critical event of mammalian fertilization. A random one-bead one-compound combinatorial peptide library represented synthetic human egg mimics and identified a previously unidentified ligand as Fc receptor–like 3, named MAIA after the mythological goddess intertwined with JUNO. This immunoglobulin super family receptor was expressed on human oolemma and played a major role during sperm-egg adhesion and fusion. MAIA forms a highly stable interaction with the known IZUMO1/JUNO sperm-egg complex, permitting specific gamete fusion. The complexity of the MAIA isotype may offer a cryptic sexual selection mechanism to avoid genetic incompatibility and achieve favorable fitness outcomes.
MAIA, Fc receptor–like 3, is a crucial human egg receptor to elicit tight membrane adhesion for gamete fusion at fertilization.
INTRODUCTION
A key event of vertebrate fertilization is the membrane fusion of the gametes, enabling the spermatozoon to enter the ooplasm and trigger the resumption of meiosis (1). Molecular recognition between the plasma membranes in the mouse involves the egg receptor on the sperm, Izumo1, a member of the immunoglobulin (Ig) superfamily, and the sperm receptor on the egg, Juno, a glycosylphosphatidylinositol-anchored nonfunctional folate receptor family member (2–4). Binding of these receptors is facilitated by a tetraspanin network (CD9 and CD81) in the egg membrane (1), such that deletion of either receptor or the tetraspanins substantially inhibits gamete fusion despite an apparently normal sperm-egg attachment (5–7). Juno/Izumo1 interaction is conserved in mammals including marsupials (8); however, gamete fusion displays a species-specific interaction that is particularly stringent for human fertilization (9). This species specificity and the observation that Juno is lost early in gamete contact before completion of membrane fusion indicate that additional membrane recognition mechanisms are likely to be involved. We sought to identify new human sperm-egg binding/fusion epitopes by using a random one-bead one-compound (OBOC) combinatorial peptide library (10, 11) to represent synthetic egg mimics.
RESULTS
FcRL3 is identified as candidate human sperm-egg fusion epitope and coevolved with IZUMO1
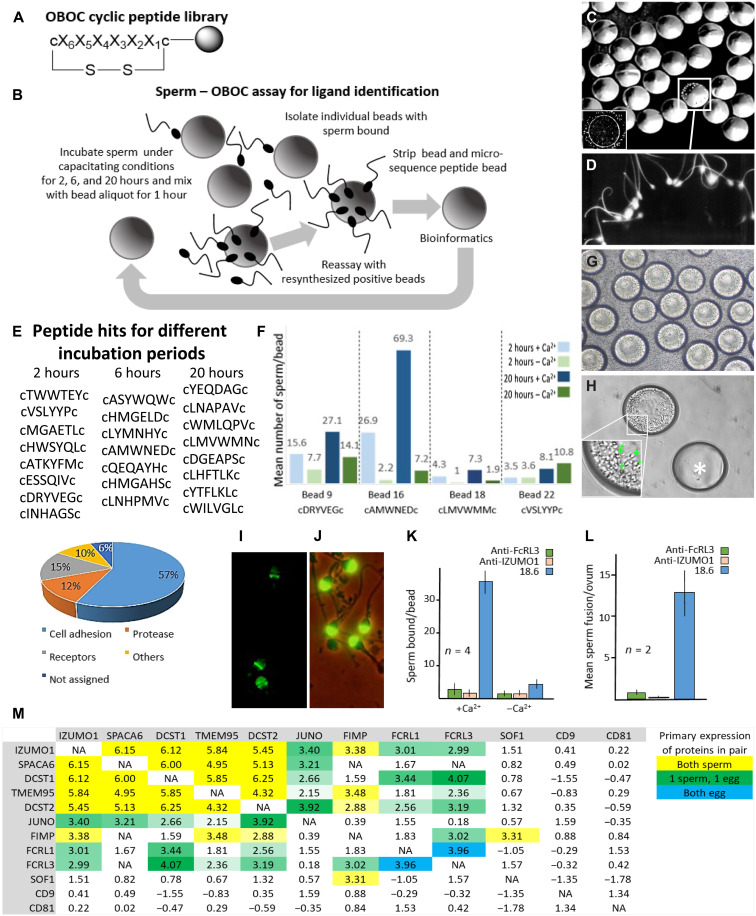
Microspheres as three-dimensional (3D) substitutes for oocytes (12) were used in combination with a cyclic OBOC library (11, 13) comprising ≥47 million permutations, with each bead displaying a unique 6-mer random l-amino acid peptide flanked by d-cysteine residues, making them resistant to proteolysis (Fig. 1A). Library beads (~90 μm in diameter) were comparable in size to human eggs (~100 to 120 μm in diameter) and used in an in vitro sperm binding assay (Fig. 1B).
Fig. 1. OBOC assay for human sperm binding and FcRL3 ERC with gamete recognition proteins.
(A) OBOC library structure (c, d-cysteine; X, 19 l-eukaryotic amino acids except l-cysteine). (B) Sperm-OBOC assay. (C) Sperm-bead binding (inset: human egg). (D) Sperm-bead binding (higher magnification). (E) Peptide hits and proportions assigned to functions. (F) Sperm (five donors) consistently bound to four specific bead aliquots incubated under specific conditions (fig. S1). Bead 16 hit for FcRL3. (G) Sperm bound to resynthesized cAMWNEDc peptide–beads during incubation. (H) Sperm-bound bead (higher magnification, bottom left corner; representative sperm highlighted by green cross) and “naked” bead (*). (I) IZUMO1 on human sperm. (J) Acrosomal antigen 18.6 on human sperm (control). (K) Antibody inhibition of sperm binding to resynthesized beads (n = repetitions) (16). (L) Antibody inhibition of sperm fusion to zona-free hamster eggs (n = animals, each of 10 to 15 oocytes). (M) Gamete interaction proteins on sperm (IZUMO1, SPACA6, DCST1, TMEM95, DCST2, and FIMP) and egg (JUNO, FcRL1, and FcRL3) experienced similar gene evolutionary histories, as shown by ERC sequence analysis. Color intensity reflects the strength of ERC value for that pair of genes. NA, not available.
The few beads with sperm bound (≤1 in 1000) were readily identified by their light-refracting sperm heads and bead rotation due to sperm motility (Fig. 1, C and D). Significantly, this sperm adhesion imitated sperm bound to oocytes in vitro with acrosome-reacted sperm pivoting about their equatorial region where sperm-egg fusion occurs (14). Washed beads with >25 sperm attached were recovered for amino acid sequencing (85% success rate), with the majority of identified peptides associated with cell adhesion properties (Fig. 1E). Another round of sperm binding with homogeneous bead aliquots identified peptide sequences that consistently bound sperm from all donors (fig. S1). Calcium is required for sperm-egg fusion, and therefore, depleting calcium from the medium can indicate whether the sperm binding assay with beads was representing a potential physiological mechanism. A particular bead aliquot (bead 16) bound 50 to 100 sperm per bead (mean, 69) with calcium, but it had much lower sperm binding without calcium (mean, 7), potentially indicating a post-acrosomal adhesion mechanism in keeping with gamete fusion (Fig. 1, F to H). The sequence cAMWNEDc showed homology with the conserved Ig domain of Fc receptor–like 3 (FcRL3), a membrane receptor of the Ig superfamily (15, 16).
Human oocytes express FcRL3 mRNA (17) and Fcγ receptors (closely related to FcRL3) on the oolemma (18). Monoclonal antibody to FcRL3 inhibited sperm adhesion to cAMWNEDc beads (±calcium), while the control antibody [mab18.6 (19)] only inhibited sperm binding in the absence of calcium (Fig. 1, I and K). Spermatozoa preincubated with monoclonal antibody to FcRL3 (and IZUMO1) and incubated with zona-free hamster oocytes [used as a clinical assay of sperm fertilizing capacity (20)] blocked both sperm attachment and fusion with zona-free hamster eggs in vitro compared with the control mab18.6 monoclonal antibody (Fig. 1, J and L). An antibody against IZUMO1 also blocked sperm attachment to the cAMWNEDc beads, suggesting a relationship between IZUMO1 and the FcRL3 binding motif (Fig. 1, I and K). Therefore, we performed evolutionary rate covariation (ERC) to analyze coevolution between proteins (21) known to be involved in mammalian gamete fusion. ERC is a sequence analysis technique that identifies genes with correlated changes in rate across species (21). ERC was previously shown to find correlations between cofunctional genes across diverse functional groups (22–26). We evaluated ERC-based functional relations between not only FcRL1/3 and IZUMO1 but also DCST1/2 (27), FIMP (28), TMEM95, SOF1, and SPACA6 (29), as male reproductive tissue (sperm and testes)–specific proteins, except SPACA6, in which minor mRNA expression was also detected in the ovary but not oocytes (30). As expected, recognized fertilization genes showed highly elevated ERC values (Fig. 1M). FcRL3 also displayed significantly elevated ERC values with IZUMO1, SPACA6, DCST1, TMEM95, DCST2, and JUNO, comprising a coevolving group within the top 5% of genome-wide values (Fig. 1M). FcRLs are expressed primarily on B cells and have an immune-modulating function, although extracellular ligands for the receptors are unknown. FcRL3 displays an Fc-homologous extracellular domain, a transmembrane domain, and cytoplasmic immunoreceptor tyrosine-based activation and inhibitory motifs (ITAM and ITIM) (15). In contrast, FcRL1 is the only FcRL expressing two ITAM-like, but no ITIM, sequences and is assumed to be a coactivation receptor (31). Anti-FcRL1 antibody failed to inhibit sperm-oocyte fusion; therefore, our investigations focused on the FcRL3 receptor. Furthermore, TMEM71, which displays homology with the selected cAMWNEDc peptide, is not known to be expressed on gametes and, moreover, does not contain Ig-like domains necessary for fusion.
