Abstract
Whipple's disease is a systemic disorder associated with a cultivation-resistant, poorly characterized actinomycete, Tropheryma whippelii. We determined a nearly complete rRNA operon sequence of T. whippelii from specimens from 3 patients with Whipple's disease, as well as partial operon sequences from 43 patients. Variability was observed in the 16S-23S rRNA spacer sequences, leading to the description of five distinct sequence types. One specimen contained two spacer sequence types, raising the possibility of a double infection. Secondary structure models for the primary rRNA transcript and mature rRNAs revealed rare or unique features.
Whipple's disease was described in 1907 (as intestinal lipodystrophy) and is a multisystem disorder of humans involving the intestinal tract as well as various other organs (3). A constant feature of the disease is the presence in affected tissues of uniform bacteria that are approximately 0.2 by 1.5 to 2.5 μm. These bacteria have a characteristic morphology when viewed with electron microscopy (25). However, numerous attempts to cultivate this bacterium have failed (3). In 1997, propagation in human macrophage cell cultures was reported (24), and recently, propagation in human fibroblasts (20), but these findings have not yet been confirmed by other groups. Thus, this organism remains poorly characterized.
Molecular data from the Whipple's disease bacterium became available through broad-range 16S ribosomal DNA (rDNA) PCR and sequence analysis (21, 31). A phylogenetic assessment based on 1,321 bp of 16S rDNA established the bacterium (Tropheryma whippelii) as an actinomycete (21). Taxon-specific 16S rDNA primers have since been used to detected the bacterium in additional patients with Whipple's disease (19, 21, 28, 29), as well as in sewage effluent (16). The determination of a nearly complete 16S rDNA sequence and the 16S-23S ribosomal RNA intergenic spacer sequence has provided additional information (16). A reassessment of its phylogeny revealed a position between the actinomycetes with group B peptidoglycan and the family Cellulomonadaceae.
The 16S-23S rDNA spacer has been used for strain differentiation in a variety of bacterial species (4). Two recent studies, both from the same group in Switzerland, addressed the potential variability of the 16S-23S ribosomal intergenic spacer of T. whippelii (5, 6). In the first study, the spacer region was found to be homogenous in specimens from 9 Swiss individuals; in the second study, three types of intergenic spacers were detected in specimens from 28 individuals. In contrast to the 16S-23S rDNA spacer, little information has been published on the intraspecies variability of the 23S-5S rDNA spacer and of regions immediately upstream and downstream of the rRNA operon.
Because of the paucity of information concerning T. whippelii and the usefulness of the rRNA operon as a source of data for microbial characterization, we determined and analyzed a nearly complete rRNA operon sequence directly from tissues from several patients with Whipple's disease. In addition, we analyzed partial operon sequences from 43 patients.
Patients and clinical specimens.
A total of 56 clinical specimens from 43 patients with Whipple's disease who originated from the United States (11 patients), Germany (28 patients), Switzerland (3 patients), and Austria (1 patient) were examined. The specimens comprised 37 intestinal biopsy specimens, 10 cerebrospinal fluid (CSF) samples, 2 brain tissue specimens, 2 lymph node tissue specimens, 1 tissue specimen associated with prosthetic hip material, 1 colon tissue specimen, 1 ocular lens capsule specimen, 1 blood sample, and 1 synovial fluid specimen. All patients, except three patients from the United States, had Whipple's disease confirmed by histology. These three patients had positive PCR results by CSF analysis, and information on histology was not available. The specimens were prepared for PCR as previously described (20, 29, 30).
Previously uncharacterized regions of the rRNA operon were amplified by PCR with primers designed from known T. whippelii sequences in combination with primers designed from highly conserved regions of other bacteria (Table 1). To obtain sequence material downstream of the 5S rRNA gene, the restriction site PCR method of Sarkar et al. (23) was used (Table 1). PCRs were performed according to standard protocols using AmpliTaq Gold polymerase (PE Applied Biosystems, Foster City, Calif.). Sequences were determined either directly from PCR products or after cloning, as described previously (15).
