Abstract
Methanosarcina barkeri 227 possesses two clusters of genes potentially encoding nitrogenases. We have previously demonstrated that one cluster, called nif2, is expressed under molybdenum (Mo)-sufficient conditions, and the deduced amino acid sequences for nitrogenase structural genes in that cluster most closely resemble those for the Mo nitrogenase of the gram-positive eubacterium Clostridium pasteurianum. The previously cloned nifH1 from M. barkeri shows phylogenetic relationships with genes encoding components of eubacterial Mo-independent eubacterial alternative nitrogenases and other methanogen nitrogenases. In this study, we cloned and sequenced nifD1 and part of nifK1 from M. barkeri 227. The deduced amino acid sequence encoded by nifD1 from M. barkeri showed great similarity with vnfD gene products from vanadium (V) nitrogenases, with an 80% identity at the amino acid level with the vnfD gene product from Anabaena variabilis. Moreover, there was a small open reading frame located between nifD1 and nifK1 with clear homology to vnfG, a hallmark of eubacterial alternative nitrogenases. Stimulation of diazotrophic growth of M. barkeri 227 by V in the absence of Mo was demonstrated. The unusual complement of nif genes in M. barkeri 227, with one cluster resembling that from a gram-positive eubacterium and the other resembling a eubacterial V nitrogenase gene cluster, suggests horizontal genetic transfer of those genes.
The nitrogenase enzyme complex typically found in eubacteria contains two components, component 2, which contains a single iron-sulfur center and is encoded by nifH, and component 1, which contains an iron-sulfur center and an iron-molybdenum cofactor and is encoded by nifD and nifK. There also exist alternative nitrogenases that contain vanadium instead of molybdenum in their component 1 and are encoded by vnf genes or that contain only iron in their component 1 and are encoded by anf genes (3). Component 1 of alternative nitrogenases also contains a third subunit type encoded by vnfG or anfG. This subunit has been found in all alternative nitrogenases studied and appears to play a role in cofactor processing and function (6, 29). The vnfG/anfG genes are situated between the vnfD/anfD and the vnfK/anfK genes, and in the case of Anabaena variabilis (28), the vnfD and vnfG genes form a single open reading frame (ORF).
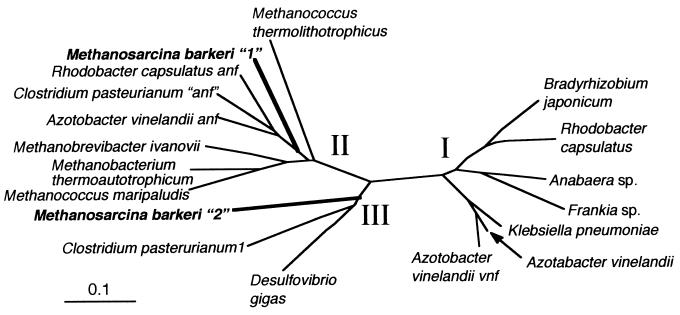
Sibold and colleagues (25) demonstrated that the methanogenic archaeon Methanosarcina barkeri 227 possesses two sets of nifH genes (designated nifH1 and nifH2), and our studies have expanded upon this finding. Phylogenetic analysis of predicted gene products (Fig. 1) shows that nifH1 from M. barkeri clusters with several other methanogen nifH genes as well as anfH genes from several eubacteria in a clade we have termed cluster II (7). The nifH2 gene product from M. barkeri clusters with part of a Mo nitrogenase from Clostridium pasteurianum and those of other anaerobes, such as Desulfovibrio gigas, in a clade called cluster III. Most common eubacterial nitrogenase nifH gene products are in cluster I. Not shown in Fig. 1 are more distantly related nifH homologues found in methanoarchaea which apparently serve purposes other than nitrogen fixation (7, 18, 21) and nifH homologues in eubacteria which play a role in chlorophyll synthesis (5).
FIG. 1.
