Abstract
During nervous system development, axons must navigate to specific target areas. In Caenorhabditis elegans, the cadherin CDH-4 is required for ventral nerve cord axonal navigation, and dorsal nerve cord fasciculation. How CDH-4 mediates axon navigation and fasciculation is currently unknown. To identify genes acting together with cdh-4, we isolated mutants suppressing the axon guidance defects of cdh-4 mutants. These suppressors showed partial suppression of axonal defects in the dorsal and ventral nerve cords seen in cdh-4 mutants. We identified one suppressor gene, prp-6, which encodes a component of the spliceosome. Complete loss-of-function alleles of prp-6 are lethal, suggesting that the mutation isolated in our suppressor screen is a partial loss-of-function allele. A previous study found that RNAi-induced suppression of prp-6 leads to changes in the expression of several 100 genes including the cadherin cdh-5. We found that overexpression of cdh-5 mimics the suppression seen in prp-6 mutants, suggesting that CDH-5 can partially compensate for the loss of CDH-4.
Keywords: Cadherin, central nervous system, nervous system development, ventral nerve cord, axon guidance
Introduction
The C. elegans nervous system is relatively simple, with a small number of neurons 1 and invariant axon trajectories. 2 This simplifies the identification of developmental defects in the nervous system. The major longitudinal axon tracts in the nervous system are the dorsal (DNC) and ventral (VNC) nerve cords. The DNC consists of a single tightly fasciculated bundle of motoneuron axons, which innervate the dorsal body wall muscles. The VNC consists of 2 axon bundles flanking the ventral midline. About 50 axons are found in the right axon bundle, whereas the left bundle contains only 4 axons. 3 The VNC contains interneuron axons in the motor circuit, which form synapses with motoneurons within the VNC. Since synapses are formed “en passant” between adjacent neuronal processes, the precise location of neuronal processes within the VNC is essential for proper neuronal circuit formation. This means that even minor navigation defects along the VNC, such as crossing from the right into the left axon tract, can potentially lead to defects in neuronal circuit formation.
Several genes controlling axon navigation in the VNC have been identified. Among these is the Fat-like cadherin, cdh-4. 4 cdh-4 mutants are characterized by fasciculation defects in the DNC, midline crossing defects in the VNC and migration defects of the Q neuroblasts. Outside the nervous system, cdh-4 mutants have epidermal and pharyngeal cell attachment defects leading to incompletely penetrant embryonic and larval lethality. While most of the defects seen in cdh-4 mutants can be interpreted as adhesion defects, the neuroblast migration defects and the midline crossing defects in the VNC cannot be fully explained by simple adhesion defects. These more complex defects point to a potential role for CDH-4 in cell migration and axon guidance.
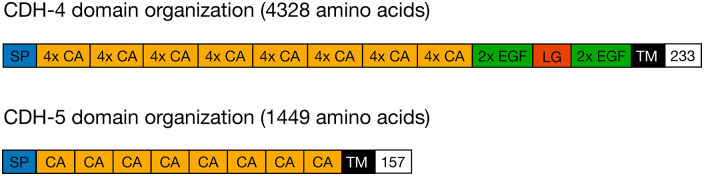
Fat-like cadherins are characterized by an unusually large number of cadherin repeats in their extracellular domain 5 (Figure 1). Fat cadherins in Drosophila control cell proliferation in imaginal discs and planar cell polarity (PCP) in multiple tissues.6-8 Different signaling pathways are used by Fat cadherins in PCP and cell proliferation.9,10 The signaling pathway used by CDH-4 in C. elegans in the context of axon guidance and fasciculation is currently unknown and is likely very different from the signaling pathways used in controlling PCP and proliferation in Drosophila.
Figure 1.
Protein domain organization of CDH-4 and CDH-5.
Abbreviations: CA single cadherin domain, “4x CA”, 4 cadherin domains; “2x EGF”, 2 epidermal growth factor domains; LG, laminin G domain; SP, signal peptide; TM, transmembrane domain; white box with number, size of intracellular domain in amino acids (contains no recognizable domain).
We performed a suppressor screen to identify genes that act together with cdh-4 to guide axons. We identified 13 suppressors that partially suppress the axonal defects of cdh-4 mutants. These suppressors only marginally suppress other defects seen in cdh-4 mutants, suggesting that they specifically interact with cdh-4 in the context of axon guidance. We identified one of these suppressor genes, prp-6, a core spliceosome component. Partial loss-of-function of prp-6 causes misregulation of several 100 genes, including the cadherin cdh-5. 11 We found that overexpression of cdh-5 can partially suppress axonal defects in cdh-4 mutants, suggesting that cdh-5 can partially compensate for the loss of cdh-4.
