Abstract
Background
Throughout the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, healthcare workers (HCWs) have faced risk of infection from within the workplace via patients and staff as well as from the outside community, complicating our ability to resolve transmission chains in order to inform hospital infection control policy. Here we show how the incorporation of sequences from public genomic databases aided genomic surveillance early in the pandemic when circulating viral diversity was limited.
Methods
We sequenced a subset of discarded, diagnostic SARS-CoV-2 isolates between March and May 2020 from Boston Medical Center HCWs and combined this data set with publicly available sequences from the surrounding community deposited in GISAID with the goal of inferring specific transmission routes.
Results
Contextualizing our data with publicly available sequences reveals that 73% (95% confidence interval, 63%–84%) of coronavirus disease 2019 cases in HCWs are likely novel introductions rather than nosocomial spread.
Conclusions
We argue that introductions of SARS-CoV-2 into the hospital environment are frequent and that expanding public genomic surveillance can better aid infection control when determining routes of transmission.
Keywords: COVID-19, SARS-CoV-2, genomic epidemiology, infection control, nosocomial infection
Outbreak analyses using genomic sequences spuriously identify transmission when genomically similar pathogens in the outside community infect cohort members. We show how to avoid this by example--leveraging community sequences within a SARS-CoV-2 outbreak at Boston Medical Center during early 2020.
At the outset of the coronavirus disease 2019 (COVID-19) pandemic, it was unclear which infection control practices would most effectively prevent viral transmission within medical centers [1]. Masks and personal protective equipment can be barriers to transmission for respiratory pathogens, but, in March 2020, how and where they would be optimally deployed was yet to be established [2, 3]. In response to this uncertainty in control and the overall uncertainty in disease severity, many communities including the greater Boston, Massachusetts, area locked down such that only workplaces deemed “essential” remained open. Healthcare workers (HCWs), as essential workers, faced an increased force of infection both at work and during public transit to work relative to those able to shelter. Treating patients with COVID-19 or congregating with other HCWs who did particularly increased the risk [4]. A first step when assessing infection control is identifying where infections likely occurred. Potential transmission events are usually identified by linking pairs of cases deemed to have been in close contact for sufficient periods of time. However, contact tracing alone is underpowered when individuals interact with multiple potential sources of infection in multiple settings.
Genomic surveillance can complement traditional contact tracing by providing an independent source of evidence that links pairs of cases as potential transmission when their sequences are sufficiently similar [5–10]. Transmission can also be ruled out if the genomes are sufficiently divergent. However, finding similar sequences in multiple individuals can indicate infection by a common source or by a dominant circulating strain, rather than direct transmission; this uncertainty is compounded by the lack of sequence diversity early in an epidemic [11]. Similarly, a lack of complete sampling of cases means that even unique pairs of putatively linked cases in any one study may not be accurately resolved. Not accounting for these possibilities can lead to incorrect inferences; thus, it is important to identify which transmission chains within an analysis are most susceptible to bias.
Public genome sequence databases provide opportunities to contextualize genomic surveillance. By focusing on sequences sampled from the same community as a study population, one can estimate how common genotypes are and assess how likely we are to see similar or identical genotypes by chance rather than as a result of direct transmission. The GISAID database [12], as of this writing, holds >7 million severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) sequences and, importantly, contains associated metadata necessary to filter for any relevant study.
In the current study, we used GISAID to refine a genomic surveillance analysis of SARS-CoV-2 at Boston Medical Center (BMC) from March to May 2020, shortly after the virus arrived in Boston. We show that cases among HCWs decreased following sequential implementation of overlapping infection control measures, despite an increased number of hospitalized patients with COVID-19. We sequenced 187 cases from a total 271 of HCWs who tested positive. Next, we show that contextualizing these samples with concurrent Massachusetts sequences deposited in GISAID informs the outbreak analysis. After linking sequences into putative transmission pairs, we show that the largest and most persistent transmission clusters consist of commonly found sequences in the community. This knowledge allows us to focus our analysis on transmission clusters with sequences unique to the medical center, showing that importations from the outside community are frequent and sustained nosocomial transmission is limited to <2 weeks. This high frequency of introductions underscores the potential magnitude of bias when identifying transmission clusters and the importance of public surveillance in mitigating this bias.
