Abstract
The Bacillus subtilis sbo-alb operon contains sboA, the structural gene for the bacteriocin subtilosin, and the alb genes required for subtilosin production. Transcription from the sbo-alb promoter is highly induced by oxygen limitation. The transcriptional regulation of the sbo-alb operon is under dual control involving the transition state regulator AbrB and the two-component regulatory proteins ResD and ResE.
The production of bacteriocins is usually induced by a combination of high cell density and low nutrient availability (12, 13, 19). The production of antimicrobial compounds will allow the bacterium to compete for scarce sources of carbon, nitrogen, and energy. Bacteriocins are often produced at high cell density to ensure that a concentration of antibiotic high enough to have an impact on the local environment is achieved. The spore-forming soil bacterium Bacillus subtilis produces an assortment of antimicrobial compounds, including the antilisterial bacteriocin subtilosin (1, 36). The structural gene for subtilosin, sboA, resides at the 5′ end of an operon that contains the alb genes (14, 34; T. Stein, S. Düsterhaus, A. Stroh, and K.-D. Entian, Abstr. 10th Int. Conf. Bacilli, abstr. P103, p. 65, 1999), which are believed to function in subtilosin chemical modification, processing, and export. The regulation of sbo-alb operon expression is complex, since it is induced in late growth cultures apparently in response to starvation and is also dramatically induced by oxygen limitation (29, 34; Stein et al., Abstr. 10th Int. Conf. Bacilli). Many of the factors governing gene expression in response to starvation (6, 8, 20, 28) and oxygen limitation (4, 25, 26, 31) have been identified in B. subtilis. Several genes that are normally expressed in response to nutritional stress are subject to repression by the transition state regulatory protein AbrB (29). The product of the abrB gene binds directly to promoter DNA of the genes that are induced by starvation and prevents their transcription during robust culture growth. We have found that sbo-alb is one of the operons that are negatively controlled by AbrB (34). The abrB gene is negatively controlled by the key transcriptional regulator of starvation-induced genes Spo0A (7, 30).
While several studies of the control of bacteriocin production in response to high cell density and nutritional stress have been reported, there are few that have uncovered induction of bacteriocin synthesis in response to anaerobiosis. Colicin E1 produced by Escherichia coli is the product of the cea gene that is transcriptionally induced under anaerobic conditions (5, 18). Activation of cea transcription requires the FNR protein, an iron-binding transcriptional activator of many anaerobically induced genes (5, 18).
In this paper, we report that anaerobic induction of the sboA-alb genes is only conditionally dependent on FNR but absolutely requires the ResDE signal transduction complex. An examination of the epistatic relationship of the various genes that function in sbo-alb control shows that both the Spo0-AbrB and ResDE systems function independently in the control of sbo-alb transcription.
Transcription of sbo-alb is induced by oxygen limitation.
The sbo-alb operon contains nine genes (sboAX-albABCDEFG) and is transcribed from a ςA-type promoter residing upstream of the sboA gene (34). Although transcription proceeds through the alb genes, there exists a sequence overlapping the end of sboA and internal to the sboX coding sequence that can potentially form a stable hairpin-loop structure which would be expected to impede transcription from the sbo-alb promoter. To study the regulation of sbo-alb expression, two lacZ transcriptional fusions were constructed as previously described (34). One fusion, sboΔBH-lacZ, carries a fragment of the 5′ half of the sboA gene, along with its promoter region, upstream of a promoterless lacZ gene that bears a B. subtilis ribosome-binding site. The plasmid containing the sboΔBH-lacZ fusion was integrated into an SPβ specialized transducing phage. The other lacZ fusion was constructed by inserting a fragment containing a segment of the sbo-alb operon from the middle of albA to the middle of albC into the integrative plasmid pMUTIN2 (33), thereby placing the fragment upstream of the promoterless lacZ gene. Integration of this plasmid, pMUPE1 (34), into the sbo-alb operon creates an (sbo-alb)-lacZ transcriptional fusion that does not disrupt any of the open reading frames within the operon but does disrupt the sbo-alb transcription unit (34).
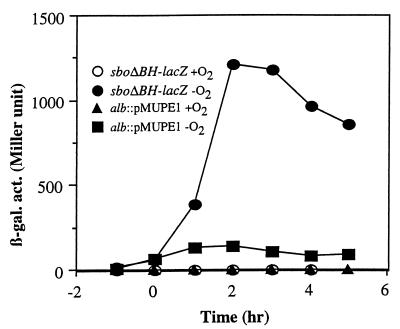
The fusion-bearing derivatives of strain JH642 were grown aerobically and anaerobically in 2xYT medium (23) supplemented with 1% glucose and 0.2% KNO3 as previously described (22). Both fusions were poorly expressed in aerobic cultures (Fig. 1) as previously shown (34), but a dramatic, 600-fold induction of phage-borne sboΔBH-lacZ was observed in anaerobically grown cultures. The alb::pMUPE1 fusion was also induced 40-fold late in the growth curve under anaerobic conditions.
