Objective:
We investigated the prevalence of preexisting M184V/I and associated risk factors among clinical trial participants with suppressed HIV and evaluated the impact of M184V/I on virologic response after switching to bictegravir/emtricitabine/tenofovir alafenamide (B/F/TAF).
Design:
Participant data were pooled from six clinical trials investigating the safety and efficacy of switching to B/F/TAF in virologically suppressed people with HIV.
Methods:
Preexisting drug resistance was assessed by historical genotypes and/or baseline proviral DNA genotyping. Virologic outcomes were determined by last available on-treatment HIV-1 RNA. Stepwise selection identified potential risk factors for M184V/I in a multivariate logistic regression model.
Results:
Altogether, 2034 participants switched treatment regimens to B/F/TAF and had follow-up HIV-1 RNA data, and 1825 of these participants had baseline genotypic data available. Preexisting M184V/I was identified in 182 (10%), mostly by baseline proviral DNA genotype (n = 167). Most substitutions were M184V (n = 161) or M184V/I mixtures (n = 10). Other resistance substitutions were often detected in addition to M184V/I (n = 147). At last on-treatment visit, 98% (179/182) with preexisting M184V/I and 99% (2012/2034) of all B/F/TAF-treated participants had HIV-1 RNA less than 50 copies/ml, with no treatment-emergent resistance to B/F/TAF. Among adult participants, factors associated with preexisting M184V/I included other resistance, black race, Hispanic/Latinx ethnicity, lower baseline CD4+ cell count, advanced HIV disease, longer duration of antiretroviral therapy, and greater number of prior third agents.
Conclusion:
M184V/I was detected in 10% of virologically suppressed clinical trial participants at study baseline. Switching to B/F/TAF demonstrated durable efficacy in maintaining viral suppression, including in those with preexisting M184V/I.
Keywords: antiretroviral therapy, bictegravir, drug resistance, HIV-1, M184V
Introduction
For people with HIV (PWH) receiving antiretroviral treatment (ART) that includes nucleoside reverse transcriptase inhibitors (NRTIs) lamivudine (3TC) or emtricitabine (FTC), the M184V and M184I substitutions in reverse transcriptase occur rapidly with incomplete suppressive therapy [1]. M184V/I confers high-level resistance to 3TC and FTC, decreases susceptibility to abacavir (ABC) and didanosine, and increases susceptibility to tenofovir and zidovudine [2]. In PWH who experienced virologic failure on 3TC, M184I typically emerges first, because of a more common nucleotide substitution, only to be replaced by M184V, which results in virus with higher replicative fitness [3–6]. Consequently, in clinical practice, M184V is more often detected after virologic failure on 3TC-containing or FTC-containing regimens than M184I, and has been observed in up to 71% of PWH who fail their first line therapy [7–9]. However, in cases of rilpivirine-based therapy failure, emergent M184I is more common [10], highlighting the clinical relevance of both M184V and M184I substitutions.
As 3TC and FTC have been part of most recommended regimens for decades, the prevalence of M184V/I among PWH is high. Nonetheless, M184V/I is often underestimated in clinical practice. In the absence of drug pressure, M184V/I variants are replaced by wild-type virus in the circulating quasispecies and are no longer able to be detected by routine plasma HIV genotyping [11,12]. The replacement of M184V/I with wild-type also occurs in the context of transmission: M184V/I is detected in ∼4% of acute HIV infections [7,13–21], but only ∼1% of newly diagnosed PWH by Sanger sequencing-based genotyping [22,23]. Despite this decline in circulating virus, drug-resistant variants are archived in the latent viral reservoir and can reemerge under therapeutic selective pressure [24,25]. Given the nature of HIV integration into CD4+ T cells, mutations are likely present for life.
The single tablet regimen bictegravir/FTC/tenofovir alafenamide (B/F/TAF) is a guideline-recommended initial therapy for HIV [26–28]. It is also approved as a replacement regimen when switching therapies in virologically suppressed PWH who have no known substitutions associated with resistance to its individual components [29]. Given the high prevalence of M184V/I and widespread use of B/F/TAF in clinical practice, whether suppressed PWH with this substitution can be switched to B/F/TAF is of interest. We summarize here the results of a comprehensive review of PWH enrolled in clinical trials of switching therapy to B/F/TAF, with a focus on those with M184V/I.
Methods
Participants and study design
Participants included in this analysis were enrolled in one of the following six clinical trials: studies 4030, 4580 (BRAAVE 2020), 1844, 1878, 4449, or 1474 (ClinicalTrials.gov NCT03110380, NCT03631732, NCT02603120, NCT02603107, NCT03405935, NCT02881320, respectively) [30–35]. At screening, participants were virologically suppressed (HIV-1 RNA <50 copies/ml for 3 or 6 months) on a three-drug antiretroviral regimen (not counting pharmacoenhancing drugs). Additional details on trial design and population age groups can be found in Table 1. Studies 4030 and 4580 permitted the enrollment of participants with documented M184V/I, whereas studies 1844, 1878, 4449, and 1474 excluded those with known or suspected M184V/I. All studies excluded substitutions associated with bictegravir resistance, and all but Study 4030 excluded other substitutions associated with FTC or TAF resistance, if known prior to enrollment. All trials were undertaken in accordance with the Declaration of Helsinki and approved by review boards or ethics committees. All participants provided written informed consent.
Table 1.
Overview of bictegravir/emtricitabine/tenofovir alafenamide switch studies in virologically suppressed people with HIV.
