Abstract
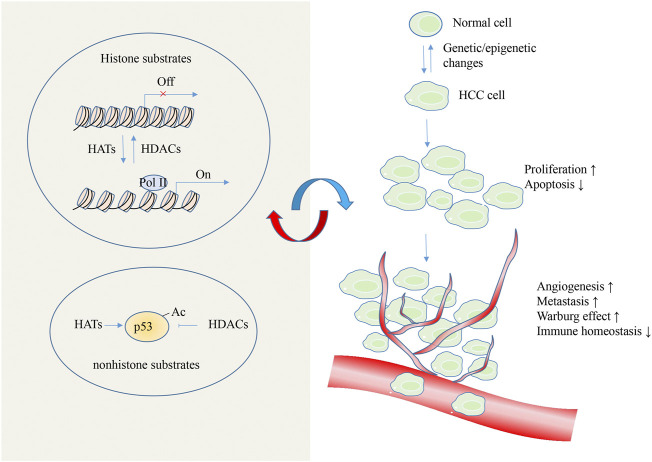
Hepatocellular Carcinoma (HCC) is the most frequent malignant tumor of the liver, but its prognosis is poor. Histone acetylation is an important epigenetic regulatory mode that modulates chromatin structure and transcriptional status to control gene expression in eukaryotic cells. Generally, histone acetylation and deacetylation processes are controlled by the opposing activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs). Dysregulation of histone modification is reported to drive aberrant transcriptional programmes that facilitate liver cancer onset and progression. Emerging studies have demonstrated that several HDAC inhibitors exert tumor-suppressive properties via activation of various cell death molecular pathways in HCC. However, the complexity involved in the epigenetic transcription modifications and non-epigenetic cellular signaling processes limit their potential clinical applications. This review brings an in-depth view of the oncogenic mechanisms reported to be related to aberrant HCC-associated histone acetylation, which might provide new insights into the effective therapeutic strategies to prevent and treat HCC.
Keywords: hepatocellula carcinoma, histone (de)acetylation, HDAC inhibition, epigenetic modfication, anticancer
1 Introduction
Hepatocellular carcinoma (HCC), the most common form of primary liver cancer and accounts for ∼90% of cases, is a severe neoplastic disease and the average 5-years survival for HCC patients is less than 15% (Erstad et al., 2019; Llovet et al., 2021). Currently, Hepatitis B virus (HBV) and hepatitis C virus (HCV) are considered as the most important pathogenic factors for HCC, but their significance will possibly decline in the coming years (Sagnelli et al., 2020). Unfortunately, the incidence rates of metabolic risk factors for HCC, including metabolic syndrome, obesity, type II diabetes and non-alcoholic fatty liver disease (NAFLD) are increasing and may jointly become the leading cause of HCC worldwide (Mcglynn et al., 2020). Despite great advances in prevention, diagnosis and therapeutic strategies, most are diagnosed at advanced stages where therapeutic options are limited, and the overall survival of patients with HCC has not improved significantly in recent decades (European Association for the Study of The et al., 2012). Sorafenib has long been the treatment strategy for advanced HCC patients; however, sorafenib resistance is considered a serious obstacle that must be overcome for HCC therapy (Zhang et al., 2012). Therefore, it is necessary to comprehend the cellular mechanisms of hepatocarcinogenesis in order to develop new and effective therapeutic targets.