Material and Methods
Strains and transgenes
The following alleles were used to isolate the suppressors and to confirm their identities: cdh-4(hd40) III, prp-6 (gk527875). The following GFP transgenes were used to evaluate nervous system defects. evIs111[rgef-1::GFP] V, hdIs36 [rgef-1::DsRed2]. The following suppressors were isolated: sup(hd158); sup(hd160); sup(hd161); sup(hd162); sup(hd163); sup(hd167); sup(hd168); sup(hd169); sup(hd170); sup(hd171); sup(hd172); sup(hd173); sup(hd174). A complete list of all strains used is in Supplemental Table S1. Animals were grown at 15°C or 20°C under standard conditions. 12
Screening, mapping, and sequencing of the suppressors
To identify suppressors of cdh-4(hd40), animals were mutagenized with 50 mM EMS for 4 hours. After mutagenesis animals were grown for 2 generations. L4 stage F2 animals were placed on individual plates. A population of cdh-4(hd40) mutant animals would take 14 days (3 generations) to exhaust the food on a plate due to the small survival rate (24%) of mutant animals. Mutagenized animals that exhausted the food on the plate in 10 days or less were considered candidate suppressors. Those candidate strains were grown further and assayed for DNC fasciculation defects. Strains with less than 35% fasciculation defects were kept as suppressors of axonal defects.
Suppressors were outcrossed 4 times with the parental strain cdh-4(hd40); evIs111 prior to whole genome sequencing. Rescue of growth rate was used to identify the suppressors after each round of outcrossing. We noticed during outcrossing that all suppressors exhibited a recessive Mendelian segregation pattern. The presence of the suppressor mutation was confirmed by evaluating DNC fasciculation defects. Whole Genome Sequencing (WGS) was performed as described. 13 Briefly, sequencing was performed on an Illumina HiSeq sequencer with approximately 20-fold sequence coverage. The reference genome (WS240) was compared to the sequences of the cdh-4(hd40) suppressor strains to identify SNPs. SNPs found in more than 1 suppressor were considered background SNPs and eliminated. Only unique mutations in each suppressor strain were considered further. Single nucleotide polymorphism (SNP) mapping was done using the Hawaiian strain CB4856. 14
Transgenic strains
To generate a strain overexpressing cdh-5, we generated transgenic animals expressing cdh-5 from a multi-copy extrachromosomal array using the fosmid WRM0613cD01, which contains the genomic region of cdh-5. The cdh-5 transgene hdEx628 was crossed into cdh-4(hd40); evIs111 to evaluate suppression effects.
Phenotypic characterization of suppressors
Axonal defects and Q cell migration defects
DNC fasciculation defects, VNC midline crossing defects, and Q cell migration defects were analyzed in late-stage larvae and adult animals expressing the pan-neuronal fluorescent marker evIs111.
DNC fasciculation defects are defined as axons splitting off the tightly fasciculated DNC axon bundle creating a visible second bundle and extending separately for at least 3 cell diameters (~15 μm) before rejoining the main bundle. VNC midline crossing defects are defined as axons crossing from the right axon bundle into left bundle (or vice versa). Q cells are 2 neuroblasts initially located near the midbody region. The Q cell on the left side (QL) migrates to the posterior, divides during the migration, and produces the SDQL and PVM neurons. The Q cell on the right side (QR) migrates to the anterior, divides during the migration and produces the SDQR and AVM neurons. Migration defects are defined as QL migrating anteriorly, so that the SDQL and PVM neurons end up in the anterior half of the animal or QR migrating posteriorly so that the SDQR and AVM neurons end up in the posterior half of the animal.
For imaging, animals were incubated with 10 mM NaN3 in M9 buffer for 1 hour and mounted on agar pads prior to analysis. Defects were scored using a 40x objective lens on a Zeiss Axioscope (Carl-Zeiss AG, Germany). Images were acquired on a Zeiss Axioplan II microscope connected to a Quorum WaveFX spinning disc system (Quorum Technologies, Canada). Stacks of confocal images with 0.2 to 0.5 μm distance between focal planes were recorded. Image acquisition and analysis was carried out using Volocity software (Perkin-Elmer, Waltham, MA). Images in the figures are maximum intensity projections of all focal planes. Figures were assembled using Keynote (Apple).
Lethality and brood size
Five to 12 L4 stage animals were placed on a plate to allow them to lay eggs. Animals were transferred daily to a fresh plate for 2 days. The number of eggs was counted to determine the brood size. The number of animals reaching adulthood 2 days after larvae hatched in relation to the number of eggs laid was used as a measure of lethality.
Movement assay
L4 stage animals were placed in an eyeglass filled with buffer solution. The number of body bends within 30 seconds was recorded. In addition, the direction of bending (ventral vs dorsal) was observed. Wild type animals alternate between ventral and dorsal body contractions in a regular fashion. cdh-4 mutants frequently fail to bend in either dorsal or ventral direction, or do not bend at all. Thirty to 100 animals were evaluated for each strain.
Animals with “normal movement” had only “normal” body bends for 30 seconds. Animals with 1 or more abnormal body bends were scored as “abnormal.” “Percent normal movement” is the fraction of animals with “normal movement,” that is, animals with “normal movement” divided by total number of animals.
Statistical analysis
We used the following tests (depending on the type of data) to determine the statistical significance of differences in phenotype penetrance: chi-square test (χ2), unpaired t-test, and 2-sample Poisson test.
