Abstract
Processing of pro-ςK in the mother cell compartment of sporulating Bacillus subtilis involves SpoIVFB and is governed by a signal from the forespore. SpoIVFB has an HEXXH motif characteristic of metalloproteases embedded in one of its transmembrane segments. Several conservative single amino acid changes in the HEXXH motif abolished function. However, changing the glutamic acid residue to aspartic acid, or changing the isoleucine residue that precedes the motif to proline, permitted SpoIVFB function. Only one other putative metalloprotease, site 2 protease has been shown to tolerate aspartic acid rather than glutamic acid in its HEXXH sequence. Site 2 protease and SpoIVFB share a second region of similarity with a family of putative membrane metalloproteases. A conservative change in this region of SpoIVFB abolished function. Interestingly, SpoIVFA increased the accumulation of certain mutant SpoIVFB proteins but was unnecessary for accumulation of wild-type SpoIVFB.
The sporulation process of Bacillus subtilis has proven to be an excellent system for discovering novel gene regulatory mechanisms and elucidating their role in temporal and spatial control of gene expression (14, 34). Sporulation involves the formation of an asymmetrically positioned septum that divides the developing cell into a larger mother cell compartment and a smaller forespore. Both cell types receive a copy of the genome, and differential gene expression, controlled primarily by the activation of new ς subunits of RNA polymerase (RNAP), drives a series of morphological changes. The mother cell side of the septum engages in a phagocytic-like process called engulfment, which results in the forespore being wholly surrounded by two membranes within the mother cell (Fig. 1). Completion of engulfment triggers activation of ςG in the forespore (12, 16). Transcription by ςG RNAP of the spoIVB gene initiates a signaling pathway that governs processing of pro-ςK to its active form in the mother cell (3, 22) (Fig. 1). ςK directs transcription of genes whose products drive further morphogenesis, including the formation of cortex and coat layers, which encapsulate the forespore, and lysis of the mother cell to release the mature spore (3, 9, 11, 13, 42–44).
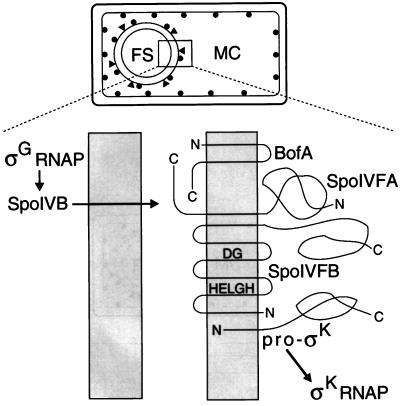
FIG. 1.
Model showing proteins involved in the signal transduction pathway that governs pro-ςK processing. The top part shows a sporangium in which engulfment of the forespore (FS) has been completed. Dots represent pro-ςK associated with the mother cell (MC) membrane and the outermost membrane surrounding the FS (39). Triangles represent SpoIVFA and SpoIVFB associated with the outermost membrane surrounding the FS (26). The bottom part shows an expanded view of the two membranes surrounding the FS. The topologies depicted for BofA, SpoIVFA, and SpoIVFB are based on analysis of lacZ and phoA fusions in E. coli (8, 35). The HELGH and DG sequences of SpoIVFB subjected to mutational analysis appear to be in transmembrane segments. The model depicts the hydrophobic prosequence of pro-ςK (13, 33) inserted into the outermost membrane surrounding the FS, but it is not known how pro-ςK associates with membranes (39). Cleavage of pro-ςK releases ςK into the MC, where it binds core RNAP and directs transcription.
