Abstract
Aechmea fasciata is one of the most popular bromeliads and bears a water-impounding tank with a vase-like rosette. The tank habit is a key innovation that has promoted diversity among bromeliads. To reveal the genomic basis of tank habit formation and ethylene-induced flowering, we sequenced the genome of A. fasciata and assembled 352 Mb of sequences into 24 chromosomes. Comparative genomic analysis showed that the chromosomes experienced at least two fissions and two fusions from the ancestral genome of A. fasciata and Ananas comosus. The gibberellin receptor gene GID1C-like was duplicated by a segmental duplication event. This duplication may affect GA signalling and promote rosette expansion, which may permit water-impounding tank formation. During ethylene-induced flowering, AfFTL2 expression is induced and targets the EIN3 binding site ‘ATGTAC’ by AfEIL1-like. The data provided here will serve as an important resource for studying the evolution and mechanisms underlying flowering time regulation in bromeliads.
Subject terms: Plant evolution, Flowering, Genome
The genome of <i>Aechmea fasciata <i > , a common house plant and popular bromelioid provides insights into its evolution.
Introduction
Aechmea fasciata, also called the urn plant or silver vase, is a common house plant and popular bromeliad. Bromeliaceae contains more than 75 genera and more than 3500 species and is one of the largest families of flowering plants found in the neotropics1–3. The chromosome number of bromeliads varies little, with most species of Bromelioideae, Puyoideae, Tillandsioideae and Pitcairnioideae being diploid at 2n = 50 and some Tillandsioideae being diploid at 2n = 484,5.One of the most diverse clades from Bromeliaceae, core bromelioids often use CAM photosynthesis and have absorptive foliar trichomes and ‘tanks’ formed by closely overlapping bases of rosette leaves to impound water and detritus, allowing these plants to absorb water and nutrients on epiphytic perches and rocks and adapt to arid regions1,6–8.
The tank habit, CAM photosynthesis, absorptive leaf trichomes, epiphytism and avian pollination are thought to be the key innovations that have allowed bromeliads to spread into the treetops of rainforests, semiarid and arid regions and microsites and become highly diversified2,9,10. The tank habit and CAM photosynthesis arose several times independently within the family; for Bromelioideae, CAM photosynthesis arose at the base of Bromelioideae-Puyoideae ca. 10.7 Mya in the Andes/central Chile;11,12 later, the tank habit coincided with epiphytism, which arose in Bromelioideae ca. 5.6 Mya in the Atlantic Forest region8. The tank epiphytic bromelioids had the highest net diversification in Bromeliaceae because CAM photosynthesis was associated with a twofold increase in speciation rate in the tank habit that was five times lower than that in tank-less habits9. Whereas Bromelioideae contains 33 genera and ca. 800 species, tank-less Ananas contains 7 species, but tank-habit Aechmea contains 260 species13. However, the genomic basis of these functional traits is poorly understood.
Ethylene-induced flowering is a remarkable feature of Bromeliaceae species and is exploited worldwide to promote flowering synchronization14. The flowering synchronization is of critical importance in Ananas comosus var. comosus and other ornamental bromeliads cultivation due to the occurrence of natural flowering out of season can cause serious scheduling problems for growers. Although ethylene promotes flower induction in Bromeliaceae species, ethylene commonly delay flowering in many plant species, including rice15, pharbitis16 and Arabidopsis17. Ethylene signalling modulates GA–DELLA signalling pathways that delay flowering by repressing the expression of flowering integrator genes such as FLOWERING LOCUS T (FT) and SUPPRESSOR OF CONSTANS OVEREXPRESSION 1 (SOC1) and the floral meristem identity gene LFY18,19. As an exceptional case, ethylene signalling activated the expression of FT in Ananas comosus var. comosus20 and A. fasciata21. But the molecular mechanism underlying ethylene-induced flowering in bromeliads remains unexplored.
The publishing of the genome of the famous fruit crop A. comosus var. comosus and A. comosus var. bracteatus provides the possibility to study the genomic basis of divergence between A. fasciata and A. comosus and the rise of tank epiphytic habits22,23. Here, we report the sequencing of A. fasciata. Ancestral genome construction and comparative genome analysis show the evolution of chromosome numbers and the loss of orthologous sets. In A. fasciata, the gibberellin receptor gene GID1C-like was duplicated; along with the insertion of 14 and 27 amino acids and multiple nonsynonymous mutations in the duplicated gene pairs relative to AcGID1C-like due to a segmental duplication event followed by mutation, it may affect GA signalling and promote rosette expansion, which allow water-impounding tank formation. Through analysis of the evolution of the FT family, coupling ChIP-seq, ChIP-qPCR and Y1H, we hypothesized that AfEIL1 can directly bind upstream of AfFTL2 proteins and that AfFTL2 may be the key gene of ethylene-induced flowering. The resulting genomic information will be helpful for studying the evolution and mechanisms of flowering time regulation in bromeliads.
Results
Genome assembly and annotation
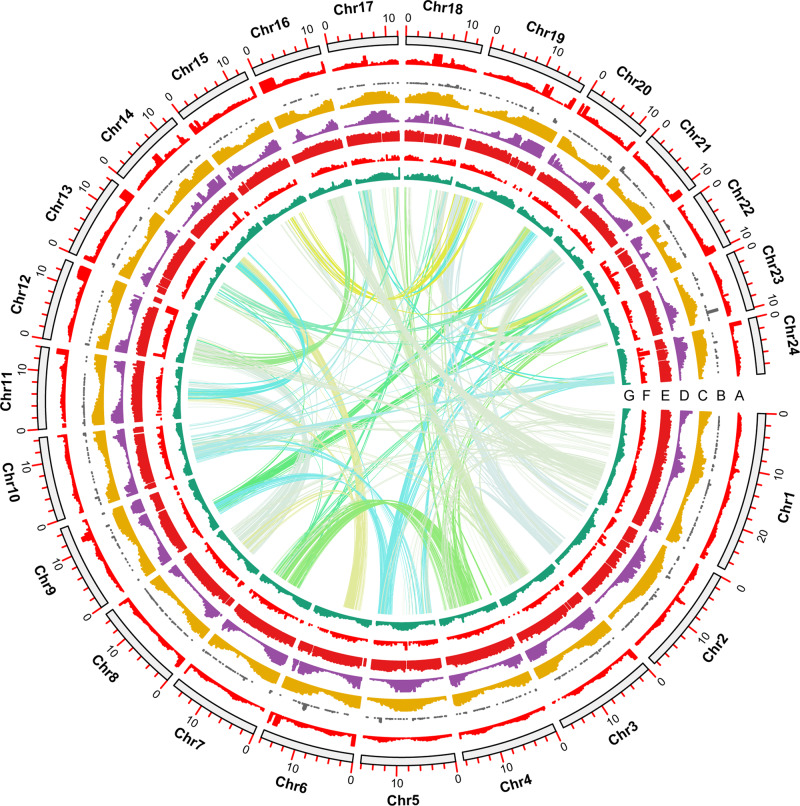
K-mer analysis of the A. fasciata genome estimated that the genome size was 359 Mb, with 1.15% heterozygosity (Supplementary Fig. 1). To overcome the impact of its high heterozygosity, we generated 40 Gb PacBio reads, 60 Gb Nanopore reads, and 158 Gb Illumina short reads for genome assembly. Using Nextdenovo (https://github.com/Nextomics/NextDenovo) and Nextpolish24, we constructed 528.98 Mb scaffold sequences, with an N50 of 4.68 Mb (Supplementary Table 1). After purging redundant sequences, 352 Mb sequences were used for Hi-C mapping (Supplementary Fig. 2). Then, we constructed 24 chromosomes with 347 Mb genome sequences, which was 96.66% of the estimated genome size (Fig. 1).
Fig. 1. Overview of the A. fasciata genome.
a DNA Retrotransposon coverage. b Pseudogene distribution density. c Repeat sequence coverage. d RNA retrotransposon coverage. e Gene expression level. f Gene density. g GC content. The genome features are shown in 1 Mb intervals. The gene expression levels were normalized to the RPKM values.
We processed RNA-seq data from roots, flowers, central leaves, mature leaves and central leaves under ethylene treatment to provide transcriptional evidence supporting the annotation and to obtain reliable gene structure annotation. Using the Marker2 pipeline25, we predicted the genes of the A. fasciata genome. Gene prediction produced 26,126 protein-coding genes (Supplementary Table 2), 348 tRNA genes and 104 rRNAs. BUSCO26 analysis shows that 93.4% of completed BUSCO orthologues were predicted (Supplementary Table 3). We also identified 1473 pseudogenes and predicted 1,239 transcription factors.