TABLE 1.
PCR primers and amplified regions within the rRNA operon sequence of T. whippelii
| Primer pair | Region amplifieda | Source and/or reference(s) for primers |
|---|---|---|
| 16Spro1 (5′-TTGAGAACTCAABAGYGTG-3′) and tw318r (5′-CGAAGTTATCCCAAAGTTAG-3′) | 1–298 | This work |
| fD1mod (5′-AGAGTTTGATCYTGGYTYAG-3′) and whip2 (5′-ATTCGCTCCACCTTGCGA-3′) | 193–1404 | 21, 28, 30 |
| whip1 (5′-AGAGATACGCCCCCCGCAA-3′) and 23S rDNA primer 2 (5′-GGTACCTTAGATGTTTCAGTTC-3′) | 1176–2182 | 12, 21, 28 |
| tw1581f (5′-GTGACTTAACCTTTTTGGAGA-3′) and tw2015r (5′-GCATCCACCATTTGCTCTTAAA-3′) | 1602–1993 | This work |
| tw1974f (5′-GTATTTGTGATTCAAGCTAC-3′) and ms37ar (5′-CTGCTTCTAAGCCAACATCCT-3′) | 1994–3136 | This work; 13 |
| tw3002f (5′-TGCCGGTAAGTTAGAGCGCA-3′) and ms38a (5′-GACAAGGAATTTCGCTACCTTA-3′) | 3022–4102 | This work; 13 |
| tw3887f (5′-GCCTGAGGCGTGACGAG-3′) and co5189r (5′-GCTTCCGGGTTCGGAATG-3′) | 3904–5171 | This work |
| tw5104f (5′-CTTGATGTGCGGCCCTTTGC-3′) and tw5745r (5′-AAGATCCCACTGCACTGACATCG-3′) | 5124–5722 | This work |
| tw5068f (5′-ATTGCTTGAAACACATTTTG-3′) or tw5104f and NheI-RSO (5′-AATACGACTCACTATAGGN10GCTAGC-3′) | 5124–5747 | This work; 23 |
In sequence AF190686. Primers are excluded.
Primary rRNA operon sequence and phylogeny.
A nearly complete rRNA operon sequence, spanning 5,747 bp, was assembled from PCR products from the intestinal biopsy specimen of a patient with Whipple's disease (patient 1 [17]). Sequences from at least three independent clones of each PCR product (Table 1) were determined. The sequence begins in the box C element, which is 50 to 380 bp downstream of the start of transcription in other actinomycetes (7, 9, 10, 18), and extends from 162 bp upstream of the 16S rRNA gene to 493 bp downstream of the 5S rRNA gene. The arrangement of rRNA genes is in the order 5′-16S-23S-5S-3′. tRNA genes were not identified.
Phylogenetic analyses were performed as described previously (14) for 16S, 23S, and 5S rRNA, utilizing 1,365, 3,033, and 120 masked nucleotide positions, respectively. Results of 16S rRNA phylogeny were consistent with those of a previous analysis (15). Both 23S rRNA- and 5S rRNA-based phylogenies consistently placed T. whippelii within the Actinobacteria, while a more detailed resolution of relationships between this organism and the actinomycetes with group B peptidoglycan and the cellulomonads was not possible, due to a lack of a sufficient number of related sequences in both 23S rRNA (2) and 5S rRNA (27) databases.
Variability of rRNA operon sequences.
Two additional nearly complete operon sequences of 5,722 bp each, ending with primer tw5745r (Table 1), were determined from a postmortem brain tissue specimen (patient 2) and from a CSF sample (patient 3). The sequence in the sample from patient 2 differed from that of patient 1 at 1 position within the 16S rRNA leader region, 1 position within the 16S rRNA gene, and 10 positions clustered within a region downstream of the 5S rRNA gene (between nucleotides 5322 and 5513). The sequence in the sample from patient 3 had one difference from that of patient 1 within the 16S rRNA gene, two in the 16S-23S ribosomal RNA intergenic spacer, and two in the 23S rRNA gene.