FIG. 1.
Anaerobic expression of sbo-alb and subtilosin production. Expression of sboA-lacZ and alb-lacZ fusions in strains ORB3147 (alb::pMUPE1) and ORB3162 (SPβsboΔBH-lacZ) under aerobic (+O2) and anaerobic (−O2) growth conditions. β-gal. act., β-galactosidase activity.
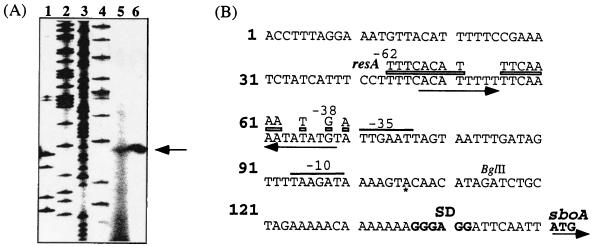
The accelerated expression of sbo-alb in anaerobically grown cells prompted us to determine if transcription from the sbo-alb promoter is stimulated under oxygen limitation and if the same promoter is utilized in anaerobic growth as in aerobic growth. Total RNA was purified, as described in a previous report (24), from JH642 cells collected at T2 of the growth curve (2 h after the end of exponential phase). Cultures were grown aerobically and anaerobically at 37°C in 2 × YT medium supplemented with 1% glucose and 0.2% KNO3. Primer extension was performed as previously described using oligonucleotide osboP4 (34). An at least 10-fold greater amount of primer extension product was produced in reaction mixtures containing RNA from anaerobically grown cells (Fig. 2A, lane 6) than in reaction mixtures containing RNA from aerobically grown cells (lane 5). It was also observed that the same transcriptional start site was utilized under both aerobic and anaerobic conditions.
FIG. 2.
(A) Utilization of the same transcription start site in the sbo-alb promoter region under aerobic and anaerobic conditions. Tenfold less primer extension reaction mixture was applied to lane 6 (RNA from anaerobic cultures) than was applied to lane 5 (RNA from aerobic cultures). Sequencing reactions G, A, T, and C are in lanes 1, 2, 3, and 4, respectively. (B) Nucleotide sequence of the sbo-alb promoter region and similarity to the regulatory region of the resA operon. The ATG start site of the sboA gene and its ribosome-binding site (SD) are shown. The −10 and −35 regions are indicated, as is the transcription start site (∗), and a region of approximate dyad symmetry upstream of the −35 sequence is marked with arrows beneath the nucleotide sequence. A broken bar above the nucleotide sequence extending from −36 to −62 marks the sequence identity between the resA and sbo-alb regulatory regions. The sequence above the bar indicates the nucleotide residues of the corresponding region in the resA promoter region.
Anaerobic induction of sbo-alb requires the ResDE signal transduction system.
The involvement of the known regulators of anaerobic gene control was next examined by measuring the expression of sboΔBH-lacZ in resDE (26, 31), fnr (4, 26), and narGH (21) mutant cells. The narGHJ genes encode subunits of respiratory nitrate reductase (9, 15) and are required for the expression of some anaerobically induced genes in the absence of nitrite (15, 26). These genes also require fnr, since narGHJI transcription is activated by the FNR protein (15, 22, 26). As shown in Table 1, all three mutations abolish sboΔBH-lacZ expression in 2xYT-glucose-nitrate-grown cells under anaerobic conditions. If nitrite (10 mM KNO2) is substituted for nitrate, then expression of sboΔBH-lacZ no longer requires fnr and narGH but still is absolutely dependent on the ResDE system (Table 1).
TABLE 1.
Expression of sboΔBH-lacZ
| Strain | Relevant genotype | β-Galactosidase activity (Miller units)a
|
|
|---|---|---|---|
| Nitrate | Nitrite | ||
| ORB3162 | Wild type | 1,462 | 2,096 |
| ORB3349 | fnr::spc | 42 | 2,182 |
| ORB3350 | ΔnarGH::phleo | 52 | 1,777 |
| ORB3348 | ΔresDE::tet | 7 | 13 |
| ORB3437 | spo0A::erm | 220 | 391 |
| ORB3241 | abrB::neo | 1,681 | NTb |
| ORB3438 | spo0A::erm abrB::neo | 2,042 | NT |
| ORB3384 | ΔresDE::tet abrB::neo | 14 | NT |
Cells were grown anaerobically in 2xYT medium with 1% glucose supplemented with 0.2% KNO3 or 10 mM KNO2. The maximal activity of at least two independent experiments is shown.
