Abstract
Panicum miliaceum L. was domesticated in northern China at least 7000 years ago and was subsequentially adopted in many areas throughout Eurasia. One such locale is Areni-1 an archaeological cave site in Southern Armenia, where vast quantities archaeobotanical material were well preserved via desiccation. The rich botanical material found at Areni-1 includes P. miliaceum grains that were identified morphologically and14C dated to the medieval period (873 ± 36 CE and 1118 ± 35 CE). To investigate the demographic and evolutionary history of the Areni-1 millet, we used ancient DNA extraction, hybridization capture enrichment, and high throughput sequencing to assemble three chloroplast genomes from the medieval grains and then compared these sequences to 50 modern P. miliaceum chloroplast genomes. Overall, the chloroplast genomes contained a low amount of diversity with domesticated accessions separated by a maximum of 5 SNPs and little inference on demography could be made. However, in phylogenies the chloroplast genomes separated into two clades, similar to what has been reported for nuclear DNA from P. miliaceum. The chloroplast genomes of two wild (undomesticated) accessions of P. miliaceum contained a relatively large number of variants, 11 SNPs, not found in the domesticated accessions. These results demonstrate that P. miliaceum grains from archaeological sites can preserve DNA for at least 1000 years and serve as a genetic resource to study the domestication of this cereal crop.
Subject terms: Plant genetics, Archaeology
Introduction
Millet is a generic term that refers to a group of small seeded grasses that were important crops in the past and remain a significant source of food and fodder in many areas of the world today1,2. The majority of cultivated millets belong to the Paniceae tribe of the Poaceae family and include Panicum miliaceum L., Cenchrus americanus (L.) R.Br., and Setaria italica (L.) P. Beuvois, while Eleusine coracana Gaertn. belongs to the Chlorideae clade of Poaceae3. Millet, along with other C4 pathway species such as Zea mays L., Saccharum officinarum L. and Sorghum bicolor (L.) Moench, are characterised by their ability to be cultivated in a wide range of environments, requiring modest amounts of water, and having short growing seasons (60–90 days).
Two millets commonly grown throughout much of Eurasia in antiquity were P. miliaceum and S. italica. Both of these millets were domesticated in Northern China with archaeological evidence indicating that P. miliaceum was being farmed as early as 5880 BCE4–8, while S. italica was first domesticated in the same region at approximately 7000–4000 BCE9. From domestication centres in Northern China, the cultivation of P. miliaceum and S. italica spread to much of Eurasia where these cereals became important food sources2. The timing and mechanism by which the cultivation of P. miliaceum and S. italica moved towards Europe is not well understood but was likely driven by mobile pastoralists who migrated along the mountain foothill ecotone of Central Asia, a path that later became one of the Silk Road trade routes2,10.
Areni-1 is a three-chambered karstic cave (also known as Birds’ Cave: 39° 43′ 53″ N, 45° 12′ 13″ E), located on the left bank of the Arpa River basin, a tributary of the river Araxes, within the eastern portion of the modern village of Areni in southern Armenia. Excavations at Areni-1 have uncovered a long history of occupations spanning approximately 6000 years. Plant remains recovered from Areni-1 include vast quantities of well-preserved desiccated and charred seeds, fruits, stones (endocarps), and stems of both wild and cultivated plants11. Among the cereals recovered were grains that were identified morphometrically as P. miliaceum11,12.
The Armenian Highlands, which includes Areni-1, and the Caucasus in general are interesting locations where wild relatives of many cereals are widely represented. The proximity to both the Fertile Crescent a region important for the development of agriculture13 and to Silk Road exchange routes14 underlies the importance of these locations to processes of domestication, adaptation, and timing of distribution events. Because of this unique relationship of the Armenian Highlands and changes in plant use, the millet grains from Areni-1 provide important information relating to the demographic and evolutionary history of P. miliaceum. The excellent preservation of the Areni-1 grains suggested that these samples would contain high levels of DNA and genetic studies of these grains were initiated using the chloroplast genome as the marker15.
Chloroplasts are semi-autonomous organelles that provide plants with energy through photosynthesis and contain an independent circular genome. Chloroplast genomes are typically present as double stranded DNA in the range of 107–218 kb that includes two inverted repeat (IR) regions that separate a large and a small single-copy regions16. It is widely accepted that the chloroplast evolved from a photosynthetic prokaryote that assimilated with a eukaryotic host more than a billion years ago17,18. Owing to various characteristics, chloroplast genes and genomes are extensively used in studies of species identification, evolutionary analyses, and phylogenetics. These attributes include primarily maternal inheritance and thus nonrecombinant19, a moderate mutation rate that lies between the mitochondrial and nuclear genomes20, and a conserved gene structure and content21,22. A particular benefit for ancient DNA (aDNA) studies, is that chloroplast genomes are present as multiple copies in cells which increases the likelihood that chloroplast DNA (cpDNA) will survive in archaeobotanical material.
Using ancient aDNA extraction techniques, hybridization capture enrichment23, and high throughput sequencing, we generated three chloroplast genomes from five Areni-1 grains assayed. First, the Areni-1 chloroplast genomes were compared to 11 members of the Paniceae tribe of grasses to verify the morphological identification of the Areni-1 grains and to validate our laboratory and analytical methods. Next, the Areni-1 chloroplasts were compared to similar genomes from 50 modern accessions of P. miliaceum. These comparisons to modern accessions sought to provided information on the route cultivated P. miliaceum travelled from domestication centres is Northern China eastward and to determine the extent to which the Areni-1 P. miliaceum had been taken through the domestication process.
Results
Areni-1 millet
The Areni-1 cave located in southern Armenia produced a vast array botanical material from occupations dating to 4000 BCE (Fig. 1 and Table 1). Exceptionally well preserved, intact Panicum miliaceum L. florets were identified from both Trench 1 and 3 based on their morphology (Table 2 and Fig. S1; methods described more fully below). Florets are the basic threshing unit of P. miliaceum, exhibiting an indurate, smooth, and shiny palea and lemma tightly enclosing an inner caryopsis, with a pointed apex and relatively blunt proximal end (Fig. 2). The vast majority of ancient millet remains across southwest Asia tend to have been preserved via charring, which frequently damages the palea and lemma, leaving only the caryopsis/seed24; the desiccated remains from Areni-1 differ in that the colour and husk (palea and lemma) were maintained (Fig. 2). The lack of charring undoubtedly enhanced the preservation of DNA. The P. miliaceum florets from Areni-1 measure roughly 3.3 × 2.2 mm. These dimensions are slightly larger than charred caryopses reported from Mongolia, Afghanistan, Turkey, Iran, and elsewhere25–27, but this not surprising given that the medieval remains from Areni-1 represent a long history of selection post-dating earlier domestication events and were preserved via desiccation with the palea and lemma of the floret intact.
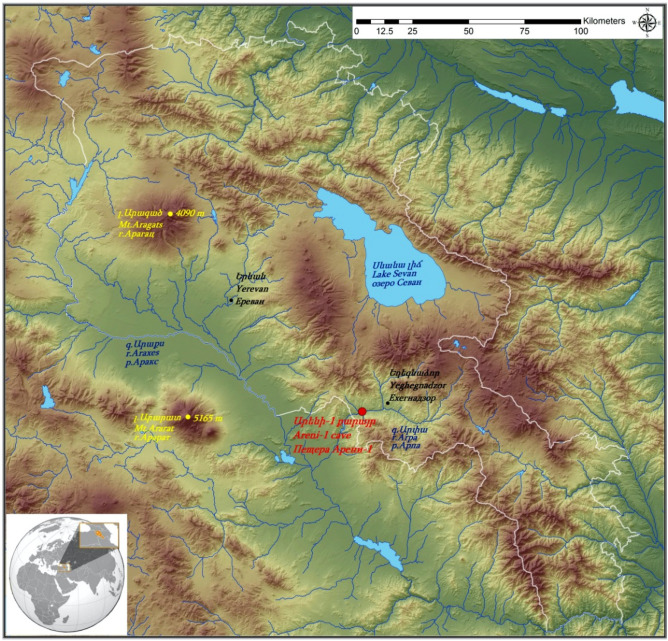
Figure 1.
Location of the Areni-1 cave. Small insert at bottom of figure shows location of Armenia within Eurasia. In the large insert, Armenia is shown with the white outline and the location of Areni-1 is indicated in red.
Table 1.