| All studies | Study 4030 | Study 4580 | Study 1844 | Study 1878 | Study 4449 | Study 1474 | |
| Screening resistance criteria: M184V/I | – | Allowed | Allowed | Excluded | Excluded | Excluded | Excluded |
| Screening resistance criteria: bictegravir-associated | – | Excluded | Excluded | Excluded | Excluded | Excluded | Excluded |
| Screening resistance criteria: TAF-associated | – | Allowed | Excluded | Excluded | Excluded | Excluded | Excluded |
| Baseline antiretroviral regimena | – | DTG + either FTC/TDF or FTC/TAF | Any 3rd agent + 2 NRTIs | DTG/ABC/3TC (single or multiple tablets) | Boosted DRV or ATV + either FTC/TDF or ABC/3TC | EVG/COBI/ FTC/TAF or any 3rd agent + FTC/TDF | Any 3rd agent + 2 NRTIs |
| Trial design | – | Double-blind placebo-controlled randomized 1 : 1 switch to B/F/TAF or DTG + FTC/TAF | Open-label randomized 2 : 1 switch to B/F/TAF or stay on baseline regimen | Double-blind placebo-controlled randomized 1 : 1 switch to B/F/TAF or DTG/ABC/3TC | Open-label randomized 1 : 1 switch to B/F/TAF or stay on baseline regimen | Open-label single arm switch to B/F/TAF | Open-label single arm switch to B/F/TAF |
| Participants enrolled (n) | 2386 | 565 | 495 | 563 | 577 | 86 | 100 |
| Median age (criteria for study) (years) | 48 | 51 (≥18) | 49 (≥18) | 46 (≥18) | 48 (≥18) | 69 (≥65) | 12 (6-<18) |
| Median time since ART initiation (IQR) (years) | 8.3 (4.3–15.4) | 10.1 (4.4–18.7) | 10.4 (5.9–17.3) | 5.5 (2.7–10) | 7.7 (4.1–14.0) | 14.9 (6.9–19.3) | 10.1 (7.4–11.4) |
| Participants switched to B/F/TAF (n) | 2044 | 284 | 493b | 547b | 534b | 86 | 100 |
| Median B/F/TAF treatment duration (IQR) (weeks) | 72 (51–102) | 59 (53–63) | 71 (48–72) | 96 (49–119) | 101 (72–120) | 96 (95–96) | 50 (30–52) |
| Participants included in LOCF analysisc (n) | 2034 | 283 | 489 | 545 | 532 | 85 | 100 |
| Timepoint for LOCF analysis | - | Week 48 | Week 72/48d | End of study | End of study | Week 96 | Week 48/24e |
| HIV-1 RNA <50 copies/ml at last visit by LOCF, % (n/N) | 99% (2012/2034) | >99% (282/283) | 99% (486/489) | 98% (535/545) | 99% (525/532) | 100% (85/85) | 99% (99/100) |
| Baseline PR/RT genotype available, % (n/N) | 90% (1825/2034) | 84% (237/283) | 98% (468/489) | 96% (522/545) | 94% (498/532) | 98% (83/85) | 17% (17/100) |
| Baseline M184V/I, % (n/N) | 10% (182/1825) | 20% (47/237) | 11% (50/468) | 3% (17/522) | 12% (62/498) | 4% (3/83) | 18% (3/17) |
| Baseline M184V/I + ≥1 other resistance substitution, % (n/N) | 81% (147/182) | 72% (34/47) | 88% (44/50) | 82% (14/17) | 79% (49/62) | 100% (3/3) | 100% (3/3) |
| M184V/I HIV-1 RNA <50 copies/ml at last visit by LOCF, % (n/N) | 98% (179/182) | 100% (47/47) | 100% (50/50) | 100% (17/17) | 95% (59/62) | 100% (3/3) | 100% (3/3) |
| Treatment emergent resistance to B/F/TAF (n) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
ART, antiretroviral therapy; B/F/TAF, bictegravir/emtricitabine/tenofovir alafenamide; IQR, interquartile range; PR/RT, protease/reverse transcriptase.
Baseline antiretroviral regimens consisted of two nucleoside reverse transcriptase inhibitors (NRTIs), such as abacavir (ABC) and lamivudine (3TC) or emtricitabine (FTC) and tenofovir disoproxil fumarate (TDF) or tenofovir alafenamide (TAF), and a third agent, such as dolutegravir (DTG), darunavir (DVR), atazanavir (ATV) or elvitegravir boosted by cobicistat (EVG/COBI).
Participants switched to B/F/TAF at baseline (4580: n = 330, 1844: n = 282, 1878: n = 290) or at weeks 24 (4580: n = 163) or 48 (1844: n = 265, 1878: n = 244).
Virologic outcomes based on last available on-treatment postswitch HIV-1 RNA using last observation carried forward (LOCF) imputation were determined for participants who switched to B/F/TAF and had at least one postswitch on-treatment HIV-1 RNA measurement.
Participants included in the LOCF analysis had outcomes determined at week 72 (n = 327 switched at baseline) or week 48 (n = 162 switched at week 24).
Participants included in the LOCF analysis had outcomes determined at week 48 (n = 75) or week 24 (n = 25) based on duration of B/F/TAF treatment at time of analysis.
Resistance analyses
Baseline resistance was assessed using two methods. First, historical HIV-1 genotypes from prior plasma HIV-1 RNA population sequencing (or from prior proviral DNA sequencing in a minority of reports) were collected if available at enrollment, and resistance-associated substitutions in protease, reverse transcriptase and integrase were recorded. Second, HIV-1 proviral DNA genotyping using the GenoSure Archive (Monogram Biosciences, South San Francisco, CA, USA) assay was performed retrospectively on baseline/day 1 samples from all adult participants with available samples and from select pediatric participants. For the GenoSure Archive assay, cell-associated HIV DNA was deep-sequenced, then bioinformatics filters removed APOBEC-mediated hypermutated sequences, and a consensus sequence was reported (hereafter referred to as BL-DNA genotype). Composite baseline sequences were derived from cumulative historical and/or BL-DNA genotypic data. As data on preexisting resistance were obtained at or after enrollment in the trials, some participants had exclusionary drug resistance substitutions detected after study drugs were initiated. All participants found to have preexisting resistance to any component of B/F/TAF were allowed to continue B/F/TAF and were included in all efficacy analyses.
Testing for resistance development was performed using the PhenoSenseGT, GeneSeq Integrase, and PhenoSense Integrase assays (Monogram Biosciences) for participants with HIV-1 RNA 200 copies/ml or greater at study endpoints, last on-treatment visit, or the visit following HIV-1 RNA 50 copies/ml or greater (confirmed virologic failure), without resuppression of HIV-1 RNA to less than 50 copies/ml. Drug resistance substitutions were adapted from IAS-USA [36].
Outcomes
All studies had postbaseline visits at weeks 4 and 12 and then every 12 weeks thereafter. Plasma HIV-1 RNA levels were measured using Roche TaqMan 2.0 (Roche Diagnostics, Rotkreuz, Switzerland). Efficacy was assessed for all participants who switched to B/F/TAF. The proportion of participants with plasma HIV-1 RNA less than 50 copies/ml at each timepoint was calculated by imputing missing as excluded (M = E). Additionally, the proportions of participants with last available on-treatment HIV-1 RNA less than 50 copies/ml and greater than or equal to 50 copies/ml were determined for all participants with at least one postswitch HIV-1 RNA measurement using the last observation carried forward imputation.
Statistical analysis
Potential factors associated with preexisting M184V/I were assessed by a multivariate logistic regression model with stepwise significance levels for entry and retention specified as 0.20 and 0.05, respectively, and adjusted for study-specific effects. All participants in the B/F/TAF and comparator treatment groups with baseline genotypic data from the adult trials (studies 4030, 4580, 1844, 1878, and 4449) were included. In the model, the dependent variable was baseline M184V/I status (yes/no) and independent baseline variables were intrinsic factors (groups of age, sex at birth, race, ethnicity, region, BMI category, and chronic kidney disease stage), HIV-specific factors (CD4+ cell count category, HIV-1 RNA category, HIV acquisition risk factor, HIV disease status, time since ART start, prior antiretroviral third agent class and number, and duration of baseline ART), and other preexisting resistance categories [NRTI other than M184V/I, protease inhibitor (PI), nonnucleoside reverse transcriptase inhibitor (NNRTI), or integrase strand transfer inhibitor (INSTI)].
