Abstract
Gene silencing mediated by small noncoding RNAs (sRNAs) is a fundamental gene regulation mechanism in eukaryotes that broadly governs cellular processes. It has been established that sRNAs are critical regulators of plant growth, development, and antiviral defence, while accumulating studies support positive roles of sRNAs in plant defence against bacteria and eukaryotic pathogens such as fungi and oomycetes. Emerging evidence suggests that plant sRNAs move between species and function as antimicrobial agents against nonviral parasites. Multiple plant pathosystems have been shown to involve a similar exchange of small RNAs between species. Recent analysis about extracellular sRNAs shed light on the understanding of the selection and transportation of sRNAs moving from plant to parasites. In this review, we summarize current advances regarding the function and regulatory mechanism of plant endogenous small interfering RNAs (siRNAs) in mediating plant defence against pathogen intruders including viruses, bacteria, fungi, oomycetes, and parasitic plants. Beyond that, we propose potential mechanisms behind the sorting of sRNAs moving between species and the idea that engineering siRNA‐producing loci could be a useful strategy to improve disease resistance of crops.
Keywords: host‐induced gene silencing (HIGS), plant immunity, small RNA, trans‐species RNAi
We summarize current advances of plant small interfering RNAs in mediating plant immunity and propose potential mechanisms behind the sorting and translocation of small noncoding RNAs during trans‐species RNA interference.
1. INTRODUCTION
Plants are in a constant battle against different kinds of parasites throughout their life cycles (e.g., viruses, bacteria, fungi, oomycetes, insects, and parasitic plants). To maintain surveillance, plants have evolved complex but fine‐tuned defence mechanisms (Jones & Dangl, 2006; Wu et al., 2018; Yuan et al., 2021). Small noncoding RNAs (sRNAs), as major modulators of gene expression, precisely regulate plant immunity. MicroRNAs (miRNAs) and small interfering RNAs (siRNA) are two major classes of plant sRNAs. miRNAs, in particular, have well‐documented roles in regulating plant immunity, including switching plant growth and immunity, regulating immune signal transduction, and buffering transcript dosage of immune receptors (Qiao, Xia, et al., 2021; Song et al., 2021; Weiberg & Jin, 2015). siRNAs, on the other hand, are primarily known for their roles in silencing viral RNAs. However, recent discoveries of trans‐species RNA interference (RNAi) have uncovered the essential role of siRNAs in repressing cellular pathogens. Emerging evidence supports a novel mode of action for plant endogenous sRNAs, particularly siRNAs, in repressing fungal and oomycete infection via the silencing of pathogen genes. These studies suggest that siRNAs, usually consisting of a mixture of diverse sequences, are deployed as a “shotgun” approach to target pathogen genes in a random, yet efficient manner. Many aspects of trans‐species RNAi, however, remain unclear. In this review, we summarize recent studies uncovering the roles of sRNAs in defence against various plant pathogens. We also discuss possible mechanisms of sRNA communication between species and the sorting of sRNAs in this process.
2. CLASSIFICATION AND FUNCTION OF PLANT sRNAs
In plants, sRNAs are classified into two major types based on their precursors and biogenesis pathways: miRNAs and siRNAs (Axtell, 2013). microRNA (MIR) genes are transcribed by RNA polymerase II to produce long hairpin‐structured pri‐miRNAs that are subsequently processed primarily by DICER‐LIKE1 (DCL1) into mature 21–22 nucleotides (nt) long miRNAs. Mature miRNAs are loaded into Argonaute (AGO) proteins to form RNA‐induced silencing complexes (RISCs) that induce posttranscriptional gene silencing (PTGS) of endogenous target genes via sequence‐complementarity by mRNA cleavage or translation repression (Axtell, 2013). Many mature miRNAs are evolutionarily conserved across different species along with their gene targets. For example, miR393, conserved among seed plants, targets TIR1‐like F‐box genes and modulates trade‐off between plant growth and immunity (Navarro et al., 2006; Ruiz‐Ferrer & Voinnet, 2009). miR482/2118, a conserved miRNA superfamily, is broadly present from gymnosperms to angiosperms and functions as essential suppressors of intracellular nucleotide‐binding/leucine‐rich repeat (NB‐LRR) receptors (Zhang, Xia, et al., 2016; Zhang et al., 2022). These studies show that interactions between miRNAs and their target genes were maintained over millions of years, providing strong selective forces to shape the conserved sequences and regulatory function of miRNAs (Zhang, Xia, et al., 2016).
siRNAs, by contrast, are generated from double‐stranded RNA (dsRNA) precursors derived from noncoding RNAs, inverted repeats, aberrant transcripts, and exogenous RNAs. These dsRNA precursors are formed by hairpin‐structured RNAs, antisense RNAs or through RNA‐dependent RNA polymerases (RDRs). The dsRNAs are processed into mature 21–24 nt siRNAs by various DCLs and are loaded into AGOs to form RISCs. siRNAs can be subdivided into two major classes: RDR6‐dependent secondary siRNAs and RNA polymerase IV‐dependent siRNAs (P4‐siRNAs) (Hudzik et al., 2020). Secondary siRNAs derive from transcripts of noncoding genes, for example TAS (trans‐acting siRNA) loci, and protein‐coding genes within large gene families, for example NB‐LRRs and pentatricopeptide repeats (PPRs). They are considered as amplifiers of the silencing effect triggered by primary sRNAs (usually 22 nt in length). The dsRNA precursors of secondary siRNAs are usually processed by DCLs in a head‐to‐tail arrangement, producing phased siRNAs with diverse sequences. These siRNAs not only silence the same loci where they originate in cis but also spread the silencing signal in trans to homologues in the same gene family (Axtell, 2013). P4‐siRNAs, predominantly 24 nt in length, are generated from heterochromatic regions and transposable elements. P4‐siRNAs are associated with RNA‐directed DNA methylation (RdDM), a process involving deposition of de novo chromatin modifications (e.g., cytosine DNA methylation and H3K9 histone methylation) at target loci to induce transcriptional gene silencing (Borges & Martienssen, 2015; Havecker et al., 2010; Mosher et al., 2008; Wu et al., 2012). DNA methylation and demethylation are also involved in fine‐tuning the expression of defence genes (Deleris et al., 2016). Additionally, groups of plant siRNAs are generated from transcription of repeats (repeat‐associated siRNAs) and convergent transcriptions (natural antisense transcripts siRNAs, nat‐siRNAs) (Axtell, 2013; Mi et al., 2008; Rajagopalan et al., 2006). Owning to the rapid increase of intriguing studies about RDR6‐dependent siRNAs in plant immunity, in this review, we mainly focus on secondary siRNAs.
It is well established that siRNA‐induced RNAi contributes to resistance to pathogen infection in eukaryotes, including invertebrates and mammals (Guo et al., 2019). While the early observation of RNAi in plant immunity was associated with antiviral defence, numerous recent studies have uncovered the crucial roles of siRNAs in gene silencing and RNAi in broad classes of plant parasites.