Relevant periods and dates associated with the occupations of the Aren-1 cave complex.
| Ages/periods | Inner division | Chronologies, ca. |
|---|---|---|
| Chalcolithic | Early/Middle | 5200–4300 BCE |
| Late | 4300–3500 BCE | |
| Bronze | Early | 3500–2400 BCE |
| Middle | 2400–1500 BCE | |
| Late | 1500–1200 BCE | |
| Iron | Early | 1200–900 BCE |
| Middle | 900–700 BCE | |
| Late | 700–600 BCE | |
| Classical | Early | 600–200 BCE |
| Middle | 200–1 BCE | |
| Late | 1–450 CE | |
| Medieval | Early | 450–900 CE |
| High | 900–1400 CE | |
| Late | 1400–1700 CE |
Periods with occupations at the Areni-1cave are given in bold.
Table 2.
Archaeological context and associated radiocarbon dates of broomcorn millet grains.
| Sample | Location in Areni-1 Cave | Dating laboratory number | Material dated | Date | Calibrated date 1 sigma (68.2%) |
Calibrated date 2 sigma (95.4%) |
|---|---|---|---|---|---|---|
| 11294a 11294b | Trench 1, Unit 3, Locus 50 (10.01.2009) |
AA-107539 X30053 |
Vitis sp. pedicel | 873 ± 36 BP | 1054–1218 CE | 1042–1248 CE |
|
11292a 11293a 11295a |
Trench 3, Sq O29/O30 Locus 2, Spit 5 |
AA-104139 X27645 |
Panicum miliaceum (pool of 3 grains) | 1118 ± 35 BP | 890–980 CE | 780–1020 CE |
Figure 2.

Areni-1 and modern millet grains. Paired photographs of 4 different grains in dorsal (left) and ventral (right) views depicting (A and B) representative archaeological Panicum miliaceum florets from Areni-1; (C) modern comparative P. miliaceum subsp. miliaceum floret (USDA PI 170588, Turkey); and (D) modern comparative Setaria italica subsp. italica floret (USDA PI 173103, Turkey). NPGS: U.S. National Plant Germplasm System (https://www.ars-grin.gov/npgs/index.html)
To confirm the contextual dating of the Areni-1 P. miliaceum,14C dating was performed on botanical material associated with the grains. The size of the Arerni-1 P. miliaceum did not provide sufficient mass to perform14C dating and extraction of aDNA from a single seed, so14C dating needed to be conducted indirectly using associated botanical material. The14C dating determined the Areni-1 grains were ≈ 1000-years-old and belonged to the Early and High medieval periods (Tables 1 and 2).
Mapping Areni-1 shotgun data to chloroplast reference genome
In the shotgun libraries, 11292a and 11295a were the only two grains that yielded a significant level of endogenous DNA and produced a large number of unique reads mapping to the chloroplast reference (> 90,000). The remaining shotgun libraries produced more than a thousand-fold fewer unique chloroplast mapped reads (Table 3). The extraction blanks and certain grains (11293a and 11294b) contained a comparatively small number of unique mapped reads in both the shotgun and cpDNA enriched libraries, suggesting that contamination from laboratory sources or between libraries during the processing of the Areni-1 samples was not a substantial issue. In the shotgun libraries, grains 11293a and 11294b both produced fewer mapped reads than the extraction blanks, but these results are likely very stochastic as these libraries contained a very small fraction of mappable reads.
Table 3.
Mapping statistics of shotgun and cpDNA enriched libraries.
| Sample | # Collapsed reads | # Mapped reads | # Unique mapped reads | % Reference covered by least 1 read (sans IRs) |
|---|---|---|---|---|
| Shotgun libraries | ||||
|
19885 (extraction blank) |
6702745 | 22 | 14 | |
| 11292a | 31367680 | 155496 | 92560 | |
| 11293a | 150049 | 2 | 2 | |
| 11294b | 6404935 | 0 | 0 | |
|
18335 (extraction blank) |
6493567 | 15 | 9 | |
| 11294a | 4635555 | 72 | 72 | |
|
18324 (extraction blank) |
6771883 | 7 | 7 | |
| 11295a | 42796618 | 444954 | 137838 | |
| cpDNA enriched libraries | ||||
|
19885 (extraction blank) |
9896218 | 1451964 | 67 | |
| 11292a | 36092254 | 21953919 | 163494 | 99.77 |
| 11293a | 35345608 | 88165 | 232 | |
| 11294b | 32245568 | 269895 | 1532 | |
|
18335 (extraction blank) |
77604223 | 270461 | 36 | |
| 11294a | 26076965 | 7472857 | 19420 | 91.28 |
|
18324 (extraction blank) |
42353298 | 128226 | 32 | |
| 11295a | 41491510 | 19512189 | 171867 | 99.95 |
Mapping statistics of libraries made from Areni-1 millet grains and mapped to the Panicum miliaceum chloroplast reference genome (Genbank#: KU343177.1). Inverted repeat regions (IRs) are two nearly identical loci in the chloroplast genome where the short fragmented aDNA sequencing data could not accurately map (Fig. 2) and were excluded from any analysis. The IRs loci excluded represent bp: 81851 to 104489 and 117101 to 139826 of the reference, which is 139,826 bp in length.
Mapping of Areni-1 enriched cpDNA data to chloroplast reference genome
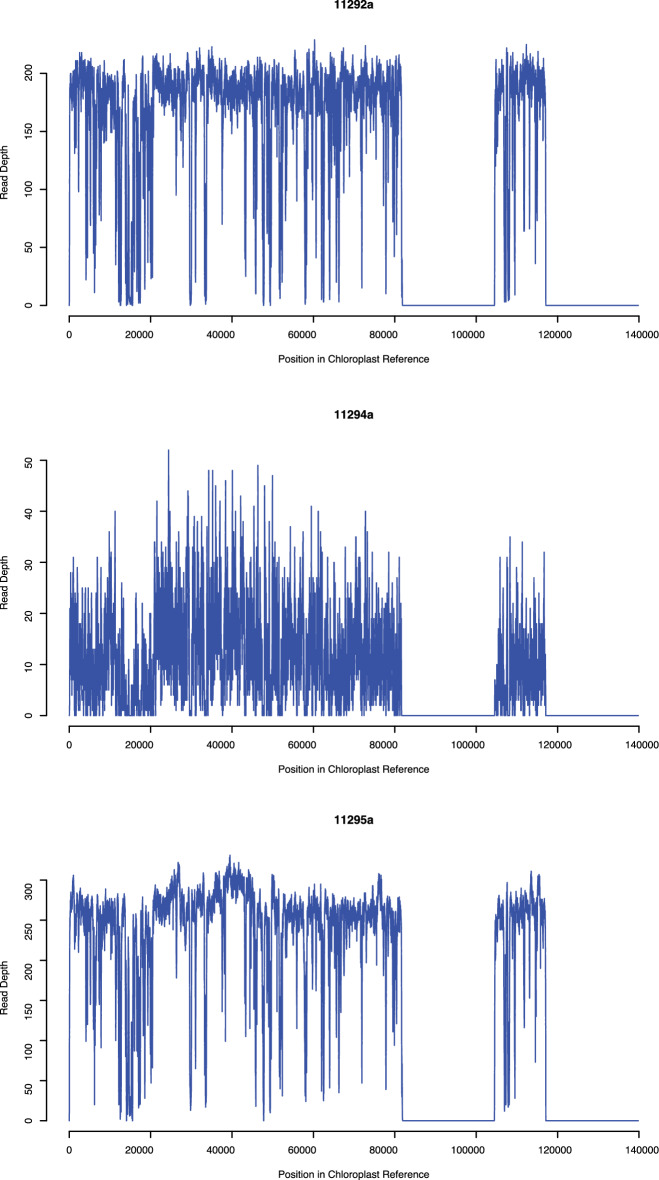
All ancient shotgun libraries were taken through two rounds of cpDNA hybridization capture regardless of endogenous DNA content in order to treat all samples in a similar manner and maximize the recovery of target sequences. Of the Areni-1 P. miliaceum enriched for cpDNA, only three grains (11292a, 11294a, and 11295a) produced libraries that contained a sufficient number of reads that mapped to the chloroplast reference genome (> 19,000) that allowed reconstruction of the Areni-1 chloroplast genomes (Table 3). No reads from the ancient libraries could be mapped to the IRs of the chloroplast genome, because the short-fragmented sequences in this data could not be accurately placed in these nearly identical loci (Fig. 3). As the IRs do not contain any mapped reads, these loci were removed from all further analysis. Libraries 11292a and 11295a covered > 99.7% of the reference (sans IRs) with at least one read, whilst the fraction of the reference covered by at least one read in library 11294a was approximately 8% less than the other two libraries.
Figure 3.
