Abstract
Background:
GATA4, a zinc-finger containing DNA-binding transcription factor, is essential for normal cardiac development and homeostasis in mice and humans, and mutations in this gene have been reported in human heart defects. Defects in alternative splicing are associated with many heart diseases, yet relatively little is known about how cell type- or cell state-specific alternative splicing is achieved in the heart. Here, we show that GATA4 regulates cell type-specific splicing through direct interaction with RNA and the spliceosome in human induced pluripotent stem cell derived cardiac progenitors (iPS-CPs).
Methods:
We leveraged a combination of unbiased approaches including affinity purification of GATA4 and mass spectrometry, enhanced cross-linking with immunoprecipitation (eCLIP), electrophoretic mobility shift assays (EMSA), in vitro splicing assays, together with unbiased transcriptomic analysis to uncover GATA4’s novel function as a splicing regulator in human iPS-CPs.
Results:
We found that GATA4 interacts with many members of the spliceosome complex in human iPS-CPs. eCLIP demonstrated that GATA4 also directly binds to a large number of mRNAs through defined RNA motifs in a sequence-specific manner. In vitro splicing assays indicated that GATA4 regulates alternative splicing through direct RNA binding, resulting in functionally distinct protein products. Correspondingly, knockdown of GATA4 in human iPS-CPs resulted in differential alternative splicing of genes involved in cytoskeleton organization and calcium ion import, with functional consequences associated with the protein isoforms.
Conclusions:
This study shows that in addition to its well-described transcriptional function, GATA4 interacts with members of the spliceosome complex and regulates cell type-specific alternative splicing via sequence-specific interactions with RNA. Several genes that have splicing regulated by GATA4 have functional consequences and many are associated with dilated cardiomyopathy, suggesting a novel role for GATA4 in achieving the necessary cardiac proteome in normal and stress-responsive conditions.
Keywords: GATA4, RNA Splicing, RNA binding, Cardiac Cells
Introduction
Regulation of the cardiac proteome is essential for normal cardiac development and function. The proteome is determined by selective transcription as well as alternative splicing of transcripts that generate specific isoforms. While mutations in transcription factors are widely known to cause human cardiac disease, mutations in the RNA splicing regulators RBFOX2 or RBM20 can also cause congenital heart defects (CHD)1–3 or dilated cardiomyopathy4–7, respectively. These proteins bind to pre-mRNA and result in alternatively spliced forms of transcribed RNA, leading to a range of protein isoforms with distinct functions8–10. Within the heart, alternative splicing results in over 1700 altered protein isoforms during the critical shift from fetal to neonatal physiology, and nearly 900 transcripts are alternatively spliced after pathologic stresses that result in hypertrophy or heart failure11,12. RBM20 is a cardiac-specific RNA-binding protein, yet it only accounts for a small subset of the known cardiac-specific splicing events, while RBFOX2 and members of the spliceosome complex are ubiquitously expressed4–7. Although tissue-specific splicing events are often involved in cell fate decisions, developmental transitions, and response to various stresses13–15, and disruption of this process can lead to many human diseases16, the mechanism by which cell type- or cell state-specific alternative splicing is achieved remains poorly understood.
Recent reports demonstrated rare examples of transcription factors that can bind to RNA, yet the functional consequences of this binding activity are unclear17–19. One possibility is co-transcriptional splicing, where the transcription factor helps “prime” regions for splicing during transcription17,20,21. In such cases, the transcriptional regulators, like CTCF and YY1, have been relatively ubiquitously expressed and thus do not address how cell-type specific splicing may be achieved19,22–24. Transcription factor estrogen receptor α (ERα), which is a key driver for over 70% of breast cancers, was recently reported to also function as an RNA-binding protein and controls steps of RNA metabolism including RNA splicing and translation25. The potential for a tissue-enriched transcription factor to interact with RNA in a sequence-specific manner and also associate with the spliceosome complex to generate alternative spliced isoforms needed for specific cell types is another intriguing model that is yet to be examined in the heart.
Here, we show that GATA4, a cardiac-enriched zinc-finger containing DNA-binding transcription factor essential for normal cardiac development in mice and humans, and for mediation of the transcriptional response to cardiac stress 26–28, also functions as a regulator of alternative RNA splicing. An unbiased search for GATA4-interacting proteins in human iPS-CPs revealed interaction with many members of the spliceosome complex. GATA4 also bound to pre-mRNAs in a sequence-specific manner that resulted in generation of alternatively spliced isoforms in human iPS-CPs. Many of these GATA4-dependent isoforms had distinct functional properties, illustrating the importance of the splicing regulation to cardiac function. These results uncover a previously unrecognized function for GATA4 in regulating alternative splicing through direct RNA interaction, and suggest a more general mechanism by which tissue-specific splicing may be achieved via cell type-enriched transcription factors that not only control mRNA production, but also the isoforms generated from them.
Methods:
The data, analytical methods, and study materials will be made available to other researchers for purposes of reproducing the results or replicating the procedure. All relevant reagents will be maintained within the Srivastava laboratory and will be supplied on request.
hiPS cell culture and cardiac differentiation
hiPS (Wtc11) were maintained on plates coated with hESC-Qualified matrigel (BD, #354277) with Essential 8 Medium (Thermo Fisher, #A1517001) at 37°C, 5% CO2, and 21% O2. Cells were passaged every 4–5 d at ~80% confluency by using Accutase (Stem Cell Technologies, #07920). Y-27632 (ROCK inhibitor, Stem Cell Technologies, #72302) was included in the culture medium for 24 hours after dissociation.
The procedure used for cardiac differentiation from hiPS was based on a previously described WNT modulation protocol with modifications29. In brief, on day 0, the WNT agonist, CHIR99021 (Stemgent Incorporated, #04–0004) was added at 6 uM in RPMI (Thermo Fisher, #21870–076) with B27 supplement minus insulin (Thermo Fisher, #A1895601) for 48 hours. Then, 5uM IWP4 (Stemgent Inc, #04–0036) in RPMI with B27 supplement minus insulin was added to activate Wnt signaling for 48 hours. At day 5, IWP4 was removed, cells were dissociated with Accutase and replated on fibronectin (12.5 ug/ml; Sigma, #F1141) coated plate at a 1:3 dilution. Y-27632 (ROCK inhibitor) was included in the culture medium for 24 hours after dissociation.
Co-immunoprecipitation
Unless stated otherwise, all biochemical steps were performed on ice or at 4 °C, and ice-cold buffers were supplemented with complete protease inhibitors (Sigma, #11836170001), PhosSTOP Phosphatase Inhibitor Cocktail (Sigma, #4906845001). For co-immunoprecipitation, cells were washed with cold PBS three times, scraped in PBS and pelleted at 300g for 4 min. The cell pellets were resuspended with 3 volumes of Hypotonic buffer (20 mM Tris-HCl PH 8, 85 mM KCl, 0.5% NP-40) for 10 min to induce cell swelling. The resulting cell suspension was homogenized using a dounce homogenizer for 12 strokes. The nuclei were pelleted at 4,300 g for 5 mins, washed once with Hypotonic buffer and resuspended with Nuclear extract buffer (20 mM HEPES-KOH, PH7.4, 0.5 M NaCl2, 2 mM MgCl2, 1 mM CaCl2, 0.5% NP-40) with Benzonase, followed by 30 min of incubation with rotation. Nuclear lysate was collected as supernatant after centrifugation at 18,000 g for 30 min. The nuclear lysate was diluted in 2 volumes of IP dilution buffer (20mM HEPES-KOH, 1 mM EDTA, 0.02% CA-630). The protein concentration was measured using Broadford reagent (Bio-Rad Laboratories).
Immunoprecipitations were performed by incubating 0.5 mg of protein with magnetic M2 Flag beads (Sigma, #M8823) for 2h at 4 °C with rotation. Beads were washed three times with IP dilution Buffer and processed for western blot or mass spectrometry.
Protein extraction and Western Blot
Cell pellets were lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with complete protease inhibitors (Sigma, #11836170001) and PhosSTOP Phosphatase Inhibitor Cocktail (Sigma, #4906845001). 30–150 μg of protein lysate was separated on a 10% SDS-polyacrylamide gel and transferred to a PVDF membrane. The membranes were blotted with the Primary antibodies (Table 1) at 4°C overnight. Secondary antibodies (GE Healthcare) and chemi-luminescence reagents (Perkin Elmer) were used as per the manufacturer’s instructions.
Table 1.
Antibodies
| Antibody Name | Cat. No |
|---|---|
| Anti-Flag antibody | Sigma/#F1804 |
| Anti-GATA4 antibody (for IP; eCLIP) | Santa Cruz/#SC-25310 |
| Anti-GATA4 antibody (for Western Blot) | Thermo Fisher/#A303-503A |
| Anti-SF3A3 antibody | Bethyl Laboratories/#A302507A |
| Anti-HNRNPH1 antibody | Bethyl Laboratories/#A300511A |
| Anti-Vinculin antibody | Sigma/#V9131 |
Human cardiac fibroblast culture
The immortalized human cardiac fibroblast line was cultured in high glucose DMEM (Life Technologies) with 10% fetal bovine serum (FBS) (Hyclone, GE Healthcare), 1x Non-Essential Amino Acid (NEAA), 10U/ml penicillin/streptomycin and 1mM sodium pyruvate (all from Life Technologies). The medium was changed every other day and the cells were split every 2–3 days.
Cell lines and eCLIP assay
Day 7 human iPS derived cardiac progenitors were used for GATA4 eCLIP experiments. HepG2 cells were used for RbFOX2 eCLIP experiments.
eCLIP experiments were performed as previously described in a detailed standard protocol30 with minor modifications. All cells were UV crosslinked on ice at 400 mJoules/cm2 with 254 nm radiation. In brief, 20 million crosslinked cells were lysed and sonicated, followed by treatment with RNase I (Thermo Fisher, AM2295) to fragment RNA. Antibodies were pre-coupled to species-specific (anti-rabbit IgG or anti-mouse IgG) Dynabeads (Thermo Fisher), added to lysate, and incubated overnight at 4 °C. Prior to IP washes, 2% of sample was removed to serve as the paired input sample. For IP samples, high- and low-salt washes were performed, after which RNA was dephosphorylated with FastAP (Thermo Fisher, EF0652) and T4 PNK (NEB, M0201L) at low pH, and a 3′ RNA adaptor was ligated with T4 RNA ligase (NEB, M0437M). IP and input samples were run on an analytical PAGE Bis-Tris protein gel and transferred to nitrocellulose membranes, after which the region from the protein size to 75 kDa above protein size was excised from the membrane, treated with proteinase K (NEB, P8107S) to release RNA, and concentrated by column purification (Zymo). Input samples were then dephosphorylated with FastAP and T4 PNK at low pH, and a 3′ RNA adapter (RiL19) was ligated with T4 RNA ligase to synchronize with IP samples. Reverse transcription was then performed with AffinityScript (Agilent, 600107), followed by ExoSAP-IT (Affymetrix, 78201) treatment to remove unincorporated primer. RNA was then degraded by alkaline hydrolysis, and a 3′ DNA adaptor was ligated with T4 RNA ligase. qPCR was then used to determine the required amplification, followed by PCR with Q5 (NEB) and gel electrophoresis to size-select the final library. Libraries were sequenced using PE100 on the HiSeq platform (Illumina).
eCLIP-seq data analysis
The Illumina adapters were trimmed from the raw reads using cutadapt version 1.1831. To control for occasional double ligation of adapters in eCLIP-seq library preparation, cutadapt were run twice as previously recommended30. Next, 10 nt unique molecular identifiers (UMI) were extracted from the beginning of second mates using UMI-tools (version 1.0.0), and appended to the read names32. Also, 10 nt from the end of first mates were removed using cutadapt under the assumption that these could be UMIs. The trimmed reads longer than 18 nt were mapped to hg19 reference using STAR version 2.5.2b33. Then, Multi-mapping reads were removed, and the remaining reads were deduplicated using UMI-tools. Next, omniCLIP version 0.1.0 (with the optional argument --bck-var) was used to call peaks of mapped reads in a CLIP sample with respect to its matched input34. The peaks that had fewer than 10 mapped fragments in total, which were counted using featureCounts v1.5.1 (with optional arguments -p -O) were removed35. The remaining peaks were extended by 30 nt in each direction, and peaks in one sample that did not overlap a peak in any of the replicate samples were removed. Finally, the remaining peaks in individual replicates were integrated by merging all overlapping peaks. To identify sequence motifs enriched in the peaks, program findMotifsGenome.pl from homer2 with the arguments “-p 10 -rna -S 10 -len 5,6,7,8,9” were used, which instructs HOMER to only search motifs for the positive strand36.
