Abstract
Yuan-Zhi Decoction (YZD) is a traditional Chinese medical formulation with demonstrated clinical benefits in Alzheimer’s disease (AD). We used liquid chromatography coupled with mass spectrometry to identify 27 unique chemical components of YZD. Analyzing these using network pharmacology and molecular docking models identified 34 potential interacting molecular targets involved in 26 biochemical pathways. When tested in an animal model of AD, the APP/PS1 transgenic mice showed measurable improvements in spatial orientation and memory after the administration of YZD. These improvements coincided with significantly reduced deposition of Aβ plaques and tau protein in the hippocampi in the treated animals. In addition, a decreased BACE1 and beta-amyloid levels, a downregulation of the p-GSK-3β/GSK-3β, and an upregulation of the PI3K and p-AKT/AKT pathway was seen in YZD treated animals. These in vivo changes validated the involvement of molecular targets and pathways predicted in silico analysis of the chemical components of YZD. This study provides scientific support for the clinical use of YZD and justifies further investigations into its effects in AD. Furthermore, it demonstrates the utility of network pharmacology in elucidating the biochemical mechanisms underlying the beneficial effects of traditional Chinese medicines (TCM).
Keywords: network pharmacology, molecular docking, molecular mechanism, Alzheimer’s disease, Yuan-Zhi decoction
Introduction
Alzheimer’s disease (AD) is an irreversible, chronic, and relentless neurodegenerative disease, characterized by memory loss, psychoses, affective disorders, and behavioral alterations (Gong et al., 2015; Fang et al., 2017; Cai F. F. et al., 2018; Pang et al., 2018; Sun et al., 2018). Numerous details of AD-related neuropathological changes have been described, including the formation of amyloid plaques, neurofibrillary tangles, and loss of neurons and synapses. Nonetheless, key events leading to the pathogenesis and progression of the disease remain poorly understood (Kumar et al., 2015).
Considering the multifactorial causation of AD, pharmacotherapy approaches targeting a single molecule may not optimal. And As an example, although the anti-amyloid antibody, aducanumab, developed by Biogen, has been shown to lower the number of amyloid plaques in the brain, there is no evidence that it improves cognitive outcomes. Thus, the accelerated approval of aducanumab by the FDA remains controversial (Mullard, 2021). Given the worldwide impact of AZ effective novel approaches to the management of the disease are urgently needed (Luo et al., 2020). TCM has been in clinical use for thousands of years, particularly in the management of chronic diseases. Numerous Chinese herbal formulations show efficacy in the treatment of AD. These are gaining gradual worldwide acceptance due their holistic approach acting simultaneously of multiple symptoms, while exhibiting limited side effects (Wang and Zhang, 2017).
Yuan-Zhi Decoction (YZD) is a natural plant-derived TCM formulation. It can be traced back to the book “Qi xiao liang fang” written by Fang Xian during the Ming Dynasty. YZD is a mixture of eight Chinese herbs, namely: Polygala tenuifolia Willd. (yuan-zhi in Chinese), Panax ginseng C. A. Mey. (ren-shen in Chinese), Acorus tatarinowii Schott (shi-chang-pu in Chinese), Notopterygium incisum Ting ex H. T. Chang (qiang-huo in Chinese), Asrum heterotropoides Fr. Schmidt var. mandshuricum (Maxim.) Kitag. (xi-xin in Chinese), Ephedra sinica Stapf (ma-huang in Chinese), Paeonia lactiflora Pall. (chi-shao in Chinese), Atractylodes macrocephala Koidz. (bai-zhu in Chinese). This formulation has been in use to treat a variety of medical issues. Recent pharmacological studies have demonstrated that many ingredients of YZD have beneficial effects in AD. Yuanzhi (Polygala tenuifolia Willd.) is an ingredient of multiple formulas use to managed age related memory impairment (May et al., 2016; Howes et al., 2017; Li B. et al., 2021). Shichangpu (Acori Tatarinowii Rhizoma), has pronounced neuro-pharmacological effects (Oh et al., 2004; Chang and Teng, 2018; Song et al., 2018; Wang L. et al., 2019; Deng et al., 2020). Xixin decoction was shown to reduce phosphorylation toxicity at specific sites of the tau protein in the brains of rats (Diwu et al., 2013). However, due to the complex composition of YZD, a comprehensive elucidation of the molecular mechanisms involved in tits beneficial effects in AD remains incomplete.
Network pharmacology, first proposed by Andrew Hopkins in 2007 is a relatively novel approach in elucidating the mechanism of action of multicomponent drugs (Luyen et al., 2015; Lee et al., 2018; Wang N. et al., 2019). Its core concept is to optimize treatment strategies based on the biological network of disease features, bioactive agents, and drug targets that are connected to each other (Hopkins, 2007). With rapid progress in bioinformatics and systems biology, network pharmacology is becoming an increasingly viable approach in identifying active compounds and elucidating the underlying pharmacological mechanisms involved in their beneficial effects (Li et al., 2011; Li and Zhang, 2013; Cai H. et al., 2018; Zhou et al., 2020). Through the integration of chemical component identification, target prediction, and network construction, this approach can effectively predict the molecular mechanisms affected by the active compounds in TCM formulations (Luo Z et al., 2020). As such network pharmacology represents a promising tool in the study of the mechanism of action of complex formulations containing a multitude of active ingredients (Li et al., 2020).
Molecular docking studies use computer simulations to study the interactions between receptors and small molecules potentially binding to them. By predicting certain features, such as the location and affinity of these interaction it is an invaluable tool in drug design and screening of chemical compounds (Chen et al., 2014; Lionta et al., 2014; Saikia and Bordoloi, 2019; Reddyrajula and Dalimba, 2020). Recently the combination of network pharmacology and molecular docking analysis was successfully applied to study the active component of TCM formulations (Wang Y. et al., 2019).
In the study presented here we systematically identified chemical constituents of YZD using liquid chromatography/mass spectrometry and explored possible targets and related pathways with potential relevance to the treatment of AD. The validity of this approach was confirmed by behavioral evaluation, microscopic analysis of the brains, and by Western blotting.
Materials and methods
Chemicals, reagents and materials
HPLC-grade acetonitrile was obtained from Merck KgaA (Darmstadt, Germany). HPLC-grade formic acid was supplied by Macklin Biochemical Co., Ltd. (Shanghai, China). Ultra-pure water (18 MΩ cm) was prepared via a Milli-Q water purification system (Milford, United States). 3,6′-disinapoyl sucrose, Ginsenoside Re, β-Asarone, Asarinin, Notopterol, Ephedrine, Paeoniflorin, and Atractylenolide II were purchased from National Institutes for Food and Drug Control (Beijing, China). The purity of each reference standard was above 98%. All eight herb constituents of YZD, including Polygala tenuifolia Willd., Panax ginseng C. A. Mey., Acorus tatarinowii Schott, Notopterygium incisum Ting ex H. T. Chang, Asrum heterotropoides Fr. Schmidt var. mandshuricum (Maxim.) Kitag., Ephedra sinica Stapf, Paeonia lactiflora Pall., and Atractylodes macrocephala Koidz. were purchased from the Hebei Renxin Pharmaceutical Co., LTD., and authenticated by Deputy Chief Pharmacist Jing He (Department of Pharmacy, General Hospital of Northern Theater Command). Their voucher specimens were deposited at the author’s laboratory.
Preparation of YZD extract and standard solution
Raw herbs contained in the formula (yuan-zhi 9.3 g; ren-shen 18.6 g; shi-chang-pu 18.6 g; qiang-huo 18.6 g; xi-xin 18.6 g; ma-huang 18.6 g; chi-shao 37.3 g; bai-zhu 37.3 g) were soaked for 2 h and extracted by reflux extraction using a 10-fold mass of 70% ethanol aqueous solution. This process was repeated twice. Raw extracts were filtered with a 200-mesh sieve, combined, and concentrated under vacuum, resulting in 60 g thick paste. The extract was obtained with a yield of 33.7% and kept frozen at −20°C until use. A 1.0 g of this paste was sonicated in 50 ml of methanol/water mixture (1:1, v/v) for 5 min, centrifuged at 1,500×g for 10 min at 4°C, the supernatant was filtered through a 0.22 μm membrane filter, and used for further analysis.
Chromatography/mass spectroscopy equipment and parameters
Chromatographic separation was carried out on a Thermo Ultimate 3000LC, Q Exactive HF LC-MS system (Thermo, United States) utilizing an Agilent Zorbax Eclipse C18 column (1.8 μm, 2.1 × 100 mm) at 30°C with a flow rate of 0.3 ml/min. The mobile phase was 0.1% formic acid in water (solvent A) and acetonitrile (solvent B). The gradient elution program was performed as follows: 0–2 min, 5% B; 2–6 min, 5–30% B; 6–7 min, 30% B; 7–12 min, 30–78% B; 12–14 min, 78% B; 14–17 min, 78–95% B; 17–20 min, 95% B; 20–21 min, 95–5% B; 21–25 min, 5% B. 2 μL of sample solution was injected for analysis. Mass detection was performed using an Thermo Ultimate 3000LC Q Exactive HF mass spectrometer (Thermo, United States) equipped with a ESI source operating in both positive and negative mode with the following operating parameters: drying gas (N2) temperature: 325°C, sheath gas flow rate: 45 arb; auxiliary gas flow rate: 15 arb, purge gas flow rate: 1 arb, electrospray voltage: 3.5 KV, capillary temperature: 330°C, S-Lens RF Level: 55% under positive or negative mode. The nozzle voltage was set at +500 V or −1000 V, respectively. The quasimolecular ions [M-H]− and [M + H]+ were selected as precursor ions and subjected to target-MS/MS analyses with different collision energies ranging from 10 to 60 V to acquire sufficient product ions. MS spectra were recorded over the m/z range of 50–1,000. The Compound Discoverer 3.1 platform was used to perform retention time correction, peak identification, peak extraction, and other tasks. Data was processed using the Thermo mzCloud online database and Thermo mzVault local database to identify substances based on the secondary mass spectrum information.
Network pharmacology analysis
Establishing the database of YZD components
Briefly, all available Chinese medicine network pharmacology research databases (TCMSP, TCMID, ETCM) were searched for previously identified chemical components of YZD. We also prepared a list a list of all chemical components identified by combined LC-MS analysis. This comprehensive list of YZD components was used to search the PubChem (https://www.ncbi.nlm.nih.gov/pubmed/), SEA (https://sea.bkslab.org), STITCH (https://stitch.embl.de/), PharmMapper (https://lilab-ecust.cn/pharmmapper/), and SwissTarget Prediction databases (http://www.swisstargetprediction.ch/) to create a comprehensive list of molecules present in YZD that could mediate beneficial effects in AD.
Establishing the AD therapeutic target database
To identify potential AD-related therapeutic targets we conducted multiple database searches with the search term “Alzheimer’s Disease” and limiting searches to “Homo sapiens”. The following databases were queried: Drugbank database (https://www.drugbank.ca/); GeneCards database (https://www.genecards.org/); OMIM database (http://omim.org/); DisGeNET database (http://www.disgenet.org/), and GEO database (https://www.ncbi.nlm.nih.gov/geo/). Potential AD-related molecular targets identified from these searches were merged into a single list and duplicates were removed, resulting in a list of potential drug targets in AD.
Protein-protein interaction network analysis
The list of potential drug targets in AD (described in 2.4.2) was intersected with a list of known or presumed molecular constituents of YZD (described in 2.4.1) using the Omic Share Tools. Molecules present in both lists represented molecules in YZD that were likely to be responsible for its beneficial effects in AD. The Omic Share Tools were used to draw Venn diagrams.
To clarify relationships between the proteins identified in this process their protein-protein interaction (PPI) network was constructed and analyzed using the STRING database (https://string-db.org/), with the species limited to Homo sapiens.