Read depth of mapped cpDNA. Read depth of cpDNA enriched libraries from the three Areni-1 millet grains mapped to the chloroplast reference genome: Genbank# KU343177.1. The IRs are shown in the large tracks of the graph which lack mapped reads.
Read length distribution and mapDamage analysis of enriched cpDNA libraries
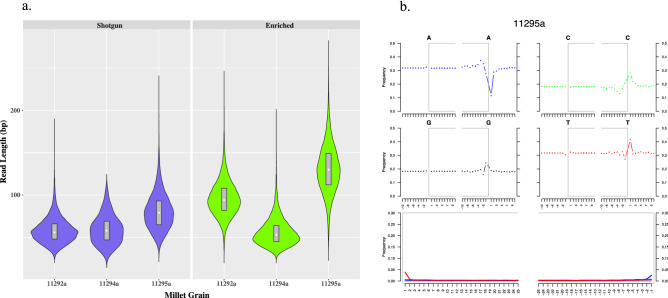
To support the ≈1000-year-old14C dating of the Areni-1 grains, sequences from the 11292a, 11294a, and 11295a cpDNA enriched libraries were examined for characteristics of authentic aDNA: short read lengths and the presence of deamination miscoding lesions28,29. All libraries from the three grains contained reads that were fragmented to a short length, generally < 150 bp (Fig. 4a). Grain 11294a produced the shortest reads, suggesting that this sample was the most degraded of the three. In the cpDNA enriched libraries, the read length of grains 11292a and 11295a was shifted towards longer sequences, which is expected as larger DNA fragments form more stable complexes with enrichment probes and are thus recovered more efficiently in hybridization capture procedures23. In contrast, the enriched library for 11294a experienced a downward shift in read length, which likely stems from the stochastic effect of having only 72 unique mapped reads in the shotgun library for this sample. In mapDamage analysis of the cpDNA enriched libraries, the ‘5 ends of reads exhibited the increased levels of C→T misincorporations caused by deamination of cytosine typical of aDNA (Fig. 4b and Fig. S2)30,31. MapDamage results are not presented for shotgun data because the library from grain 11294a contained an insufficient number of reads for this analysis. Since direct14C dating of the Areni-1 broomcorn millet used for sequencing was not possible due to the small mass of the grains, the fragmentation patterns and miscoding profiles are important data that support the medieval dating of the samples.
Figure 4.
Read length distribution and mapDamage analysis. (a) Read length distributions of the shotgun and cpDNA enriched libraries made from three Areni-1 millet grains. All libraries were mapped to the Panicum miliaceum chloroplast reference genome: Genbank# KU343177.1. The shape of the violin displays the frequency distribution of the read lengths in each library. Inside each violin, the grey rectangle represents the interquartile range of the read lengths, while the white circle is the median87. (b) The nucleotide damage pattern of the cpDNA enriched library from the 11295a grain was assayed using mapDamage 231. The enriched library exhibited the C→T nucleotide misincorporations pattern typical of authentic aDNA. The cpDNA libraries from grains 11292a and 112924a gave similar results and are presented in Fig. S2.
Mapping statistics for Areni-1 enriched cpDNA data
In the mapping of cpDNA enriched libraries, grains 11292a and 11295a produced similar numbers of unique reads (≈ 170,000) that mapped to the chloroplast genome reference, while sample 11294a generated approximately a tenth of the mapped reads observed in the other two grains. The maximum read depth was greater with grains 11292a and 11295a at 229 and 331 reads respectively, compared to 52 reads for 11294a (Fig. 3). Despite the differences in coverage of the chloroplast reference genome and read depth, all three samples produced chloroplast data that allowed for phylogenetic and haplotype network analysis.
Generation of chloroplast genomes from 50 modern accessions of broomcorn millet
For comparative purposes, chloroplast genomes were generated from 50 accessions of modern P. miliaceum (Table S1). These accessions had a mainly Eurasian distribution and were weighted towards Chinese accessions with 29 of the samples originating from various locations within China. Included in these accessions were two wild (undomesticated) broomcorn millet (SampleID 31 and 167) and two accessions that have previously been used to produce nuclear reference genomes (SampleID 69 and T296)32,33. The sequencing of these modern accessions generated average read depths of ≥ 1356 reads across the reference chloroplast genome.
Paniceae phylogenetic analysis
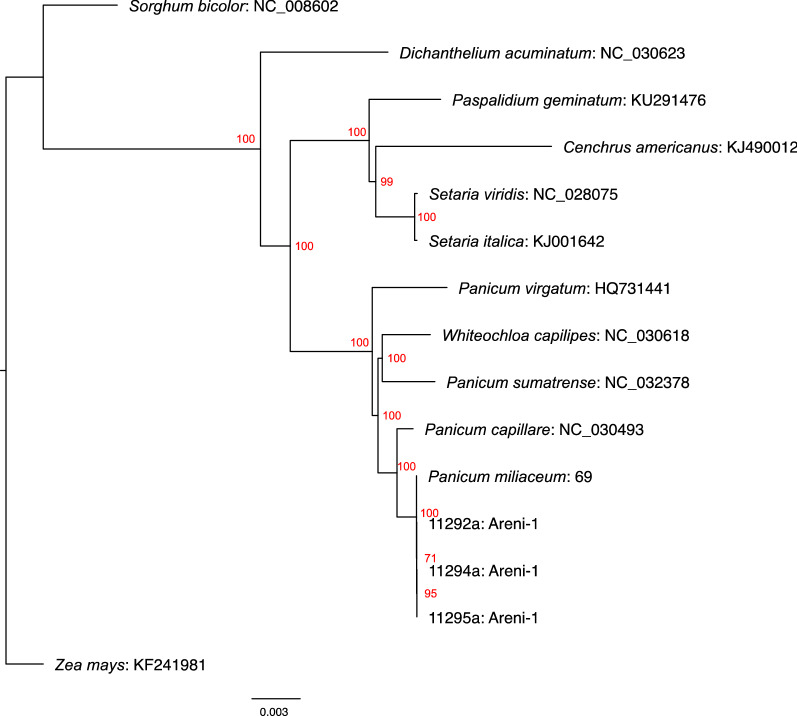
To confirm the visual identification and the taxonomic placement of the Areni-1 P. miliaceum and validate our laboratory and analytical methods, the chloroplast genomes from these medieval grains were compared to similar sequences from 11 members of the Paniceae tribe, including a modern P. miliaceum (Table S2). A phylogeny was generated of the Paniceae and Areni-1 chloroplast genomes with maximum likelihood method (Fig. 5). In the tree, the Areni-1 samples and broomcorn millet were placed together on a branch with a bootstrap support of 100, which indicated that the medieval grains were closely related to broomcorn millet and confirmed the taxonomic placement and visual identification of the medieval grains. The placement of the Panicum and Setaria species in the tree suggested the topography produced in this analysis was correct. All the Panicum spp. were placed on the same branch within the tree and domesticated S. italica formed a clade with the wild antecedent of this cereal, Setaria viridis9.
Figure 5.
Paniceae chloroplast phylogeny. A phylogeny generated with chloroplast genomes (sans inverted repeat regions) from 11 modern Paniceae species and 3 Areni-1 millet grains using the RAxML program with the GTRGAMMA model81. The tree was rooted on the outgroup Zea mays (maize). The scale bar at the bottom of the phylogeny represents the substitutions per site.