Generation of GATA4-knockout HEK293 cell line
For generating the GATA4-knockout line, Cas9/gRNA RNPs were used. SpCas9-NLS ‘HiFi’ protein (from MacroLab, UC Berkeley [40 uM, 6.4 mg/ml]) was used. gRNA was purchased from Synthego (GATA4-gRNA: GAGGCCCACUCGGCGGGAGG). We transfected the HEK293 cells with SpCas9-NLS ‘HiFi’ protein and gRNA mixture by using the nucleofection platform (Neon Transfection System). We then seeded a single cell per well in 96 well plates and generated a clonal GATA4-knock out line. The targeted genomic region of GATA4 was amplified by PCR and sequenced to confirm the deletion (GATA4-F: AGAGATCTCATGCAGGGTCG; GATA4-R: TCATGATGCCTGGCCTTACT). The expression of GATA4 were checked by qRT-PCR and Western Blot.
Electrophoretic mobility shift assays
EMSA experiments were performed using the Lightshift chemiluminescent RNA EMSA Kit (Thermo Fisher, #20158) according the manufacturer’s instruction with modifications. RNA probes were labeled with 5’ IRDye 800 CW (IDT) to be directly observed on the gel. The probes (Table 2) were mixed with proteins, binding buffers (10 mM HEPES [PH 7.3], 20 mM KCl, 1mM MgCl2, 1mM DTT), 5% Glycerol, 2ug tRNA at RT for 20 min. Samples were applied to 4–12% polyacrylaminde Gel (Thermo Fisher, #EC62352BOX) under nondenaturing conditions with 0.5x Tris-borate-EDTA buffer (8.9 mM Tris base, 8.9 mM boric acid and 0.25 mM EDTA) at 4 °C for 1h. Gels were imaged using Odyssey® Fc Imaging System.
Table 2.
EMSA probes
| Probe Name | Sequences |
|---|---|
| Probe contains Motif1 | CUCACACACUCUUCCGAUCUGGG |
| Probe contains Motif2 | CUCACACACGUGCUCUUCGGG |
| Probe contains Motif3 | CCCAGAUCGGAAGUGUGUGAG |
| Probe contains Motif4 | CCCGAAGAGCACGUGUGUGAG |
| Probe with mutant Motif1 | CUCACACACUCUGCCAAAAUGGG |
| Probe with mutant Motif2 | CUCACACACGUGCUAAAAGGG |
| Probe with mutant Motif3 | CCCAUUUUAAAAGUGUGUGAG |
| Probe with mutant Motif4 | CCCUUUUAGCACGUGUGUGAG |
RNA pull-down assay
All cells were UV crosslinked on ice at 400 mJoules/cm2 with 254 nm radiation. In brief, 20 million crosslinked cells were lysed and sonicated, followed by treatment with RNase I (Thermo Fisher, AM2295) to fragment RNA. Antibodies were pre-coupled to species-specific (anti-rabbit IgG or anti-mouse IgG) Dynabeads (Thermo Fisher), added to lysate, and incubated overnight at 4 °C. Prior to IP washes, 2% of sample was removed to serve as the paired input sample. For IP samples, high- and low-salt washes were performed, after which samples were treated with proteinase K (NEB, P8107S). RNA was extracted and concentrated by column purification (Zymo). Reverse transcription was then performed with SuperScript IV (Thermo Fisher, #18090010). qPCR was then used to determine the RNA targets of interest (Table 3). The fold enrichment of pull down RNA in each samples were normalized to the related input.
Table 3.
Primers for RNA pull down assays
| Gene | Forward |
|---|---|
| CESR2-eCLIP Peak-F | GCCTATACCTCAACCGCAGC |
| CESR2-eCLIP Peak-R | GATGTTGTCGTCGAAGGGCA |
siRNA knockdown
siRNAs against GATA4 protein were purchased from Life Technologies (Invitrogen) (Silencer® Select Pre-Designed siRNA to human GATA4 #4392420, siRNA ID: s535120, Negative Control No. 1 siRNA #4390843). siRNA transfections were performed using Lipofectamine™ RNAiMAX transfection reagent (Thermo Fisher) according to the manufacturer’s instructions. 48 hours after transfection, cells were harvested for protein and RNA extraction.
RNA extraction, real-time PCR analysis
Total RNA was extracted using Zymo Direct-zol RNA kit (Zymo Research, #R2051) with on-column DNase I digestion according to the manufacturer’s instruction. 500 ng of RNA was converted to cDNA using SuperScript IV (Thermo Fisher, #18090010). For Taqman real-time PCR, 1/50 cDNA was applied for quantitative PCR reaction using Taqman Universal PCR master mix (Life technologies). The PCR was conducted in 7900HT Fast Real-Time system (Applied Biosystem). All gene expressions were normalized with GAPDH gene (Table 4).
Table 4.
Taqman Probes
| Gene | ThermoFish Ref. # |
|---|---|
| GATA4 | Hs01034629_m1 |
| GAPDH | Hs02786624_g1 |
Bulk RNA sequencing
Total RNA was extracted using Zymo Direct-zol RNA kit (Zymo Research, #R2051) according to the manufacturer’s instruction. Nanodrop and bioanlyzer Agilent 2100 were used to check the quantity and quality of RNA. The RNA libraries were prepared with the NuGEN Ovation Ultralow System V2 (Tecan, #0344NB) according to the manufacturer’s instruction. High-throughput sequencing was done using a PE100 run on a HiSeq instrument (Illumina).
Alternative splicing analysis using rMATS
Input sequences were provided in FASTQ format. After adapters from the reads were removed, fastx_trimmer from the fastx_toolkit version 0.0.14 was used to trim all reads to 97 nt and removed the reads shorter than 97 nt37. The reads were mapped to hg19 reference using STAR (with optional argument --alignEndsType EndToEnd). Then, rMATS was used to model the counts mapping across splice junctions and the pairing of samples38. rMATS computed the p-values for significance of alternative splicing events based on the null hypothesis that the magnitude of splicing difference is less than or equal to 0.05 (Δψ≤0.05). Splicing events where there were not at least two samples with at least 5 raw reads mapping to the splicing junctions were filtered out from further analysis. FDR-adjusted p-values for the remaining events were calculated using the Benjamini-Hochberg procedure39.
Relative exon usage analysis using Dexseq
To detect the alternative splicing of specific exons relative exon usage analysis was performed using Dexseq. Reads were assigned to exons using “featureCounts,”35 part of the Subread suite (http://subread.sourceforge.net/). Differential exon usage was estimated using the DEXSeq40,41. Bioconductor package in R. The DEXSeq function outputs an adjusted p-value42 per exon that is the result of testing the hypothesis whether the fold-change (GATA4 siRNA versus control) of normalized counts for this exon is the same as the fold-changes of the other exons belonging to the same gene.
Differential gene expression analysis
Input sequences were provided in FASTQ format. Trimming of known adapters and low-quality regions of reads was performed using Fastq-mcf43. Sequence quality control was assessed using the program FastQC44 and RseQC45. The reads were mapped to hg19 reference using STAR 2.5.2a33. Reads were assigned to genes using “featureCounts,”35 part of the Subread suite (http://subread.sourceforge.net/). Gene-level counts were arrived at using Ensembl gene annotation, in GTF format. Differential expression (GATA4 siRNA treatment vs control conditions) was assessed using edgeR46, a R package available through bio-conductor. Genes with less than 5 raw reads in at least 2 replicates were filtered out from further analysis. The reads counts of remaining ones are normalized using calcNormFactors in edgeR47. The mean gene expression was modeled as a function of siRNA status (siRNA treatment versus scramble control). Genes whose expression is associated with siRNA treatment were determined by the likelihood ratio test implemented in edgeR48–50 using a FDR< 0.05 and FC≥1.5threshold42.
NRVM isolation, culture and transfection
All rat manipulations were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee following guidelines described in the US National Institutes of Health’s (NIH’s) Guide for the Care and Use of Laboratory Animals.
Neonatal rat ventricular myocytes (NRVMs) were isolated from 2- to 3-day-old Sprague Dawley rats as described previously51,52. NRVMs were cultured in growth medium (DMEM, 5% fetal bovine serum, 100 U/ml penicillin-streptomycin) for 24 h. BrdU was added in the growth medium to remove non-myocytes. The cells were then cultured in non-serum media (DMEM, 0.1% BSA, 1% insulin-transferring selenium, 100 U/ml penicillin-streptomycin) and infected with an adenovirus encoding GFP or CACNA1C (exon21+exon22). 24 hours later, phenylephrine (PE, 100 uM) was added for 48 hours.
Cellular Electrophysiology
48 hours after PE stimulation, L-type Ca2+ currents (ICa2+-L) were recorded from GFP+ NRVMs via an Axopatch-700B amplifier in whole-cell voltage-clamp mode. pCLAMP10.3 (Molecular Devices/Axon Instruments) was used for data acquisition and the pCLAMP 10.6 Clampfit module was used for offline data analysis. Cells were perfused with the bath solution (in mmol/L, pH 7.4 adjusted with CsOH): TEA-Cl 137, CaCl2 4, MgCl2 0.5, CsCl 5.4, glucose 10, and HEPES 10. Patch electrodes were pulled to a resistance of 4.0 MΩ. Pipette solution contained (in mmol/L, pH 7.2 adjusted with CsOH): CsCl 125, Mg-ATP 5, MgCl2 1, TEA-Cl 20, EGTA 10, and HEPES 10. Recordings were digitized at 20 KHz and filtered at 1 kHz. To elicit L-type calcium current, cells were held at −50 mV to inactive T-type Ca2+current, and a series of 200-ms test pulses was applied from −40 mV to +50 mV at 10-mV increments 3 second intervals. The current amplitude was defined as the difference between the peak and steady-state current at a given test potential. The current density was calculated by normalizing the current amplitude to the cell capacitance, which was measured with a 10-mV test pulse from a holding potential of −70mV.
In vitro splicing assay
In vitro splicing experiments were performed as previously described in a detailed standard protocol53 with minor modification. The in vitro splicing reporter was constructed by synthesis in IDT, including the specific alternative exon, its flanking introns, as well as its constitutive exons (Table 5). In brief, the in vitro splicing assays performed in a volume of 20 ul contained 1.5 mM ATP, 5 mM creatine phosphate, 5 mM DTT, 3 mM MgCl2, 2.6% PVA, 30 units of RiboLock RNase inhibitor (Thermo Scientific), 20 ng of splicing substrate, 50–60 μg of splicing extract, and up to 12 μL of splicing buffer (20 mM HEPES-KCl pH 7.9, 100 mM KCl, 0.2 mM EDTA, 10% glycerol, 1 mM DTT) with or without the addition of protein. Reactions were incubated at 30 °C for one hour. RNA was extracted using TRI Reagent (Sigma) and then resuspended in 10 μL of DEPC-treated water. Spliced products were amplified by RT-PCR assays using 2 μL of the recovered RNA and primers specific for upstream and downstream constitutive exons. RT-PCR products were resolved on a 3% agarose gel (Table 6).
Table 5.