METHODS
SARS-CoV-2 Sequencing
We collected all available discarded diagnostic SARS-CoV-2–positive nasopharyngeal swab samples between 16 March and 5 May 2020, from HCWs (n = 271) for genomic analysis. HCWs include all BMC employees, including both those were patient facing and those who were not. In addition, we refer to all transmission within the medical center as nosocomial, including HCW-HCW and patient-HCW transmissions. After filtering isolates based on sample adequacy and quality metrics, we were left with 233 samples for sequencing. Total RNA was isolated, and SARS-CoV-2 genomic RNA was selectively amplified using the ARTIC v3 primer set and deep sequenced using Illumina short-read technology. Reads were aligned to the Wuhan-Hu-1 reference sequence (MN908947.3), after which coverage was assessed with SAMtools 1.10 [13], and single-nucleotide variants were called using LoFreq software [14]. Single-nucleotide variations at >50% occurrence and >10× alignment depth were incorporated into a consensus sequence. We successfully sequenced 180 isolates from HCWs used in this analysis (Supplementary Figure 1).
Community Contextual Sequences
We incorporated contemporaneous SARS-CoV-2 genomes from the state of Massachusetts into this analysis to contextualize the viral landscape within the medical center with that of the outside community. We obtained 1069 sequences from GISAID (downloaded 11 July 2021) using the following filters: complete genomes, high coverage, low coverage excluded; collection dates for these samples ranged from 29 January to 6 June 2020 (GISAID agreement in the Supplementary Materials). We define a BMC consensus sequence isogenic to one in GISAID when it shares the same nucleotides at every nonambiguous site (masking N's for each compared consensus sequence).
Genotype Enrichment
We used a 1-sided binomial test to assess whether a genotype occurred more frequently within the medical center environment relative to the community contextual sequences, which we used as a proxy for circulating genomic diversity in Massachusetts. We note that the genotype frequencies in GISAID may not accurately reflect actual frequencies in the community because of a bias toward sampling other outbreak clusters by practitioners that submit sequences (Supplementary Table 1).
Clustering
We aligned samples using MAFFT 7.490 software (via Nextclade) [15], masked the 5’ and 3’ ends of each genome and generated a pairwise distance matrix of genomes using the K80 model of evolution using the ape R package [16, 17]. We considered pairs of samples to be “isogenic” if they shared the same observed mutations. That is, we ignored differences in genomic positions where one sequence contained an N, signifying inability to call the correct nucleotide owing to issues with sequencing depth and quality. Initially, we naively clustered isogenic samples into putative transmission clusters (Figure 3A). Later in the results, we refined our clustering methods and chained together pairs of sequences that differ by ≤1 SNP and co-occur within 2 weeks of each other (Figure 4). We did not chain pairs of sequences when the latter observed sequence was isogenic to any found in GISAID.
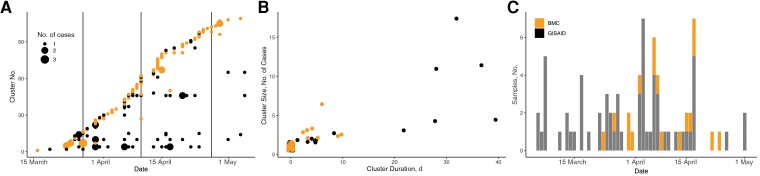
Figure 3.