Panicum miliaceum phylogenetic and haplotype network analysis
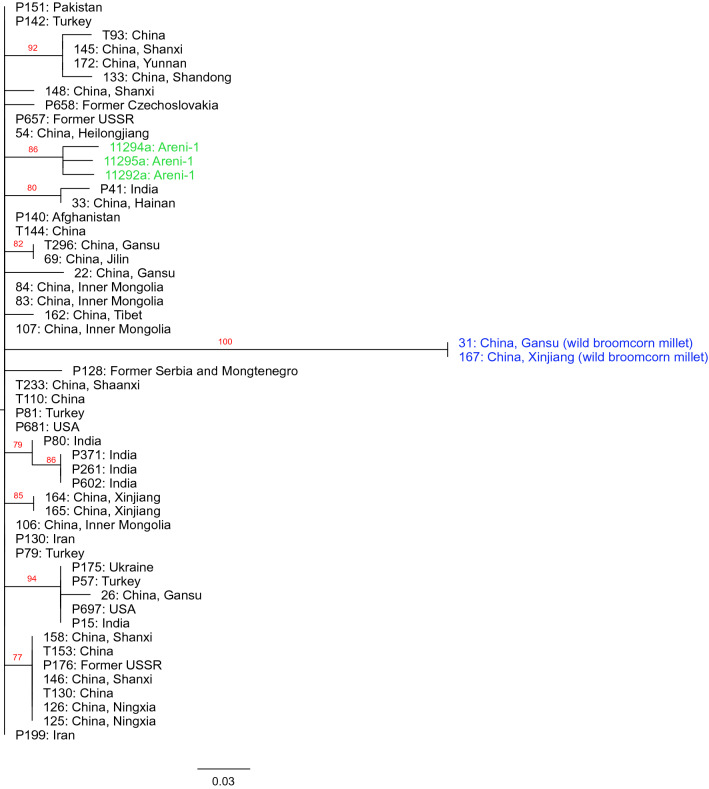
At the time the current study was started little was known about P. miliaceum chloroplast diversity or what demographic or evolutionary information the chloroplast genome from this cereal could contained. To address these deficiencies, we compared the Areni-1 chloroplast genomes to similar organelle sequences from 50 modern varieties of P. miliaceum including two wild accessions. Between the modern accessions and the Areni-1 grains a total of 63 SNPs were identified in the chloroplast genomes and these genotypes were used to generate phylogenies using maximum likelihood, Bayesian, and neighbor joining methods (Fig. 6 and Fig. S3). Generally, across the trees there was low bootstrap support and no geographic structure, so no inference on P. miliaceum demographics could be made. However, in the trees produced with the maximum likelihood and Bayesian methodologies, the domesticated accessions were split into two large clades and in each of these phylogenies the Areni-1 chloroplast genomes grouped together as a clade.
Figure 6.
Panicum miliaceum maximum likelihood phylogeny. A RaxML maximum likelihood phylogeny generated using the chloroplasts genomes from the three Areni-1 grains and 50 modern accessions of P. miliaceum with rooting on the branch containing the two wild accession (31,167). The scale bar at the bottom of the phylogenies represents the substitutions per site. Accessions highlighted in green are the Areni-1 millet and the accession in blue are the wild P. miliaceum. Additional Bayesian and neighbour joining phylogenies of this data are given in Fig S3.
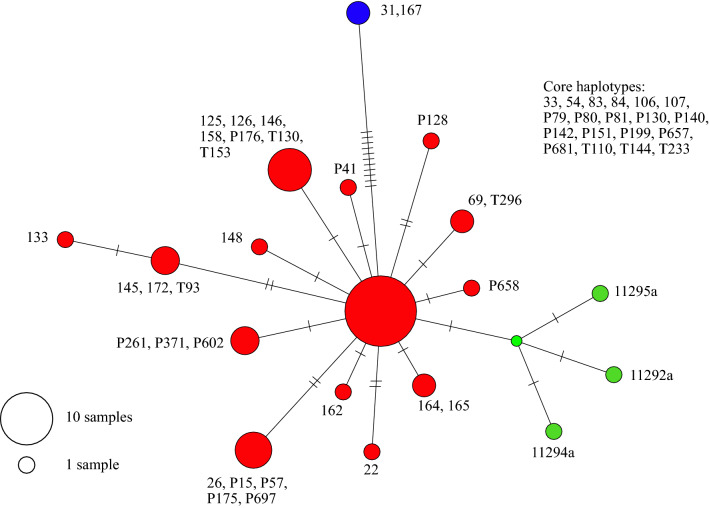
The relationship between the modern and Areni-1chloroplast genomes was further investigated in a haplotype network using the PopArt software with the median spanning network algorithm (Fig. 7). In this network, all the domesticated accessions, including the Areni-1 samples, were separated by only 1–5 SNPs. A core haplogroup, represented by the large node at the centre of the network, contained 19 accessions with a wide geographic range across Eurasia. The Areni-1 chloroplast genomes were separated from the core haplogroup by two mutations, one of which was common to all three medieval samples. The largest number of variations was observed in the wild P. miliaceum, which were separated from the domesticated accessions by 11–14 SNPs.
Figure 7.
Panicum miliaceum haplotype network. A chloroplast genome haplotype network from the three medieval Areni-1 grains and 50 modern accessions of P. miliaceum was generated using the PopArt program (http://popart.otago.ac.nz/index.shtml) with the median joining network algorithm. The hash marks on the lines connecting the circles indicate the number of mutations separating the haplotypes. The accessions contained in the core haplotype, the large node at centre of network, are listed in the upper right of the figure. Modern domesticated accessions are given in red, wild modern accessions are given in blue, and the Areni-1 millet are given in green.
Discussion
In this study, DNA extraction and hybridization capture enrichment techniques were used to recover aDNA from medieval broomcorn millet grains found in Areni-1, a cave site located in the Armenian Highlands a region that has an important history in the domestication and dispersal of many crops. Despite variable quality of aDNA found in the Areni-1 grains, we were able to recover three chloroplast genomes from five grains assayed.
Despite a similar well-preserved physical appearance, the endogenous DNA content varied among the Areni-1 grains used in this study as did the quality of the aDNA. For example, in the millet enriched for cpDNA, grain 11295a contained the highest level of endogenous DNA and the longest read length, while grain 11294a contained the least amount of endogenous DNA with the shortest read length (Table 2, Fig. 3a). The environment of the cave appears to be suited to prevent decomposition of biological material and macromolecules as excavations at Areni-1 have unearthed vertebrate soft tissues including a mummified goat34 and brain tissue in a human skull35, demonstrating that the conditions in the cave can preserve ephemeral biological material. It is not clear why the Areni-1 millet exhibited different levels of DNA preservation. Grains with poor preservation may have undergone different harvesting, threshing, or other anthropogenic treatments that influenced DNA stability. Post excavation factors, such as storage or handling conditions, may have also influenced the different levels of DNA preservation. Further, the micro-environment has been shown to impact the survival of DNA in mammalian bones36 and similar processes may have influenced the decay of aDNA in the Areni-1 millet grains. Local variations in pH, ion concentration, moisture, or other abiotic factors could have effected DNA stability in the Areni-1 millet37. Variable preservation of aDNA from grains is not unique to the Areni-1 millet as a study of 6000 year old barley found large differences in recoverable endogenous DNA in grains excavated from the same occupation layer of a cave38. Regardless of the varying integrity of the aDNA, the recovery of chloroplast genomes from the Areni-1 samples demonstrates that broomcorn millet grains can, under the right conditions, preserve DNA for at least 1000 years. Even with a poorly preserved sample (11294a), hybridization capture enrichment was able to recover sufficient cpDNA to perform a chloroplast phylogenetic and haplotype analysis.
In the Paniceae grass chloroplast phylogeny (Fig. 5), the Areni-1 and P. miliaceum reference clustered with several species native to North America (Panicum virgatum, switchgrass; Panicum capillare, witchgrass) and Whiteochloa capillipes, which is from Australia, New Guinea, and Indonesia. Panicum sumatrense, a cereal crop originally from India also clustered with the Areni-1/ P. miliaceum. Panicum is the largest genera of the grass family Paniceae and has a cosmopolitan distribution so the clustering of the Areni-1/ P. miliaceum with widely distributed species is not unusual39. While it is unlikely that the specimens from Areni-1 are directly related to these species, it is possible that some of them share a common ancestor from Southeast Asia, further supporting the east to west introduction of P. miliaceum in Eurasia. It should be noted that the chloroplast phylogeny indicates a close relationship between broomcorn millet and P. capillare (bootstrap support = 100) an observation that supports a previous study of Panicum nuclear DNA, which found that diploid P. capillare or a close relative is the likely maternal genome donor to tetraploid broomcorn millet40.
To investigate the demographic history of P. miliaceum, chloroplast phylogenies were generated for the Areni-1 samples and modern accessions using maximum likelihood, Bayesian, neighbour joining methodologies. In these analyses only 63 SNPs were identified in a group that included 48 modern domesticated accessions, two wild accessions, and the three ≈1000-year-old grains from the Areni-1 cave. Accordingly, there was little differences between these chloroplast genomes which was reflected in the phylogenies with low bootstrap support values through much of the trees. No conclusions to be drawn on the demographics of broomcorn millet form these analyses (Fig. 6 and Fig. S3). The inability of chloroplast genomes to resolve intraspecies relationships is not unique to the current study. Research on the genera Amaranthus and Euphrasia found that whole chloroplast genomes were highly conserved and lacked the power to discriminate between some intraspecies accessions41,42. Yet, in all these trees the Areni-1 chloroplast genomes fell within the diversity of domesticated accession, indicating the Areni-1 grains are a fully domesticated variety of P. miliaceum and not a wild or partially domesticated form of the cereal.