In vitro splicing templates
| Minigene Name | Sequence |
|---|---|
| SLX4IP-minigene | GTCTGTAGTTACTGTGGAATCAATAAGCCATGGCATCTAAGA AATTTGCTGTTAAAgtaagtaatgtttaatagctttttaaaaagaggctaatcactttctaatat aactgagctctgtgttaattgaaaatcgaggttcttatgtagaggcaaagcccatgccctcaggccttcc gatctctcttctctgtgcatgtgcatgcacactcacatgtccccgctcacaaacccactcactttcaacct cacaggacttcttcgaagatgccagcctccttttcacagcctttgtgtttactactcctgctgcctacaatg ctcttcccccagatattgctgggcatgagccgcacagacactgactctgccattaatgtctttcagTG TGGGAATTTTGCTGTCCTCGTGGATCTTCATATCTTGCCACAA GGTTCAAACAAAGATACAAGCTGGTTTTCTGAACAGAAGAAA GAGgtacaacaaaactttctactatttaattgcaagtagctgctgctgctataagaaatatcagctctttc cagctttcatttctgttagtgggaaaaaaatgttgcgtatttaatccttgaaaatttcccagatcacacagtg agccaacatcatggtttcgagcaaacttcagtagcaaatttaaatttgcggtgttattcatgagcacatatg acattcctttcaacattcacagctaaattaaaaagcaaaatgaaatgaaatgttcattgttaactttttaatat atatgttaatttttttgcattaaatttgaaggttatatctaattttttatttgctttagGAAGTCTGTTT ACTGTTAAAAGAAACCATTGATTCAAGAGTTCAGGAGTACTT GGAAGTTCGCAAACAGCACAGGCCATCAAATGCAGAATTCAC AAGATCCAATCCCTTGTCCTTAAAAG |
| SLX4IP-Motif-Mutant-minigene | GTCTGTAGTTACTGTGGAATCAATAAGCCATGGCATCTAAGA AATTTGCTGTTAAAgtaagtaatgtttaatagctttttaaaaagaggctaatcactttctaatat aactgagctctgtgttaattgaaaatcgaggttcttatgtagaggcaaagcccatgccctcaggcatca gtagaacttccgatctgactcttctctgtgcatgtgcatgcacactcacatgtccccgctcacaaaccca ctcactttcaacctcacaggacttcttcgaagatgccagcctccttttcacagcctttgtgtttactactcct gctgcctacaaatcagtagcccccagatattgctgggcatgagccgcacagacactgactctgccatt aatgtctttcagTGTGGGAATTTTGCTGTCCTCGTGGATCTTCATATC TTGCCACAAGGTTCAAACAAAGATACAAGCTGGTTTTCTGAA CAGAAGAAAGAGgtacaacaaaactttctactatttaattgcaagtagctgctgctgctataa gaaatatcagctctttccagctttcatttctgttagtgggaaaaaaatgttgcgtatttaatccttgaaaattt cccagatcacacagtgagccaacatcatggtttcgagcaaacttcagtagcaaatttaaatttgcggtgt tattcatgagcacatatgacattcctttcaacattcacagctaaattaaaaagcaaaatgaaatgaaatgtt cattgttaactttttaatatatatgttaatttttttgcattaaatttgaaggttatatctaattttttatttgctttagG AAGTCTGTTTACTGTTAAAAGAAACCATTGATTCAAGAGTTC AGGAGTACTTGGAAGTTCGCAAACAGCACAGGCCATCAAATG CAGAATTCACAAGATCCAATCCCTTGTCCTTAAAAG |
| GATA4-minigene | ACAACGTCTCCCTCCCACCCGGCTGAGAACAGCCTGGAATCC CTGTGCAGAGTTTGCCTCACACCAGGCCTTGAGCAGCCGCAG GGACGCCGCTCCCCACCGCGGgtgagtcccctcctggccctcctggtgacccctct gccgtgggaggctgggatgctgggggaatggaaggcttggggagtctccttgctcaccacaatccg gaggctctacgtccactccgtatccccaagaaagagtgtccgaggcacggaccatagcaagtgaag gaaggtaggtcgacgtggccttgcagctgaattcgttctccatttttccttcagcagGGACGCAT CCTGCTCTGCACCCTGGTTCTCGGCGCTGCGCCCGCGGAGGCT CGTGCAGGGCAGGCTGCCCGTGCGGGTGAGGACTGAGTGCCG CGCAGGGAAGGAGTATCGCAGACCGGCGCCCAGGCCCAGCG GGGGAATCCAAGGGCCGTGTTGCAGGACTCGGCATTCGTTCT GCGCGGGTCACCTTGAATGTCTGTCCGGATCCCTCGCGGCAG GGCCGCAGAGGCGCGTCCATATCTTGGAGGAATTCGTTCCAT AGAATGAGgtttgattctctctggggtttcttgttttccattataagactctggcgaccttggtggcg ccagattttttcagatgttgcttttgttccgggtgtagcgcccggctgcggcgaggaggaaggagccag cctagcagcttctgcgcctgtggccgcgggtgtcctggaggcctctcggtgtgacgagtgggggacc cgaaggctcgtgcgccacctccaggcctggacgctgccctccgtcttctgcccccaataggtgcgcc ggaccttcaggccctggggtgaattcagctgctcctacatcagcttccggaaccaccaaaaattcaaat tgggattttccggagtaaacaagagcctagagccctttgctcaatgctggatttaatacgtatatatttttaa gcgagttggttttttcccctttgatttttgatcttcgcgacagttcctcccacgcatattatcgttgttgccgtc gttttctctccccgcgtggctccttgacctgcgagggagagagaggacaccgaagccgggagctcgc agggaccatgtatcagagcttggccatggccgccaaccacgggccgccccccggtgcctacgagg cgggcggccccggcgccttcatgcacggcgcgggcgccgcgtcctcgccagtctacgtgcccaca ccgcgggtgccctcctccgtgctgggcctgtcctacctccagggcggaggcgcgggctctgcgtcc ggaggcgcctcgggcggcagctccggtggggccgcgtctggtgcggggcccgggacccagcag ggcagcccgggatggagccaggcgggagccgacggagccgcttacaccccgccgccggtgtcgc cgcgcttctccttcccggggaccaccgggtccctggcggccgccgccgccgctgccgcggcccgg gaagctgcggcctacagcagtggcggcggagcggcgggtgcgggcctggcgggccgcgagcag tacgggcgcgccggcttcgcgggctcctactccagcccctacccggcttacatggccgacgtgggc gcgtcctgggccgcagccgccgccgcctccgccggccccttcgacagcccggtcctgcacagcctg cccggccgggccaacccggccgcccgacaccccaatctcggtgagtaggagcgcgagggctggg gcgcgtgagggccggggcaggggccgtcttgagccctgtcgagggcctcttgtttttccaccaacgc cttcgttgggctggggatggtgcttcactacctcgagtttctagggaaggcagaagccagtgcggggc tggcgacatcacagccccagaagaccggcttctgtggaaggggccgggcctgcccgccggggcct cttctgagatggtgtcagggtcggagtgaggtggtcttctctttccaaggaaagggcattgtttctgtgcg ctctagattctcagatgtgagagctgggcataaacaaagaattaatcctctgtgtcttttcttgtctgttccc cccaactcagTAGATATGTTTGACGACTTCTCAGAAGGCAGAGAGT GTGTCAACTGTGGGGCTATGTCCACCCCGCTCTGGAGGCGAG ATGGGACGGGTCACTATCTGTGCAACGCCTGCGGCCTCTACC ACAAGATGAACGGCATCAACCGGCCGCTCATCAAGC |
| TTYH3-minigene | GCCCTGCTGCTCCTGGGGGCCGCCGCCCTGGCCTGCCTCGCCC TGGACCTCCTCTTCCTGCTCTTCTACTCCTTCTGGCTGTGCTGC CGGCGGCGCAAGAGCGAGGAGCACCTGGACGCCGACTGCTG CTGCACGGCCTGGTGTGTCATCATCGCCACGCTGGTGTGCAGg tgagcgcggtggggcggggacggagggcggggccagagggggcggcccgagggggcggggc tggagtggggtgtgtgtgggtgggcggggctacatctcacgggcccgccctctccagCGCCGG CATCGCAGTGGGATTCTACGGCAACGGGGAGACCAGTGATGG CATCCATAGGGCCACCTACTCGCTCCGCCACGCCAACCGCAC GGTGGCCGGGGTCCAGGACCGCgtgagtggccgcggagttggggccaggagc actcttgctgctccggagccccacgtctgcgcgaggacgggcggggcagccgggactggggctgt gcaggaggggcccagactctggggcgaggcgggcgcgctcccagcctcacgcccgcgggtccg gcgcagGTGTGGGACACGGCGGTGGGGCTGAACCACACGGCGG AGCCCAGCCTGCAGACCCTGGAGCGGCAGCTGGCCGGGCGG CCCGAGCCCCTGCGAGCCGTACAGAGGCTGCAGGGCCTGCTG GAGACGCTGCTGGGCTACACGGCCGCCATCCCCTTTTGGAGG AACACGGCGGTGTCGCTGGAGGTGCTGGCGGAGCAGGTGGA TCTCTACGACTGGTACAG |
Table 6.
Primers for in vitro splicing products
| Gene | Forward | Reverse |
|---|---|---|
| SLX4IP | GAGTCTGTAGTTACTGTGGAATCAA TAAGCCATGGC |
TTTGATGGCCTGTGCTGTTTGCG AACTTCC |
| GATA4 | GGCTGAGAACAGCCTGGAAT | AAGATATCGCTTGATGAGCGGC C |
| TTYH3 | GCTCTTCTACTCCTTCTGGC | GATATCCTGTACCAGTCGTAG |
Statistics and reproducibility
For statistical analysis GraphPad Prism software (version 9, GraphPad Software Inc) was used. All values are shown as means ± SEM. All experiments were carried out in at least three biological replicates. The number of replicates, statistical test used and test results are described in the figure legends. For all statistical tests, the 0.05 confidence level was considered statistically significant. In all figures, * denotes P < 0.05, ** denotes P < 0.001.
Data Availability
All source data, including sequencing reads and processed files have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE173162 (eCLIP-Seq), and GSE173163 (RNA-Seq).
Results:
GATA4 interacts with RNA splicing proteins in human iPS-derived cardiac progenitor cells (hiPS-CPs)
As most transcription factors function in complexes to regulate gene expression, our recent work investigated GATA4’s protein interactome by using an antibody to the endogenous GATA4 for affinity purification followed by mass spectrometry (AP-MS) in hiPS-CPs treated with RNase and DNase in triplicates (Figure 1A)54. Human GATA4-null iPS-CPs were used as negative control for non-specific interactions. Over 250 proteins were identified as potential GATA4 interacting partners, including several previously reported GATA4 interactors and novel interactors54. Surprisingly, there were many significant interactions between GATA4 and RNA binding proteins belonging to the spliceosome complex (e.g. SNRPF, SF3A1, SF3A3), RNA splicing regulators (e.g. HNRNPF, HNRNPR) and mRNA processing regulators (e.g. DHX15, SRRT) (Figure 1B). Co-immunoprecipitation (Co-IP) analysis of the GATA4 interaction with RNA-binding proteins for which there are high quality antibodies confirmed that interaction with SF3A3, HNRNPH1, and SF3A120 can indeed occur, even in the presence of RNAse (Figure 1C, Figure S1A).
Fig. 1: GATA4 protein interacts with RNA splicing proteins in cardiac progenitors.
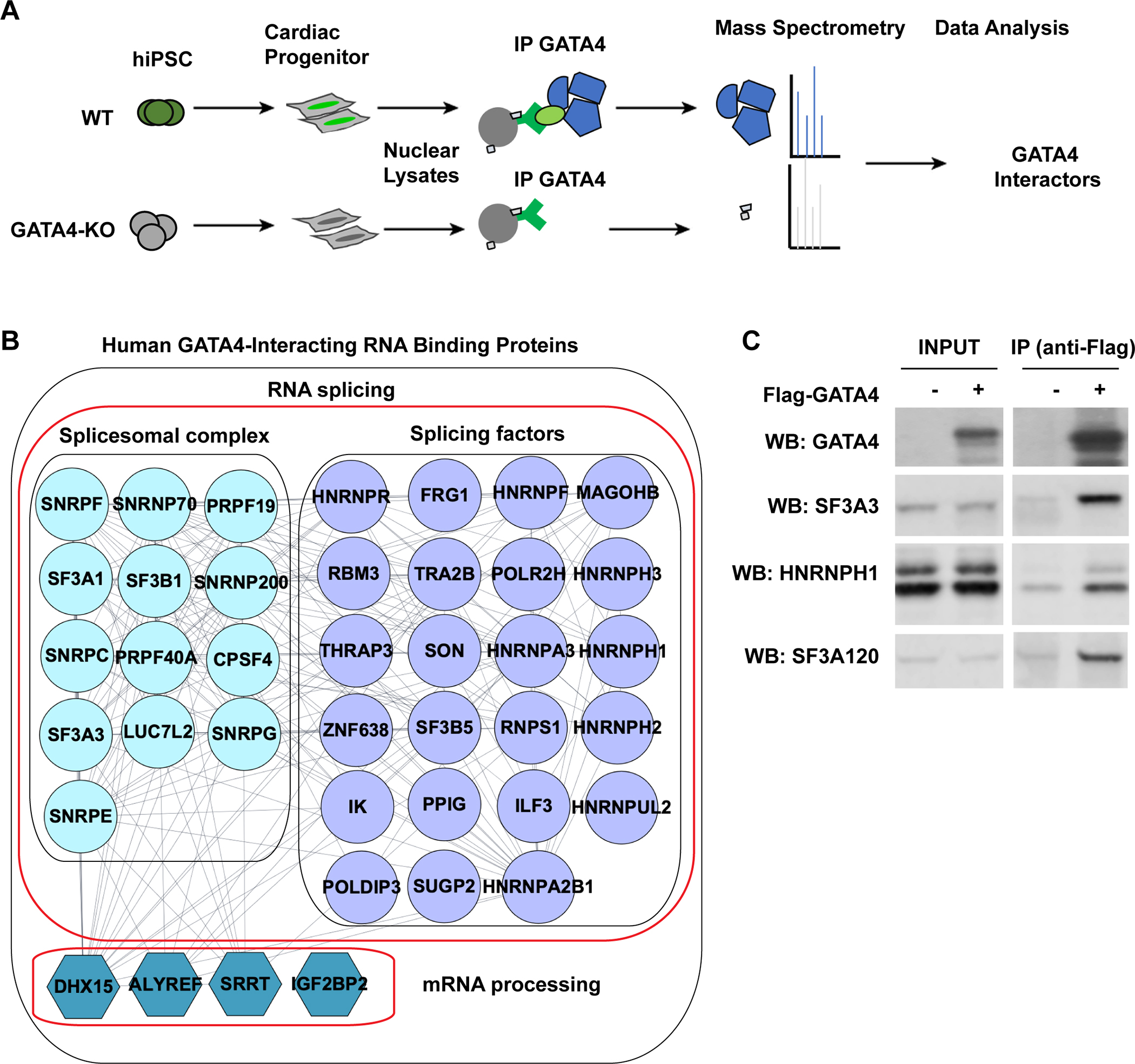
(A) Schematic of the experimental approach to identify human GATA4-interacting proteins in iPSC-derived cardiac progenitor cells (CPCs) by AP-MS using antibody to endogenous GATA4. Proteins isolated in an iPSC line lacking GATA4 were subtracted. (B) RNA binding proteins that were isolated with GATA4 from human iPS-CPs. The interaction network was realized using String. (C) Western blot (WB) of splicing factors indicated after Flag immunoprecipitation (IP) using anti-Flag antibody in HEK293T cells expressing Flag-GATA4. Protein expression in input lysate indicated. Cell lysate were treated with RNase and DNase before IP.