FcRL3 interacts with JUNO on human oolemma and was named MAIA
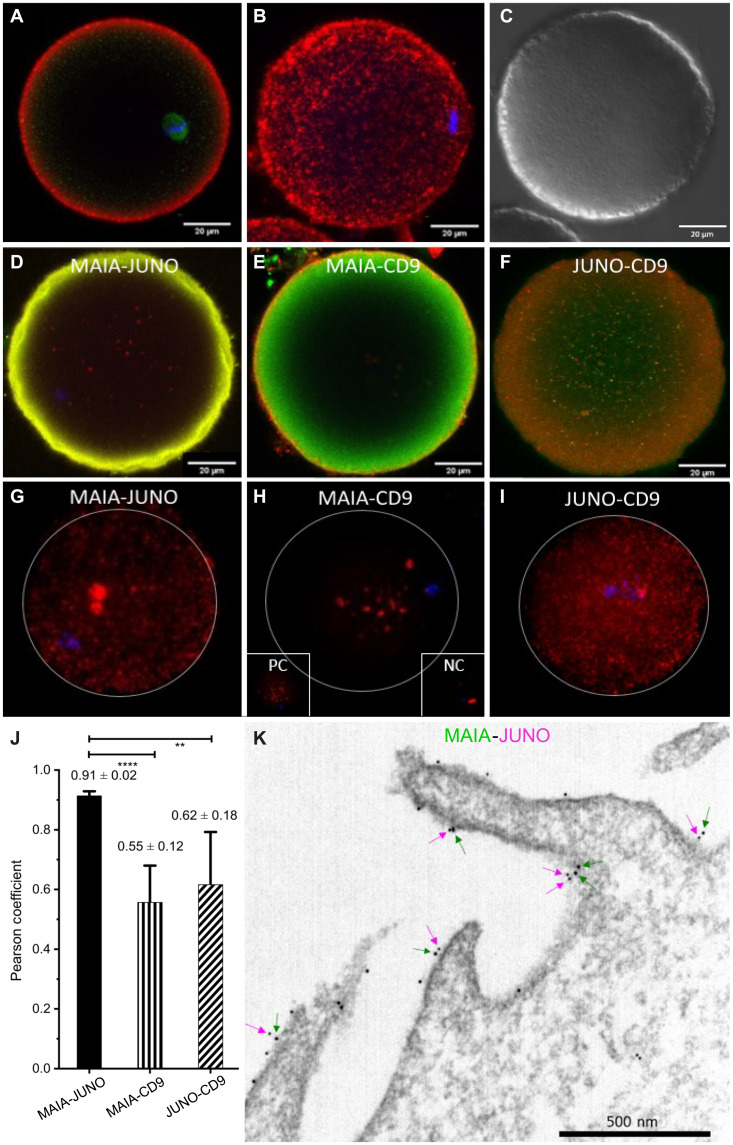
FcRL3 protein is localized within the oolemma of unfertilized human oocytes (Fig. 2, A to C, and movie S1), and we addressed its close interaction with candidate binding proteins (JUNO and CD9) with colocalization (Fig. 2, D to F, and fig. S2) and proximity ligation assays (PLAs) (Fig. 2, G to I). The strongest interaction between FcRL3-JUNO reflected a high Pearson correlation coefficient (Fig. 2J). Therefore, we named the FcRL3 on human oocytes MAIA, after the mythological goddess intertwined with JUNO. Double immunogold labeling with gold particles showed close association between JUNO and MAIA along the oolemma microvilli (Fig. 2K).
Fig. 2. MAIA immunolocalization and interaction in human oocytes.
(A to C) MAIA on human mature metaphase II (MII) stage oocyte (red), β-tubulin (green), nuclear counterstain (Hoechst 33342), confocal microscopy; (A) single plane, (B) a maximum intensity projection visualization of MAIA localization and (C) differential interference contrast (DIC). (D to F) Colocalization between protein pairs (D) MAIA-JUNO, (E) MAIA-CD9, and (F) JUNO-CD9 on human MII oocyte explained by preferential localization of the proteins in, or close to, the cell membrane (fig. S2). (G to I) PLA on human MII oocyte for protein pairs (G) MAIA-JUNO, (H) MAIA-CD9, and (I) JUNO-CD9 with appropriate negative control (NC) α-tubulin–β1 integrin and positive control (PC) α-tubulin–β-tubulin. (J) Pearson correlation coefficient for the protein pairs corresponding to colocalization analysis (fig. S2). (K) Representative transmission electron microscopy image of MAIA (big dots and green arrows) and JUNO (small dots and magenta arrows) localized on the microvilli/oolemma of MII oocyte.
MAIA induces sperm fusion with transfected CHO and HEK293T cells, and paralog Fcrl5−/− mice suggest a potential role of the FcRL protein family in reproductive processes
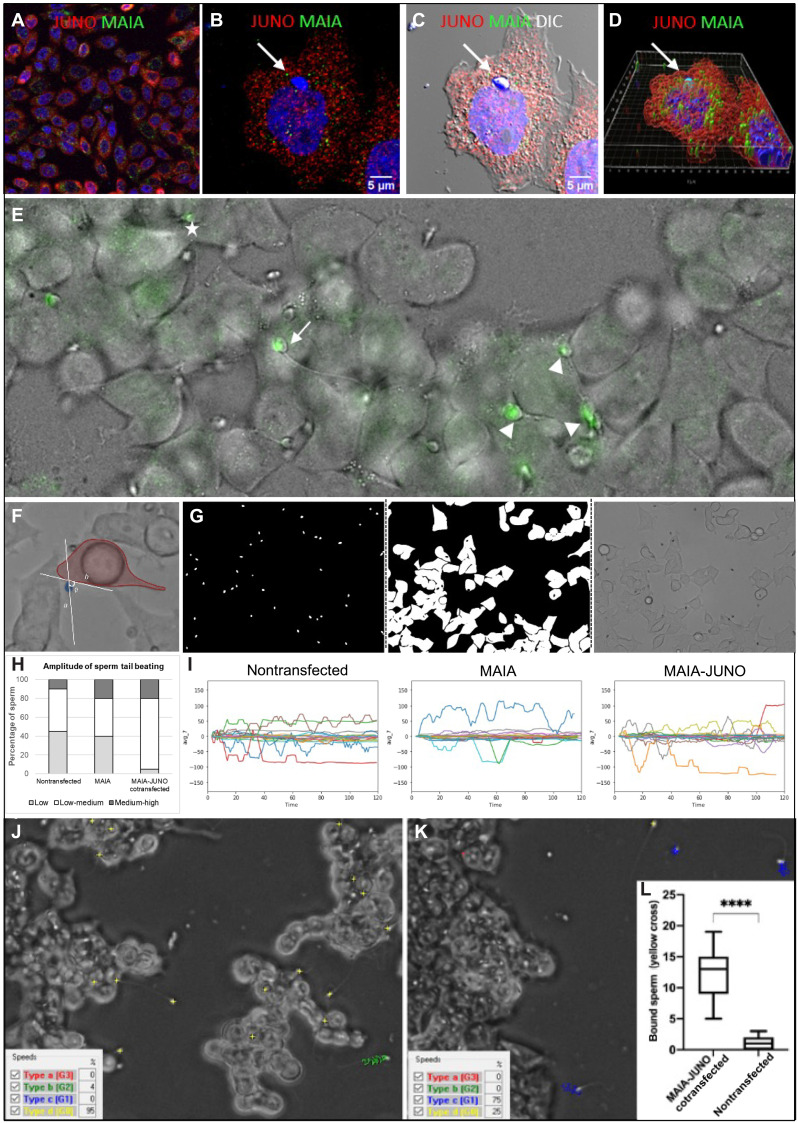
Chinese hamster ovary (CHO) cells were transfected with an FcRL3- and JUNO-carrying plasmid (Fig. 3A) and coincubated with capacitated human sperm (37°C) (Fig. 3, B to D), which adhered avidly (15 to 30 sperm per field; control, 1 per field) and displayed characteristic oscillations of the sperm head (movies S2 and S3). Double-transfected CHO cells with sperm bound in this manner also had expression of membrane-bound MAIA and JUNO, with the sperm nuclei engulfed within the cytoplasm, indicating possible cell-cell fusion (Fig. 3D, fig. S4, and movie S4). Sperm incubated with cotransfected human embryonic kidney (HEK) 293T cells displayed a range of tail beating observed previously (14), which can be reflected in the status of sperm-cell binding as a prerequisite of fusion with their viability remaining (Fig. 3E and movie S5). Using artificial intelligence (AI) object identification, we developed novel neuronal network training masks for human sperm tail beating pattern quantification (Fig. 3, F to I). On the basis of the AI findings, the difference between HEK cells—nontransfected, transfected with MAIA, or cotransfected with MAIA and JUNO—was significant in the amount of sperm displaying low and medium amplitude of tail beating. In particular, 75% of sperm incubated with cotransfected cells showed medium-amplitude beating pattern, which we interpret as the pattern of sperm binding and the most favorable for future possible fusion. We emphasize that our system took into account the data distribution between individual human sperm tail movement amplitudes, reflecting those observed in green fluorescent protein (GFP) fusion assay. The majority (P ≤ 0.0001) of these sperm later stop with a hardly noticeable movement or straighten flagellum, a behavior reminiscent of the moment of sperm/egg fusion in vitro (Fig. 3, J to L, and movies S5 to S7) (1).
Fig. 3. Sperm binding assessment in relation to cell fusion.
(A) Immunostaining of MAIA (green) and JUNO (red) cotransfected CHO cells; (B) with fused human sperm head (white arrow); nuclear counterstain (Hoechst); (C) merged with DIC; (D) Imaris Surface Render analysis (fig. S4 and movie S4). (E) Representative image of sperm captured by lifetime confocal microscopy (green) displaying a range of tail beating reflecting the status of sperm-cell fusion (movie S5); white arrow, fusing sperm (slow tail beating); white asterisks, nonfusing sperm (fast tail beating); white tips (fused sperm, static tail without beating). (F to I) AI object identification. (F) Representative image of captured tail beating pattern used for AI training. (G) Example of novel neuronal network training masks for human sperm tail beating pattern quantification. (H) Quantification of sperm tail beating amplitude calculated from the degree angle as low, medium, and high. (I) Representative charts of degree angle change. (J and K) Representative snapshot of a video recording of sperm kinematic parameter analysis assessed by CASA with phase contrast (yellow cross, static sperm–bound sperm with low tail beating; blue track, medium-progressive swimming sperm; green track, rapid-swimming sperm; red track, rapid-progressive swimming sperm); (J) MAIA-JUNO cotransfected HEK293T (HEK) cells (movie S6); (K) nontransfected HEK cells (movie S7). (L) Quantification of video recordings (ncotransfected = 17 and nnontransfected = 14) of bound sperm (yellow cross) assessed by CASA, ****P ≤ 0.0001.
