Abstract
Aims
Severe acute respiratory syndrome coronavirus-2 infection causes COVID-19, which in severe cases evokes life-threatening acute respiratory distress syndrome (ARDS). Transcriptome signatures and the functional relevance of non-vascular cell types (e.g. immune and epithelial cells) in COVID-19 are becoming increasingly evident. However, despite its known contribution to vascular inflammation, recruitment/invasion of immune cells, vascular leakage, and perturbed haemostasis in the lungs of severe COVID-19 patients, an in-depth interrogation of the endothelial cell (EC) compartment in lethal COVID-19 is lacking. Moreover, progressive fibrotic lung disease represents one of the complications of COVID-19 pneumonia and ARDS. Analogous features between idiopathic pulmonary fibrosis (IPF) and COVID-19 suggest partial similarities in their pathophysiology, yet, a head-to-head comparison of pulmonary cell transcriptomes between both conditions has not been implemented to date.
Methods and results
We performed single-nucleus RNA-sequencing on frozen lungs from 7 deceased COVID-19 patients, 6 IPF explant lungs, and 12 controls. The vascular fraction, comprising 38 794 nuclei, could be subclustered into 14 distinct EC subtypes. Non-vascular cell types, comprising 137 746 nuclei, were subclustered and used for EC-interactome analyses. Pulmonary ECs of deceased COVID-19 patients showed an enrichment of genes involved in cellular stress, as well as signatures suggestive of dampened immunomodulation and impaired vessel wall integrity. In addition, increased abundance of a population of systemic capillary and venous ECs was identified in COVID-19 and IPF. COVID-19 systemic ECs closely resembled their IPF counterparts, and a set of 30 genes was found congruently enriched in systemic ECs across studies. Receptor–ligand interaction analysis of ECs with non-vascular cell types in the pulmonary micro-environment revealed numerous previously unknown interactions specifically enriched/depleted in COVID-19 and/or IPF.
Conclusions
This study uncovered novel insights into the abundance, expression patterns, and interactomes of EC subtypes in COVID-19 and IPF, relevant for future investigations into the progression and treatment of both lethal conditions.
Keywords: SARS-CoV-2, COVID-19, IPF, Transcriptomics, Single-nucleus RNA-seq, Lung, Endothelial cells
Graphical Abstract
Graphical Abstract.
Time for primary review: 57 days
1. Introduction
The pandemic caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) continues to progress and flare up. COVID-19, caused by SARS-CoV-2 infection, manifests as acute respiratory distress syndrome (ARDS)1 in severe cases, too often with life-threatening consequences. Despite ongoing vaccination programmes and therapeutic improvements,2,3 mortality of a fraction of acutely-ill severe COVID-19 patients and chronic morbidity of severe COVID-19 survivors remain unacceptably high, and SARS-CoV-2 mutants continue to threaten healthcare, economic welfare and quality of life in multiple countries. As single-cell studies provide an unbiased and comprehensive characterization of cellular landscapes, they represent a suitable strategy for increasing our understanding of the cell phenotypes and transcriptomic underpinnings of COVID-19. Previous single-cell studies profiled the response to SARS-CoV-2 in peripheral blood mononuclear cells and bronchoalveolar lavage fluid, primarily focusing on immune cells.4–12 Subsequent single-cell studies then also compiled inventories of the pulmonary cell heterogeneity in organs from deceased COVID-19 patients (‘lethal COVID-19’).13–15
Endothelial cells (ECs) have been proposed to contribute to vascular inflammation, recruitment/invasion of immune cells, vascular leakage, hypercoagulability, vascular thrombotic occlusion, and resultant hypoxia in the lungs of severe COVID-19 patients.16–19 Moreover, non-vascular cells, in particular immune cells, in the pulmonary micro-environment can render ECs dysfunctional.20 Nevertheless, the vascular landscape, as well as its interplay with non-vascular cells, remains underexplored at in-depth single-cell resolution in COVID-19.
Important to consider when interrogating the lung vasculature, the lung harbours two circulatory systems: the pulmonary circulation, important for gas exchange, and the systemic/bronchial vascular supply, providing oxygenated blood to the entire lung.21 In fact, in addition to the more established pulmonary EC subtypes, so-called peri-bronchial venous ECs were identified as a transcriptomically distinct vascular subcluster in a single-cell RNA-sequencing (scRNA-seq) study of lungs from healthy, idiopathic pulmonary fibrosis (IPF; a lung disease characterized by progressive lung scarring and irreversible lung dysfunction) and chronic obstructive pulmonary disease patients.22 In healthy lungs, this EC subtype is restricted to the bronchial vasculature surrounding large proximal airways, while in IPF lungs, peri-bronchial venous ECs expand and are observed in areas of bronchiolization and fibrosis.22 In subsequent lung EC scRNA-seq studies, this EC subtype was specifically localized to the systemic vasculature of the bronchial vascular plexus and visceral pleura in healthy lungs, and ultimately coined as ‘systemic venous’.15,23
IPF shares a number of major risk factors and molecular characteristics with COVID-19,24–26 and patients with COVID-19-associated ARDS often develop severe pulmonary fibrosis,27,28 which can be life-threatening in the acute stage and often results in incapacitating sequelae later on.29 A population of systemic venous ECs has also been recently detected in COVID-19 lungs,15 but its putative role in fibrotic lung disease, as well as additional heterogeneity within this EC subpopulation, either within or between conditions, remains elusive to date.
Since a direct comparison of COVID-19-related vs. non-COVID-19-related pulmonary fibrosis has not been conducted to date, we performed a single-nucleus RNA-sequencing (snRNA-seq) study on frozen lung tissue from 7 COVID-19 decedents, 6 IPF patients (who required lung transplantation), and 12 controls (for detailed patient information, see Supplementary material online, Supplementary Methods and Table S1), and derived transcriptomes of 38 794 single ECs, distributed over 14 distinct subclusters. Whereas COVID-19 and IPF samples largely resembled each other in terms of subcluster distribution and differentially expressed genes, we detected notable differences in transcriptome signatures and subcluster abundances when comparing both conditions with control lungs, including an increased abundance of systemic venous, and newly discovered systemic capillary ECs in both COVID-19 and IPF lungs. By taking advantage of the 137 746 nuclei of non-vascular cell types, we performed EC-interactome analyses and identified a perturbed cross-talk between vascular and non-vascular compartments in lethal COVID-19. We moreover identified a congruent set of 30 genes, selectively enriched in systemic ECs across multiple COVID-19 and IPF studies, comprising different patient cohorts and sequencing strategies. Altogether, we highlight key transcriptomic changes and interactions perturbed in COVID-19 with focus on the endothelium, partially overlapping with IPF, and with potential importance for future therapeutic development.
2. Methods
2.1. Patient samples
Informed consent was obtained from all research subjects. Sample collection and use were approved by the local ethics committee (Medical Ethics Committee UZ/KU Leuven, see Supplementary material online, Supplementary Methods for specific ethical protocols). The study complied with the principles outlined in the Declaration of Helsinki. All SARS-CoV-2-positive samples were handled and processed in a biosafety Level-3 laboratory, according to the biocontainment procedures associated with processing of SARS-CoV-2-positive samples. For more detailed patient information see Supplementary material online, Supplementary Methods and Table S1. All non-COVID-19 control patient tissues were collected before the 2020 pandemic, and therefore the tissues were negative for SARS-CoV-2.
2.2. Single nuclei isolation from control, COVID-19, and IPF lung tissues
For snRNA-seq, after collection, freezing of lung post-mortem/explant samples was performed as quickly as possible by placing the samples in cryo-tubes, which were subsequently snap-frozen with liquid N2 (5 min). Afterwards, the tubes were placed on dry ice and stored at –80°C. The nuclei isolation protocol was adapted from Slyper et al.30 (see Supplementary material online, Supplementary Methods for more details).
2.3. snRNA-seq
Nuclei were counted using an automated cell counter (Luna, Logos Biosystems, Gyeonggi-do, South Korea), and converted to barcoded Drop-seq libraries by using the Chromium Single 3′ Library, Gel Bead & Multiplex Kit and Chip Kit (10X Genomics; Pleasanton, CA, USA), aiming for an estimated number of 10 000 nuclei per library. Libraries were sequenced on an Illumina NovaSeq 6000. Demultiplexing according to the sample barcodes, and subsequent read alignment were done using Cell Ranger (v3.1.0). A human reference genome (GRCh38) was used, including intron sequences for mapping of reads obtained from snRNA-seq data. Three Fastq files were generated per sample: The I1 Fastq contains the sample barcode, R1 Fastq contains the cell barcode and UMI, and the R2 Fastq contains the cDNA (88 nt).
2.4. snRNA-seq data analysis
After generation of the gene expression matrices, raw data were processed further in R (Version 4.0.1). The following quality control steps were performed: (i) genes expressed by <10 nuclei were removed; (ii) nuclei that expressed fewer than 250 genes (low quality), and with a detected number of genes >2 standard deviations above the mean (potential doublets) were excluded from further analysis; (iii) nuclei with a detected fraction of mitochondrial genes >20% were removed. The resulting data (259 297 nuclei) were first normalized using the NormalizeData function as implemented in the Seurat package (v3.1). We next identified the top 2000 highly variable genes (FindVariableFeatures function), followed by scaling of the data (ScaleData function). The data were then summarized by principal component analysis (PCA; RunPCA function). The top 35 PCs were used to construct a shared nearest-neighbor (SNN) graph (FindNeighbors function) used to cluster the dataset (FindClusters function, resolution = 0.5), followed by visualization using uniform manifold approximation and projection (UMAP; runUMAP function). Marker genes for each cluster were calculated using FindAllMarkers(), and clusters were annotated and subsetted (using the subset() function) into major cellular lineages based on the expression of canonical marker genes, including PECAM1 and CDH5 for ECs, COL1A1, ACTA2, DCN, and LUM for stromal cells, EPCAM and SFTPC for epithelial cells, and PTPRC for immune cells (which could be further divided into NK/T cells (CD3E, NKG7), myeloid cells (MARCO, CD163, FCN1), B cells (MS4A1), mast cells (MS4A2, KIT), and plasma cells (JCHAIN)). Individual subclustering was then performed for the epithelial, stromal, immune (NK/T and myeloid subsets only) and endothelial subsets (see Supplementary material online, Supplementary Methods for further details).
2.5. Histological and immunohistochemical analysis
For details, see Supplementary material online, Supplementary Methods.
2.6. Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism (GraphPad Software, USA). Comparison of changes between two groups was performed using an unpaired, two-tailed t-test (in case of normally distributed data, as determined by performing a Shapiro–Wilk test) or a Mann–Whitney U test (unpaired; two-tailed; in case data were not normally distributed). In case of unequal variance (F-test), a Welch t-test was used. Comparison of changes between multiple groups was performed using a Kruskal–Wallis test and Dunn’s test for multiple comparisons. All immunofluorescence or histochemical analyses were repeated in a minimum of three patients per group and representative images are displayed.