To identify which domains of GATA4 mediate GATA4:RNA binding protein interactions, a series of expression plasmids encoding GATA4 deletion mutants fused to the FLAG epitope (Figure S1B) were transfected into 293T-GATA4 knockout (KO) cells. Proteins bound to GATA4 deletion mutants were immunoprecipitated using FLAG antibody. Full length GATA4 co-immunoprecipitated with RNA binding proteins (e.g. SF3A3) as expected. Mutants lacking the C-terminal zinc finger or harboring a GATA4 G296S disease-causing missense variant near the C-terminal zinc finger were comparable with full-length GATA4 in interacting with SF3A3. In contrast, deletion of (N)-terminal amino acids or the N-terminal zinc finger resulted in diminished interaction, while absence of the (C)-terminal 119 amino acids abolished the interaction with SF3A3 (Figure S1. C; D). These findings revealed an unexpected interaction between GATA4 and RNA splicing proteins.
GATA4 binds to RNA through consensus RNA motifs
As GATA4 interacted with multiple RNA binding proteins, we tested whether GATA4 might exist in a complex with RNA. To identify the genome-wide RNA targets of GATA4, we used an antibody to endogenous GATA4 and performed enhanced crosslinking and immunoprecipitation (eCLIP)30 in triplicate using human iPS-CPs as done for AP-MS above. By normalizing the IP signal with size-matched input eCLIP libraries, we identified 1740 reproducible GATA4 binding sites over 1164 putative GATA4 target mRNAs (Table S1). Many genes had more than one identified peak (Figure 2A), as illustrated in GATA4 RNA (Figure 2B). Mapping the location of eCLIP peaks across the gene bodies revealed that the majority of GATA4 binding sites were located in the coding sequence and intron regions, similar to what was observed for other splicing proteins such as RBFOX2, which we used as a positive control for our eCLIP studies (Figure 2C). KEGG analysis showed GATA4-bound RNAs were enriched for focal adhesion, extra-cellular matrix (ECM)-receptor interaction, arrhythmogenic right ventricular cardiomyopathy pathways and cardiac contraction (Figure 2D). In summary, these data indicate that GATA4 is capable of specifically interacting with a subset of RNAs in human iPS-CPs.
Fig. 2: eCLIP demonstrates sequence-specific binding of GATA4 to RNA in human iPS-CPs.
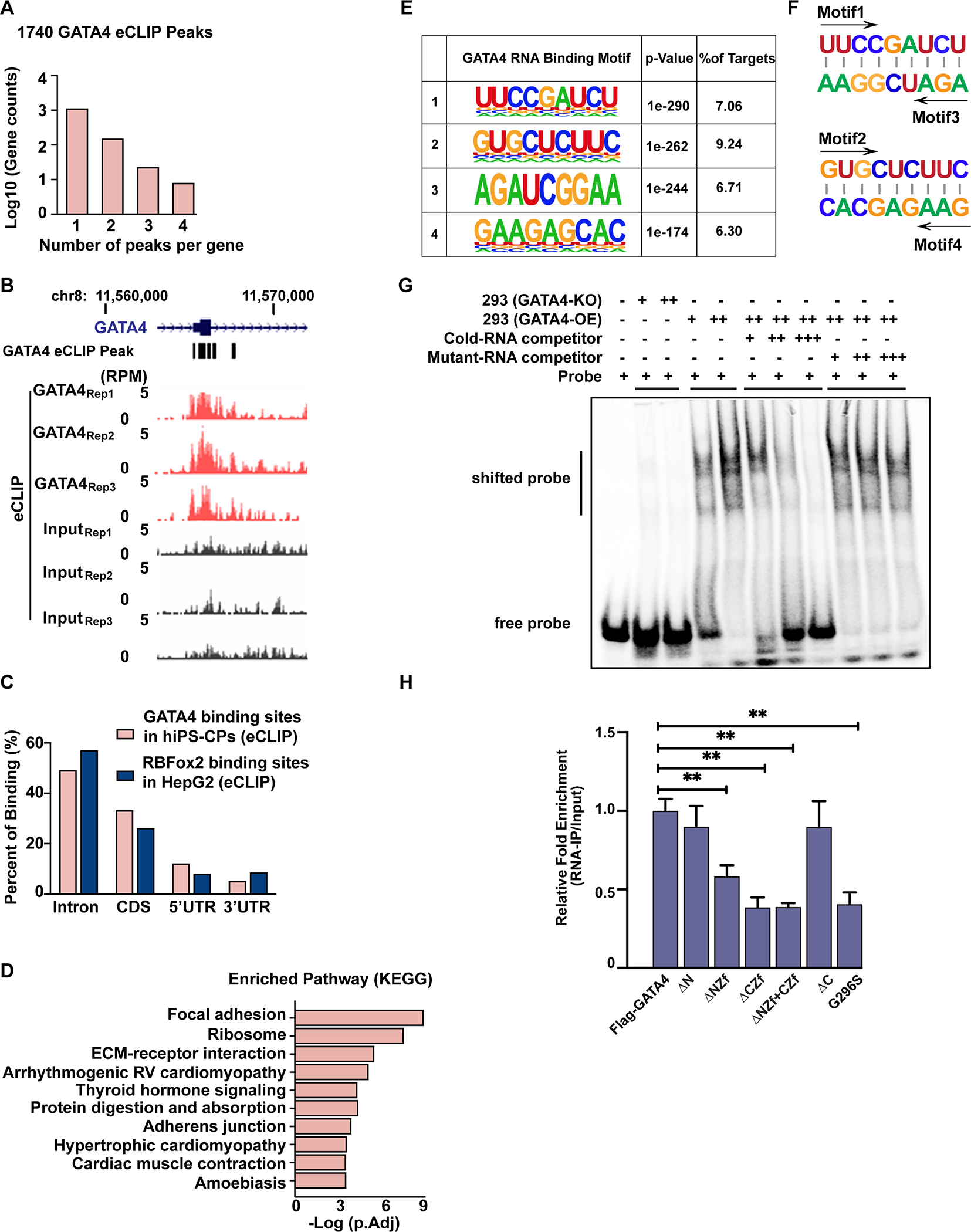
(A) Numbers of GATA4-eCLIP peaks in human iPS-CPs. (B) Integrated genome viewer tracks centered on the GATA4 gene. OmniCLIP identified peak regions indicated as black boxes above. (C) Distribution of GATA4-eCLIP (hCPCs) binding sites or RbFOX2-eCLIP (HepG2) binding sites across different regions of gene bodies. (D) KEGG pathway analysis of GATA4-bound RNAs. (E) Weblogos depicting the most significantly enriched GATA4-eCLIP-binding motifs by eCLIP in human iPS-CPs. (F) The motifs 1 or 2 were the reverse complementary sequences of motifs 3 or 4, respectively. (G) EMSAs were performed by adding GATA4-knockout (KO) or GATA4-overexpressing (OE) HEK293T cell lysate to an IRDye 800 labeled RNA probe containing the GATA4-binding motif 4 sequence. EMSA was performed by incubating HEK293T GATA4-OE lysate with labeled probe in the presence of 200(+)-, 1000(++)- and 2000(+++)-fold molar excess of the unlabeled competitor RNA or unlabeled mutant-RNA competitor. (H) RNA-pull down assay of GATA4 with the GATA4 binding site on CESR2 mRNA for GATA4. Immunoprecipitated mRNA were reverse-transcribed and amplified by qRT-PCR. One-way ANOVA coupled with a Tukey test was used to assess significance. **P < 0.001.
To determine if the GATA4-RNA interaction occurs in a sequence-specific manner, which would give greater confidence of direct interaction specificity, we used HOMER for de novo strand specific motif analysis of the GATA4-bound eCLIP peak regions. As a positive control, we did the same analysis on eCLIP results for RBFOX2 and found, as expected, that the canonical RBFOX2 motif “UGCAUG” was present in 40% of bound RNAs55. Four highly-enriched and related motifs were identified in the GATA4 eCLIP data, present in about 30% of GATA4-bound transcripts (Figure 2E). Interestingly, motifs 1 (UUCCGAUCU) or 2 (GUGCUCUUC) were the reverse complementary sequences of motifs 3 or 4, respectively, suggesting the possibility of interaction with hairpin structures (Figure 2F); indeed, a statistically significant number of peaks containing motif 2 or 4 also had a complementary sequence (motif 4 or 2) within the same transcript (Figure S2, p<0.001). To determine whether these motifs facilitate GATA4’s interaction with RNA, we performed an electromobility shift assay (EMSA) using RNA probes containing sequences derived from GATA4-eCLIP peaks with motif 4 (Figure 2G) or motifs 1, 2, or 3 (Figure S3, A to C). Specific shifted bands were detected in GATA4-expressing HEK293T nuclear extracts, and those bands were absent in GATA4-KO HEK293T nuclear extracts (Figure S3, Figure 2G and Figure S4, A to C); these bands were competed away with excess unlabeled wild-type, but not GATA4 site mutant probe (Figure 2G and Figure S4, A to C). These results suggested that GATA4 directly binds to defined mRNA motifs in a sequence-specific manner.
To identify which domains of GATA4 mediate GATA4:RNA interactions, a series of expression plasmids encoding GATA4 deletion mutants fused to the FLAG epitope (Figure S1B) were transfected into 293T-GATA4-KO cells. RNA species bound to GATA4 deletion mutants were immunoprecipitated using FLAG antibody. Consistent with our eCLIP results, full length GATA4 was able to bind to RNA targets (e.g. CESR2). GATA4 mutants lacking the (N)-terminal or (C)-terminal amino acids were comparable with full-length GATA4 in binding these RNAs. In contrast, mutants lacking the N-terminal or C-terminal zinc finger domains resulted in significant reduction of RNA binding, with most binding lost by deletion of the C-terminal zinc finger. The GATA4 G296S missense variant, which disrupts the C-terminal zinc finger and causes heart disease in children, disrupted RNA binding, demonstrating the importance of this zinc finger for GATA4:RNA interaction (Figure 2H).