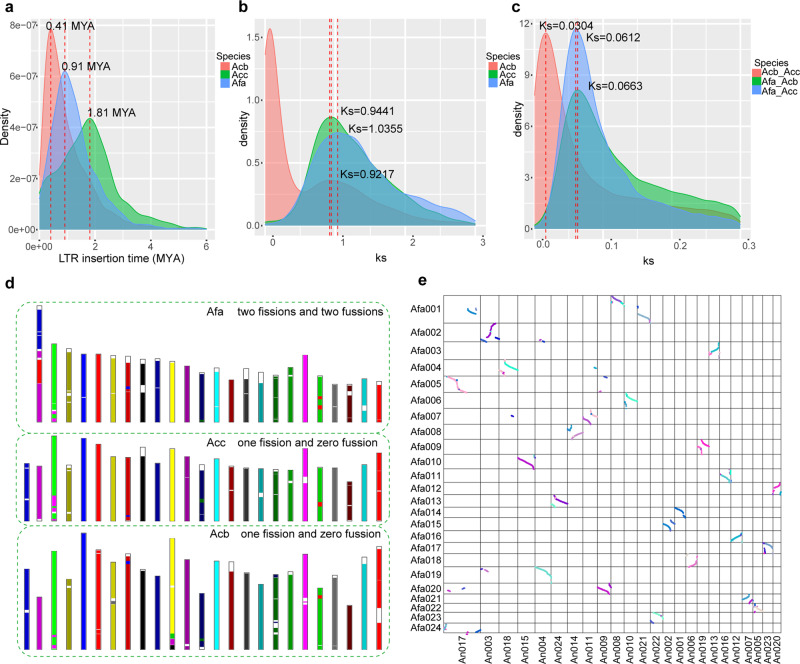
We used LTR-Retriver27 and BUSCO to estimate genome completeness. The LAI value was 13.94, and 98.4% of the BUSCO orthologues were completed, suggesting a highly complete and reference quality draft genome assembly (Supplementary Table 4). The repeat sequences consisted of 61.72% of the genome (Supplementary Table 5). A total of 0.63% consisted of transposon elements (TEs), and 58.15% of the genome consisted of retrotransposons. The LTR family Gypsy accounted for 24.31% of the genome. LTRs are thought to play a major role in the evolution of chromosome structure, specifically in repeat-dense regions such as centromeres. The LTR distribution of each chromosome was analysed, showing that LTRs were enriched near the ends of most chromosomes (18) and that a few (6) were enriched in the centre of the chromosomes (Supplementary Fig. 3). The LTR insertion time was analysed using LTR_retriever, suggesting that there was a burst of LTR insertions ca. 0.91 Mya (Fig. 2a).
Fig. 2. Genome evolution of A. fasciata, A. comosus var. comosus and A. comosus var. bracteatus.
a LTR insertion time estimation. b Ks distribution of Afa, Acc and Acb. c Ks distribution of Afa-Acc, Afa-Acb and Acb-Acc. d Bar plots of Afa, Acc and Acb with respect to the ancient genome. e Dot plot of the Afa and ancestral genomes. Afa: A. fasciata; Acc: A. comosus var. comosus; and Acb: A. comosus var. bracteatus.
Ancestral genome construction
We constructed the ancestral genomes of A. fasciata, A. comosus var. comosus and A. comosus var. bracteatus based on homologous genes detected by Orthofinder28 according to the BLAST results for proteins (Fig. 2d). The ancestral genome contains 24 ancestral chromosomes and 13,837 ordered protogenes, including 13,119 genes of A. comosus var. comosus and 12,311 genes of A. fasciata and 11,445 genes of A. comosus var. bracteatus. The 24 A. fasciata chromosomes had experienced at least two fissions and two fusions from the ancestral chromosomes, and A. comosus var. comosus and A. comosus var. bracteatus had experienced one fission and formed the current 25 chromosomes. Chr17 of the ancestral chromosomes fissioned into chr5 and chr10 in A. comosus var. comosus and into chr3 and chr11 in A. comosus var. bracteatus, while in A. fasciata, the ancestral chromosome chr17 fissioned into three parts, two of which formed chr5 and chr24, and the last part fused with the ancestral chromosomes chr8 and chr21 into the current chromosome chr1 (Fig. 2e, Supplementary Fig. 4 and Supplementary Fig. 5). Multiple translocations, inversions, and shifts were identified in the three genomes through synteny alignment with the ancestral genome. In particular, we identified three translocations of ancestral chr9, chr14 and chr21 into the 5’ of chr15, forming the current chr23 of A. comosus var. bracteatus.
Gene family expansion and contraction
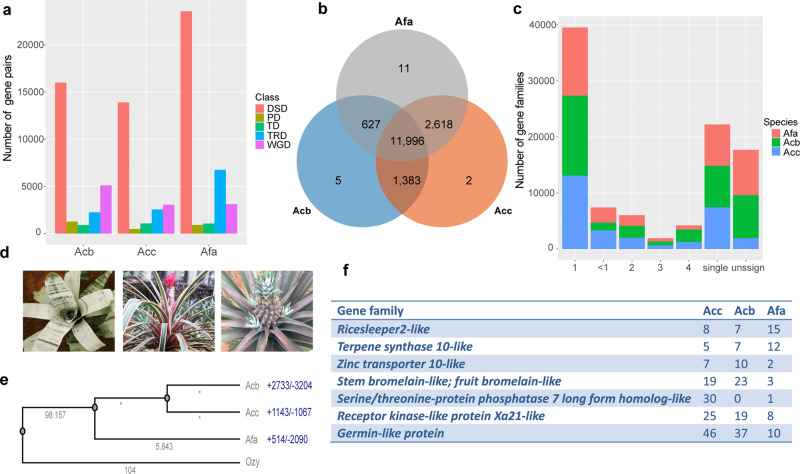
We identified 15,252 gene families (orthologue gene set) in A. fasciata, 15,999 in A. comosus var. comosus and 14,011 in A. comosus var. bracteatus, including 18,138, 20,235 and 20,699 genes, respectively. They shared 11,996 gene families, with 627, 1383 and 2618 gene families lost in A. comosus var. comosus, A. comosus var. bracteatus, and A. fasciata, respectively (Fig. 3b); of the gene families, 99.21%, 98.97% and 99.36% had gene family sizes less than 5 (Fig. 3c). There were 8,092 single copy genes in the three genomes. Compared with those of A. comosus var. comosus and A. comosus var. bracteatus, the number of gene families of size more than 1 was decreased significantly in A. fasciata. Using DupGeneFinder29, we predicted the duplicated gene pairs of the genome. As shown in Fig. 3a, A. fasciata harboured more duplicate gene pairs than A. comosus var. comosus and A. comosus var. bracteatus, with 3,112, 1,069, 914, 6757 and 23,537 whole genome duplication (WGD), tandem (TD), proximal (PD), transposed (TRD) and dispersed (DSD) gene pairs and 3062, 1072, 489, 2506 and 13,892 such pairs in A. comosus var. comosus, respectively.
Fig. 3. Gene family expansion and contraction estimation in A. fasciata, A. comosus var. comosus and A. comosus var. bracteatus.
a Duplicate gene pair distribution. b Venn plot of gene families. c Distribution of gene family size. d The tank-habit A. fasciata and tank-less pineapple and red pineapple. e Estimation of divergence time and gene family expansion and contraction. f significantly expanded or contracted gene families.
We also calculated the synonymous substitutions per synonymous site (Ks) of the three genomes. There were similar WGD peaks in the three genomes, corresponding to WGD event occurred 100– 120 million years ago;23 and the divergence peaks between A. comosus var. comosus and A. comosus var. bracteatus were significantly different from the peaks between A. fasciata and A. comosus var. comosus (Fig. 2b, c). Using the single-copy genes of A. fasciata, A. comosus var. comosus and A. comosus var. bracteatus and rice, we estimated the divergence time. Analysis showed that A. comosus var. comosus and A. fasciata diverged ca. 5.843 MYA (Fig. 3e), when the core-Bromeliaceae appeared in the Atlantic rainfall forest8. According to the estimated divergence time, we used Café30 to predict gene family expansion and contraction. There were 514, 1143, and 2733 expanded and 2090, 1067, and 3204 contracted gene families in A. fasciata, A. comosus var. comosus and A. comosus var. bracteatus, respectively.
