Abstract
The presence of the heat stress response-related ATPases ClpC and ClpX or the peptidase ClpP in the cell is crucial for tolerance of many forms of stress in Bacillus subtilis. Assays for detection of defects in protein degradation suggest that ClpC, ClpP, and ClpX participate directly in overall proteolysis of misfolded proteins. Turnover rates for abnormal puromycyl peptides are significantly decreased in clpC, clpP, and clpX mutant cells. Electron-dense aggregates, most likely due to the accumulation of misfolded proteins, were noticed in studies of ultrathin cryosections in clpC and clpP mutant cells even under nonstress conditions. In contrast, in the wild type or clpX mutants such aggregates could only be observed after heat shock. This phenomenon supports the assumption that clpC and clpP mutants are deficient in the ability to solubilize or degrade damaged and aggregated proteins, the accumulation of which is toxic for the cell. By using immunogold labeling with antibodies raised against ClpC, ClpP, and ClpX, the Clp proteins were localized in these aggregates, showing that the Clp proteins act at this level in vivo.
In bacteria, as in eukaryotic cells, heat shock proteins are part of the cellular machinery for protein folding, repair, and degradation. Important energy-dependent and heat response-related proteases in Escherichia coli are the Clp proteases, ClpAP and ClpXP, consisting of separately encoded ATPase and peptidase subunits. Different substrate specificity is determined by association of the proteolytic component ClpP with either ClpA or ClpX as a regulatory ATPase (for a review, see references 9 and 11). The resulting complexes exhibit a native molecular architecture of two rings of a ClpP heptamer, stacking back to back. A hexamer of the Clp ATPase is located either on one or on both sides of the ClpP rings. For this complex, a structural similarity to the eukaryotic proteasome has been discussed (12, 16, 41, 52).
It has been accepted that the conserved and ubiquitous Clp ATPases can function as either proteolysis regulators or molecular chaperones (for recent reviews, see references 10, 11, 39, and 43). Chaperone or disaggregase function has been shown or suggested for the ClpA and ClpB, as well as for the ClpX members of the HSP100 family of Clp ATPases (33, 44, 53, 54). Participation in overall proteolysis of misfolded proteins has also been demonstrated for the ClpYQ (HalUV) protease. ClpQ, the proteolytic subunit, shares a very high degree of similarity with members of the β-type subunit constituting the catalytic core of the eukaryotic 20S proteasome, whereas ClpY also belongs to the Hsp100 ATPase family (1, 27, 35, 36). Besides Clp in E. coli, the ATP-dependent protease Lon plays an important role in cellular processes by modulating the availability of certain regulatory proteins or degrading abnormally folded proteins (for a review, see reference 9). Regulatory proteins, such as the cell division inhibitor SulA or RcsA, involved in capsule synthesis, were shown to be degraded by the Lon protease (4, 48).
In the gram-positive soil bacterium Bacillus subtilis, deletion or disruption of either clpC, clpP, or clpX causes a very pleiotropic phenotype. The presence of ClpC, ClpP, or ClpX in the cell is essential for stress tolerance, because clp mutants cannot grow under several stress conditions (7, 21, 29). Furthermore, B. subtilis Clp proteins were found to be required for cell division and several stationary-phase phenomena, such as motility and degradative enzyme synthesis, as well as the development of sporulation and genetic competence (7, 17, 21, 29, 30, 31, 49, 50).
Our experiments on the role of Clp proteins in protein degradation revealed a direct participation of ClpC, ClpX, and ClpP in overall proteolysis of heat-damaged proteins in B. subtilis. The decreased breakdown of damaged proteins, evidenced by accumulation of protein aggregates in clpC and clpP mutants, occurred even under nonstress conditions. By immunocytochemical methods, we could localize Clp proteins at these protein aggregates, suggesting that they most likely act there in vivo in resolubilizing and/or degrading damaged proteins.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. E. coli and B. subtilis cells were routinely cultivated under vigorous agitation at 37°C in Luria-Bertani medium. The different stress conditions were induced as described earlier (51). The culture was divided during exponential growth, and one half of the culture was grown at 37°C (control), whereas the other half of the culture was exposed to heat shock at 50°C or treated with puromycin. Since clp mutant cells showed impaired growth in minimal medium (7, 21), the culture was supplemented with 0.05% (wt/vol) yeast extract. The media were supplemented with the following antibiotics if necessary: ampicillin (100 μg/ml), chloramphenicol (5 μg/ml for B. subtilis or 25 μg/ml for E. coli), kanamycin (10 μg/ml), spectinomycin (100 μg/ml), erythromycin (1 μg/ml), and lincomycin (25 μg/ml).