Data availability
Strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Results
Isolation of cdh-4 suppressors
cdh-4 mutants are characterized by a variety of defects, including substantial embryonic and larval lethality, with only 24% of animals reaching reproductive age. Embryonic lethality is due to epidermal defects during morphogenesis. Additional larval lethality is mainly caused by a detachment of the pharynx from the mouth preventing larvae from feeding. 4 cdh-4 mutants also produce fewer progeny than wild type. Together, this results in a very slow population growth, where the progeny of an individual take more than 16 days to exhaust the food on a 60 mm culture plate. This slow growth phenotype was the basis for the suppressor screen. We mutagenized cdh-4 mutant animals and subcloned individual F2 animals to determine how long it took them to exhaust the food. Populations that starved within 10 days or less were considered to carry a suppressor mutation that improved either survival or increased the number of progeny (or both). Identified suppressors were then evaluated for suppression of DNC fasciculation defects. We identified 13 suppressors that significantly improved growth and suppressed DNC fasciculation defects.
Identification of prp-6 as one of the suppressors
The suppressors were outcrossed 4 times to remove background mutations. Whole genome sequencing was used to identify unique point mutations in each strain by elimination mutations found in several strains. SNP mapping was used to narrow down the interval containing the suppressor. For sup(hd170) the gene prp-6 was left as the only candidate after mapping. We therefore focused on sup(hd170). prp-6 encodes a component of the spliceosome, specifically a component of the U5 subunit, which is part of the active spliceosome. hd170 is a missense mutation that changes glutamic acid to lysine at amino acid position 110. An independently isolated allele from the Million Mutation Project, 13 gk527875, causes a similar glutamic acid to lysine change at amino acid position 93. cdh-4(hd40); prp-6(gk527875) double mutants have 25% DNC fasciculation defects and 25% VNC defects, showing suppression of axonal defects very similar to prp-6(hd170), which has 27% DNC fasciculation defects and 24% VNC defects. cdh-4(hd40) mutants have 53% DNC fasciculation defects and 50% VNC defects. This confirms that the suppression of axonal defects is caused by a mutation in prp-6. Complete loss-of-function of prp-6 is lethal, 11 since splicing is an essential cellular process. Therefore, prp-6(hd170) is likely to be a partial loss-of-function allele.
Phenotypic characterization of suppressors
Most suppressors increased the number of progeny produced but not the fraction of surviving progeny (Table 1). Only sup(hd161), sup(hd163), sup(hd167), and sup(hd174) significantly reduced overall lethality. In other words, the majority of the mutants we isolated do not suppress defects causing lethality. This observation supports the hypothesis that lethality in cdh-4 mutants is caused through a pathway distinct from the one controlling axon navigation and fasciculation.
Table 1.
Lethality and brood size in the suppressors.
| Genotype | Brood size a | % lethality |
|---|---|---|
| cdh-4(hd40) | 62 | 76 |
| cdh-4(hd40); prp-6(hd170) | 166*** | 67 |
| cdh-4(hd40); sup(hd158) | 129*** | 80 |
| cdh-4(hd40); sup(hd160) | 126*** | 86 |
| cdh-4(hd40); sup(hd161) | 57 | 53 # |
| cdh-4(hd40); sup(hd162) | 99** | 68 |
| cdh-4(hd40); sup(hd163) | 119*** | 54 ## |
| cdh-4(hd40); sup(hd167) | 154*** | 44 ### |
| cdh-4(hd40); sup(hd168) | 163*** | 90 |
| cdh-4(hd40); sup(hd169) | 135*** | 86 |
| cdh-4(hd40); sup(hd171) | 123*** | 91 |
| cdh-4(hd40); sup(hd172) | 81 | 65 |
| cdh-4(hd40); sup(hd173) | 99** | 82 |
| cdh-4(hd40); sup(hd174) | 105** | 58 # |
The number of parental animals is 10 for hd40 and 5 for the suppressors. The experiment was done once.
P < .01 compared to cdh-4(hd40).
P < .001 compared to cdh-4(hd40); 2-sample Poisson test.
P < .05 compared to cdh-4(hd40).
P < .01 compared to cdh-4(hd40).
P < .001 compared to cdh-4(hd40); (χ2 test).