Candidate genes involved in adaptation to new environments
Of the expanded families, several may be involved in the adaptation of A. comosus to new environments, including metabolism genes Bromelain and transporter-like genes zinc transporter 10 (ZIP10)-like and disease resistance proteins Receptor kinase-like protein Xa21 (XA21)-like, Germin-like protein (GLP) like and serine/threonine-protein kinase 7 long form homologue (MAIL3)-like genes (Fig. 3f). GLPs contribute to broad-spectrum diseases, and XA21 promotes plant innate immunity31,32. MAIL3 may be involved in the regulation of root and shoot development by maintaining cell division activity33.
The transcription factor families RICESLEEPER2-like and terpene synthase 10 (TPS10)-like were expanded in A. fasciata (Supplementary Fig. 6 and Supplementary Fig. 7). RICESLEEPER2 is essential for normal plant growth and development34. There were 8 RICESLEEPER2-like loci in the ancient genome of A. fasciata. Compared with A. comosus and A. comosus var. bracteatus, two clades of Ricesleeper2-like were expanded in A. fasciata due to the dispersed duplication event after speciation with A. comosus var. comosus. TPS10 is involved in defence against oomycete infection and indirect defence against herbivores by producing mixed signalling to attract natural enemies to plants35,36. The TPS10-like gene clusters contained 5, 7 and 12 genes in A. comosus var. comosus, A. comosus var. bracteatus and A. fasciata, respectively. One of the loci, a homologue of rna23024 of A. comosus var. comosus, was expanded to form a cluster with 9 genes.
Candidate genes for tank formation
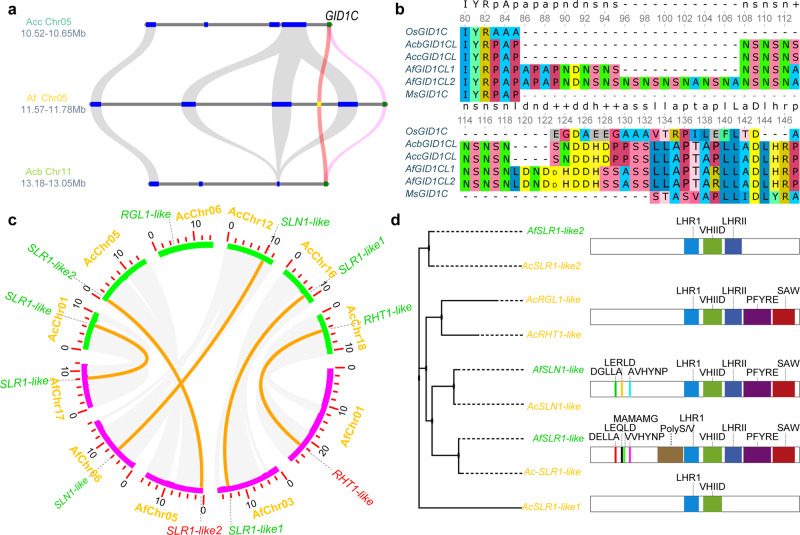
A. fasciata is tank-forming core bromelioids, while A. comosus var. comosus and A. comosus var. bracteatus are tank-less (Fig. 3d). The gibberellin (GA) receptor gene GID1C was duplicated in A. fasciata due to segmental tandem duplication in a region of chr05, which is a conserved syntenic region with that of chr05 of A. comosus var. comosus and chr11 of A. comosus var. bracteatus (Fig. 4a). In this region, the pentatricopeptide repeat-containing proteins At2g30780-like Af03915 and Af03914 and GID1C-like AfGID1CL1 (Af03916) and AfGID1CL2 (Af03917) were tandemly duplicated gene pairs. However, the duplicated GID1C gene pairs had experienced different mutations in sequence. There were 14 and 27 amino acid insertions in 86–130 amino acids of AfGID1CL2 and AfGID1CL1, respectively, and nonsynonymous mutations at positions 10, 139, 202, 226, 229, 249, 278, 355 and 391 relative to AcGID1C-like (Supplementary Fig. 8 and Fig. 4b). Gene expression analysis revealed that AfGID1CL1 and AfGID1CL2 were expressed in all plant organs and higher in the roots, and the expression level of AfGID1CL1 was one-fold higher than that of AfGID1CL2 (Supplementary Fig. 9). Overexpression of OsGID1C resulted in a GA hypersensitivity phenotype in transgenic rice37. Overexpression of GID1C-like in Arabidopsis resulted in the expansion of rosettes38,39. The tandem duplication of the GID1C-like gene increased its expression level and may improve GA sensitivity and promote rosette expansion in A. fasciata, which allows the formation of tanks with closely overlapping leaves. In addition, there were 3 and 6 homologues of DELLA in A. fasciata and A. comosus var. comosus, respectively, which were distributed in conserved syntenic blocks among the chromosomes (Fig. 4c). AfSLR1-like and AcSLR1-like have complete DELLA and GRAS domains but unique motifs, such as ‘LEQLD’, ‘MAMAMG’ and ‘VVHYNP’ (Fig. 4d), different from the previously described ‘LEQLE’,’MAMGM’ and ‘TVHYNP’ of angiosperm DELLA40. AfSLN1-like and AcSLN1-like have a ‘DGLLA’ motif and a unique ‘LERLD’ motif, which is also different from the described ‘LERLE’ motif of angiosperm ‘DGLLA’40. There is a specific ‘AAAAEVEEEGEEAAEE’ insertion in the GRAS domains of AfSLR1-like and AcSLR1-like (Supplementary Fig. 10). Moreover, four homologues lacked DELLA domains and partial GRAS domains in A. comosus var. comosus, including AcRGL1-like, AcRTH1-like, AcSLR1-like1 and AcSLR1-like2. In A. fasciata, the homologues of AcRGL1-like and AcRTH1-like became pseudogenes, and homologues of AcRGL1-like were lost.
Fig. 4. Duplication of GID1C-like and contraction of DGLLA-like in A. fasciata.
a Microsynteny block containing the duplicated GID1-likes among A. fasciata (Af), A. comosus var. comosus (Acc) and A. comosus var. bracteatus (Acb). b Portion of the multiple alignment of GID1C-like proteins; Os, Oryza sativa subsp. Japonica; Ms, Musa acuminata subsp. malaccensis; Acb, A. comosus var. bracteatus; Acc, A. comosus var. comosus; Af, A. fasciata. c The distribution and synteny of DELLA and DGLLA-like proteins between A. fasciata (Af) and A. comosus var. comosus (Acc). d The domain and polygenetic tree of A. fasciata (Af) and A. comosus var. comosus (Acc) DELLA and DGLLA-like proteins.
The FT family and ethylene-induced flowering
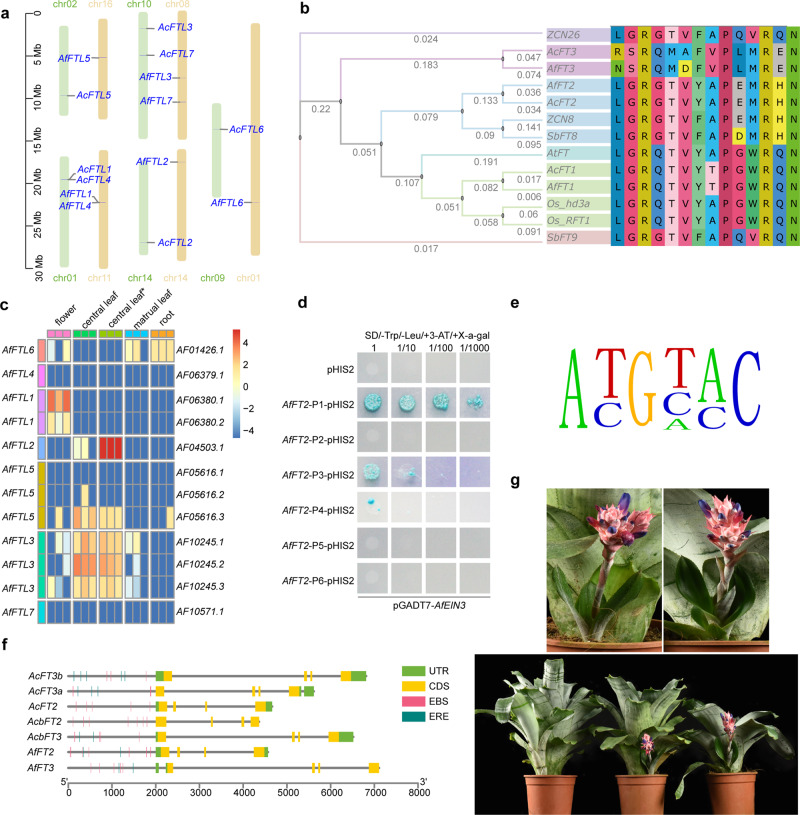
As shown in Fig. 5a, there are 7 FT-like genes in A. fasciata, distributed on 5 chromosomes. Compared with those in A. comosus var. comosus, the locations and gene structures of FT-likes in A. fasciata were conserved; FTL1 and FTL4 are tandemly duplicated genes in both A. fasciata and A. comosus var. comosus. The phylogenetic analysis of FTLs and analysis of amino acid residues showed that AfFTL2 and AcFTL2 are highly homologous to Zcn841,42 and SbFT843, which act as florigens in Zea mays and Sorghum bicolor, and harbour similar amide residues in segment B, which is the key amide residue for functioning as a florigen (Fig. 5b). FTL1 shares high similarity with HD3a and RFT1 but does not respond to ethylene treatment. FTL3 is a homologue of ZCN26, and Segment B of the FT proteins varies in different FTs. The two lines of 35 S::AfFTL2 plants flowered early, with one of the lines flowering only with three rosette leaves (Fig. 5g). Using RNA-seq, we analysed the expression of FT-like genes in roots, central leaves, mature leaves, flowers and central leaves under ethylene treatment: AfFTL4 and AfFTL7 were not expressed; AfFTL4 was mainly expressed in flowers; only AfFTL6 was expressed in roots; AfFTL2, AfFTL3, AfFTL5 and AfFTL6 were expressed in central leaves; and only AfFTL2 was dramatically induced by ethylene (Fig. 5c). To investigate the mechanism by which ethylene induces AfFTL2 expression, we conducted AfEIL1-like ChIP-seq and identified the core EIN3 binding motif ‘ATGTAC’ (Fig. 5e). ChIP-qPCR and Y1H assays validated the binding of EIN3 to AfFTL2 0–500 bp upstream of the promoters (Fig. 5d and Supplementary Table 6). We predicted the ethylene-related cis-elements EBS and ERE in the 2000 bp promoters of the genes (Fig. 5f). Both the FTL3 and FTL2 promoters harbour EBS, but AcFTL2 does not have ERE sites, suggesting that EBS is a conserved element upstream of the FTL2 promoters of A. fasciata and A. comosus var. comosus.