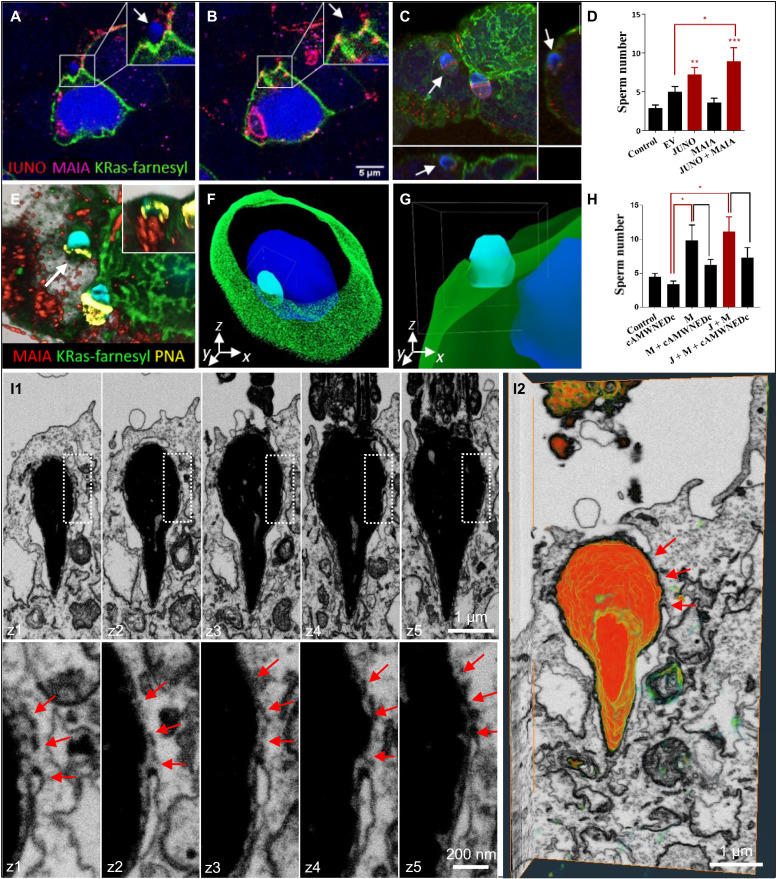
Previously, it was shown that cells expressing JUNO alone bind spermatozoa but do not fuse with them (32). Therefore, to monitor the trajectory of this sperm-cell entry, HEK293T cells were cotransfected with MAIA/FcRL3, JUNO, and a plasmid encoding with a GFP-tagged farnesylated K-Ras to visualize the cell membrane (Fig. 4, A to C, and movie S8). First, we detected significantly elevated sperm binding to these cotransfected HEK cells (Fig. 4D and fig. S3, A and B), and second, we verified the ability of sperm to fuse with these cells as visualized by segmentation and volume rendering of fluorescence using Imaris 3D (Fig. 4, E to G, and movie S9). Notably, when human sperm were preincubated with the synthetic cAMWNEDc peptide, sperm binding to HEK293T cells decreased (Fig. 4H), indicating a possible interference within the Ig domain interaction. Third, we combined correlative light and electron microscopy (CLEM; to target the site of sperm binding to the cotransfected cell) with focus ion beam scanning electron microscopy (FIB-scanning electron microscopy) volumetric imaging and found the sperm head to be exposed to the cytoplasm of this cotransfected cell by FIB-scanning electron microscopy (Fig. 4, I1 and I2, and movie S10). The reconstruction of FIB-scanning electron microscopy dataset showed membrane fusion over the equatorial region of the sperm head extending back over the post-acrosomal region as at the beginning of sperm-egg fusion. Human sperm will fuse with lymphocytes (33), which also highly express FcRL3 (fig. S3, C and D), and inhibition of FcRL3 gene expression by FcRL3 short interfering RNA (siRNA) led to decreased sperm fusion with JeKo-1 B lymphocytes (fig. S3, E to J). Furthermore, a soluble extracellular domain of FcRL3 protein bound to IZUMO1, which was relocated to the equatorial segment after the acrosome reaction (fig. S3, K and L).
Fig. 4. Sperm binding/fusion with MAIA and JUNO cotransfected cells.
(A to C) Membrane disruption and sperm head fusion (white arrows) with cotransfected HEK293T (HEK) by MAIA (purple), JUNO (red), and cell membrane–incorporated GFP-tagged farnesylated K-Ras (green) (movie S8). (D) Quantification of sperm binding to cotransfected HEK293T (HEK) with MAIA, JUNO, and MAIA + JUNO. (E to G) Immunofluorescence data postprocessing. (E) Acrosome-reacted sperm (white arrow) equatorial segment stained with PNA (yellow) fused with cell membrane (green); the apical part of the sperm head within the cell cytoplasm is shown in the top right corner (visualized by Imaris). (F and G) Sperm-HEK penetration and fusion 3D model (Imaris), MAIA (purple), JUNO (red), and AF488-phalloidine–stained F-actin (green) (movie S9). (H) Sperm-HEK binding decrease after sperm-cAMWNEDc coincubation. (H and D) Image Xpress screening system; ImageJ/FIJI; error bars (SEM); EV, empty vector; J, JUNO; and M, MAIA. (I1 and I2) 3D visualization of the sperm head within the cytoplasm of MAIA-JUNO cotransfected HEK cell using FIB-scanning electron microscopy. (I1) Consecutive FIB-scanning electron microscopy sections showing fusion (red arrows) between the plasma membrane of MAIA-JUNO cotransfected HEK cell and the inner acrosomal membrane (IAM) of the sperm head around the equatorial segment. Overview of an internalized sperm head (top) and detail of the membrane fusion (bottom) demarcated by dotted rectangle (above). Spacing between frames, 250 nm. (I2) 3D volume rendering of the whole FIB-scanning electron microscopy dataset (x-y-z projection side view) showing partial exposure of the sperm head to the cytoplasm of the HEK cell and the site of membrane fusion (red arrows) (movie S10).
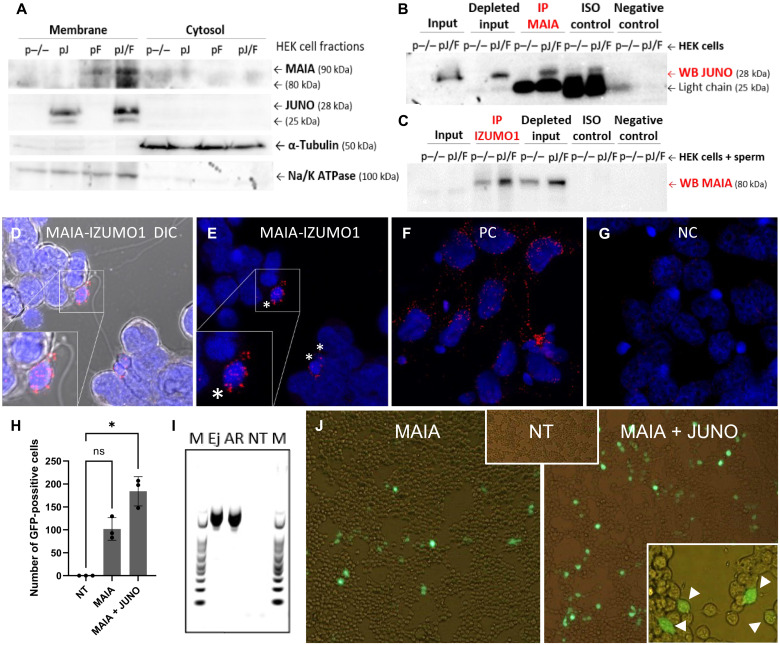
We confirmed the expression of transfected proteins (MAIA and JUNO) (Fig. 5A), along with the cis-protein interaction between MAIA and JUNO within the cell membrane (Fig. 5B and fig. S3M) and the trans-protein interaction between MAIA and IZUMO1 on the sperm (Fig. 5C and fig. S3N). IZUMO1 protein in human sperm was used as a control (fig. S3O). PLA performed on JUNO-MAIA double-transfected cells with bound sperm revealed an interaction between the MAIA and IZUMO1 proteins (Fig. 5, D and E, and fig. S5, A to D), indicating a reciprocal binding activity of these two proteins with respect to controls (Fig. 5, F and G, and fig. S5). A sperm-cell GFP fusion assay was designed, and human sperm transfected with GFP plasmid were added to MAIA or MAIA and JUNO cotransfected HEK cells. When sperm-cell fusion occurred, the GFP signal expressed by the cells was detectable (Fig. 5, H to J). The sperm fusion with cells cotransfected with MAIA and JUNO was significant (P < 0.5) compared to the single-transfected MAIA group (Fig. 5H), supporting our proposal that MAIA facilitates sperm fusion in the presence of JUNO. Transfected cells fused with sperm still had a detectable sperm tail outside the cell (Fig. 5J), similar to sperm-oocyte fusion.
Fig. 5. MAIA-IZUMO1 protein-protein interaction and sperm-cell GFP fusion assay.
(A) Recombinant MAIA and JUNO protein localization in HEK plasmatic membrane; both proteins are shown with two isoforms (two bands) because of protein glycosylation; α-tubulin was used as a loading control for cytosolic fraction; Na/K adenosine triphosphatase (ATPase) was used as a reference protein for membrane fraction. (B and C) Co-IP of (B) MAIA-JUNO and (C) MAIA-IZUMO1 using cotransfected HEK (B) without or (C) with sperm; ISO control, isotype IgG control antibody (IgG light chain, 25 kDa; IgG heavy chain, 50 kDa). (D to G) PLA assay for MAIA-IZUMO1 on JUNO + MAIA cotransfected HEK with sperm; the positive signal is designated exclusively to the point of sperm head–cell attachment (white asterisks) (fig. S5). (D and E) MAIA-IZUMO1; (F) positive control: α-tubulin–β-tubulin; (G) negative control: MAIA–β-tubulin. (H to J) GFP fusion assay of sperm with HEK cells. (H) Number of GFP-positive cells (n) per well, nontransfected (NT), MAIA, and MAIA + JUNO cotransfected HEK cells with fused GFP-transfected sperm; *P ≤ 0.05. (I) Polymerase chain reaction (PCR) showing GFP in transfected ejaculated (Ej) and acrosome-reacted (AR) sperm, nontransfected, and marker (M). (J) Representative fluorescent images showing analyzed groups (H and I), positively transfected cells (white arrows) fused with sperm with visible tail are shown in the bottom right corner, and fluorescent signal is merged with bright field. ns, not significant.
The MAIA/Fcrl3 gene is not present in mice (34); however, its protein shows 40% amino acid identity with the extracellular domain of mouse Fcrl5, which is also the most closely related paralog of FcRL3 in mice and could share functional features common to both species (fig. S6). Therefore, an Fcrl5 gene knockout (KO) mouse was prepared by deletion of the sequence between exon 4 and exon 7 (E4-E7) of the Fcrl5 gene using the bioinformatic prediction of the Fcrl5−/− (KO) allele and the allele sequencing result (fig. S7, A and B). One of the interesting reproductive traits was the inability to establish the Fcrl5−/− line from the maternal lineage (see raw data mouse model summary in the Biobox repository). In vivo mating outcome of KO female and Fcrl5+/+ [wild-type (WT)] male then resulted in 30% KO/WT pairs without fertilization output, despite the successful mating evaluated by the presence of a vaginal plug (fig. S7C), which points to fertilization defects. The average litter size was decreased in KO/WT pairs (P = 0.14), and no significant changes in sex ratio were observed (fig. S7D). In mice, even minor changes in their reproduction fitness are of importance, given their high reproductive ability as a species compared to human.