Results
Study population and pooled bictegravir/emtricitabine/tenofovir alafenamide efficacy
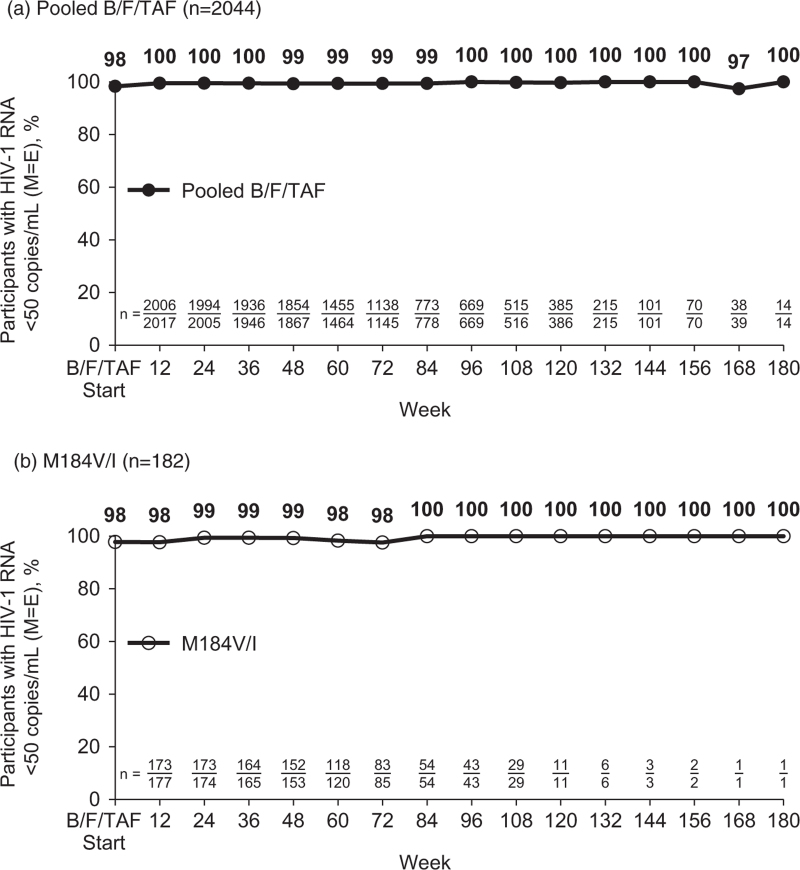
In these six clinical trials evaluating switching therapy to B/F/TAF, a total of 2386 participants were randomized and treated, and 2044 (86%) switched to B/F/TAF (1372 at baseline/day 1 and 672 after 24 or 48 weeks of continuing their baseline regimen) (Table 1). Median B/F/TAF treatment duration was 72 weeks (IQR 51–102 weeks); 92% (1888/2044) received B/F/TAF for at least 48 weeks and 33% (674/2044) for at least 96 weeks. The proportion of participants with HIV-1 RNA less than 50 copies/ml ranged from 97 to 100% at all study visits through a maximum of 180 weeks after B/F/TAF switch by M = E analysis (Fig. 1a). A total of 2034 B/F/TAF-treated participants had at least one postswitch on-treatment HIV-1 RNA measurement, and 99% (2012/2034) were virologically suppressed at their last study visit. Seven participants met criteria for resistance testing, and none had treatment emergent resistance to B/F/TAF.
Fig. 1.
Virologic suppression on bictegravir/emtricitabine/tenofovir alafenamide by missing = excluded (M = E).
(a) All B/F/TAF-treated participants (n = 2044), including those in the last observation carried forward (LOCF) analysis (n = 2034) and those with baseline/day 1 HIV-1 RNA data only (n = 10). (b) B/F/TAF-treated participants with baseline M184V/I (n = 182). B/F/TAF, bictegravir/emtricitabine/tenofovir alafenamide.
Baseline resistance: pooled bictegravir/emtricitabine/tenofovir alafenamide
Historical genotypes were available for 47% (956/2034) of B/F/TAF-treated participants with postswitch on-treatment viral load data (97% were from plasma RNA, 3% were from proviral DNA), and baseline DNA genotypes were available for 84% (1712/2034). Altogether, cumulative baseline genotypic data from historical and/or BL-DNA genotypes were obtained from 90% (1825/2034) of participants for protease/reverse transcriptase and from 85% (1731/2034) for integrase. Preexisting primary NRTI, NNRTI, PI, and INSTI resistance substitutions were detected in 16% (288/1825), 22% (397/1825), 11% (201/1825), and 2% (30/1731), respectively (Supplemental Table 1).
Preexisting M184V and/or M184I was detected in 10% (182/1825) of B/F/TAF-treated participants. Of those with M184V/I, M184V was far more common than M184I: 88% (161/182) had a V substitution only, 6% (11/182) had an I substitution only, and 5% (10/182) had a mixture of V and I (Table 2). M184V/I was the only drug resistance substitution detected in 19% (35/182) and was present with at least one other resistance substitution in 81% (147/182). Resistance (-R) substitutions detected in addition to M184V/I were other NRTI-R in 47% (86/182), including K65R in 4% (8/182) and at least one thymidine analog mutation (TAM) in 40% (72/182), NNRTI-R in 53% (97/182), and PI-R in 27% (50/182). M184V/I with a primary INSTI-R substitution (E92G, Y143H, Q148H, or N155H) was detected in 2% (4/182).
Table 2.
Preexisting M184V/I and presence with other resistance substitutions in bictegravir/emtricitabine/tenofovir alafenamide-treated participants.
| Category | Baseline genotype of participants with preexisting M184V/I | Pooled B/F/TAF (n = 182) |
| M184 substitutions | M184V only | 88% (161) |
| M184I only | 6% (11) | |
| M184V and M184I mixture | 5% (10) | |
| Other resistance | M184V/I alone (no other resistance substitution) | 19% (35) |
| M184V/I + ≥1 other resistance substitution | 81% (147) | |
| Other NRTI-R | M184V/I + other NRTI-Ra | 47% (86) |
| M184V/I + K65R/N | 4% (8) | |
| M184V/I + ≥1 TAMb | 40% (72) | |
| M184V/I + 1–2 TAMs | 18% (33) | |
| M184V/I + ≥3 TAMs | 21% (39) | |
| M184V/I + ≥3 TAMs including M41L and/or L210W | 14% (26) | |
| M184V/I + K70E, L74I/V, Y115F, and/or Q151Mc | 15% (27) | |
| NNRTI-R | M184V/I + NNRTI-Rd | 53% (97) |
| M184V/I + K103N/S | 37% (67) | |
| M184V/I + RPV-Re | 27% (50) | |
| M184V/I + E138A/K/Rf | 7% (12) | |
| PI-R | M184V/I + PI-Rg | 27% (50) |
| INSTI-R | M184V/I + primary INSTI-Rh | 2% (4) |
Data is % (n).
