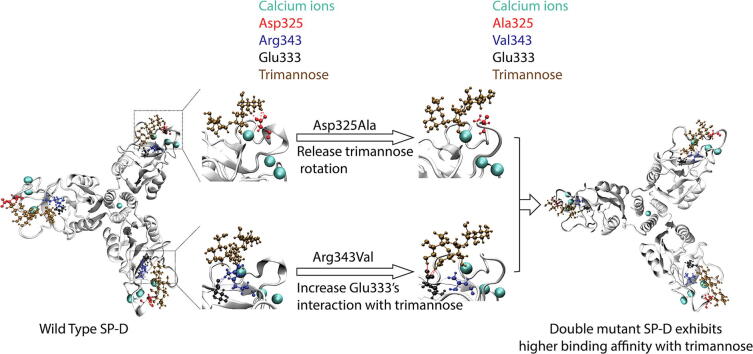
Graphical abstract
Abbreviations: SP-D, Surfactant Protein D; IAV, Influenza A Viruses; WT, Wild-type; DM, Double mutant; MD, Molecular Dynamics; FEP, Free Energy Perturbation; PS, Pulmonary Surfactant; PAP, Pulmonary Alveolar Proteinosis; SP-B, Surfactant Protein B; SP-C, Surfactant Protein C; SP-A, Surfactant Protein A; HA, Hemagglutinin; CRD, Carbohydrate Recognition Domain; PME, Particle Mesh Ewald; RMSD, Root Mean Square Deviation; RMSF, Root Mean Square Fluctuation; λ-REMD, λ-Replica-Exchange Molecular Dynamics
Keywords: Surfactant Protein D, Protein-Glycan Complexes, Molecular Dynamics Simulation, Free Energy Perturbation
Highlights
-
•
The Asp325Ala mutation in SP-D promotes a trimannose conformational change to a more stable state.
-
•
The Arg343Val mutation in SP-D reduces its interaction with Glu333 to increase the binding affinity with trimannose.
-
•
The Arg343Val mutation contributes more to the increase of SP-D’s binding affinity with trimannose than Asp325Ala.
Abstract
Surfactant protein D (SP-D) is an essential component of the human pulmonary surfactant system, which is crucial in the innate immune response against glycan-containing pathogens, including Influenza A viruses (IAV) and SARS-CoV-2. Previous studies have shown that wild-type (WT) SP-D can bind IAV but exhibits poor antiviral activities. However, a double mutant (DM) SP-D consisting of two point mutations (Asp325Ala and Arg343Val) inhibits IAV more potently. Presently, the structural mechanisms behind the point mutations’ effects on SP-D’s binding affinity with viral surface glycans are not fully understood. Here we use microsecond-scale, full-atomistic molecular dynamics (MD) simulations to understand the molecular mechanism of mutation-induced SP-D’s higher antiviral activity. We find that the Asp325Ala mutation promotes a trimannose conformational change to a more stable state. Arg343Val increases the binding with trimannose by increasing the hydrogen bonding interaction with Glu333. Free energy perturbation (FEP) binding free energy calculations indicate that the Arg343Val mutation contributes more to the increase of SP-D’s binding affinity with trimannose than Asp325Ala. This study provides a molecular-level exploration of how the two mutations increase SP-D binding affinity with trimannose, which is vital for further developing preventative strategies for related diseases.
1. Introduction
Pulmonary surfactant (PS) plays an important role in human health. PS is essential for maintaining lung function including the expansion and contraction of the alveoli needed for healthy breathing. Furthermore, PS defends against airborne pathogens such as viruses, bacteria, molds, pollen, and fungi that enter our lungs as we breathe. An imbalance of PS can lead to many diseases. When premature infants do not produce enough PS, they lack the ability to breathe and are often treated with PS taken from porcine and bovine sources. People with too much PS face diseases such as pneumonia and congenital pulmonary alveolar proteinosis (PAP). Individuals with too little PS can have emphysema, chronic bronchitis, and cystic fibrosis [1].
PS is made up of both lipids and proteins. Four main types of lipids comprise 90 % of PS and include zwitterionic saturated phospholipids, zwitterionic unsaturated phospholipids, negative unsaturated phospholipids, and cholesterol. In addition, two major types of proteins comprise 10 % of the pulmonary surfactant system: the small, hydrophobic surfactant proteins B and C (SP-B and SP-C, respectively) buried inside the lipid bilayer for the interfacial adsorption of pulmonary surfactants which is essential for reducing surface tension enabling the lungs to expand and contract [2], [3], and the large, hydrophilic surfactant proteins A and D (SP-A and SP-D respectively) that interact with airborne pathogens acting as a host defense in the lungs [4]. While not directly relevant to this study, it is interesting to note that surfactant proteins are not only found in the lungs but throughout the body. Surfactant proteins have been identified in areas that include the female reproductive tract, urinary tract, gastrointestinal tract, the eye, ear, nasal compartment, central nervous system, the coronary artery, and the skin.
SP-A and SP-D are immunoproteins belonging to the collagenous C-type lectin (collectin) subgroup and acting as our first line of defense against airborne pathogens through binding glycans on the pathogen’s surface and flagging them for phagocytosis. A variety of experimental studies have investigated immunoproteins' interactions with viruses and bacteria. One study, for example, demonstrated that both SP-A and SP-D can inhibit influenza A virus (IAV) activity [4], [5]. SP-D is a more potent inhibitor of IAV activity than SP-A through the high-level aggregation and neutralization of the IAV particles [4], [6], [7]. SP-D binds viruses such as SARS-CoV-2, playing a critical role as a first-line inhibitor [8]. Previous studies also show that both SP-A and SP-D can inhibit the activity of Histoplasma capsulatum yeast, which can cause serious long-term lung infections. SP-D can also enhance neutrophil uptake of bacteria, such as Staphylococcus aureus and Escherichia coli, commonly found in staph and bacterial infections [9]. While these studies demonstrate the critical role of immunoproteins in responding to microbial invasion, further investigations are needed to identify their molecular-level binding mechanisms.
Computational techniques such as Monte Carlo and full-atomistic molecular dynamics (MD) simulations can provide molecular-level insight into the biomaterial offering a powerful tool for understanding the immunoproteins' interactions. These simulations have successfully explored several immunoproteins with atomic-level insights. For example, Zhang et al. used MD simulations to investigate the dynamic interaction between different carbohydrates and human SP-D as well as steered MD simulations to study the unbinding pathway of glucose from human SP-D [10], [11]. Goh et al. used X-ray crystallography and mutagenesis combined with MD simulations to reveal the interaction between SP-A and lipids [12].
Herein, we aim to elucidate the molecular mechanism by which SP-D can bind high mannose oligosaccharides on the surface of pathogens, such as influenza hemagglutinin (HA). HA is found on the surface of influenza viruses, reducing viral uptake into lung epithelial cells [13], [14]. A previous study found that the antiviral activity of WT SP-D depends on the presence of high mannose oligosaccharides on the viral surface. For example, SP-D inhibits the influenza A virus (IAV) strains of H1N1, H2N2, and H3N2 less efficiently due to its HA surface’s relatively low glycosylation sites [15]. By contrast, the Asp325Ala and Arg343Val double mutant SP-D (DM SP-D) inhibits high glycosylation and low glycosylation IAV strains more efficiently than the WT [16].
More computational studies are needed to understand how specific mutations enhance the antiviral activity of SP-D. Although Crouch et al. experimentally probed the efficacy of DM SP-D’s inhibition of IAV and supported this with molecular mechanics minimization and analysis, they did not fully investigate each mutation’s contribution at the molecular level with more comprehensive computational techniques. It is critical to fully understand the molecular mechanism that details how the specific mutations modulate the antiviral activity of SP-D. Knowledge of the molecular mechanism will inform the development of specific mutant SP-D proteins with high antiviral activity to antagonize glycosylated viruses, such as IAV and SARS-CoV-2.
Based on this experimental study [16], we computationally investigate the interactions of WT SP-D with trimannose using microsecond-scale, full-atomistic MD simulations. Then, in order to thoroughly study the effect of the point mutations on binding, we analyzed simulations of each mutation separately and then the DM SP-D interacting with the glycan trimannose. This permitted an evaluation of each mutation’s contribution to SP-D’s binding affinity with trimannose along with the influence of the DM SP-D by comparing the structural dynamics and other attributes between the four SP-D models. The simulations considered for the four SP-D variants (WT, two single mutants (Asp325Ala and Arg343Val), and the DM) invoked the unbound state as well as two different binding poses. This generates a total of 12 different models employed in evaluating the protein-glycan interactions.
2. Background
2.1. Structure of SP-D
Surfactant protein D (SP-D) is an immunoprotein designed to recognize and interact with glycans found on the surface of different foreign entities such as viruses, bacteria, pollen, and fungi. SP-D is a collagenous C-type lectin (collectin). Human SP-D exhibits different oligomeric forms, among which the most common is the dodecameric SP-D (cruciform) consisting of four SP-D trimers. Each SP-D trimer is made up of three identical intertwined monomers. Each monomer contains 355 residues. Each SP-D monomer can be divided into four major structural domains: N-terminal domain, collagen-like domain, neck region, carbohydrate recognition domain (CRD):
-
•
N-terminal domain (residues 1 to 25) contains two conserved cysteines (C15 and C20) that are essential for SP-D oligomerization by forming disulfide bonds [17].
-
•
Collagen-like domain (residues 26 to 202) is a helix and twists with the repeating pattern of 59 Gly-X-Y.
-
•
Neck region (residues 203 to 232) is an α-helical coiled-coil domain mediating SP-D trimerization.
-
•
C-terminal carbohydrate recognition domain (CRD) (residues 233 to 355) recognizes and interacts with diverse cell surface glycans of various pathogens containing galactose, glucose, or mannose, and is crucial to SP-D’s antiviral function [9], [18], [19], [20], [21], [22], [23].