Fig. 5. FT-like genes are the key genes involved in ethylene-induced flowering in A. fasciata.
a, FT-like distribution in A. fasciata and A. comosus var. comosus. b Neighbour-joining tree and key amino acid residues in segment B of the fourth exon of FT-like proteins from Os (Oryza sativa subsp. japonica), Sb (Sorghum bicolor (L.) Moench), ZCN (Zea mays), At (Arabidopsis thaliana), Af (Aechmea fasciata) and Ac (Ananas comosus). c Heatmap of AfFTL expression levels in flowers, roots, central leaves, and central leaves* (central leaves 24 h after ethylene treatment). d Yeast one-hybrid (Y1H) assay showing AfEIL1-like binding to the promoter of AfFTL2. e EBS motif detected by ChIP-seq in A. fasciata. f Gene structures of AfFTL2, AfFTL3 and their paralogous genes in Acc (Ananas comosus (L.) Merr.) and Acb (Ananas comosus var. bracteatus); the EIN3 binding sites (EBS) and ethylene response elements (ERE) were detected within 2000-bp upstream of transcription initiation sites; UTR, untranslated region; CDS coding sequence. g Two lines of 35 S::AfFTL2 A. fasciata plants flowered early; bottom left, wild plants; bottom centre, 35 S::AfFTL2 line 1; bottom right, 35 S::AfFTL2 line 2; upper left, flower of 35 S::AfFTL2 line 1; and upper right, flower of 35 S::AfFTL2 line 2.
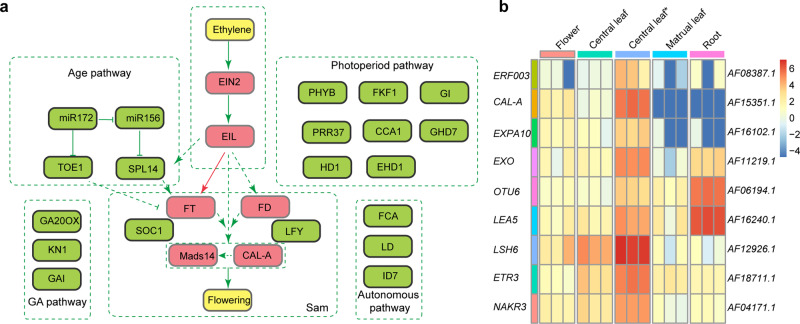
The A. fasciata homologues of five main flowering pathway genes in Arabidopsis and rice were identified by protein alignment. The flowering pathway genes were conserved in bromeliads relative to rice (Fig. 6a). The age pathway genes AfmiR156, AfmiR172, AfTOE1 and AfSPL14 were identified to be involved in A. fasciata flowering time44,45. However, in contrast to Arabidopsis and rice, there was an EIL1-dependent pathway to induce FTL2 expression and then downstream signalling genes. In addition, ethylene signalling also induced the expression of homologues of genes involved in plant development, including CAL-A, ERF3, ETR3, EXPA10, LEA5, NAKR3, and LSH6, which shared similar expression with AfFTL2 and synchronized flowering (Fig. 6b). LSH6 plays a critical role in synchronizing flowering46. NakR3 may play a role in the transport of FT proteins47. CAL-A functions as a floral meristem identity gene48. ETR3, ERF03, OTU6, and EXPA10 play roles in plant growth and varied developmental processes15,49,50.
Fig. 6. A putative flowering pathway in A. fasciata.
a Putative flowering pathway in A. fasciata. b Heatmap of putative genes synchronizing flowering in A. fasciata.
Discussion
Bromeliaceae arose in the Guayana Shield ca. 100 Mya1. The subfamily Bromelioideae is one of the most diverse clades in Bromeliaceae. The ancestral bromelioids were tank-less, coinciding with the formation of epiphytism, and the core-Bromeliodeae tank habit arose in the Atlantic Forest ca. 5.64 Mya8. Using rice as an outlier, we estimated that the divergence time between A. fasciata and rice was 104 Mya and that the divergence time between A. fasciata and A. comosus var. comosus was 5.843 Mya, which is close to the time of the tank-forming core-Bromeliodeae arising in the Atlantic Forest8. Although A. comosus var. bracteatus is a variety of pineapple, the introgression of A. macrodontes genes into A. comosus led to genetic differentiation between A. comosus var. bracteatus and A. comosus var. comosus51,52. This phenomenon allowed the construction of the ancestral genomes of A. comosus, A. comosus var. bracteatus and A. fasciata. Synteny alignment with the ancestral genome showed that A. comosus var. bracteatus experienced specific translocations on chr23. In addition to multiple translocations, inversions and shifts of chromosomes, A. comosus var. comosus and A. comosus var. bracteatus experienced one fusion, and A. fasciata experienced at least two fissions and two fusions, indicating that changes in genomic structure play key roles in tank epiphytism habit formation. Due to the chromosome fusions of ancestry chromosomes, A. fasciata has 48 (2n = 48) chromosomes and was validated by the root tip squash method (Supplementary Fig. 11). We also observed a karyotype with 2n = 50 due to breakage of chromosome 1 near the centromere, which may be the reason why previous reports described a karyotype with 2n = 50 chromosomes53. Transposon elements play key roles in the instability of the genome and genome evolution. In A. comosus var. comosus, 44% of the genome assembly consisted of TEs, and 33% consisted of LTRs, but 69% and 52% of the genome was composed of TEs and LTRs, respectively; in A. comosus var. bracteatus, 64.19% and 48.20% of the genome consisted of TEs and LTRs, respectively; and in A. fasciata, only 58.78% and 33.9% of the genome consisted of TEs and LTRs, respectively. According to the estimation of genome size, A. comosus var. comosus, A. comosus var. bracteatus and A. fasciata were 526, 591 and 359 Mb, respectively. Therefore, the main cause for the significantly smaller size of the A. fasciata genome relative to the A. comosus var. comosus and A. comosus var. bracteatus genomes was the lower insertion of TEs and LTRs. The loss of numerous genes, specifically duplicated genes, would be another reason for the significantly smaller size of the A. fasciata genome relative to that of A. comosus var. comosus and A. comosus var. bracteatus. A total of 1894 orthologue sets were lost in A. fasciata, and coupled with the loss of duplicated genes, the number of predicted protein-coding genes was 7,552 less than that of A. comosus var. bracteatus. It is supposed that WGD pairs were lost in the subsequent millions of years. The duplication of a single gene has a short half-life and occurs continuously, playing roles in key adaptations to environments54. According to the expansion and contraction analysis by Café30, 1975 gene families were contracted, and only 28 gene families were expanded. The expanded gene families may play key roles in the formation of tank-epiphytic habits in A. fasciata.