Incidence of isogenic sequences at Boston Medical Center (BMC) and their presence in GISAID. A, Transmission cluster incidence using a naive approach (only 0-single-nucleotide polymorphism connections), with the size of the point reflecting the number of transmission cluster cases sampled on the same day. Colors denote whether a cluster was seeded by a sequence unique to BMC (orange/grey in print) or a sequence identical to one found in the GISAID database (black). Vertical lines delineate timing of sequentially introduced infection control measures as described for Figure 1. B, Scatterplot of putative cluster size versus duration. Colors denote whether a cluster was seeded by a sequence unique to BMC (orange/grey in print) or by a sequence identical to one found in GISAID database (black). C, Incidence over time of the genotype characterizing the largest clusters in BMC and isogenic samples in GISAID.
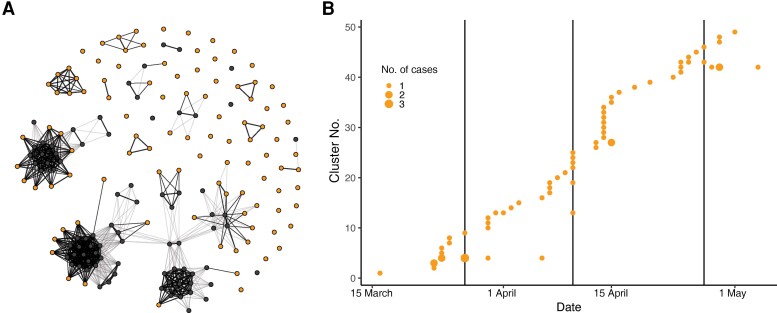
Figure 4.
Transmission clusters with sequences unique to Boston Medical Center (BMC). A, Network representation of all sequences (nodes) with edges representing potential transmission. Starting with the earliest sequence as a cluster, subsequent sequences join into a cluster if they are within 1 single-nucleotide polymorphism (SNP) of a cluster member, occur within 14 days of a cluster member, and, for 1-SNP connections, are not isogenic to any sequences observed in GISAID. Dark edges show surviving connections; lighter edges, connections between genomically similar sequences that do not satisfy the other conditions. Thicker and thinner edges reflect 0-SNP and 1-SNP differences between sequences, respectively. Healthcare worker sequences unique to BMC are shown in orange; those isogenic to sequences in GISAID, in black. B, Incidence of transmission clusters over time.
Phylogenetics
Maximum likelihood phylogenetic trees were inferred using IQ-TREE 2.2.0 software [18]. An appropriate model of rate heterogeneity (GTR + F + I + G4) was chosen using the ModelFinder feature [19], and branch supports were added using ultrafast bootstrap support [20] with 10 000 replicates. A time-resolved phylogeny was generated and rerooted using TreeTime software [21]. Tree annotation was conducted in R using the ggtree and ape packages [16, 17, 22].
RESULTS
COVID-19 Transmission Among HCWs at a Large Medical Center
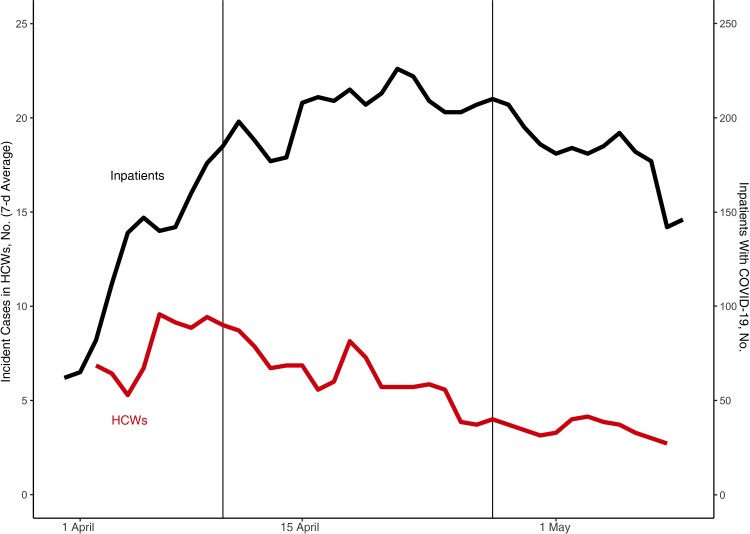