MAIA supersedes JUNO to interact with IZUMO1 during gamete binding
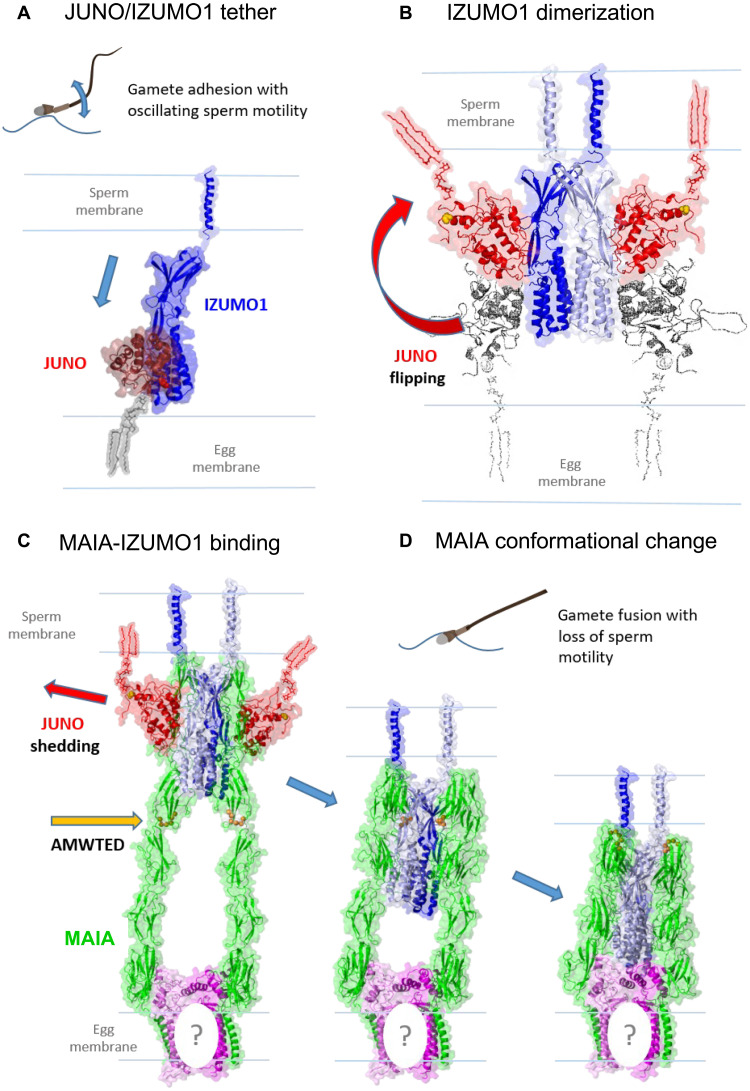
We propose a molecular mechanism for potential sperm-egg membrane recognition with MAIA (Fig. 6). JUNO is recognized and pulled from the egg membrane by IZUMO1 (Fig. 6A) driven by increasing affinity to the most stable flipped orientation (4) and creating a binding pocket for MAIA (Fig. 6, A and B, and fig. S8, A to C). Modeling CD9 (fig. S9A1) showed that both binding interfaces of the dimer occupied by domain 6 of MAIA formed a stable interaction (Fig. 6C and fig. S9A2); however, the colocalization assay (Fig. 2E) with a Pearson coefficient of 0.55 ± 0.12 (Fig. 2J) and PLA assay (Fig. 2H) suggested only very moderate interaction. This experimental detection of weak CD9-MAIA interaction could be due to the presence of JUNO in the oolemma, which could be changed after JUNO shedding. In addition, another member of the tetraspanin family could be partially involved in the interaction with MAIA in the presence of JUNO. Significantly, the 2:2 IZUMO1/JUNO complex displayed a pair of deep grooves formed between the surface of JUNO and both IZUMO1 proteins serving as a preferred binding site for all of the MAIA domains (Fig. 6B and fig. S9B). Removing JUNO from this complex and docking another MAIA domain further improved binding, using the original IZUMO1/JUNO binding interface (Fig. 6D) stabilized in bend conformation at neutral pH (fig. S9, C and D) favorable for fertilization (35). The affinity of each MAIA domain to the IZUMO1/JUNO complex leads to a stable membrane adhesion, allowing for shortening of the cell-cell distance by a molecular “ratchet-like” mechanism. Loss of JUNO at this stage offers a larger binding interface for MAIA, further stabilizing the structure for an even closer proximity for membrane fusion. Shedding of JUNO may result in the formation of extracellular vesicles in the perivitelline space that act as a “decoy egg” to prevent polyspermy (3). A graphical representation depicts the proposed interaction of binding and fusion between the egg microvillus and the sperm membrane facilitated by the key molecules including MAIA (fig. S10, A to G). The model predicts that the sperm membrane and egg microvillus apex are in close proximity during sperm attachment to the oolemma (fig. S10A). Sperm IZUMO1 and egg JUNO located in apical parts of both gametes interact with the apico-laterally located proteins IZUMO1 and MAIA (fig. S10B), followed by sperm and egg membrane budding mediated by the close apical IZUMO1-JUNO membrane interaction and convergence facilitated by the contraction of the IZUMO1-MAIA protein complex (fig. S10C). This gamete membrane proximity leads to their hybridization (fig. S10D) and surface destabilization (fig. S10E), followed by a formation of a new sperm/egg hybrid membrane (fig. S10F) and, lastly, a fertilization cone (fig. S10G).
Fig. 6. Outline of human gamete binding.
(A) Fertilizing sperm display oscillatory motility tethered to the oolemma due to the initial JUNO/IZUMO1 interaction. (B) IZUMO1 dimerization triggers the transfer of JUNO from the egg into the sperm membrane. (C) Tight binding of MAIA in the created JUNO/IZUMO1 binding pocket. (D) MAIA conformational change to extracellular Fc domains, leading to close membrane proximity enabling gamete fusion with the loss of sperm motility (figs. S8 and S9). The question mark (C and D) may represent an unknown or CD9 protein, as proposed in fig. S9 (A1 and A2).
DISCUSSION
Gamete fusion is a critical stage of vertebrate fertilization, enabling the fertilizing sperm to enter the ooplasm and trigger the resumption of meiosis (1). This is initiated in the mouse by the egg receptor on the sperm membrane, Izumo1, and the sperm receptor on the oolemma, Juno, facilitated by a tetraspanin network of the egg membrane. Juno/Izumo1 interaction is conserved in mammals; however, specificity of the human sperm for the egg remains particularly stringent, indicating that additional membrane recognition mechanisms are likely to play a role. Besides IZUMO1, several proteins located on the sperm were found to be essential for mammalian gamete fusion. Consistent with their roles in gamete interaction, all DCST1, DCST2, FIMP, TMEM95, and SPACA6, but not SOF1, showed very strong ERC values with IZUMO1, and DCST1/2 and TMEM95 exceeded the values for JUNO. These ERC correlations, which signal functional relationships and originate from the changing evolutionary pressures on the process of fertilization, strengthen the implication that FcRL3’s high score (>2) reflects a consistent signal of coevolution between gamete interaction proteins despite FcRL3 being expressed in many nonreproductive tissues.
The classical approach to identify putative receptors on scarce and ethically contentious human eggs is to inhibit sperm egg binding in vitro with antibodies and other probes. However, such methods are subject to confounding factors (i.e., quality and quantity of donor eggs and nonspecific steric hindrance), leading to considerable experimental bias. Moreover, human sperm do not readily attach or fuse to oocytes of other species in vitro, excepting those of great apes (9) and the zona-free hamster oocyte (20). The latter can indicate human sperm fertilizing potential but is otherwise an atypical model of homologous gamete fusion. Here, we used a random screening approach with microspheres as 3D substitutes for oocytes (12) and the OBOC combinatorial method as previously used to identify novel cancer cell surface markers (13). It is important to recognize that 6-mer peptides have limited specificity and do not necessarily exhibit correct conformation. However, rounds of assays eliminated inconsistent/nonspecific binding peptides, while consistent sperm-microbead binding under physiological conditions for natural fertilization (e.g., sperm capacitation and acrosome reaction) selected candidate moieties for further analysis to verify functionality during fertilization. Hence, we identified that the extracellular domain of human-specific FcRL3/MAIA provided tight attachment to tether hyperactivated motile sperm to beads and the human oolemma to allow sperm-egg fusion. Moreover, double-transfected somatic cells expressing JUNO and MAIA evoked sperm-cell fusion. These in vitro findings are all consistent with a role for MAIA during human fertilization, although other additional moieties are likely to be involved (1). The homology of the FcRL family of genes between species is variable, perhaps indicating relative species specificity (36, 37). Fcrl5 gene is the most closely related paralog of FcRL3 in mice, and our Fcrl5−/− mouse model suggests a potential role of the FcRL protein family in the reproductive process, although it remains to be determined whether Fcrl5 or another FcRL is involved in mouse fertilization.
A vigorous oscillatory sperm movement occurs during gamete binding and stops at the time of fusion (14), which was observed in sperm binding and fusing with transfected cells (Fig. 3) and might induce the molecular ratchet-like interaction of membrane receptors (14). We propose that the shedding of JUNO upon sperm-oolemma primary attachment (3, 8, 38) elicits tight apposition of membranes and permits specific gamete fusion facilitated by MAIA binding to IZUMO1 protein, which was supported by sperm-cell GFP fusion assay. Interaction between MAIA and JUNO on the oolemma was indicated, and the close association of both proteins on microvilli was confirmed. Trans-protein interaction between MAIA and IZUMO1 in vitro revealed a reciprocal binding activity of these two proteins. Significantly, in our modeling, the 2:2 IZUMO1/JUNO complex displayed a pair of deep grooves formed between the surface of JUNO and both IZUMO1 proteins serving as a preferred binding site for all of the MAIA domains. The structural modeling suggested the protein conformational change of IZUMO1 after the initial binding to JUNO, which enabled the secondary binding of IZUMO1 to FcRL3. This IZUMO1-FcRL3 bond was not proven to be feasible without the initial JUNO-IZUMO1 interaction and JUNO shedding from the oolemma. Crystallography data will be needed to challenge the proposed hypothesis further.
Fertilization (or gamete recognition) is a process that is carried out by many genes, all of which would change their rates of sequence evolution in response to changes in the evolutionary pressure on fertilization. Examples of how those pressures change include changes in the intensity of male-male competition, changes in sexual conflict between females and males, or reinforcement of reproductive isolation between species. The wide diversification of the MAIA/FcRL3 receptor family (34) during evolution is likely to confer species specificity during gamete fusion either during sperm-egg recognition or via downstream intracellular ITIM/ITAM signaling mechanisms. Recently, an ITIM motif was implicated in cell-cell fusion induced by the varicella-zoster virus (39). Moreover, FcRL isotype complexity at fertilization offers a cryptic sexual selection mechanism to avoid genetic incompatibility and achieve favorable fitness outcomes as previously postulated (40, 41). MAIA/FcRL3 receptor may reflect the ancestral traits used in both cell immunological response and gamete fusion underlying the importance of interindividual compatibility during fertilization.
MATERIALS AND METHODS
Synthesis of OBOC peptide libraries and resynthesis of individual positive peptides on beads
The OBOC libraries were synthesized on TentaGel S NH2 resin (0.26 mmol/g; Rapp Polymere GmbH, Tübingen, Germany) with 9-fluorenylmethoxycarbonyl (Fmoc) chemistry using a “split-mix” strategy using N-hydroxybenzotriazole/N,N-diisopropylcarbodiimide as a coupling reagent (13, 42). A fourfold molar excess of Fmoc-protected amino acids to resin was used with reactions at room temperature (RT) for 2 to 6 hours. We confirmed the completion of coupling with a ninhydrin test. The Fmoc group was removed with 20% piperidine in N,N′-dimethylformamide (first 5 min and then 15 min), and side-chain–protecting groups were removed with a trifluoroacetic acid (TFA) cocktail containing 82.5% TFA, 5% phenol, 5% thioanisole, 5% H2O, and 2.5% triisopropylsilane. The disulfide formation was achieved with 20% dimethyl sulfoxide in 0.1 M ammonium acetate buffer (pH 6.2) for 2 days. Ellman test was negative. Resynthesis of individual positive peptides on beads used a similar approach but without the split-mix procedure.