Several operon regions were sequenced from additional patient specimens. These included the 16S rRNA gene leader region (2 specimens from 2 patients; the region between primers 16Spro1 and tw318r) (Table 1), the 16S-23S ribosomal intergenic spacer (53 specimens from 40 patients; primers tw1581f and tw2015r), the region of the insertion in the 23S rRNA gene (1 specimen from 1 patient; primers tw3002f and ms38a), and the 23S-5S ribosomal intergenic spacer (2 specimens from 2 patients; primers tw3887f and co5189r). No differences between these additional sequences and that determined from patient 1 were found in the 16S rRNA gene leader region, the 23S rRNA gene region containing the insertion (positions 3022 to 4102), and the 23S-5S intergenic spacer region.
In previous work, three 16S-23S rDNA spacer sequence types were identified (6, 15). In our study, five types of 16S-23S ribosomal RNA intergenic spacer sequences were found. Type 1 was defined as the original sequence determined for this spacer region (15); it corresponded to the sequence of patients 1 and 2 as described above and was found in a total of 16 specimens from 14 patients. Type 2 (6) was most frequently detected; it corresponded to that of patient 3 and was found in 36 specimens from 26 patients. Type 3, as defined by Hinrikson et al. (6), was not found in any specimen. Types 4 through 6 have not been previously described and were each found in single patients (designated patients 4 to 6). There were a total of 10 variable nucleotide positions and two to six differences with the type 1 sequence. The differences, with reference to type 1 and numbered according to this type's 294-bp spacer sequence, were as follows: type 2, T56C and Δ86; type 4, A94G, C98T, G115A, and G200A; type 5, A94G, G115A, and G200A; type 6, T56C, Δ86, C124T, C143T, T148C, and A157G. To confirm the existence of distinct spacer types by an independent method, PCR products (primers tw1581f and tw2015r) from each of these types were subjected to digestion with the restriction endonucleases HaeII, HinfI, MseI, MslI, XmnI, and TaaI. Distinct and expected band patterns were obtained for each spacer type.
There were 11 patients from whom more than one specimen was examined. These comprised five patients from Germany and one from Switzerland with more than one intestinal biopsy specimen, four patients from Germany with intestinal biopsy and CSF specimens, and one patient from the United States (patient 6) with an intestinal biopsy and a blood specimen. Multiple clones from individual PCR products (range, 2 to 10 clones) were sequenced for 19 patients. In all instances (except for patient 5, whose case is discussed below), 16S-23S spacer types were consistent among different clinical samples from the same patient and between different clones of the same PCR product. This is consistent with the hypothesis that a single bacterial clone undergoes systemic dissemination (29) and is further supported by the results from patient 3, who had an intestinal biopsy taken in 1987 and two CSF specimens taken in 1998 and 1999, all with identical spacer types, and by the results of patient 6, whose samples from different sites yielded the same, unique type 6 spacer sequence. Nucleotide differences observed elsewhere in the operon sequence did not correlate with 16S-23S spacer type.
Geographic analysis of 16S-23S spacer types showed that 10 of 28 German patients had type 1, 17 had type 2, and 1 had a mixed type 2 and 5 infection (discussed below). Of 11 patients from the United States, 3 had type 1, 7 had type 2, and 1 had type 6. Of three patients from Switzerland, two had type 2, and one had type 1. The single patient from Austria had a type 4 sequence. While the number of specimens from each area is relatively small, it appears clear that the two common types (types 1 and 2) are observed at similar frequencies in patients from both the United States and Europe, suggesting that a differential distribution of T. whippelii between the Old World and the New World, such as has been described for Borrelia burgdorferi sensu lato and its genospecies (1), is unlikely.