3. Results
3.1. Atlas of pulmonary subtypes in lethal COVID-19 and IPF
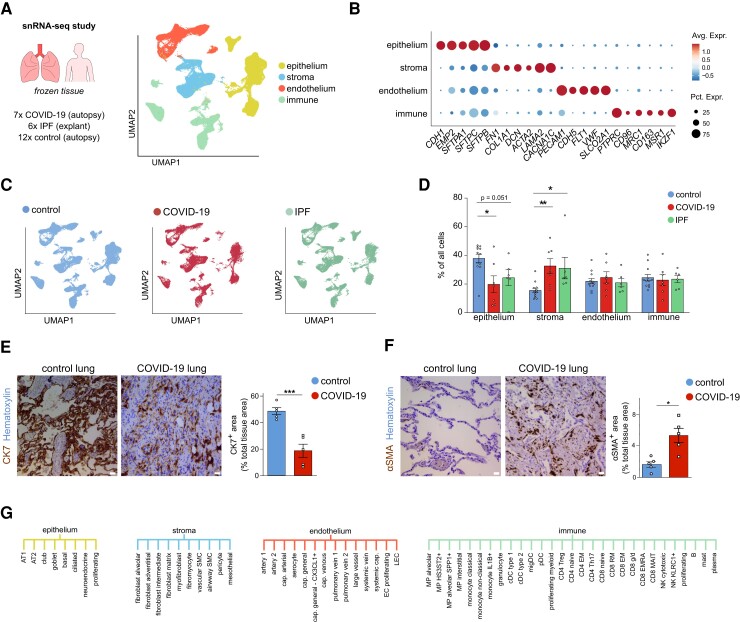
The goal of this study was to analyze pulmonary cell transcriptomic heterogeneity in lethal COVID-19 at cellular resolution, and to compare it with the single-cell transcriptome signature of IPF. To as much as possible avoid confounding study design differences introduced by comparing existing datasets of COVID-19 and IPF lung tissues, we compared head-to-head both lung diseases in a single study, by performing snRNA-seq on frozen lung tissues from 7 COVID-19 decedents, 6 IPF patients requiring lung transplantation, and 12 controls who died of causes unrelated to lung disease (Figure 1A, for clinical metadata see Supplementary material online, Table S1).
Figure 1.
Pulmonary cell types in COVID-19, IPF, and control lungs. A, UMAP plot of lung cells from 7 deceased COVID-19 patients, 6 IPF patients who required lung transplantation, and 12 SARS-CoV-2-uninfected controls (who died of causes unrelated to lung disease), colour-coded by major cellular lineage. B, Dot plot heatmap of expression of representative marker genes of major cellular lineages. The size and colour intensity of each dot represent, respectively, the percentage of cells within each cell type expressing the marker gene and the average level of expression of the marker in this cell type. Colour scale: top (red), high expression; bottom (blue), low expression. C, UMAP plot of lung cells, colour-coded for the indicated conditions. D, Fractions of major cell types in COVID-19, IPF, and control samples. Mean ± SEM, Kruskal–Wallis, and Dunn’s test for multiple comparisons, *P < 0.05, **P < 0.01. n = 7, 6, and 12 for COVID-19, IPF, and control, respectively. E, Representative images of lung sections from COVID-19 and control subjects, immunostained for the epithelial marker cytokeratin-7 (CK7; brown (dark staining)). Quantifications of the CK7-positive area (% of the total tissue area) are provided to the right of the images. Scale bar: 25 μm. Mean ± SEM, unpaired t-test, two-tailed, ***P < 0.001, n = 5 and n = 5 for COVID-19 and control, respectively. F, Representative images of lung sections from COVID-19 and control subjects, immunostained for the stromal marker alpha-smooth muscle actin (αSMA; brown (dark staining)). Scale bar: 25 μm. Quantifications of the αSMA-positive area (% of the total tissue area) are provided to the right of the images. Data are mean ± SEM, unpaired t-test with Welch correction, two-tailed, *P < 0.05, n = 5 and n = 5 for COVID-19 and control, respectively. G, Overview of the 61 different subclusters identified in epithelial, stromal, endothelial, and immune lineages (for a description of all subclusters and their marker genes, see Supplementary material online, Supplementary Methods).
We profiled a total of 176 540 nuclei, distributed over different cellular lineages, detected in every sample and condition: ECs (PECAM1), stromal cells (defined as a mix of fibroblasts (COL1A2, FN1), pericytes (PDGFRB), and smooth muscle cells (ACTA2), according to a previously published lung taxonomy31), epithelial cells (EMP2, EPCAM), and immune cells (mix of myeloid cell types (MRC1, ITGAX, FCN1), T cells (CD3E), NK cells (NKG7), B cells (MS4A1), mast cells (MS4A2) and plasma cells (JCHAIN)) (Figure1A and B; Supplementary material online, Figure S1A and Table S2). Epithelial cells were under-represented in lethal COVID-19 lungs, whereas stromal cells were enriched (Figure1C and D), a finding corroborated by immunostaining for the epithelial marker cytokeratin-7 (CK7) (Figure 1E), and the stromal cell marker alpha-smooth muscle actin (αSMA) (Figure 1F), in line with previous findings.15 Similar trends were observed in IPF, as previously reported.22,32–34 Further subclustering of the different cellular lineages revealed 61 subclusters (Figure 1G), in line with reported single-cell human lung taxonomies,22,31,35,36 and detected in all three conditions.
3.2. Phenotypic heterogeneity of ECs in COVID-19, IPF, and control lung tissue
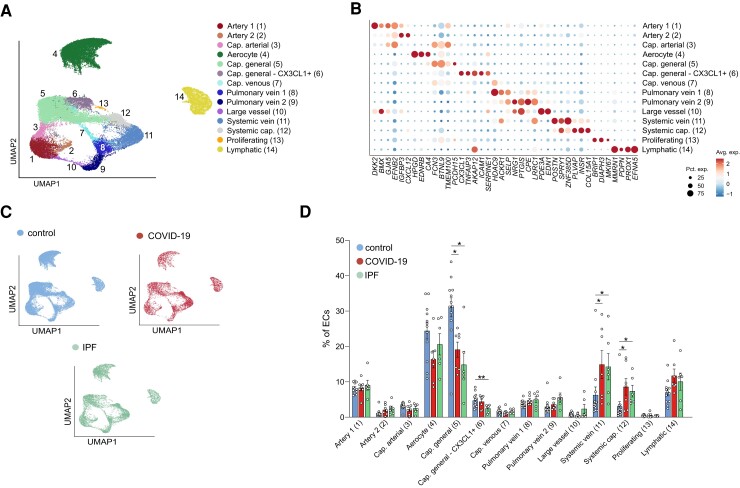
Given the increasing availability of single-cell analyses of non-vascular cell types in COVID-19 and IPF lungs,13–15 and the underexplored nature of ECs at single-cell resolution in both diseases, we focused primarily on the EC cohort in our dataset (n = 38 794 nuclei across all three conditions). Using previously published vascular bed marker genes and annotations,22,23,35 14 transcriptionally distinct EC subclusters could be identified (Figure2A and B; Supplementary material online, Figure S1B and C and Table S2). To enable accurate comparisons with other lung single-cell studies, we based our chosen EC subtype nomenclature on a comprehensive integrated single-cell atlas of human lung ECs23 as much as possible. Specifically, we uncovered: two clusters of arterial ECs (1–2; GJA5, ARL15, DKK2), of which Artery 2 distinguished itself by increased expression of IGFBP3 and CXCL12; capillary arterial ECs (3), expressing both arterial (GJA5), and capillary marker genes (FCN3) (representing arteriolar ECs); aerocytes (4; CA4, ACE, EDNRB); two clusters of general capillary ECs (5–6; NOSTRIN, FCN3, BTNL9), of which Cluster 6 additionally expressed inflammatory marker genes (CX3CL1, ICAM1); capillary venous ECs (7; FCN3, ACKR1, SELP) (representing venular ECs) and two clusters of pulmonary venous ECs (8–9; ACKR1, SELP), of which Cluster 9 specifically showed elevated expression of CPE, PTGIS and NRG1; large vessel ECs (10), expressing both arterial and venous marker genes (BMX, SELP, EDN1); the recently described COL15A1+ peri-bronchial22 or systemic venous23 ECs (11; COL15A1, SPRY1, ZNF385D, POSTN), as well as a COL15A1+ capillary EC population that we coined ‘systemic capillary’ ECs (12; COL15A1, INSR, ZNF385D) (see below for details); proliferating ECs (13; MKI67) and lymphatic ECs [14; PROX1, MMRN1 (Figure2A and B)].
Figure 2.
Vascular subclusters in COVID-19, IPF, and control lungs. A, UMAP plot of EC transcriptomes, colour- and number-coded for the 14 subtypes identified by graph-based clustering. B, Dot plot heatmap of the expression of EC subtype-specific marker genes used for subcluster annotation. The size and colour intensity of each dot represent, respectively, the percentage of cells within each cell type expressing the marker gene and the average level of expression of the marker in this cell type. Colour scale: top (red), high expression; bottom (blue), low expression. C, UMAP plots of ECs, colour-coded per condition D, Fraction of EC subtypes in COVID-19, IPF, and control samples. Data are mean ± SEM, Kruskal–Wallis, and Dunn’s test for multiple comparisons, n = 7 (COVID-19), 6 (IPF), 12 (controls), *P < 0.05, **P < 0.01.
We next explored differences in abundance of certain EC subtypes across control, COVID-19, and IPF samples, to investigate whether a differential abundance of certain EC subtypes can be associated with any of these conditions. As a population, ECs were similarly abundant in control, COVID-19, and IPF lungs (Figure1C and D), but at the subcluster level, we observed an underrepresentation of general capillaries, while systemic venous and capillary EC populations were expanded in both COVID-19 and IPF lungs (Figure2C and D).
3.3. ECs in lethal COVID-19: transcriptome signatures of increased stress, altered immune signalling, and perturbed barrier integrity
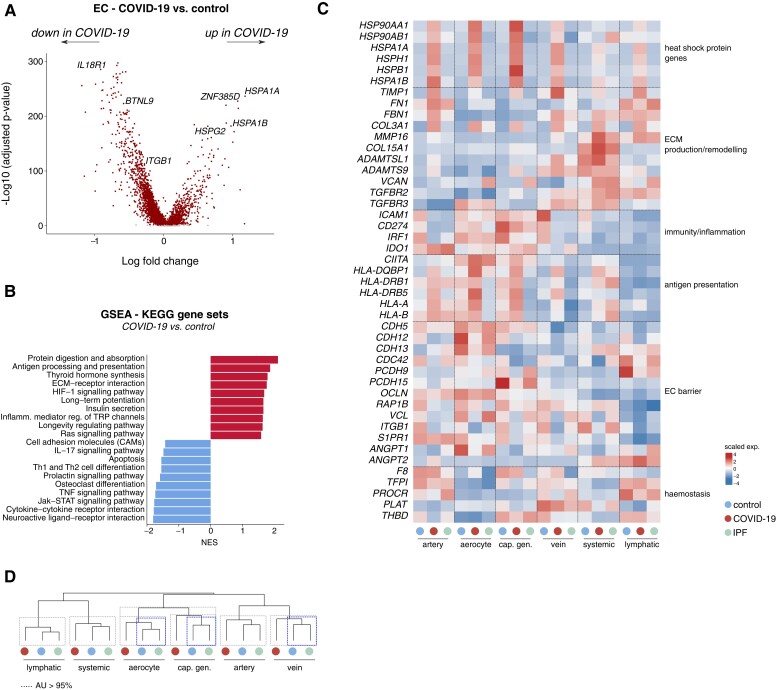
To characterize the global gene expression signatures of the vascular compartment across conditions, we performed differential gene expression analysis (DGEA) and gene set enrichment analysis of all (pooled) COVID-19 or IPF vs. control ECs (see Supplementary material online, Table S3). These analyses revealed, among others, an enrichment for genes involved in antigen presentation, hypoxia signalling and extracellular matrix (ECM) interactions in COVID-19, while several gene sets related to immune system regulation, inflammation, and cell–cell adhesion were negatively enriched (Figure3A and B). To explore whether the enrichment/depletion of these gene sets was selective to a specific EC subtype, we analyzed the expression of representative genes belonging to these enriched pathways across the major identified EC subtypes (see Supplementary material online, Supplementary Methods for details on pooling of the different subclusters). We observed that genes encoding heat shock proteins (HSP90AA1, HSPA1A) involved in cellular stress were particularly enriched in COVID-19 pulmonary microvascular ECs (Figure 3C), presumably evoked by the harsh microenvironmental conditions in these lungs and the reported endothelialitis.37 Genes involved in ECM production/remodelling and associated matrix/receptor signalling (TIMP1, FBN1, MMP16, COL15A1) were enriched in both COVID-19 and IPF ECs (Figure 3C). This enrichment was observed in arterial and systemic ECs, but was particularly prominent in venous ECs (Figure 3C), and suggests a potential involvement of the vasculature in creating a pro-fibrotic environment in both conditions.