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| E. coli | ||
| RR1 | F−mcrB mrr hsdS20(rB− mB−) ara-14 proA2 lacY1 leuB2 galK2 rpsL20 (Smr) xy1-5 mtl-l supE44 | 2 |
| DH5α | F− φ80dlacZΔ M15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA96 | 12 |
| BL21(DE3)/pLysS | F−lon hsdSB (rB− mB−) with DE3, a λ prophage carrying the T7 RNA polymerase gene, and pLysS plasmid containing the T7 phage lysozyme gene | 45 |
| B. subtilis | ||
| IS58 | trpC2 lys-3 | 42 |
| BGWLON1 | trpC2 lys-3 lonA::pJH101 | 34 |
| BUG1 | trpC2 lys-3 ΔclpP::spec | 6 |
| BUG2 | trpC2 lys-3 clpX::pMUTIN4 | 6 |
| BEK4 | trpC2 lys-3 ΔclpC::spec | 19 |
General methods.
DNA manipulations and transformation of E. coli were done according to standard protocols (37, 38). Some oligonucleotides used for PCR included mismatches, allowing creation of restriction sites. Chromosomal DNA from B. subtilis was isolated using the Wizard genomic DNA purification kit (Promega, Inc.). Transformation of B. subtilis with plasmid or chromosomal DNA was carried out by using a two-step protocol (14). Analysis of transcription by mRNA slot blotting has been described previously (21). Protein extracts were electrophoresed with standard sodium dodecyl sulfate-polyacrylamide gels (24). The protein concentrations of crude extracts were determined by the Bio-Rad protein assay (3). Western blotting was performed by transferring the proteins to polyvinylidene difluoride membranes (Bio-Rad Laboratories). For immunodetection, the membranes were blocked for 1 h in BLOTTO buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 2 mM NaN3, 2.5% [wt/vol] skim milk powder, 0.5% [vol/vol] Tween 20); incubated overnight with the polyclonal antisera for ClpC (1:8,000) (17), ClpX (1:20,000), and ClpP (1:10,000) diluted in BLOTTO; washed twice for 20 min in BLOTTO; and processed with a goat anti-rabbit or a goat anti-guinea pig alkaline phosphatase conjugate (Sigma). Cross-reacting material was visualized by chemiluminescense with CDP-Star as a substrate of the alkaline phosphatase. The Lumi-Imager system (Roche Diagnostics) was used for documentation and quantitation.
Purification of proteins and antibody production.
For overproduction and purification of ClpP and ClpX in E. coli, the entire genes were amplified by PCR using primers PRCLPPF (GGAGGATCCATGAATTTAATACCTACAGTC) and PRCLPPR (CGGAATTCTTACTTTTTGTCTTCTGTGTG), as well as PRXFOR (GGAGGATCCATGTTTAAATTTAACGAGGA) and PRXREV (CGGGGTACCTTATGCAGATGTTTTATCTT), and cloned as a BamHI/EcoRI or BamHI/KpnI fragment into pRSETA (Invitrogen, Inc.). This plasmid allowed an in-frame fusion of the clpP and the clpX gene to six histidine codons at the N terminus and transcription from a T7 promoter. Overproduction of His6-ClpP and His6-ClpX proteins with T7 RNA polymerase in the E. coli strain BL21(DE3) (45) and purification under native conditions by Ni-nitriloacetic acid affinity chromatography (Qiagen, Inc.) was done as previously described (19). The His6-ClpP and His6-ClpX proteins were used for custom antibody production in rabbits (Eurogentec, Liege, Belgium).
Measurement of degradation of puromycyl peptides.
B. subtilis wild type and the isogenic clpC, clpP, clpX, and lonA mutant strains were grown at 37°C in synthetic medium (46) until the optical density reached 0.4. Puromycin was added to a final concentration of 40 μg/ml, while the control culture contained no puromycin. After 15 min of incubation at 37°C, [3H]leucine was added to a final concentration of 20 μCi/ml. After 5 min, the cells were collected by centrifugation and washed twice. They were then grown at 37°C for 1 h in synthetic medium supplemented with leucine. Samples were taken at intervals, applied to filter disks, and precipitated with 10% trichloroacetic acid. The radioactivity of the acid-insoluble fraction was measured by liquid scintillation counting (8).
Preparation of B. subtilis cells for cryosectioning and immunocytochemistry.
Cells were harvested at an optical density of 0.3 before (control) and after heat shock at 50°C. After a fixation step (30 min in 0.2% glutaraldehyde, 2% [wt/vol] paraformaldehyde, 100 mM cacodylate buffer [pH 7.4], 1 mM CaCl2, 1 mM MgCl2, and 25 mM NaN3), the cells were washed for 5 min in the same buffer without aldehydes, quenched for 15 min in glycine–Tris-buffered saline (TBS) (50 mM glycine, 20 mM Tris-HCl [pH 8.0], 2.5 mM KCl, 135 mM NaCl, 20 mM NaN3), washed for 5 min in TBS, and soaked in a mixture of 25% (wt/vol) polyvinylpyrrolidone (Mr, 10,000; Sigma-Aldrich) and 1.6 M sucrose according to the method of Tokuyasu (47). Samples were mounted on specimen holders, frozen in liquid nitrogen, and sectioned with a diamond knife at −100°C with an ultracut S/FCS cryoultramicrotome (Leica). Ultrathin thawed cryosections were placed on Formavar-carbon-coated copper grids (400 mesh), floated sections down six times for 10 min each time on drops with glycine-TBS, for 15 min on 5% (vol/vol) goat serum in incubation buffer (1% skim milk powder [wt/vol], 0.01% Tween 20 in TBS), for 16 h on polyclonal antiserum against ClpC, ClpX, and ClpP (1:125, 1:1,000, and 1:1,000, respectively) diluted in incubation buffer, six times for 2 min each time on incubation buffer, and for 60 min on goat anti-rabbit or goat anti-guinea pig 10-nm-diameter gold conjugates (British BioCell International) diluted 1:25 in incubation buffer. After extensive washes with TBS and double-distilled water, the sections were stabilized with 2% methyl cellulose (25 cps) containing 0.3% uranyl acetate and analyzed with a Zeiss EM 906 electron microscope at 60 kV. Incubations with primary antibodies took place at 4°C; all other incubation steps were carried out at room temperature. The specificities of immune reactions were demonstrated by omitting the primary antibodies. No gold particles were detected in the negative controls.