Over the past decades, the development of epigenetics (e.g., microRNA, DNA methylation, and histone modification) has provided a fresh view to uncover the mechanisms of liver carcinogenesis (Bayo et al., 2019; Ganai, 2020b; Liu et al., 2021). Epigenetic phenomena refer to heritable adaptive reversible changes in gene expression that are not induced by changes in the DNA sequence (Cavalli and Heard, 2019). In eukaryotic cells, histone modifications, such as acetylation, phosphorylation, methylation, SUMOylation, and ubiquitination are a major source of molecular functional diversity, and their aberrant regulation is a common feature of many diseases (Bhat et al., 2021). Histone acetylation represent a prevalent event in epigenetic regulation and manipulates oncogenes and tumor suppressor genes during cancer progression (Lawrence et al., 2016). Generally, histone acetylation and deacetylation processes are catalyzed by the opposing activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs) (Verza et al., 2020). In human cells, HATs mainly include three subfamilies: the MYST family, the GNAT family, and the p300/CBP family, and all subfamilies include transcription factor and steroid receptor co-activators with catalytic activity (Gajer et al., 2015). According to the specialized functions of HDACs, they are divided into 4 major classes of 18 members, namely class I (HDAC1, 2, 3, 8), class II (IIa 4, 5, 7, 9, and IIb 6,10), class III [Sirtuin1-7 (SIRT1-7)], and class IV (HDAC11) (Mcclure et al., 2018). Group I HDACs (Class I, II, and VI) are zinc-dependent amidohydrolases. The majority of class I HDACs exist in the nucleus, except for HDAC3 and HDAC8, which can shuttle between the nucleus and cytoplasm (Ganai, 2019). The distribution of Class I HDACs show highly specific tissue expression. Class II HDACs are mostly located in both the nucleus and cytoplasm and require Class I HDACs to obtain catalytic activity (Martin et al., 2007). The second group of mammalian HDACs, Sirtuins, are named for their homology to the yeast silent information regulator 2 (Sir2) gene. Sirtuins are structurally and functionally distinct from Group I HDACs in that their deacetylase activity is NAD + dependent (Bheda et al., 2016). HATs are involved in histone acetylation by the transfer of acetyl groups from acetyl-CoA to lysine residues located on the histones, leading to an open state of chromatin and allowing access of transcription factors and promoting gene transcription. Conversely, HDACs erase the acetyl groups from the lysine residues located on N-terminal ends of histone proteins and recover the positive charge of lysine, resulting in a closed state of chromatin and silencing gene expression (Gray and Dangond, 2006; Falkenberg and Johnstone, 2014; Ganai, 2020c). Histone acetylation has been described to be capable of post-transcriptionally modulating various biochemical pathways that are essential for tumorigenesis (Biswas and Rao, 2017). Because HATs/HDACs can reversibly control the modifications (Tomaselli et al., 2020), it is appealing to develop epigenetic drugs as one of many tools in the fight against liver cancer.
HCC progression is a complex process with dysregulated cellular and molecular events driven by aberrant genetic and epigenetic activities. In particular, the pathogenesis of Hepatitis B virus X protein (HBx)/hepatitis C virus/nonalcoholic steatohepatitis-mediated HCC is tightly related to HATs/HDACs activities (Tsukiyama-Kohara, 2012; Liu et al., 2015; De Conti et al., 2017). Recently, quantitative acetylome analysis and lysine acetylome study revealed that abnormal histone modifications may predict prognosis in HCC patients (Zhao et al., 2020; Chai et al., 2021). Furthermore, several liver-targeting HDAC inhibitors potently suppress HCC growth and animal and preclinical studies with HDAC inhibitors suggest survival benefits (Yeo et al., 2012; Afaloniati et al., 2020; Tapadar et al., 2020). However, the role of acetylated proteins and the precise mechanism of individual HDACs in HCC progression is still not clear. In the present review, we provide an explicit summary of the roles and the underlying regulatory mechanisms of histone acetylation modification in HCC, which will provide us with new strategies for the treatment of HCC.