Unrooted phylogenetic tree for nifH gene products prepared using the neighbor-joining method as described in Materials and Methods. Sequences in clusters II and III are more highly represented than are those in cluster I, and the two nifH sequences from M. barkeri 227 are in boldface.
We cloned and sequenced nifD2 and nifK2 (7) and nifE2 and part of nifN2 (8) from M. barkeri 227 and demonstrated that all of these genes clustered most closely with their corresponding nif homologues in C. pasteurianum. Moreover, we demonstrated that the nif2 genes were expressed during growth in typical Mo-sufficient medium (7, 8). We had previously demonstrated that Mo stimulated diazotrophic growth in M. barkeri 227 (16), so it appears likely that the nif2 gene cluster contains genes involved in the synthesis of a Mo nitrogenase.
The role of the M. barkeri nifH1 gene described by Sibold et al. (25) (GenBank accession no. X56072) is not clear. Although the eubacterial members of cluster II are part of anf-type alternative nitrogenase genes, evidence has been obtained that at least in the cases of Methanococcus thermolithotrophicus (17) and Methanococcus maripaludis (15) these genes encode proteins that are components of Mo nitrogenases. One possibility is that the nif1 genes encode an alternative nitrogenase, possibly of the vanadium (V) type. In our initial attempts to detect stimulation of diazotrophic growth by V (16), inconclusive results were obtained. Scherer (23) showed stimulation of diazotrophic growth by either Mo or V in M. barkeri Fusaro and mentioned that similar but less convincing results could be obtained with strain 227.
To obtain a better understanding of the function of the nif1 genes in M. barkeri, we used PCR to clone and sequence the remaining part of nifD1 from M. barkeri, only 94 nucleotides of which had been sequenced by Sibold et al. (25), as well as part of nifK1. In this publication, we describe these genes and their phylogenetic similarity to vnf-type alternative nitrogenases, as well as a homologue of vnfG/anfG found between nifD1 and nifK1 in M. barkeri 227. We also describe experiments demonstrating that V stimulated diazotrophic growth in M. barkeri 227.
MATERIALS AND METHODS
Cloning and sequencing studies.
M. barkeri 227 (ATCC 43241, DSM 1538, and OCM 35) was obtained from our own culture collection, and DNA was extracted and purified from it as described previously (7). Two primers were designed for the amplification of the nifDK segment. The forward primer (5′-GAAGAAGGAGAAGACAC-3′) was designed from the published sequence (25) of the M. barkeri nifD1 gene. The degenerate, reverse primer [5′(AG)CA(AGTC)CC(TC)G(AGTC)CC(AGTC)CC(AG)TG-3′] was designed from the alignment of 12 different nifK sequences. A Hybaid OMN-E thermal cycler (Labnet, Woodbridge, N.J.) was used for the PCRs. The solutions were subjected to 32 cycles of PCR (denaturing, 1 min at 92°C; annealing, 40 s at 50°C; and extension, 1 min at 72°C). Ligation was performed overnight using an Invitrogen (San Diego, Calif.) TA cloning kit and pCR2.0 plasmid and subsequent transformation of the plasmid into competent Escherichia coli cells. DNA sequencing was performed at the Cornell Biotechnology Institute using an ABI 373A automatic sequencing apparatus and vector-specific and internal primers.
Phylogenetic analysis was performed on amino acid sequences aligned using Clustal X software (12) by using the PHYLIP 3.5 package (10), specifically the PROTDIST, PROTPARS, NEIGBOR, and FITCH programs as previously described (7). Phylogenetic trees were generated using TreeView software (20). Bootstrap analyses were performed on 100 replicates using the SEQBOOT, NEIGHBOR, and CONSENSE programs.
Media and conditions for examining effect of vanadium on growth.