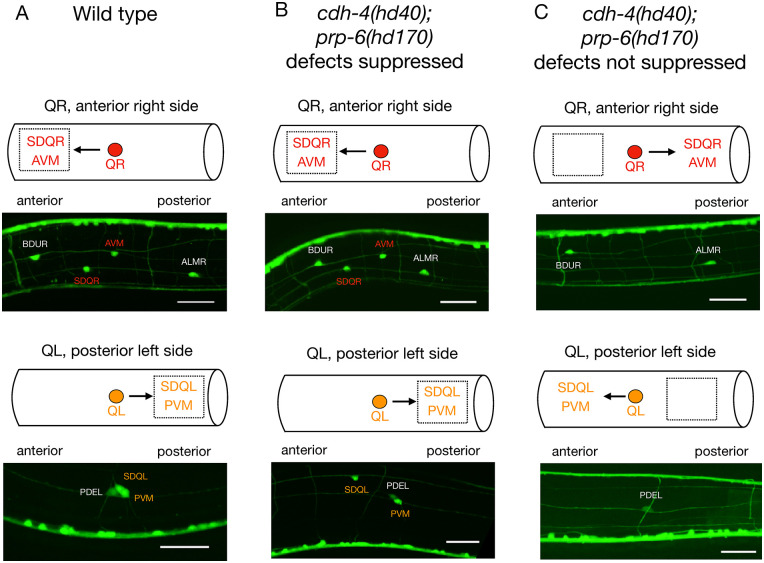
cdh-4 mutants show migration defects of the QL neuroblast, which normally migrates posteriorly in wildtype animals, but frequently migrates anteriorly in cdh-4 mutants. We sampled six suppressors for a suppression of QL migration defects and found that only two of them, prp-6(hd170) and sup(hd161), marginally suppressed the QL migration defects to 78%-83% (Table 2, Figure 2). In four of the six suppressors we found that QR neuroblasts showed migration defects with 20% to 25% penetrance, substantially higher than what is observed in cdh-4 mutants, where only 5% of QR neuroblasts have migration defects. prp-6(hd170) mutants in a wildtype background show no Q cell migration defects (Table 2). Since there are no Q cell migration defects in wild type, all the suppression effects are partial effects. Two of the descendants of the Q cells (AVM and PVM) are mechanosensory neurons. Migration defects of these cells could lead to mechanosensory defects.
Table 2.
Q cell migration defects in the suppressors.
| Genotype | % Q cell migration defects (n) | |
|---|---|---|
| QR | QL | |
| cdh-4(hd40) | 5** (100) | 95** (105) |
| cdh-4(hd40); prp-6(hd170) | 20** (100) | 83*** (150) |
| prp-6(hd170) | 0* (100) | 0*** (100) |
| cdh-4(hd40); sup(hd158) | 25*** (107) | 89** (101) |
| cdh-4(hd40); sup(hd160) | 11** (101) | 89** (101) |
| cdh-4(hd40); sup(hd161) | 22*** (102) | 78*** (103) |
| cdh-4(hd40); sup(hd162) | 9** (55) | 95** (58) |
| cdh-4(hd40); sup(hd163) | 24*** (100) | 84*** (100) |
P < .05 compared to cdh-4(hd40).
P < .01 compared to cdh-4(hd40).
P < .001 compared to cdh-4(hd40); (χ2 test).
Figure 2.
Q cell migration defects in cdh-4(hd40); prp-6(hd170). (A) In wild type animals QR migrates anteriorly and its descendants (SDQR and AVM) are located in the anterior part of the animal. QL migrates posteriorly and its descendants (SDQL and PVM) are in the posterior part of the animal. (B) In some cdh-4(hd40); prp-6(hd170) animals Q cell migration defects are suppressed, that is, the location of the Q cell descendants is as in wild type. (C) In some cdh-4(hd40); prp-6(hd170) animals Q cell migration defects are not suppressed, that is, either QR descendants are not in their normal anterior location or QL descendants are not in their normal posterior location. Scale bar is 20 μm. Marker used: evIs111.
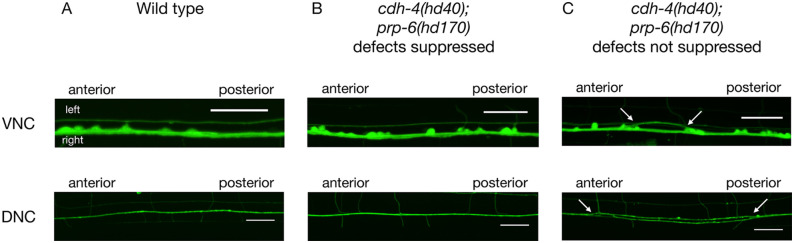
In wild type DNC and VNC defects are in the range of 4% to 5%. 4 DNC fasciculation defects were partially suppressed in all suppressors (Table 3, Figure 3), with defects ranging from 18% to 37% penetrance compared to 53% in cdh-4(hd40). VNC crossover defects were also partially suppressed in all suppressors (Table 3, Figure 3), ranging from 19% to 33% penetrance compared to 50% in cdh-4(hd40). In 5 suppressors, sup(hd162), sup(hd168), sup(hd169), prp-6(hd170), and sup(hd172), DNC and VNC defects were suppressed to a similar extent. For the other suppressors, there was no clear correlation between suppression of DNC and VNC defects. For example, VNC defects are strongly suppressed in sup(hd174) but DNC defects are not. Conversely, DNC defects are strongly suppressed in sup(hd161) but VNC defects are not. This suggests that cdh-4 might act through different pathways in the DNC and VNC and that some suppressors only affect one of the 2 pathways strongly.
Table 3.