This study was aimed at elucidating the mechanism of pro-ςK processing. Pro-ςK has 20 N-terminal amino acids that are removed by processing during sporulation (13). This processing step serves as a developmental checkpoint, coupling events in the forespore to ςK activation in the mother cell (4, 18). It delays the appearance of active ςK by about 1 h. If the checkpoint is circumvented and ςK accumulates earlier than normal, sporulation efficiency drops (4), perhaps owing to a ςK-dependent feedback loop that inhibits production of early mother cell transcription factors (40, 41). Genetic studies identified components involved in the checkpoint and led to the following model: SpoIVFB was proposed to be the protease that processes pro-ςK, or a positive regulator of such a protease (5), and SpoIVFA and BofA appeared to be negative regulators of processing until a signal from the forespore relieved this inhibition (5, 27, 28). (Fig. 1). SpoIVFB and SpoIVFA are membrane proteins that appear to be localized to the outermost membrane surrounding the forespore (8, 26). BofA is also a membrane protein (35), and recent results suggest that it stabilizes SpoIVFA, which may be the primary inhibitor of SpoIVFB (25). Consistent with the idea that SpoIVFB is the processing enzyme, it enhanced processing when coproduced with pro-ςK in growing B. subtilis or Escherichia coli, although the amount of processing was small (17). It was noted that SpoIVFB has a sequence (VLIHELGHAA) matching a motif found in zinc-dependent endopeptidases (17), but hydropathy analysis had suggested this sequence is embedded in a transmembrane domain (5) (Fig. 1), and no metalloproteases with an HEXXH motif in a membrane-spanning segment had been described.
Several findings motivated us to test the idea that SpoIVFB is a metalloprotease. First, Rawson et al. (24) described the human site 2 protease (S2P), which has an essential HEIGH sequence embedded within or near the surface of the endoplasmic reticulum (38) and is proposed to mediate intramembrane cleavage of sterol regulatory element-binding proteins (6), releasing these transcription factors from the endoplasmic reticulum and permitting regulation of genes involved in cholesterol and fatty acid metabolism (2). Second, pro-ςK was shown to be membrane associated (39) (Fig. 1). This suggested that pro-ςK, like sterol regulatory element-binding proteins, might be cleaved within a membrane or near its surface and that SpoIVFB might be functionally homologous to S2P (14). Third, a SpoIVFB-green fluorescent protein chimera appeared to accumulate to a higher level than native SpoIVFB when produced in growing B. subtilis, and more pro-ςK processing was observed, strengthening the hypothesis that SpoIVFB is the processing enzyme (27).
Here, we present mutational evidence that SpoIVFB is a metalloprotease. While this work was in progress, computer database searching and protein multiple sequence alignment identified SpoIVFB and S2P as members of a novel clan of putative zinc metallopeptidases (15, 29). Also, using a strategy different than the one we employed, Rudner et al. (29) analyzed the effect of mutations in spoIVFB. Our results confirm and extend those of Rudner et al. (29), providing additional insights into the requirements for SpoIVFB function.
Rescue of pro-ςK processing and sporulation by a plasmid bearing spoIVFB.
To develop a system for mutational analysis of spoIVFB, an autonomously replicating plasmid containing the spoIVFB gene fused to the isopropyl-β-d-thiogalactopyranoside-inducible spac promoter was constructed. A 1.0-kbp HindIII-SalI fragment containing the spoIVFB gene from pSC227 (19) was ligated to HindIII-SalI-digested pDG148 (32), resulting in pSL16. This plasmid was transformed (1) into B. subtilis OR745 (26), which lacks the chromosomal spoIVFB gene, and into B. subtilis BSL51 (19), which lacks both spoIVFA and spoIVFB. Transformants were selected on Luria-Bertani agar (30) containing kanamycin sulfate (5 μg/ml). Figure 2 shows that pSL16, but not the empty vector from which it was derived (pDG148), allowed production of ςK during sporulation. The level of ςK that accumulated was comparable in the strains with (lane 5) or without (lane 3) spoIVFA but significantly less than in wild-type PY79 (37) (lane 1). The level of ςK did not increase in pSL16-containing strains harvested 1 h later during sporulation (i.e., at T6 [data not shown]), suggesting that pro-ςK processing was not merely delayed relative to that in wild-type cells. Since it was known that very little ςK is needed for spore formation (19), we measured the sporulation efficiency of the pSL16-containing strains. Table 1 shows that OR745/pSL16 and BSL51/pSL16 formed heat-resistant spores nearly as efficiently as wild-type cells and several orders of magnitude more efficiently than negative control strains containing pDG148. These results demonstrate that pSL16 can partially rescue pro-ςK processing and sporulation of spoIVFB mutant cells in the presence (OR745) or absence (BSL51) of spoIVFA and that the sporulation assay in particular is a sensitive test for SpoIVFB function.
