Abstract
Based upon the nucleotide sequence of the relA gene from Escherichia coli, a gene fragment corresponding to the homologous gene from the pathogenic oral bacterium Porphyromonas gingivalis 381 was isolated by PCR and utilized to construct a relA mutant. The mutant, KS7, was defective in ribosome-mediated ppGpp formation and also in the stringent response.
Porphyromonas gingivalis is a black-pigmented, gram-negative bacterium which has been strongly implicated in chronic destructive periodontitis (19). Several potential virulence factors of these organisms have been characterized, including fimbriae (10). The fimbriae of P. gingivalis mediate oral colonization by serving as an adhesin for attachment to saliva-coated tooth surfaces, antecedent gram-positive bacteria, and host cells (3, 4, 11). Mutants defective in the major fimbrial subunit, FimA, have also been demonstrated to be less virulent in a rat model system (14).
Although environmental factors have been shown to alter the expression of the fimA gene in P. gingivalis (2), the molecular basis for such modulation has not yet been determined. Therefore, this investigation was initiated to examine the potential role of a growth-related regulatory mechanism, the stringent response, on virulence gene expression in these organisms. Bacteria undergoing nutritional stress, such as amino acid deprivation, synthesize ppGpp and pppGpp as part of a phenomenon termed the stringent response (7). These nucleotides have been shown to play a role in growth phase-regulated gene expression either directly or indirectly (9). Therefore, it was of interest to determine if the stringent response plays a role in regulating the expression of important virulence factors in P. gingivalis.
Construction of a P. gingivalis relA mutant.
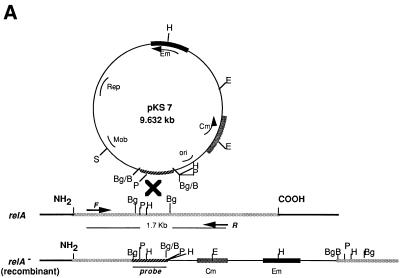
In order to examine the role of the stringent response in modulating the physiology of P. gingivalis, a strategy was utilized to construct a relA mutant of these organisms. The incomplete P. gingivalis W83 genome sequence database of The Institute for Genomic Research was searched with the BLAST program (1) using the Escherichia coli relA gene for comparison, and two open reading frames (ORFs) with significant homologies were identified. ORF1 on fragment 155 showed 29% identity and 40% amino acid similarity with the E. coli RelA over 767 amino acids, while ORF2 on fragment 40 revealed 28% identity and 41% similarity over 781 amino acids. A domain characteristic of SpoT enzymes (5) was also present in ORF2. Both ORFs exhibited several of the conserved regions found in relA genes of different bacteria (15, 16) (Fig. 1). This observation was not unexpected, as several gram-negative bacteria contain two genes, relA and spoT, which are homologous and can each synthesize ppGpp (18). To amplify an internal fragment from ORF1, the primers 5′-AAGAAGTACGCAGTCTG-CTG-3′ (F1) and 5′-GTTGTCGATAACCAATACGT-3′ (R1) were synthesized; a pair, F2 and R2, for amplifying a fragment from ORF2 were also constructed. Standard PCR conditions were utilized for each amplification. The 1.7-kb PCR product from ORF1 was digested with BglII to generate an 862-bp fragment, which was inserted into the BamHI site of the suicide plasmid pKDCMZ (17), yielding plasmid pKS7. Plasmid pKS7 was transformed into E. coli HB101 containing the helper plasmid R751. This strain was then mated with P. gingivalis 381 as previously described (13), and the transconjugants were selected on Trypticase soy broth agar plates containing both gentamicin (50 μg/ml) and erythromycin (10 μg/ml). Southern blot analysis (Fig. 2A) confirmed that 2 of 12 transconjugants examined had undergone the predicted single crossover recombination event. When digested with HindIII, the DNA of the transconjugants displayed two additional bands of 534 bp and 4.162 kbp (Fig. 2B, lanes 2 and 3; sizes determined from the P. gingivalis W83 genome database). The identity of the latter band was not clear, as a similar-size band was also produced in parental strain 381. This band most likely arose from one of the flanking regions which was recognized by the probe and masked the similar-size fragment from the transconjugants. The chromosomal DNA from one of the transconjugants, KS7, was therefore digested with a second restriction enzyme, PstI. Digestion with this enzyme should give rise to two bands of 695 bp and 8.667 kbp if a single crossover event had occurred (Fig. 2A). Two positive bands corresponding to the predicted sizes were observed in the transconjugant (Fig. 2B, lane 2) but not for the wild-type strain (Fig. 2B, lane 1), confirming that ORF1 had been disrupted. However, none of the transconjugants resulting from the use of the ORF2 amplicon yielded insertions in ORF2, and they were not further analyzed. It should be noted that attempts to inactivate the spoT gene in wild-type E. coli have also been unsuccessful (20).
FIG. 1.