However, in the maximum likelihood and Bayesian phylogenies, the domesticated accessions were split into two clades with no geographical structure and each clade containing accessions from locations throughout Eurasia (Fig. 6 and Fig. S3a). These results are similar those found in a study by Hunt et al.43 which used 16 microsatellite loci across 98 P. miliaceum landraces and divided modern broomcorn millet into two clades, one composed solely of Chinese accessions and the other clade contained accessions from throughout Eurasia. It is not clear why the chloroplast phylogenies in the current study lacked a clade of solely Chinese accessions as observed by Hunt et al. 2011. Previous studies have reported incongruent chloroplast and nuclear phylogenies and have attributed these differences to incomplete lineage sorting and introgression/hybridization44,4545. Similar processes may be involved with the discrepancies between the P. miliaceum nuclear and chloroplast genomes, however the low levels of diversity in the chloroplast genomes are likely the main contributing factor for these differences. The P. miliaceum chloroplast genomes contained sufficient information for gross scale separation of the different accessions but was insufficient to accurately resolve fine scale relationships.
In the haplotype network (Fig. 7), the majority of the haplotypes were separated by 1–5 SNPs, which did not allow for any conclusions to be drawn about the demographic history of P. miliaceum. Wild accessions contained a relatively high number of variants, 11 SNPs, that were not present in the domesticated accessions. Several scenarios that are not mutually exclusive could explain this observation. One possibility is that the wild accessions sampled may not be directly related to the broomcorn lineages that were domesticated. Another possible scenario is the SNPs observed in the wild accessions may represent diversity lost in the chloroplast genome during a domestication bottle neck. Such a loss of diversity has been reported in a study by Leigh et al.46, which described a reduction in both alleles and haplotypes in the chloroplast genomes of different wheat species after domestication. However, a much larger sampling of chloroplast genomes from wild accessions will be needed to resolve the relationship of undomesticated and domesticated P. miliaceum.
Domestication of crops involve the selection of plant traits that better meet the needs of humans. Chloroplasts are not only responsible for photosynthetic energy production, but are also involved with other cellular functions including synthesis of amino acids, nucleotides, fatty acids, phytohormones, vitamins, and the production of metabolites that are important for plant interaction with biotic and abiotic factors in the environment16. Since the chloroplast regulates such a broad range of cellular functions that are essential to P. miliaceum for survival and success as a cereal, traits under the control of this organelle will come under selection by humans. The similarity of the Areni-1 chloroplast genomes to those from modern domestications may represent a limited range of genotypes that human find beneficial to P. miliaceum.
The chloroplast inheritance pattern will aid in maintaining chloroplast genotypes that are beneficial as a crop. Although unproven in P. miliaceum, as far as the authors can determine, maternal inheritance of chloroplast in P. miliaceum is the mostly likely scenario because maternal inheritance occurs in most higher plants and maternal inheritance has been shown to be the primary inheritance pattern in Panicum virgatum, a relative of P. miliaceum47,48. With maternal inheritance there is no meiosis or recombination so chloroplast genotypes beneficial to humans will not be broken up and can be more readily maintained in a cultivated plant49.
It is possible that mapping of promiscuous DNA that has broken off the chloroplast genome and become incorporated in either the nuclear or mitochondrial genome has influenced the Areni-1 chloroplast results50,51. Effort was made to minimize the impact of promiscuous cpDNA on the Areni-1 results by requiring a relatively high read depth (≥ 10) and at least 95% of the reads to call a mutation as an alternative allele. However, as it will be difficult to distinguish between promiscuous cpDNA and DNA from the chloroplast, proving that promiscuous cpDNA has not influenced the current results is extremely challenging. Regardless, the agreement of the morphological identification of the Areni-1 millet as broomcorn millet and the placement of the Areni-1 millet with P. miliaceum in the Paniceae chloroplast phylogeny (Fig. 5) indicate that promiscuous cpDNA did not have a significant impact on the interspecies analysis.
In conclusion, here we generated 53 P. miliaceum chloroplast genomes, which includes three that are ≈1000-yeaers-old that can be used as resources to study this cereal crop and other C4 plants. A phylogeny generated from these chloroplast genomes divided the domesticated forms of broomcorn millet into two clades, an observation that parallels a report using nuclear DNA. The chloroplast genomes from domesticated accessions displayed low diversity and wild accessions sampled in this study contained SNPs that were outside the diversity of the domesticated forms of P. miliaceum. Lastly, this study demonstrates that broomcorn millet grains can preserve significant levels of DNA for hundreds if not thousands of years and can serve as an important genetic resource to investigate the evolution and domestication of this cereal.
Materials and methods
Areni-1 archaeology of botanical material
To identify the plant material at Areni-1, a comprehensive archaeobotanical sampling strategy was adopted for the cave sediment. Sediment samples with a volume of 5 L were collected throughout the excavation and separated using dry sieving instead of flotation. The majority of plant remains found at Areni-1 were desiccated and contact with water was likely to damage this material. After passing each sediment sample through a 1 mm sieve, material > 1 mm was kept for study and < 1 mm was discarded, which allowed for some small weed seeds and plant parts to be lost. Similar approaches to sieving have been used at other sites that contained copious amounts of desiccated plant material52. Sieved material was hand sorted into categories of plant remains, bones, and pottery fragments. Millet grains were among the plant material recovered from Areni-1 excavations, which also included other domesticated cereals including wheat and barley. To date, all millet found at Areni-1 came from Trench 1 and 3 within the cave site and in stratigraphic horizons attributed to medieval occupations11.
Areni-1 millet grain morphology and identification
Identification of the Areni-1 millet grains were made by comparing morphological features of the ancient florets with modern Panicum, Echinochloa, and Setaria specimens accessioned from Turkey and Armenia (curated within the Archaeobotany reference collection at the University of Connecticut). To help further hone the identification, a number of images, keys, and textual descriptions of morphological features and species distributions were also consulted, including reference manuals27,53–56, the Flora of Armenia57, Flora of Turkey58, Flora of Iraq59, and archaeobotanical publications detailing various additional Paniceae species found across Asia and Europe25,60–64. Textual descriptions within flora typically provide size ranges for spikelets rather than florets or caryopses. These descriptions remained helpful, however, given that the spikelet lengths of many taxa (particularly within the Setaria genus), were shorter than the florets observed here, and could easily be eliminated. Length to breadth ratios were also considered along with the surface texture of the florets. The Areni-1 millet grains were photographed and measured using a Nikon AZ microscope with associated NIS Elements software.
Carbon dating of botanical material from Areni-1
A single Vitis sp. pedicel from Trench 1 and a pool of three millet grains from Trench 3 (Fig. S1) were sent to the NSF Arizona AMS Laboratory (University of Arizona) for radiocarbon dating. Fraction Modern Carbon and Radiocarbon Age were calculated as weighted averages of combined machine runs to reduce overall error.
Areni-1 millet library construction and cpDNA enrichment
For genetic studies, five millet grains were chosen at random for the extraction and enrichment of cpDNA (Table 2). Extraction of aDNA and construction of Illumina libraries from the Areni-1 millet was performed in a dedicated low-DNA laboratory where no work had previously been done with millet. Isolation of aDNA from the Areni-1 grains and negative blank controls was performed with a two-step protocol, which has been previously described65–67. Shotgun and cpDNA enriched libraries were generated using standard aDNA methods and a detailed description of these protocols is given in the Supplemental Methods.
Mapping of Areni-1 millet sequencing data
Raw FASTQ files demultiplexed on the i7 adapter index were obtained directly from the sequencer. Run-specific FASTQ files were demultiplexed by the internal barcodes using Sabre 1.0 (https://github.com/najoshi/sabre). AdapterRemoval v2.2.168 was used to trim adapters, collapse reads, discard reads < 25 bp, and remove reads of low quality (< 4). Collapsed reads were mapped to a P. miliaceum chloroplast reference genome (Genbank #: KU343177.1) using BWA aln (v0.5.11-foss-2016b) with parameters recommended for aDNA (-l 1024 -n 0.01 -o 2) and a mapping quality (MAPQ) 3069,70. Duplicate reads were removed from the mapped data using Picard Tools (v 2.2.4: https://broadinstitute.github.io/picard/index.html). Levels of nucleotide misincorporation caused by deaminated cytosine were assayed using mapDamage 2.0 in cpDNA enriched libraries31. Read depths were extracted from cpDNA enriched mapped data using SAMTools (v1.3.1-foss-2016a)71 and plotted using R (v3.4.2)72. Read length distributions for the shotgun and cpDNA enriched libraries were produced using SeqKit (v 0.7.2)73 and SAMTools and plotted with R using the ggplot2 package. For all libraries, except the shotgun library for 11294a, read length analysis was performed using 19,000 reads, a number based on the smallest number of mapped reads produced by a library in this dataset. The shotgun library for 11294a produced 72 mapped reads and this number was used for the read length analysis.