Figure 3.
Transcriptomic rewiring of COVID-19 and IPF ECs. A, Volcano plot comparing differentially expressed genes in ECs from COVID-19 vs. control lungs. Representative genes are indicated; each dot represents a single gene. Red (dark) and grey (light) dots indicate up- or downregulated genes with a false discovery rate (q-value) <0.05 or >0.05, respectively. B, Gene set enrichment analysis in ECs derived from COVID-19 lungs, compared with those from controls, using KEGG gene sets. Red bars (to the right, with a value above 0) indicate upregulated gene sets, blue bars (to the left, with value below 0) indicate downregulated gene sets in COVID-19 ECs. C, Gene expression heatmap of individual genes included in the KEGG gene sets presented in (B), in the indicated cell types and conditions. Colour scale: top (red), high expression; bottom (blue), low expression. EC subtypes were pooled into major artery (EC1–3), aerocyte (EC4), general capillary (cap; EC5–6), vein (EC7–9), systemic (EC11–12), and lymphatic (EC14) subgroups. D, Hierarchical clustering analysis of major (pooled) EC subtypes. Dashed boxes indicate clusters that were resolved by multiscale bootstrapping [approximately unbiased (AU) P-value ≥ 95%]. Transcriptomes of COVID-19 aerocytes, general capillaries and veins were statistically separable from their counterparts in control and IPF lungs (blue (dark) dashed boxes); this was not observed for lymphatic, arterial and systemic ECs. GSEA, gene set enrichment analysis; NES, normalized enrichment score.
ECs in COVID-19 lungs (and similarly in IPF lungs) furthermore showed decreased expression of certain genes and gene sets involved in immunity/inflammation (chemokines/cytokines, TNF and JAK/STAT signalling), possibly contributing to dampening of the immune response (Figure3B and C). For instance, transcript levels of immunostimulatory genes including ICAM1 (leucocyte recruitment/adhesion, and a known marker of EC activation) and IRF1 (pro-inflammatory EC activation) were downregulated in both COVID-19 and IPF, as observed in tumour ECs,35,38 across the majority of EC subtypes (Figure 3C). Conversely, levels of IDO1, which positively correlates with SARS-CoV-2 viral load in COVID-19 autopsy samples,39 were upregulated in COVID-19 and IPF (arterial/microvascular) ECs (Figure 3C). An increased abundance of IDO1+ ECs (CD31+) was also observed by immunostaining of lung sections in COVID-19 (see Supplementary material online, Figure S1D). While in vitro studies suggested immunosuppressive roles of endothelial IDO1, the roles of IDO1 in ECs in the in vivo setting are yet to be determined and may be context-dependent. Overall, the immune gene signature in lethal COVID-19 (and IPF) seemed to be complex. For instance, in general capillary ECs, which are considered semi-professional antigen presenting cells40 and reduced in numbers as a population in COVID-19 (Figure2C and D), levels of genes involved in antigen processing and presentation were upregulated in COVID-19 (Figure 3C), possibly in an attempt to mount a compensatory immune response. Altogether, ECs in COVID-19 and IPF exhibit an immunosuppressive transcriptome signature, though the relevance of other immunostimulatory gene signatures requires further study.
Consistent with the reported vascular activation and leakage in COVID-19 lungs,37 gene sets involved in cell–cell adhesion were decreased in COVID-19 ECs (Figure3B and C). For instance, expression of CDH5, important for endothelial junction stability, as well as of other genes involved in EC barrier maintenance and vessel wall integrity (ITGB1, RAP1B, CDC42, OCLN, VCL)41–43 or vascular quiescence and homeostasis (S1PR1) was generally decreased in COVID-19 EC subtypes, most strikingly in aerocytes (Figure 3C). Despite reports of vascular damage in IPF,44 such transcriptome changes were not clearly detected in IPF lung ECs (Figure 3C), raising the question whether impaired EC barrier integrity is a trait more selective to lethal COVID-19. Notably, transcripts of ANGPT1 (known to tighten the vessel wall and to lower vascular permeability)45 were reduced only in COVID-19 ECs (mainly in aerocytes), while levels of ANGPT2, a context-dependent regulator of vascular leakage, pro-inflammatory signaling and a predictive biomarker of intensive care unit admission of COVID-19 patients,46 were upregulated (mainly in systemic and lymphatic ECs) in both diseases (Figure 3C).
Gene sets involved in regulating haemostasis, a process derailed in COVID-19 and modulated by ECs,47 were generally not significantly altered across conditions in our dataset (see Supplementary material online, Table S3). In line with this, key genes involved in these processes showed a mixed expression pattern in COVID-19 and IPF ECs, with the expression of the pro-coagulation gene F8 predominantly being decreased in IPF only, whereas certain anti-coagulation genes (PROCR, THBD) were more dominantly decreased in COVID-19 ECs (Figure 3C). Prominent differential expression of TFPI (anti-coagulation) and PLAT (clot dissolution), on the other hand, could not be clearly detected in COVID-19 and IPF ECs (Figure 3C).
Unbiased hierarchical clustering complemented with multiscale bootstrapping revealed that transcriptomes of microvascular and venous ECs seemed to be most prominently rewired in COVID-19 (Figure 3D), in line with our findings described above. Besides the abovementioned genes and gene sets, gene set variation analysis revealed additional genes and processes, for instance vascular smooth muscle contraction, glycolysis, HIF-1α signalling and others, specifically altered in COVID-19 and/or IPF, warranting further exploration (see Supplementary material online, Figure S2A).
To assess robustness of our findings, we next compared our data with an independent lung snRNA-seq dataset of COVID-19 and control patients.15 Using unbiased hierarchical clustering, we revealed that most COVID-19-derived EC subtypes from both studies clustered together, and separate from control (or IPF) samples, when analyzing the same set of EC-enriched genes as shown in Figure 3C (see Supplementary material online, Figure S2B). When unbiasedly calculating the set of genes enriched (adjusted P-value ≤ 0.05; log-fold change ≥ 0.25) in the EC-compartment of both studies, we found a set of 127 genes congruently enriched (see Supplementary material online, Figure S2C and Table S3). To evaluate enrichment of these congruent genes in ARDS in a non-COVID-19 context, we generated a bulk RNA-sequencing (RNA-seq) dataset of post-mortem lung tissue from severe COVID-19 and influenza A (H1N1) patients, as well as non-COVID-19 controls (see Supplementary material online, Figure S2D, for clinical information of COVID-19 and influenza patients in this cohort, see37). Bulk deconvolution, using our snRNA-seq dataset as a reference, predicted the presence of all major cellular lineages (epithelial, stromal, endothelial and immune cells) in the bulk RNA-seq dataset (see Supplementary material online, Figure S2E). Hierarchical clustering, using the congruent 127-gene signature, revealed that COVID-19 patients clustered separately from control and influenza patients (see Supplementary material online, Figure S2F). Nonetheless, gene expression patterns were largely similar in influenza and COVID-19 patients, but most pronounced in COVID-19 (see Supplementary material online, Figure S2F). Finally, when analyzing the same set of genes in a bulk RNA-seq dataset of lung tissue from a human ACE2-expressing transgenic (K18-hACE2) mouse model of severe COVID-1948 (see Supplementary material online, Figure S2G), we observed enriched expression of about half of the EC-enriched end-stage COVID-19 genes, suggesting that the enrichment of at least part of our observed signatures may be COVID-19-associated, and not merely a consequence of general ARDS or cohort-related confounders (e.g. ventilation, treatment regimens).
Altogether, ECs in COVID-19 lungs thus selectively exhibited a signature involved in cellular stress and perturbed barrier maintenance/integrity, and (partially) shared signatures with IPF indicative of increased ECM deposition/remodelling and altered immunomodulation, with on one hand the downregulation of pro-inflammatory genes and adhesion molecules, while on the other hand upregulating multiple genes involved in antigen presentation. These signatures seem, at least in part, specific to and/or more pronounced in late-stage COVID-19.
3.4. EC cross-talk with other pulmonary cell types in COVID-19 and IPF
Given the prominent transcriptomic changes in COVID-19 and IPF ECs, we next explored with which other pulmonary cells they were predicted to interact, and which of such interactions might likely explain the altered vascular gene expression landscape. We therefore used all non-vascular cell types in our snRNA-seq data to characterize their cross-talk with ECs in every condition. Given the recently published landscapes of stromal, epithelial, and immune cell subtypes in healthy, COVID-19, and IPF lung tissue,13–15,22 we refer to Supplementary material online, Figures S3–S6 and Tables S2 and S3 and the Supplementary Methods for a detailed overview of their unsupervised clustering analyses and annotation, as well as their differential abundance in COVID-19 and IPF vs. control lung tissue. Using CellPhoneDB,49 we characterized the cross-talk between ECs (all subtypes pooled) and other major pulmonary cell types by assessing their predicted receptor–ligand interaction landscape. Whereas the full interactome analysis is provided in Supplementary material online, Table S4, we specifically focused on the interactions between the pulmonary cellular environment and the endothelium (Figure 4A–C; Supplementary material online, Figure S7A and B). For subtype-specific expression of EC-expressed interaction partners, we refer to Supplementary material online, Figure S7C. Our analyses revealed several interactions, previously not yet implicated in COVID-19 or IPF pathobiology.
Figure 4.
Predicted endothelial—non-endothelial cell interactions in COVID-19 and IPF lungs. A–C, Heatmaps, visualizing the interaction score for the predicted receptor–ligand pairs (P ≤ 0.05) within the (A) vascular compartment itself (EC–EC interactions), (B) between ECs and epithelial cells, or (C) between ECs and stromal cells in control, COVID-19 and IPF lungs. Only interactions enriched or reduced in COVID-19 and/or IPF vs. control lungs are plotted. In bold indicated and boxed interactions are enriched in COVID-19 or COVID-19 and IPF lungs compared with controls.