A surge of COVID-19 cases among HCWs at BMC first emerged in late March 2020 and was sustained through mid-May, overlapping a case surge in the surrounding communities. Weekly time-averaged cases among HCWs peaked in early April, followed by a steady decline (Figure 1) (Supplementary Table 2 contains raw counts). By the end of the surge on 9 May, 271 HCWs had COVID-19. Diagnosed. Infection control implemented a universal masking policy for HCWs at all times in the medical center, including N95 masks for patient-facing HCWs beginning 27 March 2020, and subsequently including the use of enhanced personal protective equipment (beginning 10 April); diagnostic testing of all new medical center admissions and symptomatic HCWs began 27 April. HCW cases plateaued and then decreased following the implementation of masking, despite an elevated risk of infection owing to increasing numbers of inpatient cases. Next, we compared genome sequences to resolve transmission chains and estimate the total rate of transmission between HCWs relative to introductions. We successfully sequenced 180 of 271 SARS-CoV-2 samples isolated from discarded diagnostic tests (Supplementary Figure 1).
Figure 1.
Severe acute respiratory syndrome coronavirus 2 case surveillance at Boston Medical Center and the implementation of infection control measures. The rolling 7-day time-average of incident coronavirus disease 2019 (COVID-19) cases among healthcare workers (HCWs) (lower red line) is shown, along with the daily inpatient COVID-19 census (upper black line). Data were left truncated to the first date of mandatory masking (27 March). First vertical line (10 April) indicates the addition of enhanced personal protective equipment; second vertical line (27 April), the addition of universal testing for inpatients and symptomatic HCWs. Plotting begins later than our earliest data point owing to the exclusion of backlogged reporting and the use of a centered 7-day average.
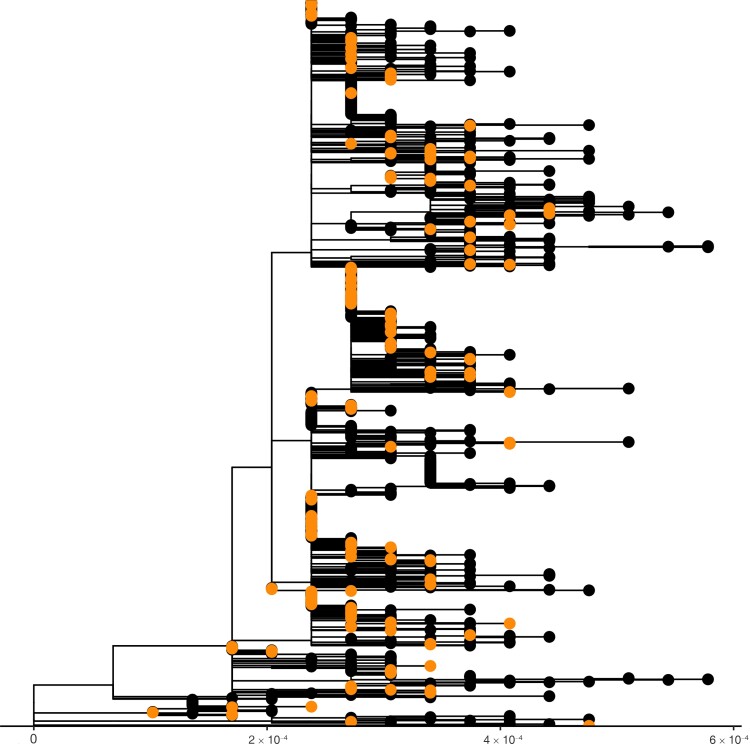
Figure 2 shows a maximum likelihood phylogenetic tree scaled by genetic divergence of our samples contextualized with all contemporaneous sequences in GISAID from the broader Massachusetts community (see Supplementary Figure 2 for a time scaled phylogeny). BMC samples fall across the tree, consistent with multiple, distinct introductions into the HCW population. Some subclades contain only BMC samples, which are hallmarks of sustained transmission clusters contained within the medical center, yet we cannot exclude cryptic reintroductions from the outside community. To further illustrate this point, we compare genotype frequencies between BMC and the Massachusetts community and find significant enrichment of some genotypes within the medical center (Supplementary Table 1). This enrichment analysis provides preliminary evidence for sustained transmission between HCWs but should not be overinterpreted. Nonrandom sequencing in GISAID biases results when comparing frequencies.