Sequence alignments of ORF1 (PgORF1), ORF2 (PgORF2), E. coli relA (EcrelA), and E. coli spoT (EcspoT). The sequences were aligned with the aid of the Pileup program of the Genetics Computer Group software package. Amino acid 1 is the first amino acid of ORF2. The amino acids in bold letters are the conserved amino acids found in at least three of the sequences. F1 and R1 represent regions from the corresponding nucleotide sequence of ORF1 from which the primers were designed to generate the 1.7-kb fragment by PCR. F2 and R2 are the primers designed from the nucleotide sequence of ORF2.
FIG. 2.
Southern blot analysis of chromosomal DNA from the transconjugants. (A) Schematic representation of the relA region of KS7 and the single crossover integration event using the suicide plasmid pKS7. (B) The chromosomal DNAs of strains 381 and KS7 were digested with HindIII or PstI and subjected to agarose gel electrophoresis and Southern blot hybridization with the relA probe (shown in panel A). Lane 1, DNA from strain 381 in each blot; lanes 2 and 3, DNA from two transconjugants which were digested with HindIII (left blot). DNA from one of the transconjugants, KS7, was further digested with PstI (right blot, lane 2). Arrowheads with molecular sizes point to the positions of the unique bands observed for the transconjugants.
Properties of mutant KS7.
In order to determine if ORF1 is the relA homolog in P. gingivalis, the RelA activity of mutant KS7 was directly determined. Ribosomes were prepared from E. coli or P. gingivalis cells essentially as previously described (6). RelA activity, determined from (p)ppGpp synthesis, was carried out as described by Krohn and Wagner (12). Routinely, 50-μl reaction mixtures contained an amount of ribosomes equivalent to an optical density at 260 nm of 3.0, 2.0 mM ATP, and 1.3 mM GTP. Either [γ-32P]ATP or [γ-32P]GTP (3.0 μCi) was used to label the ppGpp and pppGpp. The P. gingivalis ribosome mixtures also contained 2.0 mM dithiothreitol. The reactions were carried out at 30°C for E. coli ribosomes for 1 h, or at 37°C for 4 h anaerobically for the P. gingivalis ribosomes. After precipitation of the proteins with formic acid the supernatant fluids were analyzed by thin-layer chromatography with polyethylene cellulose plates (EM Sciences, Gibbstown, N.J.). After development with 1.5 M potassium phosphate, pH 3.4, the plates were dried and exposed to X-ray film overnight.
Since RelA activity is associated with ribosomes while SpoT is a cytoplasmic protein, ppGpp synthesis by the P. gingivalis ribosomes would be a measure of RelA activity. RelA activity was observed for the P. gingivalis ribosomes incubated anaerobically, since ppGpp, but not pppGpp, could be readily detected (Fig. 3). It is not clear whether the absence of the latter guanosine nucleotide indicates that this molecule is not synthesized in P. gingivalis under these conditions or that the molecule is unstable. As a positive control, ribosomes were isolated from E. coli CF3120, which harbors a plasmid containing the E. coli relA gene (kindly provided by M. Cashel, National Institutes of Health, Bethesda, Md.), and synthesizes large amounts of both ppGpp and pppGpp. However, neither ppGpp nor pppGpp was synthesized in detectable amounts by mutant KS7 under these conditions. These results suggested that ORF1 corresponds to the relA gene of P. gingivalis since SpoT would not be ribosome associated. It is also tempting to speculate that ORF2 detected in the present study corresponds to the spoT gene homolog, but this proposal will require additional investigation.
FIG. 3.
Synthesis of phosphorylated guanosine nucleotides by purified ribosomes from P. gingivalis and E. coli. Ribosomes from E. coli and P. gingivalis were isolated and incubated with GTP and ATP as described in the text. The reaction products were subjected to thin-layer chromatography analysis. Lane 1, no ribosomes; lanes 2, 3, and 4, ribosomes of E. coli CF 3120, P. gingivalis 381, and P. gingivalis KS7, respectively. The positions of the phosphorylated nucleotides are marked.
In order to further confirm that ORF1 codes for a RelA homolog, stable RNA synthesis was compared in wild-type 381 and mutant KS7 (Fig. 4). Under amino acid-limiting conditions induced by the addition of serine hydroxamate (8), RNA synthesis was inhibited in strain 381 but not significantly in mutant KS7. Inhibition of stable RNA synthesis by serine hydroxamate in P. gingivalis was not as pronounced as in E. coli (8) due to the necessity of utilizing a complex medium containing peptides for the former organisms as well as their relatively slower growth. Nevertheless, the differences in stable RNA accumulation in the two strains is consistent with the identification of the relA gene in P. gingivalis. Preliminary data further suggest that inactivation of this gene also resulted in reduced fimbrial expression in these organisms. These, as well as other phenotypic changes related to virulence, are currently under investigation.
FIG. 4.
Stringent response of P. gingivalis 381 and mutant KS7. Cells were grown in Trypticase soy broth to the end of the logarithmic phase in the presence of 32PO4 (25 μCi/ml). The arrows indicate the time of addition of serine hydroxamate (1.0 mg/ml) to each of duplicate cultures. The alkali-labile, trichloroacetic acid-precipitable radioactivity was measured as RNA, as previously described (8). (A) Wild-type 381; (B) relA mutant KS7. Symbols: □, no serine hydroxamate; ▵, serine hydroxamate. Each measurement is the average of triplicate samples.