3. FUNCTION OF siRNAs IN PLANT IMMUNITY
3.1. RNAi‐based antiviral defence in plants
siRNA‐induced PTGS was the earliest strategy investigated in antiviral immunity (Hamilton & Baulcombe, 1999; Wingard, 1928). In 1928, S. A. Wingard discovered that tobacco plants inoculated with tobacco ringspot virus acquired immunity in symptomless new leaves that were resistant to secondary infection (Wingard, 1928). However, the antiviral factor was not known at that time. In 1999, virus‐derived siRNAs (vsiRNAs) were discovered in tobacco infected by potato virus X (Hamilton & Baulcombe, 1999). vsiRNAs are derived from dsRNA precursors produced as RNA virus replicative intermediates or from the bidirectional transcription of circular DNA viruses (Guo et al., 2019). Viral dsRNAs can be directly recognized by plant DCLs to produce 21–24 nt primary vsiRNAs. To amplify the silencing signal, secondary vsiRNAs are processed by DCLs from long dsRNAs synthesized by RDRs (Brodersen & Voinnet, 2006; Hamilton & Baulcombe, 1999). Secondary vsiRNAs can be loaded into AGOs to induce the degradation of single‐stranded viral RNAs. Studies have demonstrated that the 21‐nt vsiRNAs, primarily produced by DCL4, are the major class of vsiRNAs that specifically silence viral RNAs through PTGS (Garcia‐Ruiz et al., 2010; Wang et al., 2010). DCL2, which was confirmed to have redundant roles as DCL4, can also produce vsiRNAs when DCL4 is inhibited by viruses (Bouché et al., 2006; Qin et al., 2017). In contrast, circular DNA viruses produce DCL3‐dependent 24‐nt vsiRNAs through bidirectional transcription to induce RdDM and transcriptional gene silencing (Aregger et al., 2012; Yang et al., 2011) (Figure 1a).
FIGURE 1.
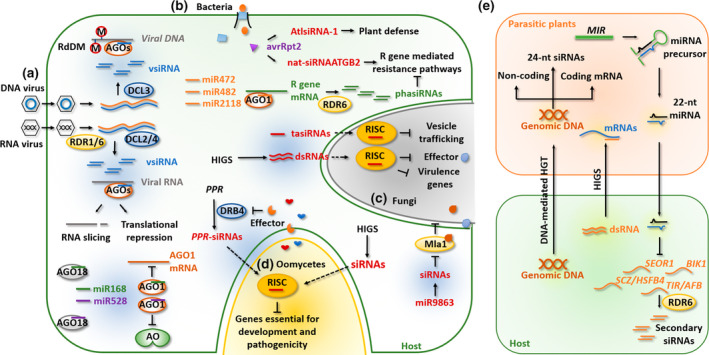
Summary of small interfering RNA (siRNA)‐mediated defence in plant–parasite interactions. (a) Bidirectional transcripts of DNA viruses are sources of virus‐derived siRNA (vsiRNA) processed by DCL3. DCL3‐processed vsiRNAs mediate RNA‐directed DNA methylation (RdDM) of the viral genome. RNA viruses generate vsiRNA derived from viral replicates or transcripts depending on RDR1/6 and DCL2/4. These vsiRNAs are associated with Argonautes (AGOs) to cleave viral transcripts or mediate translational repression. AGO18 sequesters miR168 and miR528, resulting in derepression of AGO1 and the l‐ascorbate oxidase (AO) gene required for reactive oxygen species (ROS) production, respectively. (b) Plants combat bacteria by elaborate regulation of plant resistance (R) genes. In Arabidopsis, nat‐siRNAATGB2, AtlsiRNA‐1, and miRNA‐RDR6 pathways modulate R gene‐mediated resistance pathways. NB‐LRR genes are suppressed by primary microRNA (miRNA) (miR472/482/2118) triggered‐ and RDR6‐dependent siRNAs. (c) In plant–fungi interactions, both designed double‐stranded (ds) RNAs and endogenous trans‐acting siRNAs (tasiRNAs) induce trans‐species RNA interference of pathogen genes. miR9863 modulates MLA1‐mediated resistance by triggering MLA‐siRNAs. (d) Both host‐induced gene silencing (HIGS)‐produced siRNAs and PPR gene‐derived siRNAs induce the silencing of oomycete pathogenicity genes to confer resistance. As a counterdefence strategy, oomycete effectors suppress the plant RNA silencing pathway by interfering with key components in the pathway, such as DRB4. (e) RNA translocation between parasitic plants and host. Parasitic plant‐derived components are shaded blue, and host‐derived components are shaded orange. The host genomic DNA is integrated into the genome of parasitic plants through horizontal gene transfer (HGT) and participates in 24‐nt siRNA production. Host‐produced siRNAs induce gene silencing in parasitic plants through HIGS. Endogenous 22‐nt miRNAs of the parasitic plants can be transported to the host cells and target plant mRNAs to trigger secondary siRNA production. DCL, Dicer‐like protein; RDR, RNA‐dependent RNA polymerase; RISC, RNA‐induced silencing complex.
While vsiRNAs are important for plant cells to degrade the viral genome and enhance plant antiviral immunity, some vsiRNAs can also silence host gene expression or regulate host resistance towards viral infection. One example is from a recent study on tomato yellow leaf curl virus (TYLCV) (Yang, Liu, et al., 2019). TYLCV, a geminivirus with a circular single‐stranded DNA genome, produces vsiRNAs through bidirectionally transcribed RNA from a short intergenic region. These vsiRNAs are deployed by TYLCV to silence SlLNR1, a tomato long noncoding RNA (lncRNA) involved in antiviral defence, and further enhance TYLCV symptoms. In another example, vsiRNAs derived from wheat yellow mosaic virus (WYMV) can activate broad‐spectrum plant immunity by down‐regulating host genes (Liu et al., 2021). Transgenic wheat expressing vsiRNA1, a highly abundant vsiRNA produced in the early stages of WYMV infection, showed enhanced resistance to viral infection. Further investigation revealed that vsiRNA1 silences a wheat thioredoxin‐like gene (TaAAED1) specifically to up‐regulate reactive oxygen species (ROS) production in a dose‐dependent manner (Liu et al., 2021).
In addition to vsiRNAs, another group of siRNAs involved in antiviral defence is virus‐activated siRNAs (vasiRNAs), which were first identified in Arabidopsis (Cao et al., 2014). These vasiRNAs are predominantly 21 nt in length, genetically distinct from the endogenous host siRNAs, and require DCL4 and RDR1 for their production (similar to vsiRNAs). However, unlike vsiRNAs, vasiRNAs are generated from exon regions of host genes, although their roles in modulating antiviral immunity remain unknown (Cao et al., 2014). A recent study discovered that two brassicaceous crops, turnip and oilseed rape, also produce vasiRNAs (Leonetti et al., 2021). However, the newly found vasiRNAs were predominantly 22 nt in length, which is slightly different from the 21‐nt vasiRNAs identified in Arabidopsis. They were identified to target genes involved in photosynthesis and stress response. Collectively, these studies indicate a potential role of vasiRNAs in regulating plant immunity.
In contrast to vsiRNAs and vasiRNAs, which are positively engaged in antiviral defence, most plant miRNAs negatively regulate plant immunity machineries. For example, miR319, an ancient and conserved miRNA in plants (Talmor‐Neiman et al., 2006), is induced by rice ragged stunt virus infection and silences TCP21, leading to the suppression of the jasmonic acid‐mediated defence pathway (Zhang, Ding, et al., 2016). Another miRNA, miR528, suppresses plant immunity by silencing the l‐ascorbate oxidase (AO) gene required for ROS production. In rice, miR528 is sequestered by AGO18, which restricts its loading by AGO1, thereby reducing the silencing of AO and elevating ROS production. AGO18 also decoys miR168 to alleviate repression on rice AGO1 to confer broad‐spectrum viral resistance (Wu et al., 2015).
The crucial role of RNAi in antiviral defence is also demonstrated by the production of viral suppressors of RNA silencing (VSRs) during virus–host coevolution, as a major counterdefence mechanism of viruses. Since the discovery of the first VSR (CMV2b) (Ding et al., 1995), a large number of studies have revealed that VSRs targets components involved in almost every step of the RNAi pathway (Csorba et al., 2015; Derrien et al., 2018; Kørner et al., 2018). Using VSRs to suppress host RNA silencing pathways has been well accepted as a common strategy to prevent viral genome silencing and promote viral infection.