FIG. 2.
Rescue of pro-ςK processing by pSL16 and mutant derivatives. Extracts were prepared from cells (10) collected 5 h after the onset of sporulation in DSM medium (21). Proteins (10 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subjected to Western blot analysis with polyclonal anti-pro-ςK antibodies diluted 1:10,000 (39). Lane 1, PY79; lane 2, BSL51/pDG148; lane 3, BSL51/pSL16; lane 4, OR745/pDG148; lane 5, OR745/pSL16; lane 6, OR745/pSL16 I42P; lane 7, OR745/pSL16 E44D.
TABLE 1.
Sporulation rescue by plasmids
| Plasmid | Sporulation efficiencya
|
|
|---|---|---|
| OR745 | BSL51 | |
| pDG148 | 0.00087 | 0.0003 |
| pSL16 | 54 | 48 |
| pSL16 I42P | 77 | 0.00025 |
| pSL16 H43F | 0.00029 | <0.001 |
| pSL16 E44A | 0.0013 | 0.0051 |
| pSL16 E44D | 72 | 0.00017 |
| pSL16 E44Q | 0.015 | 0.00045 |
| pSL16 H47F | 0.00052 | <0.001 |
| pSL16 D137A | 0.00033 | 0.0018 |
| pSL16 D137E | 0.00033 | 0.0013 |
| pSL16 D137H | 0.014 | 0.00088 |
| pSL16 D137N | 0.00096 | 0.00016 |
Sporulation was induced at 37°C by nutrient exhaustion in DSM medium (21). IPTG (1 mM) was added at the onset of sporulation. Sporulation efficiency is expressed as a percentage of that observed for wild-type PY79 (typically 92%) and was determined by measuring the ratio of the number of colonies formed at 37°C on Luria-Bertani agar after heat treatment (80°C for 20 min) to the number before heat treatment (21). Similar results were observed for BSL51 derivatives when sporulation was induced by resuspension in SM medium (21).
Mutational analysis of the HEXXH motif.
To test the hypothesis that SpoIVFB is a metalloprotease (17), we made mutations in and around the HELGH sequence at positions 43 to 47 (Fig. 1). Plasmids with a mutation resulting in an I42P, E44A, E44D, or E44Q substitution in SpoIVFB were derived from pSL16 using the QuickChange site-directed mutagenesis kit (Stratagene); such plasmids are referred to herein as, e.g., pSL16 I42P. Plasmids with a mutation resulting in an H43F or H47F substitution were created using a sequence overlap extension strategy to synthesize full-length spoIVFB with the desired mutation, followed by subcloning into HindIII-SalI-digested pDG148 to construct plasmids otherwise identical to pSL16. Details of the sequence overlap extension procedure and primer sequences can be found at our web site (http: //www.bch.msu.edu/faculty/kroos.htm). Each mutant spoIVFB allele was completely sequenced to ensure that no additional mutations had been introduced.
In zinc metallopeptidases, the two histidine residues of the HEXXH motif coordinate zinc, and the glutamic acid residue promotes nucleophilic attack by a water molecule on the carbonyl atom of the substrate peptide bond (23). If SpoIVFB is a metalloprotease, then changing the histidine residues to phenylalanine was predicted to prevent metal binding and cause loss of function. Changing the putative catalytic glutamic acid residue to glutamine or alanine was also expected to cause loss of function. Table 1 shows that these predictions were met. The H43F, E44A, E44Q, and H47F changes each abolished the ability of pSL16 to restore sporulation of OR745 or BSL51.