M. barkeri 227 (ATCC 43241, DSM 1538, and OCM 35) cultures were grown in molybdenum-free minimal salts medium based on that of Scherer (23), which contained the following (in millimolar): imidazole, 50; NaH2PO4, 0.5; Na2HPO4, 0.5; MgCl2, 1.7; CaCl2, 2; KCl, 5.4; NaCl, 34; and trace elements consisting of (in micromolar) H3BO3, 1.45; CoCl2, 0.25; NaSeO3, 0.1; ZnCl2, 0.1; MnCl2, 0.045; NiCl2, 0.025; and CuCl2, 0.017. All glassware used in molybdenum starvation experiments was prepared by washing twice with 1 M H2SO4 followed by copious rinsing in MilliQ water (Millipore Corp., Bedford, Mass.). Moreover, all chemicals used in the molybdenum starvation experiments were of the highest grade obtainable. V was added in the form of vanadyl sulfate (Gold Label; Aldrich, Milwaukee, Wis.) containing less than 1 ppm of molybdenum. This medium was prepared anaerobically and dispensed in 50-ml amounts into 126-ml serum bottles under a pure N2 atmosphere (16). After autoclaving, each bottle was supplemented with the following: methanol (8 M), 1 ml; sodium acetate (2.5 M), 1 ml; H2S (gas), 1 ml; NaHCO3 (1%), 0.1 ml; ferric citrate (50 mM), 0.05 ml; titanium(III) citrate (83 mM) (30), 0.1 ml; and the relevant metal addition or ammonia addition. H2S gas was prepared by the acidification (5 ml of 1 M HCl) of 5 ml of anoxic 20% (wt/vol) Na2S in a 126-ml nitrogen-sparged sealed vial.
Mo-starved starter cultures were prepared in the Mo-free medium with the addition of limiting amounts of ammonium chloride (2 mM). After three transfers (2%, vol/vol), cultures grew more slowly and were suitable for metal addition experiments. All bottles were inoculated with 1 ml of M. barkeri grown in ammonia-supplemented Mo-free medium at 37°C. Growth was determined principally by methane production using a thermal conductivity gas chromatograph (16).
Nitrogenase assays.
Whole-cell nitrogenase enzyme activity was determined by the acetylene reduction assay reaction in sealed 36-ml argon-filled acid-washed serum vials as previously described by Lobo and Zinder (16), except that a 1.5-m by 3.2-mm stainless steel column packed with 80/100 mesh alumina F-1 (Supleco, Bellefonte, Pa.) was used to resolve methane, ethane, ethene, and acetylene.
Nucleotide sequence accession number.
The accession number for the sequences reported in this paper is AF254784.
RESULTS
Analysis of the nifHDK1 gene cluster in M. barkeri 227.
Figure 2 presents a map of the M. barkeri nif1 region cloned and sequenced by Sibold et al. (25) and ourselves. Like all functional nif gene clusters in methanogens studied to date, there are two small ORFs, both resembling eubacterial glnB genes (25), located between nifH1 and nifD1. The deduced nifD1 gene was 1,398 bp long (including a TGA encoding a translation stop), has approximately 41 mol% G+C, and encodes a polypeptide of 465 amino acids with a predicted molecular mass of 52,807 Da. The predicted polypeptide lacked the 50-amino-acid insert present in the M. barkeri nifD2 gene product and the nifD1 gene product from C. pasteurianum.
FIG. 2.
Physical map of a 3.4-kb DNA fragment from M. barkeri 227 containing nifHDGK1 genes. The original clone of Sibold et al. (25) contained nifH1, ORF105, ORF 123, and a small part of nifD1. The PCR oligonucleotide primers (termed oligo1 and oligo2), used for PCR cloning of the nifDGK1 genes, are indicated by arrows. The direction of transcription for the nif genes is shown by a large arrow. H, HindIII; E, EcoRI; P, PstI; EV, EcoRV.
Immediately following the TGA encoding the translation stop of the nifD1 gene is an ATG encoding a translation start of another ORF which, as described in more detail below, shows clear homology to vnfG and anfG genes of alternative nitrogenases. Eight base pairs upstream of this ATG is an AGGAA encoding a potential Shine-Dalgarno translation initiation signal within the putative nifD1 gene. This “nifG” ORF is 336 bp long, including a TAA encoding a translation stop, and encodes a predicted polypeptide with 111 amino acids and a molecular mass of 12,897 Da. Nineteen base pairs downstream from nifG1 is the sequence encoding the potential translation start site of an ORF containing nifK1, of which there are 146 bp of data until the end of the cloned fragment.