With the formation of tank habits, bromeliads invaded the treetops of rainforests. Especially in Atlantic forests of Brazil, they presented with extraordinary biodiversity. The tank habit allows bromeliads to impound and absorb water and nutrients through their leaves, not requiring the uptake of water and nutrients by roots, leading to their degeneration into anchorage. Leaves of most bromeliads are organized in stemless rosettes that allow the development of aquatic and terrestrial ecosystems55,56. The duplication of GID1C-like genes and contraction of DELLA-like genes in A. fasciata may promote rosette expansion by affecting GA signalling, which allowed the rosettes to overlap closely and impound water. GA is a hormone that regulates various developmental processes, such as stem elongation, leaf expansion, flowering, seed germination and pollen development57–59. The effect of GA on controlling leaf expansion has been studied widely60–64. In the classical GA signalling pathway, GA regulates the gene expression of various pathways by interacting with the receptor GID1 and destabilizing the DELLA protein, which are master regulators of the GA signalling pathway37,65. DELLA proteins have conserved eponymous DELLA motifs, followed by LEQLE, TVHYNP and MAMGM motifs in the N-terminus40. The GRAS domain is in the C-terminus and connects to these motifs by a poly S/T/V motif, which consists of five subdomains, including LHRI, LHRII, VHIID, PFYRE and SAW66. All the motifs and subdomains, except LHRI, were associated with the affinity of DELLA to GA preceptor GID140. The motifs in the N-terminus function in the transactivation of plant growth regulators;67 the motifs in the C-terminus are essential for the formation of the transactivation complex and sequestration of DELLA-interacting proteins; specifically, the SAW motif plays a key role in the suppressive activity of DELLA68,69. DELLA functions as a master regulator in the trade-off between plant growth and various adverse environmental conditions. AfSLR1-like and AcSLR1-like proteins have unique LEQLD, MAMAMG and VVHYNP motifs in the N-terminus and AAAAEVEEEGEEAAEE insertions in the GRAS domain, which are different from previously reported conserved motifs in DELLA proteins, suggesting that AfSLR1-like and AfSLR1-like may regulate different growth suppressors and DELLA-interacting proteins and play a pivotal role during adaptation to specific environments in bromeliads. In A. comosus var. comosus and A. fasciata, there were 5 and 2 DGLLA genes, respectively, which were DELLA-related proteins with DGLLA motifs instead of DELLA motifs69. DGLLA homologues function as back-up GA-insensitive negative regulators of plant growth under specific conditions70. The contraction of DGLLA homologues may promote the plant growth in A. fasciata. In conclusion, the duplication of GID1-like genes and contraction of DGLLA-like genes may play key roles in the formation of tank habits. Transposase-like genes of the HAT (hobo, activator, Tam3) family are essential for plant growth in rice and Arabidopsis71. Ricesleeper-overexpressing Arabidopsis plants showed delayed inflorescence development, increased rosette leaf number and irregularity, dwarfism and fasciation. Ricesleeper-overexpressing rice plants were also dwarfed34. Compared with A. comosus var. comosus and A. comosus var. bracteatus, A. fasciata contained 8 original Ricesleeper2 loci, 2 of which had expanded. The expansion of ricesleeper2-like may also be associated with tank epiphytic habit formation in A. fasciata. In addition, Key innovations could significantly decrease the extinction rate by improving the dispersion ability, helping the plants to invade new regions or increasing defences against herbivores9. TPS10 is involved in the indirect defence against lepidopteran larvae by producing a mixture that attracts natural enemies to plants being fed upon35. TPS10 is also induced by oomycete infection in Medicago truncatula36. The expansion of AfTPS10-like gene clusters improved the defence against herbivores and pathogens and may play a critical role in adaptation to environments. Tank-less bromeliads often inhabit semiarid or arid regions, where they lie at the limit of physiological tolerance, and most other plants fail to survive. The expansion of AcMAIL3-like may play a key role in the development of pineapple roots. The expansion of ZIP10-like may help pineapple adapt to infertile environments.
FT genes are highly conserved in sequence and function in angiosperms. FT genes regulate plant growth and multiple developmental processes, including flowering, expansion of storage organs, root development, flower development, stomatal movement and so on. In previous studies, AFTL1 was cloned and promoted flowering in Arabidopsis. As homologues of HD3A, FTL1 was expressed in flowers in both pineapple and A. fasciata and may play a role in flower development. FTL3 was induced by ethylene in pineapple but not in A. fasciata. FTL3 is a homologue of SbFT9, is expressed in leaves and may play a role in leaf development. AfFTL6 was expressed in the roots of A. fasciata, indicating that FTL6 may be involved in root development. Of the FTLs, only FTL2 was induced by ethylene and had the same amino acids in segment B as ZCN8 and SbFT8, indicating that FTL2 regulates flowering in pineapple and A. fasciata. Plants need to flower under optimal conditions to ensure successful reproduction. Therefore, plants have evolved a complex mechanism to integrate endogenous and environmental cues to flower at optimal times. Pineapple flowers naturally under cool night temperatures and short days72, as does A. fasciata. It is thought that pineapple flowering is induced by a burst of endogenous ethylene controlled by environmental cues73. The ethylene synthase inhibitor amino vinylglycine could delay the flowering of pineapple74. Silencing AcAcs2, a critical ethylene biosynthesis gene, also delayed pineapple flowering73. In A. fasciata, AfEIL1-like could induce the expression of AfFTL2 directly, indicating that there is an EIL1-dependent flowering pathway in pineapple and A. fasciata. Moreover, the homologues of CAL-A, LSH6, ERF3, NAKR3, etc. were induced by ethylene, similar to pineapple, indicating that there is a complex and conserved mechanism regulating flowering in bromeliads. DELLA functions as a flowering repressor and plays essential roles in ethylene delayed flowering by reducing bioactive GA levels and increasing DELLA accumulation18. In pineapple, ethylene treatment also reduced the bioactive GA level75. Ethylene-induced flowering was independent of DELLA-mediated flowering repression, which may be due to the unique motifs of AfSLR1-like and AcSLR1-like. Further research on the mechanism by which ethylene induces flowering is needed to facilitate plantation management and culture of new varieties.
In summary, we reported the genome sequences of A. fasciata, a popular ornamental tropical plant, using Nanopore, PacBio, Illumina and Hi-C sequencing to reach a high level of completeness and accuracy of the genome. The genome sequences of A. fasciata will facilitate further research on bromeliad evolution and the genomic basis of tank habit formation of tank bromeliads and ethylene-induced flowering.
Methods
Sampling, sequencing and assembly
The A. fasciata plants for genome sequencing were sampled in a greenhouse of the national gene bank of tropical crops in Danzhou, Hainan, China. The genomic DNA of seedlings was extracted for genomic library construction. For Illumina sequencing, libraries with 350-bp insertions were constructed; for PacBio sequencing, 5 libraries with 20k-bp insertions were constructed and sequenced on the PacBio RS II system with P6-C4 chemistry; for Nanopore single-molecule sequencing, libraries with high molecular weight genomic DNA were constructed on PromethION. In total, 60,191,465,203 bp and 2,177,689 reads were produced by Nanopore single-molecule sequencing, with an average length of 27,640.06, a maximum length of 265,723 and an N50 of 33,856. There were 158 Gb Illumina short reads produced. There were 42,180,646,315 bp and 4,498,100 PacBio reads with an average length of 9377.4 bp and a maximum length of 53,937 bp.
Hi-C libraries were created from tender leaves of A. fasciata at BioMarker Technologies Company as described previously. Briefly, the leaves were fixed with formaldehyde and lysed, and then the cross-linked DNA was digested with HindIII overnight. Sticky ends were biotinylated and proximity-ligated to form chimeric junctions that were enriched and then physically sheared to a size of 500–700 bp. Chimeric fragments representing the original cross-linked long-distance physical interactions were then processed into paired-end sequencing libraries, and 1,001 million 150-bp paired-end reads were produced on the Illumina HiSeq X Ten platform.
For RNA-seq, total RNA was extracted from central leaves, roots, flowers and central leaves under ethylene treatment for 24 h. After removing genomic DNA using DNase I (Takara), mRNAs were obtained using oligo (dT) beads and broken into short fragments, followed by cDNA synthesis. Paired-end sequencing was conducted on the HiSeq X Ten platform (Illumina, CA, USA).
Genome assembly and annotation
Using GenomeScope76 for k-mer analysis, the genome size of A. fasciata was estimated. PacBio reads were first self-corrected with the parameter corOutCoverage = 100. The corrected reads, along with NanoPore long reads, were imported for assembly by Nextdenovo v2.3.1 (https://github.com/Nextomics/NextDenovo) and NextPolish (https://github.com/Nextomics/NextPolish) using the default parameters. Then, the redundant sequences of the polished contig sequences were eliminated by Purge Haplotigs77. Chromosomal assembly was performed based on proximity-guided assembly using ALLHIC78.