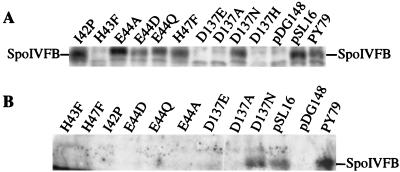
To determine whether the mutant proteins accumulate, we performed Western blot analysis with antibodies against SpoIVFB. Figure 3A shows that the mutant SpoIVFB proteins, with the exception of the H43F mutant, accumulated during sporulation of OR745 to a level comparable to that of wild-type SpoIVFB produced from pSL16 or from the chromosome (PY79); however, in BSL51 (lacking spoIVFA) the mutant SpoIVFB proteins did not accumulate to a detectable level, whereas wild-type SpoIVFB was detected (Fig. 3B). OR745 containing pSL16 with any of the mutant spoIVFB alleles failed to process pro-ςK (data not shown). These results demonstrate the critical nature of the HEXXH motif for SpoIVFB function and strongly support the idea that SpoIVFB is a metalloprotease, in agreement with the findings of Rudner et al. (29). The absence of detectable mutant SpoIVFB proteins in BSL51 indicates that SpoIVFA is more critical for accumulation of the mutant proteins than it is for the wild-type protein.
FIG. 3.
Detection of SpoIVFB produced from pSL16 and mutant derivatives. Extracts were prepared from cells (10) collected 3 h after the onset of sporulation. Proteins (10 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subjected to Western blot analysis (39) with affinity-purified anti-SpoIVFB antibodies diluted 1:500 (26). The generation of anti-SpoIVFB antiserum and affinity purification of antibodies is described at our web site (http://www.bch.msu.edu/faculty/kroos.htm). (A) Wild-type PY79 and spoIVFB mutant OR745 containing pDG148, pSL16, or pSL16 encoding the indicated amino acid substitution. Sporulation was done in DSM medium (21). (B) Wild-type PY79 and spoIVFAB mutant BSL51 containing pDG148, pSL16, or pSL16 encoding the indicated amino acid substitution. Sporulation was done in SM medium (21).
We tested the effect of changing glutamic acid at position 44 in the HEXXH motif of SpoIVFB to aspartic acid. The analogous change in three cytoplasmic metalloproteases abolished function (7, 20, 36), but the putative membrane metalloprotease S2P tolerated this substitution and retained partial activity (24). Table 1 shows that pSL16 E44D rescued sporulation of OR745. Figure 2 (lane 7) shows that pSL16 E44D partially restored pro-ςK processing during sporulation of OR745. These results demonstrate that SpoIVFB, like S2P but unlike cytoplasmic metalloproteases, tolerates the E-to-D change in its HEXXH sequence, supporting the idea that SpoIVFB and S2P belong to a novel class of metalloproteases (14, 15, 29). Perhaps these proteins have greater conformational flexibility in the region of the HEXXH motif owing to its presence in a membrane (8, 38). During catalysis, the acidic amino acid residue must be accessible to a water molecule if the mechanism of peptide bond cleavage by SpoIVFB and S2P is like that of well-studied metalloproteases (23). Interestingly, pSL16 E44D did not restore heat-resistant spore formation to BSL51 when sporulation was induced in liquid (Table 1), and the mutant protein was undetectable (Fig. 3B), whereas it accumulated in OR745/pSL16 E44D (Fig. 3A). Thus, SpoIVFA is more important for accumulation of SpoIVFB E44D than it is for accumulation of wild-type SpoIVFB. However, BSL51/pSL16 E44D colonies turned faint brown on Difco sporulation medium (DSM) agar, whereas BSL51 colonies remained cream colored. Brown pigmentation is caused by expression of cotA (31), a ςK-dependent gene (43), suggesting that SpoIVFB E44D can function weakly under these conditions. Microscopic examination suggested the presence of more spores in colonies of BSL51 with pSL16 E44D than without the plasmid, and this was confirmed by sporulation assays of samples from the colonies, with BSL51/pSL16 E44D and BSL51 showing about 10 and 10−4% sporulation efficiency, respectively, after 2 days at 37°C.