In COVID-19 and IPF, fewer interactions involved in angiogenesis, vascular integrity and homeostasis (EGFR-TGFB1, FGFR1-KL/FGF7, NRP2-SEMA3F, PDGFB-PDGFRA/PDGFRB) were identified within the vascular compartment itself, or between ECs and epithelial or stromal cells (Figure 4A–C). Moreover, among downregulated interactions between the same cell types in COVID-19 was DLL4/JAG1-NOTCH1 signalling (Figure 4A–C). Loss of endothelial NOTCH1 signalling has been associated with perturbed vascular remodelling and a reduction of fenestrae in hepatic sinusoidal ECs, portal hypertension and intussusceptive angiogenesis (IA),50 an alternative mode of vascularization documented in COVID-19 autopsy samples.37 Likewise, interactions potentially driving vascular leakage/permeability were also specifically detected in COVID-19, or COVID-19 and IPF. For instance, Ephrin receptor signalling in ECs (EPHA4; predominantly expressed in arterial and systemic ECs), induced by Ephrins (EFNA1, EFNA5) in epithelial and stromal cells (Figure4B and C; Supplementary material online, Figure S7C), which may increase EC permeability and vessel leakage,51 was detected in COVID-19 and IPF samples, but not in control lungs. Furthermore, signalling of anti-angiogenic SEMA3A (predicted to be secreted by the endothelium) through its receptors (NRP1, Plexins) on ECs, stromal or epithelial cells, was selectively predicted to occur in COVID-19 (Figure 4A–C), and has been implicated in increasing vascular permeability.52
On the other hand, certain gene expression signatures suggested possible compensatorily induced repair mechanisms. Indeed, various other interactions uniquely predicted in COVID-19 or in both COVID-19 and IPF were predominantly involved in maintaining vessel integrity. For instance, IGF1, expressed by stromal cells, was predicted to signal through IGF1R on ECs in COVID-19 and IPF (Figure 4C). IGF1 may exert pro-migratory effects on ECs,53 is believed to decrease permeability and may act as a vasodilator,54 all of which might be compensatorily induced to repair the vascular defects in COVID-19 and IPF. Furthermore, COVID-19-selective signalling of HGF (secreted by stromal cells) through the MET receptor on ECs (Figure 4C) may inhibit hypoxia-induced EC apoptosis,55 and is important for EC motility, proliferation and angiogenesis.56
In line with our abovementioned observations of reduced EC-specific expression of genes involved in immunity/inflammation in COVID-19, among interactions predicted between ECs and immune cells, we observed a selective reduction in cross-talk involved in leucocyte adhesion, recruitment and trans-endothelial migration (JAM2/JAM3 or ICAM1 on ECs with integrin complexes on immune cells) and T-cell activation (CD2–CD58 and CD46–JAG1 axes)57,58 (see Supplementary material online, Figure S7A and B). Interactions involved in myeloid cell recruitment and/or apoptosis [NRP1 on myeloid cells, SEMA3A on (aerocyte and lymphatic) ECs]59,60 were specifically enriched in COVID-19 lungs (see Supplementary material online, Figure S7A, C). A decrease in signalling involved in pathogen clearance (ANXA1–FPR1/FPR3) was also specifically observed in COVID-19 (see Supplementary material online, Figure S7A). Furthermore, while increased signalling through the GAS6–MERTK/AXL axis (also implicated in clearing of pathogens) was selectively observed in IPF lungs, this interaction was not detected in COVID-19 (see Supplementary material online, Figure S7A), highlighting potential differences between COVID-19 and IPF lung pathology from an immunoregulatory standpoint.
In agreement, we observed a few additional notable differences between COVID-19 EC-interactomes and those present in IPF explant lungs. For instance, the interaction of EC-secreted bone morphogenetic protein 6 (BMP6) (known to exert pro-fibrogenic effects61) with BMP receptors on almost all non-EC cell types, and vascular endothelial growth factor beta (described to contribute to hypoxia-induced vascular remodelling and hypertension in the lung62) secretion by epithelial cells, predicted to signal through FLT1/NRP1 on ECs, were both uniquely identified in IPF (Figure4B and C and Supplementary material online, Figure S7A and B). Moreover, increased secretion of IL15 by ECs was predicted to signal through IL15RA on stromal cells specifically in the context of COVID-19 (Figure 4C). The expression of IL15 was predominantly present in systemic ECs (see Supplementary material online, Figure S7C), whereas IL15RA was mainly detected in fibroblast subclusters of the stromal cell compartment (see Supplementary material online, Table S2), and complementary NicheNet analysis (see Supplementary material online, Figure S8A), to explore the putative downstream effects of systemic EC-mediated IL15 signalling in COVID-19 fibroblasts, revealed that the glycolytic genes PKM and PGK1 were among downstream target genes regulated by IL15 (see Supplementary material online, Figure S8B). Glycolysis is known to be important for ECM production and the fibrogenic phenotype of fibroblasts,63 and indeed, compared with control or IPF, several members of the glycolysis pathway were upregulated specifically in COVID-19 stromal cells (see Supplementary material online, Figure S8C). These findings may suggest that, despite (partial) commonalities regarding EC-interactomes involved in impaired barrier integrity in lethal COVID-19 and IPF, different drivers of the fibrogenic response and vascular remodelling may underlie both conditions.
3.5. Increased abundance of the systemic vasculature in lethal COVID-19 and IPF
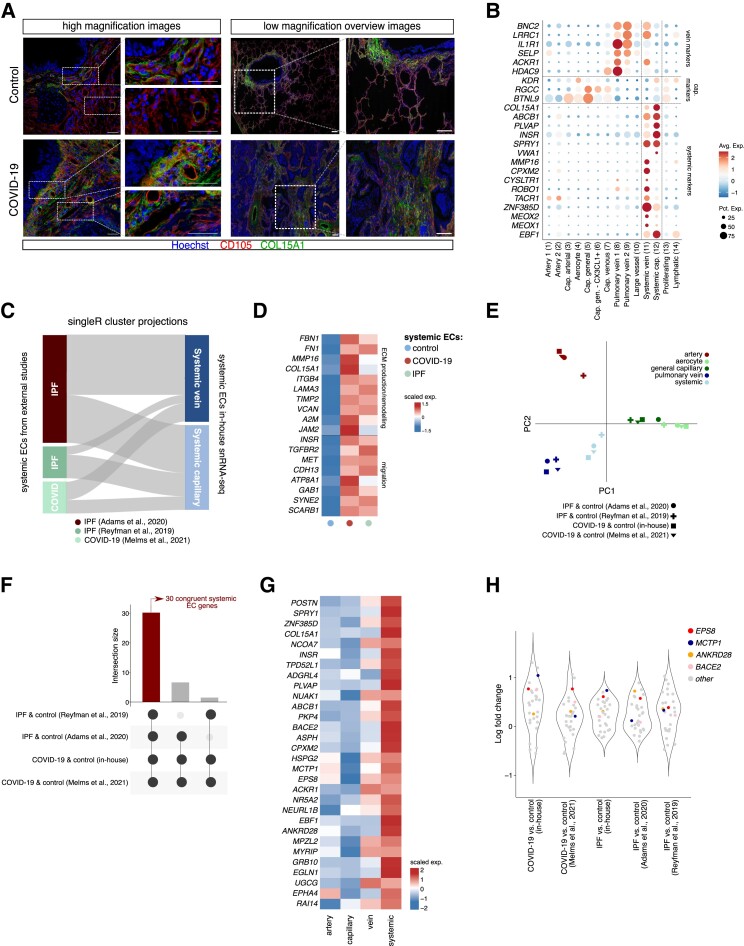
As mentioned above, we observed a selective expansion of the systemic (venous and capillary) vasculature in both COVID-19 and IPF lungs, while general capillaries significantly decreased in abundance in both conditions (Figure2C and D). This observed shift on one hand likely reflects damage of the pulmonary circulation, yet on the other hand may suggest a possible compensatory expansion of the systemic circulation to secure sufficient blood supply, as seen in IPF and other pulmonary diseases.22 Immunostainings for COL15A1 (used as a canonical marker for peri-bronchial/systemic venous ECs22,23) and CD105 confirmed the bronchial localization of systemic venous/peri-bronchial ECs in healthy lungs, opposed to a predominant presence in fibrotic regions in COVID-19 lungs (Figure 5A), verifying our snRNA-seq findings, and in line with previous observations (see Introduction) in IPF lungs.22
Figure 5.
Systemic vasculature in COVID-19 and IPF lungs. A, Representative immunofluorescent images of lung sections from COVID-19 and control subjects, immunostained for CD105 and COL15A1. Hoechst labels nuclei. High magnification (left) and low magnification overview images (right) are shown. Smaller images to the right of larger images are magnifications of the respective boxed areas. Scale bar: 50 μm in high magnification images and their zoom-in areas. Scale bar: 250 μm in low magnification overview images and their zoom-in areas. B, Dot plot heatmap of the expression of systemic, capillary, and venous EC marker genes. The size and colour intensity of each dot represent, respectively, the percentage of cells within each subcluster expressing the marker gene and the average level of expression of the marker in this subcluster. Colour scale: top (red), high expression; bottom (blue), low expression. C, SingleR annotation of systemic ECs extracted from the indicated publicly available single-cell/nucleus studies, visualized as cluster projections. The top-50 most highly ranking markers of systemic capillary and venous subclusters in our in-house snRNA-seq dataset were used as a reference. D, Gene expression heatmap of individual genes involved in ECM production/remodelling and migration, in the indicated cell types and conditions. Genes were selected from Gene Ontology enrichment analysis, as presented in Supplementary material online, Table S5. Colour scale: top (red), high expression; bottom (blue), low expression. E, PCA of pairwise Jaccard similarity coefficients of top-50 marker genes enriched in different EC subclusters extracted from indicated single-cell studies. Symbols indicate studies, colours indicate EC subclusters. F, UpSet plot of systemic EC-enriched genes across the four different datasets included in the meta-analysis. Black connected dots beneath the graph indicate which studies are intersected. Left (red) bar: 30 intersecting genes commonly enriched in systemic ECs in all studies [false discovery rate (q-value) <0.05]. G, Gene expression heatmap of genes (n = 30) commonly enriched in systemic ECs across studies (see left (red) bar in F), in the indicated EC subtypes identified in our snRNA-seq atlas. Colour scale: top (red), high expression; bottom (blue), low expression. EC subtypes were pooled into major artery (EC1–3), capillary (cap; EC4–6), vein (EC7–9) and systemic (EC11–12) subgroups. H, Violin plots, visualizing the log-fold change distribution of the 30-gene congruent systemic EC signature obtained in (F). Coloured dots indicate genes congruently enriched in COVID-19 vs. control and/or IPF vs. control lungs across all studies included in the analysis; grey dots indicate all other genes in the 30-gene signature.
Notably, and unlike previously reported work,22,23 we not only identified a subpopulation of systemic venous ECs in our dataset, but also a second population of ECs expressing reported markers of the systemic vasculature (ZNF385D, SPRY1, COL15A1, EBF1),22,23 but lacking clear expression of venous marker genes (ACKR1, HDAC9, SELP) (Figure 5B). Instead, these ECs more strongly resemble microvascular ECs, based on their expression of markers commonly detected in general capillary ECs (KDR, RGCC, BTNL9)23,35 (Figure 5B). We hypothesized that the discrepancy between our findings and other studies, in which such a microvascular systemic EC subtype was not identified as a transcriptomically separate cluster, is likely due to the substantially higher number of ECs captured in our dataset, allowing us to chart vascular heterogeneity to a larger extent. To explore this further, we extracted single systemic ECs from external, publicly available datasets of either COVID-1915 or IPF lungs,22,64 and used SingleR65 to annotate these cells using our systemic EC subclusters as a reference. Indeed, systemic ECs in all three datasets could be separated into venous and microvascular subsets (Figure 5C). This analysis verified the presence of a transcriptomically distinct subcluster of systemic capillary ECs, thereby adding a thus far overlooked, but additional layer of transcriptional heterogeneity within the pulmonary systemic vascular population. Notably, immunostainings confirmed the presence of both systemic venous ECs (COL15A1 + CD105+ in direct proximity of αSMA+ smooth muscle cells) and systemic capillary ECs (COL15A1 + CD105+, distant from αSMA+ smooth muscle cells) in COVID-19 (and healthy) lungs (Figure S9).
Considering that cellular subtypes, which are congruently altered across different conditions, may represent interesting therapeutic targets, the common enrichment of the systemic vasculature in both COVID-19 and IPF lungs may open interesting avenues for further translational investigation. We therefore performed DGEA, comparing pooled COVID-19 or IPF systemic venous and capillary EC subclusters (jointly referred to as ‘systemic ECs’) to their control counterparts (using only our in-house generated dataset), to explore robust systemic EC marker genes enriched in both conditions. This analysis revealed a set of 107 common genes, of which several are involved in ECM production/remodelling and associated matrix/receptor signalling (TIMP2, FBN1, FN1, MMP16, COL15A1, ITGB4, LAMA3, A2M, JAM2) and cellular migration (INSR, TGFBR2, MET, CDH13) (Figure 5D; Supplementary material online, Table S5), suggesting that systemic ECs in lethal COVID-19 and IPF may be endowed with increased migratory and fibrogenic properties.