GATA4 knockdown induces alternative splicing changes in human iPS-derived cardiac progenitors
As GATA4 interacts with numerous RNA binding proteins involved in splicing, and interacts with RNA targets, we tested whether knockdown of GATA4 in human iPS-CPs would alter the profile of alternatively spliced mRNAs. In comparison with human iPS-CPs treated with non-specific control siRNA (siNC), siGATA4 treatment for 48 hours significantly reduced GATA4 mRNA and protein levels (Figure S5, A and B). siRNA against GATA4 in iPS-CPs was utilized rather than CRISPR-based knockout so that emergence of CPs would not be affected and immediate consequences could be determined rather than potential secondary effects of GATA4 loss. RNA sequencing data from four biological replicates of control siRNA or siRNA targeting GATA4 in day 6 human iPS-CPs revealed 1599 differential and reproducible alternative splicing (AS) events occurring over 1247 genes upon GATA4 knockdown (Table S2). The differential AS genes included those related to sarcomere formation or calcium handling (e.g., TTN, SLC8A1, CACNA1C), the MAPK pathway (e.g., ATF2, RAP1B, SKT1, FGFR1), and transcriptional regulation (e.g., TEAD2, MEF2C, GATA4). The differential splicing events were deconvoluted into different types of splicing with a discrete range of percent spliced-in (PSI) values, including skipped exon (SE), retained intron (RI), mutually exclusive exon (MXE), alternative 5′ splice site (A5SS), and alternative 3′ splice site (A3SS). Changes most often involved exon inclusion or exclusion, representing nearly two-thirds of differential splicing events (937 events) (Figure 3A). GATA4 affected exon cassette inclusion and exclusion equally, with 458 and 489 events, respectively. Sashimi plots demonstrate differences in exon usage of EIF4A2 (Figure 3, B and C) and RYR2 (Figure S5, C and D) between control and GATA4-KD as examples. Relative exon usage plot also indicates differentially spliced exons of the GATA4 gene upon GATA4-knockdown, suggesting autoregulation of GATA4 isoforms (Figure 3D). Interestingly, GATA4 knockdown in cardiac fibroblasts also affected alternative splicing of an overlapping, yet distinct, set of RNAs (Figure S6, Table S3).
Fig. 3: GATA4 knockdown in human iPS-CPs induces alternative splicing changes.
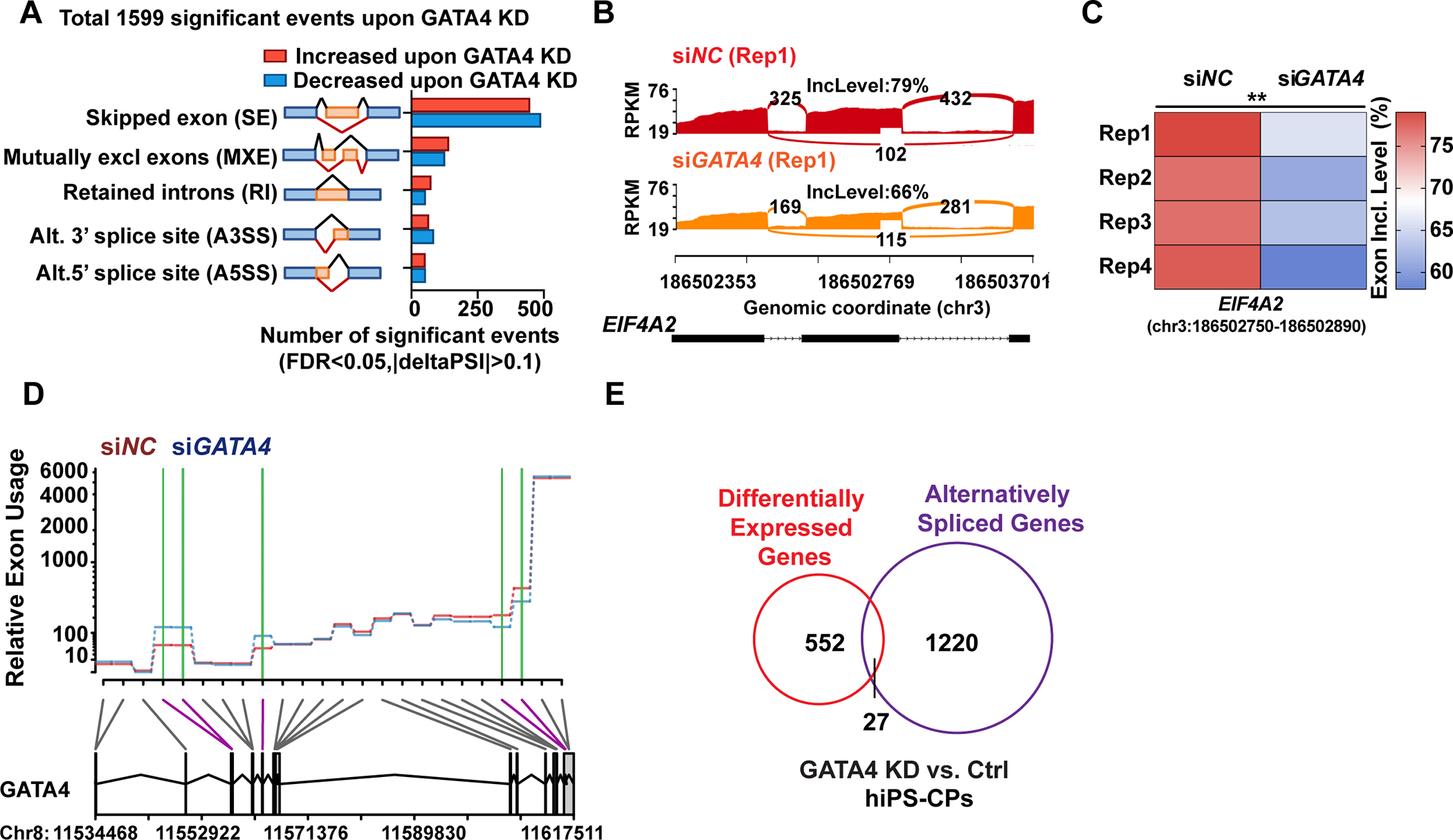
(A) Quantification of the alternative splicing events (n=1599) upon GATA4-knockdown (KD) by event type (FDR <0.05 from rMATS, junction counts ≥5) obtained from RNA-seq (n = 4). (B and C) Representative Sashimi plots depicting alternative splicing pattern of EIF4A2 in hiPS-CPs with siRNA targeting GATA4 (siGATA4) or negative control siRNA (siNC) and exon inclusion levels in each condition across four replicates (Rep). (D) Representative relative exon usage plots depicting alternative GATA4 splicing patterns in human iPS-CPs with green lines indicating change in exon inclusion upon GATA4 knockdown with siRNA compared to negative control siRNA. (E) Intersection of alternatively spliced genes upon GATA4-KD defined by rMATs (FDR <0.05, junction counts ≥5) and differentially expressed gene upon GATA4-KD (FDR<0.05, FC≥1.5).
We also investigated whether the GATA4 G296S disease-causing missense variant would affect the RNA splicing profile in human iPS-CPs. Human iPS cells with or without a homozygous G296S mutation were differentiated to day 6 cardiac progenitors for RNA sequencing. We observed 1580 differential and reproducible alternative splicing (AS) events (n=3) occurring over 1203 genes in GATA4 G296S iPS-CPs compared with wild-type isogenic cells (Table S4). Of these, 363 alternative splicing events over 152 genes were also observed upon GATA4KD. Gene ontology (GO) biological process analysis showed that these genes, which included RYR2, SLC8A1 and CACNA1C (Figure S7A), were enriched in regulating cytosolic calcium ion transport, sarcomere organization, and cardiac muscle contraction (Figure S7B), suggesting a role in titrating cardiac function.
Because GATA4 is a transcriptional regulator, we also analyzed the consequences on gene expression levels upon GATA4 knockdown and compared those to the alternatively spliced species. Differential gene expression analysis across four replicates revealed that GATA4 knockdown consistently dysregulated expression of 579 genes (FC>2, FDR<0.05) (Table S5). While many differential alternatively spliced genes were also regulated at the mRNA abundance levels upon GATA4 knockdown (27 genes), more often alternatively spliced genes were not changed in their mRNA level (Figure 3E). This suggests that while GATA4 can regulate gene splicing for some direct transcriptional targets providing a second level of isoform-specific control, it can also respond to cellular needs by regulating necessary isoforms independent of its transcriptional regulation. Importantly, we also did not observe significant changes in the levels of genes known to be involved in RNA binding or splicing upon acute knockdown of GATA4 for 48 hours (>400, Table S6)56, including RBM20 which can be affected with more permanent disruption of GATA4 (Figure S8, A to C) 28, and therefore splicing changes observed in GATA4 knockdown are unlikely to reflect direct GATA4-dependent transcriptional regulation of proteins involved in splicing.
GATA4 regulates mRNA splicing through direct interaction with mRNA
To determine if any GATA4-dependent AS events also involved direct interaction of GATA4 with pre-mRNA or were related to its transcriptional role, we integrated data sets that identified specific splicing pattern alterations upon GATA4 knockdown with GATA4-eCLIP data, and GATA4-chromatin immunoprecipitation (ChIP) data57 in human iPS-CPs. We identified 89 differentially spliced RNAs that were bound by GATA4 based on the eCLIP data (Figure 4A, p<0.001, Table S7). Only a fraction of the GATA4 bound RNA were alternatively spliced, which may suggest GATA4 function in regulating other RNA processing events, such as RNA polyadenylation or stability, similar to other RNA binding proteins that regulate splicing including RBM20 (Figure 4A)58. GATA4 was also bound to the gene body by ChIP-seq at 28 of these 89 sites (p<0.001), suggesting the possibility of co-transcriptional splicing regulation at some of these loci. The set of RNAs bound by GATA4 was enriched for genes involved in muscle stretch and calcium ion transport. Among the 89 RNAs bound by GATA4 and differentially spliced upon GATA4 knockdown, the majority of differential events involved exon inclusion or exclusion events, with far fewer intron inclusion or exclusion scenarios (Figure 4B and Figure S9A).
Fig. 4: GATA4 regulates mRNA splicing through direct interaction with mRNAs.
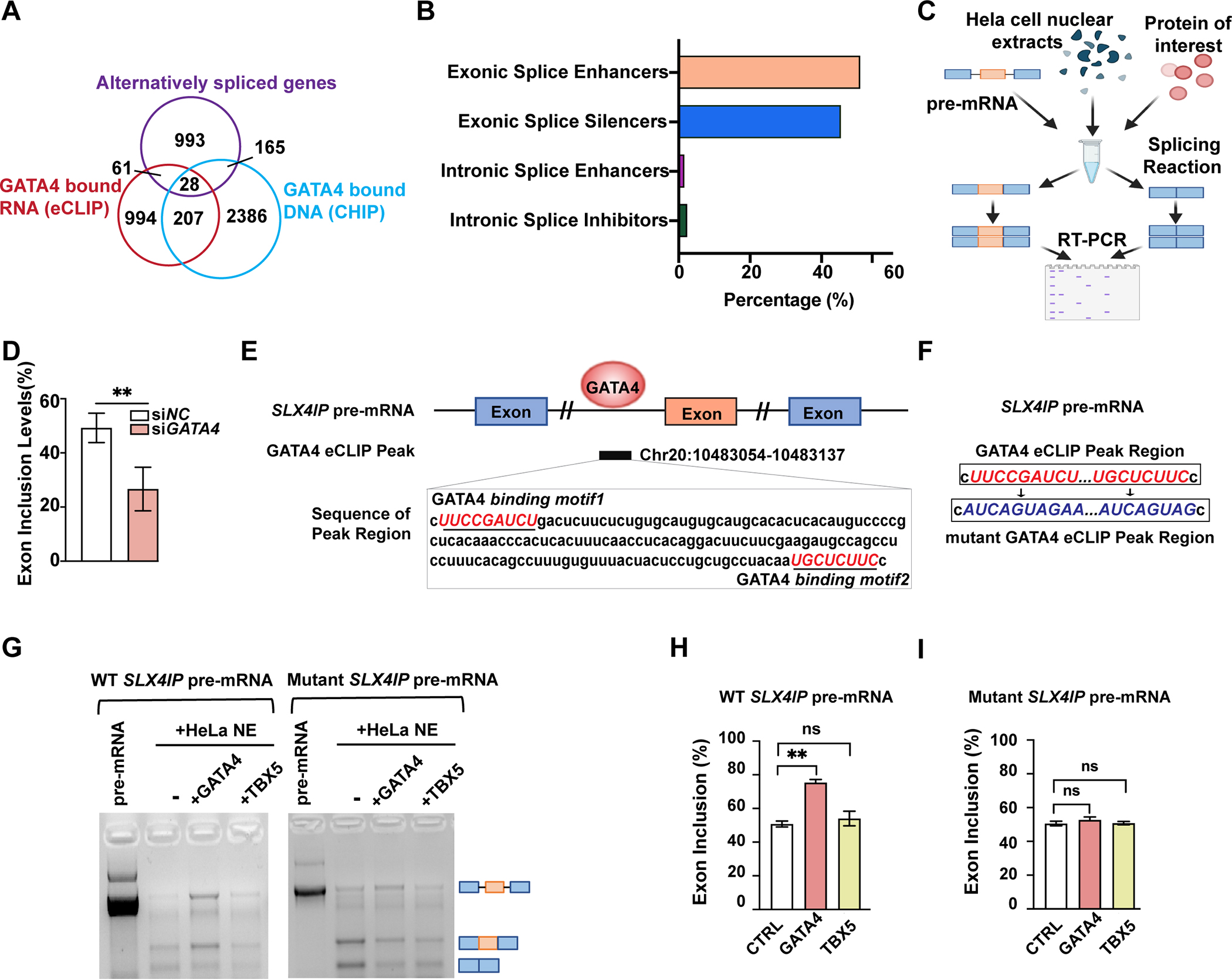
(A) Intersection of alternatively spliced genes upon GATA4-knockdown (KD) defined by rMATs (FDR <0.05, junction counts ≥5), genes with GATA4-eCLIP tag, and genes with GATA4-CHIP tag. (B) Quantification of GATA4 function in regulating splicing of 89 genes that were differentially spliced upon GATA4 KD and were bound to GATA4 based on an eCLIP binding peak. (C) Schematic representation of the in vitro splicing assay. (D) Exon inclusion level of significantly changed exon 3 of SLX4IP upon use of GATA4 siRNA compared to negative control (NC) siRNA. Data are shown as means ± SEM (n=4). Statistical significance was assessed using Student’s t test (**P < 0.001). (E) Integrated genome viewer tracks centered on gene SLX4IP. OmniCLIP identified peak regions indicated as black box. GATA4 binding motifs indicated in red. (F) Sequence information of the WT or mutant SLX4IP pre-mRNA template used in in vitro splicing assay. (G) In vitro splicing of SLX4IP minigene reporter transcripts in Hela nuclear-extracts (NE), with or without the addition of GATA4 or TBX5 protein. Bands indicate sizes of splicing events schematized with pre-mRNA length versus two alternatively spliced events. (H and I) Quantification of relative exon inclusion level in (G). Data are shown as means ± SEM (n=3). One-way ANOVA coupled with a Tukey test was used to assess significance. ns., non-significant; **P < 0.001.