2 Regulatory mechanism underlying the development of hepatocellular carcinoma
2.1 Histone acetylation is implicated in hepatocellular carcinoma metastasis and angiogenesis
Cancer metastasis is the primary obstacle to successful treatment of HCC. Epithelial-mesenchymal transition (EMT), which is characterized by the loss of epithelial cell markers epithelial-cadherin (E-cadherin), as well as the increased expression of the mesenchymal proteins such as N-cadherin, Vimentin, α-smooth muscle actin (α-SMA), and the EMT-transcription factors Snail, Slug, Twist, ZEB (Wang et al., 2018), is an essential process in invasion and metastasis of cancer cell (Dong et al., 2019). Recent evidence has suggested that aberrant acetylated activity of EMT-related genes and EMT upstream genes were tightly associated with tumorigenicity and HCC metastasis (Han et al., 2018). To drive HCC metastasis from primary tumors, HDACs-mediated histone acetylation restrain E-cadherin expression or prompt mesenchymal proteins transcription, thereby facilitating migration and invasion in HCC (Han et al., 2019; Hu et al., 2019). On the contrary, EMT process and cell migration in HCC was suppressed by overexpression of the non-acetylation Vimentin (Guo et al., 2018). β-catenin pathway is one of the critical regulatory pathways in EMT process and cancer metastasis, and the acetylated status of β-catenin or the upstream signal protein kinase B (PKB) might mediate the canonical Wnt pathway in HCC (Chen et al., 2013; Yuan et al., 2020; Han et al., 2021). Interestingly, as the substrate for acetylation reactions, acetyl-CoA plays an important role in epigenetic modifications due to its dynamic association with histone acetylation; and acyl-CoA thioesterase 12 have been reported to epigenetically inducing TWIST2 expression and the promotion of EMT in HCC (Lu et al., 2019). Therefore, biological products that interact with HDAC to correct aberrant acetylated activities provide an attractive approach for cancer therapy (Huo et al., 2021; Zhang et al., 2021). Panobinostat, a new hydroxamic acid-derived histone deacetylase inhibitor (HDACI) has shown promising anticancer effects by inhibiting HCC growth and metastasis recently (Song et al., 2013). However, opposite results highlighted that HDACI promote the expression of Snail and induce EMT in hepatoma cells, thus limiting the clinical outcome of HDACI-based therapies in HCC (Xu et al., 2018; Xiao et al., 2020).
Angiogenesis has a key role in the formation of a new vascular network and HCC is largely dependent on angiogenesis for its energy supply during metastasis process (Morse et al., 2019). Angiogenic gene, such as vasohibin 2 and integrin αV subunit gene, were transcriptionally activated by histone modification and promotes angiogenesis in HCC (Xue et al., 2013; Cai et al., 2018; Cai et al., 2019). Angiogenesis is driven by hypoxic microenvironment, and the cellular response to hypoxia is triggered by the transcription factor hypoxia-inducible factors (HIFs) (Pugh and Ratcliffe, 2003). HIFs play a critical role in the adaptation of cancer cells to hypoxic conditions by activating the transcription of several pro-oncogenic genes (Albadari et al., 2019). Increasing evidence showed that the stability and activity of HIF-1α and HIF-2α were precisely regulated by acetylation modification, thereby contributing to the subsequent EMT process and HCC metastasis (Yoo et al., 2008; Liu et al., 2013; Sun et al., 2017; Cao et al., 2020). In contrast, HDACI destabilizes HIF-1α and diminishes its transcriptional activity during hypoxic microenvironment (Lee et al., 2016). Although the antiangiogenic activity of HDACI has been determined to be associated with decreased expression of proangiogenic genes, the specific effect of individual HDAC enzymes on HCC angiogenesis is still controversial (Lv et al., 2016). Therefore, more selective novel HDACIs might improve the prognosis of patients to a greater extent.
In addition, histone acetyltransferase p300 and hMOF are involved in inducing HCC migration and vascular invasion by mediating the acetylation of some oncogenes and enhancing their transcriptions (Niu et al., 2020; Pote et al., 2020; Liang et al., 2021). Epigenetic activation of the microRNAs by histone acetylation also contributes to EMT process and HCC metastasis (Zhang et al., 2013; Wen et al., 2020). Hence, targeting aberrant acetylation present a promising new class of compounds for anticancer therapy. Given the diverse molecular targets and downstream cellular pathways of HDACs (Figure 1), understanding of the context-dependent roles of individual HDACs on HCC metastasis might give us an advantage to treat cancers by exploiting this field in a specifically targeted manner.
FIGURE 1.