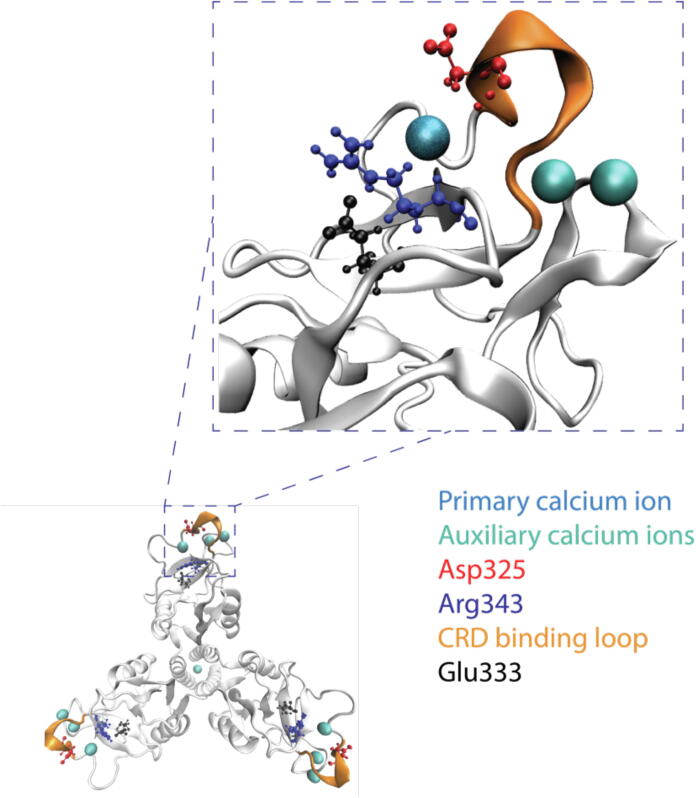
In this study, we look at both the WT SP-D and the DM SP-D with the focuses on the residues in the neck and CRD regions (residues 203 to 355). To understand the significant components relevant to this study, we have highlighted the calcium ions, the residues involved in the two point mutations (Asp325 and Arg343), Glu333, and the CRD binding loop in Fig. 1. Three calcium ions are highlighted in cyan, one primary (dark cyan) and two auxiliaries (slight cyan). The primary calcium ion is directly involved in binding with trimannose. A previous computational study has shown that without the primary calcium ion in the CRD, the glycan quickly unbinds from the CRD (7 ns), indicating the significance of the primary calcium ion for the antiviral function of SP-D [24]. The other two auxiliary calcium ions can stabilize the CRD’s conformation to enhance binding with trimannose. The two residues involved in the point mutations, Asp325 and Arg343, are highlighted in red and blue. Glu333, highlighted in black, plays a role in interacting with the residue Arg343 that is mutated. Finally, highlighted in gold, is the CRD binding loop, a fluctuating region of amino acids between the primary calcium ion and the auxiliary calcium ions. These components all have significance in SP-D’s CRD interaction with trimannose and the protein components serve as the focus of efforts described herein. While important, the calcium ion contributions are not focused upon here and will be described elsewhere.
Fig. 1.
SP-D structure focused on a portion of the CRD. Major components of the CRD involved in trimannose binding are highlighted in different colors.
3. Methods
3.1. Molecular dynamics
3.1.1. Constructing different SP-D models
In this study we used the following PDB structures to build our models of the SP-D•Trimannose complexes:
-
•
WT SP-D trimer structure from 3G83 (PDB ID) with a resolution of 1.90 Å contains two monomers consisting of residues 205–355, and a third monomer consisting of residues 206–355 [25].
-
•
WT SP-D trimer structure from 3IKQ (PDB ID) with a resolution of 2.25 Å contains two monomers consisting of residues 210–355, and a third monomer consisting of residues 211–355 [26].
-
•
DM SP-D trimer structure of 4 M18 (PDB ID) with a resolution of 3.20 Å consists of residues 211–355 for each monomer [24].
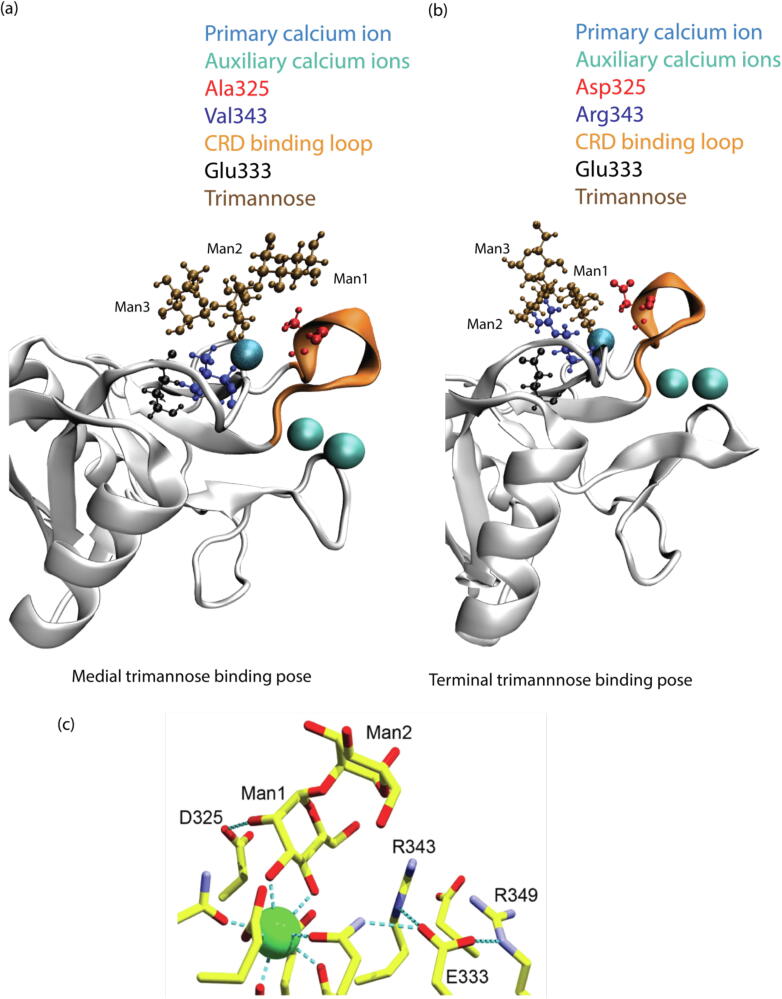
To fully understand how trimannose is interacting with SP-D’s CRD we need to understand the different possible trimannose binding poses. Because these possibilities are almost endless, we began by using the poses already captured in the crystallographic image as referenced in the PDB database. There are two major trimannose-binding poses: (1) the medial binding pose where the center mannose chelates with the primary calcium ion of SP-D, and (2) the terminal binding pose where the terminal mannose chelates with the primary calcium ion of SP-D. Fig. 2(a) shows the comparison between the medial and terminal trimannose binding poses of SP-D’s CRD. In the trimannose, three alpha-d-Mannopyranose (Man1, Man2, and Man3) connect to each other through 1–2 linkage.
Fig. 2.
(a) Medial trimannose binding pose of DM SP-D•Trimannose, and (b) terminal trimannose binding pose of WT SP-D•Trimannose. (c) Crystallographic representation of WT SP-D interacting with dimannose [25].
The medial trimannose binding pose was obtained from 4 M18 and is the only SP-D crystal structure in the PDB database containing the double mutation Asp325Ala and Arg343Val while binding with trimannose in the CRD. The 4 M18 crystal structure was produced in the experimental studies in which they found that DM SP-D has higher antiviral activities than WT SP-D in relation to IAV [16], [24]. In addition, the high-mannose glycosylation type is one of the most common types found on the surfaces of viruses such as IAV and SARS-CoV-2.
In the medial binding pose, Man2 interacts with the primary calcium ion, and Man1 interacts with the CRD binding loop. Four models were constructed with this medial pose, which are DM SP-D•Trimannose (4 M18), Asp325Ala SP-D•Trimannose, Arg343Val SP-D•Trimannose, and WT SP-D•Trimannose. The last three models were obtained by point mutation from 4 M18 using AmberTools20. After mutations, we used VMD and found that there were no steric clashes between SP-D and trimannose in each model. We then optimized the complex structures.
We did not find a terminal trimannose binding pose with SP-D in the PDB database. However, we did find the terminal binding pose of dimannose with SP-D in 3G83 (see Fig. 2(c)) [25]. The dimannose in 3G83 has the same type of mannose units and linkages as that of 4 M18. In this structure, Man1 interacts with the primary calcium ion and Asp325 (see Fig. 2(c)). To build a trimannose, we extended a mannose unit, Man3, to the original dimannose using Schrodinger. Because the insertion of Man3 to Man1 will affect its original interaction with Asp325, we added Man3 to Man2 using the 3D Builder module in Schrodinger (see Fig. 2(b)). We then optimized the complex structures.
To fully understand how each mutation influences SP-D’s binding affinity with trimannose, we constructed the following twelve models for MD simulation (see Table 1). From these twelve models, we can offer molecular-level explanations of how the two point mutations influence SP-D’s binding affinity and quantify each single mutation’s effect in enhancing SP-D’s antiviral activity. Furthermore, we can compare the two binding poses to better understand how each pose correlates with SP-D’s binding affinity.
Table 1.
Simulation models in this study.
| System | Models |
|---|---|
| Medial binding pose | WT SP-D•Trimannose |
| Asp325Ala SP-D•Trimannose | |
| Arg343Val SP-D•Trimannose | |
| DM SP-D•Trimannose (4 M18 PDB ID) | |
| Terminal binding pose | WT SP-D•Trimannose |
| Asp325Ala SP-D•Trimannose | |
| Arg343Val SP-D•Trimannose | |
| DM SP-D•Trimannose | |
| Only SP-D | WT SP-D |
| Asp325Ala SP-D | |
| Arg343Val SP-D | |
| DM SP-D |
The twelve models consist of three parts. The first and second groups are SP-D binding with trimannose in two different poses: the medial trimannose binding pose and the terminal trimannose binding pose. Each pose contains four complex models, including WT SP-D•Trimannose, Asp325Ala SP-D•Trimannose, Arg343Val SP-D•Trimannose, and DM SP-D•Trimannose. The third group includes four models without trimannose binding: WT SP-D, Asp325Ala SP-D, Arg343Val SP-D, and DM SP-D.
It is noted that only the α, α -trimannose is considered in these studies, as this configuration coincides with the available crystallographic information. Instinct may instruct that one must also consider α,β - β, α and β, β possibilities as well due to anomeric equilibrium at Cα. However, as only the α, α -trimannose is observed on the surface of influenza, the other anomers are not considered here.
3.1.2. Structural preparation, solvation, and neutralization
To prepare the structures we hydrogenated using tleap of AmberTools20 to add hydrogen atoms to the structures. The Amber ff19SB forcefield was used to model the protein structure, GLYCAM06 was utilized to model the trimannose structure, and OPC was selected as the water model [27], [28], [29]. The forcefields used in this study are the latest versions as implemented in AMBER20. Each model was then solvated with a 10 Å water shell using an octahedral water box. Na + and Cl- were placed to neutralize the system. The salt (NaCl) concentration was set to 0.15 M. All molecular dynamics simulations were carried out using AMBER20 [30].
3.1.3. Minimization and equilibration
All structures were first optimized using two stages of minimization. Initially, the entire structure was fixed by applying strong harmonic restraints of 500 kcal/(mol·Å2) on all atoms in the SP-D complexes. Subsequently, we minimized the entire system without any constraints. After that, we equilibrated the structure by employing a two-stage process. In the first stage, the system temperature was raised from 0 K to 310 K over 1 × 106 steps of MD with a timestep of 2 fs in the NVT ensemble. In this stage, we applied a weak harmonic restraint of 10 kcal/(mol·Å2) to the whole SP-D structure. Finally, the system was equilibrated at a constant temperature of 310 K with 1 bar pressure (NPT ensemble) for 100 ns. 310 K was chosen to emulate body temperature. The Langevin dynamics algorithm with a collision frequency of 1.0 ps−1 was used for temperature control, and the Berendsen barostat with a pressure regulation time of 2 ps controlled the pressure. The SHAKE algorithm was used to constrain all hydrogen-containing bonds during the equilibration stage [31]. A cutoff distance of 8 Å was used to calculate the nonbonded interactions using the Particle Mesh Ewald (PME) method. There is an additional step for the terminal trimannose binding pose system. We fixed the SP-D and its original dimannose with weak restraints and equilibrated the Man3 unit using an NPT ensemble for 50 ns (1 bar, 310 K).