B/F/TAF, bictegravir/emtricitabine/tenofovir alafenamide; INSTI, integrase strand transfer inhibitor; NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; PI, protease inhiitor.
NRTI resistance (-R) substitutions were K65R/E/N, T69 insertions, K70E, L74V/I, Y115F, Q151M, M184V/I, and thymidine analog mutations (TAMs; M41L, D67N, K70R, L210W, T215F/Y, and K219E/N/Q/R) in reverse transcriptase (RT).
TAMs present with M184V/I (alone or with other substitutions) were M41L (n = 38), D67N (n = 35), K70R (n = 39), L210W (n = 21), T215Y/F (n = 36), K219E/N/Q/R (n = 28).
Other NRTI resistance (-R) substitutions present with M184V/I (alone or with other substitutions) were K70E (n = 3), L74I/V (n = 18), Y115F (n = 10), Q151M (n = 3).
NNRTI-R substitutions were L100I, K101E/P, K103N/S, V106M/A, V108I, E138A/G/K/Q/R, V179L, Y181C/I/V, Y188C/H/L, G190A/E/Q/S, H221Y, P225H, F227C and M230L/I in RT.
Rilpivirine resistance (RPV-R) substitutions were L100I, K101E/P, E138A/G/K/Q/R, V179L, Y181C/I/V, Y188L, H221Y, F227C and M230I/L in RT.
Five participants had an M184I substitution with E138A/K/R.
PI-R substitutions were D30N, V32I, M46I/L, I47V/A, G48V, I50V/L, I54M/L, Q58E, T74P, L76V, V82A/F/L/S/T, N83D, I84V, N88S and L90M in protease.
Primary INSTI-R substitutions were T66I/A/K, E92Q/G, F121Y, Y143R/H/C, S147G, Q148H/K/R, N155H/S and R263K in integrase. Primary INSTI-R substitutions present with M184V/I (alone or with other substitutions) were E92G, Y143H, Q148H, N155H (n = 1, each). Integrase data were not available for four participants and are imputed as wild-type.
Detection of M184V/I (bictegravir/emtricitabine/tenofovir, M184V/I n = 182)
Of the 182 participants with baseline M184V/I, 95% (173) had M184V/I detected either at baseline/day 1 (167 participants) or within 5 years of study enrollment (six participants), indicating that M184V/I was present in the viral archive at the time of B/F/TAF switch. M184V/I was detected in 4% (38/956) of historical genotypes and in 10% (167/1712) of BL-DNA genotypes (Supplemental Table 2). Most cases of BL-DNA M184V/I detection occurred in participants without historical data. Of the 38 participants who enrolled with historical M184V/I, BL-DNA genotyping also detected M184V/I in 61% (23/38). Of the 918 participants with historical wild-type M184 and 869 participants with no historical data, M184V/I was discovered by BL-DNA genotyping in 3% (31/918) and 13% (113/869), respectively (Supplemental Table 3).
Outcomes on bictegravir/emtricitabine/tenofovir in participants with preexisting M184V/I (bictegravir/emtricitabine/tenofovir, M184V/I n = 182)
The 182 participants with preexisting M184V/I received B/F/TAF for a median duration of 69 weeks (IQR 50–96). By M = E analysis, the proportion of participants with HIV-1 RNA less than 50 copies/ml ranged from 98 to 100% at all study visits through a maximum of 180 weeks after B/F/TAF switch (Fig. 1b). At last on-treatment visit, 98% (179/182) with M184V/I had HIV-1 RNA less than 50 copies/ml compared with 99% (1803/1825) of those with baseline data and 99% (1624/1643) of those with wild-type M184 (P = 0.49 and 0.48, respectively, by Fisher's exact test) (Fig. 2). When analyzed by the presence of M184V/I alone or in combination with other resistance substitutions, virologic suppression at last visit ranged from 97 to 100%. All 12 participants with M184V/I and K65R or primary INSTI resistance were suppressed at last visit. When analyzed by M184V/I detection type, 98% (164/167) with BL-DNA M184V/I detection and 100% (38/38) with historical M184V/I were virally suppressed at last visit.
Fig. 2.
Virologic suppression on bictegravir/emtricitabine/tenofovir alafenamide at last on-treatment visit by last observation carried forward (LOCF).
Three participants with preexisting M184V/I from study 1878 (switch from boosted PI with two NRTIs) had HIV-1 RNA greater than 50 copies/ml at their last study visit while receiving B/F/TAF. Two had preexisting M184V with another resistance substitution (K70R or K103N) and one had an M184V/I mixture only. All three cases of M184V/I were detected by BL-DNA genotyping. Two participants had HIV-1 RNA less than 100 copies/ml at last visit, which did not meet resistance testing criteria: the participant with M184V and K103N virologically suppressed to HIV-1 RNA less than 50 copies/ml on commercial B/F/TAF and the participant with M184V/I suppressed on a regimen of ritonavir-boosted atazanavir with FTC/TDF. The third participant who had M184V and K70R experienced confirmed virologic failure with HIV-1 RNA 2860 copies/ml after documented poor adherence and undetectable plasma bictegravir levels; there were no new resistance substitutions. This participant subsequently switched regimens to rilpivirine with cobicistat-boosted darunavir and achieved virologic suppression.
Baseline factors associated with preexisting M184V/I (adults only, all treatment groups, M184V/I, n = 216)
For the analysis of factors associated with M184V/I, all adult participants with baseline data, including those not treated with B/F/TAF, were included (2079 participants); preexisting M184V/I was detected in 216 (10%). At baseline, participants with M184V/I were median 52.5 years old, 78% male, 47% black, 19% Hispanic/Latinx, and had median CD4+ cell count of 638.5 cells/μl (Supplemental Table 4). For 99% (214/216), baseline regimens at study entry consisted of two NRTIs with a third agent [61% (132/216) INSTI, 33% (72/216) boosted PI, 5% (10/216) NNRTI]; one participant was on a four-drug baseline regimen (boosted PI, INSTI, FTC/TAF), and one participant had missing data. All baseline NRTI backbones consisted of FTC or 3TC: 41% (89/216) FTC/TAF, 40% (87/216) FTC/TDF, or 18% (39/216) ABC/3TC.