Figure 2.
Phylogenetic tree of Boston Medical Center (BMC) samples (orange/grey in print) and Massachusetts samples in GISAID (black) sampled between 29 January and 6 June 2020. The x-axis scale is in genetic divergence (single nucleotide polymorphisms per genome length).
Putative Transmission Clusters Identified With Genomic Data
Identifying potential transmission events requires implicitly assuming an evolutionary epidemiology model—one that putatively links pairs of sequences that are sufficiently genomically similar and are sampled sufficiently closely in time (eg, with respect to a given serial interval and mutation rate). A commonly used conservative approach is to propose that true cases linked by transmission must have identical genomes, because there has been insufficient time for mutations to occur. Because consensus genomes may differ in sequencing coverage, we use “isogenic” sequences, which we define as those with identical sequences at all positions for which data are available; doing so yielded 24 putative transmission clusters and 109 unique sequences (Figure 3A. Putative transmission clusters arose throughout the surveillance period, and 47% of samples contained unique sequences not found in any other infection. However, 3 clusters involved >10 cases and persisted for several weeks. Figure 3B shows that the largest putative transmission clusters are also those that persist longest.
Despite our naive classification of these larger groups as putative transmission clusters, they were all “seeded” by a sequence common to the Massachusetts community and therefore compatible with potential for repeated introduction. Some sequences in BMC, but not all, are isogenic to ≥1 sequence in GISAID. Labeling our transmission clusters as to whether their sequences are isogenic to any Massachusetts sequences observed in GISAID (Figure 3Aand 3B) shows that the largest clusters have been observed in the community and are thus at higher risk for repeat introductions than less common sequences unobserved in GISAID. For illustration, Figure 3C shows the incidence of sequences in GISAID and sequences at BMC for the most persistent transmission cluster. The sequence was repeatedly observed in the community and thus reflects a potential, continual source of infection from either the community or by proxy from the patients. In contrast, the isogenic clusters unique to the BMC were relatively smaller, with the largest containing 6 sequences and a vast majority containing just a single HCW sample.
A naive model connecting only isogenic sequences ignores the possibility that mutations arise between transmissions. To account for this possibility, we linked sequences which were isogenic or within 1 SNP and occurred within a 2-week serial interval of each other, but only if the later observed sequence was not isogenic to any Massachusetts isolates in GISAID. We reason that any genotype observed in GISAID would be more common in the community and at higher risk of being independently introduced. Figure 4A shows a network diagram of how the sequences were ultimately linked into transmission clusters using this model. Here, nodes represent samples and darker edges reflect links into the same transmission cluster. Lighter edges connect sequences similar within ≤1 SNP but were not ultimately connected owing either to timing or to sequence presence in GISAID. This clustering method resulted in larger transmission clusters than the naive method, because transmission clusters could include nonisogenic sequences.