3.2. siRNAs in plant defence against bacterial pathogens
In antiviral defence, vsiRNAs originate from the viral genome and protect plants by directly degrading viral RNAs. However, in plant defence against nonviral pathogens, endogenous siRNA‐mediated gene silencing is activated to reprogramme gene expression involved in plant immunity.
Components of the siRNA pathway have been identified to regulate plant immunity. For example, RDR6 is an essential factor for the activation of secondary siRNA production and amplification of silencing signals. shl2‐rol, a rice mutant of OsRDR6, shows more severe necrotic spots after inoculation with Xanthomonas oryzae pv. oryzae, supporting the positive role of RDR6‐dependent siRNAs in the defence against bacteria (Wagh et al., 2016). NB‐LRRs are intracellular immune receptors that recognize pathogen effectors and trigger immune responses. In the absence of pathogens, NB‐LRR transcripts are suppressed by miRNA‐triggered and RDR6‐dependent secondary siRNAs. In tomato, the miR482/2118 family represses NB‐LRRs. Sequestering miR482/2118 via short tandem target mimic RNAs conferred enhanced resistance against Pseudomonas syringae (Canto‐Pastor et al., 2019). Similarly, a compromised miR472‐RDR6 silencing pathway in Arabidopsis, which is required for the repression of NB‐LRRs, enhanced plant defence against P. syringae by promoting the recognition of avirulence effector AvrPphB (recognized by the NB‐LRR receptor RPS5) (Boccara et al., 2014). In the crp1 aba1 Arabidopsis mutant, SNC1, an R protein, showed overaccumulation in the nucleus, leading to the global reduction of miRNAs and NB‐LRR‐derived secondary siRNAs. In turn, this resulted in enhanced resistance against P. syringae (Cai, Liang, et al., 2018). Another group of RDR6‐dependent siRNAs are also involved in the modulation of plant defence against bacterial infection. In Arabidopsis, an endogenous siRNA, termed nat‐siRNAATGB2, that derived from the natural antisense transcripts pair ATGB2‐PPRL, was induced on infection by P. syringae carrying the avrRpt2 effector (Katiyar‐Agarwal et al., 2006). Consequently, these siRNAs attenuated PPRL mRNA and released the suppression of NB‐LRR receptor RPS2 by PPRL to trigger disease resistance (Katiyar‐Agarwal et al., 2006) (Figure 1b). Similarly, P. syringae infection induced long siRNA AtlsiRNA‐1, derived from the SRRLK‐AtRAP natural antisense transcripts pair, to silence a negative regulator of defence responses, AtRAP, and enhance plant immunity against bacterial infection (Katiyar‐Agarwal et al., 2007) (Figure 1b).
Collectively, these discoveries suggest that RDR6‐dependent siRNAs are critical regulators of intracellular immune receptors. It is also intriguing to hypothesize that plant siRNAs are involved in modulating broad‐spectrum resistance.
3.3. siRNA‐mediated host‐induced gene silencing HIGS against filamentous pathogens
Filamentous eukaryotic pathogens, including fungi and oomycetes, are major threats to crops. Based on the discovery of RNAi in animal cells, an innovative RNAi‐based approach was developed and has been applied in controlling filamentous pathogens. Host‐induced gene silencing (HIGS) is used to produce artificial siRNAs in plants to silence pathogen genes that are required for infection (Figure 1c). Several studies have highlighted the role of HIGS in plant immunity against eukaryotic pathogens. Transgenic barley and wheat expressing artificial siRNAs targeting the fungal effector gene Avra10 showed enhanced resistance to Blumeria graminis, an obligate biotrophic fungal pathogen causing powdery mildew disease (Nowara et al., 2010). Engineered Arabidopsis and barley expressing dsRNAs that targeted the CYP51 gene family conferred resistance to the head blight‐causing fungus Fusarium graminearum (Koch et al., 2013). Similar approaches have been used to treat wilt caused by Verticillium dahliae in crops such as tomato and cotton (Song & Thomma, 2018; Zhang, Jin, et al., 2016). HIGS has also proven to be effective against oomycete pathogens (Figure 1d). Stable transgenic lettuce expressing siRNAs targeting Bremia lactucae genes HAM34 or CES1 can inhibit B. lactucae growth and sporulation (Govindarajulu et al., 2015). In another example, siRNAs that were generated from hairpin RNA expressed in transgenic potato can silence PiGBP1, a gene encoding G protein β‐subunit in Phytophthora infestans. As a result, sporangia formation of P. infestans was inhibited and its virulence was compromised (Jahan et al., 2015). However, HIGS has large variations in silencing efficiency, suggesting that successful HIGS is highly dependent on the target gene (Jahan et al., 2015).
From the successful application of engineered HIGS, it is intriguing to learn whether plant endogenous siRNAs also contribute to plant immunity against eukaryotic pathogens. As expected, plant endogenous siRNAs have been shown to be involved in the defence against filamentous pathogens. For instance, in barley inoculated with B. graminis, 22‐nt miR9863 was found to trigger the production of 21‐nt siRNAs from Mla alleles encoding NB‐LRR receptors (Liu et al., 2014) (Figure 1c). Similarly, Arabidopsis mutants in the RNA silencing pathways, including rdr6, sgs3, ago7, and dcl4, all exhibited enhanced susceptibility to V. dahliae (Ellendorff et al., 2009). In addition to defence against fungi, Arabidopsis RNAi mutants showed reduced resistance to the oomycete pathogen Phytophthora parasitica (Guo et al., 2018). Guo et al. (2018) examined the contribution of siRNAs in plant defence against oomycete pathogens) using an ectopic VSR expression strategy (Figure 1d). Specifically, transiently expressed p19 of tomato bushy stunt virus in tobacco and soybean hairy roots promoted infection of P. parasitica and Phytophthora sojae, respectively, by suppressing the plant siRNA pathway (Guo et al., 2018).
Multiple studies have suggested that plant endogenous sRNAs induce trans‐species RNAi, a natural HIGS. In the first example, Zhang et al. uncovered that miR166 and miR159 were induced in cotton on infection by V. dahliae and were exported into fungal hyphae. These miRNAs targeted and suppressed virulence gene expression in V. dahliae to confer disease resistance (Zhang, Zhao, et al., 2016). This work shed light on HIGS induced by plant endogenous sRNAs. Studies in Arabidopsis showed that secondary siRNAs are also major components triggering trans‐species RNAi against filamentous pathogens. In particular, two trans‐acting siRNAs (tasiRNAs; TAS1c‐siR483 and TAS1c‐siR453) were translocated into the fungus Botrytis cinerea during its infection and subsequently attenuated fungal pathogenicity by silencing virulence genes (Cai, Qiao, et al., 2018) (Figure 1c). In another study, an siRNA pool derived from a subset of PPR gene loci accumulated during natural infection of Phytophthora capsici and served as a major arsenal to silence virulence‐related genes in Phytophthora pathogens. As an example, PPR‐derived siR1310 potentially silenced Phyca_554980, a gene encoding U2‐associated splicing factor in P. capsici, to weaken Phytophthora development and pathogenicity (Hou et al., 2019) (Figure 1d). Collectively, these data support the idea that trans‐species RNAi between plant host and pathogen is a naturally occurring and widespread phenomenon.