RESULTS
Intracellular levels of ClpC, ClpP, and ClpX.
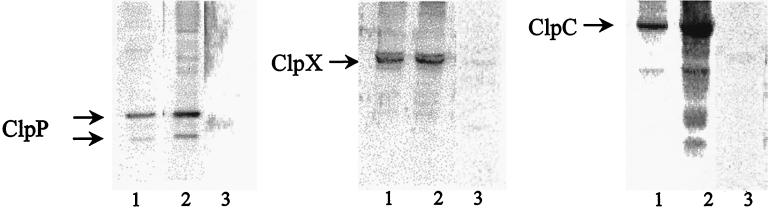
The transcription patterns of the clpC operon, as well as those of the clpP and clpX genes, showed an mRNA induction of all three genes after stress conditions producing nonnative proteins in the cell (6, 7, 20, 28). To find out more about the intracellular concentrations of the ClpC, ClpP, and ClpX proteins under stress conditions, Western blot experiments were performed with protein extracts of control and heat-shocked cells. As expected, significantly higher intracellular amounts of ClpC and ClpP were observed after heat shock in comparison to the control level before heat shock, also indicating increased synthesis of these proteins under stress conditions (Fig. 1). Interestingly, Western blots with the ClpP antibody revealed two specific signals with a mass difference of approximately 5 kDa, indicating that the ClpP protein might be present in two different forms in the cell. This has also been observed in E. coli, where the first 14 amino acids are autocatalytically processed (26). Inspection of the clpP sequence revealed a second, theoretical translation start at position 91 of the coding sequence preceded by a putative Shine-Dalgarno sequence (coding sequence position 76) (data not shown), indicating reinitiation of translation rather than processing. Surprisingly, there was no obvious difference in the level of ClpX in nonstressed and heat-shocked cells (6) (Fig. 1). This result did not agree with our transcriptional data, which showed a moderate heat shock induction of the clpX gene.
FIG. 1.
Amount of ClpC, ClpP, and ClpX in exponentially growing and heat-shocked cells. Samples were taken before (lanes 1) and 15 min after (lanes 2) 50°C heat shock and were analyzed by Western blotting using an antibody against ClpC, ClpP, or ClpX. Crude extracts of the mutants for the appropriate target proteins ClpP (BUG1), ClpX (BUG2), and ClpC (BEK4) were examined as specificity controls for the respective antibodies (lanes 3).
ClpC, ClpP, and ClpX participate in overall proteolysis of misfolded proteins.
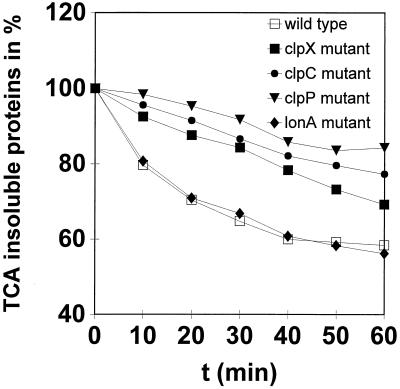
Much data is available concerning the role of ClpAP and ClpXP proteases in ATP-dependent proteolysis in E. coli (for a review, see references 9 and 11). In contrast to E. coli, little is known about the importance of Clp-mediated protein degradation in gram-positive bacteria. Genes encoding ClpA- or ClpB-type ATPases were not found in the B. subtilis genome (22). However, ClpX and ClpC appeared to be good candidates for direction of energy-dependent proteolysis in association with the ClpP peptidase subunit during stress. To look more closely at the function of putative ATP-dependent proteases in B. subtilis, we examined the abilities of the clpC, clpP, and clpX mutants as well as the lonA mutant to degrade prematurely released puromycyl polypeptides. In general, a low protein turnover was observed in the absence of puromycin (data not shown). As expected, the turnover rates rose significantly after the addition of puromycin, which induces the synthesis of abnormal proteins. The results, presented in Fig. 2, show that the clpC, clpP, and clpX mutants degraded puromycyl polypeptides both at a reduced rate and to a much lower overall extent than the wild-type strain, reflecting the important role of Clp proteases in protein degradation. Deletion of clpC and clpP affected protein breakdown more than a clpX mutation did (Fig. 2). Unexpectedly, there was no difference between the turnover rates of the wild type and the lonA mutant (Fig. 2). In contrast to E. coli lon mutants, which exhibit strongly diminished decay rates for abnormal proteins (25), the B. subtilis LonA did not participate in overall proteolysis of puromycyl polypeptides.