3.1.4. Production runs
The production MD run was obtained in the isobaric, isothermal (NPT) ensemble as described in equilibration section. Table 2 shows the information on thirteen related SP-D model systems.
Table 2.
Summary of the molecular dynamics simulation system.
| MD system | Total atoms | Simulation time (ns) | |
|---|---|---|---|
| Medial binding pose | WT SP-D•Trimannose | 67,194 | 3000 |
| Asp325Ala SP-D•Trimannose | 67,187 | 3000 | |
| Arg343Val SP-D•Trimannose | 67,279 | 3000 | |
| DM SP-D•Trimannose | 67,280 | 3000 | |
| Terminal binding pose | WT SP-D•Trimannose | 73,823 | 3000 |
| Asp325Ala SP-D•Trimannose | 73,844 | 3000 | |
| Arg343Val SP-D•Trimannose | 73,968 | 3000 | |
| DM SP-D•Trimannose | 73,981 | 3000 | |
| Without Trimannose | WT SP-D | 71,840 | 500 |
| Asp325Ala SP-D | 70,348 | 500 | |
| Arg343Val SP-D | 70,766 | 500 | |
| DM SP-D | 69,434 | 500 | |
| Simulation of Trimannose in Water | Trimannose | 3920 | 1250 |
3.2. Analysis
3.2.1. Structural analysis methods
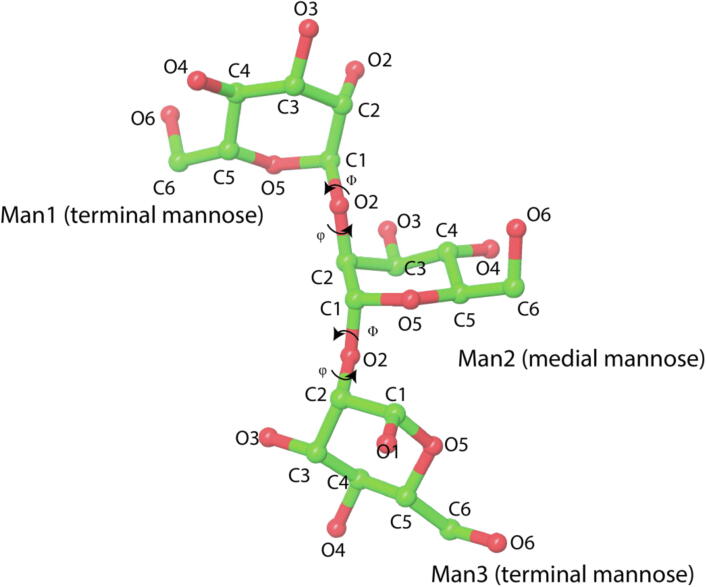
Root mean square deviation (RMSD) was calculated for the entire simulation trajectory with reference to the starting structure for each model. The root mean square fluctuation (RMSF) was calculated using the coordinates of the protein backbone atoms. We count the hydrogen bonds using the CPPTRAJ program with a default angle cutoff of 135° and a distance cutoff of 3 Å [32]. For the glycosidic torsion angles, the glycosidic ϕ and φ angles are defined as O5-C1-O2-C2 and C1-O2-C2-C1, as shown in Fig. 3. The glycosidic torsion angles are calculated using the CPPTRAJ program.
Fig. 3.
Illustration of the structure of trimannose (alpha-d-Mannopyranose-(1–2)-alpha-d-Mannopyranose-(1–2)-alpha-d-Mannopyranose (Man1-Man2-Man3)). Glycosidic torsion angles are defined as ϕ = O5-C1-O2-C2 and φ = C1-O2-C2-C1. The figure was created using Schrodinger [33].
3.2.2. Binding free energy calculation
The free energy perturbation (FEP) method was used to calculate the binding free energies of different complex systems [34]. The -Replica-Exchange Molecular Dynamics (-REMD) method was used in FEP calculation [35], [36], [37]. In the perturbation process (alchemical transformation) for each complex model, two coupling parameters, and , are used to control the nonbonded interaction (Lennard-Jones interaction and electrostatic interactions) between SP-D and trimannose. There is a total of 32 linear-spaced windows (0.0, 0.03225, 0.06451, 0.09677, 0.12903, 0.16129, 0.19354, 0.22580, 0.25806, 0.29032, 0.32258, 0.35483, 0.38709, 0.41935, 0.45161, 0.48387, 0.51612, 0.54838, 0.58064, 0.61290, 0.64516, 0.67741, 0.70967, 0.74193, 0.77419, 0.80645, 0.83870, 0.87096, 0.90322, 0.93548, 0.96774, 1.0) are used for FEP/λ-REMD calculations [36], [38], [39]. The latest CHARMM-GUI Absolute Ligand Binder Tool was used to prepare the input file for all required simulations [38]. The NAMD 3.0 was used to conduct all required simulations for the calculation of binding free energy [40]. The NPT ensemble with 310 K and 1 bar was used to equilibrate the systems, including “complex” and “ligand”, with the TIP3P water model. For each complex system, FEP/λ-REMD simulations were conducted for 6.2 ns per window, and the final 5 ns FEP/λ-REMD simulations were used for the binding free energy calculations [41].
3.2.3. Quantum calculation of trimannose
The initial trimannose structure from PDB 4 M18 was used for geometry optimization. The QM geometry optimization was conducted at the B3LYP/6–311++G** level of theory. The conformations of the most populated glycosidic torsion angles in WT SP-D•Trimannose complex and DM SP-D•Trimannose complex were used for single point energy calculation with the following geometries. The two groups of glycosidic torsion angles of WT SP-D•Trimannose are (79.219, −55.800) and (72.033, −77.362). The corresponding glycosidic torsion angles groups for DM SP-D•Trimannose are (77.030, −86.394) and (71.606, −83.132). The QM single-point energy calculations were obtained at the B3LYP/6–311++G** level of theory. All QM calculations are conducted through Gaussian 16 [42].
4. Results
To learn how each mutation in the experimental study [16] enhanced the DM SP-D’s ability to bind IAV more tightly than the WT SP-D, thus leading to enhanced virus recognition, we needed to understand how the glycan, trimannose, is interacting with SP-D. We began by running basic structural analysis techniques and then working to understand the impact of each mutation on the interaction of trimannose with SP-D in both the medial and terminal pose in the following four SP-D complex models: WT SP-D with trimannose, Asp325Ala SP-D with trimannose, Arg343Val SP-D with trimannose, and DM SP-D with trimannose.
4.1. Structural dynamics of the SP-D•Trimannose complexes
4.1.1. RMSd
The root-mean-square-deviations (RMSDs) were calculated to evaluate the structural stabilities of different SP-D complex models. The RMSDs from the starting structure of our four different SP-D complex models with trimannose binding are shown as a function of simulation time (RMSD plots for all 12 models are in the supplemental information. See Figures S1-S3.).
Upon inspection of the medial trimannose binding poses of four different SP-D complex models, significant differences in their RMSD plots are observed (see Figure S1). The RMSD plot of WT SP-D•Trimannose had a large fluctuation after 400 ns as one of the trimannose units unbound. Before the unbinding event occurred, the average RMSD of WT SP-D•Trimannose was 2.37 Å with a standard deviation of 0.32 Å. Following the unbinding event, the average RMSD increased to 6.14 Å, with a standard deviation of 1.85 Å. Another unbinding event was observed at approximately 2300 ns in Asp325Ala SP-D•Trimannose, and the average RMSD was 5.10 Å with a standard deviation of 1.65 Å.
For Arg343Val SP-D•Trimannose, the average RMSD was 4.88 Å with a standard deviation of 1.25 Å. The DM SP-D•Trimannose had a stable structural conformation without significant fluctuation in the RMSD plot. The average RMSD was 5.48 Å with a standard deviation of 0.85 Å.
4.1.2. RMSf
To explore each residue’s fluctuation within the SP-D models, the root-mean-square-fluctuation (RMSF) was produced (RMSF plots for all 12 models are in the supplemental information. See Figure S4-S6.) The RMSF plot suggests that there is a region that’s extremely flexible and we named that region the CRD binding loop region (residues 324–330).
4.1.3. Chelation
Supplementary Figures S7 to S14 illustrate the chelation between trimannose and the primary calcium ion of each SP-D model considered in this work. For the medial trimannose binding pose, we observed that the plots of the WT SP-D•Trimannose complex contain a large fluctuation after approximately 400 ns following unbinding of one of the trimannose units. The plot of Asp325Ala SP-D•Trimannose also had a significant fluctuation at about 2300 ns for the same reason, i.e. one of the trimannose units was released at that moment. The plots of Arg343Val SP-D•Trimannose and DM SP-D•Trimannose were stable without large fluctuation. For the terminal trimannose binding pose, although there were no unbinding events occurring, the Asp325Ala SP-D•Trimannose tended to switch the trimannose chelation site. The rest of the other three complex models did not observe the switching of the chelation site during the production runs. The DM SP-D•Trimannose showed stronger chelation than the WT SP-D•Trimannose.
4.2. Results of the medial trimannose binding pose
4.2.1. Impact of Asp325Ala on the medial trimannose binding pose
Asp325Ala mutation releases trimannose’s torsion angles leading to a more stable binding conformation.
In the WT SP-D•Trimannose, aspartate in 325 can form a stable hydrogen bond with trimannose (Man1). Table 3 shows how the mutation in this position significantly reduced the average number of hydrogen bonds from 1.04 to 0. Additionally, in Arg343Val SP-D•Trimannose, the Asp325 can also form a stable hydrogen bond with trimannose at an average of 1.51 hydrogen bonds. After mutation in DM SP-D•Trimannose, the hydrogen bonds dropped to 0. We found that the hydrogen bond between Asp325 and trimannose limited trimannose conformation, making the conformational change to a more optimal binding position difficult for trimannose.
Table 3.