Changing isoleucine at position 42 to proline had an effect very similar to that of the E44D substitution. pSL16 I42P rescued sporulation (Table 1) and partially restored pro-ςK processing (Fig. 2, lane 6) of OR745. These results are novel. In metalloproteases of known structure, the HEXXH motif is embedded in an α-helix (23). Proline is disruptive of α-helical structure and is not found at the position preceding the HEXXH motif in known metalloproteases (23). The ability to tolerate proline in this position may be a unique feature of metalloproteases that have the HEXXH motif in a transmembrane segment. Proline may be less disruptive of α-helical structure in the hydrophobic environment of the lipid bilayer, and in SpoIVFB the sequence preceding and including the HEXXH motif (LLCLLLIVLIHELGH) (5) strongly favors an α-helical structure, which the proline substitution may perturb only slightly. It would be very interesting to substitute proline for valine at the position preceding the HEXXH motif in S2P (24) to see whether the analogy with SpoIVFB can be extended and whether the ability to function with proline at this position might be a general feature of this family of putative membrane metalloproteases (15, 29). Like SpoIVFB E44D, SpoIVFB I42P failed to accumulate to a detectable level in BSL51/pSL16 I42P (Fig. 3B), and this strain failed to sporulate in liquid (Table 1), whereas the mutant protein accumulated to the wild-type level in OR745/pSL16 I42P (Fig. 3A). Also, BSL51/pSL16 I42P exhibited an oligosporogenous phenotype similar to that of BSL51/pSL16 E44D on DSM agar, indicative of the weak but significant function of SpoIVFB I42P in the absence of SpoIVFA.
Mutational analysis of a second conserved region of SpoIVFB.
A BLAST search with the SpoIVFB sequence identified significant similarity to several hypothetical proteins of unknown function that had been discovered as a result of archaea genome sequencing projects (data not shown). These sequences contained an HEXXH motif and a second conserved region containing the sequence DG. To test the importance of this sequence in SpoIVFB, we made mutations that changed the aspartic acid residue at position 137 (Fig. 1). While this work was in progress, Lewis and Thomas (15) published the results of iterative BLAST searches with the S2P sequence, which identified a novel clan of putative zinc metalloproteases, including SpoIVFB. They called the conserved region containing the DG sequence region C and identified a consensus sequence. Computer analysis by Rudner et al. (29) also identified this region, which they called the NPDG motif, and they performed mutational analysis of this region. We constructed plasmids with a mutation resulting in a D137H or D137N substitution in SpoIVFB using the QuickChange site-directed mutagenesis kit (Stratagene) with pSL16 as the template. Plasmids with a mutation resulting in a D137A or D137E substitution were created using a sequence overlap extension strategy as described above. Details of the sequence overlap extension procedure and primer sequences can be found at our web site (http: //www.bch.msu.edu/faculty/kroos.htm). Each mutant spoIVFB allele was completely sequenced to ensure that no additional mutations had been introduced.
Replacement of the aspartic acid at position 137 of SpoIVFB with alanine, glutamic acid, histidine, or asparagine abolished sporulation rescue of OR745 or BSL51 (Table 1). OR745 containing pSL16 with any of these mutant spoIVFB alleles failed to process pro-ςK (data not shown). Of these mutant proteins, only SpoIVFB D137N accumulated to a detectable level during sporulation, and unlike all the other mutant proteins we tested, SpoIVFB D137N accumulated to about the same level as wild-type SpoIVFB in the presence or absence of SpoIVFA (Fig. 3). We conclude, in agreement with the findings of Rudner et al. (29), that a conservative change of the aspartic acid at position 137 of SpoIVFB to asparagine abolishes function. The same change in the corresponding region of S2P has been shown to abolish function (38). Since the DG sequence is conserved in all putative metalloproteases belonging to the family that includes SpoIVFB and S2P (15, 29), it is likely to be important for the function of all members of the clan. It has been speculated that the aspartic acid residue is a zinc ligand (15, 29, 38), since metalloproteases typically coordinate zinc with the two histidine residues of the HEXXH motif and a third ligand (23).
Role of SpoIVFA in accumulation of SpoIVFB.