To more rigorously test whether GATA4 regulates RNA splicing directly, we selected several transcripts for in vitro splicing assays to determine the precise effects of GATA4 protein in promoting target exon splicing in the presence of pre-mRNA and HeLa cell nuclear extract (Figure 4C). As an example, we show the alternative splicing of SLX4IP RNA regulated by GATA4 protein. The inclusion of exon 3 of SLX4IP decreased upon GATA4 knockdown by RNAseq (Figure 4D). This splicing event is likely to be directly regulated by GATA4, as we observed eCLIP peaks at the nearby intron region that contains GATA4 RNA binding motifs (Figure 4E). In the in vitro splicing assay, addition of GATA4 protein with pre-mRNA and nuclear extract stimulated greater inclusion of the specific alternative exon in question from the SLX4IP gene compared to the spliced version without this exon (Figure 4, F to H). In contrast, comparable levels of another cardiac-enriched transcription factor, TBX5, did not affect the splicing levels significantly, demonstrating the specificity of GATA4’s effects (Figure 4, F to H). Similar to the results for SLX4IP, addition of GATA4 protein, but not TBX5, inhibited inclusion of specific alternative exons from the GATA4 gene or TTYH3 gene (Figure S9B–E), predicted to be regulated by GATA4 based on eCLIP data (Figure 2B and Figure S9F) and transcriptomes of cells with diminished levels of GATA4 (Figure 3D and Figure S9G).
To investigate whether the GATA4 RNA binding sites were necessary for GATA-dependent regulation of splicing, we made mutations in the GATA4 RNA binding motifs present in SLX4IP pre-mRNA and used the in vitro splicing assay to determine consequences on the splicing products (Figure 4, F, G and I). In contrast to the effects of GATA4 on SLX4IP pre-mRNA, mutation of the GATA4 binding site resulted in failure to enrich for the exon inclusion event stimulated by GATA4, definitively demonstrating that GATA4 regulates SLX4IP splicing through direct binding with RNA in a sequence-specific manner (Figure 4, F, G and I). The function of SLX4IP is poorly understood, so the physiologic relevance of this exon inclusion/exclusion event remains unclear, however interrogation of this gene allowed rigorous evaluation of GATA4 binding through the defined motif.
GATA4 regulated alternative splicing, resulting in functionally distinct protein products
Among differentially spliced exons induced by GATA4-KD, 40 were also significantly different between hearts of patients with dilated cardiomyopathy and healthy donors (p<0.001) 12, indicating a potential pool of GATA4-dependent alternative splicing events induced by stress (Figure 5A, and Table S8). Many exons alternatively spliced correlated with known functional modules of the encoded protein (Table 7). For example, a critical exon of RYR2 (Ryanodine Receptor 2), the major regulator of intracellular calcium ion concentration necessary for cardiac contractions, that is necessary for its phosphorylation was spliced out upon GATA4-KD59,60.
Fig. 5: GATA4 regulated physiologically-relevant alternative splicing, resulting in functionally distinct protein products.
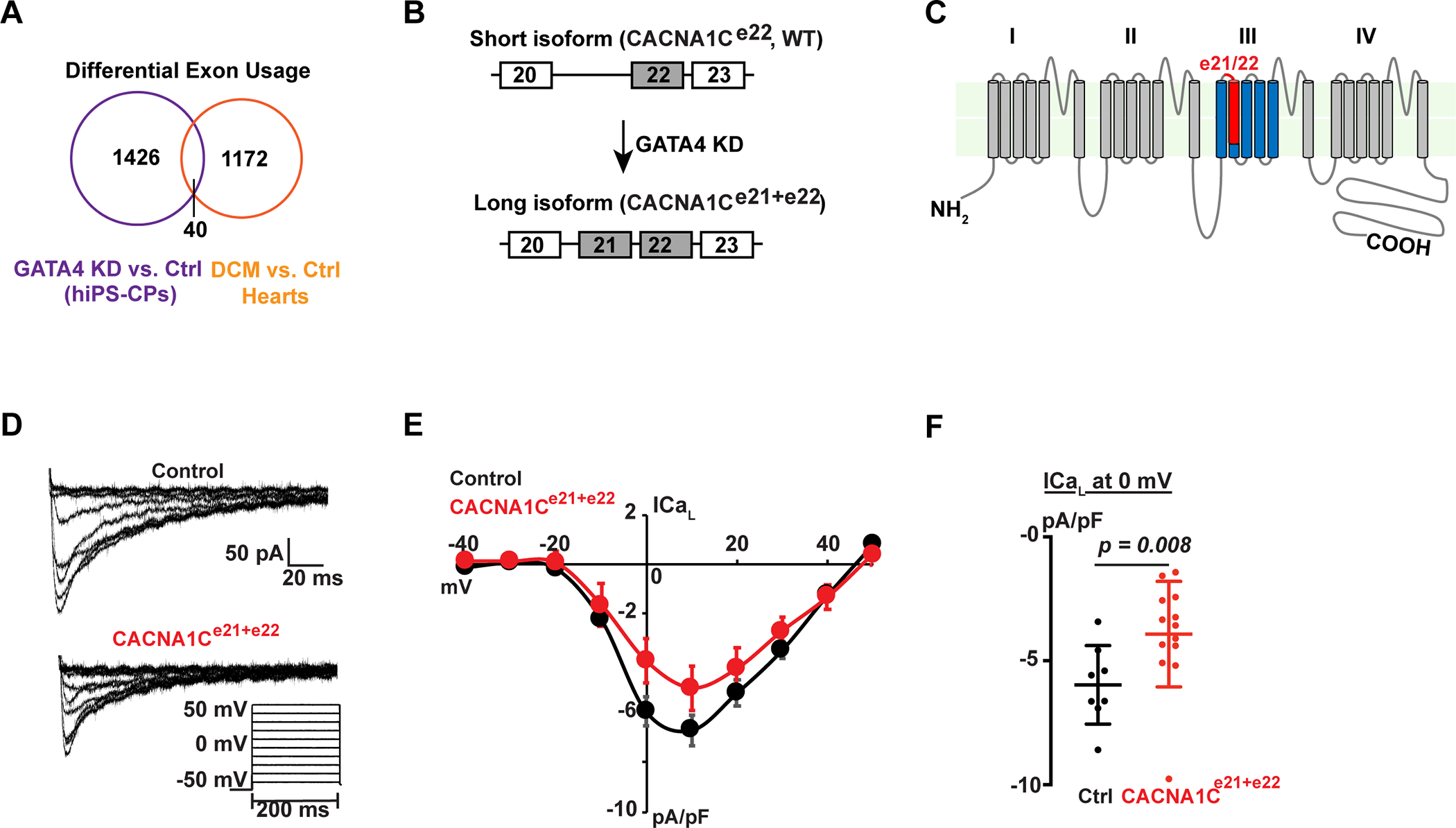
(A) Intersection of alternatively spliced exons upon GATA4-knockdown (KD) and alternatively spliced exons between human dilated cardiomyopathy (DCM) patient hearts and healthy donor hearts. (B) Schematic of short (WT) and long (CACNA1C e21+e22) isoforms of CACNA1C with differential inclusion of exon 21. (C) Predicted protein topology of the CACNA1C, showing the exon21/22 (red) localizing to the IIIS2 transmembrane segment and part of the linker region between IIIS1 and IIIS2. (D) Whole cell calcium current recordings from neonatal rat ventricular myocytes infected with adenovirus containing green fluorescent protein (GFP) or GFP + CACNA1Ce21+e22. Voltage protocol used to elicit L-type calcium current is shown at the bottom, right. (E) Current-voltage relationships for GFP infected (black, n=8) or CACNA1Ce21+e22 infected (red, n=13) neonatal rat ventricular myocytes (NRVMs), normalized to cell capacitance. Data are shown as means ± SEM. (F) Peak L-type calcium current density at 0 mV for GFP infected (black, n=8) or CACNA1Ce21+e22 infected (red, n=13) NRVMs. Error bars denote standard deviation. P=0.008 by Mann-Whitney test.
Table 7.
Examples of differentially spliced exons upon GATA4 knockdown correlated with known functional modules of the encoded protein.
| Differentially spliced region upon GATA4-KD | Protein | Affected domains | Impaired function |
|---|---|---|---|
| chr1:237850754-237850804 | RYR2 | AKAP interaction domain | RyR2 Phosphorylation |
| chr2:40404891-40404995 | SLC8A1 | Calx-beta domain | Regulatory Ca2+ binding |
| chr7:151329154-151329224 | PRKAG2 | CBS domain | Nucleotide binding |
| chr10:90770509-90770572 | FAS | Transmembrane domain | Regulation of apoptosis |
| chr11:65425757-65425970 | RELA | IPT domain | DNA binding |
In another example of GATA4-regulated isoform selection, GATA4-KD enhanced the inclusion of exon 21 of CACNA1C transcripts by 15% (p<.05), suggesting a fine-tuning function of GATA4 in regulating levels (Figure 5B, Figure S10). The CACNA1C gene codes for the α1C subunit of the Cav1.2 voltage-dependent L-type calcium channel. The CACNA1C protein consists of four repeating domains (I-IV), each with six transmembrane α helical segments (S1-S6) that are connected by intra- and extra- cellular loops as well as intercellular C-termini and N-termini (Figure 5C)61. Exon 21 encodes the IIIS2 transmembrane segment and part of the linker region between IIIS1 and IIIS2 (Figure 5C in red), and is typically spliced out; presence of exon 21 negatively affects pore formation and prevents cell surface inclusion of the functional protein derived from removal of exon 21. In HEK293 cells, overexpression of the splice variant enhanced by GATA4-KD (CACNA1Ce21+e22) decreases the whole-cell Ca2+ influx via the Cav1.2 channels62. To test whether increased transcription of the e21+22 isoform of CACNA1C can interfere with calcium channel function in cardiomyocytes, neonatal rat ventricular myocytes (NRVMs) were infected with adenovirus containing GFP alone or GFP plus CACNA1Ce21+e22, and cells were stimulated with 100 μM phenylephrine for 48hrs, which leads to increased transcription of L-type calcium channel genes63. Whole cell patch clamp was used to compare L-type calcium current levels between GFP infected and CACNA1Ce21+e22 infected NRVMs. While expression of CACNA1C did not affect the normalized current-voltage relationship, the NRVMs expressing CACNA1Ce21+e22 exhibited reduced total L-type calcium current (Figure 5, D and E). At a test potential of 0 mV, where nearly all channels are activated, NRVMs infected with CACNA1Ce21+e22 had a mean current density of −3.9 pA/pF (n = 13) of L-type calcium current compared to −6.0 pA/pF (n = 8) for NRVMs infected with GFP alone (p = 0.008 by Mann Whitney test, Figure 5F). These findings demonstrate that inclusion of the e21+e22 regions of CACNA1C results in significant reduction in L-type calcium current in cardiomyocytes and suggests that GATA4-dependent splicing, by promoting the exclusion of CACNA1C exon 21, may regulate subtle aspects of normal calcium homeostasis. Unlike transcripts above where we could rigorously demonstrate splicing regulation through direct GATA4-RNA interaction using the in vitro splicing assay, GATA4 binding is distant from the splicing site, therefore it remains to be definitively determined whether the GATA4-dependent regulation of CACNA1C is direct or indirect.