Hydrogen bonds between residue 325 and trimannose of WT SP-D•Trimannose, Asp325Ala SP-D•Trimannose, Arg343Val SP-D•Trimannose, and DM SP-D•Trimannose observed in the medial trimannose binding pose.
| System | Acceptor | Donor | Occupancy | Distance (Å) | Angle (degrees) |
|---|---|---|---|---|---|
| WT SP-D●Trimannose | Asp325@OD1 | Man1@O3 | 0.27 | 2.66 | 163.69 |
| Asp325@OD1 | Man1@O4 | 0.25 | 2.74 | 160.41 | |
| Asp325@OD2 | Man1@O3 | 0.28 | 2.66 | 163.72 | |
| Asp325@OD2 | Man1@O4 | 0.24 | 2.74 | 160.30 | |
| Total | 1.04 | ||||
| Asp325Ala SP-D●Trimannose | No Hydrogen bonds | ||||
| Arg343Val SP-D●Trimannose | Asp325@OD1 | Man1@O3 | 0.46 | 2.66 | 163.55 |
| Asp325@OD1 | Man1@O4 | 0.31 | 2.75 | 159.84 | |
| Asp325@OD2 | Man1@O3 | 0.36 | 2.66 | 163.54 | |
| Asp325@OD2 | Man1@O4 | 0.39 | 2.74 | 160.16 | |
| Total | 1.51 | ||||
| DM SP-D●Trimannose | No Hydrogen bonds | ||||
Supplemental Figures S15 and S16 show the glycosidic torsion angle (ϕ, ψ) distribution of alpha-d-Mannopyranose-(1–2)-alpha-d-Mannopyranose (α-d-Manp-(1 → 2)-α-d-Manp) linkage of Man1-Man2 and Man2-Man3 in different complex models. In the WT SP-D•Trimannose and Arg343Val SP-D•Trimannose complex models, the Asp325 formed a stable interaction through hydrogen bonding, limiting Man1′s rotation, leading to (ϕ, ψ) of Man1-Man2 populated at (79.15°, −57.60°) for WT SP-D•Trimannose and (78.66°, −57.78°) for Arg343Val SP-D•Trimannose. After mutation in this position (Asp325Ala SP-D•Trimannose and DM SP-D•Trimannose) there was no lock on the torsion angles of trimannose making (ϕ, ψ) of Man1-Man2 populated at about (77.39°, −84.59°) and (77.43°, −86.39°), respectively. To view this, refer to supplemental Figure S15 for the glycosidic torsion angles (ϕ, ψ) distribution of α-d-Manp-(1 → 2)-α-d-Manp linkage of Man1-Man2. We further calculated the single point energy of the most populated trimannose conformations of WT SP-D•Trimannose and DM SP-D•Trimannose.
We compared the glycosidic torsion angles of the linkage of Man1-Man2 and Man2-Man3 from WT SP-D•Trimannose, (79.15°, −57.60°) and (72.04°, −77.36°), to that from DM SP-D•Trimannose, (77.43°, −86.39°) and (70.91°, −80.27°), at the medial trimannose binding pose. Evaluation of the single point energies (B3LYP/6–311++G**) of trimannose in the observed conformations when bound to WT SP-D and DM SP-D revealed a 0.96 kcal/mol conformational strain is introduced by binding to the WT compared to DM SP-D. This indicates more favorable binding, based on conformational strain, for trimannose occurs when bound to DM SP-D. Furthermore, energy minimization trimannose at the B3LYP/6–311++G** level of theory reveals a conformation with (ϕ, ψ) of (80.40, −86.89) nearly identical to that observed when trimannose is bound to DM SP-D.
Säwén et al. studied a mannose disaccharide extensively with MD and NMR and derived the most probable conformation for trimannose with glycosidic angles (80.00°, −87.00°) [43]. We also ran over 1000 ns of MD simulation on free trimannose in solution to obtain the distribution of glycosidic torsion angles of the trimannose. Figure S19 shows the distributions of the two groups of glycosidic torsion angles in the trimannose-free state. Free trimannose displayed the major conformation at (ϕ, ψ) of (72.75°, −84.65°), which is consistent with previous studies [43]. The second-favored conformation has glycosidic torsion angles at approximation (ϕ, ψ) of (72.75°, −135.00°). As shown in Figure S19, the two favored glycosidic torsion angle distributions were more common.
We used quantum mechanics to locate the minimum energy trimannose structure through Gaussian. These optimized glycosidic torsion angles (ϕ, ψ) were (80.40, −86.89) consistent with our MD simulations of trimannose-free in solution in which the major glycosidic torsion angles (ϕ, ψ) were (74.32°, −84.76°).
Our results indicate that the trimannose bound at Ala325 SP-D (Asp325Ala SP-D and DM SP-D) remains in the glycosidic torsion angles favored in free solution. When trimannose is bound at Asp325 SP-D (WT SP-D and Arg343Val SP-D), the hydrogen bonding interaction between Asp325 and trimannose locks the Man1 rotation, leading to less stable torsion angles. Quantum mechanical calculations support our explanation of the molecular mechanism of Asp325Ala’s effect in enhancing SP-D’s binding affinity with trimannose.
4.2.2. Impact of Arg343Val in the medial trimannose binding pose
The mutation of Arg343Val weakens the hydrogen bonding interaction with Glu333 which frees Glu333 to interact with trimannose.
To better understand the role of the Arg343Val point mutation in binding with trimannose, the hydrogen bonding interaction between the residue in position 343 and the specific glutamate residue in position 333 within the CRD and trimannose were calculated separately. Table 4 details hydrogen bonds between the residues in position 343 and Glu333 and trimannose in different SP-D•Trimannose complex models.
Table 4.
Hydrogen bonds between residue in position 343 and Glu333 and trimannose of WT SP-D•Trimannose, Asp325Ala SP-D•Trimannose, Arg343Val SP-D•Trimannose, and DM SP-D•Trimannose at the medial trimannose binding pose.
| System | Hbond Type | Acceptor | Donor | Occupancy | Distance (Å) | Angle (degrees) |
|---|---|---|---|---|---|---|
| WT SP-D●Trimannose | Glu333-Arg343 | Glu333@OE1 | Arg343@NE | 0.13 | 2.84 | 157.52 |
| Glu333@OE2 | Arg343@NE | 0.12 | 2.84 | 156.02 | ||
| Glu333@OE1 | Arg343@NH1 | 0.25 | 2.80 | 158.00 | ||
| Glu333@OE1 | Arg343@NH2 | 0.04 | 2.82 | 156.05 | ||
| Glu333@OE2 | Arg343@NH1 | 0.27 | 2.80 | 157.57 | ||
| Glu333@OE2 | Arg343@NH2 | 0.05 | 2.81 | 156.04 | ||
| Total | 0.87 | |||||
| Glu333-Trimannose | Glu333@OE1 | Man3@O2 | 0.05 | 2.63 | 162.02 | |
| Glu333@OE2 | Man3@O2 | 0.05 | 2.64 | 162.18 | ||
| Glu333@OE1 | Man3@O3 | 0.15 | 2.67 | 163.82 | ||
| Glu333@OE2 | Man3@O3 | 0.18 | 2.67 | 163.63 | ||
| Total | 0.43 | |||||
| Asp325Ala SP-D●Trimannose | Glu333-Arg343 | Glu333@OE1 | Arg343@NH1 | 0.16 | 2.81 | 156.57 |
| Glu333@OE2 | Arg343@NH1 | 0.12 | 2.81 | 156.60 | ||
| Glu333@OE1 | Arg343@NH2 | 0.11 | 2.82 | 151.39 | ||
| Glu333@OE2 | Arg343@NH2 | 0.08 | 2.81 | 150.55 | ||
| Glu333@OE1 | Arg343@NE | 0.05 | 2.83 | 153.50 | ||
| Glu333@OE2 | Arg343@NE | 0.19 | 2.83 | 154.53 | ||
| total | 0.70 | |||||
| Glu333-Trimannose | Glu333@OE1 | Man3@O2 | 0.19 | 2.65 | 162.57 | |
| Glu333@OE2 | Man3@O2 | 0.15 | 2.68 | 161.59 | ||
| Glu333@OE1 | Man3@O3 | 0.09 | 2.70 | 163.67 | ||
| Glu333@OE2 | Man3@O3 | 0.05 | 2.71 | 163.73 | ||
| total | 0.49 | |||||
| Arg343Val SP-D●Trimannose | Glu333-Val343 | No Hydrogen bonds | ||||
| Glu333-Trimannose | Glu333@OE1 | Man3@O2 | 0.04 | 2.71 | 156.56 | |
| Glu333@OE2 | Man3@O2 | 0.04 | 2.72 | 156.49 | ||
| Glu333@OE1 | Man3@O3 | 0.48 | 2.67 | 163.77 | ||
| Glu333@OE2 | Man3@O3 | 0.43 | 2.67 | 163.68 | ||
| total | 1.00 | |||||
| DM SP-D●Trimannose | Glu333-Val343 | No Hydrogen bonds | ||||
| Glu333-Trimannose | Glu333@OE1 | Man3@O2 | 0.23 | 2.72 | 157.01 | |
| Glu333@OE2 | Man3@O2 | 0.20 | 2.73 | 156.31 | ||
| Glu333@OE1 | Man3@O3 | 0.41 | 2.68 | 164.30 | ||
| Glu333@OE2 | Man3@O3 | 0.35 | 2.68 | 164.24 | ||
| total | 1.19 | |||||
Before mutation, Arg343 is a positive residue in the WT SP-D•Trimannose complex and Asp325Ala SP-D•Trimannose complex. Arg343 formed a stable interaction through hydrogen bonding with the negatively charged Glu333, whose hydrogen bond number with the glutamate residue was larger than that in the Arg343Val SP-D•Trimannose complex and DM SP-D•Trimannose complex (see Table 4). Meanwhile, Glu333 formed fewer hydrogen bonds with trimannose in WT SP-D•Trimannose and Asp325Ala SP-D•Trimannose than with the Arg343Val SP-D•Trimannose complex and DM SP-D•Trimannose complex.