The five 16S-23S rRNA spacer types found in our 43 patients expand our understanding of T. whippelii sequence diversity (6). Whipple's disease was confirmed by histological methods in all but 3 of our 43 patients. We found a predominance of spacer type 2 (65%), whereas the Swiss study (6) described a predominance of type 1. The previously reported uniformity of spacer type 1 in Swiss individuals (5) could not be confirmed.
Sequence variability within a single host.
In the case of patient 5, four clones of the PCR-amplified 16S-23S spacer were sequenced: three yielded an identical but novel type 5 sequence, and one contained a type 2 sequence. Direct sequencing and restriction endonuclease analysis of the PCR product revealed expected sequence ambiguities corresponding to differences between sequence types 2 and 5. These findings suggest that both sequence types were present in the patient's specimen and raise two possibilities: either the bacterium has more than one rRNA operon, or the patient was infected by two different bacterial strains. While it is not possible to draw a firm conclusion between these two alternatives, there are two findings consistent with the notion of a single rRNA operon in T. whippelii. First, there was concordance of rRNA spacer type between different clinical samples and clones of PCR products from specimens from each of the other patients (discussed above). Second, amplification of genomic DNA flanking the 3′ end of the operon by restriction site PCR (24) from the specimen of patient 1 generated two fragments in the same reaction, yielding 491- and 677-bp sequences, respectively; the sequence of the 491-bp fragment extended 206 bp downstream of the 5S rRNA gene and was identical to the sequence of the 677-bp PCR product in the region of overlap. Thus, this initial analysis failed to find variability in the genome adjacent to, but outside of, the rRNA operon. For comparison, in the Mycobacterium tuberculosis genome (accession no. Z73902), a non-rRNA gene begins 120 bp downstream of the 5S rDNA. Thus, the second explanation, i.e., a dual infection by different T. whippelii strains, appears more likely.
Structural and regulatory features of the primary RNA transcript.
A secondary structure model of the primary rRNA transcript was constructed by visual comparison to published structures (7–11, 18), with some assistance from the Mfold program (Genetics Computer Group, Madison, Wisc.). The proposed model is shown in Fig. 1. This model is consistent with published models for other members of the class Actinobacteria (7–11, 19) and shows two stems, one bracketing pre-16S rRNA and consisting of the 16S rRNA leader region and the 5′ portion of the 16S-23S rRNA spacer (spacer 1a), and another bracketing pre-23S rRNA and consisting of the 3′ portion of the 16S-23S rRNA spacer (spacer 1b) and the 23S-5S rRNA spacer (spacer 2). The 5′ end of the model begins with primer 16Spro1, which is located within and between the box A and box C elements of the operon leader region. The polymorphic positions within the 16S-23S spacer were mapped on the secondary structure model (Fig. 1). According to the model, all nucleotide differences between 16S-23S spacer types either are located in loops, where they are not involved in base pairing, or create tolerated changes in paired positions that preserve the structure.
FIG. 1.
Proposed secondary structure model of the primary RNA transcript of the T. whippelii rRNA operon. Helices are numbered according to their location in the leader (L), the 5′ portion of the 16S-23S rRNA spacer (S1a), the 3′ portion of the 16S-23S rRNA spacer (S1b), and the 23S-5S rRNA spacer (S2). Helix T is a putative Rho-independent terminator. The locations of the primer 16Spro1 and the regulatory elements (box A and box C) are indicated by braces. Numbers and nucleotides in boxes indicate 16S-23S rRNA spacer types and nucleotide changes in these types, respectively. Helix S1b-1 contains a putative box B element. Asterisk, no information is available for this nucleotide in 16S-23S rRNA spacer types 4 and 5.
Structural features associated with mature rRNAs.