To more comprehensively establish a signature of systemic EC marker genes congruently enriched in IPF and COVID-19 across different patient cohorts, we again took advantage of the abovementioned publicly available COVID-1915 and IPF lung22,64 datasets to assess similarity of (systemic) ECs across all studies. First, we calculated the top-50 most highly ranking marker genes of all EC subtypes (pooled, see Methods), and used pairwise Jaccard similarity coefficients to reveal that the transcriptomes of EC subtypes (including systemic ECs) are highly conserved across studies (and thus conditions) (Figure 5E). Furthermore, a meta-analysis of marker genes specific to systemic ECs in each dataset revealed a list of 30 congruent genes, significantly enriched in systemic ECs, in all studies in health and disease (Figure5F and G; Supplementary material online, Table S5). Within this signature, obtained from independent patient cohorts, experimental setups (single-cell vs. single-nucleus RNA-seq) and conditions (COVID-19 or IPF), EPS8, ANKRD28, BACE2 and MCTP1 were also robustly enriched in COVID-19 and IPF systemic ECs (compared with their control counterparts) across studies (Figure 5H). The function of these genes in COVID-19 and IPF, or ECs in general, however remains elusive to date.
Together, ECs in lethal COVID-19 are dominantly enriched for a population of systemic ECs, with high transcriptional resemblance to their counterparts in IPF, suggesting, besides the vast differences in the cause and progression of COVID-19 and IPF, a common EC subtype may contribute to vascular remodelling observed in both conditions.
4. Discussion
We conducted this pulmonary single-nucleus analysis to identify EC phenotypes exhibiting transcriptome signature changes that might suggest a possible contribution to the vascular problems faced by lethal COVID-19 patients. Moreover, we aimed to compare EC transcriptomes between COVID-19 and IPF lungs, to pinpoint key similarities and differences between two conditions characterized by progressive fibrotic lung disease. Our study, in which we profiled >35 000 ECs extracted from post-mortem biopsies of COVID-19 and control lungs, as well as IPF explant lungs, resulted in multiple novel insights.
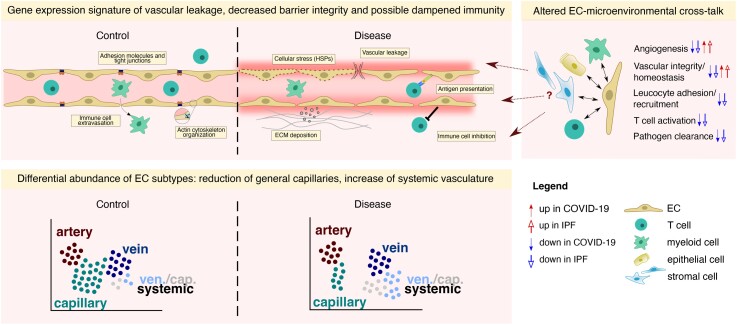
First, we observed a gene expression signature suggestive of vascular leakage, decreased barrier integrity, and possible dampened immunity in the vascular compartment of COVID-19 and (to some extent) IPF lungs (Figure 6). Of note, while changes in RNA abundance may not always be informative for inference of final protein activity, the functional roles for some of our identified barrier-associated genes may be context-dependent, the downregulation of only one tight junction protein may be compensated by others, and vascular barrier regulation by these junctional molecules also relies at levels beyond mRNA transcription, our results are based on the downregulation of a group of genes involved in these processes, not a single gene. Alongside the plethora of indications of vascular leakage in other studies of COVID-19 lung disease,17,37,66 our results are thus in line with the concept of vascular leakage as a key hallmark of end-stage COVID-19. Moreover, our data highlight prominent transcriptome rewiring of barrier-related genes in the aerocyte (important for gas exchange, and part of the blood–air barrier)67 and general capillary compartments of the pulmonary vasculature, both of which are shown to localize to the alveolar wall of the lung,23 possibly in line with the extensive alveolar damage reported in severe COVID-19 patients.68
Figure 6.
Transcriptomic changes in the COVID-19 and IPF vasculature. Schematic representation of vascular transcriptomic rewiring in COVID-19 and IPF vs. control lungs. Upper left panel: the vasculature in (lethal) COVID-19 and IPF lungs harbours a gene expression signature suggestive of vascular leakage, decreased barrier integrity, increased ECM deposition, and possible dampened immunity. Upper right panel: EC-centred interactome analysis revealed various routes of EC-microenvironmental cross-talk that could potentially drive the dysfunctional state of the vasculature in COVID-19 and IPF. Lower panel: ECs in lethal COVID-19 and IPF are dominantly enriched for systemic venous and capillary ECs, whereas general (pulmonary) capillary ECs are decreased in abundance. The transcriptomic signature of systemic ECs suggests an involvement in ECM production/deposition, possibly contributing to the overall fibrotic environment in lethal COVID-19 and IPF. ECM, extracellular matrix; HSPs, heat shock proteins.
Our EC-centred interactome analysis, based on predictions requiring further validation, confirmed these observations and further revealed various routes of EC-microenvironmental cross-talk that could potentially drive this dysfunctional state of the vasculature, predominantly driven by decreased EC–non-EC signalling involved in general vascular integrity and homeostasis (Figure 6), together revealing novel insights into and suggesting potential drivers of vascular derailment in fibrotic lung conditions. In addition, EC activation is commonly reported as a key characteristic in acute COVID-19, yet our results (e.g. decreased expression of ICAM1, IRF1) may reflect a potential dampening of EC-mediated immune responses in lethal COVID-19. While in tumour ECs, downregulation of immunostimulatory genes is considered an immune escape mechanism of the tumour, in chronic disease conditions like COVID-19 and IPF, it may potentially present a compensatory mechanism to thwart the uncontrollable inflammation in the tissue. On the other hand, the immune gene signature of ECs in lethal COVID-19 (and IPF) seemed to be complex, as we also found evidence for reduced levels of CD274 (encoding PD-L1), an immune checkpoint inhibitor,69 in both COVID-19 and IPF, while levels of genes involved in antigen processing and presentation were upregulated primarily in COVID-19. Whereas certain hallmarks of EC activation (e.g. loss of vascular integrity, upregulation of HLA genes) are thus apparent in the transcriptome signature of lethal COVID-19 ECs, other hallmarks (upregulation of leucocyte adhesion molecules) are absent, possibly highlighting important vascular differences between early (acute) and lethal disease stages in COVID-19 lungs, warranting further investigation into the (translational) relevance of our identified gene signatures.
Second, while single-cell resolution studies of post-mortem lung tissue in other types of ARDS are currently lacking, our comparative analyses suggest that the transcriptomic changes observed in COVID-19 (ECs) can in part also be found in lungs of SARS-CoV-2-infected mice, suggesting that, at least to a substantial extent, these changes are independent of potential confounding factors inherently present in patient cohorts (e.g. underlying health conditions, treatment regimen, post-mortem ischaemia, etc.). Moreover, although bulk RNA-seq informs on general transcriptional signatures and thus cannot particularly inform on EC-selective transcriptome changes, we observed similar expression patterns of a COVID-19-enriched, EC-specific gene expression signature in bulk RNA-seq data of COVID-19 and influenza-associated ARDS lung samples. However, changes were most pronounced in COVID-19.
Third, whereas the overall abundance of the COVID-19 and IPF pulmonary vascular compartment was unchanged in comparison with control lungs, we observed a significant reduction of general capillaries in both conditions. We did not specifically observe an enrichment in (regenerative) proliferating ECs as reported in the resolution phase of influenza infection in mice.70 Differences in species and disease type/severity/staging may have caused this discrepancy between the two studies. The systemic EC population, within which we identified a thus far overlooked microvascular population, was significantly enriched in both COVID-19 and IPF lungs, compared with the control setting. In line with reports on inflammatory lung disease,71 these findings suggest that the systemic EC phenotype may possibly be triggered to induce repair of the damaged pulmonary circulation in COVID-19 (Figure 6).
However, the transcriptome signature of systemic ECs presented with a notable enrichment for genes involved in ECM remodelling/organization and migration in both COVID-19 and IPF. Interactome analysis furthermore suggested a selective interaction between IL15, predicted to be secreted predominantly by systemic ECs, and IL15RA expressed on stromal cells. By possibly stimulating glycolysis, our data may suggest that systemic ECs, besides their potential intrinsic pro-fibrotic properties, could also act as a driver of the fibrogenic response of stromal cells. Altogether, these findings may indicate a possible contribution of the systemic vasculature to progressive pulmonary fibrosis, raising the question whether targeting the systemic vasculature may represent a plausible anti-fibrotic strategy. However, considering their likely contribution to tissue repair, an optimal targeting strategy could entail specific inhibition of their pro-fibrotic or potential pathological properties, instead of a complete impediment of the systemic vasculature. In that light, our integrated meta-analysis revealed a set of 30 genes robustly expressed by systemic ECs across different COVID-19 and IPF studies, patient cohorts and sequencing strategies, with 4 genes robustly upregulated in the disease context, and may thus represent a good starting point for further study into the functional and/or pathological role of these candidates in the systemic vasculature. Whether systemic venous and capillary ECs might, despite their partially overlapping transcriptomes, exhibit distinct functions during either the fibrotic response or vascular repair remains to be elucidated. Notably, the fibrotic response also involves stromal cell types [in which ECM remodelling-related genes/processes were also found upregulated in COVID-19 (see Supplementary material online, Figure S5G and H)], which have to be considered in this context as well.
We acknowledge that our findings are limited by the patient cohort size, COVID-19-associated confounders (e.g. prolonged mechanical ventilation, therapeutic regimens) and require further, functional validation. Furthermore, while in lethal/end-stage COVID-19 patients the virus is considered to no longer actively replicate,72 we did not determine active SARS-CoV-2 infection at the time of death in our patient cohort. We also cannot exclude the possibility that treatment with corticosteroids might have affected (in part) the observed transcriptomic landscape in severe COVID-19 tissues, including the decreased expression of immunoregulatory genes. Nonetheless, the analyzed samples are comparable with previous COVID-19 lung/tissue atlases,14,15 and representative of patients who received the standard-of-care treatments given in the respective clinical setting. While the therapeutic implications of our findings remain elusive, our study has nevertheless contributed to unravelling the heterogeneous composition and potential functions of the vasculature in COVID-19 and IPF lungs, and provides a rich resource for exploration of both vascular and non-vascular cell types in the context of progressive pulmonary fibrosis in these two lethal conditions. In addition, since pulmonary fibrosis is often a long-term consequence of severe COVID-19, we speculate that the abundant presence of systemic ECs may not only pose problems for acute COVID-19 patients, but also for COVID-19 survivors. This is particularly relevant in the context of long COVID, a condition in which the vasculature may play an important role as well.73 Finally, given the current scarcity of model systems that accurately reflect severe/lethal COVID-19, our results shed important novel light into the gene expression landscape of the vasculature in this affliction, and may open up future opportunities regarding screening, monitoring and therapeutic management of (long) COVID patients.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
L.P.M.H.d.R. and L.M.B. designed and analyzed experiments. B.B. and G.P. performed QC and mapping of transcriptomics data. L.-A.T., S.J., L.L, T.V.B., S.J.D., E.M, M.B., L.S., A.-C.K.T, K.R., R.B., V.G., and J.A. performed experiments and/or provided (experimental) support. S.V. performed immunohistochemistry. S.F., Sti.V., A.K.S., S.L., A.D., J.G., P.V.M., J.D.H., T.M., M.K., D.J., V.G., W.W., D.V.R., W.J., L.J.C., B.W., M.L., J.v.D., A.T., E.W., and J.W. contributed to the collection of clinical specimens. L.P.M.H.d.R., L.M.B., and P.C. wrote the manuscript. G.E., M.D., and Lu.S. supervised the study. B.T., M.M., J.N., and D.L. provided insights and/or resources. Lu.S. and P.C. contributed to project administration and funding acquisition. P.C. conceptualized the study. All authors approved the manuscript.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge Ann Manderveld, An Carton, Thomas van Brussel, Rogier Schepers, Naima Dai, Christophe Hermans, Toine Mercier, and Koen De Winne for their technical assistance.