FIG. 2.
Overall cellular proteolysis of clpC, clpP, clpX, and lonA mutants. Relative puromycylpolypeptide degradation in the wild type and the ΔclpC, ΔclpP, ΔclpX, and lonA mutants is shown. Cellular protein was labeled with l-[3H]leucine (20 μCi/ml) in the presence of puromycin (40 μg/ml) and then chased with nonradioactive l-leucine in the absence of puromycin. Radioactivity of the trichloroacetic acid (TCA)-insoluble fraction was measured by liquid scintillation counting as described in Materials and Methods.
Subcellular localization of Clp proteins.
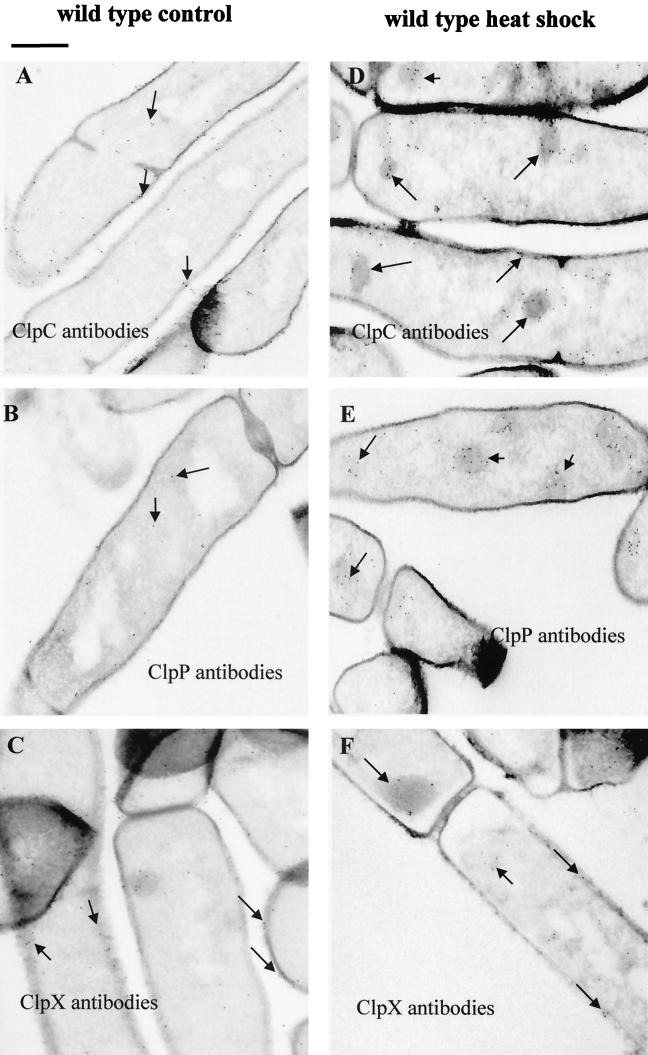
Ultrastructural analysis of heat-shocked Saccharomyces cerevisiae cells revealed electron-dense particles, which were considered to be aggregates of heat-denatured proteins (33). In order to get more information about general heat shock damage and the subcellular localization of the ClpC, ClpP, and ClpX proteins in B. subtilis cells, we performed electron microscopic experiments in combination with the immunogold labeling technique. Cryosections of exponentially growing or heat-shocked cells were processed with antibodies against one of the three Clp proteins and with a secondary antibody-gold conjugate to visualize the proteins by electron microscopy.
Consistent with the yeast data, equivalent aggregates could be observed in electron microscopic studies of wild-type cells heat shocked for 10 min at 50°C (Fig. 3). Generally, treatment with 20 μg of puromycin/ml gave similar results (not shown). Similar to S. cerevisiae, general heat damage of B. subtilis cells was accompanied by aggregation of heat-damaged proteins, which can be visualized by electron microscopy. These aggregates did not show a preferred localization but were randomly distributed in the cell. Immunocytochemical experiments with exponentially growing wild-type cells showed that ClpP was spread inside the cells, as were some of the ClpC and ClpX proteins (Fig. 3A to C). However, gold particles corresponding to the ClpC and ClpX ATPases were also found at the cell envelope, indicating that they might also have functions there (Fig. 3A and C). In heat-shocked cells, all three proteins could be detected at the electron-dense aggregates (Fig. 3D to F). Thus, it was possible to show that ClpC, ClpP, and ClpX indeed adhere to these aggregates in vivo, most likely for resolubilization and degradation of heat-damaged and aggregated proteins. Table 2 shows a quantitation of the immunogold particles in cryosections of wild-type cells before and after heat shock.