Sperm-bead binding assay
Human sperm samples were donated by healthy volunteers (University of Sheffield Research Ethics Committee no: SMBRER293) with informed consent and prepared by the swim-up method as described previously (43). Aliquots (1 ml) were coincubated with beads (105 sperm/104 beads) for 1, 6, and 20 hours (triplicate for each condition) in the presence or absence of calcium chloride (2 mM). Beads displaying sperm attachment (>25 sperm) were recovered separately by a fine glass pipette under dissecting microscope and washed by repeated aspiration. Beads exhibiting the highest sperm binding after washing were transferred in 100 μl of phosphate-buffered saline (PBS) to small Eppendorf tubes and stored at −20°C before freight transfer to the Lam laboratory for microsequencing as described previously (42). Peptide sequence hits were subjected to bioinformatic search analysis (BLAST and PANTHER).
Zona-free hamster egg penetration test
Sperm samples and zona-free hamster eggs were prepared as described previously (43, 44). Hamsters were obtained from the animal facility of the Faculty of Science, Charles University, Prague, Czech Republic. Animal procedures and the experimental protocol, approved by the Local Ethics Committee of the Faculty of Science, Charles University, were carried out in accordance with the Animal Scientific Procedure (accreditation numbers 13060/2014-MZE-17214 and 16OZ27335/2013-17214) and subjected to review by the Local Ethics Committee of the Faculty of Science, Charles University. Oocytes were collected the morning following estrus. Human sperm samples classified by World Health Organization standards (2010) as normozoospermic were donated by the Assisted Reproduction Centre ProCrea, Prague. Sperm were incubated in 20-μl drops of the Biggers, Bavister and Whitten medium (BBW) under mineral oil at 37°C in 5% CO2 in air for 3 hours with or without antibody [anti-FcRL3 and anti-IZUMO1; mab18.6 (44) at a final concentration of 1 μg/ml] before addition of 10 to 15 zona-free eggs and incubation for 1 hour. Eggs were recovered, washed by aspiration, and mounted on wax spot slides to determine sperm binding and sperm fusion (decondensed sperm heads).
Mouse model
Gene manipulation was performed using the CRISPR-Cas9 genome editing tool to generate an Fcrl5 KO C57BL/6NCRL mouse model in the Research Institute for Microbial Diseases, Osaka University, Osaka, Japan. The mouse strain was reanimated into the specific pathogen–free facility of the Czech Centre for Phenogenomics (Institute of Molecular Biology, Czech Academy of Sciences, BIOCEV). Fcrl5 gene targeting was confirmed with polymerase chain reaction (PCR) using specific primers for Fcrl5 gene (Generi Biotech, Czech Republic): GTGGTGTGCCTGATGTGTTCC (forward) and CACTGAGCAGTTAAGTAGTCTGC (reverse), and the expected PCR product size was 527 base pairs. For genotyping, we used the following primers: F1, AAGCTCCACCTGCTGTGTTT; R2, GCAGGAACCCAGGACTTACC; recognized Fcrl5 KO product was sequenced by Sanger sequencing method (SEQme, Czech Republic). An in vivo mating study of Fcrl5+/+ (WT) male and Fcrl5−/− (KO) female mice was performed and compared to WT pair reproductive success. All animal procedures and experimental protocols were approved by the Animal Welfare Committee of the Czech Academy of Sciences (Animal Ethics number 66866/2015-MZE-17214, 18 December 2015).
Immunolabeling of human oocytes for MAIA
Human samples were obtained from consenting individuals under the appropriate ethical committee approval of collaborating institutions (BIOCEV 012019, Reprofit International 1/2015, and Masaryk University 16/2016 and 1/2019). Unfertilized oocytes were obtained from young egg donors (aged 19 to 35 years) participating in an egg donation program (Reprofit International, Brno, Czech Republic). Immature oocytes retrieved in stimulated in vitro fertilization cycles were incubated in Continuous Single Culture Medium-Complete (90165, Irvine Scientific, USA) at 5% CO2 and 37°C until they reached the metaphase II (MII) stage. To remove the zona pellucida, live oocytes were briefly treated with prewarmed Tyrode’s solution (T1788, Sigma-Aldrich). Following 1 hour of incubation in fixative solution containing 2% formaldehyde (EM grade), 1 M Hepes, 0.5 EGTA, and 1 M MgSO4 at 37°C, the samples were rinsed in PBS supplemented with 1% bovine serum albumin (BSA) and incubated for 15 min in PBS with 0.1% Triton X-100 at RT. After blocking in 3% BSA, all samples were incubated with primary monoclonal anti-hFcRL3/FcRH3 (MAB3126, R&D Systems, UK) diluted 1:50 in 1% BSA for 2 hours at 37°C and 5% CO2. After washing in 1% BSA, a secondary donkey anti-mouse IgG coupled to Alexa Fluor 568 (Molecular Probes, UK) was added for 1 hour (dilution 1:500 in 1% BSA). Oocytes were washed and mounted in 4′,6-diamidino-2-phenylindole (DAPI)–containing VECTASHIELD medium (Vector Laboratories, Peterborough, UK) for confocal imaging using a Carl Zeiss LSM 880 NLO microscope (Imaging Methods Core Facility at the BIOCEV Research Centre, Vestec, Czech Republic). An open-source software, ImageJ/Fiji (45), was used for image processing. Huygens Professional (Scientific Volume Imaging, The Netherlands; http://svi.nl) software was used for 3D visualization of images.
Immunogold transmission electron microscopy to show MAIA and JUNO oolemma localization
Human oocytes were counterstained with 6- or 12-nm gold-conjugated IgG secondary antibodies (Jackson ImmunoResearch, UK) for 1 hour, washed, and fixed with 3% glutaraldehyde solution (16220, Electron Microscopy Science, USA) in 0.1% BSA (A-1933, Sigma-Aldrich) in PBS for 1 hour at RT and 72 hours at 4°C. The samples were postfixed with 1% aqueous OsO4 (19152, Electron Microscopy Science, USA), embedded in 2% low-melting agarose type II (17856, Thermo Fisher Scientific), and dehydrated with a cold ethanol series on ice and a final dehydration with acetone-anhydride for 5 min. The samples were infiltrated using Epon Embed-812 resin (14120, Electron Microscopy Science, USA) at ratios 1:2, 1:1, and 2:1 with acetone for 5 to 30 min before polymerization at 60°C for 72 hours. The 80-nm-thick sections were examined at 120 kV using the transmission electron microscope Jeol JEM 2100-Plus by the TEM Centre imaging system.
CLEM to visualize sperm fused with MAIA/JUNO cotransfected HEK cells in nanoscale resolution
Optical fluorescent microscopy sample imaging
Optical microscopy imaging was performed on a Nikon spinning disk confocal microscope equipped with Nikon Eclipse Ti2 microscope body, Yokogawa CSU-W1 spinning disk module, and two PRIME BSI sCMOS cameras. Four-color fluorescence z-stack (4.5 μm, 300-nm step size) was acquired sequentially using 405-, 488-, 561-, and 638-nm lasers and 430- to 480-nm, 500- to 550 nm, 575- to 625 nm, and 665- to 715-nm emission filters; 60xW/1.2 water objective; and pinhole sizes of 50 μm. The ROI (region of interest) was determined by localization of the target cell on the coordinates of a glass-bottom gridded MatTek dish on tile scan overview DIC image (tile scan: 5 × 5 tiles) and registered for subsequent relocation of the ROI under the FIB-scanning electron microscopy.
Sample preparation for FIB-scanning electron microscopy
Immediately after optical microscopy image acquisition, the samples were chemically postfixed with a solution containing 2.5% glutaraldehyde and 2% formaldehyde in PBS buffer for 1 hour, washed in PBS, and postfixed first in ferricyanide-reduced osmium (1% OsO4 and 1.5% K[Fe(CN)6] in PBS), followed by nonreduced osmium (1% OsO4 in PBS) for 30 min each. Samples were then washed in PBS, rinsed in distilled H2O, and contrasted with 1% uranyl acetate in distilled H2O (30 min). After rinsing in distilled H2O, samples were dehydrated in ethanol series (30, 50, 80, 95, and 100%), embedded in epoxy resin (Embed-812), and polymerized for 72 hours at 60°C. The glass bottom of each gridded MatTek dish was removed, and the resin block (with monolayer of embedded cells) was mounted on aluminum scanning electron microscopy stubs. Samples were coated with 25 nm of platinum (using High Vacuum Coater, Leica ACE600) for subsequent FIB milling and scanning electron microscopy imaging.
FIB-scanning electron microscopy imaging
Ion milling and scanning electron microscopy image acquisition were performed in a dual-beam scanning electron microscope (FEI Helios NanoLab 660 G3 UC). The target ROIs were localized by finder-grid coordinates (registered during optical microscopy imaging and etched to the surface of the resin block) visible under the Everhart-Thornley detector at 0.8 nA and 20 kV. On the top of a localized target cell, a protective layer of platinum (1000 nm) was deposited on top of the ROI using a single gas injection system, and a trench around the ROI was milled by a focused ion beam at 21 nA and 30 kV. FIB-scanning electron microscopy data collection was done by fine FIB milling at 0.79 nA and 30 kV (5-nm slice thickness), followed by scanning electron microscopy image acquisition at 0.2 nA and 2 kV using the In-Column backscattered electron detector with a pixel size of 3 nm and a pixel dwell time of 15 μs. The size (number of pixels) of acquisition area was adjusted to the size of the area of interest.
Data analysis
The FIB-scanning electron microscopy dataset was processed with Amira Software 6.2. Stacks of 2D images were first aligned and then denoised with the median filter function in the software. For 3D visualization of the whole dataset, the threshold-based segmentation and volume rendering and pseudo-color LUT were used.
Sperm binding to CHO/HEK293T cells with increased expression of JUNO, MAIA, and/or membrane-incorporating GFP-tagged farnesylated K-Ras
Both lines were useful and complementary models. The CHO cells were used in suspension contrary to HEK293T, but their transfection efficiency was lower than for HEK especially for visualization of cell membranes. HEK or CHO cells were seeded at 5 × 104 cells per well in glass-bottom 24-well plates (precoated with poly-d lysine) and cultured in Dulbecco’s modified Eagle’s medium + 10% fetal bovine serum (FBS) + 1% penicillin-streptomycin (Invitrogen) for 24 hours before transfection. Cells at 70 to 80% confluence were transfected using Lipofectamine 3000 (Invitrogen) with 0.5 μg of plasmid DNA {human FcRL3 plasmid [OriGene, MD, USA: complementary DNA (cDNA) clone MGC: 34866], human FOLR4 plasmid (OriGene, MD, USA: cDNA clone NM_001199206), and pCMV6-XL5 mammalian expression vector (EMPTY plasmid) (OriGene, MD, USA: pCMV6-XL5)} in the MEM for 16 hours. Control cells were without transfection. Human sperm samples were prepared by density gradient, after washing in PBS and Sperm Preparation Medium (Origio), and were processed by swim-up technique for 1 hour. The best sperm in appropriate concentration (5 × 104 sperm per well) were capacitated in Sperm Preparation Medium with added progesterone (10 μg/ml; Sigma-Aldrich) and then were incubated with cells (300 μl per well) for 2 hours at 37°C.