A de novo repeat library of the genome was customized using RepeatModeler79, which can automatically execute two de novo repeat finding programs, RECON (version 1.08) and RepeatScout (version 1.0.5). Consensus transposable element (TE) sequences generated above were imported to RepeatMasker (version 4.05) to identify and cluster repetitive elements. Unknown TEs were further classified using TEclass (version 2.1.3). To identify tandem repeats within the genome, the Tandem Repeat Finder (TRF) package (version 4.07) was used with the modified parameters of ‘1 1 2 80 5 200 2,000 –d –h’ to find high-order repeats.
To further investigate LTRs, we applied the LTR_retriever pipeline27, which can integrate results from public programs such as LTR_FINDER and LTRharvest and efficiently remove false positives from the initial predictions. The predicted LTRs were further classified into intact and nonintact LTRs, and the insertion time was estimated as T = K/2μ (K is the divergence rate, and μ is the neutral mutation rate; the default is 1.38 × 10 − 8 in LTR_retriever) using the scripts implemented in the LTR_retriever package. Genome completeness was assessed with BUSCO v5.2.226.
Gene annotations were processed with MAKER225. Two rounds of MAKER2 processing were used to achieve high-quality gene annotation. The RNA-seq reads were imported to Trinity, genome-guided and de novo assembled with default parameters. Then, the combined reads were imported to the PASA pipeline80. The PASA assembled transcripts were used to train the ab initio predictors SNAP81, GENEMARK82 and AUGUSTUS83. After that, MAKER2 processed the first gene annotation. After filtering the values of predicted proteins by MAKER2, the ab initio predictors were trained again. BRAKER84 were used for predicting genes using aligned RNA-seq reads as input. Then, using the transcripts processed by Stringtie85 as input and gene models predicted by BRAKER, the MAKER2 pipeline was run again. Using the GenblastA86 pipeline and Genewise87, we also predicted pseudogenes. PlantTFDB88 was used for transcription factor prediction. The A. comosus var. bracteatus genomic data were downloaded from EBI-ENA (PRJEB33121)89. The A. comosus var. comosus genomic data were downloaded from NCBI (ASM154086v1)90.
Genome structure and evolution
Orthofinder28 was used for the identification of putative paralogous and orthologous genes from A. fasciata and other species. Because the coding genes have one more transcript, we retained the longest transcripts. Based on homology and collinearity, we investigated the paleohistory of A. fasciata, A. comosus var. comosus and A. comosus var. bracteatus. After alignment of coding genes using BLAST, we identified putative protogenes (pPGs). Then, the pPGs, ordered by location on the chromosome, were imported to MGRA291. After analysis of gene gain/loss, the ancestral genome was reconstructed with an exhaustive set of ordered protogenes (oPGs). Then, the oPGs were used for collinearity analysis by MCscanX92.
ChIP-seq and ChIP-qPCR
Chromatin immunoprecipitation assays were performed as previously described93. Briefly, 3 g of sample was washed twice in cold PBS buffer, and proteins were cross-linked to DNA by incubating the samples with formaldehyde at a final concentration of 1%. Afterwards, samples were lysed, and chromatin was obtained on ice. Chromatins were sonicated to obtain soluble sheared chromatin (average DNA length of 200–500 bp). One part of the soluble chromatin was stored at –20 °C for input DNA, and the remainder was used for immunoprecipitation by the antibodies AfEIL1-like and normal rabbit IgG (CST, 2729). Immunoprecipitated DNA was amplified by PCR using specific primers. All primers are listed in Supplementary Table 6.
To construct ChIP-Seq libraries, the resulting ChIP DNA above was used to generate a sequencing library according to the Illumina ChIP-Seq manufacturer’s instructions. Then, an Illumina Genome Analyser II (Illumina, San Diego, CA) was used for sequencing according to the manufacturer’s instructions.
Y1H assay
The yeast one-hybrid assay was performed as previously described45. The AfEIL1-like sequences were inserted into the pGADT7 vector (Clontech, USA) to construct pGANDT7-AfEIL1-like vectors. For Y1H cDNA library screening, the 200-bp promoter fragment AfFTL2 was cloned into the destination vector pGADT7. The growth of the transformants on SD-His-Ura-Trp medium was considered an indicator of AfEIL1-like binding to the corresponding DNA fragments.
Transgenic plants
The coding sequence of AfFTL2 was cloned into the binary vector Cam35S under the control of the CaMV35S promoter. The confirmed construct was transformed into A. fasciata using agrobacterium strain GV3101 with the leaf base dip method. Transgenic plants were verified by PCR to detect exogenous genes and qRT–PCR to detect the expression of AfFTL2.
Statistics and reproducibility
Statistical analyses were performed using the s software R (version 3.6.3). The significance level was typically set to 0.05 for all the statistical analyses. The RNA-seq, ChIP-seq and ChIP-qPCR experiments were performed with three biological replicates and the merged tissues of three plants were sampled for each treatment.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This work is financially supported by the National Key Research and Development Plan Program (No. 2021YFC2600603), the National Natural Science Foundation of China (No. 31372106), the Natural Science Foundation of Hainan Province of China (No. 319MS082) and the Central Public Interest Scientific Institution Basal Research Fund (No. 1630032016006).
Author contributions
L.X. and Z.Y.L. designed the research; Z.Y.L., J.B.W., Y.L.J, B.L.H., Y.L.F., and X.B.W. performed the experiments, L.X., Z.Y.L. and J.B.W. analysed the data; Q.Q.Y. and C.Y.M. supplied the materials; J.B.W. and Z.Y.L. wrote the manuscript, and L.X., X.B.Z. and G.H.Z. revised the manuscript. All authors critically read and approved the final version of the manuscript.
Peer review
Peer review information
Communications Biology thanks Xingtan Zhang, Clarisse Palma-Silva and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Diane Saunders and Caitlin Karniski. Peer reviewer reports are available.
Data availability
The Aechmea fasciata genome sequences and raw sequence data from RNA-seq and genome sequencing have been deposited under BioProject accession number PRJNA74855794. The AfEIL1-like ChIP-seq data were deposited under GEO accession number GSE20579995.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhiying Li, Jiabin Wang.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-022-03918-4.
References
- 1.Givnish TJ, et al. Phylogeny, adaptive radiation, and historical biogeography in Bromeliaceae: insights from an eight-locus plastid phylogeny. Am. J. Bot. 2011;98:872–895. doi: 10.3732/ajb.1000059. [DOI] [PubMed] [Google Scholar]
- 2.Crayn DM, Winter K, Jac S. Multiple origins of crassulacean acid metabolism and the epiphytic habit in the Neotropical family Bromeliaceae. Proc. Natl Acad. Sci. 2004;101:3703–3708. doi: 10.1073/pnas.0400366101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gouda, E. J., Butcher, D. & Gouda, C. S. In Encyclopaedia of Bromeliads, Version 4. Utrecht University Botanic Gardens. http://bromeliad.nl/encyclopedia/(accessed: [19-08-2022]), (2018).
- 4.Gitaí J, Paule J, Zizka G, Schulte K, Benko-Iseppon AM. Chromosome numbers and DNA content in Bromeliaceae: additional data and critical review. Botanical J. Linn. Soc. 2015;176:349–368. doi: 10.1111/boj.12211. [DOI] [Google Scholar]
- 5.Zanella CM, et al. Genetics, evolution and conservation of Bromeliaceae. Genet. Mol. Biol. 2012;35:1020–1026. doi: 10.1590/S1415-47572012000600017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Givnish TJ, Millam KC, Berry P, Sytsma KJ. Phylogeny, Adaptive Radiation, and Historical Biogeography of Bromeliaceae Inferred from ndhF Sequence Data. Aliso A J. Syst. Evolut. Bot. 2007;23:3–26. [Google Scholar]
- 7.Schulte K, Barfuss M, Zizka G. Phylogeny of Bromelioideae (Bromeliaceae) inferred from nuclear and plastid DNA loci reveals the evolution of the tank habit within the subfamily. Mol. Phylogenetics Evolution. 2009;51:327–339. doi: 10.1016/j.ympev.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Givnish TJ, et al. Adaptive radiation, correlated and contingent evolution, and net species diversification in Bromeliaceae. Mol. phylogenetics evolution. 2014;71:55–78. doi: 10.1016/j.ympev.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 9.Silvestro D, Zizka G, Schulte K. Disentangling the effects of key innovations on the diversification of Bromelioideae (Bromeliaceae) Evolution. 2014;68:163–175. doi: 10.1111/evo.12236. [DOI] [PubMed] [Google Scholar]
- 10.Crayn DM, Winter K, Schulte K, Smith JAC. Photosynthetic pathways in Bromeliaceae: phylogenetic and ecological significance of CAM and C3 based on carbon isotope ratios for 1893 species. Botanical J. Linn. Soc. 2015;178:169–221. doi: 10.1111/boj.12275. [DOI] [Google Scholar]
- 11.Kürschner WM, Kvaček Z, Dilcher DL. The impact of Miocene atmospheric carbon dioxide fluctuations on climate and the evolution of terrestrial ecosystems. Proc. Natl Acad. Sci. 2008;105:449–453. doi: 10.1073/pnas.0708588105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winter, K. & Smith, J. Crassulacean Acid Metabolism: Current Status and Perspectives, (Springer, Berlin, Heidelberg, 1996).