Limiting our analysis to transmission clusters containing only sequences unique to BMC reduces bias due to repeat introductions from genotypes common in the community. While such viruses could be present in the wider community and unsampled, their likely reduced frequency to those observed means there is a smaller chance of repeated introductions. Figure 4B shows the incidence of the resulting transmission clusters over time, containing only sequences unique to BMC. All persisted for <2 weeks, and the largest contained 7 samples, occurring early. The number of clusters relative to the total number of samples sets a minimum bound on the total number of importations in the sample. We estimate that 73% (95% confidence interval, 63%–84%) of the infections were independent introductions, based on the transmission clusters that were found to be unique to BMC. Conservatively focusing only on sequences unique to BMC is expected to induce a selection bias against earlier transmission clusters, owing to the reduced diversity at the beginning of the pandemic, which may increase the estimated importation rate due to changes in infection control. Yet using all transmission clusters with our prior clustering still gives a minimum importation rate of 43% (95% confidence interval, 36%–51%). Overall, these high rates suggest that introductions remain a continual source of infection in addition to HCW-HCW transmissions.
DISCUSSION
We retrospectively analyzed SARS-CoV-2 transmission dynamics at BMC, primarily using genomic surveillance in the months after the virus arrived in Boston. After grouping sequences into potential transmission clusters using genomic similarity and sampling time, we found low rates of transmission between HCWs relative to rates of transmission from unobserved sources, similar to what was observed at another medical center [23]. Importantly, we aimed to reduce bias when analyzing transmission cluster dynamics by leveraging contemporary Massachusetts SARS-CoV-2 sequences deposited in GISAID. Focusing on less common sequences reduces bias by ignoring sequences more likely to be repeatedly introduced, yet it may increase bias in the opposite direction by selectively removing earlier transmission clusters when there were fewer infection control interventions implemented. Furthermore, we can never fully distinguish transmission events between HCWs from repeated introductions as the source of any transmission index case could transmit to multiple HCWs. Nevertheless, we aimed to minimize the overall bias, and the retained transmission clusters were smaller and persisted for <2 weeks, suggesting effective infection control interventions, though some HCW-HCW spread remained.
Repeated introductions bias surveillance efforts to detect clusters, particularly at the outset of an epidemic, when the overall pathogen diversity is low. With a small enough mutation rate, multiple transmissions may occur before genomes diverge at a consensus level. Deeper analysis using subconsensus variation increases the power to link samples as transmission events, and could be particularly useful [9, 24, 25]. However, formal methods linking genomics and contact tracing using subconsensus variation remain to be developed and face considerable statistical challenges, such as the large amount of genetic drift due to small transmission bottlenecks and recurring mutations not associated with transmission [26, 27]. As such, we simplified comparisons and used consensus sequences.
Our study faced several limitations, particularly in sampling. It is possible that early cases identified in BMC and the community were missed, given the imperfect state of surveillance at the time. Furthermore, our genomic surveillance was not robust to missing intermediate steps along a transmission chain, such as a HCW testing at a site outside of the hospital. Similarly, steps in transmission chains may have been missed owing to undetected, asymptomatic SARS-CoV-2 infections which have been estimated to range between 1.6% and 56.5% of cases [10, 28, 29]. Because our sample contains only HCWs, it is impossible to conclude whether any of the identified putative clusters were a result of patient-HCW or HCW-HCW transmission. Note, patient-HCW transmissions represent introductions into the study cohort. As such, we were unable to address how infection control measures altered routes of transmission, but even those effects are contingent on widespread adoption among HCWs, and individual behavior could undermine these efforts.
We leveraged public data from GISAID, which is populated with data collected for other projects, and thus the sampling is likely not completely random and instead biased toward other outbreaks to some degree. Some of these other outbreaks may be other nosocomial transmission clusters, such as nursing facility outbreak investigations, which may be more likely to spread to other medical centers like BMC. In response to these complex factors affecting rates of introduction, we adapted our analysis toward qualitative presence of sequences in GISAID as opposed to quantitatively comparing frequencies, which may not accurately reflect frequencies of introduction to our study cohort. Yet the overall power to observe any one sequence also depends on the overall sequencing effort, which was particularly low early in the pandemic [30].