For analysis of sperm-cell binding in the presence of cAMWNEDc peptide (corresponding to the FcRL3), human sperm were mixed with this synthetic peptide (200 μM) and then coincubated with transfected cells as mentioned above. After washing, cells were fixed with 4% paraformaldehyde in PBS [dilution 1:300; sperm stained with peanut agglutinin (PNA)–fluorescein isothiocyanate (FITC) for 15 min] (Invitrogen) and incubated overnight at 4°C in the dark with primary antibodies rat monoclonal anti-hFOLR4 (JUNO) (MAB6328, R&D Systems) diluted 1:50 in 1.5% BSA (100 μl per well) and/or mouse monoclonal anti-hFcRL3/FcRH3 (MAB3126, R&D Systems) diluted 1:50 in 1.5% BSA. Cells were counterstained for cytoskeletal F-actin using phalloidine-AF488 (150 nM) (20 min at RT) and followed by Alexa Fluor 568 goat anti-rat IgG or Alexa Fluor 647 goat anti-mouse (Molecular Probes) secondary antibodies (1:500 in 1.5% BSA; 1 hour of incubation at RT in the dark). Nuclei were stained with Hoechst 33342 (Sigma-Aldrich) (0.1 μg/ml for 10 min at RT in the dark). Cells were examined with the Carl Zeiss LSM 880 NL0 confocal microscope and/or ImageXpress Micro Confocal High-Content Imaging System and analyzed by the open-source software ImageJ/FIJI.
MAIA extracellular protein domain binding to human sperm
Human sperm samples were capacitated by swim-up for 2 hours in Sperm Preparation Medium (Origio) and acrosome-reacted by calcium ionophore for 30 min at 37°C. Acrosome-reacted sperm were checked by PNA-FITC (1:300), for 15 min at RT after fixation by acetone:methanol for 5 min. Diluted MAIA/FcRL3 protein extracellular domain with His-tag (R&D Systems, 3126-FC) (1 μg/ml in PBS) was incubated with sperm for 1 hour at 37°C. Sperm suspension was fixed by acetone, and bound protein was visualized by His-tag antibody (Sigma-Aldrich) diluted 1:500 for 1 hour.
Sperm binding to human JeKo-1 B lymphocytes
The human JeKo-1 mantle cell lymphoma cell line was cultured in RPMI medium + 20% inactivated FBS for 24 hours before use for binding assay with human sperm. Human sperm or JeKo-1 cells were preloaded and stained with Hoechst 33342 (Sigma-Aldrich) (0.1 μg/ml for 10 min at RT in the dark) before binding to cells for 2 hours. Then, knockdown of FcRL3 gene in JeKo-1 was done by siRNA transfection. For these experiments, cells were plated at a density of 500,000 cells per well in a 24-well plate. Transfections were performed 24 hours later using human FcRL3 siRNA (sc-45791) (25, 50, and 100 nM) or control scrambled off-target siRNA-GFP (sc-3686) (25, 50, and 100 nM). All siRNAs were provided by Santa Cruz Biotechnology. Transfections were performed with Lipofectamine 2000 (Invitrogen), according to the manufacturer’s protocol for 72 hours. Efficiency of transfection was checked by immunofluorescence signal in cells transfected by scrambled siRNA-GFP and by Western blot. Transfected cells were washed in fresh medium, capacitated and acrosome-reacted sperm were added to the cell pellet (500,000 cells) for 2 hours, and JeKo-1 cell–bound sperm were visualized by light microscopy. For quantification of GFP-transfected sperm fusing with JeKo-1 cells, samples were scanned by an Etaluma Microscope LS720.
SDS–polyacrylamide gel electrophoresis, immunoblotting, and detection
Nonreducing lysis buffer (2× SDS) was used for whole-cell lysates. Using the Mem-PER Plus kit, membrane and cytosol protein fraction were isolated. Protein was quantified using a NanoDrop 3000 spectrophotometer (Thermo Fisher Scientific). Molecular masses were estimated with prestained Precision Plus Protein Dual Color Standards (Bio-Rad). Tris-glycine buffer (pH 9.6) with 20% methanol was used for transfer of proteins onto a polyvinylidene difluoride (PVDF) membrane (Immobilon-P, Millipore, Darmstadt, Germany) for electroblotting carried out for 1 hour at 500 mA. PVDF membranes blocked with 5% dry milk (Bio-Rad) in PBS-Tween were washed and incubated with primary antibody mouse monoclonal anti-hFcRL3/FcRH3 (MAB3126, R&D Systems) or rat monoclonal anti-hFOLR4 (JUNO) (MAB6328, R&D Systems), both diluted 1:500 in 5% dry milk (Bio-Rad) in PBS-Tween, overnight at 4°C. A mouse monoclonal anti–α-tubulin (TU02) (sc8035, Santa Cruz Biotechnology) diluted 1:5000 in 5% dry milk (Bio-Rad) in PBS-Tween was used as a loading control. Incubation with secondary antibody anti-mouse or anti-rat IgG conjugated to horseradish peroxidase (HRP; Bio-Rad), both diluted 1:3000 in PBS-Tween, was performed for 1 hour at RT. After washing, membranes were developed using the SuperSignal Chemiluminescence Substrate (Thermo Fisher Scientific).
Coimmunoprecipitation from cell lysates
Transfected cell lysate supernatants were incubated with mouse monoclonal anti-hFcRL3/FcRH3 (MAB3126, R&D Systems), rat monoclonal anti-hFOLR4 (JUNO) (MAB6328, R&D Systems), or rabbit polyclonal anti-hIZUMO1 ABIN6059408 (antibodies online) primary antibodies overnight at 4°C on a rocking platform. Isotype controls used appropriate antibodies of mouse, rat, or rabbit IgGs (Sigma-Aldrich). With added agarose, protein A/G beads (Thermo Fisher Scientific) were incubated at RT for 2 hours. A negative control of protein A/G agarose beads alone was also prepared. Coimmunoprecipitates (Co-IPs) were eluted from protein A/G agarose beads by incubation in the reducing sample buffer for 5 min at 95°C. After electrophoretic separation in 10% polyacrylamide gel and transfer onto a PVDF membrane, the FcRL3 Co-IP was incubated with rat monoclonal anti-hFOLR4 (JUNO) antibody (MAB6328, R&D Systems); IZUMO1 Co-IP was incubated with mouse antibody against FcRL3/FcRH3 (MAB3126, R&D Systems) diluted 1:500 in 5% milk overnight at 4°C. After washing and incubation with secondary anti-rat or anti-mouse antibody conjugated with HRP (Bio-Rad) diluted 1:3000 in 5% milk, the antibody reaction was visualized using the SuperSignal Chemiluminescence Substrate (Thermo Fisher Scientific).
Proximity ligation assay
The interaction of experimental proteins was studied using the Duolink In Situ Red Starter Kit, DUO92101 (Sigma-Aldrich), with primary antibodies directed against each of the target proteins. Fixed ZP free human mature oocytes (MII) were acquired in 1% BSA in PBS. Oocytes were transferred into 100 μl of mouse monoclonal anti-hFcRL3/FcRH3 (dilution 1:50; MAB3126, R&D Systems) and rat monoclonal anti-hFOLR4 (dilution 1:50; MAB6328, R&D Systems) or rabbit polyclonal anti-CD9/MRP-1 (dilution 1:100; bs-2489R) and rat monoclonal anti-hFOLR4 (dilution 1:50; MAB6328, R&D Systems) primary antibodies diluted in 1% BSA and incubated in a wet chamber at 4°C overnight. Washed oocytes were incubated with PLUS and MINUS PLA probes and amplified following the manufacturer’s protocol and transferred into 2 μl of VECTASHIELD Mounting Medium with DAPI. A known interaction between the α-tubulin (1:1000; TU02 sc8035, Santa Cruz Biotechnology) and β-tubulin (1:1000; ab15568, Abcam) was used as a positive control (DUO92101 Duolink, In Situ Red Starter Kit Mouse/Rabbit, Sigma-Aldrich). A known absent interaction between α-tubulin and β1-integrin (1:200; M106, sc8978, Santa Cruz Biotechnology) was used as a negative control. Fluorescence was detected with a confocal microscope (Carl Zeiss LSM 880 NLO; 100 to 120 z-stacks per sample). Similar methods were used with unfertilized human sperm-egg samples and with HEK293T cells. Experimental setting of PLA assay in cell-to-cell interaction model was performed in agreement with previously published methodology (46) with a slight modification for application on gametes.
Sperm binding and fusion assessment
HEK293T cells were seeded and transfected as mentioned previously in Materials and Methods. Sperm were stained by SYBR Green from the LIVE/DEAD Sperm Viability Kit (L-7011, Thermo Fisher Scientific) according to the manufacturer’s protocol to mark only live sperm; propidium iodide from the kit was not used in these settings. After washing, sperm were incubated with JUNO + FcRL3 cotransfected cells for 1 hour, and unbound sperm were washed thoroughly three times. Live sperm (green) displaying a range of tail beating reflecting the status of sperm-cell binding/fusion were scanned under a confocal microscope (Nikon CSU-W1 Spinning Disk Confocal Microscope). In parallel, the same experiment was prepared, and sperm kinematic parameters were analyzed, assessed, and quantified by computer-assisted sperm analysis (CASA) with phase contrast (yellow cross, static sperm–bound sperm with low tail beating; blue track, medium-progressive swimming sperm; green track, rapid-swimming sperm; red track, rapid-progressive swimming sperm).
Sperm-cell GFP fusion assay
Sperm and cell transfection
On the basis of the split-GFP complementation approach (47), all cells were transfected using Lipofectamine 2000 (Thermo Fisher Scientific, 11668030). Sperm cells (5 × 106 cells) were transfected with 2 μg of GFP plasmid (Addgene, no. 101865) in OptiPRO serum-free medium (Thermo Fisher Scientific, 12309019) for 1 hour. Then, cells were washed with PBS three times, pelleted, resuspended in SpermPrep capacitation medium (Origio), and used for fusion reaction. Transfection efficacy of sperm cells was analyzed by PCR reaction using DNA lysis buffer [1 M tris (pH 8.5), 1 M NaCl, 0.5 M EDTA, and 10% SDS] for isolation of DNA (samples were heated for 30 min at 55°C and then for 10 min at 95°C) and DreamTaq Green PCR Master Mix 2× for PCR reaction (Thermo Fisher Scientific, K1081). HEK293T cells were transfected with FcRL3 plasmid (Origene, SC125617), IZUMO 1R (Origene, SC329497), or a combination of both. In all cases, 2 μg of plasmid was used per 5 × 105 cells in combination with Lipofectamine in Opti-MEM medium (Thermo Fisher Scientific, 31985062). List of primers used for detection of plasmid DNA after transfection is as follows: IZUMO1, FCRL3, and IZUMO1R (JUNO): VP1.5, 5′-GGACTTTCCAAAATGTCG-3′ (forward); XL39, 5′-ATTAGGACAAGGCTGGTGGG-3′ (reverse): pKAM GFP: RRE, 5′-CAGGAAGCACTATGGGCGCAGC-3′ (forward); EGFPC5, 5′-CATGGTCCTGCTGGAGTTCGTG -3′ (reverse).