- 13.Evans TM, et al. Phylogenetic Relationships in Bromeliaceae Subfamily Bromelioideae based on Chloroplast DNA Sequence Data. Syst. Bot. 2015;40:116–128. doi: 10.1600/036364415X686413. [DOI] [Google Scholar]
- 14.Poel BVD, Ceusters J, Proft MPD. Determination of pineapple (Ananas comosus, MD-2 hybrid cultivar) plant maturity, the efficiency of flowering induction agents and the use of activated carbon. Sci. Horticulturae. 2009;120:58–63. doi: 10.1016/j.scienta.2008.09.014. [DOI] [Google Scholar]
- 15.Wuriyanghan H, et al. The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice. Plant Cell. 2009;21:1473–1494. doi: 10.1105/tpc.108.065391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kęsy J, et al. The possible role of PnACS2 in IAA-mediated flower inhibition in Pharbitis nil. Plant growth Regul. 2010;61:1–10. doi: 10.1007/s10725-010-9443-3. [DOI] [Google Scholar]
- 17.Zemlyanskaya EV, Levitsky VG, Oshchepkov DY, Ivo G, Mironova VV. The Interplay of Chromatin Landscape and DNA-Binding Context Suggests Distinct Modes of EIN3 Regulation in Arabidopsis thaliana. Front. Plant Sci. 2016;7:2044. doi: 10.3389/fpls.2016.02044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Achard P, et al. The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl Acad. Sci. 2007;104:6484–6489. doi: 10.1073/pnas.0610717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukazawa J, Ohashi Y, Takahashi R, Nakai K, Takahashi Y. DELLA degradation by gibberellin promotes flowering via GAF1-TPR-dependent repression of floral repressors in Arabidopsis. Plant Cell. 2021;33:2258–2272. doi: 10.1093/plcell/koab102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, J. et al. Integrated DNA methylome and transcriptome analysis reveals the ethylene-induced flowering pathway genes in pineapple. Sci. Rep.7, 17167 (2017). [DOI] [PMC free article] [PubMed]
- 21.Li Z, et al. Transcriptome sequencing determined flowering pathway genes in Aechmea fasciata treated with ethylene. J. Plant Growth Regul. 2016;35:316–329. doi: 10.1007/s00344-015-9535-4. [DOI] [Google Scholar]
- 22.Chen LY, Vanburen R, Paris M, Zhou H, Ming R. The bracteatus pineapple genome and domestication of clonally propagated crops. Nat. Genet. 2019;51:1–10. doi: 10.1038/s41588-018-0328-0. [DOI] [PubMed] [Google Scholar]
- 23.Ming R, et al. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 2015;47:1435–1442. doi: 10.1038/ng.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu J, Fan J, Sun Z, Liu S. NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics. 2020;36:2253–2255. doi: 10.1093/bioinformatics/btz891. [DOI] [PubMed] [Google Scholar]
- 25.Cantarel BL, et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 27.Ou S, Jiang N. LTR_retriever: A highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 2018;176:1410–1422. doi: 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:1–14. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiao X, et al. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 2019;20:1–23. doi: 10.1186/s13059-019-1650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han MV, Thomas GWC, Jose LM, Hahn MW. Estimating Gene Gain and Loss Rates in the Presence of Error in Genome Assembly and Annotation Using CAFE3. Mol. Biol. Evolution. 2013;30:1987–1997. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
- 31.Manosalva PM, et al. A Germin-Like Protein Gene Family Functions as a Complex Quantitative Trait Locus Conferring Broad-Spectrum Disease Resistance in Rice. Plant Physiol. 2009;149:286–296. doi: 10.1104/pp.108.128348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, et al. A conserved threonine residue in the juxtamembrane domain of the XA21 pattern recognition receptor is critical for kinase autophosphorylation and XA21-mediated immunity. J. Biol. Chem. 2010;285:10454–10463. doi: 10.1074/jbc.M109.093427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.ühlken C, Horvath B, Stadler R, Sauer N, Weingartner M. MAIN-LIKE1 is a crucial factor for correct cell division and differentiation in Arabidopsis thaliana. Plant J. 2014;78:107–120. doi: 10.1111/tpj.12455. [DOI] [PubMed] [Google Scholar]
- 34.Knip M, de Pater S, Hooykaas PJ. The SLEEPER genes: a transposase-derived angiosperm-specific gene family. BMC plant Biol. 2012;12:1–15. doi: 10.1186/1471-2229-12-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Köllner TG, Gershenzon J, Degenhardt J. Molecular and biochemical evolution of maize terpene synthase 10, an enzyme of indirect defense. Phytochemistry. 2009;70:1139–1145. doi: 10.1016/j.phytochem.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 36.Yadav H, Dreher D, Athmer B, Porzel A, Hause B. Medicago TERPENE SYNTHASE 10 is involved in defense against an oomycete root pathogen. Plant Physiol. 2019;180:1598–1613. doi: 10.1104/pp.19.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueguchi-Tanaka M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- 38.Griffiths J, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell. 2006;18:3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mauriat M, Moritz T. Analyses of GA20ox- and GID1-overexpressing aspen suggest that gibberellins play two distinct roles in wood formation. Plant J. 2009;58:989–1003. doi: 10.1111/j.1365-313X.2009.03836.x. [DOI] [PubMed] [Google Scholar]
- 40.Van De Velde K, Ruelens P, Geuten K, Rohde A, Van Der Straeten D. Exploiting DELLA Signaling in Cereals. Trends Plant Sci. 2017;22:880–893. doi: 10.1016/j.tplants.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 41.Meng X, Muszynski MG, Da Nilevskaya ON. The FT-Like ZCN8 Gene Functions as a Floral Activator and Is Involved in Photoperiod Sensitivity in Maize. Plant Cell. 2011;23:942–960. doi: 10.1105/tpc.110.081406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo L, et al. Stepwise cis-regulatory changes in ZCN8 contribute to maize flowering-time adaptation. Curr. Biol. 2018;28:3005–3015. doi: 10.1016/j.cub.2018.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, et al. Three FLOWERING LOCUS T-like genes function as potential florigens and mediate photoperiod response in sorghum. N. Phytologist. 2016;210:946–959. doi: 10.1111/nph.13834. [DOI] [PubMed] [Google Scholar]
- 44.Lei M, et al. AfAP2-1, An age-dependent gene of Aechmea fasciata, responds to exogenous ethylene treatment. Int. J. Mol. Sci. 2016;17:303. doi: 10.3390/ijms17030303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ming L, et al. Constitutive Expression of Aechmea fasciata SPL14 (AfSPL14) Accelerates Flowering and Changes the Plant Architecture in Arabidopsis. Int. J. Mol. ences. 2018;19:2085. doi: 10.3390/ijms19072085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macalister C, et al. Synchronization of the flowering transition by the tomato TERMINATING FLOWER gene. Nat. Genet. 2012;44:1393–1398. doi: 10.1038/ng.2465. [DOI] [PubMed] [Google Scholar]
- 47.Zhu Y, Liu L, Shen L, Yu H. NaKR1 regulates long-distance movement of FLOWERING LOCUS T in Arabidopsis. Nat. Plants. 2016;2:1–10. doi: 10.1038/nplants.2016.75. [DOI] [PubMed] [Google Scholar]
- 48.Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development. 2000;127:725–734. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- 49.Marowa P, Ding A, Kong Y. Expansins: roles in plant growth and potential applications in crop improvement. Plant cell Rep. 2016;35:949–965. doi: 10.1007/s00299-016-1948-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Radjacommare R, Usharani R, Kuo C-H, Fu H. Distinct phylogenetic relationships and biochemical properties of Arabidopsis ovarian tumor-related deubiquitinases support their functional differentiation. Front. plant Sci. 2014;5:84. doi: 10.3389/fpls.2014.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.d’Eeckenbrugge GC, Govaerts R. Synonymies in Ananas (Bromeliaceae) Phytotaxa. 2015;239:273–279. doi: 10.11646/phytotaxa.239.3.8. [DOI] [Google Scholar]
- 52.Duval M-F, et al. Relationships in Ananas and other related genera using chloroplast DNA restriction site variation. Genome. 2003;46:990–1004. doi: 10.1139/g03-074. [DOI] [PubMed] [Google Scholar]