It is noteworthy that both endogenous miRNAs and siRNAs contribute to HIGS. Generally, plant miRNAs harbour conserved and unique sequences, usually in high abundance, and confer RNA silencing efficiently, making them advantageous for silencing specific genes (Hou & Ma, 2020). By contrast, most siRNAs are generated from nonprotein‐coding loci, which have fewer constraints on sequence diversification, and protein‐coding genes, such as NB‐LRR and PRR, which exhibit high rates of diversifying selection (Bergelson et al., 2001; Fujii et al., 2011). These siRNAs typically consist of a population of diverse sequences that, collectively, could silence multiple targets simultaneously (Hou et al., 2019; Hou & Ma, 2020). The relentless arms race between plants and pathogens drives constant dynamic variation in pathogen genes. However, miRNAs are under conserved constraints to ensure accurate regulation of plant endogenous genes and formation of the stem‐loop structure of primary transcripts (Alonso‐Peral et al., 2010; Hou & Ma, 2020; Yan et al., 2016). On the other hand, sequences of secondary siRNA loci or PHAS loci have diverged rapidly, except for the miRNA target site required for the initiation of the RDR6‐dependent siRNA pathway, which is under biased selection for conservation (Fujii et al., 2011; Tian et al., 2021). Diversification of flanking sequences of miRNA target site makes them able to generate siRNAs to silence fast‐evolving pathogen genes, undergoing coevolution with target sites in the pathogens. This shotgun approach confers efficient resistance by targeting multiple pathogen genes or even a broad‐spectrum resistance by targeting diversified pathogens (Axtell, 2019). Thus, in this regard, siRNAs are beneficial for HIGS (Hou & Ma, 2020).
3.4. siRNAs play a critical role during plant–plant parasite interaction
Parasitic plants cause major problems and affect global crop yield. They lead a unique lifestyle, depending on stolen nutrients from their host plants. For example, Cuscuta campestris, an obligate parasite on a wide range of herbaceous plants, forms a direct connection with the host plant through a specialized feeding structure termed the haustorium, which is the channel for nutrients, water, metabolites, and biological molecules. As a result of parasitism, host growth is severely reduced.
Engineered dsRNA‐mediated HIGS has been demonstrated to confer resistance to parasitic plants (Figure 1e). Transgenic tobacco expressing dsRNAs against STM‐like, a transcription factor that controls haustorial development, showed decreased vigour of Cuscuta pentagona (Alakonya et al., 2012). A similar strategy has been used to control Orobanche aegyptiaca on tomatoes. Transgenic tomatoes producing dsRNA targeting M6PR, a key enzyme required for the accumulation of mannitol during parasitism, caused the death of O. aegyptiaca tubercles (Aly et al., 2009). These studies highlight the essential role of host‐produced artificial siRNAs in defeating parasitic plants.
Recent research also verified trans‐species sRNA transportation from parasites to host plants (Shahid et al., 2018) (Figure 1e). During the process of parasitism, a group of C. campestris‐derived 22‐nt miRNAs accumulated in the haustoria. These miRNAs were found to target specific Arabidopsis genes involved in plant defence, including TIR1, AFB2, AFB3, BIK1, SEOR1, and HSFB4. As a result, secondary siRNAs were triggered from these loci to silence defence genes and promote parasitism. These observations suggest that trans‐species miRNA‐triggered secondary siRNAs affect the outcome of parasitism.
In addition to sRNAs, both DNA and mRNAs are also cargos that are trafficked between host and parasites (LeBlanc et al., 2013). Such broad exchange of nucleic acids can lead to horizontal gene transfer (HGT), probably through reverse transcription and genomic integration (Yang et al., 2016). A recent study identified 108 HGT events between parasites and host plants (Yang, Wafula, et al., 2019). Interestingly, HGT sequences seemed to be the source of 24‐nt siRNAs in Cuscuta, indicating a potential role of these siRNAs to silence host gene expression and thus facilitate parasitism. This study not only provided new insights into the origination of siRNA loci in parasites but highlighted the importance of siRNAs during plant–plant parasite interactions.
4. TRANS‐SPECIES siSIRNA MOVEMENT
sRNAs are highly mobile small molecules that traffic intercellularly and systemically in a noncell‐autonomous manner (Liu & Chen, 2018). The mobility of plant sRNAs is a prerequisite for carrying out their vital functions. How sRNAs move within and between organisms has been widely studied. Plants sRNAs, either naked, associated with RNA‐binding proteins, or encased by vesicles, can travel for short distances via the plasmodesmata and for long distances through the phloem system and even between species (Wang & Dean, 2020). For example, primary siRNAs can move 10–15 cells without producing secondary siRNAs, while long‐distance sRNA transport via the phloem involves the amplification of an RDR‐mediated silencing signal (Kim, 2005). Moreover, plant sRNAs can be translocated to invading fungi, oomycetes, and parasitic plants and subsequently silence virulence genes in the invaders and thereby confer resistance. It is noteworthy that delivery of sRNAs between species is a frequent and bidirectional process (Wang & Dean, 2020; Weiberg & Jin, 2015).
These molecules can be transported through the symplastic or apoplastic pathway. In cell‐to‐cell movement, sRNAs can move through three mechanisms: (1) move through the smooth endoplasmic reticulum (ER)‐derived desmotubule of two adjacent cells, (2) spread through the spaces between the plasma membrane and desmotubules, or (3) be secreted directly from the plasma membrane and cross the cell wall to the extracellular matrix, where they can be taken up and absorbed by adjacent cells (Wang & Dean, 2020). In systemic movement, sRNAs are trafficked from source cells to companion cells through the plasmodesmata and arrive at the sieve tube elements. From there, they travel over long distances through the sieve plates of the phloem or are secreted from the plasma membrane and cell wall into the extracellular matrix and are absorbed by other cells directly (Wang & Dean, 2020).
Trans‐species sRNA movement has been demonstrated in several plant–pathogen interactions and been shown to participate in trans‐species RNAi. Both necrotrophic and biotrophic pathogens have been found to absorb plant sRNAs (Cai et al., 2018b; Hou et al., 2019). Both dsRNAs and exosome‐carried sRNAs could be detected in fungal hyphae, suggesting direct uptake of sRNAs by the hyphae (Cai, Qiao, et al., 2018; Qiao, Lan et al., 2021). Haustoria, the specialized intimate structures formed by biotrophic/hemibiotrophic filamentous eukaryotic pathogens such as Phytophthora that interact with plant cells, provide an integrated portal into plant cells for material exchanges, including nutrients, virulence effectors, and antimicrobial agents (Micali et al., 2011; Wang et al., 2017). Based on these observations, two potential secretion routes could be proposed. One is dependent on the conventional protein secretion pathway. siRNAs produced on rough ER are internalized into budding vesicles, probably associated with RNA‐binding proteins, and then the cargos are released to the extrahaustorial matrix through the ER–Golgi route and absorbed by the haustoria. Alternatively, plant sRNAs are encapsulated into intraluminal vesicles and multivesicular bodies, which migrate to and fuse with the plasmamembrane to unload extracellular vesicles (EVs) to the apoplast. Apoplastic EVs fuse with the haustorial or hyphal membrane through endocytosis and release functional sRNAs into cells of pathogenic organisms, which subsequently assemble the RISC complexes to silence target genes (Ding et al., 2014; Hou & Ma, 2020). Exosomes or EVs have been well documented as essential vehicles of extracellular sRNAs (Valadi et al., 2007). It has been reported that Arabidopsis cells send sRNAs into B. cinerea by secreting EVs and so silences pathogen genes (Cai, Qiao, et al., 2018). However, this study did not rule out other possibilities, such as involvement of a nonvesicle ribonucleoprotein complex (Rutter & Innes, 2020).