FIG. 3.
Subcellular localization of Clp proteins using the immunogold labeling technique. Cryosections of exponentially growing (A, B, and C) or heat-shocked (D, E, and F) wild-type cells were treated with antibodies against ClpC (A and D), ClpP (B and E), and ClpX (C and F) with a secondary antibody-gold conjugate to visualize the localization of the proteins. The localization of gold particles corresponding to Clp proteins is indicated by arrows. Bar, 0.5 μm.
TABLE 2.
Quantitation of immunogold particles in ultrathin cryosections of B. subtilis wild-type cells before and after heat shock
| Parameter | Valuea
|
|||||
|---|---|---|---|---|---|---|
| ClpP antibody
|
ClpC antibody
|
ClpX antibody
|
||||
| Controlb | Heat shockc | Controlb | Heat shockc | Controlb | Heat shockc | |
| % Cells with inclusions bodies | 0 | 99.2 | 0 | 100 | 0 | 98.3 |
| No. (± SD) of gold particles in cytoplasm per cell | 12.6 ± 6.6 | 17.0 ± 6.9 | 26.4 ± 9.2 | 39.4 ± 11.5 | 15.9 ± 5.1 | 11.9 ± 4.9 |
| No. (± SD) of gold particles at protein aggregates per cell | 0 | 14.0 ± 6.7 | 0 | 30.4 ± 10.2 | 0 | 7.2 ± 4.1 |
At least 100 cells were evaluated.
Cells were harvested during exponential growth.
Cells were harvested 10 min after heat shock at 50°C.
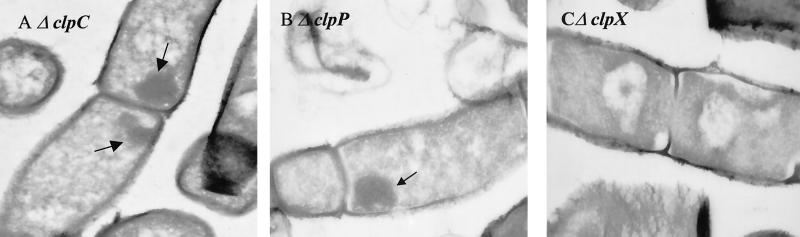
clpC and clpP mutants accumulate aggregates of denatured proteins under nonstress conditions.
Yeast with mutations in the hsp104 gene, encoding a B-type Hsp100 ATPase, failed to recover from aggregation damage (33). Since clpC, clpP, and clpX mutants exhibit heat-sensitive phenotypes and defects in overall protein breakdown (see above), electron microscopic studies with these mutants were performed in the next step. Ultrathin sections of heat-shocked wild-type cells revealed that aggregation damage was significantly decreased or completely disappeared after 30 min of incubation at 50°C, whereas after 30 min clpC and clpP mutants were as damaged as immediately after stress. This phenomenon was less pronounced in clpX mutant cells (not shown). No aggregation damage was observed in nonstressed exponentially growing wild-type and clpX mutant cells (Fig. 3A to C and 4C). In contrast, in clpC and clpP mutant cells, accumulation of electron-dense material could also be detected under nonstress conditions (Fig. 4A and B). These observations support the conclusion that B. subtilis ClpC and ClpP play a crucial role in protein turnover under nonstress as well as under stress conditions.
FIG. 4.
Ultrastructural analysis of clp mutants. Ultrathin sections of exponentially growing cells of ΔclpC (A), ΔclpP (B), and clpX (C) mutants are shown. Accumulations of electron-dense material in the clpC and clpP mutants are indicated by arrows. Bar, 0.5 μm.
Changes in the localization of ClpP were observed in clpC mutant cells. Gold particles corresponding to ClpP found at electron-dense aggregates were considerably diminished in exponentially growing clpC mutant cells, whereas deletion of clpP had no effect on localization of ClpC at electron-dense material (data not shown). The distribution of ClpX in clpP mutant cells and that of ClpP in clpX mutants resembled the wild-type situation (not shown). In summary, these results suggest that ClpC could direct ClpP in the degradation of denatured proteins.
DISCUSSION
Energy-dependent degradation of misfolded proteins in E. coli is assigned to the ClpAP, the ClpYQ (HslUV), and the Lon proteases (for a review, see reference 9). Despite the conservation and ubiquity of Clp proteins in bacteria and higher organisms, little is known about the importance of Clp-mediated proteolysis in organisms other than E. coli. Lactococcus lactis cells lacking ClpP had a reduced ability to degrade puromycyl-containing peptides (5). No A-type Clp ATPases were apparent from the complete sequence of the B. subtilis genome (22). Instead, a direct participation in the overall proteolysis of misfolded proteins was shown for a member of the ClpC subfamily in B. subtilis, most prominently found in gram-positive bacteria and plants. Besides its global function in removal of damaged proteins, CtsR, the global repressor of clp gene expression in gram-positive bacteria, has been proved to be a specific substrate of the ClpCP protease (our unpublished observation).