Sequencing of modern broomcorn millet accessions
DNA was extracted from seedling leaves of 50 modern broomcorn millet accessions (Table S1) using the DNAeasy Plant Mini Kit (QIAGEN) and the provided instructions. Shotgun libraries were constructed from the isolated DNA using a NEBNext Ultra II FS DNA Library Prep Kit for Illumina (New England Biolabs) using the protocol provided by the manufacture. Sequencing of the modern libraries was performed at Core Facility for Genomics at Shanghai Center for Plant Stress Biology (Shanghai, China) on an Illumina HiSeq 2500 platform using 2 × 125 bp (250 cycle) chemistry.
Mapping of modern broomcorn millet sequencing data
Fastq reads from the modern broomcorn millet accessions were firstly mapped to a broomcorn millet nuclear reference genome (NCBI Assembly: GCA_003046395.2) using BWA mem (v0.5.11-foss-2016b: https://arxiv.org/abs/1303.3997v2). From the resulting bam file, reads that did not map to the nuclear reference were extracted using SAMTools (v1.12) and remapped to the broomcorn millet chloroplast genome (Accession: KU343177.1) using BWA mem. Duplicates were marked with Picard Tools to prevent processing of these reads in downstream analysis. Mapping to the chloroplast reference genome produced average read depths of between 1356 and 4548 across the chloroplast genome (Table S2).
Variant calling in the Areni-1 and modern millet sequencing data
Variants were called in parallel in the Areni-1 and modern millet using freebayes (v 1.0.2-GCC-4.9.3-binutils-2.25: Garrison and Marth, 2012, arXiv:1207.3907) with the following parameters: --min-base-quality 30 --min-coverage 10 --hwe-priors-off --ploidy 1 --pooled-continuous --genotype-qualities --min-alternate-fraction 0.95 to generate vcf files. The value for the --min-alternate-fraction was chosen to mask polymorphic sites caused by promiscuous cpDNA (chloroplast DNA that has migrated to the nuclear or mitochondrial genomes)74. Freebayes automatically disregards reads marked as duplicate, so only unique reads from the modern millet were analysed. Chloroplast genome consensus sequences were generated for the Areni-1 millet using the FastaAlternateReferenceMaker function of GATK (v 3.5-Java-1.8.0_71)75 using the freebayes vcf files and the KU343177.1 reference. Two sections of the Areni-1 consensus sequences, bp 81851 to 104489 and 117101 to 139826, were removed with Geneious Prime (Build 2019-04-26) using 11295a as a guide76, because these regions represent the IRs of the chloroplast genome where ancient reads could not be mapped (Fig. 3).
Paniceae phylogenetic analysis
Chloroplast reference genomes for 10 members of the Paniceae tribe of grasses (which includes members of the Panicum genus) were downloaded from the NCBI website (Table S1). The reference sequence for the Zea mays chloroplast genome was also downloaded for use as an out-group. Although the Grass Phylogeny Working Group II, which used three genetic markers, placed Whiteochloa capillipes in the Cenchrinae subtribe77, this species was included in the analysis because a more recent study of complete chloroplast genomes placed W. capillipes in the Paniceae subtribe78. The Areni-1 consensus sequences and the grass references were aligned using MAFFT (v 7.130-foss-2016b-with-extensions) and the default settings79.
P. miliaceum variety 69 from the modern accessions (Table S2) was included the MAFFT alignment to serve as the broomcorn millet chloroplast genome. After alignment, the Paniceae genomes were trimmed of the IRs with Geneious Prime using sample 11295a as before. Poorly aligned regions were removed from the alignment using the trimAl program with the default parameters on the Phylemon 2.0 website (http://phylemon2.bioinfo.cipf.es)80. A maximum likelihood tree was built for the alignment with RAxML81 (v. 8.2.10) with 1,000 rapid bootstraps and the “GTRGAMMA” model. Tree visualization was performed with FigTree (v 1.4.3; http://tree.bio.ed.ac.uk/software/figtree/) with rooting on the branch containing the maize chloroplast genome.
Broomcorn millet phylogenetic and haplotype analysis
The translated genotypes of the Areni-1 and modern millet were extracted from the freebayes vcf file and exported as a multiple sequence file using bcftools (v1.12)71. An MAFFT alignment with the IRs and poorly aligned regions removed was produced as before. Phylogenies were then produced for the chloroplast genomes using three methodologies: (1) maximum likelihood using RAxML as describe above; (2) Bayesian using MrBayes (v3.2.7a, https://nbisweden.github.io/MrBayes/download.html with 5 million generations82; (3) neighbor joining MEGA11 (https://megasoftware.net/) with 1000 bootstrap replicates83. Trees were generated for the phylogenies using FigTree as above. A PHYLIP format file generated from the MAFFT alignment using Geneious Prime was used to produce a haplotype network in the PopART program (http://popart.otago.ac.nz/index.shtml) with the median joining network algorithm84.
Use of plant material
The collection and use of the medieval and modern millet grains complied with all relevant institutional, national, and international guidelines and legislation on the study of plant material. No permission was needed for any of the millet grains used in this study.
Supplementary Information
Acknowledgements
The excavations at Areni-1 were directed by Boris Gasparyan (Institute of Archaeology and Ethnography, National Academy of Sciences, Armenia and Gfoeller Renaissance Foundation, USA) and co-directed by Ron Pinhasi (School of Archaeology, University College Dublin, Ireland) and Gregory Areshian (Cotsen Institute of Archaeology at UCLA, USA). The sorting of biological material was primarily supervised by Tamara Bagoyan (Institute of Archaeology and Ethnography, National Academy of Sciences, Armenia).
Author contributions
S.M.R. and H.Z. initiated and designed the study. N.H., B.G., and A.S. provided the ancient broomcorn millet. H.Z. and L.L. provided the modern accessions of broomcorn millet. A.S. performed the morphology analysis. S.M.R. and L.L. generated the data, whilst all authors contributed to analysis and interpretation of data. S.M.R. wrote the initial draft of the manuscript and Supplementary Information. All authors reviewed and edited the manuscript and Supplementary Information.
Funding
Laboratory work for the Areni-1 millet was funded by the following grant:LP130100648: Identifying the diversity and evolution of loci associated with adaptation to aridity/heat and salinity in ancient cereal crops awarded to M.G. and A.C. from the Australian Research Council. Archaeobotany work was partly funded by a Curtiss T. and Mary G. Brennan Award to A.S. Sequencing of the 50 modern broomcorn millet accessions was funded by a grant from the National Natural Science Foundation of China (31900189) awarded to L.L. and grants from Shanghai Science and Technology Committee (17391900200), National Natural Science Foundation of China (31922008), and CAS Youth Innovation Promotion Association (Y201844) awarded to H.Z.
Data availability
Sequencing data for the Areni-1 samples can be found at the NCBI website under BioProject: PRJNA575768 and the chloroplast sequencing data for the 50 modern broomcorn millet accessions are deposited at figshare: https://doi.org/10.6084/m9.figshare.14914605.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Stephen M. Richards, Leiting Li, Alan Cooper and Heng Zhang.
Contributor Information
Stephen M. Richards, Email: smr3434@gmail.com
Heng Zhang, Email: hengzhang@psc.ac.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-17931-4.