NT, not tested.
Transcription of sbo-alb is controlled by two independent regulatory pathways.
The transition state regulator AbrB exerts negative control over transcription from the sbo-alb promoter (34). This form of regulation was reexamined in anaerobically grown cells bearing the sboΔBH-lacZ fusion. A mutation in the spo0A gene (10) which results in overproduction of the abrB product causes a sharp reduction in sboΔBH-lacZ expression in cells grown anaerobically (Table 1). The effect of a spo0A mutation is overcome by introducing a mutation in abrB (34) (strain ORB3438, Table 1). Since the temporal regulation of sbo-alb expression is still observed in the abrB and spo0A abrB mutants (data not shown), ResD might be activated only after exponential growth, probably as a result of nitrite accumulation. However, we cannot rule out the possibility that another regulator is involved in the postexponential induction of sboA-alb.
ResDE, like the Spo0 system, could positively regulate sbo-alb transcription by repressing the abrB gene or indirectly affect AbrB concentration and/or activity. If this were the case, then we would expect an abrB null mutation to result in sbo-alb expression in the absence of ResDE. Examination of sboΔBH-lacZ expression in the resDE abrB double mutant showed that the ResDE system is still required for sbo-alb transcription in the absence of AbrB protein (Table 1).
To further investigate the relationship among Spo0A, AbrB, and ResDE, the expression of ResDE-controlled genes was examined in spo0A, abrB, and spo0A abrB mutant cells bearing either an fnr (26)-, an hmp (flavohemoglobin) (15)-, or an nasD (nitrite reductase) (22)-lacZ construct, each of which requires ResDE for expression. If expression of resDE or the activity of their products required spo0A or was repressed by AbrB, then a spo0A mutation would have resulted in reduced expression of the resDE-controlled lacZ fusions. No effect of a spo0A or an abrB mutation on lacZ expression was detected (data not shown), indicating that Spo0A and AbrB do not influence the activity of ResDE in anaerobically grown cells. The addition of nitrite, which is thought to be necessary for ResDE-dependent activation of anaerobically induced genes, did not significantly overcome the inhibition of sbo-alb transcription caused by AbrB overproduction in a spo0A mutant background (Table 1). These results show that the Spo0-AbrB and ResDE systems of control operate independently of each other. An earlier observation that membrane-bound nitrate reductase was hyperactive in Spo0 mutants (3) was explained by possible membrane alteration in the mutant strains; however, an alternative possibility that AbrB is a positive regulator of the narGHJI operon is worth examining.
The results presented herein and those from previously published studies (34) provide compelling evidence that Spo0-AbrB- and ResDE-dependent mechanisms of control are exerted at the level of sbo-alb transcription initiation. Inspection of the sbo-alb regulatory region reveals a sequence extending from −37 to −62 that contains a region of approximate dyad symmetry and sequences identical to the −37 to −62 sequence within the regulatory region of the resA operon which is also controlled by ResDE (Fig. 2B) (2, 31). Access to the sbo-alb promoter region may be blocked by the AbrB protein early in the growth curve. Following Spo0A repression of abrB when cells approach stationary phase, ResD, activated as nitrite accumulates in the cell, will gain access to the region upstream of the promoter −35 sequence, where it will interact with RNA polymerase to stimulate transcription initiation.
Under aerobic growth conditions, Spo0A-P will accumulate as cultures approach stationary phase due to the activities of KinA, -B, -C, and -D, all of which are histidine protein kinases that donate high-energy phosphate to the Spo0 phosphorelay (6, 7, 11, 16, 17, 27, 32). The initial phosphorylation of Spo0A is thought to depend on KinC (16, 17). This results in a level of Spo0A-P sufficient for repression of the abrB gene. KinC has been shown to phosphorylate Spo0A in the absence of other Spo0 phosphorelay components, but its preferred target is believed to be Spo0F (11, 17). It seems that the KinC-Spo0 system is functional in anaerobically grown B. subtilis cells in which sbo-alb expression is induced. However, B. subtilis cells do not readily undergo sporulation late in the growth curve when grown anaerobically in sporulation medium (data not shown; 9). It is not known if the other kinases that supply the Spo0 phosphorelay with high-energy phosphate are functional in anaerobically grown cells.