DNC and VNC axonal defects in the suppressors.
| Genotype | % DNC fasciculation defects (n) | % VNC cross-over defects (n) |
|---|---|---|
| cdh-4(hd40) | 53 (100) | 50 (100) |
| cdh-4(hd40); prp-6(hd170) | 27*** (109) | 24*** (100) |
| cdh-4(hd40); sup(hd158) | 23*** (134) | 29** (112) |
| cdh-4(hd40); sup(hd160) | 26*** (135) | 29** (101) |
| cdh-4(hd40); sup(hd161) | 18*** (101) | 30** (100) |
| cdh-4(hd40); sup(hd162) | 28*** (120) | 25*** (106) |
| cdh-4(hd40); sup(hd163) | 25*** (106) | 33* (100) |
| cdh-4(hd40); sup(hd167) | 32** (105) | 23*** (128) |
| cdh-4(hd40); sup(hd168) | 19*** (105) | 21*** (109) |
| cdh-4(hd40); sup(hd169) | 24*** (105) | 19*** (109) |
| cdh-4(hd40); sup(hd171) | 35** (114) | 25** (100) |
| cdh-4(hd40); sup(hd172) | 29*** (103) | 25** (107) |
| cdh-4(hd40); sup(hd173) | 37* (124) | 28** (109) |
| cdh-4(hd40); sup(hd174) | 39 (120) | 21*** (113) |
P < .05 compared to cdh-4(hd40).
P < .01 compared to cdh-4(hd40).
P < .001 compared to cdh-4(hd40); (χ2 test).
Figure 3.
Ventral and dorsal nerve cord phenotypes in cdh-4(hd40); prp-6(hd170). (A) In wild type animals axons do not cross between the right and left VNC axon tracts. The DNC consists of a tightly fasciculated axon bundle. (B) In some cdh-4(hd40); prp-6(hd170) animals VNC or DNC defects are suppressed, that is, VNC and DNC look like wild type. (C) In some cdh-4(hd40); prp-6(hd170) animals VNC or DNC defects are not suppressed, that is, axons cross the midline in the VNC (arrows) or DNC axons are defasciculated (arrows). Scale bar is 20 μm. Marker used: evIs111.
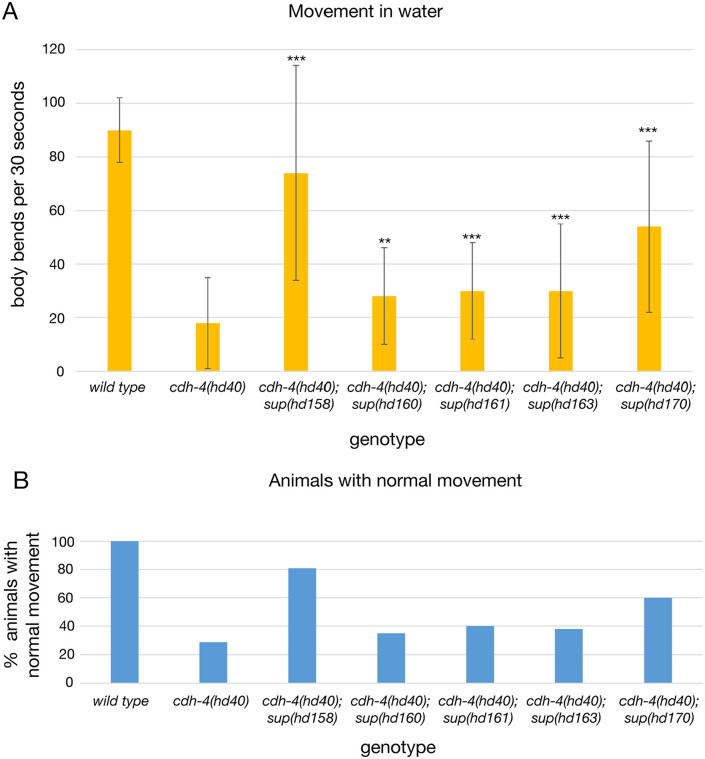
cdh-4(hd40) mutants also show movement defects. These could be the consequence of axonal defects 4 or defects seen in synapse formation in D-type motoneurons. 15 When put in liquid media, wildtype animals bend regularly and rapidly by alternate and coordinated contractions of dorsal and ventral body wall muscles with a frequency of around 180 bends per minute. This frequency is reduced to about 40 bends per minute in cdh-4(hd40) mutant animals. Furthermore, while all wildtype animals show regular and complete bending in both directions, only about 30% of cdh-4(hd40) mutant animals show normal movement behavior. We tested 5 suppressors that strongly suppressed axon guidance defects for a potential suppression of movement defects by qualitatively and quantitatively evaluating movement in liquid. Two of the suppressors, sup(hd158) and prp-6(hd170), showed partial improvement of movement both qualitatively and quantitatively (Figure 4). The three other suppressors, sup(hd160), sup(hd161), and sup(hd163) showed movement defects similar to cdh-4(hd40) (Figure 4). Since all five suppressors show comparable suppression of axonal defects, this indicates that suppression of movement defects is likely not due to suppression of axonal defects.
Figure 4.
Movement defects in suppressor mutants. (A) Movement in water: Shown are body bends per 30 seconds (n = 31-100). **P < .01 compared to cdh-4(hd40); ***P < .001 compared to cdh-4(hd40); unpaired t-test. (B) Animals with normal movement: shown is the fraction of animals showing only normal body bends within 30 seconds of scoring (n = 31-100).