- 53.Marchant CJ. Chromosome Evolution in the Bromeliaceae. Kew Bull. 1967;21:161–168. doi: 10.2307/4108461. [DOI] [Google Scholar]
- 54.Lynch M, Conery JS. The Evolutionary Fate and Consequences of Duplicate Genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 55.Gonçalves AZ, Mercier H, Oliveira RS, Romero GQ. Trade-off between soluble protein production and nutritional storage in Bromeliaceae. Ann. Bot. 2016;118:1199–1208. doi: 10.1093/aob/mcw174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ngai JT, Srivastava DS. Predators Accelerate Nutrient Cycling in a Bromeliad Ecosystem. Science. 2006;314:963–963. doi: 10.1126/science.1132598. [DOI] [PubMed] [Google Scholar]
- 57.Olszewski N, Sun TP, Gubler F. Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell. 2002;14:S61–S80. doi: 10.1105/tpc.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hedden P, Sponsel V. A Century of Gibberellin Research. J. Plant Growth Regul. 2015;34:740–760. doi: 10.1007/s00344-015-9546-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Daviere JM, Achard P. Gibberellin signaling in plants. Development. 2013;140:1147–1151. doi: 10.1242/dev.087650. [DOI] [PubMed] [Google Scholar]
- 60.Wheeler AW, Humphries EC. Separation of the Effects of Gibberellic Acid on Leaf and Stem Growth of Dwarf French Bean. Nature. 1964;202:616–616. doi: 10.1038/202616a0. [DOI] [Google Scholar]
- 61.Li J, et al. Tomato SlDREB gene restricts leaf expansion and internode elongation by downregulating key genes for gibberellin biosynthesis. J. Exp. Bot. 2012;63:6407–6420. doi: 10.1093/jxb/ers295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miceli A, Moncada A, Sabatino L, Vetrano F. Effect of Gibberellic Acid on Growth, Yield, and Quality of Leaf Lettuce and Rocket Grown in a Floating System. Agronomy. 2019;9:382. doi: 10.3390/agronomy9070382. [DOI] [Google Scholar]
- 63.Prasetyaningrum P, et al. Nocturnal gibberellin biosynthesis is carbon dependent and adjusts leaf expansion rates to variable conditions. Plant Physiol. 2021;185:228–239. doi: 10.1093/plphys/kiaa019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gallego-Giraldo L, et al. Gibberellin homeostasis in tobacco is regulated by gibberellin metabolism genes with different gibberellin sensitivity. Plant cell Physiol. 2008;49:679–690. doi: 10.1093/pcp/pcn042. [DOI] [PubMed] [Google Scholar]
- 65.Thomas SG, Blázquez MA, Alabadí D. Della Proteins: Master Regulators Gibberellin-Responsive Growth Dev. 2016;49:189–228. [Google Scholar]
- 66.Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. Cell Mol. Biol. 2010;18:111–119. doi: 10.1046/j.1365-313X.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- 67.Hirano K, et al. The suppressive function of the rice DELLA protein SLR1 is dependent on its transcriptional activation activity. Plant J. 2012;71:443–453. doi: 10.1111/j.1365-313X.2012.05000.x. [DOI] [PubMed] [Google Scholar]
- 68.Ikeda A, et al. slender Rice, a Constitutive Gibberellin Response Mutant, Is Caused by a Null Mutation of the SLR1 Gene, an Ortholog of the Height-Regulating Gene GAI/RGA/RHT/D8. Plant Cell. 2001;13:999–1010. doi: 10.1105/tpc.13.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Itoh H, et al. Overexpression of a GRAS protein lacking the DELLA domain confers altered gibberellin responses in rice. Plant J. 2005;44:669–679. doi: 10.1111/j.1365-313X.2005.02562.x. [DOI] [PubMed] [Google Scholar]
- 70.Fukao T, Bailey-Serres J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl Acad. Sci. 2008;105:16814–16819. doi: 10.1073/pnas.0807821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bundock P, Hooykaas P. An Arabidopsis hAT-like transposase is essential for plant development. Nature. 2005;436:282–284. doi: 10.1038/nature03667. [DOI] [PubMed] [Google Scholar]
- 72.Bartholomew, D. P., Paull, R. E. & Rohrbach, K. G. The pineapple: botany, production and uses, (CABI Publishing, 2003).
- 73.Trusov Y, Botella JR. Silencing of the ACC synthase gene ACACS2 causes delayed flowering in pineapple [Ananas comosus (L.) Merr.] J. Exp. Bot. 2006;57:3953–3960. doi: 10.1093/jxb/erl167. [DOI] [PubMed] [Google Scholar]
- 74.Kuan C-S, et al. Foliar application of aviglycine reduces natural flowering in pineapple. HortScience. 2005;40:123–126. doi: 10.21273/HORTSCI.40.1.123. [DOI] [Google Scholar]
- 75.Ávila M, et al. Early histological, hormonal, and molecular changes during pineapple (Ananas comosus (L.) Merrill) artificial flowering induction. J. Plant Physiol. 2016;209:11–19. doi: 10.1016/j.jplph.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 76.Vurture GW, et al. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics. 2017;33:2202–2204. doi: 10.1093/bioinformatics/btx153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roach MJ, Schmidt SA, Borneman AR. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinforma. 2018;19:1–10. doi: 10.1186/s12859-018-2485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang X, Zhang S, Zhao Q, Ming R, Tang H. Assembly of allele-aware, chromosomal-scale autopolyploid genomes based on Hi-C data. Nat. Plants. 2019;5:833–845. doi: 10.1038/s41477-019-0487-8. [DOI] [PubMed] [Google Scholar]
- 79.Flynn JM, Hubley R, Rosen J, Clark AG, Smit AF. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl Acad. Sci. 2020;117:9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haas BJ, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Korf I. Gene finding in novel genomes. BMC Bioinforma. 2004;5:1–9. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lukashin AV, Borodovsky M. GeneMark. hmm: new solutions for gene finding. Nucleic acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 84.Hoff, K. J., Lomsadze, A., Borodovsky, M. & Stanke, M. Whole-genome annotation with BRAKER. in Gene prediction 65-95 (Springer, 2019). [DOI] [PMC free article] [PubMed]
- 85.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.She R, Chu JS-C, Wang K, Pei J, Chen N. GenBlastA: enabling BLAST to identify homologous gene sequences. Genome Res. 2009;19:143–149. doi: 10.1101/gr.082081.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Birney E, Clamp M, Durbin R. GeneWise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jin J, et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016;45:D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen, L. Y., Vanburen, R., Paris, M., Zhou, H. & Ming, R. The Bracteatus Pineapple Genome and Domestication of Clonally Propagated Crops. European Nucleotide Archive. https://www.ebi.ac.uk/ena/browser/view/PRJEB33121 (2019). [DOI] [PubMed]
- 90.Ming, R. et al. The pineapple genome and the evolution of CAM photosynthesis. National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/assembly/GCF_001540865.1 (2016).
- 91.Avdeyev P, Jiang S, Aganezov S, Hu F, Alekseyev MA. Reconstruction of ancestral genomes in presence of gene gain and loss. J. Computational Biol. 2016;23:150–164. doi: 10.1089/cmb.2015.0160. [DOI] [PubMed] [Google Scholar]
- 92.Wang Y, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49–e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li N, et al. OsASR2 regulates the expression of a defence‐related gene, Os2H16, by targeting the GT-1 cis-element. Plant Biotechnol. J. 2018;16:771–783. doi: 10.1111/pbi.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li, Z. et al. The genome of Aechmea fasciata provides insights into the evolution of tank epiphytic habits and ethylene-induced flowering. National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA748557 (2022). [DOI] [PMC free article] [PubMed]
- 95.Li, Z. et al. Ethylene-induced flowering in Aechmea fasciata. National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE205799 (2022).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Aechmea fasciata genome sequences and raw sequence data from RNA-seq and genome sequencing have been deposited under BioProject accession number PRJNA74855794. The AfEIL1-like ChIP-seq data were deposited under GEO accession number GSE20579995.