Contributor Information
Laura P M H de Rooij, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, VIB, Leuven 3000, Belgium.
Lisa M Becker, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, VIB, Leuven 3000, Belgium.
Laure-Anne Teuwen, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, VIB, Leuven 3000, Belgium.
Bram Boeckx, Laboratory of Translational Genetics, Center for Cancer Biology, VIB & Department of Genetics, KU Leuven, Leuven 3000, Belgium.
Sander Jansen, Laboratory of Virology & Chemotherapy, KU Leuven, Leuven 3000, Belgium.
Simon Feys, Medical Intensive Care Unit, UZ Gasthuisberg & Laboratory for Clinical Infectious and Inflammatory Disorders, Department of Microbiology, Immunology and Transplantation, KU Leuven, Leuven 3000, Belgium.
Stijn Verleden, Laboratory of Respiratory Diseases and Thoracic Surgery (BREATHE), KU Leuven, Leuven 3000, Belgium.
Laurens Liesenborghs, Laboratory of Virology & Chemotherapy, KU Leuven, Leuven 3000, Belgium.
Anna K Stalder, Institute of Medical Genetics and Pathology, University Hospital Basel, Basel 4031, Switzerland.
Sasha Libbrecht, Department of Pathology, Ghent University Hospital, Ghent University, Ghent 9000, Belgium.
Tina Van Buyten, Laboratory of Virology & Chemotherapy, KU Leuven, Leuven 3000, Belgium.
Gino Philips, Laboratory of Translational Genetics, Center for Cancer Biology, VIB & Department of Genetics, KU Leuven, Leuven 3000, Belgium.
Abhishek Subramanian, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, VIB, Leuven 3000, Belgium.
Sébastien J Dumas, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, VIB, Leuven 3000, Belgium.
Elda Meta, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, VIB, Leuven 3000, Belgium.
Mila Borri, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, VIB, Leuven 3000, Belgium.
Liliana Sokol, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, VIB, Leuven 3000, Belgium.
Amélie Dendooven, Department of Pathology, Ghent University Hospital, Ghent University, Ghent 9000, Belgium; University of Antwerp, Faculty of Medicine, Wilrijk 2610, Belgium.
Anh-Co K Truong, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, VIB, Leuven 3000, Belgium.
Jan Gunst, Laboratory of Intensive Care Medicine, Department of Cellular and Molecular Medicine, KU Leuven, Leuven 3000, Belgium.
Pierre Van Mol, Laboratory of Translational Genetics, Center for Cancer Biology, VIB & Department of Genetics, KU Leuven, Leuven 3000, Belgium.
Jasmin D Haslbauer, Laboratory of Tumor Inflammation and Angiogenesis, Center for Cancer Biology, VIB, Leuven 3000, Belgium.
Katerina Rohlenova, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, VIB, Leuven 3000, Belgium.
Thomas Menter, Institute of Medical Genetics and Pathology, University Hospital Basel, Basel 4031, Switzerland.
Robbert Boudewijns, Laboratory of Virology & Chemotherapy, KU Leuven, Leuven 3000, Belgium.
Vincent Geldhof, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, VIB, Leuven 3000, Belgium.
Stefan Vinckier, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, VIB, Leuven 3000, Belgium.
Jacob Amersfoort, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, VIB, Leuven 3000, Belgium.
Wim Wuyts, Department of Respiratory Medicine, Unit for Interstitial Lung Diseases, UZ Gasthuisberg, Leuven 3000, Belgium.
Dirk Van Raemdonck, Laboratory of Respiratory Diseases and Thoracic Surgery (BREATHE), KU Leuven, Leuven 3000, Belgium.
Werner Jacobs, Medical CBRNe unit, Queen Astrid Military Hospital, Belgian Defense, Neder-Over-Heembeek 1120, Belgium; Department of Forensic Pathology, ASTARC Antwerp University Hospital and University of Antwerp, Antwerp 2610, Belgium.
Laurens J Ceulemans, Laboratory of Respiratory Diseases and Thoracic Surgery (BREATHE), KU Leuven, Leuven 3000, Belgium.
Birgit Weynand, Translational Cell & Tissue Research, Department of Imaging & Pathology, KU Leuven, Leuven 3000, Belgium.
Bernard Thienpont, Laboratory for Functional Epigenetics, Department of Human Genetics, KU Leuven, Leuven 3000, Belgium.
Martin Lammens, Department of Pathology Antwerp University Hospital, Edegem 2560, Belgium; Center for Oncological Research, University of Antwerp, Antwerp 2000, Belgium.
Mark Kuehnel, Medizinische Hochschule Hannover (MHH), Institut für Pathologie, D-30625 Hannover, Germany; Biomedical Research in Endstage and Obstructive Lung Disease Hannover (BREATH) Member of the German Centre for Lung research (DZL), Hannover 30625, Germany.
Guy Eelen, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, VIB, Leuven 3000, Belgium.
Mieke Dewerchin, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, VIB, Leuven 3000, Belgium.
Luc Schoonjans, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, VIB, Leuven 3000, Belgium; Laboratory of Translational Genetics, Center for Cancer Biology, VIB & Department of Genetics, KU Leuven, Leuven 3000, Belgium.
Danny Jonigk, Medizinische Hochschule Hannover (MHH), Institut für Pathologie, D-30625 Hannover, Germany; Biomedical Research in Endstage and Obstructive Lung Disease Hannover (BREATH) Member of the German Centre for Lung research (DZL), Hannover 30625, Germany.
Jo van Dorpe, Department of Pathology, Ghent University Hospital, Ghent University, Ghent 9000, Belgium.
Alexandar Tzankov, Institute of Medical Genetics and Pathology, University Hospital Basel, Basel 4031, Switzerland.
Els Wauters, Laboratory of Respiratory Diseases and Thoracic Surgery (BREATHE), KU Leuven, Leuven 3000, Belgium; Respiratory Oncology Unit, University Hospital KU Leuven, Leuven 3000, Belgium.
Massimiliano Mazzone, Laboratory of Tumor Inflammation and Angiogenesis, Center for Cancer Biology, VIB, Leuven 3000, Belgium; Laboratory of Tumor Inflammation and Angiogenesis, Center for Cancer Biology, Department of Oncology, KU Leuven, Leuven 3000, Belgium.
Johan Neyts, Laboratory of Virology & Chemotherapy, KU Leuven, Leuven 3000, Belgium.
Joost Wauters, Medical Intensive Care Unit, UZ Gasthuisberg & Laboratory for Clinical Infectious and Inflammatory Disorders, Department of Microbiology, Immunology and Transplantation, KU Leuven, Leuven 3000, Belgium.
Diether Lambrechts, Laboratory of Translational Genetics, Center for Cancer Biology, VIB & Department of Genetics, KU Leuven, Leuven 3000, Belgium.
Peter Carmeliet, Laboratory of Angiogenesis and Vascular Metabolism, Department of Oncology and Leuven Cancer Institute (LKI), KU Leuven, VIB Center for Cancer Biology, VIB, Leuven 3000, Belgium; Laboratory of Angiogenesis and Vascular Heterogeneity, Department of Biomedicine, Aarhus University, Aarhus 8000, Denmark; Center for Biotechnology, Khalifa University of Science and Technology, Abu Dhabi, United Arab Emirates.
Funding
This work was supported by the ‘Fonds Wetenschappelijk Onderzoek’ (FWO: P.C., L.P.M.H.d.R., L.M.B, A.-C.K.T., K.R., M.B., J.A., S.F., S.V., and P.V.M.); KU Leuven (L.-A.T.); a Marie Skłodowska-Curie-individual fellowship (HORIZON EUROPE Marie Sklodowska-Curie Actions; L.M.B., J.A., and S.J.D.), Strategisch Basisonderzoek FWO (SB-FWO: V.G.); UZ Antwerp (S.V.); The University Research Fund by the Flemish Government (BOF-UA, S.V.); Broere Charitable Foundation (D.V.R.); the clinical research and education council of the University Hospitals Leuven (J.G.); Stichting tegen Kanker (Mandate for basic & clinical oncology research; E.W.); COVID-19-Fund KU Leuven/UZ Leuven (J.W., J.N.); FWO (G0G4820N; J.N.); FWO Fundamental Clinical Mandate (1833317N; J.W.); European Research Council (ERC) Consolidator Grant (XHale) (771883; D.J.); the Botnar Research Foundation Grant (FTC-2020-10; A.K.S.); the Botnar Research Centre for Child Health (BRCCH) Basel (A.T.); Methusalem funding ("Bijzonder Onderzoeksfonds Methusalem" by the Flemish Government, P.C.), the NNF Laureate Research Grant from Novo Nordisk Foundation (Denmark, P.C.), and Advanced ERC Research Grant (EU-ERC743074; P.C.). The computational resources used in this work were partly provided by the Flemish Supercomputer Center (VSC), funded by the Hercules Foundation and the Flemish Government, Department of Economy, Science and Innovation (EWI).
Data availability
All raw and processed sequencing data generated during this study are available at GEO, under accession code GSE159585. This study did not generate any new code.
Translational perspective.
While assessing clinical and molecular characteristics of severe and lethal COVID-19 cases, the vasculature’s undeniable role in disease progression has been widely acknowledged. COVID-19 lung pathology moreover shares certain clinical features with late-stage idiopathic pulmonary fibrosis (IPF)—yet an in-depth interrogation and direct comparison of the endothelium at single-cell level in both conditions is still lacking. By comparing the transcriptomes of endothelial cells (ECs) from lungs of deceased COVID-19 patients to those from IPF explant and control lungs, we gathered key insights into the heterogeneous composition and potential roles of ECs in both lethal diseases, which may serve as a foundation for development of novel therapeutics.