The techniques used here could be used in future outbreak investigations involving other pathogens. Generally, public databases and community surveillance are paramount for contextualizing local outbreaks, as seen previously with GISAID and influenza and now with SARS-CoV-2. The better curated these databases are with respect to unbiased, random sampling and overall coverage, the better practitioners can distinguish importation from outbreak chains. We argue, given our high rates of importation, that outbreaks in the outside community regularly enter medical centers. It is important that this surveillance infrastructure is maintained and expanded to combat SARS-CoV-2 or future emerging infectious diseases as well as endemic pathogens such as influenza.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Jacquelyn Turcinovic, National Emerging Infectious Diseases Laboratories, Boston University, Boston, Massachusetts, USA; Bioinformatics Program, Boston University, Boston, Massachusetts, USA.
Beau Schaeffer, Center for Communicable Disease Dynamics, Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, Massachusetts, USA.
Bradford P Taylor, Center for Communicable Disease Dynamics, Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, Massachusetts, USA.
Tara C Bouton, Section of Infectious Diseases, Boston University School of Medicine and Boston Medical Center, Boston, Massachusetts, USA.
Aubrey R Odom-Mabey, Bioinformatics Program, Boston University, Boston, Massachusetts, USA; Division of Computational Biomedicine, Boston University School of Medicine, Boston, Massachusetts, USA.
Sarah E Weber, Section of Infectious Diseases, Boston University School of Medicine and Boston Medical Center, Boston, Massachusetts, USA.
Sara Lodi, Department of Biostatistics, Boston University School of Public Health, Boston, Massachusetts, USA.
Elizabeth J Ragan, Section of Infectious Diseases, Boston University School of Medicine and Boston Medical Center, Boston, Massachusetts, USA.
John H Connor, National Emerging Infectious Diseases Laboratories, Boston University, Boston, Massachusetts, USA; Bioinformatics Program, Boston University, Boston, Massachusetts, USA; Department of Microbiology, Boston University School of Medicine, Boston, Massachusetts, USA.
Karen R Jacobson, Section of Infectious Diseases, Boston University School of Medicine and Boston Medical Center, Boston, Massachusetts, USA.
William P Hanage, Center for Communicable Disease Dynamics, Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, Massachusetts, USA.
Notes
Acknowledgments. The authors thank Boston Medical Center (BMC) for providing financial support for this study; and all the patients and staff involved in the BMC coronavirus disease 2019 (COVID-19) efforts. The biologic samples used for the analyses presented here were obtained from the BMC/Boston University COVID-19 Biorepository.
Disclaimer. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Financial support. This work was supported by the National Center for Advancing Translational Sciences, National Institutes of Health (NIH), through Clinical and Translational Science Institute, Boston University and Genome Science Institute, Boston University (grant 1UL1TR001430 to T. C. B. and J. H. C.), by the NIH (grant NIAID K23 AI152930-01A1 to T.C.B.), and by the National Institute of General Medical Sciences, NIH (grant T32GM100842 to A. R. O. M.). Funding to pay the Open Access publication charges for this article was provided by NIH R01AI128344.