We performed a phylogenetic analysis of the predicted gene product of the nifD1 gene, and the results are presented in Fig. 3. It clearly clustered with the vnfD gene products from Azotobacter vinelandii and Anabaena variabilis, with a closer relationship with the latter, and was distant from the other methanogen nifD gene products. The identity at the amino acid level with the A. variabilis vnfD gene product was 80%, whereas it was 69, 58, and 35% with the A. vinelandii vnfD, anfD, and nifD gene products, respectively. Thus, there was a high degree of similarity with V nitrogenases. The eubacterial anfD gene products clustered with the vnfD gene products to the exclusion of the three methanogen nifD gene products in cluster II, which, it could be argued, form a separate cluster. As previously demonstrated (8), the nifD2 gene product from M. barkeri was most closely related to that of C. pasteurianum.
FIG. 3.
Unrooted phylogenetic tree for nifD gene products prepared using the neighbor-joining method. The nifD gene products from M. barkeri 227 are in boldface.
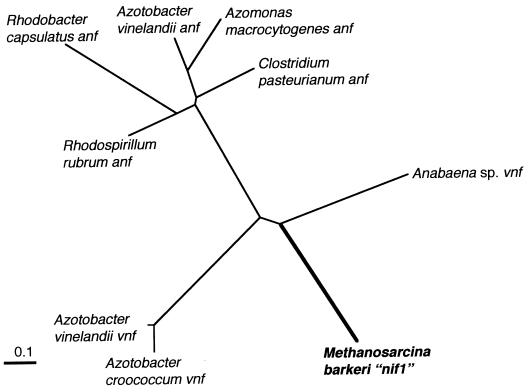
Figure 4 shows the phylogenetic relationship between the M. barkeri “nifG1” gene product and the corresponding predicted sequences of various vnfG and anfG gene products. Similar to the case for the nifD gene products, the “nifG” gene product from M. barkeri had the greatest similarity with the vnfG gene product from A. variabilis. However, the nifG gene product amino acid sequences were not nearly as conserved as were those for nifD. The M. barkeri nifG1 predicted gene product had 44, 38, and 35% amino acid identity with the A. variabilis vnfG gene product and the A. vinelandii vnfG and anfG gene products, respectively. The vnf and anf genes clearly formed distinct phylogenetic clusters.
FIG. 4.
Unrooted tree prepared using the neighbor-joining method showing phylogeny of vnfG/anfG gene products. The “nifG1” sequence from M. barkeri 227 is in boldface.
Diazotrophy of M. barkeri 227 grown with V in the absence of Mo.
We have found it difficult to obtain reproducible Mo limitation of growth using our standard growth medium and conditions. In our original studies on trace element requirements for diazotrophy in M. barkeri 227, growth in the absence of added Mo was 60% of that with Mo. Examining the procedures developed by Scherer (23) for metal limitation of growth of M. barkeri 227, we found that one particularly important source of variability is sodium sulfide, which, despite our rinsing the crystals with distilled water, seems to be a major source of contamination. Replacing it with H2S gas generated by acidification of sodium sulfide appeared to limit the amount of variability. Indeed, we were eventually able to detect Mo limitation of growth with ammonia, which is expected since M. barkeri uses Mo as a prosthetic group in the enzyme formylmethanofuran dehydrogenase (13, 24), which carries out a reaction analogous to that of formate dehydrogenase. We also found that it took at least two transfers in medium lacking added Mo from our standard medium containing ca. 0.4 μM Mo before we detected growth limitation. Similar results were obtained in studies on C. pasteurianum by Hinton and Mortenson (11), who detected molybdenum storage proteins in that organism.