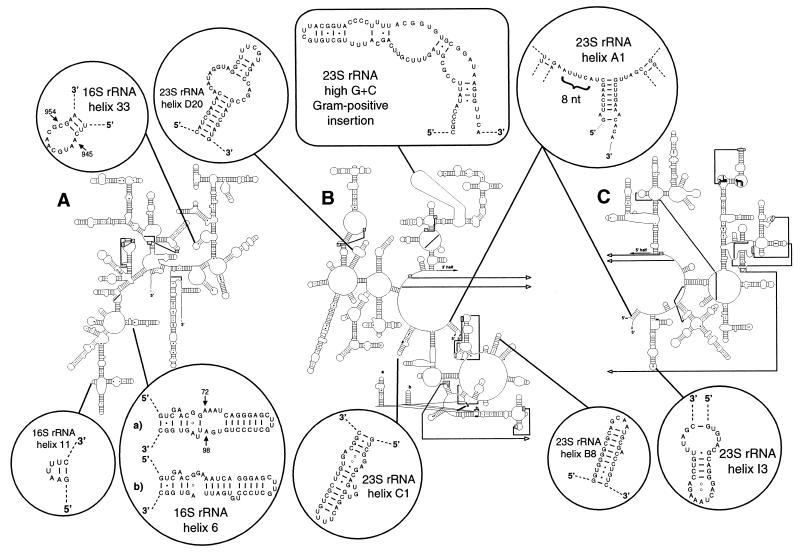
Putative secondary structures of 16S, 23S, and 5S rRNAs were inferred by manual alignment to published structures (18; http://www.rna.icmb.utexas.edu/). The proposed model for 5S rRNA (data not shown) was identical to a previously-published model for Streptomyces ambofaciens (18), except for a paired set of nucleotides in T. whippelii 5S rRNA (G20 and C63). Several regions in the T. whippelii predicted 16S and 23S rRNA secondary structures were found to differ from those of other actinomycetes (http://www.rna.icmb.utexas.edu/) (Fig. 2). Most of these variable regions involved differences in helix length and unpaired or unilateral bulges along helix stems. Three features of the 16S rRNA and 23S rRNA, however, were found to be unique to T. whippelii, after comparison with a larger set of structures and sequences (10,073 16S rRNA and 503 23S rRNA bacterial sequences). One of these features is the absence of an otherwise conserved pairing between 16S rRNA positions 945 and 954 in helix 33 (Fig. 2). The second was observed in the vicinity of the stem formed by the 5′ and 3′ ends of mature 23S rRNA (Fig. 2). Here, the central loop of 23S rRNA consists of eight nucleotides, whereas six are present in nearly all other bacterial 23S rRNAs. The third feature was located in helix I3 of 23S rRNA; it consisted of two noncanonical G-A pairings at the end of the helix, closed by a pentaloop. Except for Micrococcus luteus, which has the same constellation, all other bacteria in the database have a shorter helix without G-A pairings, closed by a tetraloop. G-A pairings are only observed in archaea and eukaryotes.
FIG. 2.
Variable and distinguishing structures of T. whippelii rRNAs. (A) 16S rRNA; (B) 5′ half of 23S rRNA; (C) 3′ half of 23S rRNA. 16S rRNA helix 33 and 23S rRNA helices A1 and I3 are considered unique to T. whippelii (see text); the others have helix length and bulge variabilities relative to the structures of other actinomycetes (http://www.rno.icmb.vtexas.edu/). Arrows, with positions in mature rRNAs, indicate unusual nucleotides. There are two possible structures (a and b) for helix 6 in the 16S rRNA. The 23S rRNA insertion of the high-G+C-content gram-positive bacteria is located at positions 3522 to 3601 of the sequence AF190686.