The ClpX-type ATPase of B. subtilis is also involved in degradation of damaged proteins, but to a lesser extent (Fig. 2). Recently, potential specific substrates have been determined by a two-dimensional-gel approach (7), suggesting that ClpX has a regulatory function. Interestingly, homologous proteins of components of the ClpYQ protease, found in E. coli (27, 35) and other eubacteria, also exist in B. subtilis (22). However, nothing is known about the contribution to energy-dependent proteolysis of this proteolytic system in B. subtilis.
Lon was reported to be the primary protease in E. coli for degrading abnormally folded proteins (9). Although Lon is very well conserved, B. subtilis LonA was obviously not involved in degradation of misfolded proteins. A second lon gene, lonB, has been identified upstream of B. subtilis lonA and shown to increase total cellular ATP-dependent protease activity, but only after overproduction (R. Ye and S.-L. Wong, Abstr. Proc. 8th Conf. Bacilli, abstr. T31, p. 78, 1995). Possibly, the Lon proteins can compensate for one another. However, our data strongly suggest that the dominant portion of energy-dependent proteolysis in vivo is executed by the ClpCP and ClpXP proteases in B. subtilis (Fig. 2). It should be mentioned, however, that the interaction of ClpP with ClpX has not yet been proven. Whereas mutations in the lon gene of E. coli exhibit various phenotypes, such as filamentation or mucoidy (4, 48), no growth defect has been described so far for clpA, clpX, or clpP mutants (15, 26). Conversely, a single mutation in any of the clp genes in B. subtilis has profound effects on cell morphology and growth even under standard conditions (7, 18, 21, 23, 29). This phenomenon can be due to the deficiency in the ability of the clpC and clpP mutants to remove damaged and aggregated proteins and may cause the extreme sensitivity of those mutants, not only during stress (7, 21, 29, 30). Consequently, they are unable to recover from stress (7, 21, 29) (Fig. 2 and 4). From a functional point of view, ClpC seems to combine properties of ClpA as well as ClpB ATPase in the direction of proteolysis while also protecting the cell from stress by resolubilization of protein aggregates (for a review, see references 9, 10, 11, and 43). In this context, it is interesting to note that double clp gene mutants in any combination do not appear viable (our unpublished observation).
Our experiments indicate that ClpC, ClpP, and ClpX adhere to aggregates of damaged proteins generated by heat shock, presumably acting as disaggregases and/or proteases in vivo (Fig. 3 and 4). So far, however, we cannot exclude the possibility that the Clp proteins themselves aggregate under stress conditions, thus explaining their presence in denatured protein aggregates, but this is probably not the main reason for their localization at these aggregates. Similarly, Clp proteins were detected at inclusion bodies caused by an overproduction of a foreign protein in B. subtilis (B. Jürgen, M. Hecker, and T. Schweder, unpublished observation). Immunocytochemical experiments with clpP mutants showed that both ATPases, ClpC and ClpX, are located alone at these aggregates. This may support the assumption of disaggregase capacity that has also been suggested for the yeast Hsp104 ATPase (33). Localization of the ATPases at the cell envelope implies further specific and possibly chaperone functions, e.g., protein transport or translocation, as already shown in eukaryotic systems for a chloroplastic ClpC and for yeast Hsp78 in mitochondria (32, 40).
In summary, these data provide evidence of synergistic roles of the ClpCP and ClpXP proteases of B. subtilis in energy-dependent protein degradation. In contrast to E. coli, the primary proteolytic activity in degrading heat-damaged and abnormally folded proteins of B. subtilis can be assigned not to the Lon protease but to the ClpCP protease. Our future projects will focus on investigation of specific substrates of the Clp proteases in B. subtilis under different physiological conditions to determine the participation of proteolysis in regulatory pathways.
ACKNOWLEDGMENTS
We thank Kürsad Turgay and David Dubnau for helpful comments and valuable discussions and R. Bednarsky for critical reading of the manuscript. Furthermore, we are grateful to Jörg Mosterz and Ulf Gerth for overproduction and purification of ClpP and ClpX. Renate Gloger, Annette Meuche, and Hartmut Fischer are acknowledged for excellent technical assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie to M.H.