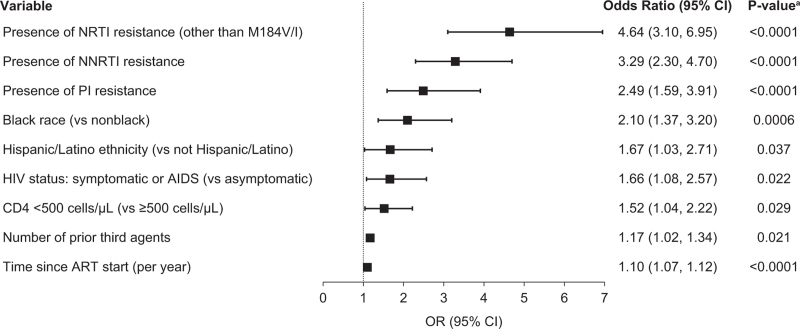
By multivariate logistic regression model (adjusted for study effect), the presence of NRTI-R (other than M184V/I), NNRTI-R, and PI-R substitutions were associated with the greatest odds of also having preexisting M184V/I [odds ratio (OR) 4.64, 3.29, 2.49, respectively) (Fig. 3). Black race and Hispanic/Latinx ethnicity were also associated with preexisting M184V/I [OR 2.10 (black versus nonblack), 1.67 (Hispanic/Latinx versus non-Hispanic/Latinx)], as was symptomatic HIV or AIDS at baseline and CD4+ cell counts less than 500 cells/μl [OR 1.66 (symptomatic/AIDS versus asymptomatic), 1.52 (CD4+ <500 versus ≥500 cells/μl)]. Finally, a greater number of prior third agents and a longer duration since ART initiation were also associated with presence of M184V/I, with each third agent increasing the odds of preexisting M184V/I by 17% and each year since initiation of ART increasing the odds by 10%.
Fig. 3.
Risk factors associated with preexisting M184V/I by multivariate logistic regression model.
Odds ratios (OR), confidence intervals (CI), and P values were derived from multivariate logistic regression modeling that included baseline resistance and characteristics as potential risk factors and was adjusted for study specific effects.
Discussion
Preexisting M184V/I was common among the virologically suppressed clinical trial participants in B/F/TAF switch studies, all of whom had prior FTC or 3TC exposure. The overall frequency of baseline M184V/I was 10%; however, individual study frequency ranged from 3 to 20% depending on population characteristics and entry criteria. Although high M184V/I frequencies were expected in the studies that permitted M184V/I at enrollment (20% in study 4030 and 11% in study 4580), it was surprising to retrospectively find preexisting M184V/I in 12% of participants in study 1878, which excluded known M184V/I. Comparing studies 1878 and 1844, 1844 had similar population size and demographics as study 1878 but only 3% M184V/I frequency. This highlights the factors associated with M184V/I presence identified by multivariate regression: longer time on ART (median 7.7 years in study 1878 versus median 5.5 years in study 1844: P < 0.0001 by Wilcoxon rank sum test) and higher prevalence of other preexisting antiretroviral resistance (44% in study 1878 versus 31% in study 1844: P < 0.0001 by Fisher's exact test) [31,35]. Additionally, participants switched from a PI-based regimen in study 1878 (versus from DTG/ABC/3TC in study 1844), and a 2019 observational study of five European cohorts found significant associations between M184V/I and prior PI treatment, longer ART duration, and higher prevalence of at least two TAMs [37]. In the TANGO study, which evaluated switching to DTG/3TC in virologically suppressed participants, broad resistance exclusion criteria and a short duration of prior ART (median 2.9 years) that was primarily INSTI-based likely contributed to the low 1% (7/643) detection of baseline M184V/I by retrospective GenoSure Archive [38]. Thus, extrapolation of M184V/I frequencies in clinical trials to other groups of virologically suppressed PWH should take ART history and presence of other resistance substitutions into account.
In the B/F/TAF studies, M184V/I was often discovered by BL-DNA genotyping, particularly in those without historical genotypic data. The utility of proviral DNA genotyping is often debated with limited, but growing, evidence for clinical use. Three studies have found an association between resistance detected by DNA genotype while suppressed and subsequent virologic failure [39–41]. Furthermore, a recent retrospective observational study demonstrated that DNA genotype-guided regimen changes were well tolerated with no increased risk of virologic failure [42]. In a similar study conducted in France, DNA genotype-guided regimen changes increased the probability of maintaining viral suppression; however, the inclusion of APOBEC-mediated hypermutated sequences in their dataset confounds interpretation [43]. As GenoSure Archive removes hypermutated sequences from assay results, our dataset does not over-report resistance substitutions caused by hypermutation, such as M184I, which was detected at low frequency (1%). In clinical care, DNA genotyping assays may be beneficial in certain situations, including when switching regimens is under consideration and treatment history is incomplete or unknown, past resistance tests are unavailable, or after recent low-level viremia when standard resistance testing cannot be performed, but hypermutated sequences must be removed to prevent over-reporting of drug resistance from hypermutated, non-viable genomes.
Despite the potential utility of DNA genotyping, these assays can lack standardization and are limited by the low frequency of latently infected CD4+ T cells. Only a fraction of the HIV reservoir is circulating in peripheral blood; therefore, the small sample volumes assayed are unlikely to be representative of the entire HIV archive. DHHS guidelines note that DNA resistance tests must be interpreted with caution as they often fail to detect all substitutions that were previously reported by RNA genotypes [26,44,45]. M184V/I was detected by GenoSure Archive in only 61% (23/38) of participants with historical M184V/I in our study, and in only 48% in another recent study [46]. Although this may suggest decay of mutated viruses within the reservoir over time [47], it is important to note that the reproducibility of substitution detection within the same sample by DNA testing is variable [48,49]. Furthermore, the average length of time between historical and BL-DNA M184V/I detection was 10 years in our study (data on file), indicating that M184V/I was not meaningfully lost over time.
High levels of virologic suppression were maintained in the 182 participants with preexisting M184V/I who switched to B/F/TAF, regardless of how M184V/I was detected or the presence of additional resistance substitutions, and we identified several factors associated with the presence of M184V/I at baseline. Our study has limitations, however. Only two of the studies prospectively allowed participants with documented M184V/I to enroll, and one of them included only participants who self-identified as black race (Study 4580), which could have biased the analysis of M184V/I prevalence and risk factors. Other enrollment criteria, including age and baseline regimen, may also have affected the analysis of M184V/I risk factors; however, study-specific effects were included in the multivariate model. Furthermore, potentially incomplete ART histories could have affected our model's accuracy. Finally, we only studied switching to B/F/TAF in participants with suppressed HIV. Additional studies in those with M184V/I who are viremic are necessary to further our understanding of the efficacy of B/F/TAF against HIV harboring M184V/I.