Although sbo-alb undergoes significant transcriptional induction when cells encounter anaerobic conditions, this is not accompanied by a proportional increase in antilisterial activity, at least not under the growth conditions used in this study (data not shown). However, an increase in the anaerobic production of subtilosin has been reported (Stein et al., 10th Int. Conf. Bacilli). Mutations in sboA or in any of the alb genes do not significantly impair anaerobic growth (data not shown). It is not obvious why the cell would benefit from the massive increase in sbo-alb expression during anaerobiosis. It is possible that we have not used the appropriate anaerobic growth conditions required for active subtilosin production. It is also possible that the cell accumulates inactive precursors of subtilosin, which then undergo oxygen-dependent modifications to yield an active peptide when an aerobic environment is encountered. A greater knowledge of sbo-alb gene products and their function, as well as an examination of other antimicrobial production systems in anaerobically grown Bacillus species, may provide clues as to the role bacteriocins play in the anaerobic life of bacteria.
Acknowledgments
Support for this work was provided by grant GM45898 from the National Institutes of Health (to P.Z.), grant MCB-9996014 from the National Science Foundation (to M.M.N.), and a grant from The Oregon Research Foundation. P.Z. gratefully acknowledges support from E. I. du Pont de Nemours, Inc.
REFERENCES
- 1.Babasaki K, Takao T, Shimonishi Y, Kurahashi K. Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: isolation, structural analysis, and biogenesis. J Biochem. 1985;98:583–603. doi: 10.1093/oxfordjournals.jbchem.a135315. [DOI] [PubMed] [Google Scholar]
- 2.Birkey S M, Liu W, Zhang X, Duggan M F, Hulett F M. Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol Microbiol. 1998;30:943–953. doi: 10.1046/j.1365-2958.1998.01122.x. [DOI] [PubMed] [Google Scholar]
- 3.Bohin J-P, Bohin A, Schaeffer P. Increased nitrate reductase A activity as a sign of membrane alteration in early blocked asporogenous mutants of Bacillus subtilis. Biochimie. 1976;58:99–108. doi: 10.1016/s0300-9084(76)80360-4. [DOI] [PubMed] [Google Scholar]
- 4.Cruz-Ramos H, Boursier L, Moszer I, Kunst F, Danchin A, Glaser P. Anaerobic transcription activation in Bacillus subtilis: identification of distinct FNR-dependent and -independent regulatory mechanisms. EMBO J. 1995;14:5984–5994. doi: 10.1002/j.1460-2075.1995.tb00287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eraso J M, Weinstock G M. Anaerobic control of colicin E1 production. J Bacteriol. 1992;174:5101–5109. doi: 10.1128/jb.174.15.5101-5109.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grossman A D. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu Rev Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- 7.Hoch J A. Control of cellular development in sporulating bacteria by the phosphorelay two-component signal transduction system. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 129–144. [Google Scholar]
- 8.Hoch J A. spo0 genes, the phosphorelay, and the initiation of sporulation. In: Hoch J A, Sonenshein A L, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 747–755. [Google Scholar]
- 9.Hoffmann T, Troup B, Szabo A, Hungerer C, Jahn D. The anaerobic life of Bacillus subtilis: cloning of the genes encoding the respiratory nitrate reductase system. FEMS Microbiol Lett. 1995;131:219–225. doi: 10.1111/j.1574-6968.1995.tb07780.x. [DOI] [PubMed] [Google Scholar]
- 10.Ireton K, Rudner D Z, Siranosian K J, Grossman A D. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 1993;7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- 11.Jiang M, Tzeng Y L, Feher V A, Perego M, Hoch J A. Alanine mutants of the Spo0F response regulator modifying specificity for sensor kinases in sporulation initiation. Mol Microbiol. 1999;33:389–395. doi: 10.1046/j.1365-2958.1999.01481.x. [DOI] [PubMed] [Google Scholar]
- 12.Katz E, Demain A L. The peptide antibiotics of Bacillus: chemistry, biogenesis, and possible functions. Bacteriol Rev. 1977;41:449–474. doi: 10.1128/br.41.2.449-474.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleerebezem M, Quadri L E N, Kuipers O P, de Vos W M. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 14.Kunst F, Ogasawara N, Moszer I, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 15.LaCelle M, Kumano M, Kurita K, Yamane K, Zuber P, Nakano M M. Oxygen-controlled regulation of the flavohemoglobin gene in Bacillus subtilis. J Bacteriol. 1996;178:3803–3808. doi: 10.1128/jb.178.13.3803-3808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ledeaux J R, Grossman A D. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J Bacteriol. 1995;177:166–175. doi: 10.1128/jb.177.1.166-175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ledeaux J R, Yu N, Grossman A D. Different roles for KinA, KinB, and KinC in the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1995;177:861–863. doi: 10.1128/jb.177.3.861-863.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malkhosyan S R, Panchenko Y A, Rekesh A N. A physiological role for DNA supercoiling in the anaerobic regulation of colicin gene expression. Mol Gen Genet. 1991;225:342–345. doi: 10.1007/BF00269868. [DOI] [PubMed] [Google Scholar]