References
- 1.Habiyaremye C, et al. Proso Millet (Panicum miliaceum L.) and Its potential for cultivation in the Pacific Northwest, U.S.: A review. Front. Plant Sci. 2017;7:5. doi: 10.3389/fpls.2016.01961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller NF, Spengler RN, Frachetti M. Millet cultivation across Eurasia: Origins, spread, and the influence of seasonal climate. The Holocene. 2016;26:1566–1575. doi: 10.1177/0959683616641742. [DOI] [Google Scholar]
- 3.Cannarozzi G, et al. Genome and transcriptome sequencing identifies breeding targets in the orphan crop tef (Eragrostis tef) BMC Genomics. 2014;15:581. doi: 10.1186/1471-2164-15-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motuzaite-Matuzeviciute G, Staff RA, Hunt HV, Liu X, Jones MK. The early chronology of broomcorn millet (Panicum miliaceum) in Europe. Antiquity. 2013;87:1073–1085. doi: 10.1017/S0003598X00049875. [DOI] [Google Scholar]
- 5.Lu H, et al. Earliest domestication of common millet (Panicum miliaceum) in East Asia extended to 10,000 years ago. Proc. Natl. Acad. Sci. 2009;106:7367–7372. doi: 10.1073/pnas.0900158106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunt HV, et al. Genetic evidence for a western Chinese origin of broomcorn millet (Panicum miliaceum) The Holocene. 2018 doi: 10.1177/0959683618798116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leipe C, Long T, Sergusheva EA, Wagner M, Tarasov PE. Discontinuous spread of millet agriculture in eastern Asia and prehistoric population dynamics. Sci. Adv. 2019;5:6225–62215. doi: 10.1126/sciadv.aax6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevens CJ, Shelach-Lavi G, Zhang H, Teng M, Fuller DQ. A model for the domestication of Panicum miliaceum (common, proso or broomcorn millet) in China. Veg. Hist. Archaeobotany. 2020 doi: 10.1007/s00334-020-00804-z. [DOI] [Google Scholar]
- 9.Diao, X. & Jia, G. In Plant Genetics and Genomics: Crops and Models Vol. Volume 19 (ed Richard, A. J.) Ch. 4, 61–72 (Springer International Publishing, Berlin, 2017).
- 10.Spengler R, et al. Early agriculture and crop transmission among Bronze Age mobile pastoralists of Central Eurasia. Proc. R. Soc. B Biol. Sci. 2014;281:5. doi: 10.1098/rspb.2013.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith, A., Bagoyan, T., Gabrielyan, I., Pinhasi, R. & Gasparyan, B. In Stone Age of Armenia, A Guide-book to the Stone Age Archaeology in the Republic of Armenia (eds Boris, G. & Makoto, A.) 233–260 (Center for Cultural Resource Studies, Kanazawa University, 2014).
- 12.Zohary, D., Hopf, M. & Weiss, E. Domestication of Plants in the Old World : The Origin and Spread of Domesticated Plants in Southwest Asia, Europe, and the Mediterranean Basin. Fourth edition edn, (Oxford University Press, Oxford, 2013).
- 13.Zeder MA. Domestication and early agriculture in the Mediterranean Basin: Origins, diffusion, and impact. Proc. Natl. Acad. Sci. 2008;105:11597. doi: 10.1073/pnas.0801317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christian D. Silk roads or steppe roads? The silk roads in world history. J. World Hist. 2000;11:1–26. doi: 10.1353/jwh.2000.0004. [DOI] [Google Scholar]
- 15.Wang Y, et al. Chloroplast genome variation and phylogenetic relationships of Atractylodes species. BMC Genomics. 2021;22:103–103. doi: 10.1186/s12864-021-07394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniell H, Lin C-S, Yu M, Chang W-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134–134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sánchez-Baracaldo P, Raven-John A, Pisani D, Knoll-Andrew H. Early photosynthetic eukaryotes inhabited low-salinity habitats. Proc. Natl. Acad. Sci. 2017;114:E7737–E7745. doi: 10.1073/pnas.1620089114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Llorente B, et al. Homecoming: rewinding the reductive evolution of the chloroplast genome for increasing crop yields. Nat. Commun. 2021;12:6734–6734. doi: 10.1038/s41467-021-26975-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birky CW., Jr Uniparental inheritance of mitochondrial and chloroplast genes: Mechanisms and evolution. Proc. Natl. Acad. Sci. USA. 1995;92:11331–11338. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith DR. Mutation rates in plastid genomes: They are lower than you might think. Genome Biol. Evol. 2015;7:1227–1234. doi: 10.1093/gbe/evv069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mower, J. P. & Vickrey, T. L. In Advances in Botanical Research Vol. 85 (eds Shu-Miaw, C. & Robert, K. J.) 263–292 (Academic Press, Hoboken, 2018).
- 22.Gao L-Z, et al. Evolution of Oryza chloroplast genomes promoted adaptation to diverse ecological habitats. Commun. Biol. 2019;2:278–278. doi: 10.1038/s42003-019-0531-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brotherton P, et al. Neolithic mitochondrial haplogroup H genomes and the genetic origins of Europeans. Nat. Commun. 2013;4:1764. doi: 10.1038/ncomms2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filipović D, et al. New AMS 14C dates track the arrival and spread of broomcorn millet cultivation and agricultural change in prehistoric Europe. Sci. Rep. 2020;10:13698. doi: 10.1038/s41598-020-70495-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korolyuk EA, Krasnikov AA, Polosmak NV. Panicoids in Xiongnu burial ground (Mongolia, First Century AD): Problems of identification. Turczaninowia. 2018;21:145–159. doi: 10.14258/turczaninowia.21.4.14. [DOI] [Google Scholar]
- 26.Nesbitt M, Summers GD. Some recent discoveries of millet (Panicum miliaceum L. and Setaria italica (L.) P. Beauv) at excavations in Turkey and Iran. Anatol. Stud. 1988;38:85–97. doi: 10.2307/3642844. [DOI] [Google Scholar]
- 27.Motuzaite-Matuzeviciute G, Hunt HV, Jones MK. Experimental approaches to understanding variation in grain size in Panicum miliaceum (broomcorn millet) and its relevance for interpreting archaeobotanical assemblages. Veg. Hist. Archaeobotany. 2012;21:69–77. doi: 10.1007/s00334-011-0322-2. [DOI] [Google Scholar]
- 28.Dabney J, Meyer M, Pääbo S. Ancient DNA damage. Cold Spring Harb. Perspect. Biol. 2013;5:1–6. doi: 10.1101/cshperspect.a012567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armbrecht L, Hallegraeff G, Bolch CJS, Woodward C, Cooper A. Hybridisation capture allows DNA damage analysis of ancient marine eukaryotes. Sci. Rep. 2021;11:3220. doi: 10.1038/s41598-021-82578-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brotherton P, et al. Novel high-resolution characterization of ancient DNA reveals C > U-type base modification events as the sole cause of post mortem miscoding lesions. Nucleic Acids Res. 2007;35:5717–5728. doi: 10.1093/nar/gkm588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi J, et al. Chromosome conformation capture resolved near complete genome assembly of broomcorn millet. Nat. Commun. 2019;10:464. doi: 10.1038/s41467-018-07876-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou C, et al. The genome of broomcorn millet. Nat. Commun. 2019;10:436. doi: 10.1038/s41467-019-08409-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zarikian N, Gasparyan B. Micromammal remains from Areni-1 Cave, Armenia. IOSR J. Human. Soc. Sci. 2016;21:20–25. doi: 10.9790/0837-2110042025. [DOI] [Google Scholar]
- 35.Wilkinson KN, et al. Areni-1 Cave, Armenia: A Chalcolithic-Early Bronze Age settlement and ritual site in the southern Caucasus. J. Field Archaeol. 2012;37:20–33. doi: 10.1179/0093469011Z.0000000002. [DOI] [Google Scholar]
- 36.Campana MG, McGovern T, Disotell T. Evidence for differential ancient DNA survival in human and pig bones from the Norse North Atlantic. Int. J. Osteoarchaeol. 2014;24:704–708. doi: 10.1002/oa.2255. [DOI] [Google Scholar]
- 37.Pääbo S, et al. Genetic analyses from ancient DNA. Annu. Rev. Genet. 2004;38:645–679. doi: 10.1146/annurev.genet.37.110801.143214. [DOI] [PubMed] [Google Scholar]
- 38.Mascher, M. et al. Genomic analysis of 6,000-year-old cultivated grain illuminates the domestication history of barley. Nature Genet.48, 1089–1093 10.1038/ng.3611, http://www.nature.com/ng/journal/v48/n9/abs/ng.3611.html#supplementary-information (2016). [DOI] [PubMed]
- 39.Aliscioni SS, Giussani LM, Zuloaga FO, Kellogg EA. A molecular phylogeny of Panicum (Poaceae: Paniceae): Tests of monophyly and phylogenetic placement within the Panicoideae. Am. J. Bot. 2003;90:796–821. doi: 10.3732/ajb.90.5.796. [DOI] [PubMed] [Google Scholar]