Acknowledgments
We thank J. Hernandez for helpful advice and discussions.
This investigation was supported in part by National Institutes of Health grant DE08293.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J H, Zhang Z, Miller W, Lipman D J. GAPPED BLAST and PSI-BLAST—a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3390–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amano A, Sharma A, Sojar H T, Kuramitsu H K, Genco R J. Effects of temperature stress on expression of fimbriae and superoxide dismutase by Porphyromonas gingivalis. Infect Immun. 1994;62:4682–4685. doi: 10.1128/iai.62.10.4682-4685.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amano A, Shizukuishi S, Horie H, Kimura S, Morisaki I, Hamada S. Binding of Porphyromonas gingivalis fimbriae to proline-rich glycoproteins in parotid saliva via a domain shared by major salivary components. Infect Immun. 1998;66:2072–2077. doi: 10.1128/iai.66.5.2072-2077.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amano A, Fujiwara T, Nagata H, Kuboniwa M, Sharma A, Sojar H T, Genco R J, Hamada S, Shizukuishi S. Porphyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralis through specific domains. J Dent Res. 1997;76:852–857. doi: 10.1177/00220345970760040601. [DOI] [PubMed] [Google Scholar]
- 5.Avarind L, Koonin E V. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23:469–472. doi: 10.1016/s0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- 6.Cammack K A, Wade H E. The sedimentation behavior of ribonuclease: active and inactive ribosomes from bacteria. Biochem J. 1965;96:671–680. doi: 10.1042/bj0960671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cashel M, Rudd K E. The stringent response. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C.: American Society for Microbiology; 1987. pp. 1410–1438. [Google Scholar]
- 8.Gentry D, Xiao H, Burgess R, Cashel M. The omega subunit of Escherichia coli K-12 RNA polymerase is not required for stringent RNA control in vivo. J Bacteriol. 1991;173:3901–3903. doi: 10.1128/jb.173.12.3901-3903.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gentry D R, Hernandez V J, Nguyen L H, Jensen D B, Cashel M. Synthesis of the stationary-phase sigma factor ςS is positively regulated by ppGpp. J Bacteriol. 1993;175:7982–7989. doi: 10.1128/jb.175.24.7982-7989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holt S C, Bramanti T E. Factors in virulence expression and their role in periodontal disease pathogenesis. Crit Rev Oral Biol Med. 1991;2:177–281. doi: 10.1177/10454411910020020301. [DOI] [PubMed] [Google Scholar]
- 11.Kontani M, Ono H, Shibata H, Okamura Y, Tanaka T, Fujiwara T, Kimura S, Hamada S. Cysteine protease of Porphyromonas gingivalis 381 enhances binding of fimbriae to cultured human fibroblasts and matrix proteins. Infect Immun. 1996;64:756–762. doi: 10.1128/iai.64.3.756-762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krohn M, Wagner R. A procedure for the rapid preparation of guanosine tetraphosphate (ppGpp) from Escherichia coli ribosomes. Anal Biochem. 1995;225:188–190. doi: 10.1006/abio.1995.1138. [DOI] [PubMed] [Google Scholar]
- 13.Lynch M C, Kuramitsu H K. Role of superoxide dismutase activity in the physiology of Porphyromonas gingivalis. Infect Immun. 1999;67:3367–3375. doi: 10.1128/iai.67.7.3367-3375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malek R, Fisher J G, Caleca A, Stinson M, van Oss C J, Lee J-Y, Cho M-I, Genco R J, Evans R T, Dyer D W. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J Bacteriol. 1994;176:1052–1059. doi: 10.1128/jb.176.4.1052-1059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mechold U, Cashel M, Steiner K, Gentry D, Malke H. Functional analysis of a relA/spoT gene homolog from Streptococcus equisimilis. J Bacteriol. 1996;178:1401–1411. doi: 10.1128/jb.178.5.1401-1411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metzger S, Sarubbi E, Glaser G, Cashel M. Protein sequences encoded by the relA and spoT genes of Escherichia coli are interrelated. J Biol Chem. 1989;264:9122–9125. [PubMed] [Google Scholar]
- 17.Nakayama K. Rapid viability loss on exposure to air in a superoxide dismutase-deficient mutant of Porphyromonas gingivalis. J Bacteriol. 1994;176:1939–1943. doi: 10.1128/jb.176.7.1939-1943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarubbi E, Russ K E, Xiao H, Ikehara K, Kalman M, Cashel M. Characterization of the spoT gene of Escherichia coli. J Biol Chem. 1989;264:15074–15082. [PubMed] [Google Scholar]
- 19.Slots J, Genco R J. Black-pigmented Bacteroides species, Capnocytophaga species, and Actinobacillus actinomycetemcomitans in human periodontal disease: virulence factors in colonization, survival, and tissue destruction. J Dent Res. 1984;63:412–421. doi: 10.1177/00220345840630031101. [DOI] [PubMed] [Google Scholar]
- 20.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutation. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]