SpoIVFA has been proposed to stabilize thermolabile SpoIVFB during sporulation, based on the finding that an in-frame deletion in spoIVFA greatly reduces sporulation at 37°C but not at 30°C (5, 8). We observed no significant difference in the sporulation efficiency at 37°C of OR745 (expressing spoIVFA from the chromosome) and BSL51 (lacking spoIVFA) containing pSL16 (Table 1), and the sporulation efficiency of BSL51/pSL16 did not increase at 30°C (data not shown). The altered timing and/or the increased level of SpoIVFB production from pSL16 appears to make SpoIVFA dispensable for sporulation at 37°C. However, production of SpoIVFB from pSL16 was not optimal for rescue of pro-ςK processing (Fig. 2). Perhaps spoIVFB must be expressed in cis with spoIVFA and/or the two proteins must be produced at a certain stoichiometric ratio to function optimally. Resnekov (25) recently reported that spoIVFA must be expressed in cis with spoIVFB-gfp in order to allow a SpoIVFB-green fluorescent protein chimera to accumulate more abundantly in B. subtilis engineered to produce these proteins during growth (27).
Although expression in cis may be required for optimal function, our results clearly show that SpoIVFA produced in trans (from the chromosome of OR745) caused increased accumulation during sporulation of certain mutant SpoIVFB proteins produced from plasmids (Fig. 3). On the other hand, wild-type SpoIVFB and one mutant protein (SpoIVFB D137N) accumulated in the absence of SpoIVFA (Fig. 3B). In E. coli, SpoIVFB adopts a six-transmembrane-segment configuration (Fig. 1) when SpoIVFA is also produced, but when SpoIVFA is absent, the third and fourth transmembrane segments appear to be exposed in the cytoplasm (8). Perhaps wild-type SpoIVFB and SpoIVFB D137N produced in B. subtilis BSL51 achieve the six-transmembrane-segment configuration despite the absence of SpoIVFA and are therefore more resistant to proteolysis, allowing these proteins to accumulate to a detectable level (Fig. 3B), whereas certain mutant SpoIVFB proteins require SpoIVFA to achieve the six-transmembrane-segment configuration and accumulate detectably (Fig. 3A).
Acknowledgments
We thank S. Lu for constructing pSL16 and pSL28a. We thank D. Green, S. Cutting, D. Rudner, P. Fawcett, R. Losick, and O. Resnekov for communicating results prior to publication. We are grateful to D. Rudner for encouraging us to use OR745 and for sending the strain, and we are grateful to O. Resnekov for the SpoIVFB antibody affinity purification protocol.
This research was supported by the Michigan Agricultural Experiment Station and by grant GM43585 from the National Institutes of Health.
REFERENCES
- 1.Bron S. Plasmids. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. New York, N.Y: John Wiley & Sons; 1990. pp. 75–174. [Google Scholar]
- 2.Brown M S, Goldstein J L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 3.Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. Forespore-specific transcription of a gene in the signal transduction pathway that governs pro-ςK processing in Bacillus subtilis. Genes Dev. 1991;5:456–466. doi: 10.1101/gad.5.3.456. [DOI] [PubMed] [Google Scholar]
- 4.Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. A forespore checkpoint for mother-cell gene expression during development in Bacillus subtilis. Cell. 1990;62:239–250. doi: 10.1016/0092-8674(90)90362-i. [DOI] [PubMed] [Google Scholar]
- 5.Cutting S, Roels S, Losick R. Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis. J Mol Biol. 1991;221:1237–1256. doi: 10.1016/0022-2836(91)90931-u. [DOI] [PubMed] [Google Scholar]
- 6.Duncan E, Dave U, Sakai J, Goldstein J, Brown M. Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junction as determined by cysteine panning. J Biol Chem. 1998;273:17801–17809. doi: 10.1074/jbc.273.28.17801. [DOI] [PubMed] [Google Scholar]