The insertion sequences of high-G+C-content gram-positive bacteria in the 23S rRNA determined by Roller et al. (22) were visually examined for possible conserved base pairings and covariation at paired positions, and the T. whippelii insertion sequence was aligned to these sequences with the ARB program package (26). The insertion of T. whippelii (80 bp) was quite different from the insertions of other actinomycetes (86 to 116 bp) with respect to both length and primary sequence. It had similarity values of only 25 to 43% with those of other actinomycetes, while the other previously determined insertion sequences (22) had values of 42 to 81% among each other. This significant divergence from the other actinomycetes seems intriguing and suggests an unusual evolutionary history for this organism.
Conclusions.
The nearly complete T. whippelii rRNA operon sequence is currently the largest body of molecular data for this medically important organism, an organism that has otherwise resisted characterization. The sequence will provide targets for new diagnostic primers and probes. Several features are particularly noteworthy, such as (i) the very low degree of nucleotide similarity of the 23S rRNA insertion with those of other actinomycetes and (ii) rare or unique rRNA secondary structures, which support a relatively distant relationship of T. whippelii to the cultivated members of the Actinobacteria. The possibility of dual infection of a host by two T. whippelii strains is raised for the first time. Our identification of new 16S-23S rRNA intergenic spacer types and downstream genomic polymorphisms may facilitate further analysis of the ecology of this organism and the epidemiology of Whipple's disease.
Nucleotide sequence accession numbers.
The rRNA operon sequence determined from patient 1 has been deposited in the GenBank and EMBL databases under the accession no. AF190686. The operon sequences of patients 2 and 3 and the 16S-23S ribosomal RNA intergenic spacer types 4 to 6 of patients 4, 5, and 6 have been assigned accession no. AF190687 through AF190691. The secondary structures of 16S rRNA and 23S rRNA have been deposited at the rRNA secondary structure website (http://www.rna.icmb.utexas.edu/).
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (Ma 1663/3-1) to M.M. and by grants from the Lucille P. Markey Charitable Trust and the Donald and Delia Baxter Foundation to D.A.R.
We thank D. H. Persing, P. S. Mitchell, and J. N. Thorvilson (Rochester, Minn.) for providing specimens from 11 patients diagnosed at the Mayo Clinic; M. Schneemann (Zürich, Switzerland) for providing specimens from 2 Swiss patients; I. J. McLaughlin (Applied Biosystems) for technical advice and support, R. R. Gutell (Austin, Tex.) for assistance with rRNA secondary structure analysis, and T. M. Schmidt (Michigan State University, East Lansing, Mich.) for advice regarding rRNA sequence analysis.
REFERENCES
- 1.Baranton G, Postic D, Saint-Girons I, Boerlin P, Piffaretti J C, Assous M, Grimont P A D. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS461 associated with Lyme borreliosis. Int J Syst Bacteriol. 1992;42:378–383. doi: 10.1099/00207713-42-3-378. [DOI] [PubMed] [Google Scholar]
- 2.De Rijk P, Robbrecht E, de Hoog S, Caers A, van de Peer Y, De Wachter R. Database on the structure of large subunit ribosomal RNA. Nucleic Acids Res. 1999;27:174–178. doi: 10.1093/nar/27.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dobbins W O., III . Whipple's disease. Springfield: Charles C Thomas, Publisher; 1987. p. 242. , III. [Google Scholar]