- 40.Hunt HV, et al. Reticulate evolution in Panicum (Poaceae): The origin of tetraploid broomcorn millet P. miliaceum. J. Exp. Botany. 2014;65:3165–3175. doi: 10.1093/jxb/eru161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viljoen E, Odeny DA, Coetzee MPA, Berger DK, Rees DJG. Application of chloroplast phylogenomics to resolve species relationships within the plant genus Amaranthus. J. Mol. Evol. 2018;86:216–239. doi: 10.1007/s00239-018-9837-9. [DOI] [PubMed] [Google Scholar]
- 42.Zhou T, et al. The complete chloroplast genome of Euphrasia regelii, pseudogenization of ndh genes and the phylogenetic relationships within Orobanchaceae. Front. Genet. 2019;10:5. doi: 10.3389/fgene.2019.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunt HV, et al. Genetic diversity and phylogeography of broomcorn millet (Panicum miliaceum L.) across Eurasia. Mol. Ecol. 2011;20:4756–4771. doi: 10.1111/j.1365-294X.2011.05318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu W-B, Huang P-H, Li D-Z, Wang H. Incongruence between Nuclear and chloroplast DNA phylogenies in pedicularis section cyathophora (Orobanchaceae) PLoS ONE. 2013;8:e74828. doi: 10.1371/journal.pone.0074828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nge FJ, Biffin E, Thiele KR, Waycott M. Reticulate evolution, ancient chloroplast haplotypes, and rapid radiation of the australian plant genus Adenanthos (Proteaceae) Front. Ecol. Evol. 2021;8:5. doi: 10.3389/fevo.2020.616741. [DOI] [Google Scholar]
- 46.Leigh FJ, et al. Using diversity of the chloroplast genome to examine evolutionary history of wheat species. Genet. Resour. Crop Evol. 2013;60:1831–1842. doi: 10.1007/s10722-013-9957-4. [DOI] [Google Scholar]
- 47.Park H-S, et al. Inheritance of chloroplast and mitochondrial genomes in cucumber revealed by four reciprocal F1 hybrid combinations. Sci. Rep. 2021;11:2506. doi: 10.1038/s41598-021-81988-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martínez-Reyna JM, Vogel KP, Caha C, Lee DJ. Meiotic stability, chloroplast DNA polymorphisms, and morphological traits of upland × lowland switchgrass reciprocal hybrids. Crop Sci. 2001;41:1579–1583. doi: 10.2135/cropsci2001.4151579x. [DOI] [Google Scholar]
- 49.Greiner S, Sobanski J, Bock R. Why are most organelle genomes transmitted maternally? BioEssays News Rev. Mol. Cell. Dev. Biol. 2015;37:80–94. doi: 10.1002/bies.201400110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang G-J, et al. Nuclear integrants of organellar DNA contribute to genome structure and evolution in plants. Int. J. Mol. Sci. 2020;21:707. doi: 10.3390/ijms21030707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roark LM, Hui AY, Donnelly L, Birchler JA, Newton KJ. Recent and frequent insertions of chloroplast DNA into maize nuclear chromosomes. Cytogenet. Genome Res. 2010;129:17–23. doi: 10.1159/000312724. [DOI] [PubMed] [Google Scholar]
- 52.Van der Veen, M. Consumption, Trade and Innovation. Exploring the Botanical Remains from the Roman and Islamic Ports at Quseir a-Qadim, Egypt. 20–33 (Africa Magna Verlag, 2011).
- 53.Fuller, D. Q. In http://www.homepages.ucl.ac.uk/~tcrndfu/Abot/Millet%20Handout06.pdf (ed University College London. Institute of Archaeology) (2006).
- 54.Nesbitt M. Identification Guide for Near Eastern Grass Seeds. University College London; 2006. [Google Scholar]
- 55.Cappers, R. T. J., Neef, R. & Bekker, R. M. Digital Atlas of Economic Plants. Volume 2b: Icacinaceae–Zygophyllaceae. (Barkhuis & Groningen University Library, 2009).
- 56.Neef, R., Cappers, R. T. J. & Bekker, R. M. Digital Atlas of Economic Plants in Archaeology. (Barkhuis & Groningen University Library, 2012).
- 57.Agababian, M. Y. et al. Flora of Armenia Vol 11. Poaceae [in Russian]. (A.R.G. Gantner Verlag KG, 2010).
- 58.Davis, P. H. (Edinburgh University Press, Edinburgh, 1985).
- 59.Bor, N. L. Flora of Iraq Vol. 9. (Ministry of Agriculture, Republic of Iraq, 1968).
- 60.Nasu H, Momohara A, Yasuda Y, He J. The occurrence and identification of Setaria italica (L.) P. Beauv (foxtail millet) grains from the Chengtoushan site (ca 5800 cal B.P.) in central China, with reference to the domestication centre in Asia. Veg. Hist. Archaeobotany. 2007;16:481–494. doi: 10.1007/s00334-006-0068-4. [DOI] [Google Scholar]
- 61.Tsang C-H, et al. Broomcorn and foxtail millet were cultivated in Taiwan about 5000 years ago. Bot. Stud. 2017;58:3. doi: 10.1186/s40529-016-0158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fukunaga K, Kawase M, Sakamoto S. Variation of caryopsis length and width among landraces of foxtail millet, Setaria italica (L.) P. Beauv. Jpn. J. Trop. Agric. 1997;41:235–240. doi: 10.11248/jsta1957.41.235. [DOI] [Google Scholar]
- 63.Stevens CJ, Shelach-Lavi G, Zhang H, Teng M, Fuller DQ. A model for the domestication of Panicum miliaceum (common, proso or broomcorn millet) in China. Veg. Hist. Archaeobotany. 2021;30:21–33. doi: 10.1007/s00334-020-00804-z. [DOI] [Google Scholar]
- 64.Jiang H, Zhang Y, Lü E, Wang C. Archaeobotanical evidence of plant utilization in the ancient Turpan of Xinjiang, China: A case study at the Shengjindian cemetery. Veg. Hist. Archaeobotany. 2015;24:165–177. doi: 10.1007/s00334-014-0495-6. [DOI] [Google Scholar]
- 65.Chomczynski P, Mackey K, Drews R, Wilfinger W. DNAzol®: A reagent for the rapid isolation of genomic DNA. Biotechniques. 1997;22:550–553. doi: 10.2144/97223pf01. [DOI] [PubMed] [Google Scholar]
- 66.Rohland N, Glocke I, Aximu-Petri A, Meyer M. Extraction of highly degraded DNA from ancient bones, teeth and sediments for high-throughput sequencing. Nat. Protoc. 2018;13:2447–2461. doi: 10.1038/s41596-018-0050-5. [DOI] [PubMed] [Google Scholar]
- 67.Richards SM, et al. Low-cost cross-taxon enrichment of mitochondrial DNA using in-house synthesised RNA probes. PLoS ONE. 2019;14:e0209499. doi: 10.1371/journal.pone.0209499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schubert M, Lindgreen S, Orlando L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC. Res. Notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schubert M, et al. Improving ancient DNA read mapping against modern reference genomes. BMC Genomics. 2012;13:178. doi: 10.1186/1471-2164-13-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria, 2019).
- 73.Shen W, Le S, Li Y, Hu F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE. 2016;11:e0163962. doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scarcelli N, et al. Intra-individual polymorphism in chloroplasts from NGS data: Where does it come from and how to handle it? Mol. Ecol. Resour. 2016;16:434–445. doi: 10.1111/1755-0998.12462. [DOI] [PubMed] [Google Scholar]
- 75.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinform. (Oxf., Engl.) 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Edwards EJ. New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol. 2012;193:304–312. doi: 10.1111/j.1469-8137.2011.03972.x. [DOI] [PubMed] [Google Scholar]
- 78.Burke SV, et al. Evolutionary relationships in Panicoid grasses based on plastome phylogenomics (Panicoideae; Poaceae) BMC Plant Biol. 2016;16:140. doi: 10.1186/s12870-016-0823-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinform.s (Oxf., Engl.) 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kong S, Sánchez-Pacheco SJ, Murphy RW. On the use of median-joining networks in evolutionary biology. Cladistics. 2016;32:691–699. doi: 10.1111/cla.12147. [DOI] [PubMed] [Google Scholar]
- 85.Ramsey CB, Lee S. Recent and planned developments of the program OxCal. Radiocarbon. 2013;55:720–730. doi: 10.1017/S0033822200057878. [DOI] [Google Scholar]
- 86.Reimer PJ, et al. IntCal13 and Marine13 radiocarbon age calibration curves 0–50,000 years cal BP. Radiocarbon. 2013;55:1869–1887. doi: 10.2458/azu_js_rc.55.16947. [DOI] [Google Scholar]
- 87.Hintze JL, Nelson RD. Violin plots: A box plot-density trace synergism. Am. Stat. 1998;52:181–184. doi: 10.1080/00031305.1998.10480559. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data for the Areni-1 samples can be found at the NCBI website under BioProject: PRJNA575768 and the chloroplast sequencing data for the 50 modern broomcorn millet accessions are deposited at figshare: https://doi.org/10.6084/m9.figshare.14914605.