Recent data have shown that in addition to B/F/TAF, other three-drug regimens, such as DTG/ABC/3TC and cobicistat-boosted elvitegravir or darunavir with FTC/TAF also have high efficacy in virologically suppressed PWH with M184V/I who switch to these regimens [37,50–52]. Factors contributing to the efficacy of three-drug FTC or 3TC-containing regimens against M184V/I include third agents with high barriers to resistance, reduced replicative fitness and higher RT fidelity of HIV carrying M184V/I [10,53–56], and/or increased activity of tenofovir against M184V/I mutants [57,58]. It is important to note that the efficacy demonstrated by B/F/TAF and other three-drug regimens in the presence of M184V/I can only be extrapolated to use of FTC or 3TC with at least two other fully active drugs, consistent with current guidelines [26].
In conclusion, preexisting M184V/I was commonly detected in participants with suppressed HIV enrolled in B/F/TAF switch studies. Overall, 182 participants (10% of those with baseline genotyping data) had M184V/I at baseline and switched to B/F/TAF. Similarly high rates of virologic suppression were maintained in B/F/TAF-treated participants with or without preexisting M184V/I for at least 1 year with no emergent resistance. M184V/I was associated with presence of other resistance substitutions, black race, Hispanic/Latinx ethnicity, symptomatic or AIDS HIV disease status at baseline, baseline CD4+ cell counts less than 500 cells/μl, greater number of prior third agents, and longer duration of ART. Our analysis of this large population demonstrates the durable efficacy of B/F/TAF in virologically suppressed PWH, including those with known or possible M184V/I.
Acknowledgements
We thank the individuals who participated in these trials and their partners and families, the principal investigators and their staff, and the Gilead study staff. This study was sponsored by Gilead Sciences, Inc.
Justification of authors’ contributions: all authors were involved in the development of the manuscript and interpretation of data and have read and approved the final version. K.A., R.A., M.L.D., S.C., and R.M. analyzed data. P.E.S., E.S.D., J.-M.M., and D.H. enrolled participants. H.L. and C.B. performed statistical analyses. K.A., P.E.S., J.-M.M., E.S.D., D.H., R.A., M.L.D., H.L., C.B., I.M., J.G., S.E.C., H.M., and K.L.W. interpreted results and edited and approved the final manuscript. The first draft was written by K.A. and P.E.S.. All authors contributed edits of the final manuscript.
Financial support: this analysis was supported by Gilead Sciences, Inc.
Conflicts of interest
P.E.S. reports grants or research support from GlaxoSmithKline (GSK)/ViiV, and Gilead Sciences and honoraria or consultation fess from GSK/ViiV, Gilead Sciences, Janssen, and Merck. J.-M.M. reports serving on advisory boards for Gilead Sciences, Merck, ViiV Healthcare, Janssen, Bristol-Myers Squibb (BMS), and Teva, and has received research grants from Gilead Sciences. E.S.D. reports grants from Gilead Sciences, Merck, and ViiV Healthcare, and serves as a consultant or an adviser for BMS, Gilead Sciences, Merck, and Viiv Healthcare. K.A., R.A., M.L.D., S.C., R.M., H.L., C.B., I.M., J.G., S.E.C., H.M., and K.L.W. are all employees and stock shareholders of Gilead Sciences. D.H. declares no competing interests.
Previously presented in part: the 17th European AIDS Conference, 6–9 November 2019, Basel, Switzerland and HIV Glasgow 2020 Virtual, 5–8 October 2020.
Supplementary Material
Paul E. Sax and Kristen Andreatta share first authorship.
Supplemental digital content is available for this article.
References
- 1.Katlama C, Ingrand D, Loveday C, Clumeck N, Mallolas J, Staszewski S, et al. Safety and efficacy of lamivudine-zidovudine combination therapy in antiretroviral-naive patients. JAMA 1996; 276:118–125. [PubMed] [Google Scholar]
- 2.Miller MD, Haddad M, Su C, Gibbs C, McColl DJ, Guyer B. Trends in HIV-1 reverse transcriptase resistance-associated mutations and antiretroviral prescription data from 2003–2010. Antivir Ther 2012; 17:993–999. [DOI] [PubMed] [Google Scholar]
- 3.Wainberg MA, Salomon H, Gu Z, Montaner JS, Cooley TP, McCaffrey R, et al. Development of HIV-1 resistance to (−)2’-deoxy-3’-thiacytidine in patients with AIDS or advanced AIDS-related complex. AIDS 1995; 9:351–357. [PubMed] [Google Scholar]
- 4.Frost SD, Nijhuis M, Schuurman R, Boucher CA, Brown AJ. Evolution of lamivudine resistance in human immunodeficiency virus type 1-infected individuals: the relative roles of drift and selection. J Virol 2000; 74:6262–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuurman R, Nijhuis M, van Leeuwen R, Schipper P, de Jong D, Collis P, et al. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC). J Infect Dis 1995; 171:1411–1419. [DOI] [PubMed] [Google Scholar]
- 6.Keulen W, Back NK, van Wijk A, Boucher CA, Berkhout B. Initial appearance of the 184Ile variant in lamivudine-treated patients is caused by the mutational bias of human immunodeficiency virus type 1 reverse transcriptase. J Virol 1997; 71:3346–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wainberg MA, Moisi D, Oliveira M, Toni TD, Brenner BG. Transmission dynamics of the M184 V drug resistance mutation in primary HIV infection. J Antimicrob Chemother 2011; 66:2346–2349. [DOI] [PubMed] [Google Scholar]
- 8.TenoRes Study Group. Global epidemiology of drug resistance after failure of WHO recommended first-line regimens for adult HIV-1 infection: a multicentre retrospective cohort study. Lancet Infect Dis 2016; 16:565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCluskey SM, Pepperrell T, Hill A, Venter WDF, Gupta RK, Siedner MJ. Adherence, resistance, and viral suppression on dolutegravir in sub-Saharan Africa: implications for the TLD era. AIDS 2021; 35:S127–S135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulkarni R, Babaoglu K, Lansdon EB, Rimsky L, Van Eygen V, Picchio G, et al. The HIV-1 reverse transcriptase M184I mutation enhances the E138K-associated resistance to rilpivirine and decreases viral fitness. J Acquir Immune Defic Syndr 2012; 59:47–54. [DOI] [PubMed] [Google Scholar]
- 11.Deeks SG, Hoh R, Neilands TB, Liegler T, Aweeka F, Petropoulos CJ, et al. Interruption of treatment with individual therapeutic drug classes in adults with multidrug-resistant HIV-1 infection. J Infect Dis 2005; 192:1537–1544. [DOI] [PubMed] [Google Scholar]