- 4.Gürtler V, Stanisich V A. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 5.Hinrikson H P, Dutly F, Altwegg M. Homogeneity of 16S-23S ribosomal intergenic spacer regions of Tropheryma whippelii in Swiss patients with Whipple's disease. J Clin Microbiol. 1999;37:152–156. doi: 10.1128/jcm.37.1.152-156.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinrikson H P, Dutly F, Nair S, Altwegg M. Detection of three different types of ‘Tropheryma whippelii’ directly from clinical specimens by sequencing, single-strand conformation polymorphism (SSCP) analysis and type-specific PCR of their 16S-23S ribosomal intergenic spacer region. Int J Syst Bacteriol. 1999;49:1701–1706. doi: 10.1099/00207713-49-4-1701. [DOI] [PubMed] [Google Scholar]
- 7.Ji Y, Colston M J, Cox R A. Nucleotide sequence and secondary structures of precursor 16S rRNA of slow-growing mycobacteria. Microbiology. 1994;140:123–132. doi: 10.1099/13500872-140-1-123. [DOI] [PubMed] [Google Scholar]
- 8.Ji Y, Kempsell K E, Colston M J, Cox R A. Nucleotide sequences of the spacer-1, spacer-2 and trailer regions of the rrn operons and secondary structures of precursor 23S rRNAs and precursor 5S rRNAs of slow-growing mycobacteria. Microbiology. 1994;140:1763–1773. doi: 10.1099/13500872-140-7-1763. [DOI] [PubMed] [Google Scholar]
- 9.Ji Y, Colston M J, Cox R A. The ribosomal RNA (rrn) operons of fast-growing mycobacteria: primary and secondary structures and their relation to rrn operons of pathogenic slow-growers. Microbiology. 1994;140:2829–2840. doi: 10.1099/00221287-140-10-2829. [DOI] [PubMed] [Google Scholar]
- 10.Kempsell K E, Ji Y, Estrada I C E, Colston M J, Cox R A. The nucleotide sequence of the promoter, 16S rRNA and spacer region of the ribosomal RNA operon of Mycobacterium tuberculosis and comparison with Mycobacterium leprae precursor rRNA. J Gen Microbiol. 1992;138:1717–1727. doi: 10.1099/00221287-138-8-1717. [DOI] [PubMed] [Google Scholar]
- 11.Kim E, Kim H, Hong S-P, Kang K H, Kho Y H, Park Y H. Gene organization and primary structure of a ribosomal RNA gene cluster from Streptomyces griseus subsp. griseus. Gene. 1993;132:21–31. doi: 10.1016/0378-1119(93)90510-a. [DOI] [PubMed] [Google Scholar]
- 12.Kostman J R, Edlind T D, LiPuma J J, Stull T L. Molecular epidemiology of Pseudomonas cepacia determined by polymerase chain reaction ribotyping. J Clin Microbiol. 1992;30:2084–2087. doi: 10.1128/jcm.30.8.2084-2087.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotilainen P, Jalava J, Meurman O, Lehtonen O-P, Rintala E, Seppälä O-P, Eerola E, Nikkari S. Diagnosis of meningococcal meningitis by broad-range bacterial PCR with cerebrospinal fluid. J Clin Microbiol. 1998;36:2205–2209. doi: 10.1128/jcm.36.8.2205-2209.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kroes I, Lepp P W, Relman D A. Bacterial diversity within the human subgingival crevice. Proc Natl Acad Sci USA. 1999;96:14547–14552. doi: 10.1073/pnas.96.25.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maiwald M, Ditton H-J, von Herbay A, Rainey F A, Stackebrandt E. Reassessment of the phylogenetic position of the bacterium associated with Whipple's disease and determination of the 16S-23S ribosomal intergenic spacer sequence. Int J Syst Bacteriol. 1996;46:1078–1082. doi: 10.1099/00207713-46-4-1078. [DOI] [PubMed] [Google Scholar]
- 16.Maiwald M, Schuhmacher F, Ditton H-J, von Herbay A. Environmental occurrence of the Whipple's disease bacterium (Tropheryma whippelii) Appl Environ Microbiol. 1998;64:760–762. doi: 10.1128/aem.64.2.760-762.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meier-Willersen H J, Maiwald M, von Herbay A. Morbus Whipple in Assoziation mit opportunistischen Infektionen. Dtsch Med Wochenschr. 1993;118:854–860. doi: 10.1055/s-2008-1059397. [DOI] [PubMed] [Google Scholar]