References
- 1. Gibson PG, Qin L, Puah SH. COVID-19 acute respiratory distress syndrome (ARDS): clinical features and differences from typical pre-COVID-19 ARDS. Med J Aust 2020;213:54–56.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tregoning JS, Flight KE, Higham SL, Wang Z, Pierce BF. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat Rev Immunol 2021;21:626–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shang L, Lye DC, Cao B. Contemporary narrative review of treatment options for COVID-19. Respirology 2021;26:745–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, Cheng L, Li J, Wang X, Wang F, Liu L, Amit I, Zhang S, Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med 2020;26:842–844. [DOI] [PubMed] [Google Scholar]
- 5. Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martinez-Colon GJ, McKechnie JL, Ivison GT, Ranganath T, Vergara R, Hollis T, Simpson LJ, Grant P, Subramanian A, Rogers AJ, Blish CA. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med 2020;26:1070–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wauters E, Van Mol P, Garg AD, Jansen S, Van Herck Y, Vanderbeke L, Bassez A, Boeckx B, Malengier-Devlies B, Timmerman A, Van Brussel T, Van Buyten T, Schepers R, Heylen E, Dauwe D, Dooms C, Gunst J, Hermans G, Meersseman P, Testelmans D, Yserbyt J, Tejpar S, De Wever W, Matthys P, Bosisio M, Casaer M, De Smet F, De Munter P, Humblet-Baron S, Liston A, Lorent N, Martinod K, Proost P, Raes J, Thevissen K, Vos R, Weynand B, Wouters C, Neyts J, Wauters J, Qian J, Lambrechts D; CONTAGIOUS collaborators . Discriminating mild from critical COVID-19 by innate and adaptive immune single-cell profiling of bronchoalveolar lavages. Cell Res 2021;31:272–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang JY, Wang XM, Xing X, Xu Z, Zhang C, Song JW, Fan X, Xia P, Fu JL, Wang SY, Xu RN, Dai XP, Shi L, Huang L, Jiang TJ, Shi M, Zhang Y, Zumla A, Maeurer M, Bai F, Wang FS. Single-cell landscape of immunological responses in patients with COVID-19. Nat Immunol 2020;21:1107–1118. [DOI] [PubMed] [Google Scholar]
- 8. Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE, Alanio C, Kuri-Cervantes L, Pampena MB, D’Andrea K, Manne S, Chen Z, Huang YJ, Reilly JP, Weisman AR, Ittner CAG, Kuthuru O, Dougherty J, Nzingha K, Han N, Kim J, Pattekar A, Goodwin EC, Anderson EM, Weirick ME, Gouma S, Arevalo CP, Bolton MJ, Chen F, Lacey SF, Ramage H, Cherry S, Hensley SE, Apostolidis SA, Huang AC, Vella LA, Betts MR, Meyer NJ, Wherry EJ. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 2020;369:eabc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wen W, Su W, Tang H, Le W, Zhang X, Zheng Y, Liu X, Xie L, Li J, Ye J, Dong L, Cui X, Miao Y, Wang D, Dong J, Xiao C, Chen W, Wang H. Immune cell profiling of COVID-19 patients in the recovery stageby single-cell sequencing. Cell Discov 2020;6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang W, Su B, Pang L, Qiao L, Feng Y, Ouyang Y, Guo X, Shi H, Wei F, Su X, Yin J, Jin R, Chen D. High-dimensional immune profiling by mass cytometry revealed immunosuppression and dysfunction of immunity in COVID-19 patients. Cell Mol Immunol 2020;17:650–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang L, Shi Y, Gong B, Jiang L, Liu X, Yang J, Tang J, You C, Jiang Q, Long B, Zeng T, Luo M, Zeng F, Zeng F, Wang S, Yang X, Yang Z. Blood single cell immune profiling reveals the interferon-MAPK pathway mediated adaptive immune response for COVID-19. medRxiv 2020. [Google Scholar]
- 12. Ren X, Wen W, Fan X, Hou W, Su B, Cai P, Li J, Liu Y, Tang F, Zhang F, Yang Y, He J, Ma W, He J, Wang P, Cao Q, Chen F, Chen Y, Cheng X, Deng G, Deng X, Ding W, Feng Y, Gan R, Guo C, Guo W, He S, Jiang C, Liang J, Li Y-m ,Lin J, Ling Y, Liu H, Liu J, Liu N, Liu S-Q, Luo M, Ma Q, Song Q, Sun W, Wang GX, Wang F, Wang Y, Wen X, Wu Q, Xu G, Xie X, Xiong X, Xing X, Xu H, Yin C, Yu D, Yu K,Yuan J, Zhang B, Zhang P, Zhang T, Zhao J, Zhao P, Zhou J, Zhou W, Zhong S, Zhong X, Zhang S, Zhu L, Zhu P, Zou B, Zou J, Zuo Z, Bai F, Huang X, Zhou P, Jiang Q, Huang Z, Bei J-X, Wei L, Bian X-W, Liu X, Cheng T, Li X, Zhao P, Wang F-S, Wang H, Su B, Zhang Z, Qu K, Wang X, Chen J, Jin R, Zhang Z.. COVID-19 immune features revealed by a large-scale single cell transcriptome atlas. Cell 2021;184:1895–1913.33657410 [Google Scholar]
- 13. Bharat A, Querrey M, Markov NS, Kim S, Kurihara C, Garza-Castillon R, Manerikar A, Shilatifard A, Tomic R, Politanska Y, Abdala-Valencia H, Yeldandi AV, Lomasney JW, Misharin AV, Budinger GRS. Lung transplantation for patients with severe COVID-19. Sci Transl Med 2020;12:eabe4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Delorey TM, Ziegler CGK, Heimberg G, Normand R, Yang Y, Segerstolpe Å, Abbondanza D, Fleming SJ, Subramanian A, Montoro DT, Jagadeesh KA, Dey KK, Sen P, Slyper M, Pita-Juárez YH, Phillips D, Biermann J, Bloom-Ackermann Z, Barkas N, Ganna A, Gomez J, Melms JC, Katsyv I, Normandin E, Naderi P, Popov YV, Raju SS, Niezen S, Tsai LT, Siddle KJ, Sud M, Tran VM, Vellarikkal SK, Wang Y, Amir-Zilberstein L, Atri DS, Beechem J, Brook OR, Chen J, Divakar P, Dorceus P, Engreitz JM, Essene A, Fitzgerald DM, Fropf R, Gazal S, Gould J, Grzyb J, Harvey T, Hecht J, Hether T, Jané-Valbuena J, Leney-Greene M, Ma H, McCabe C, McLoughlin DE, Miller EM, Muus C, Niemi M, Padera R, Pan L, Pant D, Pe'er C, Pfiffner-Borges J, Pinto CJ, Plaisted J, Reeves J, Ross M, Rudy M, Rueckert EH, Siciliano M, Sturm A, Todres E, Waghray A, Warren S, Zhang S, Zollinger DR, Cosimi L, Gupta RM, Hacohen N, Hibshoosh H, Hide W, Price AL, Rajagopal J, Tata PR, Riedel S, Szabo G, Tickle TL, Ellinor PT, Hung D, Sabeti PC, Novak R, Rogers R, Ingber DE, Jiang ZG, Juric D, Babadi M, Farhi SL, Izar B, Stone JR, Vlachos IS, Solomon IH, Ashenberg O, Porter CBM, Li B, Shalek AK, Villani AC, Rozenblatt-Rosen O, Regev A. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 2021;595:107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Melms JC, Biermann J, Huang H, Wang Y, Nair A, Tagore S, Katsyv I, Rendeiro AF, Amin AD, Schapiro D, Frangieh CJ, Luoma AM, Filliol A, Fang Y, Ravichandran H, Clausi MG, Alba GA, Rogava M, Chen SW, Ho P, Montoro DT, Kornberg AE, Han AS, Bakhoum MF, Anandasabapathy N, Suárez-Fariñas M, Bakhoum SF, Bram Y, Borczuk A, Guo XV, Lefkowitch JH, Marboe C, Lagana SM, Del Portillo A, Zorn E, Markowitz GS, Schwabe RF, Schwartz RE, Elemento O, Saqi A, Hibshoosh H, Que J, Izar B. A molecular single-cell lung atlas of lethal COVID-19. Nature 2021;595:114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Varga Z, Flammer AJ, Steiger P, Haberecker M, Andermatt R, Zinkernagel AS, Mehra MR, Schuepbach RA, Ruschitzka F, Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020;395:1417–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Teuwen LA, Geldhof V, Pasut A, Carmeliet P. COVID-19: the vasculature unleashed. Nat Rev Immunol 2020;20:389–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sarelius IH, Glading AJ. Control of vascular permeability by adhesion molecules. Tissue Barriers 2015;3:e985954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Levolger S, Bokkers RPH, Wille J, Kropman RHJ, de Vries JPPM. Arterial thrombotic complications in COVID-19 patients. J Vasc Surg Cases Innov Tech 2020;6:454–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dull RO, Garcia JGN. Leukocyte-Induced microvascular permeability. Circ Res 2002;90:1143–1144. [DOI] [PubMed] [Google Scholar]
- 21. Suresh K, Shimoda LA. Lung circulation. Compr Physiol 2016;6:897–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, Chu SG, Raby BA, DeIuliis G, Januszyk M, Duan Q, Arnett HA, Siddiqui A, Washko GR, Homer R, Yan X, Rosas IO, Kaminski N. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv 2020;6:eaba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schupp JC, Adams TS, Cosme C Jr, Raredon MSB, Yuan Y, Omote N, Poli S, Chioccioli M, Rose KA, Manning EP, Sauler M, DeIuliis G, Ahangari F, Neumark N, Habermann AC, Gutierrez AJ, Bui LT, Lafyatis R, Pierce RW, Meyer KB, Nawijn MC, Teichmann SA, Banovich NE, Kropski JA, Niklason LE, Pe'er D, Yan X, Homer RJ, Rosas IO, Kaminski N. Integrated single cell atlas of endothelial cells of the human lung. Circulation 2021;144:286–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wigén J, Löfdahl A, Bjermer L, Elowsson Rendin L, Westergren-Thorsson G. Converging pathways in pulmonary fibrosis and COVID-19—the fibrotic link to disease severity. Respir Med X 2020;2:100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lechowicz K, Drożdżal S, Machaj F, Rosik J, Szostak B, Zegan-Barańska M, Biernawska J, Dabrowski W, Rotter I, Kotfis K. COVID-19: the potential treatment of pulmonary fibrosis associated with SARS-CoV-2 infection. J Clin Med 2020;9:1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barratt S, Millar A. Vascular remodelling in the pathogenesis of idiopathic pulmonary fibrosis. QJM 2014;107:515–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grillo F, Barisione E, Ball L, Mastracci L, Fiocca R. Lung fibrosis: an undervalued finding in COVID-19 pathological series. Lancet Infect Dis 2020;21:E72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vasarmidi E, Tsitoura E, Spandidos DA, Tzanakis N, Antoniou KM. Pulmonary fibrosis in the aftermath of the COVID-19 era (review). Exp Ther Med 2020;20:2557–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ojo AS, Balogun SA, Williams OT, Ojo OS. Pulmonary fibrosis in COVID-19 survivors: predictive factors and risk reduction strategies. Pulm Med 2020;2020:6175964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Slyper M, Porter CBM, Ashenberg O, Waldman J, Drokhlyansky E, Wakiro I, Smillie C, Smith-Rosario G, Wu J, Dionne D, Vigneau S, Jané-Valbuena J, Tickle TL, Napolitano S, Su MJ, Patel AG, Karlstrom A, Gritsch S, Nomura M, Waghray A, Gohil SH, Tsankov AM, Jerby-Arnon L, Cohen O, Klughammer J, Rosen Y, Gould J, Nguyen L, Hofree M, Tramontozzi PJ, Li B, Wu CJ, Izar B, Haq R, Hodi FS, Yoon CH, Hata AN, Baker SJ, Suvà ML, Bueno R, Stover EH, Clay MR, Dyer MA, Collins NB, Matulonis UA, Wagle N, Johnson BE, Rotem A, Rozenblatt-Rosen O, Regev A. A single-cell and single-nucleus RNA-seq toolbox for fresh and frozen human tumors. Nat Med 2020;26:792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, Chang S, Conley SD, Mori Y, Seita J, Berry GJ, Shrager JB, Metzger RJ, Kuo CS, Neff N, Weissman IL, Quake SR, Krasnow MA. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 2020;587:619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harris WT, Kelly DR, Zhou Y, Wang D, Macewen M, Hagood JS, Clancy JP, Ambalavanan N, Sorscher EJ. Myofibroblast differentiation and enhanced tgf-B signaling in cystic fibrosis lung disease. PLoS One 2013;8:e70196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Habermann AC, Gutierrez AJ, Bui LT, Yahn SL, Winters NI, Calvi CL, Peter L, Chung MI, Taylor CJ, Jetter C, Raju L, Roberson J, Ding G, Wood L, Sucre JMS, Richmond BW, Serezani AP, McDonnell WJ, Mallal SB, Bacchetta MJ, Loyd JE, Shaver CM, Ware LB, Bremner R, Walia R, Blackwell TS, Banovich NE, Kropski JA. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci Adv 2020;6:eaba1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Noble PW, Homer RJ. Back to the future: historical perspective on the pathogenesis of idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol 2005;33:113–120. [DOI] [PubMed] [Google Scholar]