REFERENCES
- 1.Bochtler M, Ditzel L, Groll M, Huber R. Crystal structure of heat shock locus V (HslV) from Escherichia coli. Proc Natl Acad Sci USA. 1997;94:6070–6074. doi: 10.1073/pnas.94.12.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolivar F, Rodrigues R L, Greener P J, Betlach M C, Heyneker H L, Boyer H W, Crosa J H, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–133. [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Canceill D, Dervyn E, Huisman O. Proteolysis and modulation of the activity of the cell division inhibitor SulA in Escherichia coli lon mutants. J Bacteriol. 1990;172:7297–7300. doi: 10.1128/jb.172.12.7297-7300.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frees D, Ingmer H. ClpP participates in the degradation of misfolded protein in Lactococcus lactis. Mol Microbiol. 1999;31:79–87. doi: 10.1046/j.1365-2958.1999.01149.x. [DOI] [PubMed] [Google Scholar]
- 6.Gerth U, Wipat A, Harwood C, Carter N, Emmerson P T, Hecker M. Sequence and transcriptional analysis of clpX—a class III heat shock gene of Bacillus subtilis. Gene. 1996;181:77–83. doi: 10.1016/s0378-1119(96)00467-2. [DOI] [PubMed] [Google Scholar]
- 7.Gerth U, Krüger E, Derré I, Msadek T, Hecker M. Stress induction of the Bacillus subtilis clpP gene encoding the proteolytic component of the Clp protease and involvement of ClpP and ClpX in stress tolerance. Mol Microbiol. 1998;28:787–802. doi: 10.1046/j.1365-2958.1998.00840.x. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg A L. Degradation of abnormal proteins in Escherichia coli. Proc Natl Acad Sci USA. 1972;69:422–426. doi: 10.1073/pnas.69.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 10.Gottesman S, Maurizi M R, Wickner S. Regulatory subunits of energy-dependent proteases. Cell. 1997;91:435–438. doi: 10.1016/s0092-8674(00)80428-6. [DOI] [PubMed] [Google Scholar]
- 11.Gottesman S, Wickner S, Maurizi M R. Protein quality control: triage by chaperones and proteases. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 12.Grimaud R, Kessel M, Beuron F, Steven A C, Maurizi M R. Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J Biol Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D. Techniques for transformation of Escherichia coli. In: Glover D M, editor. DNA cloning: a practical approach. Vol. 1. Oxford, United Kingdom: IRL Press; 1985. pp. 109–135. [Google Scholar]
- 14.Hoch J A. Genetic analysis in Bacillus subtilis. Methods Enzymol. 1991;204:305–320. doi: 10.1016/0076-6879(91)04015-g. [DOI] [PubMed] [Google Scholar]
- 15.Katayama Y, Gottesman S, Pumphrey J, Rudikoff S, Clark W P, Maurizi M R. The two-component ATP-dependent Clp protease of Escherichia coli: purification, cloning, and mutational analysis of the ATP-binding component. J Biol Chem. 1988;263:15226–15236. [PubMed] [Google Scholar]
- 16.Kessel M, Maurizi M R, Kim B, Kocsis E, Trus B L, Singh S K, Steven A C. Homology in structural organization between E. coli ClpAP protease and eukaryotic 26 S proteasome. J Mol Biol. 1995;250:587–594. doi: 10.1006/jmbi.1995.0400. [DOI] [PubMed] [Google Scholar]
- 17.Kong L, Dubnau D. Regulation of competence-specific gene expression by Mec-mediated protein-protein interaction in Bacillus subtilis. Proc Natl Acad Sci USA. 1994;91:5793–5797. doi: 10.1073/pnas.91.13.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krüger E, Hecker M. Bacillus subtilis ClpC. In: Gething M J, editor. Guidebook to molecular chaperones and protein-folding catalysts. Oxford, United Kingdom: Oxford University Press; 1997. pp. 243–245. [Google Scholar]
- 19.Krüger E, Hecker M. The first gene of the clpC-operon in Bacillus subtilis, ctsR, encodes a negative regulator of its own operon and other class III heat shock genes. J Bacteriol. 1998;180:6681–6688. doi: 10.1128/jb.180.24.6681-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krüger E, Msadek T, Hecker M. Alternate promoters direct stress induced transcription of the Bacillus subtilis clpC operon. Mol Microbiol. 1996;20:713–723. doi: 10.1111/j.1365-2958.1996.tb02511.x. [DOI] [PubMed] [Google Scholar]
- 21.Krüger E, Völker U, Hecker M. Stress induction of clpC in Bacillus subtilis and its involvement in stress tolerance. J Bacteriol. 1994;176:3360–3367. doi: 10.1128/jb.176.11.3360-3367.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 23.Kunst F, Msadek T, Rapoport G. Signal transduction network controlling degradative enzyme synthesis and competence in Bacillus subtilis. In: Piggot P J, Moran C P Jr, Youngman P, editors. Regulation of bacterial differentiation. Washington, D. C.: American Society for Microbiology; 1994. pp. 1–20. [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Maurizi M R, Trisler P, Gottesman S. Insertional mutagenesis of the lon gene in Escherichia coli: lon is dispensable. J Bacteriol. 1985;164:1124–1135. doi: 10.1128/jb.164.3.1124-1135.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maurizi M R, Clark W P, Katayama Y, Rudikoff S, Pumphrey J, Bowers B, Gottesman S. Sequence and structure of ClpP, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J Biol Chem. 1990;265:12536–12545. [PubMed] [Google Scholar]
- 27.Missiakas D, Schwager F, Betton J-M, Georgopolus C, Raina S. Identification and characterization of HslV HslU (ClpQ ClpY) proteins involved in overall proteolysis of misfolded proteins in Escherichia coli. EMBO J. 1996;15:6899–6909. [PMC free article] [PubMed] [Google Scholar]