KEGG pathway enrichment analysis and Gene Ontology term performance
KEGG Pathway enrichment and Gene Ontology (GO) term performance analysis were conducted on proteins involved in the network using the Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/) (Ogata et al., 1999) database and the Database for Annotation, Visualization and Integrated Discovery (DAVID) system (http://david.abcc.ncifcrf.gov/home. jsp/, v6.7) (Dennis et al., 2003) Relevant pathways and GO terms with a p value < 0.05 were selected as significant in the development of AD.
Network construction and analysis
To facilitate the interpretation of the complex relationships between putative beneficial components of YZD and known or presumed therapeutic targets in AD, an interaction network was created using the Cytoscape software (version 3.6.0, Boston, MA, United States). This provided an approach for data integration, analysis, and visualization for complex network analysis. (Huang et al., 2019)
Molecular docking analysis of potential active YZD ingredients
To further verify the reliability of the target prediction results, a group of key molecules was selected based on their importance in the“component-target-pathway” network. Three-dimensional structures of the selected target proteins were downloaded from the RCSB (http://www.rcsb.org) database and saved in pdb format. The Maestro software package was used to create a representation of these key proteins, including the removal of repeat units, hydrogenation and charge treatment. Then the docking site of various YZD-derived chemicals on the target proteins was determined, and a grid file was prepared. Models containing the selected protein and the interacting YZD chemical were crated by molecular docking, using the docking score (GlideScore, GS) as the scoring standard.
Experimental evaluation
Animals and drug administration
Six-month-old male transgenic (APPswe PSEN1dE9) 85Dbo mice (APP/PS1) on a C57BL/6 background were obtained from Beijing HFK Bioscience Co. Ltd. These double transgenic animals express both a human-mouse chimeric APP gene (Mo/Hu APP695) carrying the Swedish mutation (Swedish KM594/595NL) and a human PS1 gene with a mutation in its ninth exon (dE9). APP/PS1 mice and wild-type littermates were genotyped by PCR analysis of genomic DNA from tail biopsies. Animals were maintained at 22 ± 2°C with a 12-h light-dark cycle (lights on at 8 a.m., lights off at 8 p.m.) with free access to food and water. All experiments were approved by the Animal Care and Use Institutional Committee of the General Hospital of the Northern Theater Command. APP/PS1 transgenic AD mice were randomly divided into five groups. Animals in the three YZD treatment groups received 0.11 g/kg, 0.32 g/kg, or 0.96 g/kg YZD. Two control groups were given either 4 mg/kg Donazepil or water. A group of wild type mice (WT), sham treated with the same volume of water was used as an additional control group. Treatment or sham treatment was administered via oral gavage for 3 months.
Morris water maze test
After the 3 months treatment period the Morris water maze (MWM) test was used to assess cognitive behavior. The test consisted of a one-day conditioning test, a five-day spatial acquisition test, and a one-day probe trial test. Tests were performed in a circular pool (120 cm in diameter and 50 cm in height) filled with water at 20–22°C. The pool was filled to a depth of 26 cm with water containing an opaque nontoxic Titanium dioxide die, and placed in a dimly lit, soundproof test room surrounded by distinct extra maze cues. The pool was equally divided into four quadrants. A white hidden circular platform (12 cm in diameter and 25 cm high) was placed randomly under a quadrant of the pool, submerged 1 cm below the surface of the water. In the conditioning test, mice swam freely in the pool for 60 s. Each mouse was placed in the pool in the four different quadrants every day. The timer was set to 60 s and automatically stopped once the mouse found the platform and remained on it for 5 s. Animals unable to locate the platform themselves withing 60 s were manually moved to the platform and allowed to stay on it for 20 s. The probe trial test was performed on the 7th day when the platform was removed. Mice were allowed to swim for 60 s, searching for the missing platform at its last know location. The escape latency, distance, speed, and the number of times animals crossed the platform were recorded using a video camera-based Supermaze System (Shanghai Xinruan Information Technology Co., Ltd., Shanghai, China).
Western blot analysis
Mice were anesthetized with sodium pentobarbital (50 mg/kg), killed, and brains were harvested for Western blot analysis. Hippocampi, isolated from the brain, were homogenized in RIPA lysis buffer (Solarbio Science & Technology Co., Beijing, China) containing a protease and phosphatase inhibitor cocktail (Thermo Scientific, Waltham, MA, United States). The homogenized tissue was centrifuged at 12,000 rpm for 10 min at 4°C to remove debris and the protein concentration of each sample was determined using a BCA protein assay (Solarbio Science & Technology Co.). The prepared homogenates were used to estimate the protein expression levels of APP, BACE1, beta-amyloid, pGSK3B/GSK3B, and pAKT/AKT. Samples were electrophoresed using SDS-10% PAGE, transferred to a PVDF membrane, and protein expression was determined using antibodies against APP, BACE1, beta-amyloid, pGSK3B/GSK3B, pAKT/AKT, and actin (1:1,000; Abcam, Boston, United States), respectively. Signal was detected using an ECL method and quantified in a ChemiDoc MP Imaging System (Bio-Rad, California, United States).
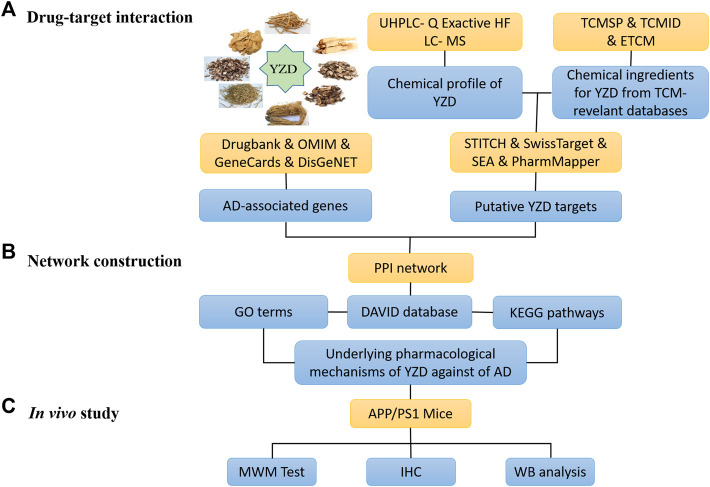
The schematic diagram depicting the steps of all experimental work is shown in Figure 1.
FIGURE 1.
Flowchart of the steps taken during the system pharmacology analysis of the therapeutic mechanisms of action of Yuan-Zhi Decoction (YZD) in Alzheimer’s disease (AD).
Immunohistochemical analyses
Hemispheres prepared for immunohistochemical analyses were immersion fixed in 5% paraformaldehyde, and paraffin embedded brain samples were cut into 3 μm slices using a microtome (Leica RM2235). Slides were deparaffinize with graded ethanol, incubated with the primary antibody overnight at 4°C and with the appropriate fluoroprobe labelled secondary antibody for 1 h at room temperature, protected from light. Antibodies used were: Anti-Amyloid beta 40 Rabbit (Servicebio, GB111197) (1:500) for the detection of Aβ plaques, and anti-Tau Rabbit (Servicebio, GB111178) (1:200) for Tau.
Statistical analysis
The experimental data were analyzed using SPSS 20.0 package (SPSS Inc., Chicago, IL, United States). Figures were created by GraphPad Prism 6.0 software (GraphPad Software Inc., San Diego, CA, United States). All results are expressed as mean ± standard error of the mean (SEM). Multigroup comparisons in the MWM test were performed using two-way ANOVA. For other experimental data, one-way ANOVA was used, using LSD to test for homogeneity of variance. A non-parametric test was used for unequal variance. A p value <0.05 was considered statistically significant.
Results
Identifying chemical compounds present in YZD
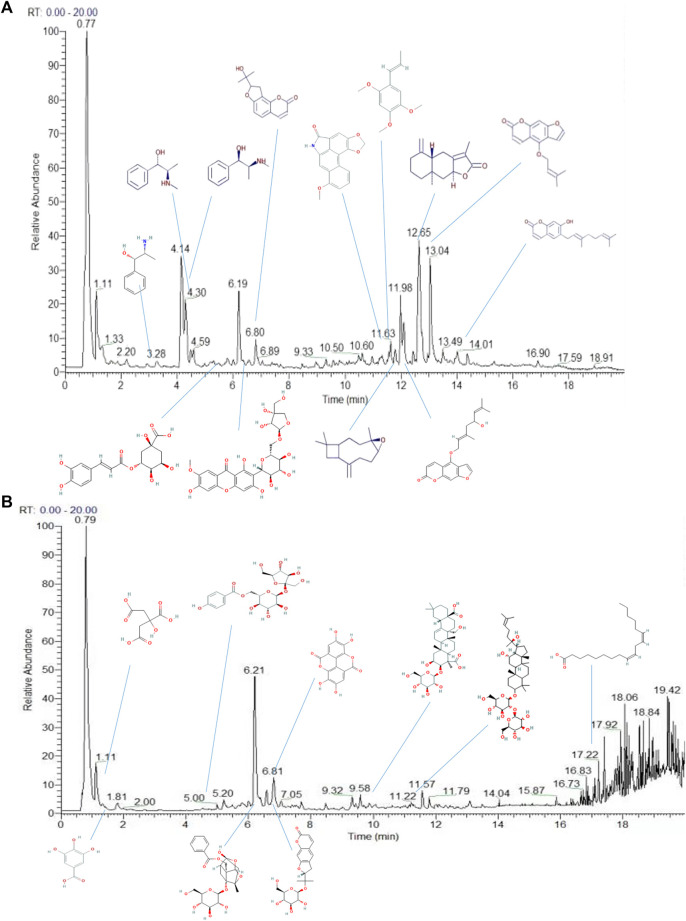
Using a combination of liquid chromatography and mass spectroscopy identified 211 constituents of YZD based on mass, chromatographic behavior, MS/MS data, and fragmentation rules (Figure 2 and Supplementary Table S1 ). Additional data mining of databases relevant to TCM revealed a total of 1026 individual compounds present in YZD (Supplementary Table S2 ). Of these, 27 represented chemical components uniquely present in YZD, listed in Table 1. It is notable that of the 27 chemical ingredients, three compounds were found to be present in more than one herb.
FIGURE 2.
Total ion chromatograms of the chemical constituents of YZD in positive (A) and negative (B) ion modes during UHPLC-Q Exactive HF analysis.
TABLE 1.