- 18.Pernodet J-L, Boccard F, Alegre M-T, Gagnat J, Guérineau M. Organization and nucleotide sequence analysis of a ribosomal RNA gene cluster from Streptomyces ambofaciens. Gene. 1989;79:33–46. doi: 10.1016/0378-1119(89)90090-5. [DOI] [PubMed] [Google Scholar]
- 19.Ramzan N N, Loftus E, Burgart L J, Rooney M, Batts K P, Wiesner R H, Fredricks D N, Relman D A, Persing D H. Diagnosis and monitoring of Whipple disease by polymerase chain reaction. Ann Intern Med. 1997;126:520–527. doi: 10.7326/0003-4819-126-7-199704010-00004. [DOI] [PubMed] [Google Scholar]
- 20.Raoult D, Birg M L, La Scola B, Fournier P E, Enea M, Lepidi H, Roux V, Piette J C, Vandenesch F, Vital-Durand D, Marrie T J. Cultivation of the bacillus of Whipple's disease. N Engl J Med. 2000;342:620–625. doi: 10.1056/NEJM200003023420903. [DOI] [PubMed] [Google Scholar]
- 21.Relman D A, Schmidt T M, MacDermott R P, Falkow S. Identification of the uncultured bacillus of Whipple's disease. N Engl J Med. 1992;327:293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 22.Roller C, Ludwig W, Schleifer K H. Gram-positive bacteria with a high DNA G+C content are characterized by a common insertion within their 23S rRNA genes. J Gen Microbiol. 1992;138:1167–1175. doi: 10.1099/00221287-138-6-1167. [DOI] [PubMed] [Google Scholar]
- 23.Sarkar G, Turner R T, Bolander M E. Restriction-site PCR: A direct method of unknown sequence retrieval adjacent to a known locus by using universal primers. PCR Methods Appl. 1993;2:318–322. doi: 10.1101/gr.2.4.318. [DOI] [PubMed] [Google Scholar]
- 24.Schoedon G, Goldenberger D, Forrer R, Gunz A, Dutly F, Höchli M, Altwegg M, Schaffner A. Deactivation of macrophages with interleukin-4 is the key to the isolation of Tropheryma whippelii. J Infect Dis. 1997;176:672–677. doi: 10.1086/514089. [DOI] [PubMed] [Google Scholar]
- 25.Silva M T, Macedo P M, Moura Nunes J F. Ultrastructure of bacilli and the bacillary origin of the macrophagic inclusions in Whipple's disease. J Gen Microbiol. 1985;131:1001–1013. doi: 10.1099/00221287-131-5-1001. [DOI] [PubMed] [Google Scholar]
- 26.Strunk, O., O. Gross, B. Reichel, M. May, S. Hermann, N. Stuckmann, B. Nonhoff, T. Ginhart, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, K.-H. Schleifer, and W. Ludwig. ARB: a software environment for sequence data. Department of Microbiology, Technische Universität München, Munich, Germany.
- 27.Szymanski M, Barciszewska M Z, Barciszewski J, Erdmann V A. 5S ribosomal RNA data bank. Nucleic Acids Res. 1999;27:158–160. doi: 10.1093/nar/27.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Herbay A, Ditton H J, Maiwald M. Diagnostic application of a polymerase chain reaction assay for the Whipple's disease bacterium to intestinal biopsies. Gastroenterology. 1996;110:1735–1743. doi: 10.1053/gast.1996.v110.pm8964398. [DOI] [PubMed] [Google Scholar]
- 29.von Herbay A, Ditton H J, Schuhmacher F, Maiwald M. Whipple's disease: staging and monitoring by cytology and polymerase chain reaction analysis of cerebrospinal fluid. Gastroenterology. 1997;113:434–441. doi: 10.1053/gast.1997.v113.pm9247461. [DOI] [PubMed] [Google Scholar]
- 30.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson K H, Blitchington R, Frothingham R, Wilson J A P. Phylogeny of the Whipple's disease-associated bacterium. Lancet. 1991;338:474–475. doi: 10.1016/0140-6736(91)90545-z. [DOI] [PubMed] [Google Scholar]