- 12.Paredes R, Sagar M, Marconi VC, Hoh R, Martin JN, Parkin NT, et al. In vivo fitness cost of the M184 V mutation in multidrug-resistant human immunodeficiency virus type 1 in the absence of lamivudine. J Virol 2009; 83:2038–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain V, Sucupira MC, Bacchetti P, Hartogensis W, Diaz RS, Kallas EG, et al. Differential persistence of transmitted HIV-1 drug resistance mutation classes. J Infect Dis 2011; 203:1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Little SJ, Holte S, Routy J-P, Daar ES, Markowitz M, Collier AC, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med 2002; 347:385–394. [DOI] [PubMed] [Google Scholar]
- 15.Metzner KJ, Scherrer AU, Preiswerk B, Joos B, von Wyl V, Leemann C, et al. Origin of minority drug-resistant HIV-1 variants in primary HIV-1 infection. J Infect Dis 2013; 208:1102–1112. [DOI] [PubMed] [Google Scholar]
- 16.Ananworanich J, Sirivichayakul S, Pinyakorn S, Crowell TA, Trichavaroj R, Weerayingyong J, et al. High prevalence of transmitted drug resistance in acute HIV-infected Thai men who have sex with men. J Acquir Immune Defic Syndr 2015; 68:481–485. [DOI] [PubMed] [Google Scholar]
- 17.Castor D, Low A, Evering T, Karmon S, Davis B, Figueroa A, et al. Transmitted drug resistance and phylogenetic relationships among acute and early HIV-1-infected individuals in New York City. J Acquir Immune Defic Syndr 2012; 61:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurt CB, McCoy SI, Kuruc J, Nelson J, Kerkau M, Fiscus S, et al. Transmitted antiretroviral drug resistance among acute and recent HIV infections in North Carolina from 1998 to 2007. Antivir Ther 2009; 14:673–678. [PMC free article] [PubMed] [Google Scholar]
- 19.Machnowska P, Meixenberger K, Schmidt D, Jessen H, Hillenbrand H, Gunsenheimer-Bartmeyer B, et al. Prevalence and persistence of transmitted drug resistance mutations in the German HIV-1 Seroconverter Study Cohort. PLoS One 2019; 14:e0209605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onywera H, Maman D, Inzaule S, Auma E, Were K, Fredrick H, et al. Surveillance of HIV-1 pol transmitted drug resistance in acutely and recently infected antiretroviral drug-naive persons in rural western Kenya. PLoS One 2017; 12:e0171124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yerly S, von Wyl V, Ledergerber B, Böni J, Schüpbach J, Bürgisser P, et al. Transmission of HIV-1 drug resistance in Switzerland: a 10-year molecular epidemiology survey. AIDS 2007; 21:2223–2229. [DOI] [PubMed] [Google Scholar]
- 22.Rhee SY, Clutter D, Fessel WJ, Klein D, Slome S, Pinsky BA, et al. Trends in the molecular epidemiology and genetic mechanisms of transmitted human immunodeficiency virus type 1 drug resistance in a large US clinic population. Clin Infect Dis 2018; 68:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhee SY, Kassaye SG, Barrow G, Sundaramurthi JC, Jordan MR, Shafer RW. HIV-1 transmitted drug resistance surveillance: shifting trends in study design and prevalence estimates. J Int AIDS Soc 2020; 23:e25611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambotte O, Chaix ML, Gubler B, Nasreddine N, Wallon C, Goujard C, et al. The lymphocyte HIV reservoir in patients on long-term HAART is a memory of virus evolution. AIDS 2004; 18:1147–1158. [DOI] [PubMed] [Google Scholar]
- 25.Turriziani O, Andreoni M, Antonelli G. Resistant viral variants in cellular reservoirs of human immunodeficiency virus infection. Clin Microbiol Infect 2010; 16:1518–1524. [DOI] [PubMed] [Google Scholar]
- 26. Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents with HIV. Available at: https://clinicalinfo.hiv.gov/sites/default/files/guidelines/documents/AdultandAdolescentGL.pdf. [Accessed 1 August 2021]. [Google Scholar]
- 27. European AIDS Clinical Society (EACS). Guidelines Version 10.1 (English). October, 2020. [Google Scholar]
- 28.Saag MS, Gandhi RT, Hoy JF, Landovitz RJ, Thompson MA, Sax PE, et al. Antiretroviral drugs for treatment and prevention of HIV infection in adults: 2020 recommendations of the International Antiviral Society-USA Panel. JAMA 2020; 324:1651–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. BIKTARVY, Gilead Sciences Inc. BIKTARVY® (bictegravir, emtricitabine, and tenofovir alafenamide) tablets, for oral use. U.S. Prescribing Information. Foster City, CA. Revised May, 2021. [Google Scholar]
- 30.Sax PE, Rockstroh JK, Luetkemeyer AF, Yazdanpanah Y, Ward D, Trottier B, et al. Switching to bictegravir, emtricitabine, and tenofovir alafenamide in virologically suppressed adults with Human Immunodeficiency Virus. Clin Infect Dis 2020; 73:e485–e493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molina JM, Ward D, Brar I, Mills A, Stellbrink HJ, López-Cortés L, et al. Switching to fixed-dose bictegravir, emtricitabine, and tenofovir alafenamide from dolutegravir plus abacavir and lamivudine in virologically suppressed adults with HIV-1: 48 week results of a randomised, double-blind, multicentre, active-controlled, phase 3, noninferiority trial. Lancet HIV 2018; 5:e357–e365. [DOI] [PubMed] [Google Scholar]
- 32.Gaur AH, Cotton MF, Rodriguez CA, McGrath EJ, Helström E, Liberty A, et al. Fixed-dose combination bictegravir, emtricitabine, and tenofovir alafenamide in adolescents and children with HIV: week 48 results of a single-arm, open-label, multicentre, phase 2/3 trial. Lancet Child Adolesc Health 2021; 5:642–651. [DOI] [PubMed] [Google Scholar]
- 33.Maggiolo F, Rizzardini G, Molina JM, Pulido F, De Wit S, Vandekerckhove L, et al. Bictegravir/emtricitabine/tenofovir alafenamide in virologically suppressed people with HIV aged >/ = 65 years: week 48 results of a phase 3b, open-label trial. Infect Dis Ther 2021; 10:775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagins D, Kumar P, Saag M, Wurapa AK, Brar I, Berger D, et al. BRAAVE2020 Investigators. Switching to bictegravir/emtricitabine/tenofovir alafenamide in black Americans with HIV-1: a randomized phase 3b, multicenter, open-label study. J Acquir Immune Defic Syndr 2021; 88:86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daar ES, DeJesus E, Ruane P, Crofoot G, Oguchi G, Creticos C, et al. Efficacy and safety of switching to fixed-dose bictegravir, emtricitabine, and tenofovir alafenamide from boosted protease inhibitor-based regimens in virologically suppressed adults with HIV-1: 48 week results of a randomised, open-label, multicentre, phase 3, noninferiority trial. Lancet HIV 2018; 5:e347–e356. [DOI] [PubMed] [Google Scholar]