The main active ingredients of YZD.
| Ingredients | CAS | Chemical formula |
|---|---|---|
| Polygalaxanthone | 162857-78-5 | C25H28O15 |
| Tenuifolin | 20183-47-5 | C36H56O12 |
| Ferulaldehyde | 458-36-6 | C10H10O3 |
| Sibiricose A3 | 139726-39-9 | C19H26O13 |
| Citric acid | 77-92-9 | C6H8O7 |
| Ginsenoside-Rg3 | 14197-60-5 | C42H72O13 |
| Ginsenoside-Rg1 | 22427-39-0 | C42H72O14 |
| Ginsenoside-Re | 52286-59-6 | C48H82O18 |
| Ginsenoside-Rb1 | 41753-43-9 | C54H92O23 |
| Caryophyllene oxide | 1139-30-6 | C15H24O |
| Aristolactam I | 13395-02-3 | C17H11NO4 |
| β-asarone | 494-40-6 | C12H16O3 |
| Ephedrine | 299-42-3 | C10H15NO |
| Pseudoephedrine | 90-82-4 | C10H15NO |
| Norephedrine | 37577-28-9 | C9H13NO |
| linoleic acid | 60-33-3 | C18H32O2 |
| Paeoniflorin | 23180-57-6 | C23H28O11 |
| Gallic acid | 149-91-7 | C7H6O5 |
| Ellagic acid | 476-66-4 | C14H6O8 |
| Albiflorin | 39011-90-0 | C23H28O11 |
| chlorogenic acid | 327-97-9 | C16H18O9 |
| Notopterol | 88206-46-6 | C21H22O5 |
| Isoimperatorin | 482-45-1 | C16H14O4 |
| Columbianetin | 3804-70-4 | C14H14O4 |
| Nodakenin | 495-31-8 | C20H24O9 |
| Ostruthin | 148-83-4 | C19H22O3 |
| Atractylenolide II | 73069-14-4 | C15H20O2 |
Putative targets of the chemical constituents of YZD
Based on database searches we found that the 27 key components of YZD had the potential to interact with a total of 949 predicted cellular targets. The contribution of the individual herbs is as follows: Polygalaxanthone: 12 targets, Tenuifolin: 43 targets, Ferulaldehyde: 238 targets, Sibiricose A3: 152 targets, citric acid: 376 targets, Ginsenoside-Rg3: 370 targets, Ginsenoside-Rg1: 102 targets, Ginsenoside-Re: 103 targets, Ginsenoside-Rb1: 114 targets, Caryophyllene oxide: 285 targets, β-asarone: 339 targets, Aristolactam I: 113 targets, Ephedrine: 109 targets, Pseudoephedrine: 48 targets, Norephedrine: 88 targets, linoleic acid: 546 targets, Paeoniflorin: 77 targets, gallic acid: 434 targets, ellagic acid: 479 targets, albiflorin: 426 targets, chlorogenic acid: 424 targets, Notopterol: 362 targets, Isoimperatorin: 419 targets, Columbianetin: 265 targets, Nodakenin 327: targets, Ostruthin: 474 targets, and Atractylenolide II: 48 targets. After combining and deduplicating the list of potential interaction partners, resulted in 949 predicted targets. Detailed information of these is given in Supplementary Table S3.
Known AD-related therapeutic targets
Database searches identified a total of 231 cellular molecular targets with presumed or proven relevance to AD. Of these 31 were found in the DrugBank database, 87 in the GeneCards database, 43 in the DisGeNET database, and 25 in the OMIM database. After removing duplicates resulted in 169 AD-related targets. The details of these, including additional information, are given in Supplementary Table S4. We also looked for genes specifically up- or downregulated in AD at mRNA level. The GSE1297 and GSE36980 chip series from the GEO database were selected for this analysis. These chipsets were submitted to the GEO2R online analysis tool that identified 65 co-expressed, differentially regulated genes that are potentially relevant to AD. These mRNAs were matched to universal gene names from the Uniprot database. The results of this analysis are shown in Supplementary Table S5.
Network and pathway analysis
YZD chemical component—Disease target intersection
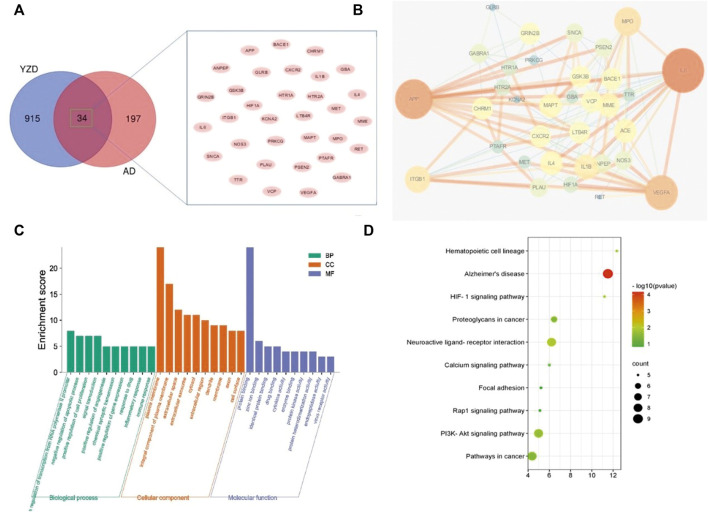
We compared the 949 know molecular targets of the active ingredients of YZD with the 231 AD-related differentially expressed mRNAs identified in Section 3.3. As shown in the Venn diagram (Figure 3A) 34 molecules were present in the overlap area. These molecules, representing targets in AD on which the chemical constituents of YZD are likely to act, are listed in Table 2.
FIGURE 3.
Analysis of proteins potentially affected by YZD treatment. (A) Venn diagram illustrating the overlap between proteins interacting with components of YZD and proteins know or predicted to be involved in the pathogenesis of AD. (B) PPI network of AD-related proteins that can interact with YZD components. (C) Analysis of the GO classification of the same set of proteins. (D) KEGG enrichment results of the interacting proteins represented as a bubble map. Red indicates greater, green a lesser enrichment of a given KEGG pathway, while the size of the bubbles corresponds to the number of proteins in each group.
TABLE 2.
The common target of drugs and diseases.
| Name | Number | Common targets |
|---|---|---|
| The common target of drugs and diseases | 34 | ACE, ANPEP, APP, BACE1, CHRM1, CXCR2, GBA, GLRB, GRIN2B, GSK3B, HIF1A, HTR1A, HTR2A, IL1B, IL4, IL6, ITGB1, KCNA2, LTB4R, MAPT, MET, MME, MPO, NOS3, PLAU, PRKCG, PSEN2, PTAFR, RET, SNCA, TTR, VCP, VEGFA, GABRA1 |
Construction of protein-protein interaction network map and selection of key targets
To investigate protein-protein interaction (PPI) relationships between the identified 34 molecules, a PPI network was built using the String platform and Cytoscape. This network included 34 nodes and 137 edges, with an average degree value of 8.06, and a PPI enrichment p value of less than 1.0e−16. Importing the interaction relationship data into the Cytoscape 3.6.0 software resulted in the PPI network map shown in Figure 3B. The calculated average shortest path length, betweenness centrality, closeness centrality, and degree of nodes in the network are shown in Table 3. According to network topology properties, the targets are sorted from high to low (degree score≥8) with larger degree values indicating the greater relevance of a node within the network. Sorting molecules accordingly identified 17 key molecules: IL6, APP, VEGFA, MPO, ITGB1, IL1B, ACE, MME, MAPT, IL4, CXCR2, GRIN2B, VCP, GSK3B, LTB4R, CHRM1, and BACE1. These proteins may represent the critical targets of YZD in the prevention and treatment of AD.
TABLE 3.
Topological parameters of the targets.
| name | Degree | Closeness centrality | Betweenness centrality |
|---|---|---|---|
| IL6 | 22 | 0.717391 | 0.174759 |
| APP | 19 | 0.702128 | 0.211938 |
| VEGFA | 16 | 0.647059 | 0.111052 |
| MPO | 12 | 0.559322 | 0.021201 |
| ITGB1 | 11 | 0.589286 | 0.064417 |
| IL1B | 10 | 0.559322 | 0.014098 |
| ACE | 9 | 0.55 | 0.008529 |
| MME | 9 | 0.55 | 0.019121 |
| MAPT | 9 | 0.568966 | 0.023683 |
| IL4 | 9 | 0.540984 | 0.013697 |
| CXCR2 | 8 | 0.559322 | 0.027057 |
| GRIN2B | 8 | 0.5 | 0.046652 |
| VCP | 8 | 0.540984 | 0.018205 |
| GSK3B | 8 | 0.568966 | 0.020185 |
| LTB4R | 8 | 0.559322 | 0.024173 |
| CHRM1 | 8 | 0.532258 | 0.031334 |
| BACE1 | 8 | 0.540984 | 0.012885 |
Functional analysis based on GO classification
To investigate further the biological process (BP), molecular function (MF), and cellular components (CC) of these key proteins, GO enrichment analysis was carried out. The top 10 GO terms (p < 0.05) were selected as significant entries based on their p value (Figure 3C). This GO category analysis indicated that the majority of hubs were involved in transcription of RNA polymerase II promoters, apoptosis, cell proliferation, and signal transduction. Classifying proteins based on molecular functions, they were involved in protein binding, zinc ion binding, and drug binding, while they were situated in various cellular locations, being associated with the plasma membrane, forming integral components of plasma membrane, or being present in the extracellular space.
KEGG pathway enrichment analysis
KEGG pathway enrichment analysis demonstrated that the 34 key targets contributed to 26 distinct pathways (p value <0.05). The top 10 of these are listed in Table 4 and Figure 3D. The most prominently represented KEGG pathways contained pathways associated with, Neuroactive ligand-receptor interaction, and the PI3K-Akt signaling pathway.
TABLE 4.
The enriched KEGG pathway and the related genes.
| Pathway | Genes | Fold enrichment | p Value |
|---|---|---|---|
| Alzheimer’s disease | BACE1, GSK3B, APP, MME, IL1B, PSEN2, MAPT, GRIN2B, SNCA | 11.52 | 0.0000 |
| Neuroactive ligand-receptor interaction | GABRA1, CHRM1, GLRB, PTAFR, HTR1A, HTR2A, GRIN2B, LTB4R | 6.21 | 0.0002 |
| PI3K-Akt signaling pathway | ITGB1, IL4, GSK3B, IL6, CHRM1, NOS3, MET, VEGFA | 4.98 | 0.0007 |
| Pathways in cancer | ITGB1, RET, PRKCG, GSK3B, IL6, HIF1A, MET, VEGFA | 4.38 | 0.0015 |
| Proteoglycans in cancer | ITGB1, PRKCG, PLAU, HIF1A, MET, VEGFA | 6.45 | 0.0018 |
| Hematopoietic cell lineage | IL4, IL6, MME, IL1B, ANPEP | 12.35 | 0.0006 |
| HIF-1 signaling pathway | PRKCG, IL6, NOS3, HIF1A, VEGFA | 11.20 | 0.0008 |
| Calcium signaling pathway | PRKCG, CHRM1, NOS3, PTAFR, HTR2A | 6.00 | 0.0081 |
| Focal adhesion | ITGB1, PRKCG, GSK3B, MET, VEGFA | 5.22 | 0.0130 |
| Rap1 signaling pathway | ITGB1, PRKCG, GRIN2B, MET, VEGFA | 5.12 | 0.0139 |
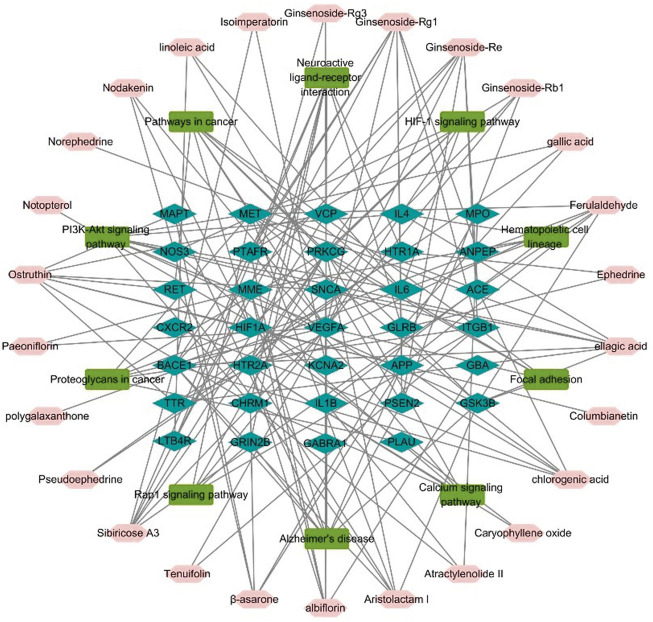
Construction of a network illustrating the roles of herb components in molecular pathways and their interacting proteins
To represent the relationship between key chemicals components of YZD, their putative molecular targets, and the molecular pathways they are involved in a herb component - putative molecular target - major pathway network map was constructed using Cytoscape 3.6.0 (Figure 4). This map illustrates clearly the relationship between the 27 key constituents of YZD, the 10 most prominent pathways identified in KEGG enrichment analysis, and the 34 AD-related proteins identified in Section 3.4. This mapping illustrated that the main pathways in the action of YZD in AD belong to KEGG pathways associated with Alzheimer’s disease, PI3K-Akt signaling, and Neuroactive ligand-receptor interactions (Table 4).