5. SORTING OF siRNA TRANSPORT BETWEEN SPECIES
The efficiency of HIGS is determined by the potential sorting and transport mechanisms of sRNAs from the donor plant to the pathogen recipient. Several studies have shown that plants can release EVs containing defence proteins, RNA‐binding proteins, and sRNA cargos in response to pathogen infection (Baldrich et al., 2019; Cai, Qiao, et al., 2018; Hou et al., 2019; Regente et al., 2017; Rutter & Innes, 2017). Given that some transferred sRNAs are low in abundance and that siRNAs originating from the same TAS loci have different fates in trans‐species movement (Baldrich et al., 2019; Cai, Qiao, et al., 2018), it can be concluded that movement of plant endogenous sRNAs into pathogens is not a simple concentration‐dependent diffusion process, but probably requires a selective sRNA sorting mechanism. Recent studies suggest that such a rigorous sorting mechanism might be dictated by sRNA biosynthetic pathways, sRNA sizes, sequence features such as 5′ nucleotide, or selective RNA‐binding protein partners (Figure 2). Uncovering the mechanism of sRNA selection for trans‐species transport will potentially enhance success in designing artificial sRNAs to control plant disease.
FIGURE 2.
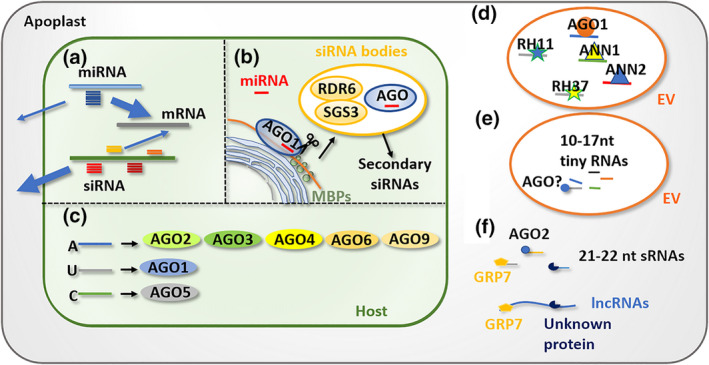
Potential mechanisms involved in small noncoding RNA (sRNA) sorting. (a) Plant microRNAs (miRNAs) with unique sequences are retained in the cytoplasm to regulate intracellular gene expression, while most small interfering RNAs (siRNAs), without intracellular targets, are more prone to be exported to the extracellular space. (b) miRNAs and siRNAs are synthesized in different cellular compartments. miRNA precursors are processed in the nucleus by Dicer‐like proteins. siRNA‐producing transcripts are cleaved on membrane‐bound polysomes (MBP) and rough endoplasmic reticulum, and further processed in siRNA bodies. The partitioning of sRNA biosynthesis potentially determines secretion of different classes of sRNAs. (c) sRNAs with the 5′ terminal nucleotides A, U, and C are preferentially recruited by AGO2/3/4/6/9, AGO1, and AGO5, respectively. sRNA loading by different AGOs may contribute to the selection of secreted sRNAs. (d) Extracellular RNAs encapsulated within extracellular versicles (EVs) need to be associated with RNA‐binding proteins for the purpose of sRNAs loading and/or stabilization. (e) Tiny RNAs (10–17 nucleotides [nt]) are enriched in EVs and could be selected by AGOs carried in EVs. (f) sRNAs (21–22 nt) and long noncoding RNAs (lncRNAs) are more probably translocated into the apoplast independent of EVs. Alternatively, AGO2, GRP7, and other RNA‐binding proteins are associated with RNAs to confer selection and/or stabilization.
sRNAs (e.g., siRNAs, miRNAs) are synthesized through distinct biosynthetic pathways and their cytoplasmic partitioning may also determine their selective secretion (Figure 2a). siRNAs, as major players in trans‐species RNAi, are enriched in the apoplast (Baldrich et al., 2019), whereas miRNAs are preferentially retained in the cytoplasm to silence endogenous genes (Hou & Ma, 2020). Interestingly, high‐frequency cleavage of secondary siRNA precursors, such as miR2118‐triggered 21‐nt phasiRNAs and miR2275‐triggered 24‐nt phasiRNAs, occurred on membrane‐bound polysomes and rough ER (Li et al., 2016; Yang et al., 2021). Furthermore, cytoplasmic “siRNA body”, a phase‐separated biomolecular condensate containing enzymes for siRNA biogenesis, accumulates adjacent to cis‐Golgi (Jouannet et al., 2012; Yu et al., 2017). Therefore, although speculative, it is reasonable to suggest that the cytoplasmic partitioning pattern of siRNA biosynthesis may confer secretion selectivity to the extracellular space through the ER–Golgi pathway (Figure 2b).
In addition, sRNA sequence features could be another factor that underpins the difference in sRNA mobility between species. A recent study found that selective loading of animal miRNAs into exosomes can be determined by specific RNA motifs (Hobor et al., 2018). A secreted AGO protein (exWAGO), which is highly conserved and abundantly expressed in nematode parasites but not in the free‐living genus Caenorhabditis, has been identified as a mediator that associated with specific sRNAs and was secreted into the host environment through nematode EVs (Chow et al., 2019). In Arabidopsis, sRNAs the assortment of rRNAs with AGOs is associated with the 5′ terminal nucleotide and origin loci (Havecker et al., 2010; Mi et al., 2008). Arabidopsis encodes 10 AGOs, with the major protein AGO1 preferentially harbouring sRNAs with 5′ terminal uridine. By contrast, the AGO2/3 and AGO4/6/9 clades and AGO5 preferentially recruit sRNAs with 5′ terminal adenosine and cytosine, respectively (Havecker et al., 2010; Mi et al., 2008; Zhang, Liu, et al., 2016) (Figure 2c). A study supporting this found that Arabidopsis AGO1 was secreted by exosome‐like EVs and selectively bound EV‐enriched sRNAs in tobacco (He et al., 2021). Therefore, 5′ terminal nucleotide selection by exAGOs could contribute to the sRNA sorting mechanism.
Several secreted RNA‐binding proteins have been identified to be involved in sRNA selection, in addition to AGOs. In Arabidopsis, RNA helicases (RH11 and RH37) and annexins (ANN1 and ANN2) were identified in EVs during B. cinerea infection (Figure 2d). ago1, rh11rh37, and ann1 ann2 mutants showed reduced translocation of EVs sRNAs and enhanced susceptibility to B. cinerea (He et al., 2021). This suggested that RNA‐binding proteins may function in loading and/or stabilizing sRNAs in EVs for transportation to pathogens. It was noted that AGO1, RH11, and RH37 specifically associated with EV‐enriched sRNAs, while ANN1 and ANN2 bound sRNAs nonspecifically (He et al., 2021).
Intriguingly, besides relatively low abundance of siRNAs and specific miRNA species, plant EVs preferentially loaded a novel class of “tiny RNAs” (10–17 nt) with broad and diverse genome origin (Baldrich et al., 2019) (Figure 2e). Tiny RNAs have been proposed to be RNA degradation products with minor function. However, a study in human AGOs revealed that human AGO3 was catalytically activated by 14‐nt tiny guide RNAs, indicating the specific selection and biological function of tiny RNAs (Park et al., 2020) (Figure 2e). Further in‐depth studies are required to determine whether these tiny RNAs are selectively loaded into EVs by their short sizes or through a specific RNA biosynthesis pathway. Note that EV content may change in response to pathogen infection, probably through the switch of RNA sorting mechanisms (Hou et al., 2019). Interestingly, Karimi et al. recently revealed that Arabidopsis apoplastic RNAs, including 21–22 nt sRNAs and lncRNAs, were mostly located outside of EVs and associated with RNA‐binding proteins (Karimi et al., 2022) (Figure 2f). Glycine‐rich RNA‐binding protein 7 (GRP7) and AGO2 were identified in the apoplast independent of EVs. Given that apoplastic sRNAs and lncRNAs in grp7 and ago2 mutants show a remarkable reduction, it is reasonable to propose that RNA‐binding proteins contribute to apoplastic RNA selection or stabilization. Collectively, partition of sRNAs in the apoplast might be determined by RNA length and associated RNA‐binding proteins.
siRNA‐induced RNA interference is a fundamental defence mechanism employed by plants. In addition to regulating cellular immunity against invading pathogens, siRNAs have been shown to act extracellularly to induce trans‐species RNAi. How sRNAs are sorted for transportation into pathogens remains unknown. One feasible strategy to illuminate the mechanism would be to produce engineered sRNA populations in plants that share common basic sequences but are characterized by different lengths, various 5′ terminal nucleotides, or are generated from distinct biogenesis pathways, which would be followed by analysis of preferentially transferred sRNAs during plant–pathogen interaction. Uncovering this sRNA sorting mechanism could improve the engineering of plant siRNAs for efficient natural HIGS against crop pathogens and ultimately contribute to reducing crop loss by conferring broad‐spectrum disease resistance.