Fusion experimental setup
All fusion experiments were done in 96-well plates, and cells were seeded and transfected with competent plasmids FcRL3 and cotransfected with FcRL3- and JUNO-carrying plasmid. The next day, capacitated and acrosome-reacted sperm transfected with GFP plasmid were added. Control cells were without transfection. All experimental groups were measured in three replicates. Cells with sperm were incubated in a 37°C and 5% CO2 incubator and scanned with Etaluma Microscope LS720 in 20-min intervals simultaneously in one time for all experimental and control groups. The captured bright-field and fluorescent signal in the well (total area captured by 10× objective) was analyzed in cells fused with GFP-transfected sperm by Lumaquant Image Analysis Software and quantified by using a created mask in ImageJ/FIJI software.
Evolutionary rate covariation
ERC is a phylogenetic signature that reflects the covariation of protein evolutionary rates over evolutionary time. Because cofunctional genes often show this covariation (21), ERC can predict new genes in pathways and protein complexes, such as for reproductive proteins (48). The calculation of ERC values was performed beginning with orthologous gene sequences from 62 mammalian species obtained from the 100-way alignment at the University of California Santa Cruz Genome Browser. Those 17,486 coding sequence alignments were used to calculate gene-specific branch lengths over the mammalian species tree topology using the PAML package (49). For each orthologous gene group/tree, the branch lengths were normalized into relative evolutionary rates (50) and then used to calculate the Pearson correlation coefficient (i.e., the ERC value) between each gene pair. To standardize values between genes and across datasets, ERC values were Fisher-transformed (51).
Phylogenetic study of FCRL family
The phylogenetic tree of the FCRL family was made with the sequences contained in data file FCRL_alignment.mfa that were then trimmed by the GBlocks server with all three options chosen for a less stringent selection, resulting in an alignment of 350 amino acid columns (52). PhyML was used to infer the phylogeny using default parameters and the LG substitution model (53). Branch support was measured using probabilities from the approximate likelihood ration test method.
Protein structural modeling
The amino acid sequences of proteins were obtained from the UniProt database (54) as deposited under the following entry names: CD9 (P21926), FcRL3 (Q96P31), IZUMO1 (Q8IYV9), and JUNO (A6ND01). The I-TASSER (55) stand-alone package version 5.0 was used to obtain homology models of CD9 (extracellular part and extracellular part with the transmembrane region) and FcRL3 (extracellular part) proteins. The CD9 model used the transmembrane region of tetraspanin CD81 [Protein Data Bank (PDB) ID 5tcx] (56) with the homology model of the CD9 extracellular region, further refined by 50-ns molecular dynamics (MD) simulation. This accounted for the structure-sequence differences between CD81 and CD9 within the transmembrane region and in the extracellular part. The set of recently available crystal structures of the IZUMO1/JUNO complex (PDB IDs 5jkc, 5jkd, and 5jke) (4) was used for the modeling of the 2:2 complex. Residues missing in the crystal structure and transmembrane helices were added by MODELLER (57) version 9.14. The prediction of protein-protein interactions used the locally available version of the ClusPro (58) 2.0 protein-protein docking server. The parameters of implicit solvation/lipid membrane model (EEF1/IMM1) (59) simulations were assigned using the web-based graphical user interface CHARMM-GUI (60). The MD simulation of suggested complexes was performed using the CHARMM (61) version c41b1 MD package. The electrostatic potential surface for various pH values was calculated using the PDB2PQR (62)/APBS (63) version 1.5. The molecular graphics was prepared using PyMOL (64) version 1.8.4.
AI object identification for sperm tail beating quantification
Cell line and sperm preparation
HEK293T cells were seeded and transfected with transfected with competent FcRL3 and cotransfected with FcRL3- and JUNO-carrying plasmids as described previously. Human sperm were capacitated and acrosome-reacted according to the defined methodology above. The data were captured after 1 hour of sperm-cell incubation.
Neural network creation
The required dataset was processed using a neural network trained specifically for the required cell type. A suitable set of training data was selected for training the network to identify objects using AI. The network was trained on 20 images composed of different experiments (HEK_Control, HEK_MAIA, and HEK_JUNO + MAIA). Manually processed images formed the basis of a neural network for her learning.
Creating a training dataset and cell segmentation
NIS Elements software AR 5.30.01 with AI module (65) was used to create the training dataset and cell segmentation. By segmenting the cells using the Binary Editor tools, two different binary layers were created, one for sperm only and the other for cells interacting with sperm. The exact boundaries of the heads of all sperm in one binary layer (“sperm”) and the exact edges of the cells in the other binary layer (“cells”) were manually defined on all images, and the planes were distinguished by different colors. The images processed in this way served as a training model for neural network training.
Neural network training
The Train segment ai module was used to train the neural network for Segment.ai of objects. The module was provided with a previously created set of input data (training pattern for sperm and for cells) with the expected output. The process of learning the neural network according to the presented data took 7 hours. The neural network trained according to the sample training sets was able to identify the required objects.
Trained neural network applications
The trained neural network was applied to the processing of the required sets of experimental images using the NIS.ai-Segment.ai module, and the objects were again detected in two binary layers (sperm and cell), as in the training dataset. Masks in the ome.tif format, a separate mask for the sperm layer for cells, and a separate mask for the original image were exported from the segmentations of precisely detected objects.
Image analysis
Segmented masks were processed in Python 3.9.4 to compute angles between cells and sperm cells automatically through all time stacks. Required packages are available in the Biobox repository (https://biobox.biocev.org/index.php/s/ND9iXitJkDGNwdB). The subset of sperm cells was selected in such a way that only sperm cells that maintain close proximity to some cell for a reasonable amount of time were taken into it. For each cell of interest, the region of interaction was defined to encircle the sperm cell by the dilation of the sperm cell mask. In this region, the part of the sperm cell overlapping with the interaction region was selected to be used in the computation of the interaction tangent line. Then, two axes were defined: the major axis of the sperm cell, and the second axis that is the line going through the two furthest points of the cell mask that are part of the interaction overlap. These two axes refer, respectively, to sperm cell orientation and tangent to the cell body in the place of cell–sperm cell interaction. The angle between these vectors was measured as . The same approach was applied to all time stacks, and results of the measurements were recorded.
Statistical evaluation of the data
Data were statistically analyzed using GraphPad Prism 7 and OriginLab Origin software. The analysis of variance (ANOVA) with Dunnett’s post hoc test was applied on data presented in Figs. 3L and 4 (D and H), Mann-Whitney test on data presented in fig. S7 (C and D), Kruskal-Wallis test on data presented in Fig. 5H, and two-tailed t test on data presented in Fig. 2J. Bars denote arithmetic means, and error bars denote SEM (Figs. 4, D and H, and 5H) and SD (Figs. 2J and 3L and fig. S7, C and D). P value equal or lower than 0.05 was considered to be significant (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001; only for Figs. 2J and 3L).
Acknowledgments
We thank S. Gregory, B. Aflatoonian, and N. Sanchez-Hernandez for technical support. We also thank the Combinatorial Chemistry and Chemical Biology Shared Resource at University of California, Davis for providing OBOC library and resynthesizing peptides on beads.
Funding: K.E. was supported by an Underwood Fellowship award from the Biotechnology and Biological Sciences Research Council, UK. N.C. and R.P. were supported by NIH grant R01 HG0009299. This work was supported by the following: BIOCEV CZ.1.05/1.1.00/02.0109; Grant Agency of the Czech Republic GA18-11275S (to K.K.), GA22-30494S (to K.K.), and GA21-04607X (to J.N.); Institute of Biotechnology of the Czech Academy of Sciences RVO 86652036; Czech-BioImaging CZ.02.1.01/0.0/0.0/16_013/0001775 and CZ.02.1.01/0.0/0.0/18_046/0016045; MEYS CR Large RI Project LM2018129; UC Davis Comprehensive Cancer Center support Grant NCI P30CA093373; MRC G0801059 / ID 87252 (to H.M.); and UKRI grant (UK Research and innovation) for open-access policy. We used services of the Czech Centre for Phenogenomics at the Institute of Molecular Genetics supported by the Czech Academy of Sciences RVO 68378050 and by the project LM2018126 Czech Centre for Phenogenomics provided by Ministry of Education, Youth and Sports of the Czech Republic.