FIGURE 4.
Herb component—putative molecular target - major pathway network map. Pink nodes represent active ingredients of YZD, blue nodes represent potential targets, and the green nodes indicate the involved KEGG pathways.
Molecular docking analysis
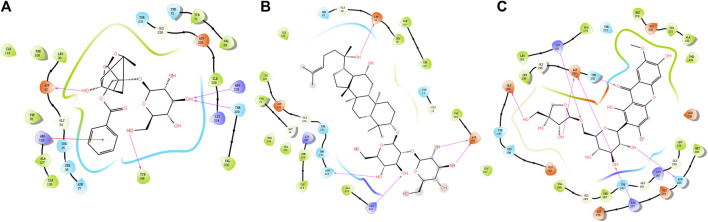
Available crystal structures of identified key molecular targets were downloaded and the Maestro platform was utilized to analyze their docking relationship with chemical components identified from YZD. We selected 8 proteins from the herb component - putative molecular target - major pathway network map for molecular docking analysis. Their interactions with the 27 key compounds of YZD were used as docking partners. In the resulting models a lower the score indicates higher binding energy, with a more stable conformation in ligand-receptor binding. This in turn increases the possibility of interaction in vitro and in vivo biological systems. The results of molecular docking (Figure 5; Table 5) and Glide energy analysis (Table 6) demonstrated strong binding between the active components of YZD and the 8 proteins selected from the herb component - putative molecular target - major pathway network map. These results provide further verification of the predicted interactions between YZD components and molecular targets. Figure 5 shows data obtained from three docking analysis models. Interactions between APP and Paeoniflorin occur through hydrogen bonds via amino acid residues ASP32, TYR198, ARG235, THR329, and a p-π conjugate is formed with Arg 128 (Figure 5A). BACE1 forms stable hydrogen bonds with Ginsenoside Rg3 via amino acid residues ASP32, ASN233, Lys321, ASP311 (Figure 5B), while there are stable hydrogen bonds between GSK3β and Polygalaxanthone via amino acid residues Glu200, ASP293, Lys181, and ASN 280 Figure 5C).
FIGURE 5.
Molecular docking models depicting interactions between YZD components and their putative molecular targets/receptors. (A) APP (Beta amyloid A4 protein Beta, PDB code: 6UWP) interacting with Paeoniflorin; (B) BACE (Beta-secretase 1, PDB code: 5CLM) interacting with Ginsenoside Rg3; (C) GSK3B (Glycogen synthase kinase-3 beta, PDB code: 1O6L) with Polygalaxanthone. Structures drawn entirely in black represent YZD components, the colored dots are amino acid residues from the interacting protein. Pink arrows represent hydrogen bonds while red lines indicate a P-π conjugate.
TABLE 5.
The docking information of 8 targets with 27 compounds of YZD.
| IL6 | APP | VEGFA | MPO | IL1B | ACE | GSK3B | BACE1 | |
|---|---|---|---|---|---|---|---|---|
| Citric acid | −5.9 | −3.1 | −2.6 | −6.6 | −4.4 | −3.9 | −4.2 | −4.7 |
| Gallic acid | −6.5 | −5.3 | −4.1 | −5.5 | −5.4 | −6.3 | −5.1 | −5.1 |
| Pseudoephedrine | −5.3 | −6.6 | −4.7 | −4.1 | −5.1 | −6.6 | −5.4 | −5.6 |
| Ephedrine | −5.4 | −6.4 | −4.3 | −4.0 | −5.3 | −7.9 | −5.7 | −5.6 |
| Ferulaldehyde | — | −4.5 | −3.2 | — | −5.0 | — | −5.8 | −4.9 |
| Norephedrine | −6.2 | −6.7 | −5.1 | −4.7 | −5.6 | −8.0 | −6.5 | −6.4 |
| Isoimperatorin | −3.7 | −4.5 | −3.0 | −3.6 | −4.6 | −6.7 | −5.1 | −4.4 |
| Nodakenin | −5.5 | −5.4 | −3.7 | −3.2 | −4.2 | −7.2 | −5.6 | −5.9 |
| Columbianetin | −5.2 | −8.4 | −6.2 | −4.4 | ||||
| Aristolactam I | −4.0 | −4.4 | −2.7 | −4.3 | −4.6 | −7.5 | −4.4 | −4.8 |
| Ginsenoside-Re | — | — | — | — | −3.5 | — | — | −5.9 |
| Ginsenoside-Rg1 | — | — | — | — | −5.9 | — | −5.6 | −4.7 |
| Paeoniflorin | −2.3 | −4.7 | −2.1 | −2.7 | −5.1 | −6.4 | −5.0 | −5.1 |
| Caryophyllene oxide | −4.6 | −3.7 | −4.4 | −6.0 | −4.3 | −4.2 | ||
| Chlorogenic acid | −5.5 | −5.4 | −4.6 | −5.4 | −4.9 | −5.9 | −7.4 | −4.3 |
| Linoleic acid | −3.0 | −0.7 | 0.9 | −1.5 | 0.7 | −2.9 | −0.5 | −1.1 |
| Ostruthin | −4.5 | −5.6 | −2.9 | −3.2 | −4.1 | −6.9 | −5.6 | −6.0 |
| β-asarone | −3.9 | −4.4 | −3.1 | −3.5 | −4.5 | −5.4 | −4.6 | −3.2 |
| albiflorin | −4.1 | −5.3 | −2.7 | −4.8 | −5.2 | −8.5 | −5.2 | −5.4 |
| Notopterol | −3.7 | −5.1 | — | −3.9 | −5.3 | −6.4 | −5.4 | −5.1 |
| Ginsenoside-Rg3 | −4.1 | — | — | — | −4.1 | −8.4 | −5.0 | |
| Polygalaxanthone | −6.5 | −1.0 | −4.2 | −5.1 | −7.9 | −7.3 | −4.6 | |
| Atractylenolide II | −3.1 | −5.3 | — | −4.1 | −4.6 | −6.6 | −6.1 | −4.0 |
| Ellagic acid | −2.6 | −3.6 | −0.9 | −2.5 | −3.5 | −6.4 | −3.8 | −2.8 |
| Ginsenoside-Rb1 | — | — | — | — | −3.6 | — | −4.8 | |
| Tenuifoli | — | — | — | — | −3.6 | — | −3.1 | −4.4 |
| Sibiricose A3 | −2.5 | −6.8 | −4.3 | −4.4 | −6.5 | −8.7 | −7.8 | −5.5 |
TABLE 6.
The Glide energy information of 8 targets with 27 compounds of YZD.
| IL6 | APP | VEGFA | MPO | IL1B | ACE | GSK3B | BACE1 | |
|---|---|---|---|---|---|---|---|---|
| Citric acid | −22.0 | −12.7 | −9.3 | −29.7 | −22.2 | −22.3 | −23.4 | −21.0 |
| Gallic acid | −21.4 | −22.5 | −17.4 | −25.3 | −23.8 | −18.1 | −27.0 | −23.0 |
| Pseudoephedrine | −20.8 | −30.0 | −18.8 | −20.9 | −24.2 | −29.7 | −27.3 | −25.9 |
| Ephedrine | −21.5 | −28.4 | −18.5 | −19.1 | −22.7 | −30.7 | −27.8 | −27.4 |
| Ferulaldehyde | −25.3 | −26.9 | −18.0 | −25.0 | −26.2 | −32.3 | −33.0 | −28.3 |
| Norephedrine | −22.3 | −25.5 | −20.7 | −21.7 | −24.6 | −28.4 | −24.9 | −24.3 |
| Isoimperatorin | −22.4 | −29.8 | −21.1 | −23.8 | −28.7 | −36.3 | −35.4 | −30.0 |
| Nodakenin | −37.7 | −46.4 | −27.5 | −31.6 | −38.9 | −46.2 | −52.4 | −48.8 |
| Columbianetin | — | — | — | — | −27.2 | −31.0 | −37.6 | −31.6 |
| Aristolactam I | −22.3 | −32.5 | −16.9 | −25.2 | −28.7 | −35.6 | −34.1 | −32.1 |
| Ginsenoside-Re | — | — | — | — | −36.2 | — | — | −56.1 |
| Ginsenoside-Rg1 | — | — | — | — | −51.9 | −43.2 | −57.2 | −49.2 |
| Paeoniflorin | −24.4 | −44.7 | −26.5 | −35.7 | −40.8 | −35.5 | −54.4 | −46.2 |
| Caryophyllene oxide | — | −25.9 | — | −22.4 | −23.3 | −27.5 | −27.3 | −25.0 |
| Chlorogenic acid | −33.0 | −42.7 | −34.3 | −43.1 | −39.3 | −40.0 | −48.8 | −40.2 |
| Linoleic acid | −26.7 | −33.2 | −21.4 | −27.3 | −25.1 | −38.6 | −33.3 | −35.7 |
| Ostruthin | −26.5 | −37.2 | −22.3 | −28.3 | −32.1 | −42.1 | −41.4 | −38.9 |
| β-asarone | −18.9 | −26.4 | −14.9 | −22.0 | −22.8 | −27.2 | −27.3 | −22.9 |
| Albiflorin | −23.1 | −36.5 | −15.1 | −26.9 | −31.6 | −24.5 | −39.0 | −31.9 |
| Notopterol | −28.5 | −39.9 | — | −31.5 | −36.9 | −42.2 | −42.4 | −40.1 |
| Ginsenoside-Rg3 | −36.4 | — | — | — | −39.4 | −48.0 | — | −51.5 |
| Polygalaxanthone | — | −66.9 | −25.8 | −44.9 | −3.9 | −53.2 | −74.2 | −55.3 |
| Atractylenolide II | −12.8 | −26.2 | −21.9 | −21.6 | −27.2 | −27.9 | −22.1 | |
| Elagic acid | −24.6 | −40.1 | −17.3 | −29.7 | −34.2 | −40.9 | −42.2 | −38.1 |
| Ginsenoside-Rb1 | — | — | — | — | −38.3 | — | — | −50.2 |
| Tenuifoli | — | — | — | — | −34.0 | −35.2 | −24.7 | −44.1 |
| Sibiricose A3 | −29.2 | −58.9 | −34.2 | −40.5 | −52.0 | −52.7 | −60.3 | −50.7 |
Studies in biological systems
YZD administration has beneficial effects on learning and memory in APP/PS1 mice
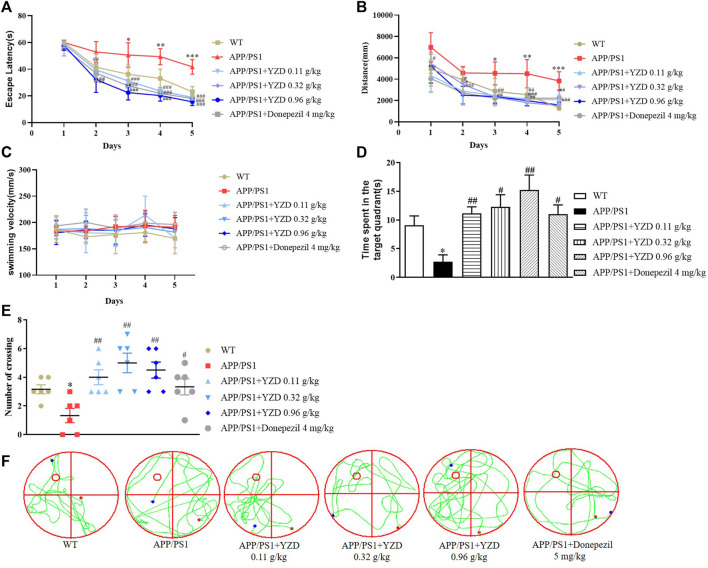
The effects of YZD on spatial learning and memory deficits in APP/PS1 transgenic mouse model were investigated in MWM trials. While there was no difference in swimming velocity (Figure 6A), APP/PS1 mice showed a significantly longer latency finding the hidden platform compared to their WT littermates. However, the administration of YZD at different doses to the transgenic animals significantly shortened the latency period (Figure 6B). Swimming distance was also reduced in the YZD treated groups (Figure 6C). Probe trials were performed to assess memory retention after the last training session. Compared to the WT group APP/PS1 mice showed a smaller number of intersections in the target quadrant and spent less time at the target area previously containing the escape platform (Figures 6D,E). YZD-treated APP/PS1 mice performed better, spending more time at the target area (Figure 6D). The number of crossovers in the YZD treated APP/PS1 mice was also greater than that of untreated controls (Figure 6E). Figure 6F shows typical swimming trajectory maps of mice passing over the platform during the test period. Taken together, these data suggest that YZD administration provided beneficial effects on learning and memory in APP/PS1 mice.