ACKNOWLEDGEMENTS
This work is sponsored by Shanghai Collaborative Innovation Center of Agri‐Seeds (ZXWH2150201/002). The authors declare that there are no conflicts of interest.
Kong, X. , Yang, M. , Le, B.H. , He, W. & Hou, Y. (2022) The master role of siRNAs in plant immunity. Molecular Plant Pathology, 23, 1565–1574. Available from: 10.1111/mpp.13250
Xiuzhen Kong and Meng Yang contributed equally to this work.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed.
REFERENCES
- Alakonya, A. , Kumar, R. , Koenig, D. , Kimura, S. , Townsley, B. , Runo, S. et al. (2012) Interspecific RNA interference of SHOOT MERISTEMLESS‐like disrupts Cuscuta pentagona plant parasitism. The Plant Cell, 24, 3153–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso‐Peral, M.M. , Li, J. , Li, Y. , Allen, R.S. , Schnippenkoetter, W. , Ohms, S. et al. (2010) The microRNA159‐regulated GAMYB‐like genes inhibit growth and promote programmed cell death in Arabidopsis . Plant Physiology, 154, 757–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aly, R. , Cholakh, H. , Joel, D.M. , Leibman, D. , Steinitz, B. , Zelcer, A. et al. (2009) Gene silencing of mannose 6‐phosphate reductase in the parasitic weed Orobanche aegyptiaca through the production of homologous dsRNA sequences in the host plant. Plant Biotechnology Journal, 7, 487–498. [DOI] [PubMed] [Google Scholar]
- Aregger, M. , Borah, B.K. , Seguin, J. , Rajeswaran, R. , Gubaeva, E.G. , Zvereva, A.S. et al. (2012) Primary and secondary siRNAs in geminivirus‐induced gene silencing. PLoS Pathogens, 8, e1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J. (2013) Classification and comparison of small RNAs from plants. Annual Review of Plant Biology, 64, 137–159. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J. (2019) Second to none: plant secondary siRNAs as defensive agents against Phytophthora . Cell Host & Microbe, 25, 7–9. [DOI] [PubMed] [Google Scholar]
- Baldrich, P. , Rutter, B.D. , Karimi, H.Z. , Podicheti, R. , Meyers, B.C. & Innes, R.W. (2019) Plant extracellular vesicles contain diverse small RNA species and are enriched in 10‐ to 17‐nucleotide “tiny” RNAs. The Plant Cell, 31, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson, J. , Kreitman, M. , Stahl, E. & Tian, D. (2001) Evolutionary dynamics of plant R genes. Science, 292, 2281–2285. [DOI] [PubMed] [Google Scholar]
- Boccara, M. , Sarazin, A. , Thiebeauld, O. , Jay, F. , Voinnet, O. , Navarro, L. et al. (2014) The Arabidopsis miR472‐RDR6 silencing pathway modulates PAMP‐and effector‐triggered immunity through the post‐transcriptional control of disease resistance genes. PLoS Pathogens, 10, e1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges, F. & Martienssen, R.A. (2015) The expanding world of small RNAs in plants. Nature Reviews Molecular Cell Biology, 16, 727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché, N. , Lauressergues, D. , Gasciolli, V. & Vaucheret, H. (2006) An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. The EMBO Journal, 25, 3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, P. & Voinnet, O. (2006) The diversity of RNA silencing pathways in plants. Trends in Genetics, 22, 268–280. [DOI] [PubMed] [Google Scholar]
- Cai, Q. , Liang, C. , Wang, S. , Hou, Y. , Gao, L. , Liu, L. et al. (2018) The disease resistance protein SNC1 represses the biogenesis of microRNAs and phased siRNAs. Nature Communications, 9, 5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Q. , Qiao, L. , Wang, M. , He, B. , Lin, F.‐M. , Palmquist, J. et al. (2018) Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science, 360, 1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto‐Pastor, A. , Santos, B. , Valli, A.A. , Summers, W. , Schornack, S. & Baulcombe, D.C. (2019) Enhanced resistance to bacterial and oomycete pathogens by short tandem target mimic RNAs in tomato. Proceedings of the National Academy of Sciences of the United States of America, 116, 2755–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, M. , Du, P. , Wang, X. , Yu, Y.‐Q. , Qiu, Y.‐H. , Li, W. et al. (2014) Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis . Proceedings of the National Academy of Sciences of the United States of America, 111, 14613–14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow, F.W.‐N. , Koutsovoulos, G. , Ovando‐Vázquez, C. , Neophytou, K. , Bermúdez‐Barrientos, J.R. , Laetsch, D.R. et al. (2019) Secretion of an Argonaute protein by a parasitic nematode and the evolution of its siRNA guides. Nucleic Acids Research, 47, 3594–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba, T. , Kontra, L. & Burgyán, J. (2015) Viral silencing suppressors: tools forged to fine‐tune host‐pathogen coexistence. Virology, 479, 85–103. [DOI] [PubMed] [Google Scholar]
- Deleris, A. , Halter, T. & Navarro, L. (2016) DNA methylation and demethylation in plant immunity. Annual Review of Phytopathology, 54, 579–603. [DOI] [PubMed] [Google Scholar]
- Derrien, B. , Clavel, M. , Baumberger, N. , Iki, T. , Sarazin, A. , Hacquard, T. et al. (2018) A suppressor screen for AGO1 degradation by the viral F‐box P0 protein uncovers a role for AGO DUF1785 in sRNA duplex unwinding. The Plant Cell, 30, 1353–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S.‐W. , Li, W.‐X. & Symons, R. (1995) A novel naturally occurring hybrid gene encoded by a plant RNA virus facilitates long distance virus movement. The EMBO Journal, 14, 5762–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Y. , Robinson, D.G. & Jiang, L. (2014) Unconventional protein secretion (UPS) pathways in plants. Current Opinion in Cell Biology, 29, 107–115. [DOI] [PubMed] [Google Scholar]
- Ellendorff, U. , Fradin, E.F. , De Jonge, R. & Thomma, B.P. (2009) RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. Journal of Experimental Botany, 60, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, S. , Bond, C.S. & Small, I.D. (2011) Selection patterns on restorer‐like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proceedings of the National Academy of Sciences of the United States of America, 108, 1723–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Ruiz, H. , Takeda, A. , Chapman, E.J. , Sullivan, C.M. , Fahlgren, N. , Brempelis, K.J. et al. (2010) Arabidopsis RNA‐dependent RNA polymerases and Dicer‐like proteins in antiviral defense and small interfering RNA biogenesis during turnip mosaic virus infection. The Plant Cell, 22, 481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajulu, M. , Epstein, L. , Wroblewski, T. & Michelmore, R.W. (2015) Host‐induced gene silencing inhibits the biotrophic pathogen causing downy mildew of lettuce. Plant Biotechnology Journal, 13, 875–883. [DOI] [PubMed] [Google Scholar]
- Guo, N. , Zhao, J. , Yan, Q. , Huang, J. , Ma, H. , Rajput, N.A. et al. (2018) Resistance to Phytophthora pathogens is dependent on gene silencing pathways in plants. Journal of Phytopathology, 166, 379–385. [Google Scholar]