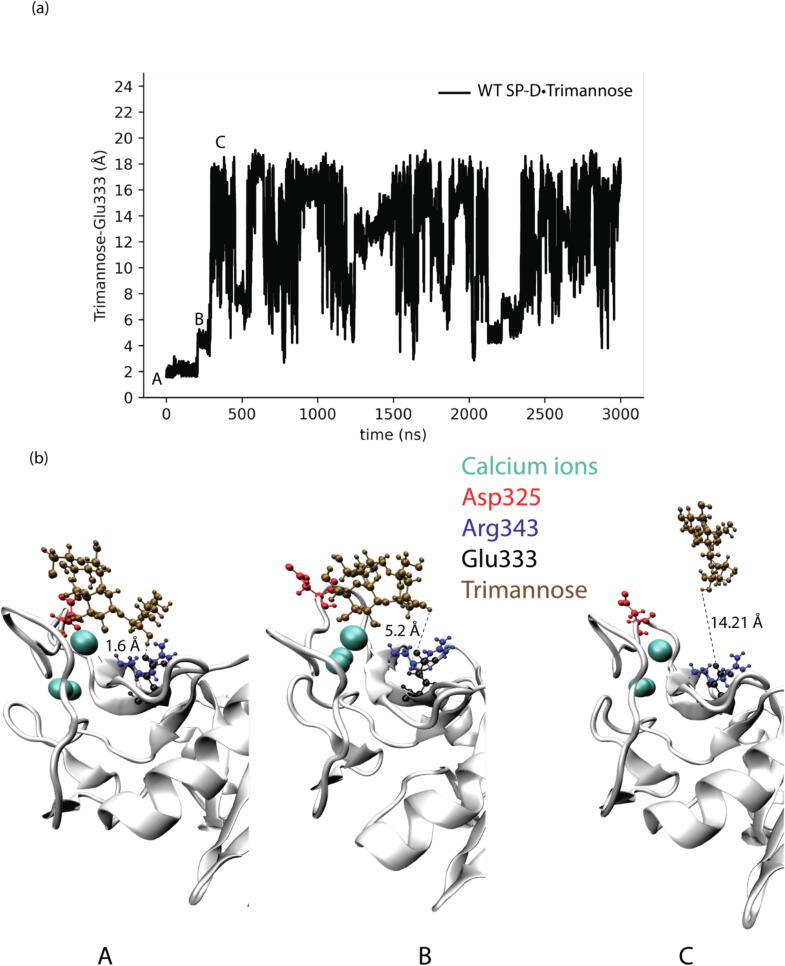
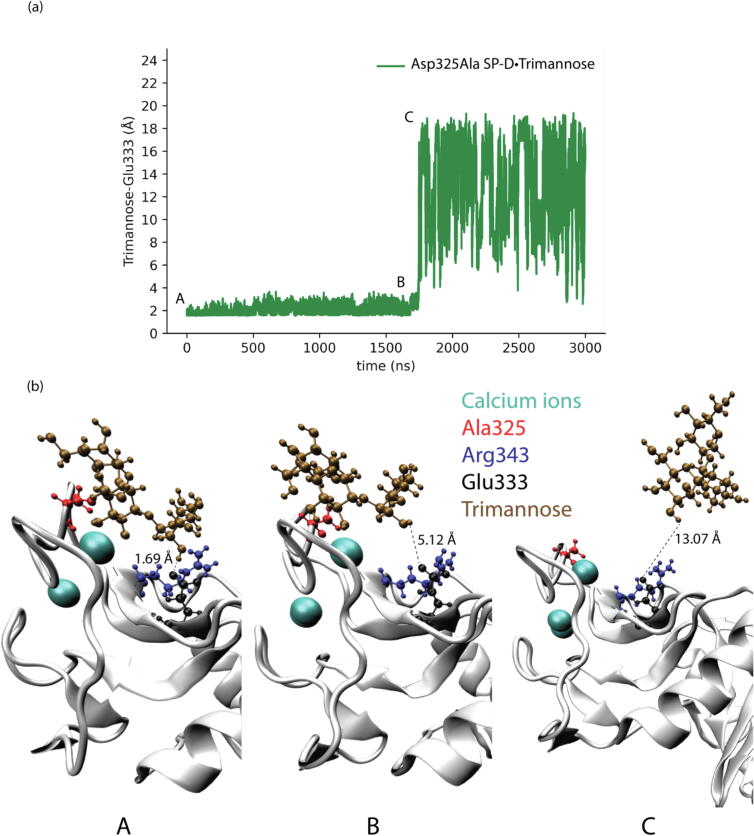
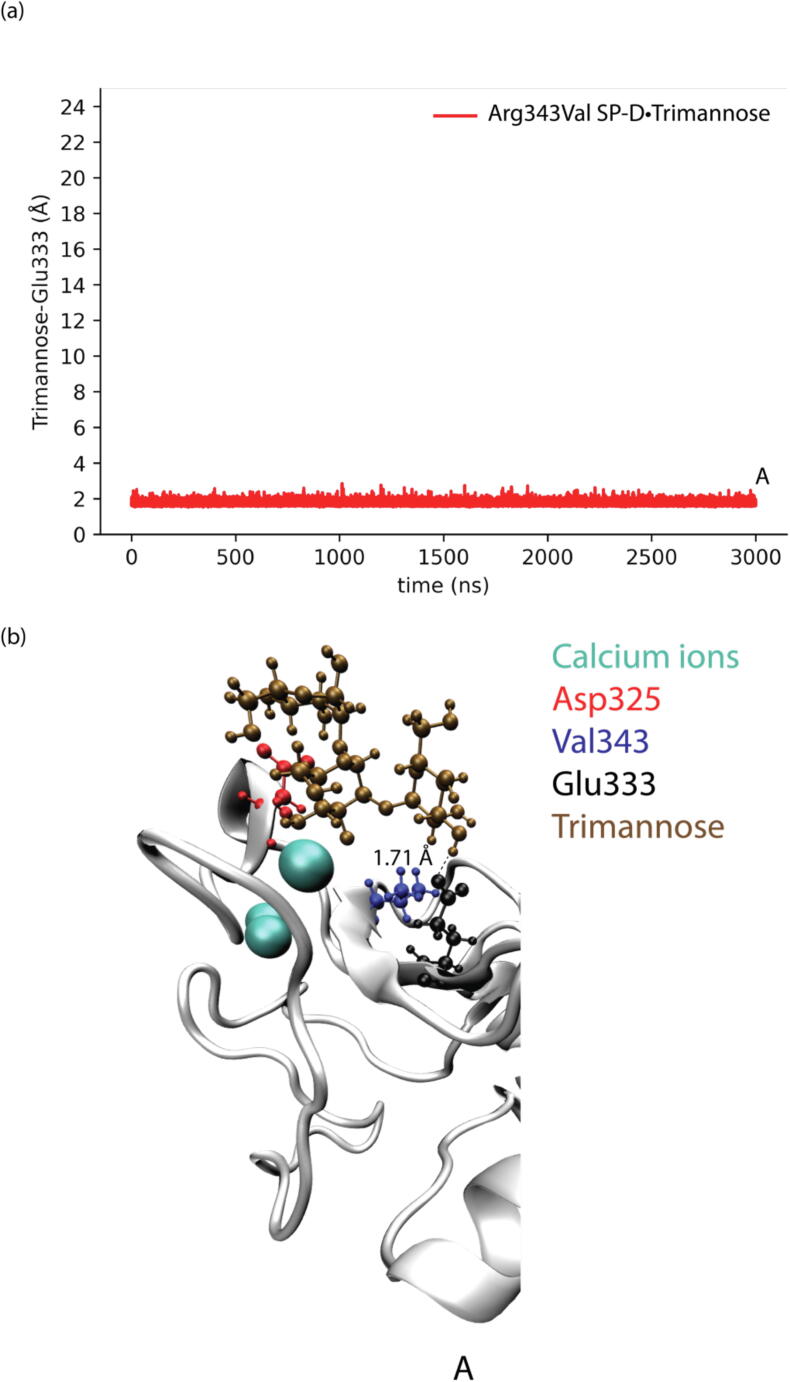
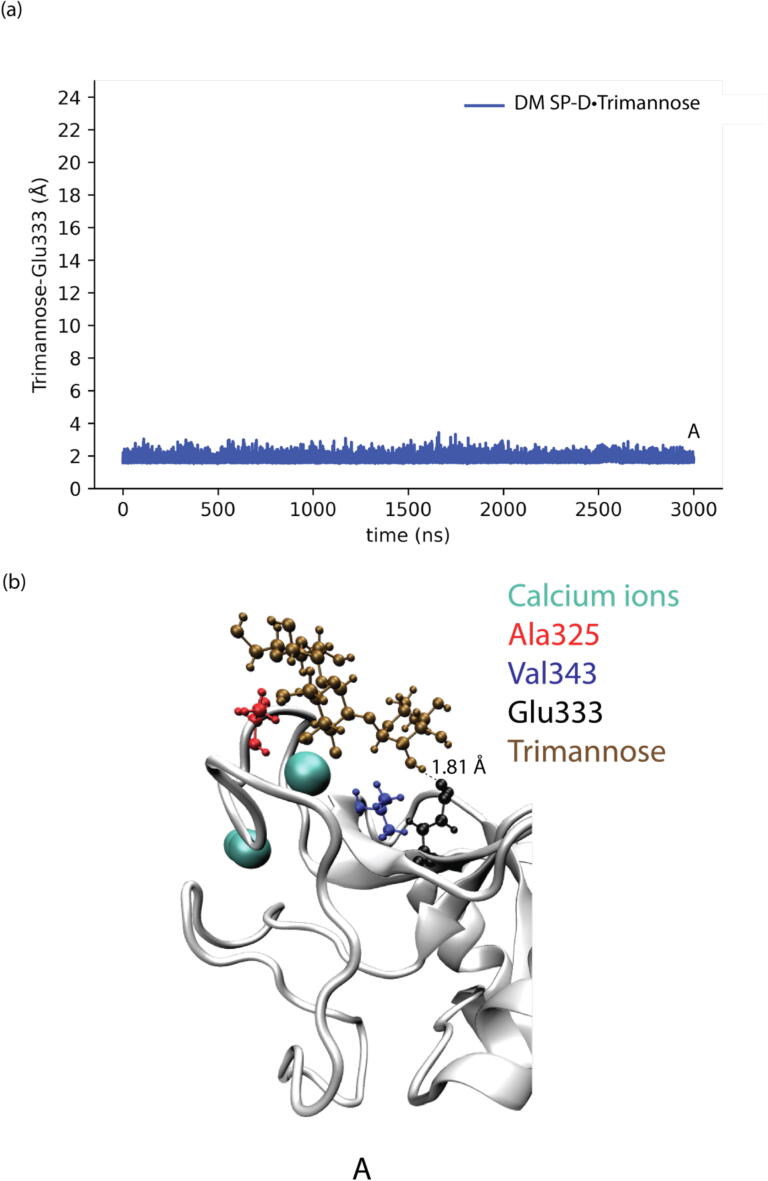
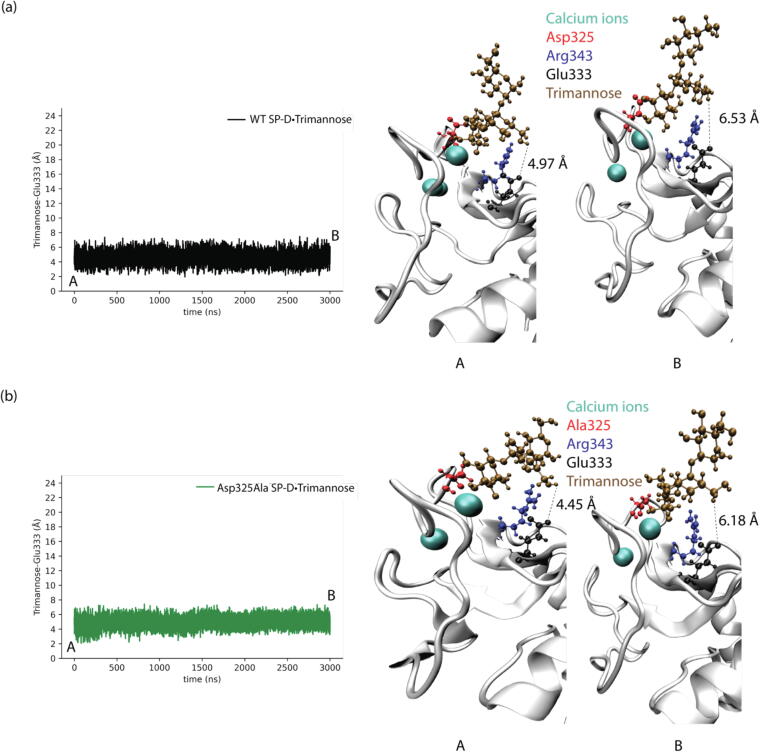
The shortest distances between trimannose and Glu333 in different SP-D•Trimannose complex models were also analyzed to explore the representative conformations of how Glu333 interacts with trimannose, as shown in Fig. 4, Fig. 5, Fig. 6, Fig. 7. For the WT SP-D•Trimannose complex (see Fig. 4), the shortest distance between trimannose and Glu333 began to greatly fluctuate after 200 ns of simulation time, as one of the trimannose units unbound here. There were three corresponding representative conformations as shown in Fig. 4(b), with the shortest distance increasing from 1.6 Å at the bound state to 14.21 Å at the unbound state. Fig. 5 shows the shortest distance plot Asp325Ala SP-D•Trimannose complex. The plot stayed constant until 2250 ns had elapsed in the simulation, and its shortest distance increased to 13.07 Å at that position. After that, it increased sharply as one of the trimannose units unbound here. Three corresponding representative conformations are shown in Fig. 5(b). The shortest plot of Arg343Val SP-D•Trimannose and DM SP-D•Trimannose stayed constant at about 2 Å without obvious fluctuation (see Fig. 6 and Fig. 7). Both had one representative conformation as shown in Fig. 6(b) and Fig. 7(b).