FIGURE 6.
YZD administration ameliorated cognitive impairment in APP/PS1 mice. WT: wild-type mice; APP/PS1: APP/PS1 mice. (A) Swimming speed of mice in the Morris water maze. (B) Changes of escape latency of mice from the first day to the fifth day in different groups. (C) Changes in swimming distance from the first day to the fifth day in different groups. (D) The time that the indicated groups of mice spent at the target area that previously contained an escape platform in the Morris water maze. (E) The times of indicated groups of mice crossing the target area that previously contained an escape platform in the Morris water maze experiment. (F) Typical swimming trajectory maps of mice passing through the platform during the test period. *p < 0.05 compared to the WT group, #p < 0.05, ##p < 0.01 compared to the APP/PS1 group. All data are presented as mean ± SE (n = 6).
Western blot analysis
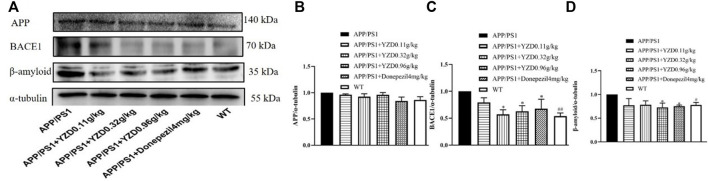
The hippocampi of animals from the MWM test were analyzed for the abundance of several AD-related proteins using Western blot analysis. As shown in Figure 7, while the abundance of APP appear different this change does not reach statistical significance (Figure 7A). However, the abundance of BACE1 (Figure 7B) and beta-amyloid (Figure 7C) expression were markedly down-regulated in APP/PS1 mice (p < 0.05). As expected, YZD and Donazepil (DON) treatment significantly decreased BACE1 expression (Figure 7B) and beta-amyloid (Figure 7C) levels compared to the untreated APP/PS1 group (p < 0.05). These results suggest that the therapeutic effect of YZD in AD may be attributed to the inhibition of BACE1 expression and the reduced production of beta-amyloid.
FIGURE 7.
YZD treatment for 3 months reduces BACE1 and beta-amyloid protein abundance in the hippocampi of APP/PS1 transgenic mice. (A) Western blot analysis of APP, BACE1, beta-amyloid levels. Densitometry data of APP (B), BACE1 (C) and beta-amyloid (D). Values represented as the mean ± SE (n = 3), *p < 0.05, **p < 0.01, #p < 0.05 was considered statistically significant. Distinct symbols indicate differences in the level of significance between groups.
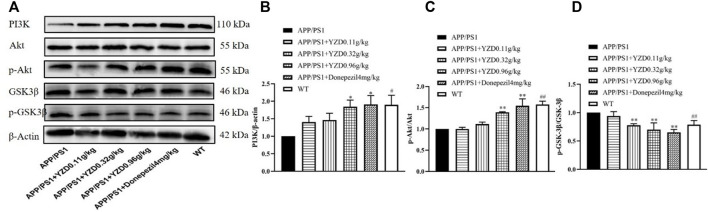
The PI3K/AKT/GSK-3β pathway correlates closely with the progression of AD. Therefore, we explored whether YZD had any effect on the expression of these proteins. Figure 8 shows a representative Western blot of PI3K, AKT, pAKT, GSK-3β and pGSK-3β levels. When compared to the WT group, the levels of PI3K (Figure 8A and p-AKT/AKT (Figure 8B) was significantly reduced in the hippocampi of untreated transgenic animals, while the levels of p-GSK-3β/GSK-3β increased (Figure 8C), with all observed differences reaching statistical significance (at least p < 0.05). Compared to untreated transgenic animals, the levels of PI3K (Figure 8A) and p-AKT/AKT PI3K (Figure 8B) increased in the groups receiving high doses YZD. Simultaneously, the levels of p-GSK-3β/GSK-3βPI3K (Figure 8C) decreased in the medium and high dose YZD treatment groups. The above results suggest that the therapeutic effect of YZD on AD may be related to the PI3K/AKT/GSK-3β signaling.
FIGURE 8.
Effects of 3-months of YZD treatment on the expression of PI3K/AKT/GSK-3β pathway related proteins in the hippocampi of APP/PS1 mice. (A) Western blot analysis of PI3K, AKT, p-AKT, GSK-3β, p-GSK-3β. (B) Densitometry of PI3K, (C) Densitometry of p-AKT/AKT, (D) Densitometry of p-GSK-3β/GSK-3β. Values show the mean ± SE (n = 3), differences in the level of significance are: *p < 0.05, **p < 0.01, ##p < 0.01.
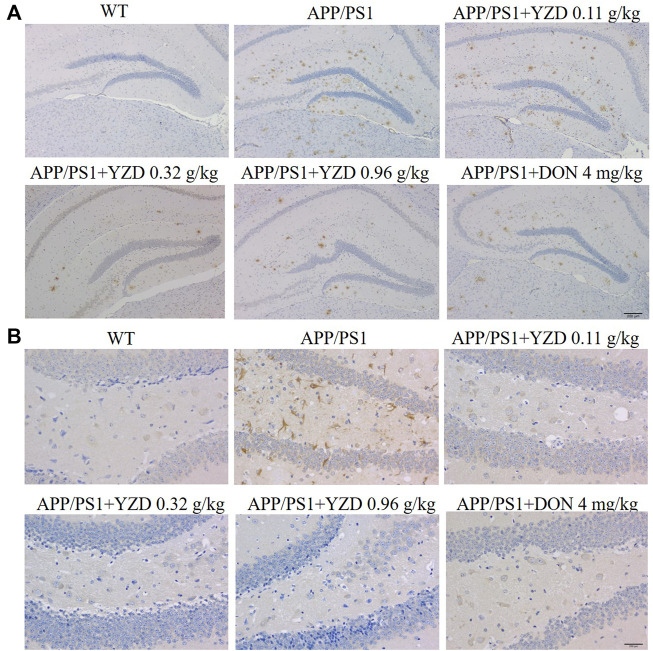
YZD treatment inhibited βAmyloid deposition and NFT in APP/PS1 transgenic mice
β Amyloid (Aβ) accumulation is a pathological feature of AD, inducing neuronal amyloidosis and even apoptosis. Therefore, we studied the effect of YZD on Aβ accumulation in the brain of APP/PS1 mice by immunohistochemistry. The resulting images shown numerous brown plaques. Tau protein, the main protein constituent of microtubule structure, maintains the stability of the microtubular system. In the pathogenesis of AD, microtubules depolymerize, resulting in the production of NFTs and the acceleration of the aging of neuronal fibers. Our results showed that the deposition of Aβ plaques was significantly decreased in the hippocampi of YZD-treated APP/PS1 transgenic mice (Figure 9). A similar reduction was seen in the deposition of Tau in the studied hippocampi.
FIGURE 9.
YZD inhibited Aβ and Tau deposition in the hippocampi of APP/PS1 mice. (A) Representative images of Aβ deposits (showing brown staining) in the hippocampi. Magnification: ×25, scale bar: 200 μm. (B) Images of Tau deposits in the hippocampi (appearing as brown deposits). Magnification: 400 × scale bar: 200 μm.
Discussion
AD is a complex disease affecting multiple biochemical pathways. Given its unclear etiology and pathogenesis, there is an increasing interest in the use of traditional medical formulations in the treatment of the condition (Poornima et al., 2016). TCM generally uses natural products, with a good safety record and low toxicity demonstrated during extensive periods of clinical use. TCM also relies on multi-compound formulations affecting several targets and multi-component pathways. Ultimately, this approach may have significant advantages over single compound medications in the treatment of complex, multifactorial diseases (Poornima et al., 2016).
To the best of our knowledge, this is the first study to systematically investigate the mechanism of action of YZD in combatting AD using network pharmacology analysis. The main chemical constituents of YZD were identified using liquid chromatography coupled to mass spectroscopy. This was followed by network pharmacology studies to assess potential interactions between the detected chemical components and putative AD-related protein targets identified in database searches. With this approach, both the protein targets and candidate active constituents of YZD were predicted in silico. By comparing the list of all potential target proteins affected by the constituents of YZD with a comprehensive list of proteins relevant to the development of AD we identified those proteins YZD is likely to act to deliver its beneficial effects in AD. Next, pathway enrichment analysis was performed to investigate the molecular pathways affected by the 27 components of YZD that were predicted act in AD. Following this computational phase of the work, the APP1/PS1 transgenic mouse model of AD was used to assess the in vivo efficacy of YZD in improving learning and memory deficits. These experiments demonstrated that YZD administration had a positive impact on spatial learning and memory, evidenced by reducing the time needed to reach a hidden platform during MWM trials. Finally, some of the network pharmacology predictions were validated by Western blot analysis and immunohistochemistry, confirming that several AD-related pathways predicted in silico indeed played a critical role in a live animal model.
In this study, data from UHPLC-Q Exactive HF mass spectrometry was complemented with TCM database searches to maximize the number of identified chemical constituents of YZD (Chu et al., 2016). In total 211 compounds were identified experimentally, including high levels of Prenol lipids, Phenol ethers, Benzopyrans, Benzene and substituted derivatives. Searches conducted in TCM-relevant databases found 1026 chemical compounds (Supplementary Table S2). Out of these two lists, a total of 27 compounds were identified that were uniquely present in YZD. Four of these were derived from the herb YuanZhi, 5 from RenShen, 2 from ShiChangPu, 6 from QiangHuo, 4 from MaHuang, 3 from XiXin, 4 from ChiShaoYao, and a single compound from BaiZhu, with 3 ingredients being present in more than one herb.
Oligosaccharide esters are a unique class of sugar esters in Polygonum consisting of a sucrose core and connect monosaccharides, mainly glucose and rhamnose, linked with different glycosidic bonds (Zhao et al., 2020). This raises the possibility that oligosaccharide esters may be biologically active substances in Polygonum japonica that could be utilized in the development of new anti-aging and neuroprotective drugs. While oligosaccharide esters exist in a variety of plants, a sugar core containing three or more monosaccharides can only be found in Polygala. It has been suggested that these unique oligosaccharide esters may have unusual pharmacologic effects (Li, 2008). Studies have shown that tenuifoliside A, tenuifoliside C (E), tenuifoliose B (D), tenuifoliside K, and tenuifoliside I (J) are reasonably well absorbed and enter the systemic circulation, exerting biological effects (Wang et al., 2017).