References
- 1. Iacobucci G. COVID-19: doctors sound alarm over hospital transmissions. BMJ 2020; 369:m2013. [DOI] [PubMed] [Google Scholar]
- 2. Heneghan CJ, Spencer EA, Brassey J, et al. . SARS-CoV-2 and the role of airborne transmission: a systematic review. F1000Res 2021; 10:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Greenhalgh T, Jimenez JL, Prather KA, Tufekci Z, Fisman D, Schooley R. Ten scientific reasons in support of airborne transmission of SARS-CoV-2. Lancet 2021; 397:1603–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sikkema RS, Pas SD, Nieuwenhuijse DF, et al. . COVID-19 in health-care workers in three hospitals in the south of the Netherlands: a cross-sectional study. Lancet Infect Dis 2020; 20:1273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lumley SF, Constantinides B, Sanderson N, et al. . Epidemiological data and genome sequencing reveals that nosocomial transmission of SARS-CoV-2 is underestimated and mostly mediated by a small number of highly infectious individuals. J Infect 2021; 83:473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stirrup O, Hughes J, Parker M, et al. . Rapid feedback on hospital onset SARS-CoV-2 infections combining epidemiological and sequencing data. eLife 2021; 10:e65828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Snell LB, Fisher CL, Taj U, et al. . Combined epidemiological and genomic analysis of nosocomial SARS-CoV-2 infection early in the pandemic and the role of unidentified cases in transmission. Clin Microbiol Infect 2022; 28:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meredith LW, Hamilton WL, Warne B, et al. . Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect Dis 2020; 20:1263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Borges V, Isidro J, Macedo F, et al. . Nosocomial outbreak of SARS-CoV-2 in a “non-COVID-19” hospital ward: virus genome sequencing as a key tool to understand cryptic transmission. Viruses 2021; 13:604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lucey M, Macori G, Mullane N, et al. . Whole-genome sequencing to track severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission in nosocomial outbreaks. Clin Infect Dis 2021; 72:e727–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Villabona-Arenas CJ, Hanage WP, Tully DC. Phylogenetic interpretation during outbreaks requires caution. Nat Microbiol 2020; 5:876–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khare S, Gurry C, Freitas L, et al. . GISAID’s role in pandemic response. China CDC Weekly 2021; 3:1049–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li H, Handsaker B, Wysoker A, et al. . The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilm A, Aw PPK, Bertrand D, et al. . LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 2012; 40:11189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aksamentov I, Roemer C, Hodcroft E, Neher R. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw 2021; 6:3773. [Google Scholar]
- 16. Paradis E, Schliep K. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019; 35:526–8. [DOI] [PubMed] [Google Scholar]
- 17. R Core Team . R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2021. Available at: https://www.R-project.org/. Accessed 20 August 2022. [Google Scholar]
- 18. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015; 32:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. Modelfinder: fast model selection for accurate phylogenetic estimates. Nat Methods 2017; 14:587–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 2018; 35:518–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sagulenko P, Puller V, Neher RA. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol 2018; 4:vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. GGTREE: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 2017; 8:28–36. [Google Scholar]
- 23. Braun KM, Moreno GK, Buys A. Viral sequencing to investigate sources of SARS-CoV-2 infections in US healthcare personnel. Clin Infect Dis 2021; 73:e1329–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lythgoe KA, Hall M, Ferretti L, et al. . SARS-CoV-2 within-host diversity and transmission. Science 2021; 372:eabg0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. San JE, Ngcapu S, Kanzi AM, et al. . Transmission dynamics of SARS-CoV-2 within-host diversity in two major hospital outbreaks in South Africa. Virus Evol 2021; 7:veab041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sobel Leonard A, Weissman DB, Greenbaum B, Ghedin E, Koelle K. Transmission bottleneck size estimation from pathogen deep-sequencing data, with an application to human influenza a virus. J Virol 2017; 91:e00171-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Valesano AL, Rumfelt KE, Dimcheff DE, et al. . Temporal dynamics of SARS-CoV-2 mutation accumulation within and across infected hosts. PLoS Pathog 2021; 17:e1009499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rivett L, Sridhar S, Sparkes D, et al. . Screening of healthcare workers for SARS-CoV-2 highlights the role of asymptomatic carriage in COVID-19 transmission. eLife 2020; 9:e58728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gao Z, Xu Y, Sun C, et al. . A systematic review of asymptomatic infections with COVID-19. J Microbiol Immunol Infect 2021; 54:12–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Davis JT, Chinazzi M, Perra N, et al. . Cryptic transmission of SARS-CoV-2 and the first COVID-19 wave. Nature 2021; 600:127–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.