Overexpression of cdh-5 partially suppresses axonal defects in cdh-4 mutants
prp-6(hd170), as a component of the spliceosome, could suppress cdh-4(hd40) defects by directly affecting splicing of the cdh-4 mRNA. Since there is only one known splice variant of cdh-4, prp-6 is unlikely to affect alternative splicing of the cdh-4 mRNA. cdh-4(hd40) is a deletion eliminating the first exon and is unlikely to produce any mRNA. To test whether the prp-6 suppression effects are specific for the cdh-4(hd40) allele, we evaluated defects in a second cdh-4 allele, rh310. cdh-4(rh310) is a point mutation creating a stop codon in exon 5 and causes defects like those seen in cdh-4(hd40) mutants. We found that prp-6(hd170) suppressed DNC defects of both cdh-4 alleles equally well (Table 4). This suggests that suppression of axonal defects by prp-6(hd170) is not specific to the hd40 allele of cdh-4.
Table 4.
DNC and VNC axonal defects in prp-6 mutants.
| Genotype | % DNC fasciculation defects (n) | % VNC cross-over defects (n) |
|---|---|---|
| cdh-4(hd40) # | 53 (100) | 50 (100) |
| cdh-4(rh310) | 50 (101) | 45 (100) |
| prp-6(hd170) | 1*** (100) | 2*** (100) |
| cdh-4(hd40); prp-6(hd170) # | 27*** (109) | 24*** (100) |
| cdh-4(rh310); prp-6(hd170) | 30** (100) | 22*** (100) |
| cdh-4(hd40); prp-6(gk527875) | 25*** (107) | 25** * (100) |
| cdh-4(hd40); cdh-5(+) | 34** (117) | 28** (101) |
| cdh-5(+) | 1** (100) | 7** (91) |
Data taken from Table 3.
P < .05 compared to cdh-4(hd40).
P < .01 compared to cdh-4(hd40).
P < .001 compared to cdh-4(hd40); (χ2 test).
Partial inactivation of prp-6 by RNAi leads to changes in the expression levels of 475 genes; 407 of these are upregulated and the rest are downregulated. 11 Among the upregulated genes is cdh-5, encoding a functionally uncharacterized member of the cadherin superfamily. 16 The domain structure of CDH-5 is similar to that of CDH-4 (Figure 1); however, CDH-5 is much smaller (1449 amino acids) than CDH-4 (4328 amino acids). This raises the possibility that overexpression of cdh-5 caused by a partial inactivation of prp-6 in the prp-6(hd170) mutant might be the cause for the suppression of cdh-4 defects. This would suggest that cdh-5 can partially replace cdh-4 for its axon guidance function.
To test whether overexpression of cdh-5 can suppress axonal defects in cdh-4 mutants, we generated transgenic animals overexpressing cdh-5 from an extrachromosomal array containing multiple copies of the wild type cdh-5 gene. cdh-5 overexpression in a wild type background has no significant DNC or VNC defects (Table 4). We crossed the transgene into cdh-4(hd40) mutants and scored DNC and VNC defects. We found that VNC crossover defects in this strain are reduced to 28% (n = 101) and DNC defects are reduced to 34% (n = 117). The suppression effects resemble those seen in cdh-4(hd40); prp-6(hd170) with 27% DNC and 24% VNC defects (Table 4). This suggests that prp-6 mediated overexpression of cdh-5 accounts for the suppression seen in the prp-6(hd170) mutant.
If overexpression of cdh-5 causes the suppression, introducing a cdh-5 mutation into the cdh-4(hd40); prp-6(hd170) strain should abolish suppression. All our attempts to generate a cdh-4(hd40); cdh-5(ok1977); prp-6(hd170) triple mutant strain failed. We concluded that this mutant combination is not viable. This is an indication that a mutation in cdh-5 enhances cdh-4 associated lethality, supporting the idea that overexpression of cdh-5 ameliorates defects of cdh-4 mutants.
Discussion
Suppressors of axon guidance defects of cdh-4 mutants
To identify genes that act together with cdh-4 to guide axons we performed a suppressor screen. In the primary screen, we selected mutants that grew substantially better than cdh-4 mutants, since it is difficult to directly screen for suppression of axon guidance defects. Two factors contribute to the slow growth of cdh-4 mutants. First, only 24% of cdh-4(hd40) mutant animals survive to adulthood. Second, the number of progeny per individual is substantially reduced in cdh-4(hd40) mutants compared to wild type. Lethality is mainly caused by hypodermal defects leading to morphogenetic defects and embryonic lethality. In addition, in some larvae the pharynx becomes detached from the mouth region leading to an inability to feed and larval lethality. 4 The basis for the reduced number of progeny is unknown. We expected the suppressors to improve survival rates, but this was not the case for most of the suppressors. Instead, the improved growth rate was predominantly due to an increased number of progeny. This suggests that most suppressors do not suppress the defects that cause lethality.