Some of the identified YZD components were previously shown to be beneficial in AD. Tenuifolin attenuates Amyloid-β42-induced neuroinflammation (Wei, 2016; Wang L. et al., 2019; Chen and Jia, 2020). This mechanism was protective against neuronal apoptosis and improved learning and memory deficits in a mouse AD model (Wang N. et al., 2019). Ginsenoside Rg1 significantly improved cognitive behavioral impairments in several animal models of AD (Nie et al., 2017; Liang et al., 2021), while ginsenoside Rg2 had a protective effect against memory impairments and neuronal death seen in Aβ25-35 transgenic rats (Cui J. et al., 2017; Cui et al., 2021). Ginsenoside Rg3 prevented cognitive impairment in a rat AD model (Zhang et al., 2019) and Notopterol effectively ameliorated cognitive deficits in APP/PS1 mice by attenuating pathology caused by the accumulation of amyloid-β and tau (Jiang X. et al., 2020; Jiang X. W. et al., 2020). Other studies have shown that Paeoniflorin exerted neuroprotective effects in a transgenic mouse model of AD (Zhang et al., 2015; Gu et al., 2016; Ma et al., 2018; Huang et al., 2020; Kong et al., 2020). Albiflorin ameliorated memory deficits in APP/PS1 transgenic mice (Xu et al., 2019) and Atractylodes macrocephala Koidz. was also used to treat Alzheimer’s disease (Zhu et al., 2018). β-Asarone, a crucial compound of YZD, was reported to prevent amyloid β-induced apoptosis by regulating the PI3K/AKT pathway in PC-12 cells (Li Z. et al., 2021; Meng et al., 2021). β-Asarone could also inhibit Amyloid deposition and reverse cognitive deficits in experimental animals (Wang et al., 2021). Nodakenin exhibited potent inhibitory activities against the β-site amyloid precursor protein cleaving enzyme 1 (BACE1) and is regarded as a promising therapeutic option in the treatment of AD (Yousof Ali et al., 2015).
Network pharmacology analysis is able to identify interactions between chemical constituents and their targets/pathways, and establish “compound-target-pathway” maps identifying chemical constituents and the mechanisms associated with their pharmacological activity (Ren et al., 2018). Our work is the first to explore the possible actions of YZD in AD using network pharmacology, laying the foundation for the use of this approach in further research. The potential pharmacological targets of the 27 YZD-derived compounds were identified with the help of the SEA, STITCH, Swiss Target Prediction database searches and PharmMapper. At the same time, protein molecules associated with the pathogenesis of AD were identified from the Drugbank, OMIM, GeneCards, and DisGeNET databases and by the analysis of differentially expressed genes in GEO datasets. This work identified 34 target molecules in AD that could be affected by ingredients in YZD.
After network topology analysis 17 of these genes, including IL6, APP, VEGFA, MPO, ITGB1, IL1B, ACE, MME, MAPT, IL4, CXCR2, GRIN2B, VCP, GSK3B, LTB4R, CHRM1, and BACE1 were considered to be the key molecules targeted by the chemical constituents of YZD. These proteins show complex interactions with each other, forming a PPI network. In such networks a change affecting a single molecule can result in a series of functional abnormalities, potentially leading to the emergence of pathologic abnormalities (Gu et al., 2020). Our results showed that APP, IL6, VEGFA, GSK3B and BACE1 could extensively interact with other members of the network. For example, APP interacts with BACE1, GSK3B, IL6, and PSEN2. APP is a widely expressed transmembrane protein playing a key role in the formation of Aβ in the pathogenesis of AD (Zhu et al., 2017). BACE1 is the main rate-limiting enzyme in the production of Aβ. Previous studies experimentally validated BACE1 as a YZD target in the treatment of AD (Ahmad et al., 2017; Prati et al., 2017). Furthermore, there is emerging evidence that BACE1 inhibitors may be potentially interesting tools in the management of AD (Rueeger et al., 2021), while the inhibition of GSK3B can reduce the phosphorylation of Tau and the conversion of APP to Aβ. These observations provide strong evidence that several components of YZD have significant association with various AD-related signaling pathways. Our molecular docking models investigating the interactions between validated targets (i.e., APP, BACE1 and GSK3B) and the 27 identified key chemical compounds found in YZD provides additional support for a role of YZD in the treatment of AD.
KEGG pathway enrichment analysis showed that the identified molecules preferentially affected the transcription of RNA polymerase II promoters, the process of apoptosis, cell proliferation, signal transduction, synaptic transmission, neuroactive ligand-receptor interactions, and the PI3K-Akt, HIF-1, and calcium signaling pathways. The PI3K/AKT signaling pathway is involved in regulating cell proliferation, differentiation, metabolism, and apoptosis. It also exerts anti-oxidative, anti-inflammatory, and neuroprotective effects. Altered regulation of this pathway has been found to play a central role in the development of AD (Cui W. et al., 2017; Razani et al., 2021). PI3K activation can act as a second messenger to transmit signals into cells, activating AKT phosphorylation. In turn, AKT could exert its anti-apoptotic effects by promoting the phosphorylation of downstream substrates, such as GSK-3β and mTOR. However, it was found that the activation of GSK-3β could lead to an abnormal increase in the level of phosphorylation of tau protein (Jimenez et al., 2011). Gene ontology analysis has also demonstrated that the molecular targets of YZD show a varied distribution within cells.
We also conducted behavioral tests to assess the efficacy of YZD in treating learning and memory deficits in aged APP/PS1 transgenic mice. These mice exhibit impairments in spatial learning and memory, as evidenced by their poor performance in MWM tests. Our results showed that YZD treatment restored learning and memory deficits in these animals, demonstrating a protective effect of this formulation in APP/PS1 transgenic AD mice.
The PI3K/AKT/GSK-3β signaling pathway plays a crucial role in cell growth, survival, apoptosis, and proliferation, and is also one of the important pathways that lead to the phosphorylation of tau protein and then the development of AD. (Kitagishi et al., 2014) Activation of PI3K will promote the phosphorylation of Thr308 within AKT, activating the protein. Thus, the activity of PI3K is reflected by the phosphorylation status of AKT. Activated AKT inhibits the action of GSK-3β downstream in this signaling pathway. GSK-3β is normally in the activated state in the cytoplasm of neurons. The activation of AKT results in the phosphorylation of the Ser9 residue of GSK-3β, inhibiting the activity of GSK-3β. GSK-3β is an important threonine/serine protein kinase, which can promote the phosphorylation of tau protein at multiple sites. (Kitagishi et al., 2014).
Our Western blot experiments indicated that treatment with YZD promoted the expression of PI3K and p-Akt, and reduced the levels of p-GSK3β, proving that YZD can regulate the activated PI3K/AKT/GSK3β pathway. The expression of BACE1 and β-amyloid were also down-regulated significantly, providing experimental confirmation of predictions reached by using network pharmacology and molecular docking analyses. Based on these findings, we propose that YZD may protect against the progression of AD by inhibiting the expression of BACE1, thereby reducing the production of beta-amyloid. The other main action of the formulation is via the regulation of the PI3K/AKT/GSK-3β pathway.
Although this work shed light on some of the therapeutic mechanisms of YZD in AD, the results need to be interpreted with caution. Additional pharmacologic experiments will be necessary to validate further mechanisms of YZD predicted to occur in our network analysis. Second, network pharmacology fails to identify dose-effect relationships of multi-component formulations. These issues will be addressed in subsequent studies. Finally, further work is needed to study global metabolic changes in other species to fully elucidate the mechanisms by which YZD improves AD. Enhanced metabolomics technologies, such as targeted metabolomics, are required to detect metabolite biomarkers (Wei et al., 2020).
In summary, multiple compounds in YZD show promise in the potential treatment of AD (Figure 10). Our experiments showed that YZD could reduce the expression of BACE1 and the production of beta-amyloid and regulate the PI3K/AKT/GSK-3β pathway. These findings are consistent with the results of our network pharmacology and molecular docking-based predictions. Thus, we propose that YZD is likely to affect a multitude of molecular mechanisms in the treatment of AD.
FIGURE 10.
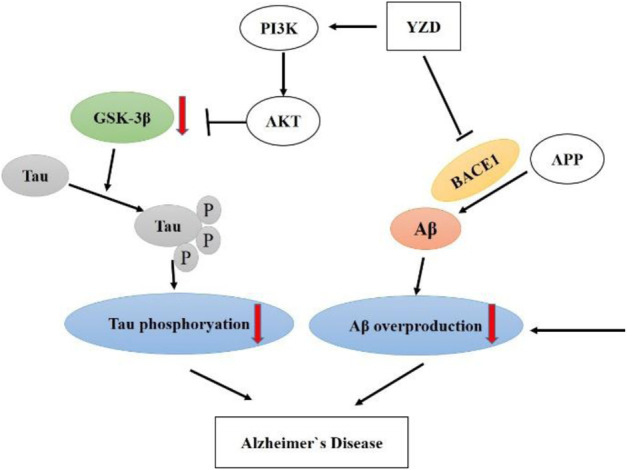
A schematic of proposed neuroprotective mechanism of YZD in AD.
Conclusion
Herbal medicines have provided valuable tools in preventing and treating AD for centuries. In this study, we have identified 27 chemical constituents of YZD. Using a network pharmacology approach combined with molecular docking analyses identified some of the mechanisms involved in the beneficial effects of YZD treatment in AD. These involve effects on neuroactive ligand-receptor interaction pathways, the PI3K/AKT signaling pathway, as well as HIF-1 and calcium signaling. The results showed that YZD has potential benefits on learning and memory deficits in APP/PS1 transgenic AD mice, and regulated the apoptotic process and signal transduction. The key proteins of related pathways were quantitatively analyzed using Western blots indicating that the extract of YZD could inhibit or activate key protein pathways. The insights provided by this preliminary work will pave the way for further studies into the potential mechanisms by which YZD acts as a therapeutic agent in AD.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal experiments were permitted by the Aminal Ethics Committee of Northern Theater General Hospital.