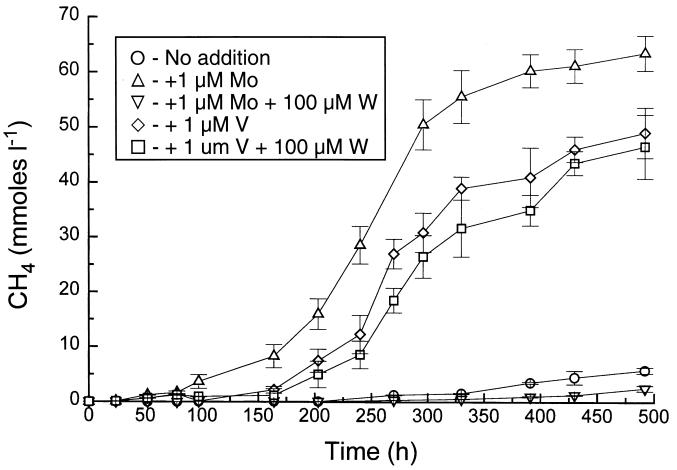
Using medium and conditions adapted from those of Scherer, we were able to demonstrate V stimulation of diazotrophic growth as measured by methane production (Fig. 5). We (16, 19) and Scherer (23) have found an excellent correlation between methanogenesis and growth in Methanosarcina spp. The culture was able to grow with ammonium in the absence of added Mo or V either because the amounts of these elements in the medium met the needs of the nondiazotrophic cells or, perhaps, because there was some contaminating Mo or V in the ammonium chloride used. Schmitz et al. (24) found that M. barkeri Fusaro could grow with methanol without added Mo, whereas growth with H2-CO2, which presumably required higher levels of the molybdoenzyme formylmethanofuran dehydrogenase, was Mo dependent. Under diazotrophic conditions, methanogenesis was severely limited in the absence of added Mo or V, and there was slight stimulation by the vitamin solution that we added, similar to results of Scherer (23). Addition of Mo greatly stimulated methanogenesis under diazotrophic conditions, and V caused a smaller but still significant effect.
FIG. 5.
Effects of additions of trace metals on methanogenesis by growing cultures of M. barkeri 227. All cultures except the culture designated No addition received a vitamin mixture (1). Except in the case of addition of NH4+, the cultures were grown diazotrophically. Error bars indicate standard deviations.
We also examined the effects of the Mo antagonist tungsten (W) (added as sodium tungstate) on methanogenesis when the organism was growing diazotrophically with Mo or V. W inhibits Mo nitrogenases but not alternative ones (3). Our previous results (16) demonstrated that W inhibited methanogenesis by M. barkeri 227 when neither Mo nor V was added to the medium under diazotrophic conditions but not when growth was with NH4+ as the nitrogen source. As shown in Fig. 6, the stimulation of methanogenesis by Mo under diazotrophic conditions was completely inhibited by adding a 100-fold excess of W. As before, adding V caused a smaller stimulation of methanogenesis under diazotrophic conditions, but addition of 100 μM W caused slight, if any, inhibition.
FIG. 6.
Effects of W on methanogenesis by M. barkeri 227 grown diazotrophically in the presence of Mo or V. Error bars indicate standard deviations.
A hallmark of V-type alternative nitrogenases is that when they reduce acetylene, they produce ethane amounting to 1 to 3% of the amount of ethene produced, whereas ethane is barely detectable from Mo-type nitrogenases (3, 9). Methanogenic nitrogenases reduce acetylene at very low rates overall compared to eubacterial nitrogenases (2, 16, 17), so that results may not be completely comparable. In the case of M. barkeri 227 growing diazotrophically in the absence of Mo and in the presence of V, the ethane produced by cells was 1.24% ± 0.14% of the ethene produced, whereas ethane was undetectable in cells growing with Mo in the growth medium.