Fig. 4.
(a) Shortest distances between trimannose and Glu333 of WT SP-D•Trimannose at the medial trimannose binding pose. (b) Representative conformations of A, B, and C show how Glu333 interacts with trimannose.
Fig. 5.
(a) Shortest distances between trimannose and Glu333 of Asp325Ala SP-D•Trimannose at the medial trimannose binding pose. (b) Representative conformations of A, B, and C show how Glu333 interacts with trimannose.
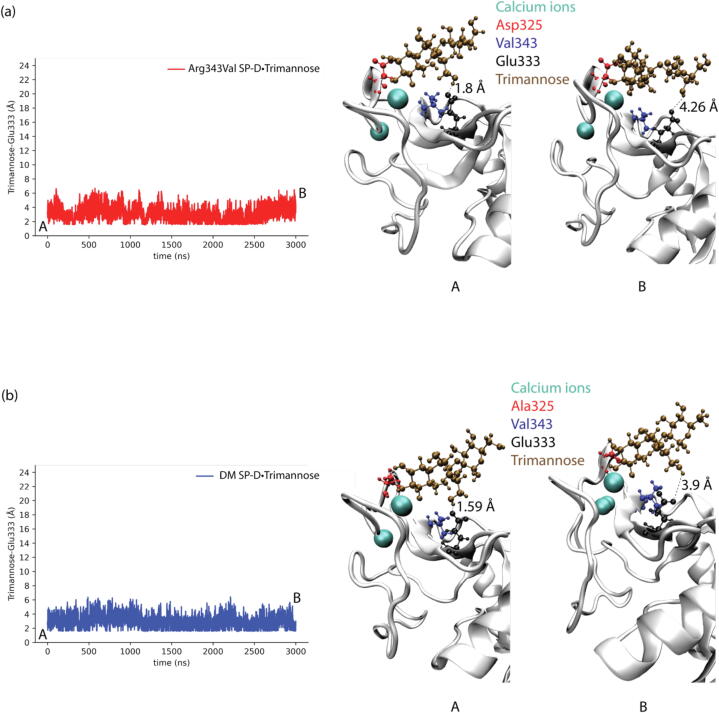
Fig. 6.
(a) Shortest distances between trimannose and Glu333 of Arg343Val SP-D•Trimannose at the medial trimannose binding pose. (b) Representative conformation of A shows how Glu333 interacts with trimannose.
Fig. 7.
(a) Shortest distances between trimannose and Glu333 of DM SP-D•Trimannose at the medial trimannose binding pose. (b) Representative conformation of A shows how Glu333 interacts with trimannose.
4.2.3. Weight SP-D and DM SP-D in the medial trimannose binding pose
Combining both mutations to form DM leads to better binding with trimannose than the WT.
In WT SP-D•Trimannose, we found that Asp325 forms a hydrogen bond with trimannose. This hydrogen bond locks the trimannose glycosidic torsion angles leading to unstable torsion angles and creating an uncomfortable binding pose for trimannose. In addition, before mutation, the positive Arg343 residue forms a significant hydrogen bond with the negative Glu333 weakening Glu333′s ability to interact with trimannose.
In contrast, DM SP-D•Trimannose, at the medial binding pose, increases the binding affinity of trimannose through changes due to each mutation. The Asp325Ala mutation releases the trimannose torsion angles enabling trimannose to relax into a more stable conformation increasing the binding ability of trimannose with SP-D. In addition, in the Arg343Val mutation, Val’s neutral charge changes by freeing negative Glu333 to instead form an enhanced bonding interaction with trimannose.
4.3. Results of the terminal trimannose binding pose
4.3.1. Impact of Asp325Ala in the terminal trimannose binding pose
The influence of the Asp325Ala mutation in the terminal trimannose binding pose does not enhance SP-D’s binding affinity with trimannose as strongly as that in the medial trimannose binding pose.
In the WT SP-D•Trimannose, aspartate in 325 forms a hydrogen bond with trimannose (Man1) but this interaction is weaker than that found in the medial trimannose binding pose. In this position, Asp325Ala decreased the average hydrogen bond number from 0.64 to 0 (see Table 5). In the other single mutant complex, Arg343Val SP-D•Trimannose, the Asp325 also formed an average 0.40 hydrogen bond with trimannose (see Table 5). After mutation in DM SP-D•Trimannose, the hydrogen number changed to 0 (see Table 5). The weak hydrogen bond makes the influence of residue 325 in locking trimannose conformation not as obvious as in the medial trimannose binding pose.
Table 5.
Hydrogen bond number between residue 325 and trimannose of WT SP-D•Trimannose, Asp325Ala SP-D•Trimannose, Arg343Val SP-D•Trimannose, and DM SP-D•Trimannose at terminal trimannose binding pose.
| System | Acceptor | Donor | Occupancy | Distance (Å) | Angle (degrees) |
|---|---|---|---|---|---|
| WT SP-D●Trimannose | Asp325@OD1 | Man1@O2 | 0.27 | 2.67 | 161.44 |
| Asp325@OD2 | Man1@O2 | 0.27 | 2.67 | 161.39 | |
| Asp325@OD1 | Man1@O6 | 0.05 | 2.70 | 161.25 | |
| Asp325@OD2 | Man1@O6 | 0.05 | 2.70 | 161.60 | |
| Total | 0.64 | ||||
| Asp325Ala SP-D●Trimannose | No Hydrogen bonds | ||||
| Arg343Val SP-D●Trimannose | Asp325@OD1 | Man1@O2 | 0.20 | 2.67 | 159.30 |
| Asp325@OD2 | Man1@O2 | 0.20 | 2.67 | 159.31 | |
| Total | 0.40 | ||||
| DM SP-D●Trimannose | No Hydrogen bonds | ||||
Figure S17 and Figure S18 show the glycosidic torsion angles (ϕ, ψ) distribution of α-d-Manp-(1 → 2)-α-d-Manp linkage (Man1-Man2 and Man2-Man3) in different complex models at the terminal trimannose binding pose. The distributions of the four SP-D complex models were similarly populated as that observed in the trimannose-free.
4.3.2. Impact of Arg343Val in the terminal trimannose binding pose
Arg343Val weakens the hydrogen bond with Glu333, therefore, enhancing Glu333′s interaction with trimannose.
The hydrogen bond between the residue in position 343 and the specific glutamate residue in position 333 within the CRD and trimannose was similar to that of the medial trimannose binding pose (see Table 6). Arg343 of WT SP-D•Trimannose complex and Asp325Ala SP-D•Trimannose complex formed a stronger hydrogen bonding interaction with the negatively charged Glu333, whose hydrogen bond number with the Glu333 was larger than that in Arg343Val SP-D•Trimannose complex and DM SP-D•Trimannose complex. Glu333, therefore, formed fewer hydrogen bonds with trimannose in WT SP-D•Trimannose and Asp325 SP-D•Trimannose than that of Arg343Val SP-D•Trimannose and DM SP-D•Trimannose. As observed in the medial pose, Arg343Val weakens the hydrogen bond with Glu333 which enhances Glu333′s interaction with trimannose.
Table 6.
Hydrogen bonds between residue in position 343 and Glu333 and trimannose of WT SP-D•Trimannose, Asp325Ala SP-D•Trimannose, Arg343Val SP-D•Trimannose, and DM SP-D•Trimannose at the terminal trimannose binding pose.
| System | Hbond Type | Acceptor | Donor | Occupancy | Distance (Å) | Angle (degrees) |
|---|---|---|---|---|---|---|
| WT SP-D●Trimannose | Glu333-Arg343 | Glu333@OE1 | Arg343@NE | 0.37 | 2.80 | 151.05 |
| Glu333@OE2 | Arg343@NE | 0.37 | 2.80 | 151.34 | ||
| Glu333@OE1 | Arg343@NH1 | 0.06 | 2.77 | 160.22 | ||
| Glu333@OE2 | Arg343@NH1 | 0.05 | 2.77 | 160.23 | ||
| Glu333@OE1 | Arg343@NH2 | 0.33 | 2.78 | 149.59 | ||
| Glu333@OE2 | Arg343@NH2 | 0.33 | 2.78 | 149.51 | ||
| Total | 1.51 | |||||
| Glu333-Trimannose | No Hydrogen bonds | |||||
| Asp325Ala SP-D●Trimannose | Glu333-Arg343 | Glu333@OE1 | Arg343@NE | 0.38 | 2.81 | 152.91 |
| Glu333@OE2 | Arg343@NE | 0.47 | 2.80 | 152.00 | ||
| Glu333@OE1 | Arg343@NH2 | 0.32 | 2.79 | 150.41 | ||
| Glu333@OE2 | Arg343@NH2 | 0.42 | 2.79 | 150.04 | ||
| Total | 1.58 | |||||
| Glu333-Trimannose | No Hydrogen bonds | |||||
| Arg343Val SP-D●Trimannose | Glu333-Val343 | No Hydrogen bonds | ||||
| Glu333-Trimannose | Glu333@OE1 | Man2@O6 | 0.18 | 2.69 | 164.06 | |
| Glu333@OE2 | Man2@O6 | 0.21 | 2.69 | 164.19 | ||
| Total | 0.39 | |||||
| DM SP-D●Trimannose | Glu333-Val343 | No Hydrogen bonds | ||||
| Glu333-Trimannose | Glu333@OE1 | Man2@O6 | 0.25 | 2.69 | 163.93 | |
| Glu333@OE2 | Man2@O6 | 0.19 | 2.69 | 164.14 | ||
| Total | 0.44 | |||||
The WT SP-D•Trimannose complex and Asp325Ala SP-D•Trimannose complex had similar shortest distance plots between trimannose and Glu333 (see Fig. 8). Both fluctuated between 3.5 Å and 7 Å, thus they both have two representative conformations. Although the Arg343Val SP-D•Trimannose complex and DM Trimannose complex also had two corresponding representative conformations of their distance plots, their plots range from 2 Å to 4 Å, which indicates the closer distance between trimannose and Glu333 (see Fig. 9).