Author contributions
ZZ, Q-CZ, and QW contributed to project design. QW wrote the paper; XL, X-WJ, and Z-HX supervised the study; DY, L-JZ, NW, and QW developed major methodologies, databases, reagents, and primary experiments; QW and XL analyzed different aspects of the results. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (Grant Nos. 81973209 and 82173716).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.893244/full#supplementary-material
References
- Ahmad A., Ali T., Park H. Y., Badshah H., Rehman S. U., Kim M. O. (2017). Neuroprotective effect of fisetin against amyloid-beta-induced cognitive/synaptic dysfunction, neuroinflammation, and neurodegeneration in adult mice. Mol. Neurobiol. 54, 2269–2285. 10.1007/s12035-016-9795-4 PubMed Abstract | 10.1007/s12035-016-9795-4 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Cai F. F., Zhou W. J., Wu R., Su S. B. (2018a). Systems biology approaches in the study of Chinese herbal formulae. Chin. Med. 13, 65. 10.1186/s13020-018-0221-x PubMed Abstract | 10.1186/s13020-018-0221-x | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Luo Y., Yan X., Ding P., Huang Y., Fang S., et al. (2018b). The mechanisms of bushen-yizhi formula as a therapeutic agent against Alzheimer's disease. Sci. Rep. 8, 3104. 10.1038/s41598-018-21468-w PubMed Abstract | 10.1038/s41598-018-21468-w | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W., Teng J. (2018). Combined application of tenuigenin and beta-asarone improved the efficacy of memantine in treating moderate-to-severe Alzheimer's disease. Drug Des. devel. Ther. 12, 455–462. 10.2147/DDDT.S155567 PubMed Abstract | 10.2147/DDDT.S155567 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Du J., Dai Q., Zhang H., Pang W., Hu J. J. E. J. O. M. C. (2014). Prediction of anti-tumor chemical probes of a traditional Chinese medicine formula by HPLC fingerprinting combined with molecular docking. Eur. J. Med. Chem. 83, 294–306. 10.1016/j.ejmech.2014.06.037 PubMed Abstract | 10.1016/j.ejmech.2014.06.037 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Chen S., Jia J. (2020). Tenuifolin attenuates amyloid-β42-induced neuroinflammation in microglia through the NF-κB signaling pathway. J. Alzheimers Dis. 76, 195–205. 10.3233/JAD-200077 PubMed Abstract | 10.3233/JAD-200077 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Chu H., Tang Q., Huang H., Hao W., Wei X. (2016). Grape-seed proanthocyanidins inhibit the lipopolysaccharide-induced inflammatory mediator expression in RAW264.7 macrophages by suppressing MAPK and NF-κb signal pathways. Environ. Toxicol. Pharmacol. 41, 159–166. 10.1016/j.etap.2015.11.018 PubMed Abstract | 10.1016/j.etap.2015.11.018 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Cui J., Shan R., Cao Y., Zhou Y., Liu C., Fan Y. (2021). Protective effects of ginsenoside Rg2 against memory impairment and neuronal death induced by Aβ25-35 in rats. J. Ethnopharmacol. 266, 113466. 10.1016/j.jep.2020.113466 PubMed Abstract | 10.1016/j.jep.2020.113466 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Cui J., Wang J., Zheng M., Gou D., Liu C., Zhou Y. (2017a). Ginsenoside Rg2 protects PC12 cells against beta-amyloid25-35-induced apoptosis via the phosphoinositide 3-kinase/Akt pathway. Chem. Biol. Interact. 275, 152–161. 10.1016/j.cbi.2017.07.021 PubMed Abstract | 10.1016/j.cbi.2017.07.021 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Cui W., Wang S., Wang Z., Wang Z., Sun C., Zhang Y. (2017b). Inhibition of PTEN attenuates endoplasmic reticulum stress and apoptosis via activation of PI3K/AKT pathway in Alzheimer's disease. Neurochem. Res. 42, 3052–3060. 10.1007/s11064-017-2338-1 PubMed Abstract | 10.1007/s11064-017-2338-1 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Deng M., Huang L., Zhong X. (2020). β-asarone modulates Beclin-1, LC3 and p62 expression to attenuate Aβ40 and Aβ42 levels in APP/PS1 transgenic mice with Alzheimer's disease. Mol. Med. Rep. 21, 2095–2102. 10.3892/mmr.2020.11026 PubMed Abstract | 10.3892/mmr.2020.11026 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G., Sherman B., Hosack D., Yang J., Gao W., Lane H., et al. (2003). David: Database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3. 10.1186/gb-2003-4-5-p3 PubMed Abstract | 10.1186/gb-2003-4-5-p3 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Diwu Y., Tian J., Shi J. (2013). Effect of Xixin decoction on phosphorylation toxicity at specific sites of tau protein in brains of rats with sporadic Alzheimer disease. J. Traditional Chin. Med. 33, 787–793. 10.1016/S0254-6272(14)60013-3 PubMed Abstract | 10.1016/S0254-6272(14)60013-3 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Fang J., Wang L., Wu T., Yang C., Gao L., Cai H., et al. (2017). Network pharmacology-based study on the mechanism of action for herbal medicines in Alzheimer treatment. J. Ethnopharmacol. 196, 281–292. 10.1016/j.jep.2016.11.034 PubMed Abstract | 10.1016/j.jep.2016.11.034 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Gong Y., Liu Y., Zhou L., Di X., Li W., Li Q., et al. (2015). A UHPLC–TOF/MS method based metabonomic study of total ginsenosides effects on Alzheimer disease mouse model. J. Pharm. Biomed. Anal. 115, 174–182. 10.1016/j.jpba.2015.07.007 PubMed Abstract | 10.1016/j.jpba.2015.07.007 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Gu L., Lu J., Li Q., Wu N., Zhang L., Li H., et al. (2020). A network-based analysis of key pharmacological pathways of Andrographis paniculata acting on Alzheimer's disease and experimental validation. J. Ethnopharmacol. 251, 112488. 10.1016/j.jep.2019.112488 PubMed Abstract | 10.1016/j.jep.2019.112488 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Gu X., Cai Z., Cai M., Liu K., Liu D., Zhang Q., et al. (2016). Protective effect of paeoniflorin on inflammation and apoptosis in the cerebral cortex of a transgenic mouse model of Alzheimer's disease. Mol. Med. Rep. 13, 2247–2252. 10.3892/mmr.2016.4805 PubMed Abstract | 10.3892/mmr.2016.4805 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Hopkins A. J. N. B. (2007). Network pharmacology. Nat. Biotechnol. 25, 1110–1111. 10.1038/nbt1007-1110 PubMed Abstract | 10.1038/nbt1007-1110 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Howes M. R., Fang R., Houghton P. J. (2017). Effect of Chinese herbal medicine on Alzheimer's disease. Int. Rev. Neurobiol. 135, 29–56. 10.1016/bs.irn.2017.02.003 PubMed Abstract | 10.1016/bs.irn.2017.02.003 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Huang J., Wang X., Xie L., Wu M., Zhao W., Zhang Y. B., et al. (2020). Extract of Danggui-Shaoyao-San ameliorates cognition deficits by regulating DHA metabolism in APP/PS1 mice. J. Ethnopharmacol. 253, 112673. 10.1016/j.jep.2020.112673 PubMed Abstract | 10.1016/j.jep.2020.112673 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Huang P., Ke H., Qiu Y., Cai M., Qu J., Leng A. (2019). Evidence-based complementary alternative medicine, 2019, 2675287. 10.1155/2019/2675287 Systematically characterizing chemical profile and potential mechanisms of qingre lidan decoction acting on cholelithiasis by integrating UHPLC-QTOF-MS and network target analysis eCAM PubMed Abstract | 10.1155/2019/2675287 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Lu H., Li J., Liu W., Wu Q., Xu Z., et al. (2020a). A natural BACE1 and GSK3β dual inhibitor Notopterol effectively ameliorates the cognitive deficits in APP/PS1 Alzheimer's mice by attenuating amyloid-β and tau pathology. Clin. Transl. Med. 10, e50. 10.1002/ctm2.50 PubMed Abstract | 10.1002/ctm2.50 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X. W., Liu W. W., Wu Y. T., Wu Q., Lu H. Y., Xu Z. H., et al. (2020b). Notopterygium incisum extract (NRE) rescues cognitive deficits in APP/PS1 Alzhneimer's disease mice by attenuating amyloid-beta, tau, and neuroinflammation pathology. J. Ethnopharmacol. 249, 112433. 10.1016/j.jep.2019.112433 PubMed Abstract | 10.1016/j.jep.2019.112433 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Jimenez S., Torres M., Vizuete M., Sanchez-Varo R., Sanchez-Mejias E., Trujillo-Estrada L., et al. (2011). Age-dependent accumulation of soluble amyloid beta (Abeta) oligomers reverses the neuroprotective effect of soluble amyloid precursor protein-alpha (sAPP(alpha)) by modulating phosphatidylinositol 3-kinase (PI3K)/Akt-GSK-3beta pathway in Alzheimer mouse model. J. Biol. Chem. 286, 18414–18425. 10.1074/jbc.M110.209718 PubMed Abstract | 10.1074/jbc.M110.209718 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagishi Y., Nakanishi A., Ogura Y., Matsuda S. (2014). Dietary regulation of PI3K-AKT-GSK-3β pathway in Alzheimer’s disease. Alzheimer's Res. Ther., 6 (3), 35. 10.1186/alzrt2650 PubMed Abstract | 10.1186/alzrt2650 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y., Peng Q., Lv N., Yuan J., Deng Z., Liang X., et al. (2020). Paeoniflorin exerts neuroprotective effects in a transgenic mouse model of Alzheimer's disease via activation of adenosine A1 receptor. Neurosci. Lett. 730, 135016. 10.1016/j.neulet.2020.135016 PubMed Abstract | 10.1016/j.neulet.2020.135016 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Kumar A., Singh A., Ekavali (2015). A review on Alzheimer's disease pathophysiology and its management: An update. Pharmacol. Rep. 67, 195–203. 10.1016/j.pharep.2014.09.004 PubMed Abstract | 10.1016/j.pharep.2014.09.004 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Lee A. Y., Park W., Kang T. W., Cha M. H., Chun J. M. (2018). Network pharmacology-based prediction of active compounds and molecular targets in Yijin-Tang acting on hyperlipidaemia and atherosclerosis. J. Ethnopharmacol. 221, 151–159. 10.1016/j.jep.2018.04.027 PubMed Abstract | 10.1016/j.jep.2018.04.027 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Li B., Xie P.-J., Hao Y.-W., Guo Y., Yu J.-R., Gong D.-Y., et al. (2021a). Yuan-zhi-san inhibits tau protein aggregation in an Aβ1-40-induced Alzheimer's disease rat model via the ubiquitin-proteasome system. Mol. Med. Rep. 23, 279. 10.3892/mmr.2021.11918 10.3892/mmr.2021.11918 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. J. (2008). The bioactivity of chemical constituents in Polygala tenuifolia and Polygala glomerata lour. Beijing: Peking Union Medical College. Google Scholar [Google Scholar]
- Li S., Zhang B. (2013). Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chin. J. Nat. Med. 11, 110–120. 10.1016/s1875-5364(13)60037-0 PubMed Abstract | 10.1016/s1875-5364(13)60037-0 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Li S., Zhang B., Zhang N. (2011). Network target for screening synergistic drug combinations with application to traditional Chinese medicine. BMC Syst. Biol. 5 Suppl 1, S10. 10.1186/1752-0509-5-s1-s10 PubMed Abstract | 10.1186/1752-0509-5-s1-s10 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Yang H., Xiao J., Zhang J., Zhang J., Liu M., et al. (2020). Network pharmacology based investigation into the bioactive compounds and molecular mechanisms of Schisandrae Chinensis Fructus against drug-induced liver injury. Bioorg. Chem. 96, 103553. 10.1016/j.bioorg.2019.103553 PubMed Abstract | 10.1016/j.bioorg.2019.103553 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Li Z., Ma J., Kuang Z., Jiang Y. (2021b). β-Asarone attenuates aβ-induced neuronal damage in PC12 cells overexpressing APPswe by restoring autophagic flux. Front. Pharmacol. 12, 701635. 10.3389/fphar.2021.701635 PubMed Abstract | 10.3389/fphar.2021.701635 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H. Y., Zhang P. P., Zhang X. L., Zheng Y. Y., Huang Y. R., Zheng G. Q., et al. (2021). Preclinical systematic review of ginsenoside Rg1 for cognitive impairment in Alzheimer’s disease. AGING 13, 7549–7569. 10.18632/aging.202619 PubMed Abstract | 10.18632/aging.202619 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionta E., Spyrou G., Vassilatis D., Cournia Z. (2014). Structure-based virtual screening for drug discovery: Principles, applications and recent advances. Curr. Top. Med. Chem. 14, 1923–1938. 10.2174/1568026614666140929124445 PubMed Abstract | 10.2174/1568026614666140929124445 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Yu G., Chen X., Liu Y., Zhou Y., Wang G., et al. (2020). Integrated phytochemical analysis based on UHPLC-LTQ–Orbitrap and network pharmacology approaches to explore the potential mechanism of Lycium ruthenicum Murr. for ameliorating Alzheimer’s disease. Food Funct. 11, 1362–1372. 10.1039/c9fo02840d PubMed Abstract | 10.1039/c9fo02840d | Google Scholar [DOI] [PubMed] [Google Scholar]