DISCUSSION
The results presented here support the hypothesis that the nif1 gene cluster from M. barkeri 227 encodes an alternative nitrogenase, most likely of the vanadium type. Evidence supporting the presence of an alternative nitrogenase includes the presence of a “nifG” gene (heretofore found only in alternative nitrogenases) in the nif1 cluster, the sequence similarity between the nifG1 and nifD1 gene products and their vnf counterparts in A. variabilis, the stimulation of diazotrophic growth by V added to the medium, the lack of inhibition by W of nitrogen-fixing cells grown with V, and the production of significant amounts of ethane from acetylene. Direct demonstration that the purified enzyme is a vanadium nitrogenase would require much more cell material than we were able to obtain using Mo-depleted medium. Thus, our evidence for a V-containing nitrogenase in M. barkeri is tentative, depending mainly on the stimulation of diazotrophic growth by V in Mo-depleted medium.
The nif1 gene cluster in M. barkeri clearly resembles those of other methanogens (4, 25–27), containing two ORFS homologous to glnB located between nifH and nifD. These ORFS are not found in eubacterial nif gene clusters and have recently been shown to modulate ammonia switch-off of nitrogenase activity in Methanococcus maripaludis (14). However, unlike other methanogen nif gene clusters, but similar to eubacterial vnf and anf clusters, there is an ORF located between nifD1 and nifK1 in M. barkeri. The “nifG1” of M. barkeri clearly clusters with vnfG genes, with closest homology to vnfG of the cyanobacterium A. variabilis.
The 80% amino acid identity between deduced nifD1 gene product from M. barkeri and the vnfD gene product from the cyanobacterium A. variabilis is one of the highest degrees of identity ever found between eubacterial and archaeal gene products (R. Doolittle, personal communication). To provide perspective, the identity between the nifD polypeptides in A. vinelandii and Klebsiella pneumoniae, two members of the gamma proteobacteria, is 72%. The moles percent G+C for the M. barkeri nifD1 gene is 41%, essentially identical with the organism's value of 42% (1), whereas that for the A. variabilis ATCC 29413 vnfDG genes is 45.3%, similar to that of the organism's genomic DNA, which is 45% (22). It is difficult to imagine how these genes could be so highly conserved in the absence of genetic transfer, but it is not clear in which direction this putative transfer occurred. The fact that the vnfD gene product from the eubacterium A. vinelandii clusters outside the M. barkeri and A. vinelandii products suggests that the transfer occurred into an ancestor of M. barkeri; however, more vnfD sequences need to be obtained from diverse organisms to clarify this situation. For example, there are no vnf genes thus far detected in clostridia.
The situation regarding vnfH genes is interesting. Whereas the vnfD gene from A. vinelandii is clearly an alternative nitrogenase gene in cluster II (Fig. 3), the A. vinelandii vnfH gene is very closely related to nifH in cluster I (Fig. 1). This has been interpreted as a recruitment of a copy of nifH by the vnfDGK genes in A. vinelandii (3). In the case of the cyanobacterium A. variabilis (28), no vnfH was found immediately upstream or downstream of the vnfDGK genes, and all four nifH-homologous genes found in this organism fall within cluster I (T. Thiel, personal communication). Thus, an authentic vnfH has not been yet identified. The M. barkeri nifH1 gene product clusters with anfH gene products to the exclusion of other methanogen nifH gene products in a neighbor-joining analysis (Fig. 1), suggesting that it may be an authentic vnfH gene. Analysis of the same data set shown in Fig. 1 using least-squares analysis (FITCH program [see Materials and Methods]) or parsimony analysis (PROTPARS program) gave similar results, but in a bootstrap analysis using neighbor joining, the M. barkeri product clustered with the anfH gene products to the exclusion of the methanogen sequences in only 59 of 100 replicates. Thus, it is possible that the M. barkeri nifH1 gene was a methanogenic nifH recruited by incoming vnf genes, similar to the situation proposed for A. vinelandii.