- 7.Fujimura-Kamada K, Nouvet F J, Michaelis S. A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-terminal processing of the yeast α-factor precursor. J Cell Biol. 1997;136:271–285. doi: 10.1083/jcb.136.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green D H, Cutting S M. Membrane topology of the Bacillus subtilis pro-ςK processing complex. J Bacteriol. 2000;182:278–285. doi: 10.1128/jb.182.2.278-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halberg R, Kroos L. Sporulation regulatory protein SpoIIID from Bacillus subtilis activates and represses transcription by both mother-cell-specific forms of RNA polymerase. J Mol Biol. 1994;243:425–436. doi: 10.1006/jmbi.1994.1670. [DOI] [PubMed] [Google Scholar]
- 10.Healy J, Weir J, Smith I, Losick R. Post-transcriptional control of a sporulation regulatory gene encoding transcription factor ςH in Bacillus subtilis. Mol Microbiol. 1991;5:477–487. doi: 10.1111/j.1365-2958.1991.tb02131.x. [DOI] [PubMed] [Google Scholar]
- 11.Ichikawa H, Halberg R, Kroos L. Negative regulation by the Bacillus subtilis GerE protein. J Biol Chem. 1999;274:8322–8327. doi: 10.1074/jbc.274.12.8322. [DOI] [PubMed] [Google Scholar]
- 12.Kellner E M, Decatur A, Moran C P. Two-stage regulation of an anti-sigma factor determines developmental fate during bacterial endospore formation. Mol Microbiol. 1996;21:913–924. doi: 10.1046/j.1365-2958.1996.461408.x. [DOI] [PubMed] [Google Scholar]
- 13.Kroos L, Kunkel B, Losick R. Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science. 1989;243:526–529. doi: 10.1126/science.2492118. [DOI] [PubMed] [Google Scholar]
- 14.Kroos L, Zhang B, Ichikawa H, Yu Y-T N. Control of ς factor activity during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1285–1294. doi: 10.1046/j.1365-2958.1999.01214.x. [DOI] [PubMed] [Google Scholar]
- 15.Lewis A, Thomas P. A novel clan of zinc metallopeptidases with possible intramembrane cleavage properties. Protein Sci. 1999;8:439–442. doi: 10.1110/ps.8.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis P J, Magnin T, Errington J. Compartmentalized distribution of the proteins controlling the prespore-specific transcription factor ςF of Bacillus subtilis. Genes Cells. 1996;1:881–894. doi: 10.1046/j.1365-2443.1996.750275.x. [DOI] [PubMed] [Google Scholar]
- 17.Lu S, Cutting S, Kroos L. Sporulation protein SpoIVFB from Bacillus subtilis enhances processing of the sigma factor precursor pro-ςK in the absence of other sporulation gene products. J Bacteriol. 1995;177:1082–1085. doi: 10.1128/jb.177.4.1082-1085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu S, Halberg R, Kroos L. Processing of the mother-cell ς factor, ςK, may depend on events occurring in the forespore during Bacillus subtilis development. Proc Natl Acad Sci USA. 1990;87:9722–9726. doi: 10.1073/pnas.87.24.9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu S, Kroos L. Overproducing the Bacillus subtilis mother cell sigma factor precursor, pro-ςK, uncouples ςK-dependent gene expression from dependence on intercompartmental communication. J Bacteriol. 1994;176:3936–3943. doi: 10.1128/jb.176.13.3936-3943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGwire B S, Chang K P. Posttranslational regulation of a Leishmania HEXXH metalloprotease (gp63). The effects of site-specific mutagenesis of catalytic, zinc binding, N-glycosylation, and glycosyl phosphatidylinositol addition sites on N-terminal end cleavage, intracellular stability, and extracellular exit. J Biol Chem. 1996;271:7903–7909. doi: 10.1074/jbc.271.14.7903. [DOI] [PubMed] [Google Scholar]
- 21.Nicholson W L, Setlow P. Sporulation, germination, and outgrowth. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. New York, N.Y: John Wiley & Sons; 1990. pp. 391–450. [Google Scholar]
- 22.Oke V, Shchepetov M, Cutting S. SpoIVB has two distinct functions during spore formation in Bacillus subtilis. Mol Microbiol. 1997;23:223–230. doi: 10.1046/j.1365-2958.1997.2091573.x. [DOI] [PubMed] [Google Scholar]