Q neuroblasts and their descendants migrate to defined positions along the anterior-posterior axis of the animal, with Q-cells on the right side (QR) migrating anteriorly and Q-cells on the left side (QL) migrating posteriorly. 17 cdh-4(hd40) mutants show prominent Q-cell migration defects, with most QL neuroblasts migrating anteriorly. Only three of the six suppressors we tested reduced QL migration defects. Four of the six suppressors showed increased QR migration defects when compared to cdh-4(hd40) mutants. Suppression of QL migration defects means more QL cells migrating posteriorly. Likewise, increased QR migration defects mean more QR cells migrating posteriorly. This suggests that some of the suppressors support posterior migration of Q cells in general.
All 13 suppressors partially suppressed DNC fasciculation as well as VNC midline crossing defects. Most suppressors suppressed DNC and VNC defects to different degrees. The lack of correlation between suppression of DNC and VNC defects in many suppressors may imply that separate pathways control guidance and fasciculation of axons in VNC and DNC, and that CDH-4 activates multiple pathways. DNC fasciculation defects could affect motoneurons that innervate dorsal body wall muscles. However, since muscle cells send muscle arm processes toward the motoneurons and are able to locate even misguided motoneuron axons, 18 the fasciculation defects in the DNC are not expected to cause significant problems in the innervation of dorsal muscle cells. Midline crossing defects in the VNC affecting interneuron axons synapsing with motoneurons in the VNC may lead to a loss of motor circuit synapses, with potentially negative consequences for the functioning of the motor circuit. cdh-4(hd40) mutants have prominent movement defects including uncoordinated and slow movement in a swimming assay. These behavioral phenotypes are partially suppressed in some but not all cdh-4 suppressing alleles analyzed for movement defects. As all suppressors show partial suppression of axonal defects, the lack of suppression of movement defects in some of these lines suggests that defects other than those affecting axon guidance are responsible for at least some of the behavioral phenotypes. cdh-4 is implicated together with fmi-1, a Flamingo-like cadherin, in the patterning of GABAergic neuromuscular junctions. 15 It is possible that synaptic defects are a major cause for the movement defects observed, and that some but not all suppressors improve synaptic development leading to a partial suppression of movement defects.
prp-6 as suppressor of axonal defects of cdh-4 mutants
We identified prp-6 as one of the suppressors. PRP-6 is an evolutionarily conserved component of the spliceosome.19 -21 The spliceosome removes introns from pre-processed mRNA. PRP-6 is a U5 component, which is part of the active core of the spliceosome that catalyzes the splicing reactions. 21 Since complete loss-of function mutations in prp-6 are lethal, 11 the missense mutation in prp-6(hd170) most likely results in a partial loss-of-function allele. A partially dysfunctional spliceosome causes inefficiencies in splicing and other associated transcriptional activities.22,23 In addition, a dysfunctional spliceosome may lead to a shift in the prevalence of specific splice variants of genes that have more than one splice variant. In mammalian cells, PRP6 is implicated in affecting alternative splice site choices when depleted.24,25 Partially inactivating prp-6 by RNAi in C. elegans also leads to changes in the expression levels of 475 genes; 407 of these are upregulated and the rest are downregulated. 11 Among the upregulated genes is cdh-5, encoding a functionally uncharacterized member of the cadherin superfamily. 16 We found that overexpression of cdh-5 in a cdh-4(hd40) mutant background suppresses DNC and VNC defects similar to those observed in prp-6 mutants. This suggests that cdh-5 can partially replace cdh-4 for its axon guidance function, and that overexpression of cdh-5 is responsible for the suppression effect rather than changes in splicing patterns.
In this study, we identified 13 alleles that partially suppress VNC and DNC axon guidance defects of cdh-4 mutants. Non-neuronal defects such as embryonic and larval lethality as well as Q cell migration defects were not, or only marginally, suppressed by these suppressors. This suggests that the different functions of cdh-4 might be mediated through different pathways, and that the suppressors we identified interact with only a few of those pathways. We identified prp-6, a key component of the spliceosome, as one of the suppressors. We found evidence that upregulation of cdh-5 in prp-6 mutants is likely responsible for this suppression effect, suggesting that cdh-5 can partially replace cdh-4 to ensure correct navigation of axons in the ventral and dorsal nerve cords.
Supplemental Material
Supplemental material, sj-docx-1-exn-10.1177_26331055221123346 for Mutations in the Spliceosome Component prp-6 and Overexpression of cdh-5 Suppress Axon Guidance Defects of cdh-4 Mutants in Caenorhabditis elegans by Zina Aburegeba, Jie Pan and Harald Hutter in Neuroscience Insights
Acknowledgments
We would like to thank members of the Hutter lab for discussions and comments on the paper and Rick Zapf for generating transgenic strains. We are grateful to members of the Moerman lab for WGS and sequence analysis. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Author Contributions: Concept/Design: HH, JP. Data acquisition: JP, ZA. Data processing/analysis: ZA, JP. Interpretation of results: ZA, JP, HH. Writing: HH.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Canadian Institute of Health research grant MOP 93719 and the National Sciences and Engineering Research Council grant RGPIN-2017-03942 both awarded to HH.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Significance Statement: The Fat-like cadherin cdh-4 controls axonal navigation and fasciculation as well as epidermal and pharyngeal cell attachment in Caenorhabditis elegans. Genes acting together with cdh-4 controlling these processes are currently unknown. In a suppressor screen we identified 13 suppressors that partially suppress the axonal defects of cdh-4 mutants. One of the suppressor genes, prp-6, encodes a core spliceosome component. In addition, we show that overexpression of the cadherin cdh-5 can suppress axonal defects in cdh-4 mutants, suggesting that CDH-5 can partially compensate for loss of CDH-4.