The genes encoding M. barkeri nitrogenases are considerably different from those in other methanogens examined thus far, which have cluster II genes apparently encoding their Mo nitrogenases and lack alternative nitrogenases. In contrast, M. barkeri contains one set of nif genes resembling those of C. pasteurianum and another resembling eubacterial vnf genes, including the presence of a vnfG-homologous ORF. Despite these differences, both nif gene clusters contain two ORFs homologous to glnB, making them similar to other methanogen nif gene clusters. It is difficult to hypothesize a scenario that can account for these similarities and differences, and perhaps more nif sequences from diverse archaea and eubacteria will clarify this situation.
Because the nif2 genes in M. barkeri are most homologous to those encoding the Mo nitrogenase from C. pasteurianum and because we have demonstrated that they are expressed in Mo-containing medium (8), we propose calling them simply nif genes. Because of their similarity to vnf genes and the evidence for V stimulation of diazotrophic growth of M. barkeri, we propose calling the nif1 genes in M. barkeri “vnf,” with the quotation marks indicating the tentative assignment.
ACKNOWLEDGMENTS
This research was supported by NSF grant 9506330 and DOE grant DE-FG02-85ER13370.
We thank Vasilios Roditis for his assistance in preliminary experiments on V stimulation of nitrogenase and P. Bishop, T. Thiel, and R. Doolittle for useful discussions.
REFERENCES
- 1.Balch W E, Fox G E, Magrum L J, Woese C R, Wolfe R S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belay N, Sparling R, Daniels L. Dinitrogen fixation by a thermophilic methanogenic bacterium. Nature. 1984;312:286–288. doi: 10.1038/312286a0. [DOI] [PubMed] [Google Scholar]
- 3.Bishop P E, Premakumar R. Alternative nitrogen fixation systems. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 736–762. [Google Scholar]
- 4.Blank C E, Kessler P S, Leigh J A. Genetics in methanogens: transposon insertion mutagenesis of a Methanococcus maripaludis nifH gene. J Bacteriol. 1995;177:5773–5777. doi: 10.1128/jb.177.20.5773-5777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke D H, Hearst J E, Sidow A. Early evolution of photosynthesis—clues from nitrogenase and chlorophyll iron proteins. Proc Natl Acad Sci USA. 1993;90:7134–7138. doi: 10.1073/pnas.90.15.7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee R, Ludden P W, Shah V K. Characterization of VNFG, the delta subunit of the vnf-encoded apodinitrogenase from Azotobacter vinelandii. Implications for its role in the formation of functional dinitrogenase 2. J Biol Chem. 1997;272:3758–3765. doi: 10.1074/jbc.272.6.3758. [DOI] [PubMed] [Google Scholar]
- 7.Chien Y-T, Zinder S H. Cloning, DNA sequencing, and characterization of a nifD-homologous gene from the archaeon Methanosarcina barkeri 227 which resembles nifD from the eubacterium Clostridium pasteurianum. J Bacteriol. 1994;176:6590–6598. doi: 10.1128/jb.176.21.6590-6598.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chien Y-T, Zinder S H. Cloning, functional organization, transcript studies, and phylogenetic analysis of the complete nitrogenase structural genes (nifHDK2) and associated genes in the archaeon Methanosarcina barkeri 227. J Bacteriol. 1996;178:143–148. doi: 10.1128/jb.178.1.143-148.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dilworth M J, Eady R R, Robson R L, Miller R W. Ethane formation from acetylene as a potential test for vanadium nitrogenase in vivo. Nature. 1987;327:167–168. [Google Scholar]
- 10.Felsenstein J. Phylogenetic inference program (PHYLIP) manual version 3.5c. Seattle: University of Washington; 1993. [Google Scholar]