- 19.Marahiel M A, Nakano M M, Zuber P. Regulation of peptide antibiotic production in Bacillus. Mol Microbiol. 1993;7:631–636. doi: 10.1111/j.1365-2958.1993.tb01154.x. [DOI] [PubMed] [Google Scholar]
- 20.Msadek T. When the going gets tough: survival strategies and environmental signaling networks in Bacillus subtilis. Trends Microbiol. 1999;7:201–207. doi: 10.1016/s0966-842x(99)01479-1. [DOI] [PubMed] [Google Scholar]
- 21.Nakano M M, Dailly Y P, Zuber P, Clark D P. Characterization of anaerobic fermentative growth of Bacillus subtilis: identification of fermentation end products and genes required for growth. J Bacteriol. 1997;179:6749–6755. doi: 10.1128/jb.179.21.6749-6755.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakano M M, Hoffmann T, Zhu Y, Jahn D. Nitrogen and oxygen regulation of Bacillus subtilis nasDEF encoding NADH-dependent nitrite reductase by TnrA and ResDE. J Bacteriol. 1998;180:5344–5350. doi: 10.1128/jb.180.20.5344-5350.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakano M M, Marahiel M A, Zuber P. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J Bacteriol. 1988;170:5662–5668. doi: 10.1128/jb.170.12.5662-5668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakano M M, Xia L, Zuber P. Transcription initiation region of the srfA operon which is controlled by the comP-comA signal transduction system in Bacillus subtilis. J Bacteriol. 1991;173:5487–5493. doi: 10.1128/jb.173.17.5487-5493.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakano M M, Zuber P. Anaerobic growth of a “strict aerobe.”. Annu Rev Microbiol. 1998;52:165–190. doi: 10.1146/annurev.micro.52.1.165. [DOI] [PubMed] [Google Scholar]
- 26.Nakano M M, Zuber P, Glaser P, Danchin A, Hulett F M. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis. J Bacteriol. 1996;178:3796–3802. doi: 10.1128/jb.178.13.3796-3802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perego M, Cole S P, Burbulys D, Trach K, Hoch J A. Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis. J Bacteriol. 1989;171:6187–6196. doi: 10.1128/jb.171.11.6187-6196.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith I. Regulatory proteins that control late-growth development. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: physiology, biochemistry, and molecular biology. Washington, D.C.: American Society for Microbiology; 1993. pp. 785–800. [Google Scholar]
- 29.Strauch M A. AbrB, a transition state regulator. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: physiology, biochemistry, and molecular biology. Washington, D.C.: American Society for Microbiology; 1993. pp. 757–764. [Google Scholar]
- 30.Strauch M A, Webb V, Speigelman B, Hoch J A. The Spo0A protein of Bacillus subtilis is a repressor of the abrB gene. Proc Natl Acad Sci USA. 1990;87:1801–1805. doi: 10.1073/pnas.87.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun G, Sharkova E, Chesnut R, Birkey S, Duggan M F, Sorokin A, Pujic P, Ehrlich S D, Hulett F M. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J Bacteriol. 1996;178:1374–1385. doi: 10.1128/jb.178.5.1374-1385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trach K, Hoch J A. Multisensory activation of the phosphorelay initiating sporulation in Bacillus subtilis: identification and sequence of the protein kinase of the alternate pathway. Mol Microbiol. 1993;8:69–79. doi: 10.1111/j.1365-2958.1993.tb01204.x. [DOI] [PubMed] [Google Scholar]
- 33.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 34.Zheng G, Yan L Z, Vederas J C, Zuber P. Genes of the sbo-alb locus of Bacillus subtilis are required for production of the antilisterial bacteriocin subtilosin. J Bacteriol. 1999;181:7346–7355. doi: 10.1128/jb.181.23.7346-7355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuber P, Nakano M M, Marahiel M A. Peptide antibiotics. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: physiology, biochemistry, and molecular biology. Washington, D.C.: American Society for Microbiology; 1993. pp. 897–916. [Google Scholar]