- 36.Wensing AM, Calvez V, Ceccherini-Silberstein F, Charpentier C, Günthard HF, Paredes R, et al. 2019 update of the drug resistance mutations in HIV-1. Topics Antiviral Med 2019; 27:111–121. [PMC free article] [PubMed] [Google Scholar]
- 37.Olearo F, Nguyen H, Bonnet F, Yerly S, Wandeler G, Stoeckle M, et al. Impact of the M184V/I mutation on the efficacy of abacavir/lamivudine/dolutegravir therapy in HIV treatment-experienced patients. Open Forum Infect Dis 2019; 6:ofz330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Wyk J, Ajana F, Bisshop F, Ajana F, Bisshop F, Portilla J, et al. Efficacy and safety of switching to dolutegravir/lamivudine fixed-dose two-drug regimen versus continuing a tenofovir alafenamide-based three- or four-drug regimen for maintenance of virologic suppression in adults with HIV-1: phase 3, randomized, noninferiority TANGO Study. Clin Infect Dis 2020; 71:1920–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmisano L, Galluzzo CM, Giuliano M. The importance of testing genotypic resistance in proviral DNA of patients fully responding to highly active antiretroviral therapy. J Acquir Immune Defic Syndr 2009; 51:233–234. [DOI] [PubMed] [Google Scholar]
- 40.Armenia D, Zaccarelli M, Borghi V, Gennari W, Di Carlo D, Giannetti A, et al. Resistance detected in PBMCs predicts virological rebound in HIV-1 suppressed patients switching treatment. J Clin Virol 2018; 104:61–64. [DOI] [PubMed] [Google Scholar]
- 41.Cutrell AG, Schapiro JM, Perno CF, Kuritzkes DR, Quercia R, Patel P, et al. Exploring predictors of HIV-1 virologic failure to long-acting cabotegravir and rilpivirine: a multivariable analysis. AIDS 2021; 35:1333–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ellis KE, Nawas GT, Chan C, York L, Fisher J, Connick E, Zangeneh TT. Clinical outcomes following the use of archived proviral HIV-1 DNA genotype to guide antiretroviral therapy adjustment. Open Forum Infect Dis 2020; 7:ofz533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meybeck A, Alidjinou EK, Huleux T, Boucher A, Tetart M, Choisy P, et al. Virological outcome after choice of antiretroviral regimen guided by proviral HIV-1 DNA genotyping in a real-life cohort of HIV-infected patients. AIDS Patient Care STDS 2020; 34:51–58. [DOI] [PubMed] [Google Scholar]
- 44.Delaugerre C, Braun J, Charreau I, Delarue S, Nere ML, de Castro N, et al. Comparison of resistance mutation patterns in historical plasma HIV RNA genotypes with those in current proviral HIV DNA genotypes among extensively treated patients with suppressed replication. HIV Med 2012; 13:517–525. [DOI] [PubMed] [Google Scholar]
- 45.Wirden M, Soulie C, Valantin MA, Fourati S, Simon A, Lambert-Niclot S, et al. Historical HIV-RNA resistance test results are more informative than proviral DNA genotyping in cases of suppressed or residual viraemia. J Antimicrob Chemother 2011; 66:709–712. [DOI] [PubMed] [Google Scholar]
- 46.Margot N, Ram R, McNicholl I, Haubrich R, Callebaut C. Differential detection of M184V/I between plasma historical HIV genotypes and HIV proviral DNA from PBMCs. J Antimicrob Chemother 2020; 75:2249–2252. [DOI] [PubMed] [Google Scholar]
- 47.Nouchi A, Nguyen T, Valantin MA, Simon A, Sayon S, Agher R, et al. Dynamics of drug resistance-associated mutations in HIV-1 DNA reverse transcriptase sequence during effective ART. J Antimicrob Chemother 2018; 73:2141–2146. [DOI] [PubMed] [Google Scholar]
- 48. D’Antoni ML, Andreatta K, Acosta R, Liu H, Shao Y, White KL. HIV-1 DNA genotyping is often variable in repeat testing from single blood draws [Poster 1749]. In: Conference on Retroviruses and Opportunistic Infections (CROI) [virtual]. [Google Scholar]
- 49.Milliere L, Bocket L, Tinez C, Robineau O, Veyer N, Wojciechowski F, et al. Assessment of intra-sample variability in HIV-1 DNA drug resistance genotyping. J Antimicrob Chemother 2021; 76:2143–2147. [DOI] [PubMed] [Google Scholar]
- 50. Perez-Valero I, Llibre JM, Lazzarin A, di Perri G, Pulido F, Molina JM, et al. A phase 3b open-label pilot study to evaluate switching to elvitegravir/cobicistat/emtricitabine/tenofovir alafenamide (E/C/F/TAF) single-tablet regimen in virologically-suppressed HIV-1 Infected adults harboring the NRTI resistance mutation M184V and/or M1841 (GS-US-292-1824): week 24 results [Presentation TUAB0104]. In: International AIDS Conference (IAC). Amsterdam, the Netherlands. [Google Scholar]
- 51.Jary A, Marcelin AG, Charpentier C, Wirden M, Lê MP, Peytavin G, et al. M184V/I does not impact the efficacy of abacavir/lamivudine/dolutegravir use as switch therapy in virologically suppressed patients. J Antimicrob Chemother 2020; 75:1290–1293. [DOI] [PubMed] [Google Scholar]
- 52.Lathouwers E, Wong EY, Brown K, Baugh B, Ghys A, Jezorwski J, et al. AMBER and EMERALD Study Groups. Week 48 resistance analyses of the once-daily, single-tablet regimen darunavir/cobicistat/emtricitabine/tenofovir alafenamide (D/C/F/TAF) in adults living with HIV-1 from the phase III randomized AMBER and EMERALD Trials. AIDS Res Hum Retroviruses 2020; 36:48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larder BA, Kemp SD, Harrigan PR. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science 1995; 269:696–699. [DOI] [PubMed] [Google Scholar]
- 54.Back NKT, Nijhuis M, Keulen W, Boucher CA, Oude Essink BO, van Kuilenburg AB, et al. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J 1996; 15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 55.Lu J, Kuritzkes DR. A novel recombinant marker virus assay for comparing the relative fitness of HIV-1 reverse transcriptase variants. J Acquir Immune Defic Syndr 2001; 27:7–13. [DOI] [PubMed] [Google Scholar]
- 56.Wainberg MA, Drosopoulos WC, Salomon H, Hsu M, Borkow G, Parniak M, et al. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science 1996; 271:1282–1285. [DOI] [PubMed] [Google Scholar]
- 57.Whitcomb JM, Parkin NT, Chappey C, Hellmann NS, Petropoulos CJ. Broad nucleoside reverse-transcriptase inhibitor cross-resistance in human immunodeficiency virus type 1 clinical isolates. J Infect Dis 2003; 188:992–1000. [DOI] [PubMed] [Google Scholar]
- 58.Wolf K, Walter H, Beerenwinkel N, Keulen W, Kaiser R, Hoffmann D, et al. Tenofovir resistance and resensitization. Antimicrob Agents Chemother 2003; 47:3478–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.