Fig. 8.
Shortest distances between trimannose and Glu333 of (a) WT SP-D•Trimannose and (b) Asp325Ala SP-D•Trimannose at the terminal trimannose binding pose. Representative conformations on the right panel show how Glu333 interacts with trimannose.
Fig. 9.
Shortest distances between trimannose and Glu333 of (a) Arg343Val SP-D•Trimannose and (b) DM SP-D•Trimannose at the terminal trimannose binding pose. Representative conformations on the right panel show how Glu333 interacts with trimannose.
4.3.3. Weight SP-D and DM SP-D in the terminal trimannose binding pose
WT SP-D•Trimannose, at the terminal binding pose, forms a weaker bonding interaction with trimannose than the DM SP-D•Trimannose complex. However, unlike the medial binding pose, in which both mutations played an important role, in the terminal pose, only one of the two mutations, Arg343Val, plays a significant role in enhancing SP-D’s interaction with trimannose. From this, we observed a benefit provided by having two mutations to enhance the binding process: when one of the mutations fails to play a significant role, the second is able to assist.
The influence of the Asp325Ala mutation in the terminal trimannose binding pose was not as strong as that in the medial trimannose binding pose to enhance the binding affinity with trimannose. Nevertheless, as seen in the medial pose, before mutation the positive Arg343 residue forms a significant hydrogen bond with the negative Glu333 weakening Glu333′s ability to interact with trimannose.
In summary, DM SP-D•Trimannose, at the terminal binding pose, increases the binding affinity of trimannose through changes largely due to the Arg343Val mutation as Val’s neutral charge frees negative Glu333 to form an enhanced bond with trimannose.
4.4. Binding free energy
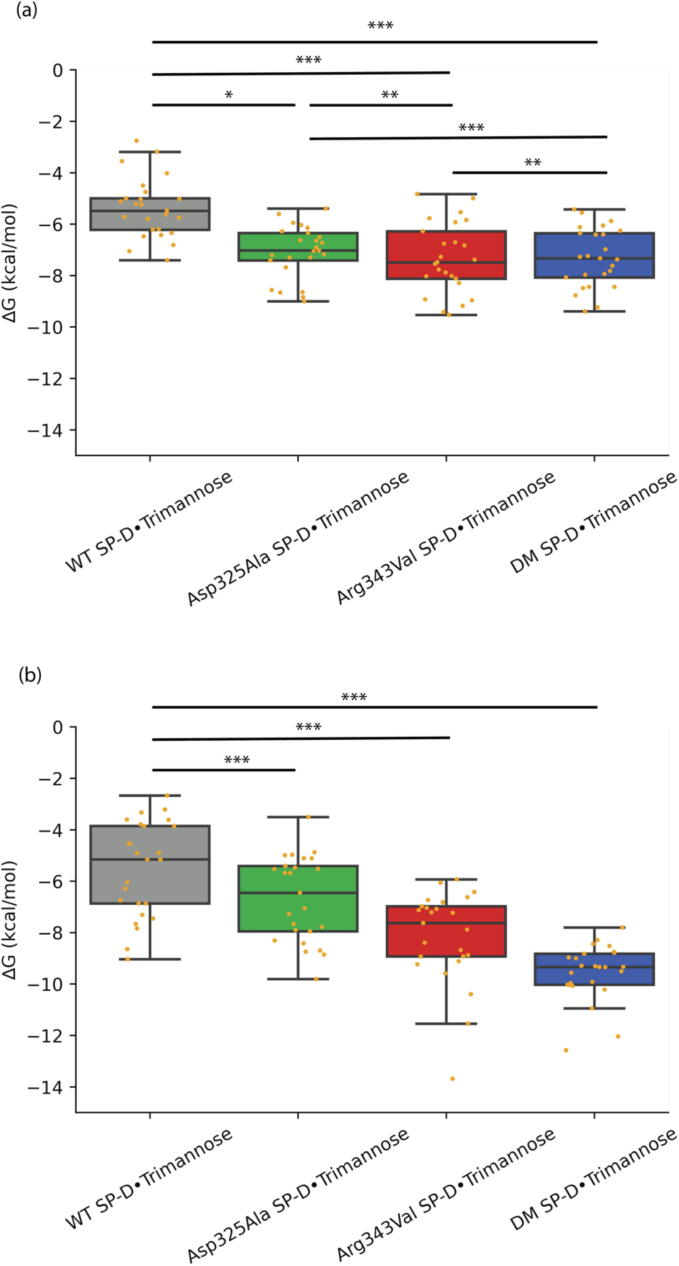
We use binding free energy to investigate the trends and understand the contributions of each of the mutations, Asp325Ala and Arg343Val, as well as how they come together in the DM SP-D from an energetic perspective. The binding free energies between different SP-D complexes at the medial and terminal binding poses were calculated in Fig. 10.
Fig. 10.
Binding free energy analysis of the medial trimannose binding pose (a) and the terminal trimannose binding pose (b). P-values are calculated by two-tailed t-tests. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
For the medial trimannose binding pose (see Fig. 10(a)), the ΔG for WT SP-D•Trimannose was −5.518 ± 1.812 kcal/mol. Each mutation increased SP-D’s binding affinity with Asp325Ala SP-D gaining an average ΔG of −6.674 ± 1.626 kcal/mol and Arg343Val SP-D obtaining an average ΔG of −8.167 ± 1.774 kcal/mol. This indicates that the Arg343Val mutation contributes more than Asp325Ala to the enhanced binding affinity with trimannose. The DM SP-D demonstrated the best binding affinity with trimannose with an average ΔG of −9.556 ± 1.066 kcal/mol. We also found that the binding free energy gains with DM SP-D are roughly the sum of that with two single mutant SP-Ds (P-value = 0.673). The effects of Asp325Ala and Arg343Val of SP-D are independent in increasing binding affinity with trimannose at the medial trimannose binding pose.
For the terminal trimannose binding pose (see Fig. 10(b)), the WT SP-D had an average ΔG of −5.390 ± 1.142 kcal/mol with trimannose. These two single mutations (Asp325Ala and Arg343Val) increased SP-D’s binding affinity. However, the difference in binding affinity with trimannose between these two single mutant SP-Ds (Asp325Ala SP-D and Arg343Val SP-D) was not obvious (P-value = 0.824), whose average ΔG was –7.082 ± 0.997 kcal/mol and −7.333 ± 1.335 kcal/mol, respectively. For the binding affinity of DM SP-D, the effect of increasing SP-D’s binding affinity was similar to either of the single mutant SP-Ds, whose average ΔG was −7.317 ± 1.120 kcal/mol.
It is worth noting that the effect of the mutations on increasing SP-D’s binding affinity is more obvious in the medial trimannose binding pose than that in the terminal trimannose binding pose. The medial trimannose binding pose of trimannose forms a much stronger connection with SP-D. In the medial trimannose binding pose, each mannose unit is in closer contact with SP-D. Man1 interacts with the CRD binding loop, and the Asp325Ala is also located within this region. Man2 chelates with the calcium ion of SP-D, and Man3 interacts with some other residues inside CRD, such as Glu333. However, for the terminal trimannose binding pose of trimannose, only Man1 chelates with the calcium ion. Some atoms inside Man1 interact with residue 325. While Man2 or Man3 can form a hydrogen bond with Glu333 after Arg343Val mutation, their interactions are not stronger than those in the medial trimannose binding pose.
These quantitative energies support our assertions regarding binding affinity based on hydrogen bonding and proximity to the trimannose. We observed that Asp325Ala does not have a large energetic impact; however, the single mutation Arg343Val is more important energetically and the DM SP-D has the highest energetic impact when both mutations act in concert. In addition, these effects contribute to the better binding affinity of the medial trimannose binding pose (ΔG of DM was −9.556 ± 1.066 kcal/mol) when compared to that of the terminal trimannose binding pose (ΔG of DM was −7.317 ± 1.120 kcal/mol).
5. Conclusions
In this study, we carried out full-atomistic MD simulations with microsecond trajectories to explore the molecular mechanism of SP-D’s mutation-induced higher binding affinity with trimannose. We systematically investigated each point mutation’s role in boosting SP-D’s binding affinity with trimannose, which is one of the most common units of viral surface glycans. We found that Asp325Ala stabilizes binding with trimannose by promoting a ligand conformational change. Although Asp325 forms a hydrogen bond with trimannose, it locks the glycosidic torsion angles, leading to unstable torsion angle distributions. The Asp325Ala mutation unlocks trimannose’s rotation, permitting relaxation to a stable conformation and further enhancing its binding with SP-D. The Arg343Val point mutation in SP-D changes its interaction with Glu333 to strengthen the binding ability with trimannose. The Arg343 residue has a more robust hydrogen bonding interaction with Glu333, which decreases the capability of the latter to bind with trimannose. In contrast, the Arg343Val mutation induces a weaker hydrogen bonding interaction with Glu333, which enhances Glu333′s hydrogen bonding interaction with trimannose. We further used the free energy perturbation (FEP) method to calculate the binding free energies between trimannose and different SP-D models to investigate each point mutation’s contribution toward enhancing SP-D binding affinity. The results show that the Arg343Val contributes more than Asp325Ala in increasing SP-D’s binding affinity with trimannose. And the DM SP-D gains the highest binding affinity with trimannose among the four different SP-Ds. The approximate order of binding affinity with trimannose is WT SP-D < Asp325Ala SP-D < Arg343Val SP-D < DM (Asp325Ala + Arg343Val) SP-D.
These findings deepen our understanding of the molecular mechanism for the enhanced binding affinity of DM SP-D to trimannose and explain the double mutant’s strong affinity for the IAV. We simultaneously captured the unbinding events of trimannose from WT SP-D and Asp325Ala SP-D in our computational generated trajectories, which is invisible through experimental techniques. The understanding of the mechanism at the molecular level can inform future investigations on the related drug design, which can target glycans precisely and yield promising clinical strategies. On the other hand, the computational modeling flow in this study can also be used to demonstrate the critical role of other immunoproteins’ behaviors at the molecular level, such as microbial invasion.