Author contributions: In vitro cell lines experiments, immunofluorescence staining, sperm-cell assays, Western blot, PLA, confocal microscopy, and data processing: J.V. Egg experiments, fluorescent staining, colocalization analysis, PLA, confocal microscopy, transmission electron microscopy, and data processing: M.F. Statistical and bioinformatic analysis, Imaris analysis, creation of sperm-egg microvillus membrane interaction model, and designing of Fcrl5 KO mice model genotyping strategy: L.D. Structural modeling and analysis: J.C. Biochemical analysis of proteins: P.P. Fcrl5 KO mouse model characterization and genotyping: V.P. and K.B. Time lapse and quantification of sperm movement and CASA: O.S. Sperm-cell GFP fusion assay: O.S. and Z.N. JeKo-1 cell line experiments: O.S. and P.P. Human egg manipulation and technical support: E.V. Statistical colocalization analysis and Huygens software resulting data evaluation: R.M. Organization and management of sperm donors and performed experimental analysis: A.P. Organization, management, and provision of human egg material: Z.H. and P.K. Plasmid isolation, cell line experiments, and sperm cell: Z.E. and Z.N. Preparation of Fcrl5 mouse model: S.P. Preparation of OBOC peptide bead libraries and performing of peptide sequencing and analysis: R.L. and K.S.L. ERC and phylogenetic analysis methods: R.P. and N.C. Supervision of the cell line analysis and editing of the manuscript: J.N. Designing and providing Fcrl5 mouse model and editing of the manuscript: M.I. Aspects performed for all other experimental analysis: K.E., K.S.L., H.M., and K.K. Direction of the research and writing the manuscript with assistance from all other authors: H.M. and K.K.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw data and other supplementary materials and movies are available in the Biobox repository: https://biobox.biocev.org/index.php/s/ND9iXitJkDGNwdB.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S10
REFERENCES AND NOTES
- 1.Klinovska K., Sebkova N., Dvorakova-Hortova K., Sperm-egg fusion: A molecular enigma of mammalian reproduction. Int. J. Mol. Sci. 15, 10652–10668 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inoue N., Ikawa M., Isotani A., Okabe M., The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature 434, 234–238 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Bianchi E., Doe B., Goulding D., Wright G. J., JUNO is the egg Izumo receptor and is essential for mammalian fertilization. Nature 508, 483–487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohto U., Ishida H., Krayukhina E., Uchiyama S., Inoue N., Shimizu T., Structure of IZUMO1–JUNO reveals sperm–oocyte recognition during mammalian fertilization. Nature 534, 566–569 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Le Naour F., Rubinstein E., Jasmin C., Prenant M., Boucheix C., Severely reduced female fertility in CD9-deficient mice. Science 287, 319–321 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Miyado K., Yamada G., Yamada S., Hasuwa H., Nakamura Y., Ryu F., Suzuki K., Kosai K., Inoue K., Ogura A., Okabe M., Mekada E., Requirement of CD9 on the egg plasma membrane for fertilization. Science 287, 321–324 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Han L., Nishimura K., Sadat Al Hosseini H., Bianchi E., Wright G. J., Jovine L., Divergent evolution of vitamin B9 binding underlies JUNO-mediated adhesion of mammalian gametes. Curr. Biol. 26, R100–R101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianchi E., Wright G. J., Sperm meets egg: The genetics of mammalian fertilization. Annu. Rev. Genet. 50, 93–111 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Bedford J., Why mammalian gametes don’t mix. Nature 291, 286–288 (1981). [DOI] [PubMed] [Google Scholar]
- 10.Lam K. S., Salmon S. E., Hersh E. M., Hruby V. J., Kazmierski W. M., Knapp R. J., A new type of synthetic peptide library for identifying ligand-binding activity. Nature 354, 82–84 (1991). [DOI] [PubMed] [Google Scholar]
- 11.Peng L., Ruiwu L., Marik J., Wang X., Takada Y., Lam K. S., Combinatorial chemistry identifies high-affinity peptidomimetics against α4β1 integrin for in vivo tumor imaging. Nat. Chem. Biol. 2, 381–389 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Hamze J., Hamze J. G., Canha-Gouveia A., Algarra B., Gómez-Torres M. J., Concepción Olivares M., Romar R., Jiménez-Movilla M., Mammalian spermatozoa and cumulus cells bind to a 3D model generated by recombinant zona pellucida protein-coated beads. Sci. Rep. 9, 17989 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao W., Wang Y., Lau E. Y., Luo J., Yao N., Shi C., Meza L., Tseng H., Maeda Y., Kumaresan P., Liu R., Lightstone F. C., Takada Y., Lam K. S., The use of one-bead one-compound combinatorial library technology to discover high-affinity αvβ3 integrin and cancer targeting arginine-glycine-aspartic acid ligands with a built-in handle. Mol. Cancer Ther. 9, 2714–2723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravaux B., Garroum N., Perez E., Willaime H., Gourier C., A specific flagellum beating mode for inducing fusion in mammalian fertilization and kinetics of sperm internalization. Sci. Rep. 6, 31886 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis R. S., Fc receptor-like molecules. Annu. Rev. Immunol. 25, 525–560 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Kochi Y., Myouzen K., Yamada R., Suzuki A., Kurosaki T., Nakamura Y., Yamamoto K., FCRL3, an autoimmune susceptibility gene, has inhibitory potential on B-cell receptor-mediated signaling. J. Immunol. 183, 5502–5510 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Kocabas A. M., Crosby J., Ross P. J., Otu H. H., Beyhan Z., Can H., Tam W. L., Rosa G. J., Halgren R. G., Lim B., Fernandez E., Cibelli J. B., The transcriptome of human oocytes. Proc. Natl. Acad. Sci. U.S.A. 103, 14027–14032 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bronson R., Fusi F., Fleit H., Monoclonal antibodies identify Fcγ receptors on unfertilized human oocytes but not spermatozoa. J. Reprod. Immunol. 21, 293–307 (1992). [DOI] [PubMed] [Google Scholar]
- 19.Moore H. D., Hartman T. D., Brown A. C., Smith C. A., Ellis D. H., Expression of sperm antigens during spermatogenesis and maturation detected with monoclonal antibodies. Exp. Clin. Immunogenet. 2, 84–96 (1985). [PubMed] [Google Scholar]
- 20.Yanagimachi R., Yanagimachi H., Rogers B. J., The use of zona-free animal ova as a test-system for the assessment of the fertilizing capacity of human spermatozoa. Biol. Reprod. 15, 471–476 (1976). [DOI] [PubMed] [Google Scholar]
- 21.Clark N., Alani E., Aquadro C., Evolutionary rate covariation reveals shared functionality and coexpression of genes. Genome Res. 22, 714–720 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolfe N. W., Clark N. L., ERC analysis: Web-based inference of gene function via evolutionary rate covariation. Bioinformatics 31, 3835–3837 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Priedigkeit N., Wolfe N., Clark N. L., Evolutionary signatures amongst disease genes permit novel methods for gene prioritization and construction of informative gene-based networks. PLOS Genet. 11, e1004967 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brunette G. J., Jamalruddin M. A., Baldock R. A., Clark N. L., Bernstein K. A., Evolution-based screening enables genome-wide prioritization and discovery of DNA repair genes. Proc. Natl. Acad. Sci. U.S.A. 116, 19593–19599 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raza Q., Choi J. Y., Li Y., O’Dowd R. M., Watkins S. C., Chikina M., Hong Y., Clark N. L., Kwiatkowski A. V., Evolutionary rate covariation analysis of E-cadherin identifies Raskol as a regulator of cell adhesion and actin dynamics in Drosophila. PLOS Genet. 15, e1007720 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kowalczyk A., Gbadamosi O., Kolor K., Sosa J., Andrzejczuk L., Gibson G., Croix C. S., Chikina M., Aizenman E., Clark N., Kiselyov K., Evolutionary rate covariation identifies SLC30A9 (ZnT9) as a mitochondrial zinc transporter. Biochem. J. 478, 3205–3220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inoue N., Hagihara Y., Wada I., Evolutionarily conserved sperm factors, DCST1 and DCST2, are required for gamete fusion. eLife 10, e66313 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujihara Y., Lu Y., Noda T., Oji A., Larasati T., Kojima-Kita K., Yu Z., Matzuk R. M., Matzuk M. M., Ikawa M., Spermatozoa lacking fertilization influencing membrane protein (FIMP) fail to fuse with oocytes in mice. Proc. Natl. Acad. Sci. U.S.A. 117, 9393–9400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbaux S., Ialy-Radio C., Chalbi M., Dybal E., Homps-Legrand M., Do Cruzeiro M., Vaiman D., Wolf J.-P., Ziyyat A., Sperm SPACA6 protein is required for mammalian sperm-egg adhesion/fusion. Sci. Rep. 10, 5335 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noda T., Lu Y., Fujihara Y., Oura S., Koyano T., Kobayashi S., Matzuk M. M., Ikawa M., Sperm proteins SOF1, TMEM95, and SPACA6 are required for sperm−oocyte fusion in mice. Proc. Natl. Acad. Sci. U.S.A. 117, 11493–11502 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leu C.-M., Davis R. S., Gartland L. A., Fine W. D., Cooper M. D., FcRH1: An activation coreceptor on human B cells. Blood 105, 1121–1126 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Kato K., Satouh Y., Nishimasu H., Kurabayashi A., Morita J., Fujihara Y., Oji A., Ishitani R., Ikawa M., Nureki O., Structural and functional insights into IZUMO1 recognition by JUNO in mammalian fertilization. Nat. Commun. 7, 12198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashida E., Scofield V., Lymphocyte major histocompatibility complex-encoded class II structures may act as sperm receptors. Proc. Natl. Acad. Sci. U.S.A. 84, 3395–3399 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fayngerts S., Najakshin A., Taranin A., Species-specific evolution of the FcR family in endothermic vertebrates. Immunogenetics 59, 493–506 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Liu Y., Wang D.-K., Chen L.-M., The physiology of bicarbonate transporters in mammalian reproduction. Biol. Reprod. 86, 99 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Guselnikov S. V., Ershova S. A., Mechetina L. V., Najakshin A. M., Volkova O. Y., Alabyev B. Y., Taranin A. V., A family of highly diverse human and mouse genes structurally links leukocyte FcR, gp42 and PECAM-1. Immunogenetics 54, 87–95 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Davis R. S., Stephan R. P., Chen C.-C., Dennis G. Jr., Cooper M. D., Differential B cell expression of mouse Fc receptor homologs. Int. Immunol. 16, 1343–1353 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Bianchi E., Wright G. J., Find and fuse: Unsolved mysteries in sperm-egg recognition. PLOS Biol. 18, e3000953 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliver S. L., Brady J. J., Sommer M. H., Reichelt M., Sung P., Blau H. M., Arvin A. M., An immunoreceptor tyrosine-based inhibition motif in varicella-zoster virus glycoprotein B regulates cell fusion and skin pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, 1911–1916 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tregenza T., Wedell N., Genetic compatibility, mate choice and patterns of parentage: Invited review. Mol. Ecol. 9, 1013–1027 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Wedekind C., Mate choice and maternal selection for specific parasite resistances before; during and after fertilization. Philos. Trans. R. Soc. Lond. B Biol. Sci. 346, 303–311 (1994). [DOI] [PubMed] [Google Scholar]
- 42.Yao N., Xiao W., Wang X., Marik J., Park S. H., Takada Y., Lam K. S., Discovery of targeting ligands for breast cancer cells using the one-bead one-compound combinatorial method. J. Med. Chem. 52, 126–133 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brewis I., Clayton R., Barratt C. L., Hornby D. P., Moore H. D., Recombinant human zona pellucida glycoprotein 3 induces calcium influx and acrosome reaction in human spermatozoa. Mol. Hum. Reprod. 2, 583–589 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Clayton R., Cooke D., Partridge L., Moore H., A combinatorial phage display library for the generation of specific Fab fragments recognizing human spermatozoa and inhibiting fertilizing capacity in vitro. Biol. Reprod. 59, 1180–1186 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sable R., Jambunathan N., Singh S., Pallerla S., Kousoulas K. G., Jois S., Proximity ligation assay to study protein–protein interactions of proteins on two different cells. Biotechniques 65, 149–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamas-Toranzo I., Hamze J. G., Bianchi E., Fernández-Fuertes B., Pérez-Cerezales S., Laguna-Barraza R., Fernández-González R., Lonergan P., Gutiérrez-Adán A., Wright G. J., Jiménez-Movilla M., Bermejo-Álvarez P., TMEM95 is a sperm membrane protein essential for mammalian fertilization. eLife 2, e53913 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Findlay G. D., Sitnik J. L., Wang W., Aquadro C. F., Clark N. L., Wolfner M. F., Evolutionary rate covariation identifies new members of a protein network required for Drosophila melanogaster female post-mating responses. PLOS Genet. 10, e1004108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Partha R., Kowalczyk A., Clark N. L., Chikina M., Robust method for detecting convergent shifts in evolutionary rates. Mol. Biol. Evol. 36, 1817–1830 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R. Partha, Evolutionary-Based Methods for Predicting Genotype-Phenotype Associations in the Mammalian Genome, thesis, University of Pittsburgh, 2019; http://d-scholarship.pitt.edu/37231/.
- 52.Talavera G., Castresana J., Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O., New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 54.The UniProt Consortium , UniProt: The universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y., The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zimmerman B., Kelly B., McMillan B. J., Seegar T. C., Dror R. O., Kruse A. C., Blacklow S. C., Crystal structure of a full-length human tetraspanin reveals a cholesterol-binding pocket. Cell 167, 1041–1051.e11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Webb B., Sali A., Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 47, 5.6.1–5.6.32 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Kozakov D., Hall D. R., Xia B., Porter K. A., Padhorny D., Yueh C., Beglov D., Vajda S., The ClusPro web server for protein-protein docking. Nat. Protoc. 12, 255–278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lazaridis T., Effective energy function for proteins in lipid membranes. Proteins 52, 176–192 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Brooks B. R., Brooks C. L. III, Mackerell A. D. Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M., CHARMM: The biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., Baker N. A., PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A., Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schrodinger LLC, The PyMOL Molecular Graphics System, Version 1.8 (2015).
- 65.NIS-Elements imaging software. www.microscope.healthcare.nikon.com/products/software/nis-elements.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Movies S1 to S10