ORCID iD: Harald Hutter  https://orcid.org/0000-0002-1162-4199
https://orcid.org/0000-0002-1162-4199
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64-119. [DOI] [PubMed] [Google Scholar]
- 2. White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1-340. [DOI] [PubMed] [Google Scholar]
- 3. White JG, Southgate E, Thomson JN, Brenner S. The structure of the ventral nerve cord of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976;275:327-348. [DOI] [PubMed] [Google Scholar]
- 4. Schmitz C, Wacker I, Hutter H. The Fat-like cadherin CDH-4 controls axon fasciculation, cell migration and hypodermis and pharynx development in Caenorhabditis elegans. Dev Biol. 2008;316:249-259. [DOI] [PubMed] [Google Scholar]
- 5. Tanoue T, Takeichi M. New insights into fat cadherins. J Cell Sci. 2005;118:2347-2353. [DOI] [PubMed] [Google Scholar]
- 6. Mahoney PA, Weber U, Onofrechuk P, Biessmann H, Bryant PJ, Goodman CS. The fat tumor suppressor gene in Drosophila encodes a novel member of the cadherin gene superfamily. Cell. 1991;67:853-868. [DOI] [PubMed] [Google Scholar]
- 7. Yang CH, Axelrod JD, Simon MA. Regulation of frizzled by fat-like cadherins during planar polarity signaling in the Drosophila compound eye. Cell. 2002;108:675-688. [DOI] [PubMed] [Google Scholar]
- 8. Cho E, Irvine KD. Action of fat, four-jointed, dachsous and dachs in distal-to-proximal wing signaling. Development. 2004;131:4489-4500. [DOI] [PubMed] [Google Scholar]
- 9. Fanto M, McNeill H. Planar polarity from flies to vertebrates. J Cell Sci. 2004;117:527-533. [DOI] [PubMed] [Google Scholar]
- 10. Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a fat tumor suppressor pathway. Nat Genet. 2006;38:1142-1150. [DOI] [PubMed] [Google Scholar]
- 11. Rubio-Peña K, Fontrodona L, Aristizábal-Corrales D, et al. Modeling of autosomal-dominant retinitis pigmentosa in Caenorhabditis elegans uncovers a nexus between global impaired functioning of certain splicing factors and cell type-specific apoptosis. RNA N Y N. 2015;21:2119-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thompson O, Edgley M, Strasbourger P, et al. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 2013;23:1749-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet. 2001;28:160-164. [DOI] [PubMed] [Google Scholar]
- 15. Najarro EH, Wong L, Zhen M, et al. Caenorhabditis elegans flamingo cadherin fmi-1 regulates GABAergic neuronal development. J Neurosci. 2012;32:4196-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pettitt J. The cadherin superfamily. In: The C. elegans Research Community, ed. WormBook. Published online December 29, 2005. doi: 10.1895/wormbook.1.50.1 [DOI] [Google Scholar]
- 17. Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110-156. [DOI] [PubMed] [Google Scholar]
- 18. Hedgecock EM, Culotti JG, Hall DH. The unc-5, unc-6, and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans. Neuron. 1990;4:61-85. [DOI] [PubMed] [Google Scholar]
- 19. Maita H, Kitaura H, Ariga H, Iguchi-Ariga SM. Association of PAP-1 and prp3p, the products of causative genes of dominant retinitis pigmentosa, in the tri-snrnp complex. Exp Cell Res. 2005;302:61-68. [DOI] [PubMed] [Google Scholar]
- 20. Liu S, Rauhut R, Vornlocher HP, Lührmann R. The network of protein–protein interactions within the human U4/U6.U5 tri-snRNP. RNA N Y N. 2006;12:1418-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fabrizio P, Dannenberg J, Dube P, et al. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell. 2009;36:593-608. [DOI] [PubMed] [Google Scholar]
- 22. Das R, Yu J, Zhang Z, et al. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867-881. [DOI] [PubMed] [Google Scholar]
- 23. Lee KM, Tarn WY. Coupling pre-mRNA processing to transcription on the RNA factory assembly line. RNA Biol. 2013;10:380-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tanackovic G, Ransijn A, Thibault P, et al. PRPF mutations are associated with generalized defects in spliceosome formation and pre-mRNA splicing in patients with retinitis pigmentosa. Hum Mol Genet. 2011;20:2116-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-exn-10.1177_26331055221123346 for Mutations in the Spliceosome Component prp-6 and Overexpression of cdh-5 Suppress Axon Guidance Defects of cdh-4 Mutants in Caenorhabditis elegans by Zina Aburegeba, Jie Pan and Harald Hutter in Neuroscience Insights
Data Availability Statement
Strains are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.