- Guo, Z. , Li, Y. & Ding, S.W. (2019) Small RNA‐based antimicrobial immunity. Nature Reviews Immunology, 19, 31–44. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J. & Baulcombe, D.C. (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Havecker, E.R. , Wallbridge, L.M. , Hardcastle, T.J. , Bush, M.S. , Kelly, K.A. , Dunn, R.M. et al. (2010) The Arabidopsis RNA‐directed DNA methylation Argonautes functionally diverge based on their expression and interaction with target loci. The Plant Cell, 22, 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, B. , Cai, Q. , Qiao, L. , Huang, C.‐Y. , Wang, S. , Miao, W. et al. (2021) RNA‐binding proteins contribute to small RNA loading in plant extracellular vesicles. Nature Plants, 7, 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobor, F. , Dallmann, A. , Ball, N.J. , Cicchini, C. , Battistelli, C. , Ogrodowicz, R.W. et al. (2018) A cryptic RNA‐binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets. Nature Communications, 9, 831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y. & Ma, W. (2020) Natural host‐induced gene silencing offers new opportunities to engineer disease resistance. Trends in Microbiology, 28, 109–117. [DOI] [PubMed] [Google Scholar]
- Hou, Y. , Zhai, Y. , Feng, L. , Karimi, H.Z. , Rutter, B.D. , Zeng, L. et al. (2019) A Phytophthora effector suppresses trans‐kingdom RNAi to promote disease susceptibility. Cell Host & Microbe, 25, 153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudzik, C. , Hou, Y. , Ma, W. & Axtell, M.J. (2020) Exchange of small regulatory RNAs between plants and their pests. Plant Physiology, 182, 51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan, S.N. , Åsman, A.K. , Corcoran, P. , Fogelqvist, J. , Vetukuri, R.R. & Dixelius, C. (2015) Plant‐mediated gene silencing restricts growth of the potato late blight pathogen Phytophthora infestans . Journal of Experimental Botany, 66, 2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. & Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jouannet, V. , Moreno, A.B. , Elmayan, T. , Vaucheret, H. , Crespi, M.D. & Maizel, A. (2012) Cytoplasmic Arabidopsis AGO7 accumulates in membrane‐associated siRNA bodies and is required for ta‐siRNA biogenesis. The EMBO Journal, 31, 1704–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, H.Z. , Baldrich, P. , Rutter, B.D. , Borniego, L. , Zajt, K.K. , Meyers, B.C. et al. (2022) Arabidopsis apoplastic fluid contains sRNA‐and circular RNA‐protein complexes that are located outside extracellular vesicles. The Plant Cell, 34, 1863–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar‐Agarwal, S. , Morgan, R. , Dahlbeck, D. , Borsani, O. , Villegas, A. , Zhu, J.‐K. et al. (2006) A pathogen‐inducible endogenous siRNA in plant immunity. Proceedings of the National Academy of Sciences of the United States of America, 103, 18002–18007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar‐Agarwal, S. , Gao, S. , Vivian‐Smith, A. & Jin, H. (2007) A novel class of bacteria‐induced small RNAs in Arabidopsis . Genes & Development, 21, 3123–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.‐Y. (2005) Regulation of short‐distance transport of RNA and protein. Current Opinion in Plant Biology, 8, 45–52. [DOI] [PubMed] [Google Scholar]
- Koch, A. , Kumar, N. , Weber, L. , Keller, H. , Imani, J. & Kogel, K.‐H. (2013) Host‐induced gene silencing of cytochrome P450 lanosterol C14α‐demethylase–encoding genes confers strong resistance to fusarium species. Proceedings of the National Academy of Sciences of the United States of America, 110, 19324–19329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kørner, C.J. , Pitzalis, N. , Peña, E.J. , Erhardt, M. , Vazquez, F. & Heinlein, M. (2018) Crosstalk between PTGS and TGS pathways in natural antiviral immunity and disease recovery. Nature Plants, 4, 157–164. [DOI] [PubMed] [Google Scholar]
- LeBlanc, M. , Kim, G. , Patel, B. , Stromberg, V. & Westwood, J. (2013) Quantification of tomato and Arabidopsis mobile RNAs trafficking into the parasitic plant Cuscuta pentagona . New Phytologist, 200, 1225–1233. [DOI] [PubMed] [Google Scholar]
- Leonetti, P. , Ghasemzadeh, A. , Consiglio, A. , Gursinsky, T. , Behrens, S.E. & Pantaleo, V. (2021) Endogenous activated small interfering RNAs in virus‐infected Brassicaceae crops show a common host gene‐silencing pattern affecting photosynthesis and stress response. New Phytologist, 229, 1650–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Le, B. , Ma, X. , Li, S. , You, C. , Yu, Y. et al. (2016) Biogenesis of phased siRNAs on membrane‐bound polysomes in Arabidopsis . eLife, 5, e22750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. & Chen, X. (2018) Intercellular and systemic trafficking of RNAs in plants. Nature Plants, 4, 869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Cheng, X. , Liu, D. , Xu, W. , Wise, R. & Shen, Q.H. (2014) The miR9863 family regulates distinct Mla alleles in barley to attenuate NLR receptor‐triggered disease resistance and cell‐death signaling. PLoS Genetics, 10, e1004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P. , Zhang, X. , Zhang, F. , Xu, M. , Ye, Z. , Wang, K. et al. (2021) A virus‐derived siRNA activates plant immunity by interfering with ROS scavenging. Molecular Plant, 14, 1088–1103. [DOI] [PubMed] [Google Scholar]
- Mi, S. , Cai, T. , Hu, Y. , Chen, Y. , Hodges, E. , Ni, F. et al. (2008) Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell, 133, 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micali, C.O. , Neumann, U. , Grunewald, D. , Panstruga, R. & O'Connell, R. (2011) Biogenesis of a specialized plant–fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cellular Microbiology, 13, 210–226. [DOI] [PubMed] [Google Scholar]
- Mosher, R.A. , Schwach, F. , Studholme, D. & Baulcombe, D.C. (2008) PolIVb influences RNA‐directed DNA methylation independently of its role in siRNA biogenesis. Proceedings of the National Academy of Sciences of the United States of America, 105, 3145–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, L. , Dunoyer, P. , Jay, F. , Arnold, B. , Dharmasiri, N. , Estelle, M. et al. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science, 312, 436–439. [DOI] [PubMed] [Google Scholar]
- Nowara, D. , Gay, A. , Lacomme, C. , Shaw, J. , Ridout, C. , Douchkov, D. et al. (2010) HIGS: host‐induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis . The Plant Cell, 22, 3130–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, M.S. , Sim, G. , Kehling, A.C. & Nakanishi, K. (2020) Human Argonaute2 and Argonaute3 are catalytically activated by different lengths of guide RNA. Proceedings of the National Academy of Sciences of the United States of America, 117, 28576–28578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, L. , Lan, C. , Capriotti, L. , Ah‐Fong, A. , Sanchez, J.N. , Hamby, R. et al. (2021) Spray‐induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnology Journal, 19, 1756–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, Y. , Xia, R. , Zhai, J. , Hou, Y. , Feng, L. , Zhai, Y. et al. (2021) Small RNAs in plant immunity and virulence of filamentous pathogens. Annual Review of Phytopathology, 59, 265–288. [DOI] [PubMed] [Google Scholar]
- Qin, C. , Li, B. , Fan, Y. , Zhang, X. , Yu, Z. , Ryabov, E. , et al. (2017) Roles of Dicer‐like proteins 2 and 4 in intra‐ and intercellular antiviral silencing. Plant Physiology, 174, 1067–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan, R. , Vaucheret, H. , Trejo, J. & Bartel, D.P. (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana . Genes & Development, 20, 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regente, M. , Pinedo, M. , San Clemente, H. , Balliau, T. , Jamet, E. & De La Canal, L. (2017) Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. Journal of Experimental Botany, 68, 5485–5495. [DOI] [PubMed] [Google Scholar]