- 11.Hinton S M, Mortenson L E. Identification of molybdoproteins in Clostridium pasteurianum. J Bacteriol. 1985;162:477–484. doi: 10.1128/jb.162.2.477-484.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeanmougin F, Thompson J D, Gouy M, Higgins D G, Gibson T J. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 13.Karrasch M, Börner G, EnBle M, Thauer R K. Formylmethanofuran dehydrogenase from methanogenic bacteria, a molybdoenzyme. FEBS Lett. 1989;253:226–230. doi: 10.1016/0014-5793(89)80964-0. [DOI] [PubMed] [Google Scholar]
- 14.Kessler P S, Leigh J A. Genetics of nitrogen regulation in Methanococcus maripaludis. Genetics. 1999;152:1343–1351. doi: 10.1093/genetics/152.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kessler P S, McLarnan J, Leigh J A. Nitrogenase phylogeny and the molybdenum dependence of nitrogen fixation in Methanococcus maripaludis. J Bacteriol. 1997;179:541–543. doi: 10.1128/jb.179.2.541-543.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobo A L, Zinder S H. Diazotrophy and nitrogenase activity in the archaebacterium Methanosarcina barkeri 227. Appl Environ Microbiol. 1988;54:1656–1661. doi: 10.1128/aem.54.7.1656-1661.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magot M, Possot O, Souillard N, Henriquet M, Sibold L. Structure and expression of nif (nitrogen fixation) genes in methanogens. In: Dubourgier H C, et al., editors. Biology of anaerobic bacteria. Amsterdam, The Netherlands: Elsevier Science Publishers B.V.; 1986. pp. 193–199. [Google Scholar]
- 18.Maupin-Furlow J A, Ferry J G. Analysis of the CO dehydrogenase/acetyl-coenzyme A synthase operon of Methanosarcina thermophila. J Bacteriol. 1996;178:6849–6856. doi: 10.1128/jb.178.23.6849-6856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray P A, Zinder S H. Nutritional requirements of Methanosarcina thermophila strain TM-1. Appl Environ Microbiol. 1985;50:49–55. doi: 10.1128/aem.50.1.49-55.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Page R D. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 21.Possot O, Henry M, Sibold L. Distribution of DNA sequences homologous to nifH among archaebacteria. FEMS Microbiol Lett. 1986;34:173–177. [Google Scholar]
- 22.Rippka R, Derruelles J, Waterbury J B, Herdman M, Stanier R Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 23.Scherer P. Vanadium and molybdenum requirement for the fixation of molecular nitrogen by two Methanosarcina strains. Arch Microbiol. 1989;151:44–48. [Google Scholar]
- 24.Schmitz R A, Bertram P A, Thauer R K. Tungstate does not support synthesis of active formymethanofuran dehydrogenase in Methanosarcina barkeri. Arch Microbiol. 1994;161:528–530. [Google Scholar]
- 25.Sibold L, Henriquet M, Possot O, Aubert J-P. Nucleotide sequence of nifH regions from Methanobacterium ivanovii and Methanosarcina barkeri and characterization of glnB-like genes. Res Microbiol. 1991;142:5–12. doi: 10.1016/0923-2508(91)90091-n. [DOI] [PubMed] [Google Scholar]
- 26.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J I, Rice P, Nolling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum Delta-H: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Souillard N, Sibold L. Primary structure, functional organization and expression of nitrogenase structural genes of the thermophilic archaebacterium Methanococcus thermolithotrophicus. Mol Microbiol. 1989;3:541–551. doi: 10.1111/j.1365-2958.1989.tb00200.x. [DOI] [PubMed] [Google Scholar]
- 28.Thiel T. Characterization of genes for an alternative nitrogenase in the cyanobacterium Anabaena variabilis. J Bacteriol. 1993;175:6276–6286. doi: 10.1128/jb.175.19.6276-6286.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waugh S I, Paulsen D M, Mylona P V, Maynard R H, Premakumar R, Bishop P E. The genes encoding the delta subunits of dinitrogenases 2 and 3 are required for Mo-independent diazotrophic growth by Azotobacter vinelandii. J Bacteriol. 1995;177:1505–1510. doi: 10.1128/jb.177.6.1505-1510.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zehnder A J B, Wuhrmann K. Titanium (III) citrate as a nontoxic oxidation-reduction buffering system for the culture of obligate anaerobes. Science. 1976;194:1165–1166. doi: 10.1126/science.793008. [DOI] [PubMed] [Google Scholar]