- 28.Mogk A, Völker A, Engelmann S, Hecker M, Schumann W, Völker U. Nonnative proteins induce expression of the Bacillus subtilis CIRCE regulon. J Bacteriol. 1998;180:2895–2900. doi: 10.1128/jb.180.11.2895-2900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Msadek T, Dartois V, Kunst F, Herbaud M-L, Denizot F, Rapoport G. ClpP of Bacillus subtilis is required for competence development, degradative enzyme synthesis, motility, growth at high temperature and sporulation. Mol Microbiol. 1998;27:899–914. doi: 10.1046/j.1365-2958.1998.00735.x. [DOI] [PubMed] [Google Scholar]
- 30.Msadek T, Kunst F, Rapoport G. MecB of Bacillus subtilis, a member of the ClpC ATPase family, is a pleiotropic regulator controlling competence gene expression and survival at high temperature. Proc Natl Acad Sci USA. 1994;91:5788–5792. doi: 10.1073/pnas.91.13.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nanamiya H, Ohashi Y, Asai K, Moriya S, Ogasawara N, Fujita M, Sadiae Y, Kawamura F. ClpC regulates the fate of a sporulation initiation sigma factor, ςH protein, in Bacillus subtilis at elevated temperatures. Mol Microbiol. 1998;29:505–513. doi: 10.1046/j.1365-2958.1998.00943.x. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen E, Akita M, Davila-Aponte J, Keegstra K. Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 1997;16:935–946. doi: 10.1093/emboj/16.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parsell D A, Kowall A S, Singer M A, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 34.Riethdorf S, Völker U, Gerth U, Winkler A, Engelmann S, Hecker M. Cloning, nucleotide sequence, and expression of the Bacillus subtilis lon gene. J Bacteriol. 1994;176:6518–6527. doi: 10.1128/jb.176.21.6518-6527.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohrwild M, Coux O, Huang H C, Moerschell R P, Yoo S J, Seol J H, Chung C H, Goldberg A L. HslV-HslU: a novel ATP-dependent protease complex in Escherichia coli related to the eukaryotic proteasome. Proc Natl Acad Sci USA. 1996;93:5808–5813. doi: 10.1073/pnas.93.12.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohrwild M, Pfeifer G, Santarius U, Muller S A, Huang H C, Engel A, Baumeister W, Goldberg A L. The ATP-dependent HslVU protease from Escherichia coli is a four-ring structure resembling the proteasome. Nat Struct Biol. 1997;4:133–139. doi: 10.1038/nsb0297-133. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schirmer E C, Glover J R, Singer M A, Lindquist S. HSP 100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 40.Schmitt M, Neupert W, Langer T. Hsp78, a Clp homologue within mitochondria, can substitute for chaperone functions of mt-hsp70. EMBO J. 1995;14:3434–3444. doi: 10.1002/j.1460-2075.1995.tb07349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin D H, Lee C S, Chung C H, Suh S W. Molecular symmetry of the ClpP component of the ATP-dependent Clp protease, an Escherichia coli homolog of 20 S proteasome. J Mol Biol. 1996;262:71–76. doi: 10.1006/jmbi.1996.0499. [DOI] [PubMed] [Google Scholar]
- 42.Smith I, Paress P, Cabane K, Dubnau E. Genetics and physiology of the rel system of Bacillus subtilis. Mol Gen Genet. 1980;178:271–279. doi: 10.1007/BF00270472. [DOI] [PubMed] [Google Scholar]
- 43.Squires C, Squires C L. The Clp proteins—proteolysis regulators or molecular chaperones? J Bacteriol. 1992;174:1081–1085. doi: 10.1128/jb.174.4.1081-1085.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Squires C L, Pederson S, Ross B M, Squires C. ClpB is the Escherichia coli heat shock protein F84.1. J Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 46.Stülke J, Hanschke R, Hecker M. Temporal activation of β-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J Gen Microbiol. 1993;139:2041–2045. doi: 10.1099/00221287-139-9-2041. [DOI] [PubMed] [Google Scholar]
- 47.Tokuyasu K T. Use of poly(vinylpyrrolidone) and poly(vinyl alcohol) for cryoultramicrotomy. Histochem J. 1989;21:163–171. doi: 10.1007/BF01007491. [DOI] [PubMed] [Google Scholar]
- 48.Torres-Cabassa A S, Gottesmann S. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J Bacteriol. 1987;169:981–989. doi: 10.1128/jb.169.3.981-989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turgay K, Hahn J, Burghorn J, Dubnau D. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 1998;17:6730–6738. doi: 10.1093/emboj/17.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turgay K, Hamoen L, Venema G, Dubnau D. Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes Dev. 1997;11:119–128. doi: 10.1101/gad.11.1.119. [DOI] [PubMed] [Google Scholar]
- 51.Völker U, Engelmann S, Maul B, Riethdorf S, Völker A, Schmid R, Mach H, Hecker M. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology. 1994;140:741–752. doi: 10.1099/00221287-140-4-741. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, Hartling J A, Flanagan J M. The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 53.Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M. The ClpX heat-shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP-ClpX protease, is a novel molecular chaperone. EMBO J. 1995;14:1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi M R. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci USA. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]