- 35. Goveia J, Rohlenova K, Taverna F, Treps L, Conradi LC, Pircher A, Geldhof V, de Rooij LPMH, Kalucka J, Sokol L, García-Caballero M, Zheng Y, Qian J, Teuwen LA, Khan S, Boeckx B, Wauters E, Decaluwé H, De Leyn P, Vansteenkiste J, Weynand B, Sagaert X, Verbeken E, Wolthuis A, Topal B, Everaerts W, Bohnenberger H, Emmert A, Panovska D, De Smet F, Staal FJT, McLaughlin RJ, Impens F, Lagani V, Vinckier S, Mazzone M, Schoonjans L, Dewerchin M, Eelen G, Karakach TK, Yang H, Wang J, Bolund L, Lin L, Thienpont B, Li X, Lambrechts D, Luo Y, Carmeliet P. An integrated gene expression landscape profiling approach to identify lung tumor endothelial cell heterogeneity and angiogenic candidates. Cancer Cell 2020;37:21–36.e13. [DOI] [PubMed] [Google Scholar]
- 36. Andreatta M, Corria-Osorio J, Müller S, Cubas R, Coukos G, Carmona SJ. Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat Commun 2021;12:2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ackermann M, Verleden SE, Kuehnel M, Haverich A, Welte T, Laenger F, Vanstapel A, Werlein C, Stark H, Tzankov A, Li WW, Li VW, Mentzer SJ, Jonigk D. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in COVID-19. N Engl J Med 2020;383:120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Griffioen AW, Damen CA, Martinotti S, Blijham GH, Groenewegen G. Endothelial intercellular adhesion molecule-1 expression is suppressed in human malignancies: the role of angiogenic factors. Cancer Res 1996;56:1111. [PubMed] [Google Scholar]
- 39. Desai N, Neyaz A, Szabolcs A, Shih AR, Chen JH, Thapar V, Nieman LT, Solovyov A, Mehta A, Lieb DJ, Kulkarni AS, Jaicks C, Xu KH, Raabe MJ, Pinto CJ, Juric D, Chebib I, Colvin RB, Kim AY, Monroe R, Warren SE, Danaher P, Reeves JW, Gong J, Rueckert EH, Greenbaum BD, Hacohen N, Lagana SM, Rivera MN, Sholl LM, Stone JR, Ting DT, Deshpande V. Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection. Nat Commun 2020;11:6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Amersfoort J, Eelen G, Carmeliet P. Immunomodulation by endothelial cells—partnering up with the immune system? Nat Rev Immunol 2022;22:576–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cerutti C, Ridley AJ. Endothelial cell-cell adhesion and signaling. Exp Cell Res 2017;358:31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dorland YL, Huveneers S. Cell-cell junctional mechanotransduction in endothelial remodeling. Cell Mol Life Sci 2017;74:279–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Glass J, Robinson R, Bloom J, Churchwell L, Lee TJ, Sharma A, Sharma S. RNA-Seq reveals IL-6 trans-signaling mediated regulation of paracellular permeability in human retinal endothelial cells. Invest Ophthalmol Vis Sci 2021;62:3122–3122. [Google Scholar]
- 44. Probst CK, Montesi SB, Medoff BD, Shea BS, Knipe RS. Vascular permeability in the fibrotic lung. Eur Respir J 2020;56:1900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stoeltzing O, Ahmad SA, Liu W, McCarty MF, Wey JS, Parikh AA, Fan F, Reinmuth N, Kawaguchi M, Bucana CD, Ellis LM. Angiopoietin-1 inhibits vascular permeability, angiogenesis, and growth of hepatic colon cancer tumors. Cancer Res 2003;63:3370. [PubMed] [Google Scholar]
- 46. Smadja DM, Guerin CL, Chocron R, Yatim N, Boussier J, Gendron N, Khider L, Hadjadj J, Goudot G, Debuc B, Juvin P, Hauw-Berlemont C, Augy JL, Peron N, Messas E, Planquette B, Sanchez O, Charbit B, Gaussem P, Duffy D, Terrier B, Mirault T, Diehl JL. Angiopoietin-2 as a marker of endothelial activation is a good predictor factor for intensive care unit admission of COVID-19 patients. Angiogenesis 2020;23:611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Teuwen LA, Geldhof V, Pasut A, Carmeliet P. COVID-19: the vasculature unleashed. Nat Rev Immunol 2020;20:389–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Winkler ES, Bailey AL, Kafai NM, Nair S, McCune BT, Yu J, Fox JM, Chen RE, Earnest JT, Keeler SP, Ritter JH, Kang LI, Dort S, Robichaud A, Head R, Holtzman MJ, Diamond MS. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol 2020;21:1327–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat Protoc 2020;15:1484–1506. [DOI] [PubMed] [Google Scholar]
- 50. Dill MT, Rothweiler S, Djonov V, Hlushchuk R, Tornillo L, Terracciano L, Meili–Butz S, Radtke F, Heim MH, Semela D. Disruption of Notch1 induces vascular remodeling, intussusceptive angiogenesis, and angiosarcomas in livers of mice. Gastroenterology 2012;142:967–977.e2. [DOI] [PubMed] [Google Scholar]
- 51. Woodruff T, Wu M, Morgan M, Bain N, Jeanes A, Lipman J, Ting M, Taylor S, Coulthard M. Epha4-Fc treatment reduces ischemia/reperfusion-induced intestinal injury by inhibiting vascular permeability. Shock 2016;45:184–191. [DOI] [PubMed] [Google Scholar]
- 52. Hou ST, Nilchi L, Li X, Gangaraju S, Jiang SX, Aylsworth A, Monette R, Slinn J. Semaphorin3A elevates vascular permeability and contributes to cerebral ischemia-induced brain damage. Sci Rep 2015;5:7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Imrie H, Viswambharan H, Sukumar P, Abbas A, Cubbon RM, Yuldasheva N, Gage M, Smith J, Galloway S, Skromna A, Rashid ST, Futers TS, Xuan S, Gatenby VK, Grant PJ, Channon KM, Beech DJ, Wheatcroft SB, Kearney MT. Novel role of the IGF-1 receptor in endothelial function and repair: studies in endothelium-targeted IGF-1 receptor transgenic mice. Diabetes 2012;61:2359–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Higashi Y, Sukhanov S, Shai SY, Danchuk S, Snarski P, Li Z, Hou X, Hamblin MH, Woods TC, Wang M, Wang D, Yu H, Korthuis RJ, Yoshida T, Delafontaine P. Endothelial deficiency of insulin-like growth factor-1 receptor reduces endothelial barrier function and promotes atherosclerosis in apoe-deficient mice. Am J Physiol Heart Circ Physiol 2020;319:H730–H743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang X, Zhou Y, Kim HP, Song R, Zarnegar R, Ryter SW, Choi AMK. Hepatocyte growth factor protects against hypoxia/reoxygenation-induced apoptosis in endothelial cells. J Biol Chem 2004;279:5237–5243. [DOI] [PubMed] [Google Scholar]
- 56. Ding S, Merkulova-Rainon T, Han ZC, Tobelem G. HGF Receptor up-regulation contributes to the angiogenic phenotype of human endothelial cells and promotes angiogenesis in vitro. Blood 2003;101:4816–4822. [DOI] [PubMed] [Google Scholar]
- 57. Mestas J, Hughes CCW. Endothelial cell costimulation of T cell activation through CD58-CD2 interactions involves lipid raft aggregation. J Immunol 2001;167:4378. [DOI] [PubMed] [Google Scholar]
- 58. Le Friec G, Sheppard D, Whiteman P, Karsten CM, Shamoun SAT, Laing A, Bugeon L, Dallman MJ, Melchionna T, Chillakuri C, Smith RA, Drouet C, Couzi L, Fremeaux-Bacchi V, Köhl J, Waddington SN, McDonnell JM, Baker A, Handford PA, Lea SM, Kemper C. The CD46-Jagged1 interaction is critical for human TH1 immunity. Nat Immunol 2012;13:1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ji JD, Park-Min KH, Ivashkiv LB. Expression and function of semaphorin 3A and its receptors in human monocyte-derived macrophages. Hum Immunol 2009;70:211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Casazza A, Laoui D, Wenes M, Rizzolio S, Bassani N, Mambretti M, Deschoemaeker S, Van Ginderachter JA, Tamagnone L, Mazzone M. Impeding macrophage entry into hypoxic tumor areas by Sema3A/Nrp1 signaling blockade inhibits angiogenesis and restores antitumor immunity. Cancer Cell 2013;24:695–709. [DOI] [PubMed] [Google Scholar]
- 61. Hong JH, Lee GT, Lee JH, Kwon SJ, Park SH, Kim SJ, Kim IY. Effect of bone morphogenetic protein-6 on macrophages. Immunology 2009;128:e442–e450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wanstall JC, Gambino A, Jeffery TK, Cahill MM, Bellomo D, Hayward NK, Kay GF. Vascular endothelial growth factor-B-deficient mice show impaired development of hypoxic pulmonary hypertension. Cardiovasc Res 2002;55:361–368. [DOI] [PubMed] [Google Scholar]
- 63. Zhao X, Psarianos P, Ghoraie LS, Yip K, Goldstein D, Gilbert R, Witterick I, Pang H, Hussain A, Lee JH, Williams J, Bratman SV, Ailles L, Haibe-Kains B, Liu FF. Metabolic regulation of dermal fibroblasts contributes to skin extracellular matrix homeostasis and fibrosis. Nat Metab 2019;1:147–157. [DOI] [PubMed] [Google Scholar]
- 64. Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, Fernandez R, Akbarpour M, Chen CI, Ren Z, Verma R, Abdala-Valencia H, Nam K, Chi M, Han S, Gonzalez-Gonzalez FJ, Soberanes S, Watanabe S, Williams KJN, Flozak AS, Nicholson TT, Morgan VK, Winter DR, Hinchcliff M, Hrusch CL, Guzy RD, Bonham CA, Sperling AI, Bag R, Hamanaka RB, Mutlu GM, Yeldandi AV, Marshall SA, Shilatifard A, Amaral LAN, Perlman H, Sznajder JI, Argento AC, Gillespie CT, Dematte J, Jain M, Singer BD, Ridge KM, Lam AP, Bharat A, Bhorade SM, Gottardi CJ, Budinger GRS, Misharin AV. Single-Cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med 2019;199:1517–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, Butte AJ, Bhattacharya M. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 2019;20:163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. D'Agnillo F, Walters KA, Xiao Y, Sheng ZM, Scherler K, Park J, Gygli S, Rosas LA, Sadtler K, Kalish H, Blatti CA III, Zhu R, Gatzke L, Bushell C, Memoli MJ, O'Day SJ, Fischer TD, Hammond TC, Lee RC, Cash JC, Powers ME, O'Keefe GE, Butnor KJ, Rapkiewicz AV, Travis WD, Layne SP, Kash JC, Taubenberger JK. Lung epithelial and endothelial damage, loss of tissue repair, inhibition of fibrinolysis, and cellular senescence in fatal COVID-19. Sci Transl Med 2021;13:eabj7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gillich A, Zhang F, Farmer CG, Travaglini KJ, Tan SY, Gu M, Zhou B, Feinstein JA, Krasnow MA, Metzger RJ. Capillary cell-type specialization in the alveolus. Nature 2020;586:785–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Carsana L, Sonzogni A, Nasr A, Rossi RS, Pellegrinelli A, Zerbi P, Rech R, Colombo R, Antinori S, Corbellino M, Galli M, Catena E, Tosoni A, Gianatti A, Nebuloni M. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: a two-centre descriptive study. Lancet Infect Dis 2020;20:1135–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity 2018;48:434–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Niethamer TK, Stabler CT, Leach JP, Zepp JA, Morley MP, Babu A, Zhou S, Morrisey EE. Defining the role of pulmonary endothelial cell heterogeneity in the response to acute lung injury. Elife 2020;9:e53072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. McCullagh A, Rosenthal M, Wanner A, Hurtado A, Padley S, Bush A. The bronchial circulation—worth a closer look: a review of the relationship between the bronchial vasculature and airway inflammation. Pediatr Pulmonol 2010;45:1–13. [DOI] [PubMed] [Google Scholar]
- 72. Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe 2021;2:e13–e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. de Rooij LPMH, Becker LM, Carmeliet P. A role for the vascular endothelium in post–acute COVID-19? Circulation 2022;145:1503–1505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed sequencing data generated during this study are available at GEO, under accession code GSE159585. This study did not generate any new code.