- Luyen B. T., Tai B. H., Thao N. P., Cha J. Y., Lee H. Y., Lee Y. M., et al. (2015). Anti-inflammatory components of Chrysanthemum indicum flowers. Bioorg. Med. Chem. Lett. 25, 266–269. 10.1016/j.bmcl.2014.11.054 PubMed Abstract | 10.1016/j.bmcl.2014.11.054 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Ma X. H., Duan W. J., Mo Y. S., Chen J. L., Li S., Zhao W., et al. (2018). Neuroprotective effect of paeoniflorin on okadaic acid-induced tau hyperphosphorylation via calpain/Akt/GSK-3β pathway in SH-SY5Y cells. Brain Res. 1690, 1–11. 10.1016/j.brainres.2018.03.022 PubMed Abstract | 10.1016/j.brainres.2018.03.022 | Google Scholar [DOI] [PubMed] [Google Scholar]
- May B. H., Feng M., Zhou I. W., Chang S. Y., Lu S. C., Zhang A. L., et al. (2016). Memory impairment, dementia, and Alzheimer's disease in classical and contemporary traditional Chinese medicine. J. Altern. Complement. Med. 22, 695–705. 10.1089/acm.2016.0070 PubMed Abstract | 10.1089/acm.2016.0070 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Meng M., Zhang L., Ai D., Wu H., Peng W. (2021). β-Asarone ameliorates β-amyloid-induced neurotoxicity in PC12 cells by activating P13K/Akt/Nrf2 signaling pathway. Front. Pharmacol. 12, 659955. 10.3389/fphar.2021.659955 PubMed Abstract | 10.3389/fphar.2021.659955 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A. (2021). FDA approval for Biogen's aducanumab sparks Alzheimer disease firestorm. Nat. Rev. Drug Discov. 20, 496. 10.1038/d41573-021-00099-3 PubMed Abstract | 10.1038/d41573-021-00099-3 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Nie L., Xia J., Li H., Zhang Z., Yang Y., Huang X., et al. (2017). Ginsenoside Rg1 ameliorates behavioral abnormalities and modulates the hippocampal proteomic change in triple transgenic mice of Alzheimer's disease. Oxid. Med. Cell. Longev. 2017, 6473506. 10.1155/2017/6473506 PubMed Abstract | 10.1155/2017/6473506 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H., Goto S., Sato K., Fujibuchi W., Kanehisa M. (1999). Kegg: Kyoto Encyclopedia of genes and Genomes. Nucleic Acids Res. 27, 29–34. 10.1093/nar/27.1.29 PubMed Abstract | 10.1093/nar/27.1.29 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M. H., Houghton P. J., Whang W. K., Cho J. H. (2004). Screening of Korean herbal medicines used to improve cognitive function for anti-cholinesterase activity. Phytomedicine 11, 544–548. 10.1016/j.phymed.2004.03.001 PubMed Abstract | 10.1016/j.phymed.2004.03.001 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Pang X. C., Kang D., Fang J. S., Zhao Y., Xu L., Lian W. W., et al. (2018). Network pharmacology-based analysis of Chinese herbal Naodesheng formula for application to Alzheimer's disease. Chin. J. Nat. Med. 16, 53–62. 10.1016/s1875-5364(18)30029-3 PubMed Abstract | 10.1016/s1875-5364(18)30029-3 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Poornima P., Kumar J. D., Zhao Q., Blunder M., Efferth T. (2016). Network pharmacology of cancer: From understanding of complex interactomes to the design of multi-target specific therapeutics from nature. Pharmacol. Res. 111, 290–302. 10.1016/j.phrs.2016.06.018 PubMed Abstract | 10.1016/j.phrs.2016.06.018 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Prati F., Bottegoni G., Bolognesi M. L., Cavalli A. (2017). BACE-1 inhibitors: From recent single-target molecules to multitarget compounds for Alzheimer’s disease. J. Med. Chem. 61, 619–637. 10.1021/acs.jmedchem.7b00393 PubMed Abstract | 10.1021/acs.jmedchem.7b00393 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Razani E., Pourbagheri-Sigaroodi A., Safaroghli-Azar A., Zoghi A., Shanaki-Bavarsad M., Bashash D. (2021). The PI3K/akt signaling axis in Alzheimer's disease: A valuable target to stimulate or suppress? Cell Stress Chaperones 26, 871–887. 10.1007/s12192-021-01231-3 PubMed Abstract | 10.1007/s12192-021-01231-3 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddyrajula R., Dalimba U. (2020). The bioisosteric modification of pyrazinamide derivatives led to potent antitubercular agents: Synthesis via click approach and molecular docking of pyrazine-1, 2, 3-triazoles. Bioorg. Med. Chem. Lett. 30, 126846. 10.1016/j.bmcl.2019.126846 PubMed Abstract | 10.1016/j.bmcl.2019.126846 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Ren D. Y., Wang P., Liu C. L., Wang J., Liu X., Liu J., et al. (2018). Hazelnut protein-derived peptide LDAPGHR shows anti-inflammatory activity on LPS-induced RAW264.7 macrophage. J. Funct. Foods. doi:doi:46, 449, 455. 10.1016/j.jff.2018.04.024 10.1016/j.jff.2018.04.024 | Google Scholar [DOI] [Google Scholar]
- Rueeger H., Lueoend R., Machauer R., Veenstra S. J., Holzer P., Hurth K., et al. (2021). Synthesis of the potent, selective, and efficacious β-secretase (BACE1) inhibitor NB-360. J. Med. Chem. 64, 4677–4696. 10.1021/acs.jmedchem.0c02143 PubMed Abstract | 10.1021/acs.jmedchem.0c02143 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Saikia S., Bordoloi M. (2019). Molecular docking: Challenges, advances and its use in drug discovery perspective. Curr. Drug Targets 20, 501–521. 10.2174/1389450119666181022153016 PubMed Abstract | 10.2174/1389450119666181022153016 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Song Z., Yin F., Xiang B., Lan B., Cheng S. (2018). Acori tatarinowii RhizomaSystems pharmacological approach to investigate the mechanism of for Alzheimer's disease. Evidence-based Complement. Altern. Med. 2018, 5194016. eCAM. 10.1155/2018/5194016 10.1155/2018/5194016 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. M., Zhu B. J., Cao H. T., Zhang X. Y., Zhang Q. C., Xin G. Z., et al. (2018). Explore the effects of Huang-Lian-Jie-Du-Tang on Alzheimer's disease by UPLC-QTOF/MS-based plasma metabolomics study. J. Pharm. Biomed. Anal. 151, 75–83. 10.1016/j.jpba.2017.12.053 PubMed Abstract | 10.1016/j.jpba.2017.12.053 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Wang L., Yu H., Lu X. H., Jin G. F., Liang J. Q., Yang H., et al. (2019a). Protective effects of tenuifolin isolated from Polygala tenuifolia Willd roots on neuronal apoptosis and learning and memory deficits in mice with Alzheimer’s disease. Food Funct. 10, 7453–7460. 10.1039/c9fo00994a PubMed Abstract | 10.1039/c9fo00994a | Google Scholar [DOI] [PubMed] [Google Scholar]
- Wang N., Ahmad H., Jia Y. M. (2017). Identification of polygala oligosaccharide esters and their metabolites in rat plasma after oral administration of ethanol extract of Kai Xin San by UHPLC-MS. Acta Pharm. Sin. 52, 7. 10.16438/j.0513-4870.2017-0373-en 10.16438/j.0513-4870.2017-0373-en | Google Scholar [DOI] [Google Scholar]
- Wang N., Wang H., Li L., Li Y., Zhang R. (2019b). β-Asarone inhibits amyloid-β by promoting autophagy in a cell model of Alzheimer's disease. Front. Pharmacol. 10, 1529. 10.3389/fphar.2019.01529 PubMed Abstract | 10.3389/fphar.2019.01529 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Wang H., Pan Q., Kang J., Liang Z., Zhang R. (2021). βThe combination of -asarone and icariin inhibits amyloid- and reverses cognitive deficits by promoting mitophagy in models of Alzheimer's disease. Oxid. Med. Cell. Longev. 2021, 7158444. 10.1155/2021/7158444 PubMed Abstract | 10.1155/2021/7158444 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. J., Zhang T. (2017). Integration of traditional Chinese medicine and Western medicine in the era of precision medicine. J. Integr. Med. 15, 1–7. 10.1016/s2095-4964(17)60314-5 PubMed Abstract | 10.1016/s2095-4964(17)60314-5 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Wang Y., Sun Y. W., Wang Y. M., Ju Y., Meng D. L. (2019c). Virtual screening of active compounds from Artemisia argyi and potential targets against gastric ulcer based on Network pharmacology. Bioorg. Chem. 88, 102924. 10.1016/j.bioorg.2019.102924 PubMed Abstract | 10.1016/j.bioorg.2019.102924 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Wei S. (2016). Potential therapeutic action of natural products from traditional Chinese medicine on Alzheimer's disease animal models targeting neurotrophic factors. Fundam. Clin. Pharmacol. 30, 490–501. 10.1111/fcp.12222 PubMed Abstract | 10.1111/fcp.12222 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Wei T. F., Zhao L., Huang P., Hu F. L., Jiao J. Y., Xiang K. L., et al. (2020). Qing-yi decoction in the treatment of acute pancreatitis: An integrated approach based on chemical profile, network pharmacology, molecular docking and experimental evaluation. Front. Pharmacol. 12, 590994. 10.3389/fphar.2021.590994 10.3389/fphar.2021.590994 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. J., Mei Y., Shi X. Q., Zhang Y. F., Wang X. Y., Guan L., et al. (2019). Albiflorin ameliorates memory deficits in APP/PS1 transgenic mice via ameliorating mitochondrial dysfunction. Brain Res. 1719, 113–123. 10.1016/j.brainres.2019.05.037 PubMed Abstract | 10.1016/j.brainres.2019.05.037 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Yousof Ali M., Jung H., Choi J. (2015). Anti-diabetic and anti-Alzheimer's disease activities of Angelica decursiva. Arch. Pharm. Res. 38, 2216–2227. 10.1007/s12272-015-0629-0 PubMed Abstract | 10.1007/s12272-015-0629-0 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Zhang H. R., Peng J. H., Cheng X. B., Shi B. Z., Zhang M. Y., Xu R. X. (2015). Paeoniflorin atttenuates amyloidogenesis and the inflammatory responses in a transgenic mouse model of Alzheimer's disease. Neurochem. Res. 40, 1583–1592. 10.1007/s11064-015-1632-z PubMed Abstract | 10.1007/s11064-015-1632-z | Google Scholar [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yang X., Wang S., Song S. (2019). Ginsenoside Rg3 prevents cognitive impairment by improving mitochondrial dysfunction in the rat model of Alzheimer's disease. J. Agric. Food Chem. 67, 10048–10058. 10.1021/acs.jafc.9b03793 PubMed Abstract | 10.1021/acs.jafc.9b03793 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Zhao X., Cui Y., Wu P., Zhao P., Zhou Q., Zhang Z., et al. (2020). Polygalae radix: A review of its traditional uses, phytochemistry, pharmacology, toxicology, and pharmacokinetics. Fitoterapia 147, 104759. 10.1016/j.fitote.2020.104759 PubMed Abstract | 10.1016/j.fitote.2020.104759 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Zhou F., He K., Guan Y., Yang X., Chen Y., Sun M., et al. (2020). Network pharmacology-based strategy to investigate pharmacological mechanisms of Tinospora sinensis for treatment of Alzheimer's disease. J. Ethnopharmacol. 259, 112940. 10.1016/j.jep.2020.112940 PubMed Abstract | 10.1016/j.jep.2020.112940 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Zhu B., Zhang Q. L., Hua J. W., Cheng W. L., Qin L. P. (2018). The traditional uses, phytochemistry, and pharmacology of Atractylodes macrocephala Koidz.: A review. J. Ethnopharmacol. 226, 143–167. 10.1016/j.jep.2018.08.023 PubMed Abstract | 10.1016/j.jep.2018.08.023 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Zhu H., Zhang F., Chen F., Xiao Z. (2017). The application and research progress of the role of APP in neurogenesis and Alzheimer's disease. Chin. J. Ethnomedicine Ethnopharmacy 026 (012), 74–79,81. Google Scholar [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.