CRediT authorship contribution statement
Deng Li: Writing – original draft. Mona S. Minkara: Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors appreciate the financial support from Northeastern University . We are grateful to the Discovery Cluster of Northeastern University for computer time and facilities. The authors thank Dr. Mike Weaver and Dr. Elizabeth Janney for meaningful discussion on this work, and support from the other members from the COMBINE lab, as well as individual support provided by Benjamin Roy Greenvall- , and Anxhela Becolli.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.08.045.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Griese M. Pulmonary surfactant in health and human lung diseases: state of the art. Eur Respir J. 1999;13(6):1455–1476. doi: 10.1183/09031936.99.13614779. [DOI] [PubMed] [Google Scholar]
- 2.Echaide M., Autilio C., Arroyo R., Perez-Gil J. Restoring pulmonary surfactant membranes and films at the respiratory surface. Bba-Biomembranes. 2017;1859(9):1725–1739. doi: 10.1016/j.bbamem.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Veldhuizen E.J.A., Haagsman H.P. Role of pulmonary surfactant components in surface film formation and dynamics. Bba-Biomembranes. 2000;1467(2):255–270. doi: 10.1016/s0005-2736(00)00256-x. [DOI] [PubMed] [Google Scholar]
- 4.Kishore U., Greenhough T.J., Waters P., Shrive A.K., Ghai R., Kamran M.F., et al. Surfactant proteins SP-A and SP-D: structure, function and receptors. Mol Immunol. 2006;43(9):1293–1315. doi: 10.1016/j.molimm.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Sano H., Kuroki Y. The lung collectins, SP-A and SP-D, modulate pulmonary innate immunity. Mol Immunol. 2005;42(3):279–287. doi: 10.1016/j.molimm.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Hartshorn K.L., Reid K.B.M., White M.R., Jensenius J.C., Morris S.M., Tauber A.I., et al. Neutrophil deactivation by influenza a viruses: Mechanisms of protection after viral opsonization with collectins and hemagglutination-inhibiting antibodies. Blood. 1996;87(8):3450–3461. [PubMed] [Google Scholar]
- 7.Reading P.C., Morey L.S., Crouch E.C., Anders E.M. Collectin-mediated antiviral host defense of the lung: evidence from influenza virus infection of mice. J Virol. 1997;71(11):8204–8212. doi: 10.1128/jvi.71.11.8204-8212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsieh M.H., Beirag N., Murugaiah V., Chou Y.C., Kuo W.S., Kao H.F., et al. Human surfactant protein D binds spike protein and acts as an entry inhibitor of SARS-CoV-2 pseudotyped viral particles. Front Immunol. 2021;12:1–11. doi: 10.3389/fimmu.2021.641360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartshorn K.L., Crouch E., White M.R., Colamussi M.L., Kakkanatt A., Tauber B., et al. Pulmonary surfactant proteins A and D enhance neutrophil uptake of bacteria. Am J Physiol. 1998;274(6):L958–L969. doi: 10.1152/ajplung.1998.274.6.L958. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J.L., Zheng Q.C., Zhang H.X. Insight into the dynamic interaction of different carbohydrates with human surfactant protein D: molecular dynamics simulations. J Phys Chem B. 2010;114(21):7383–7390. doi: 10.1021/jp9113078. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J.L., Zheng Q.C., Zhang H.X. Unbinding of glucose from human pulmonary surfactant protein D studied by steered molecular dynamics simulations. Chem Phys Lett. 2010;484(4–6):338–343. [Google Scholar]
- 12.Goh B.C., Wu H.X., Rynkiewicz M.J., Schulten K., Seaton B.A., McCormack F.X. Elucidation of lipid binding sites on lung surfactant protein A using X-ray crystallography. Mutagenesis, Mol Dyn Simulat Biochem. 2016;55(26):3692–3701. doi: 10.1021/acs.biochem.6b00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartshorn K.L., Crouch E.C., White M.R., Eggleton P., Tauber A.I., Chang D., et al. Evidence for a protective role of pulmonary surfactant protein D (SP-D) against influenza A viruses. J Clin Invest. 1994;94(1):311–319. doi: 10.1172/JCI117323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anders E.M., Hartley C.A., Jackson D.C. Bovine and mouse serum beta inhibitors of influenza A viruses are mannose-binding lectins. Proc Natl Acad Sci USA. 1990;87(12):4485–4489. doi: 10.1073/pnas.87.12.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi L., Kash J.C., Dugan V.G., Jagger B.W., Lau Y.-F., Sheng Z.-M., et al. The ability of pandemic influenza virus hemagglutinins to induce lower respiratory pathology is associated with decreased surfactant protein D binding. Virology. 2011;412(2):426–434. doi: 10.1016/j.virol.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crouch E., Nikolaidis N., McCormack F.X., McDonald B., Allen K., Rynkiewicz M.J., et al. Mutagenesis of surfactant protein D informed by evolution and X-ray crystallography enhances defenses against influenza A virus in vivo. J Biol Chem. 2011;286(47):40681–40692. doi: 10.1074/jbc.M111.300673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.BrownAugsburger P., Chang D., Rust K., Crouch E.D. Biosynthesis of surfactant protein D - Contributions of conserved NH2-terminal cysteine residues and collagen helix formation to assembly and secretion. J Biol Chem. 1996;271(31):18912–18919. doi: 10.1074/jbc.271.31.18912. [DOI] [PubMed] [Google Scholar]
- 18.Crouch E., Persson A., Chang D., Heuser J. Molecular-structure of pulmonary surfactant protein-D (Sp-D) J Biol Chem. 1994;269(25):17311–17319. [PubMed] [Google Scholar]
- 19.Watson A., Phipps M.J.S., Clark H.W., Skylaris C.K., Madsen J. Surfactant proteins A and D: trimerized innate immunity proteins with an affinity for viral fusion proteins. J Innate Immun. 2019;11(1):13–28. doi: 10.1159/000492974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pikaar J.C., Voorhout W.F., van Golde L.M., Verhoef J., Van Strijp J.A., van Iwaarden J.F. Opsonic activities of surfactant proteins A and D in phagocytosis of gram-negative bacteria by alveolar macrophages. J Infect Dis. 1995;172(2):481–489. doi: 10.1093/infdis/172.2.481. [DOI] [PubMed] [Google Scholar]
- 21.Madan T., Eggleton P., Kishore U., Strong P., Aggrawal S.S., Sarma P.U., et al. Binding of pulmonary surfactant proteins A and D to Aspergillus fumigatus conidia enhances phagocytosis and killing by human neutrophils and alveolar macrophages. Infect Immun. 1997;65(8):3171–3179. doi: 10.1128/iai.65.8.3171-3179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ofek I., Mesika A., Kalina M., Keisari Y., Podschun R., Sahly H., et al. Surfactant protein D enhances phagocytosis and killing of unencapsulated phase variants of Klebsiella pneumoniae. Infect Immun. 2001;69(1):24–33. doi: 10.1128/IAI.69.1.24-33.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen M.J., Harbeck R., Smith B., Voelker D.R., Mason R.J. Binding of rat and human surfactant proteins A and D to Aspergillus fumigatus conidia. Infect Immun. 1999;67(9):4563–4569. doi: 10.1128/iai.67.9.4563-4569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goh B.C., Rynkiewicz M.J., Cafarella T.R., White M.R., Hartshorn K.L., Allen K., et al. Molecular mechanisms of inhibition of influenza by surfactant protein D revealed by large-scale molecular dynamics simulation. Biochemistry. 2013;52(47):8527–8538. doi: 10.1021/bi4010683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crouch E., Hartshorn K., Horlacher T., McDonald B., Smith K., Cafarella T., et al. Recognition of mannosylated ligands and influenza A virus by human surfactant protein D: contributions of an extended site and residue 343. Biochemistry. 2009;48(15):3335–3345. doi: 10.1021/bi8022703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shrive A.K., Martin C., Burns I., Paterson J.M., Martin J.D., Townsend J.P., et al. Structural characterisation of ligand-binding determinants in human lung surfactant protein D: influence of Asp325. J Mol Biol. 2009;394(4):776–788. doi: 10.1016/j.jmb.2009.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian C., Kasavajhala K., Belfon K.A.A., Raguette L., Huang H., Migues A.N., et al. ff19SB: amino-acid-specific protein backbone parameters trained against quantum mechanics energy surfaces in solution. J Chem Theory Comput. 2020;16(1):528–552. doi: 10.1021/acs.jctc.9b00591. [DOI] [PubMed] [Google Scholar]
- 28.Kirschner K.N., Yongye A.B., Tschampel S.M., Gonzalez-Outeirino J., Daniels C.R., Foley B.L., et al. GLYCAM06: a generalizable Biomolecular force field. Carbohydrates. J Comput Chem. 2008;29(4):622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong Y., Shabane P.S., Onufriev A.V. Melting points of OPC and OPC3 water models. ACS Omega. 2020;5(39):25087–25094. doi: 10.1021/acsomega.0c02638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Case K.B.D.A., Ben-Shalom I.Y., Brozell S.R., Cerutti D.S., Cheatham T.E., III, Cruzeiro V.W.D., et al. University of California; San Francisco: 2020. AMBER 2020. [Google Scholar]
- 31.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23(3):327–341. [Google Scholar]
- 32.Roe D.R., Cheatham T.E., 3rd PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput. 2013;9(7):3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 33.Schrödinger L, New York, NY, Schrödinger Release 2022-1: Maestro, 2021.
- 34.Zwanzig R.W. High-temperature equation of state by a perturbation method. I. Nonpolar gases. J Chem Phys. 1954;22(8):1420–1426. [Google Scholar]
- 35.Jiang W., Chipot C., Roux B. Computing relative binding affinity of ligands to receptor: An effective hybrid single-dual-topology free-energy perturbation approach in NAMD. J Chem Inf Model. 2019;59(9):3794–3802. doi: 10.1021/acs.jcim.9b00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang W., Phillips J.C., Huang L., Fajer M., Meng Y.L., Gumbart J.C., et al. Generalized scalable multiple copy algorithms for molecular dynamics simulations in NAMD. Comput Phys Commun. 2014;185(3):908–916. doi: 10.1016/j.cpc.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang W., Thirman J., Jo S., Roux B. Reduced free energy perturbation/Hamiltonian replica exchange molecular dynamics method with unbiased alchemical thermodynamic axis. J Phys Chem B. 2018;122(41):9435–9442. doi: 10.1021/acs.jpcb.8b03277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S., Oshima H., Zhang H., Kern N.R., Re S., Lee J., et al. CHARMM-GUI free energy calculator for absolute and relative ligand solvation and binding free energy simulations. J Chem Theory Comput. 2020;16(11):7207–7218. doi: 10.1021/acs.jctc.0c00884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu N.D., Kofke D.A., Woolf T.B. Improving the efficiency and reliability of free energy perturbation calculations using overlap sampling methods. J Comput Chem. 2004;25(1):28–39. doi: 10.1002/jcc.10369. [DOI] [PubMed] [Google Scholar]
- 40.Chen H.C., Maia J.D.C., Radak B.K., Hardy D.J., Cai W.S., Chipot C., et al. Boosting free-energy perturbation calculations with GPU-accelerated NAMD. J Chem Inf Model. 2020;60(11):5301–5307. doi: 10.1021/acs.jcim.0c00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W.G., Tsirulnikov K., Zhekova H.R., Kayik G., Khan H.M., Azimov R., et al. Cryo-EM structure of the sodium-driven chloride/bicarbonate exchanger NDCBE. Nat Commun. 2021;12(1):1–12. doi: 10.1038/s41467-021-25998-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, et al. Gaussian 16 Rev. C.01, Wallingford, CT, 2016.
- 43.Sawen E., Massad T., Landersjo C., Damberg P., Widmalm G. Population distribution of flexible molecules from maximum entropy analysis using different priors as background information: application to the phi, psi-conformational space of the alpha-(1 -> 2)-linked mannose disaccharide present in N- and O-linked glycoproteins. Org Biomol Chem. 2010;8(16):3684–3695. doi: 10.1039/c003958f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.