Discussion
Proteomic diversity is central to the heart’s physiologic function during cardiogenesis, transition to the post-natal circulation, and in response to stress or disease. In this study, we show that the cardiac-enriched transcription factor GATA4 interacts with members of the spliceosome complex, directly binds to pre-mRNAs in a sequence-specific manner through defined motifs, and can dynamically regulate splicing events of bound mRNAs independent of its DNA binding in human iPS-CPs (Figure 6). These findings expand the functions of the cardiac-enriched transcription factor GATA4 to RNA binding and mRNA splicing, broadening our understanding of how the tissue specific transcriptome and its derived proteome is regulated by tissue-enriched transcription factors in the heart.
Fig. 6: Model of RNA splicing mediated by GATA4 in human cardiomyocytes through direct binding with RNA.
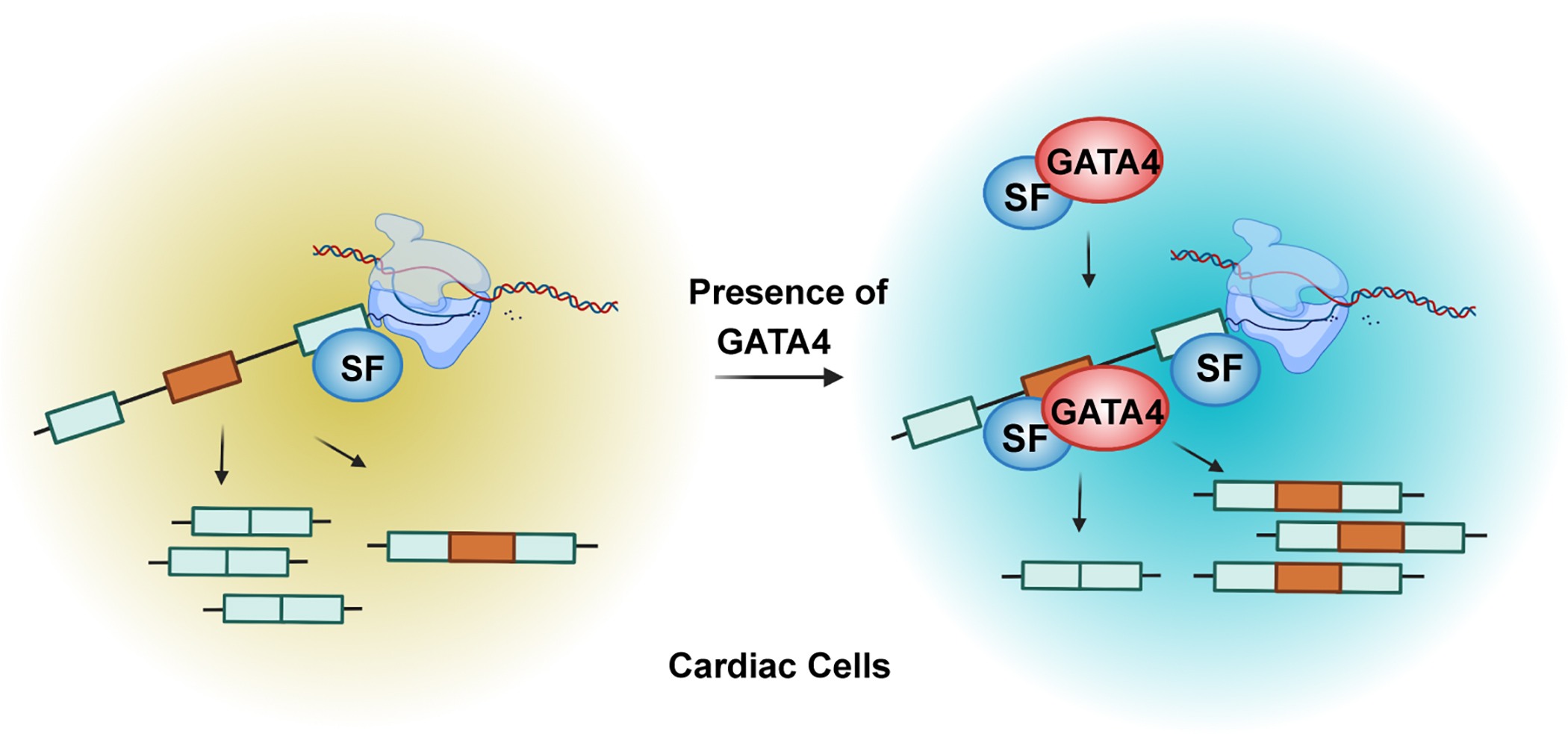
It has emerged in recent years that several transcription factors can bind RNA and some can impact alternative splicing, like the more ubiquitous factors YY1 and CTCF19,22–24. Since GATA4 is a critical transcription factor involved in cardiogenesis and human disease, and serves as a transcriptional integrator of numerous cardiac stress signaling pathways26–28, it is in a unique position to regulate both the transcriptome as well as the isoforms generated within a cell to execute specific functions. The unexpected observation in human cardiac cells that GATA4 interacts with many spliceosome proteins led us to uncover GATA4’s function in RNA splicing. By applying eCLIP-Seq, RNA-Seq and in vitro splicing, we found that GATA4 regulates tissue specific splicing through direct interaction with RNA. GATA4 modulates splicing patterns without affecting the expression of known splicing factors. GATA4 also regulates the expression levels and the alternative splicing patterns of two distinct sets of genes (Figure 3E). Furthermore, in vitro splicing assays revealed GATA4’s ability to control splicing events independent of its transcription regulation. Therefore, our data indicates that the splicing activity of GATA4 is uncoupled from its function as a transcription factor.
eCLIP of endogenous GATA4 with RNA demonstrated GATA4 in a complex with RNA transcripts, with enrichment of four specific binding motifs among interacting mRNAs, suggesting the interactions occur in a sequence-specific manner. Our EMSA results also provide evidence that GATA4 is able to bind single strand RNA through these specific motifs. Interestingly, motifs 1 or 2 were the reverse complementary sequences of motifs 3 or 4, respectively, suggesting the possibility of GATA4’s binding to RNA based on structural features.
Cardiac splicing defects cause disease in the setting of mutations of the splicing factors RBM20 and RBFOX2, highlighting the importance of alternative splicing for normal function of the heart1–3,5–7. However, current understanding of the factors regulating alternative splicing in the heart is very limited, with RBM20 only explaining a fraction of the alternative splicing that occurs in disease settings. We found that knockdown of GATA4 in human iPS-CPs resulted in >1500 differential alternative mRNA splicing events, with enrichment for genes involved in cytoskeleton organization and calcium ion import, such as CACNA1C, TTN, RYR2, and SLC8A1 (aka., NCX1). Many GATA4 regulated exons correlated with known functional modules of the protein. Indeed, we show that several genes that have GATA4-dependent splicing isoforms are associated with dilated cardiomyopathy and therefore regulation of alternative splicing might be a novel mechanism to explain how GATA4 mutations can lead to heart disease.
GATA4/5/6 are also critical for endoderm development while GATA1/2/3 are known to be essential regulators of hematopoiesis, and it will be interesting to determine if the functions associated with splicing regulation occur in a similar manner in endodermal and blood derivatives64–70. How broadly GATA family members or other transcription factors will also function to regulate splicing isoforms by directly binding to pre-mRNA and recruiting splicing factors remains to be determined, particularly for loci whose transcription is simultaneously being regulated. Further understanding of mechanisms by which tissue-enriched transcription factors like GATA4 regulate splicing to maintain homeostasis and respond to stimuli in times of stress will be important new avenues for investigation. It is clear that much remains to be characterized about the role of tissue enriched factors in mediating alternative splicing events but the novel paradigms described here deepen our understanding on how tissue-specific splicing is achieved to closely coordinate the establishment of gene programs that control specific cellular functions.
Supplementary Material
Clinical Perspective.
What is New?
GATA4, a cardiac enriched transcription factor, interacts with members of the spliceosome complex in human cardiac cells.
GATA4 is capable of directly interacting with RNA in a sequence-specific manner.
GATA4 regulates cell type specific splicing through direct interaction with RNA in human cardiac cells, independent of its transcriptional activity, including those encoding sarcomeric and calcium handling genes.
What are the Clinical Implications?
GATA4 is a novel RNA splicing factor in regulating alternative splicing, including that of many genes alternatively spliced in human heart disease.
GATA4 G296S congenital heart disease-causing missense variant affects RNA splicing profile of many critical cardiac genes in human cardiac progenitors, suggests a novel mechanism through which GATA4 mutations can lead to heart disease.
Acknowledgments:
We thank members of the Srivastava laboratory for helpful discussion and feedback. We acknowledge the Gladstone Stem Cell Core, the Gladstone Genomics Core and the Gladstone Bioinformatic Core for their support. We thank B. Taylor for her assistance with editing and graphics.
Funding:
National Institutes of Health grant DP2HL152425 (VV)
National Institutes of Health grant P01 HL146366 (DS, BGB, NJK)
National Institutes of Health grant R01 HL057181(DS)
National Institutes of Health grant R01 HL015100 (DS)
National Institutes of Health grant R01 HL127240 (DS)
Roddenberry Foundation (DS)
L.K. Whittier Foundation (DS)
Dario and Irina Sattui (DS)
Younger Family Fund (DS, BGB)
Additional Ventures (DS, BGB)
Non-standard Abbreviations and Acronyms:
- iPS-CPs
induced pluripotent stem cell derived cardiac progenitors
- eCLIP
enhanced cross-linking with immunoprecipitation
- EMSA
electrophoretic mobility shift assays
- CHD
congenital heart defects
- ERα
estrogen receptor α
- RIPA
radioimmunoprecipitation assay
- FBS
fetal bovine serum
- UMI
unique molecular identifiers
- NRVMs
Neonatal rat ventricular myocytes
- hiPS-CPs
human iPS-derived cardiac progenitor cells
- AP-MS
affinity purification followed by mass spectrometry
- Co-IP
Co-immunoprecipitation
- KO
knockout
- ECM
extra-cellular matrix
- siNC
non-specific control siRNA
- AS
alternative splicing
- PSI
percent spliced-in
- SE
skipped exon
- RI
retained intron
- MXE
mutually exclusive exon
- A5SS
alternative 5′ splice site
- A3SS
alternative 3′ splice site
- GO
Gene ontology
- ChIP
chromatin immunoprecipitation
Footnotes
Declaration of Interests:
D.S. is a co-founder and member of the board of directors of Tenaya Therapeutics and has equity in Tenaya Therapeutics; B.G.B. and B.R.C. are co-founders with equity in Tenaya Therapeutics; K.S.P. and N.K. have equity in Tenaya Therapeutics. The N.K. Laboratory has received research support from Vir Biotechnology and F. Hoffmann-La Roche. N.K. has consulting agreements with the Icahn School of Medicine at Mount Sinai, New York, Maze Therapeutics and Interline Therapeutics and has received stocks from Maze Therapeutics and Interline Therapeutics. The other authors declare no competing interests.