- 23.Rawlings N, Barrett A. Evolutionary families of metallopeptidases. Methods Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 24.Rawson R, Zelenski N, Nijhawan D, Ye J, Sakai J, Hasan M, Chang T, Brown M, Goldstein J. Complementation cloning of SP2, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 25.Resnekov O. Role of the sporulation protein BofA in regulating activation of the Bacillus subtilis developmental transcription factor ςK. J Bacteriol. 1999;181:5384–5388. doi: 10.1128/jb.181.17.5384-5388.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Resnekov O, Alper S, Losick R. Subcellular localization of proteins governing the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Genes Cells. 1996;1:529–542. doi: 10.1046/j.1365-2443.1996.d01-262.x. [DOI] [PubMed] [Google Scholar]
- 27.Resnekov O, Losick R. Negative regulation of the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Proc Natl Acad Sci USA. 1998;95:3162–3167. doi: 10.1073/pnas.95.6.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricca E, Cutting S, Losick R. Characterization of bofA, a gene involved in intercompartmental regulation of pro-ςK processing during sporulation in Bacillus subtilis. J Bacteriol. 1992;174:3177–3184. doi: 10.1128/jb.174.10.3177-3184.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudner D, Fawcett P, Losick R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci USA. 1999;96:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sandman K, Kroos L, Cutting S, Youngman P, Losick R. Identification of the promoter for a spore coat protein gene in Bacillus subtilis and studies on the regulation of its induction at a late stage of sporulation. J Mol Biol. 1988;200:461–473. doi: 10.1016/0022-2836(88)90536-0. [DOI] [PubMed] [Google Scholar]
- 32.Stragier P, Bonamy C, Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- 33.Stragier P, Kunkel B, Kroos L, Losick R. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science. 1989;243:507–512. doi: 10.1126/science.2536191. [DOI] [PubMed] [Google Scholar]
- 34.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 35.Varcamonti M, Marasco R, De Felice M, Sacco M. Membrane topology analysis of the Bacillus subtilis BofA protein involved in pro-ςK processing. Microbiology. 1997;143:1053–1058. doi: 10.1099/00221287-143-4-1053. [DOI] [PubMed] [Google Scholar]
- 36.Vazeux G, Wang J, Corvol P, Llorens-Cortes C. Identification of glutamate residues essential for catalytic activity and zinc coordination in aminopeptidase A. J Biol Chem. 1996;271:9069–9074. doi: 10.1074/jbc.271.15.9069. [DOI] [PubMed] [Google Scholar]
- 37.Youngman P, Perkins J B, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- 38.Zelenski N, Rawson R, Brown M, Goldstein J. Membrane topology of S2P, a protein required for intramembranous cleavage of sterol regulatory element-binding proteins. J Biol Chem. 1999;274:21973–21980. doi: 10.1074/jbc.274.31.21973. [DOI] [PubMed] [Google Scholar]
- 39.Zhang B, Hofmeister A, Kroos L. The prosequence of pro-ςK promotes membrane association and inhibits RNA polymerase core binding. J Bacteriol. 1998;180:2434–2441. doi: 10.1128/jb.180.9.2434-2441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang B, Kroos L. A feedback loop regulates the switch from one sigma factor to the next in the cascade controlling Bacillus subtilis mother cell gene expression. J Bacteriol. 1997;179:6138–6144. doi: 10.1128/jb.179.19.6138-6144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang B, Struffi P, Kroos L. ςK can negatively regulate sigE expression by two different mechanisms during sporulation of Bacillus subtilis. J Bacteriol. 1999;181:4081–4088. doi: 10.1128/jb.181.13.4081-4088.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Ichikawa H, Halberg R, Kroos L, Aronson A I. Regulation of the transcription of a cluster of Bacillus subtilis spore coat genes. J Mol Biol. 1994;240:405–415. doi: 10.1006/jmbi.1994.1456. [DOI] [PubMed] [Google Scholar]
- 43.Zheng L, Halberg R, Roels S, Ichikawa H, Kroos L, Losick R. Sporulation regulatory protein GerE from Bacillus subtilis binds to and can activate or repress transcription from promoters for mother-cell-specific genes. J Mol Biol. 1992;226:1037–1050. doi: 10.1016/0022-2836(92)91051-p. [DOI] [PubMed] [Google Scholar]
- 44.Zheng L, Losick R. Cascade regulation of spore coat gene expression in Bacillus subtilis. J Mol Biol. 1990;212:645–660. doi: 10.1016/0022-2836(90)90227-d. [DOI] [PubMed] [Google Scholar]