- Ruiz‐Ferrer, V. & Voinnet, O. (2009) Roles of plant small RNAs in biotic stress responses. Annual Review of Plant Biology, 60, 485–510. [DOI] [PubMed] [Google Scholar]
- Rutter, B.D. & Innes, R.W. (2017) Extracellular vesicles isolated from the leaf apoplast carry stress‐response proteins. Plant Physiology, 173, 728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter, B.D. & Innes, R.W. (2020) Growing pains: addressing the pitfalls of plant extracellular vesicle research. The New Phytologist, 228, 1505–1510. [DOI] [PubMed] [Google Scholar]
- Shahid, S. , Kim, G. , Johnson, N.R. , Wafula, E. , Wang, F. , Coruh, C. et al. (2018) MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature, 553, 82–85. [DOI] [PubMed] [Google Scholar]
- Song, Y. & Thomma, B.P. (2018) Host‐induced gene silencing compromises Verticillium wilt in tomato and Arabidopsis . Molecular Plant Pathology, 19, 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, L. , Fang, Y. , Chen, L. , Wang, J. & Chen, X. (2021) Role of non‐coding RNAs in plant immunity. Plant Communications, 2, 100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmor‐Neiman, M. , Stav, R. , Frank, W. , Voss, B. & Arazi, T. (2006) Novel micro‐RNAs and intermediates of micro‐RNA biogenesis from moss. The Plant Journal, 47, 25–37. [DOI] [PubMed] [Google Scholar]
- Tian, P. , Zhang, X. , Xia, R. , Liu, Y. , Wang, M. , Li, B. et al. (2021) Evolution and diversification of reproductive phased small interfering RNAs in Oryza species. New Phytologist, 229, 2970–2983. [DOI] [PubMed] [Google Scholar]
- Valadi, H. , Ekström, K. , Bossios, A. , Sjöstrand, M. , Lee, J.J. & Lötvall, J.O. (2007) Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 9, 654–659. [DOI] [PubMed] [Google Scholar]
- Wagh, S. , Alam, M. , Kobayashi, K. , Yaeno, T. , Yamaoka, N. , Toriba, T. et al. (2016) Analysis of rice RNA‐dependent RNA polymerase 6 (OsRDR6) gene in response to viral, bacterial and fungal pathogens. Journal of General Plant Pathology, 82, 12–17. [Google Scholar]
- Wang, M. & Dean, R.A. (2020) Movement of small RNAs in and between plants and fungi. Molecular Plant Pathology, 21, 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.‐B. , Wu, Q. , Ito, T. , Cillo, F. , Li, W.‐X. , Chen, X. et al. (2010) RNAi‐mediated viral immunity requires amplification of virus‐derived siRNAs in Arabidopsis thaliana . Proceedings of the National Academy of Sciences of the United States of America, 107, 484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Boevink, P.C. , Welsh, L. , Zhang, R. , Whisson, S.C. & Birch, P.R. (2017) Delivery of cytoplasmic and apoplastic effectors from Phytophthora infestans haustoria by distinct secretion pathways. New Phytologist, 216, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg, A. & Jin, H. (2015) Small RNAs—the secret agents in the plant–pathogen interactions. Current Opinion in Plant Biology, 26, 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingard, S.A. (1928) Hosts and symptoms of ring spot, a virus disease of plants. Journal of Agricultural Research, 37, 127–153. [Google Scholar]
- Wu, L. , Mao, L. & Qi, Y. (2012) Roles of Dicer‐like and Argonaute proteins in TAS‐derived small interfering RNA‐triggered DNA methylation. Plant Physiology, 160, 990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Yang, Z. , Wang, Y. , Zheng, L. , Ye, R. , Ji, Y. et al. (2015) Viral‐inducible Argonaute18 confers broad‐spectrum virus resistance in rice by sequestering a host microRNA. eLife, 4, e05733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C.H. , Derevnina, L. & Kamoun, S. (2018) Receptor networks underpin plant immunity. Science, 360, 1300–1301. [DOI] [PubMed] [Google Scholar]
- Yan, J. , Zhao, C. , Zhou, J. , Yang, Y. , Wang, P. , Zhu, X. et al. (2016) The miR165/166 mediated regulatory module plays critical roles in ABA homeostasis and response in Arabidopsis thaliana . PLoS Genetics, 12, e1006416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Wang, Y. , Guo, W. , Xie, Y. , Xie, Q. , Fan, L. et al. (2011) Characterization of small interfering RNAs derived from the geminivirus/betasatellite complex using deep sequencing. PLoS One, 6, e16928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. , Zhang, Y. , Wagula, E.K. , Honaas, L.A. , Ralph, P.E. , Jones, S. et al. (2016) Horizontal gene transfer is more frequent with increased heterotrophy and contributes to parasite adaptation. Proceedings of the National Academy of Sciences of the United States of America, 113, E7010–E7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Liu, T. , Shen, D. , Wang, J. , Ling, X. , Hu, Z. et al. (2019) Tomato yellow leaf curl virus intergenic siRNAs target a host long noncoding RNA to modulate disease symptoms. PLoS Pathogens, 15, e1007534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. , Wafula, E.K. , Kim, G. , Shahid, S. , Mcneal, J.R. , Ralph, P.E. et al. (2019) Convergent horizontal gene transfer and cross‐talk of mobile nucleic acids in parasitic plants. Nature Plants, 5, 991–1001. [DOI] [PubMed] [Google Scholar]
- Yang, X. , You, C. , Wang, X. , Gao, L. , Mo, B. , Liu, L. et al. (2021) Widespread occurrence of microRNA‐mediated target cleavage on membrane‐bound polysomes. Genome Biology, 22, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Jia, T. & Chen, X. (2017) The ‘how’ and ‘where’ of plant micro RNAs. New Phytologist, 216, 1002–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, M. , Ngou, B. , Ding, P. & Xin, X.F. (2021) PTI‐ETI crosstalk: an integrative view of plant immunity. Current Opinion in Plant Biology, 62, 102030. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Ding, Z. , Wu, K. , Yang, L. , Li, Y. , Yang, Z. et al. (2016a) Suppression of jasmonic acid‐mediated defense by viral‐inducible microRNA319 facilitates virus infection in rice. Molecular Plant, 9, 1302–1314. [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Jin, Y. , Zhao, J.‐H. , Gao, F. , Zhou, B.‐J. , Fang, Y.‐Y. et al. (2016) Host‐induced gene silencing of the target gene in fungal cells confers effective resistance to the cotton wilt disease pathogen Verticillium dahliae . Molecular Plant, 9, 939–942. [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Zhao, Y.‐L. , Zhao, J.‐H. , Wang, S. , Jin, Y. , Chen, Z.‐Q. et al. (2016) Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nature Plants, 2, 1–6. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Xia, R. , Kuang, H. & Meyers, B.C. (2016) The diversification of plant NBS‐LRR defense genes directs the evolution of microRNAs that target them. Molecular Biology and Evolution, 33, 2692–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Liu, X. , Guo, X. , Wang, X.J. & Zhang, X. (2016) Arabidopsis AGO3 predominantly recruits 24‐nt small RNAs to regulate epigenetic silencing. Nature Plants, 2, 16049. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Waseem, M. , Zeng, Z. , Xu, J. , Chen, C. , Liu, Y. et al. (2022) MicroRNA482/2118, a miRNA superfamily essential for both disease resistance and plant development. The New Phytologist, 233, 2047–2057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed.