References:
- 1.Wei C, Qiu J, Zhou Y, Xue Y, Hu J, Ouyang K, Banerjee I, Zhang C, Chen B, Li H, et al. Repression of the Central Splicing Regulator RBFox2 Is Functionally Linked to Pressure Overload-Induced Heart Failure. Cell Rep. 2015;10:1521–1533. doi: 10.1016/j.celrep.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verma SK, Deshmukh V, Nutter CA, Jaworski E, Jin W, Wadhwa L, Abata J, Ricci M, Lincoln J, Martin JF, et al. Rbfox2 function in RNA metabolism is impaired in hypoplastic left heart syndrome patient hearts. Sci Rep. 2016;6:30896. doi: 10.1038/srep30896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science (New York, NY. 2015;350:1262–1266. doi: 10.1126/science.aac9396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18:766–773. doi: 10.1038/nm.2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe T, Kimura A, Kuroyanagi H. Alternative splicing regulator RBM20 and cardiomyopathy. Front Mol Biosci. 2018;5:105. doi: 10.3389/fmolb.2018.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fochi S, Lorenzi P, Galasso M, Stefani C, Trabetti E, Zipeto D, Romanelli MG. The emerging role of the RBM20 and PTBP1 ribonucleoproteins in heart development and cardiovascular diseases. Genes. 2020;11. doi: 10.3390/genes11040402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider JW, Oommen S, Qureshi MY, Goetsch SC, Pease DR, Sundsbak RS, Guo W, Sun M, Sun H, Kuroyanagi H, et al. Dysregulated ribonucleoprotein granules promote cardiomyopathy in RBM20 gene-edited pigs. Nat Med. 2020;26:1788–1800. doi: 10.1038/s41591-020-1087-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harper JW, Bennett EJ. Proteome complexity and the forces that drive proteome imbalance. Nature. 2016;537:328–338. doi: 10.1038/nature19947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-Dov C, Hartmann B, Lundgren J, Valcárcel J. Genome-wide analysis of alternative pre-mRNA splicing. J Biol Chem. 2008;283:1229–1233. doi: 10.1074/jbc.R700033200 [DOI] [PubMed] [Google Scholar]
- 10.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022 [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Chen Y, Li X, Chen G, Zhong L, Chen G, Liao Y, Liao W, Bin J. Genome-wide analysis of alternative splicing during human heart development. Sci Rep. 2016;6:35520. doi: 10.1038/srep35520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heinig M, Adriaens ME, Schafer S, van Deutekom HWM, Lodder EM, Ware JS, Schneider V, Felkin LE, Creemers EE, Meder B, et al. Natural genetic variation of the cardiac transcriptome in non-diseased donors and patients with dilated cardiomyopathy. Genome Biol. 2017;18:170. doi: 10.1186/s13059-017-1286-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tazi J, Bakkour N, Stamm S. Alternative splicing and disease. Biochimica et biophysica acta. 2009;1792:14–26. doi: 10.1016/j.bbadis.2008.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803 [DOI] [PubMed] [Google Scholar]
- 16.Montes M, Sanford BL, Comiskey DF, Chandler DS. RNA splicing and disease: Animal models to therapies. Trends Genet. 2019;35:68–87. doi: 10.1016/j.tig.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herzel L, Ottoz DSM, Alpert T, Neugebauer KM. Splicing and transcription touch base: co-transcriptional spliceosome assembly and function. Nat Rev Mol Cell Biol. 2017;18:637–650. doi: 10.1038/nrm.2017.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rambout X, Dequiedt F, Maquat LE. Beyond transcription: Roles of transcription factors in pre-mRNA splicing. Chem Rev. 2018;118:4339–4364. doi: 10.1021/acs.chemrev.7b00470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han H, Braunschweig U, Gonatopoulos-Pournatzis T, Weatheritt RJ, Hirsch CL, Ha KCH, Radovani E, Nabeel-Shah S, Sterne-Weiler T, Wang J, et al. Multilayered control of alternative splicing regulatory networks by transcription factors. Mol Cell. 2017;65:539–553.e537. doi: 10.1016/j.molcel.2017.01.011 [DOI] [PubMed] [Google Scholar]
- 20.Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15:1896–1908. doi: 10.1261/rna.1714509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brugiolo M, Herzel L, Neugebauer KM. Counting on co-transcriptional splicing. F1000Prime Rep. 2013;5:9. doi: 10.12703/p5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sigova AA, Abraham BJ, Ji X, Molinie B, Hannett NM, Guo YE, Jangi M, Giallourakis CC, Sharp PA, Young RA. Transcription factor trapping by RNA in gene regulatory elements. Science (New York, NY 2015;350:978–981. doi: 10.1126/science.aad3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wai DC, Shihab M, Low JK, Mackay JP. The zinc fingers of YY1 bind single-stranded RNA with low sequence specificity. Nucleic acids research. 2016;44:9153–9165. doi: 10.1093/nar/gkw590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saldaña-Meyer R, Rodriguez-Hernaez J, Escobar T, Nishana M, Jácome-López K, Nora EP, Bruneau BG, Tsirigos A, Furlan-Magaril M, Skok J, et al. RNA interactions are essential for CTCF-mediated genome organization. Mol Cell. 2019;76:412–422.e415. doi: 10.1016/j.molcel.2019.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y, Huangyang P, Wang Y, Xue L, Devericks E, Nguyen HG, Yu X, Oses-Prieto JA, Burlingame AL, Miglani S, et al. ERalpha is an RNA-binding protein sustaining tumor cell survival and drug resistance. Cell. 2021;184:5215–5229 e5217. doi: 10.1016/j.cell.2021.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. [DOI] [PubMed] [Google Scholar]
- 27.Whitcomb J, Gharibeh L, Nemer M. From embryogenesis to adulthood: Critical role for GATA factors in heart development and function. IUBMB life. 2020;72:53–67. doi: 10.1002/iub.2163 [DOI] [PubMed] [Google Scholar]
- 28.Ang YS, Rivas RN, Ribeiro AJ, Srivas R, Rivera J, Stone NR, Pratt K, Mohamed TM, Fu JD, Spencer CI, et al. Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell. 2016;167:1734–1749. doi: 10.1016/j.cell.2016.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods. 2016;13:508–514. doi: 10.1038/nmeth.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kechin A, Boyarskikh U, Kel A, Filipenko M. cutPrimers: A new tool for accurate cutting of primers from reads of targeted next generation sequencing. J Comput Biol. 2017;24:1138–1143. doi: 10.1089/cmb.2017.0096 [DOI] [PubMed] [Google Scholar]
- 32.Smith T, Heger A, Sudbery I. UMI-tools: Modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drewe-Boss P, Wessels HH, Ohler U. omniCLIP: Probabilistic identification of protein-RNA interactions from CLIP-seq data. Genome Biol. 2018;19:183. doi: 10.1186/s13059-018-1521-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao Y, Smyth GK, Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 36.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gordon A, Hannon GJ. Fastx-toolkit. FASTQ/A short-reads preprocessing tools. http://hannonlab.cshl.edu/fastx_toolkit 5. 2010.
- 38.Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korthauer K, Kimes PK, Duvallet C, Reyes A, Subramanian A, Teng M, Shukla C, Alm EJ, Hicks SC. A practical guide to methods controlling false discoveries in computational biology. Genome Biol. 2019;20:118. doi: 10.1186/s13059-019-1716-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reyes A, Anders S, Weatheritt RJ, Gibson TJ, Steinmetz LM, Huber W. Drift and conservation of differential exon usage across tissues in primate species. Proc Natl Acad Sci USA. 2013;110:15377–11582. doi: 10.1073/pnas.1307202110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Stat Soc. 1995;57:289–300. [Google Scholar]
- 43.Aronesty E EA-Utils: Command-line tools for processing biological sequence data. http://code.google.com/p/ea-utils. 2011.
- 44.Andrews S FastQC. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 45.Wang L, Wang S, Li W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356 [DOI] [PubMed] [Google Scholar]
- 46.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smyth GK, Verbyla AP. A conditional approach to residual maximum likelihood estimation in generalized linear models. R Stat Soc B. 1996;58:565–572. [Google Scholar]
- 49.Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23:2881–2887. doi: 10.1093/bioinformatics/btm453 [DOI] [PubMed] [Google Scholar]
- 50.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9:321–332. doi: 10.1093/biostatistics/kxm030 [DOI] [PubMed] [Google Scholar]
- 51.Duan Q, McMahon S, Anand P, Shah H, Thomas S, Salunga HT, Huang Y, Zhang R, Sahadevan A, Lemieux ME, et al. BET bromodomain inhibition suppresses innate inflammatory and profibrotic transcriptional networks in heart failure. Sci Transl Med. 2017;9:eaah5084. doi: 10.1126/scitranslmed.aah5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsu A, Duan Q, McMahon S, Huang Y, Wood SA, Gray NS, Wang B, Bruneau BG, Haldar SM. Salt-inducible kinase 1 maintains HDAC7 stability to promote pathologic cardiac remodeling. J Clin Invest. 2020;130:2966–2977. doi: 10.1172/jci133753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Movassat M, Mueller WF, Hertel KJ. In vitro assay of pre-mRNA splicing in mammalian nuclear extract. Methods Mol Biol. 2014;1126:151–160. doi: 10.1007/978-1-62703-980-2_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gonzalez-Teran B, Pittman M, Felix F, Richmond-Buccola D, Thomas R, Choudhary K, Moroni E, Colombo G, Alexanian M, Cole B, et al. Integration of protein interactome networks with congenital heart disease variants reveals candidate disease genes. bioRxiv. 2021. doi: 10.1101/2021.01.05.423837 [DOI] [Google Scholar]
- 55.Lovci MT, Ghanem D, Marr H, Arnold J, Gee S, Parra M, Liang TY, Stark TJ, Gehman LT, Hoon S, et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol. 2013;20:1434–1442. doi: 10.1038/nsmb.2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR. RBPDB: A database of RNA-binding specificities. Nucleic acids research. 2011;39:D301–D308. doi: 10.1093/nar/gkq1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonzalez-Teran B, Pittman M, Felix F, Thomas R, Richmond-Buccola D, Hüttenhain R, Choudhary K, Moroni E, Costa MW, Huang Y, et al. Transcription factor protein interactomes reveal genetic determinants in heart disease. Cell. 2022;185:794–814.e730. doi: 10.1016/j.cell.2022.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fenix AM, Miyaoka Y, Bertero A, Blue S, Spindler MJ, Tan KKB, Perez-Bermejo J, Chan AH, Mayer SJ, Nguyen T, et al. Gain-of-function cardiomyopathic mutations in RBM20 rewire splicing regulation and re-distribute ribonucleoprotein granules within processing bodies. bioRxiv. 2021: 10.1101/2021.1106.1102.446820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bovo E, Huke S, Blatter LA, Zima AV. The effect of PKA-mediated phosphorylation of ryanodine receptor on SR Ca(2+) leak in ventricular myocytes. J Mol Cell Cardiol. 2017;104:9–16. doi: 10.1016/j.yjmcc.2017.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yano M, Yamamoto T, Kobayashi S, Matsuzaki M. Role of ryanodine receptor as a Ca2(+) regulatory center in normal and failing hearts. J Cardiol. 2009;53:1–7. doi: 10.1016/j.jjcc.2008.10.008 [DOI] [PubMed] [Google Scholar]
- 61.Abernethy DR, Soldatov NM. Structure-functional diversity of human L-type Ca2+ channel: perspectives for new pharmacological targets. J Pharmacol Exp Ther. 2002;300:724–728. doi: 10.1124/jpet.300.3.724 [DOI] [PubMed] [Google Scholar]
- 62.Hu Z, Wang JW, Yu D, Soon JL, de Kleijn DP, Foo R, Liao P, Colecraft HM, Soong TW. Aberrant splicing promotes proteasomal degradation of L-type Ca(V)1.2 calcium channels by competitive binding for Ca(V)β subunits in cardiac hypertrophy. Sci Rep. 2016;6:35247. doi: 10.1038/srep35247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaughan JP, Hefner CA, Houser SR. Electrophysiological properties of neonatal rat ventricular myocytes with alpha1-adrenergic-induced hypertrophy. Am J Physiol. 1998;275:H577–H590. doi: 10.1152/ajpheart.1998.275.2.H577 [DOI] [PubMed] [Google Scholar]
- 64.Tremblay M, Sanchez-Ferras O, Bouchard M. GATA transcription factors in development and disease. Development. 2018;145. doi: 10.1242/dev.164384 [DOI] [PubMed] [Google Scholar]
- 65.DeLaForest A, Quryshi AF, Frolkis TS, Franklin OD, Battle MA. GATA4 is required for budding morphogenesis of posterior foregut endoderm in a model of human stomach development. Front Med (Lausanne). 2020;7:44. doi: 10.3389/fmed.2020.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xuan S, Sussel L. GATA4 and GATA6 regulate pancreatic endoderm identity through inhibition of hedgehog signaling. Development. 2016;143:780–786. doi: 10.1242/dev.127217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pikkarainen S, Tokola H, Kerkelä R, Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovasc Res. 2004;63:196–207. doi: 10.1016/j.cardiores.2004.03.025 [DOI] [PubMed] [Google Scholar]
- 68.Tiyaboonchai A, Cardenas-Diaz FL, Ying L, Maguire JA, Sim X, Jobaliya C, Gagne AL, Kishore S, Stanescu DE, Hughes N, et al. GATA6 plays an important role in the induction of human definitive endoderm, development of the pancreas, and functionality of pancreatic β cells. Stem Cell Reports. 2017;8:589–604. doi: 10.1016/j.stemcr.2016.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fujiwara T, O’Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Katsumura KR, Bresnick EH. The GATA factor revolution in hematology. Blood. 2017;129:2092–2102. doi: 10.1182/blood-2016-09-687871 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All source data, including sequencing reads and processed files have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE173162 (eCLIP-Seq), and GSE173163 (RNA-Seq).