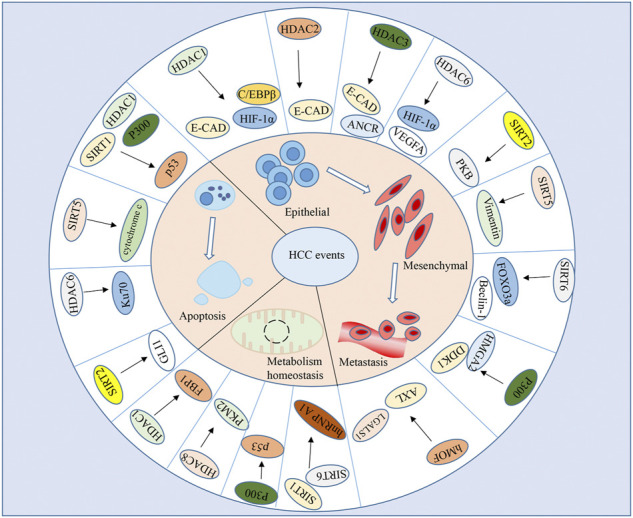
Target genes of HATs/HDACs in HCC. HATs/HDACs mediated-histone modifications affect key protein function that govern a wide array of biological processes in HCC metastasis, apoptosis, and metabolic homeostasis.
2.2 Histone acetylation is implicated in hepatocellular carcinoma metabolism
Special metabolic change, including the Warburg effect, unsaturated fatty acid biosynthesis, and so on, induces molecular changes in cancer cell, thereby allowing it to grow and proliferate in a nutrient-poor environment (Pavlova and Thompson, 2016). Cancer cells metabolize glucose to lactic acid to produce ATP and generate metabolic intermediates for the synthesis of lipids, nucleic acids, and proteins during aerobic glycolysis (Warburg effect). As such, it represents a potential therapeutic strategy for cancer. There is an amount of information indicated that histone acetylation was an important factor in cancer metabolism. More than 1000 acetylation sites in proteins were identified in human liver tissues, and metabolic enzymes accounted for a large amount (Zhao et al., 2010). More importantly, acetylation of significant enzymes in the metabolic glycolysis pathway is considered a mainly regulatory mechanism for promoting their enzymatic activities and liver cancer cell metabolism (Figure 1)(Table 1) (Hu et al., 2017; Zhang R. et al., 2020; Gao et al., 2021). The acetylation status of pyruvate kinase M2 isoform (PKM2), a key enzyme for glycolysis, affects the metabolic phenotype of HCC cells. HDAC8 reprograms the glucose metabolism of HCC cells by regulating K62 acetylation of PKM2 protein, and TSP50 promotes the Warburg effect by increasing PKM2 K433 acetylation level (Zhang R. et al., 2020; Gao et al., 2021). Sirtuin-mediated deacetylation of hnRNP A1 also suppresses glycolysis and growth in HCC by PKM2 pathway (Yang et al., 2019). Some HDACI is protective against HCC via correcting aberrant acetylated activity of fructose-1,6-bisphosphatase (FBP1) gene, thus, suppressing glucose metabolism and HCC cell growth in vitro and tumor growth in mice (Yang et al., 2017). Particularly, the tumor suppressor gene p53 was shown to revert the Warburg effect and negatively influence the oncogenic metabolic adaption of cancer cells (Gomes et al., 2018). p53 was the first non-histone protein shown to be regulated by histone acetyltransferases and histone deacetylases, and this type of modification is essential for p53 activity in HCC. The acetylated p53 is responsible for the deregulation of glycogen metabolism and represents a promising therapeutic target for the clinical management of HCC (Chen et al., 2019; Di Leo et al., 2019). Of note, the glycolytic product lactate also plays a crucial role in regulating gene transcription by inhibiting the HDAC enzymes, promoting hyperacetylation in nucleosomes and active transcriptional state (Liu and Zhang, 2018). The HAT activity of p300/CBP is often aberrantly controlled in human disease, and targeting p300/CBP has been shown to produce antitumor effects in vitro against several hematological malignancies, prostate and colorectal cancers (Du et al., 2017; Lasko et al., 2017). Increased expression of p300 has also been reported to correlate with poor survival and aggressive phenotypes in HCC, and p300 inhibitor attenuates HCC through epigenetic regulation of glycolytic function and nucleotide synthesis (Cai et al., 2021).
TABLE 1.
Key proteins modified by acetylation during HCC metabolism.
| Acetylated Proteins | upstream Regulator | metabolic Process | references |
|---|---|---|---|
| PKM2 | HDAC8 | Glucose metabolism | (Zhang et al., 2020c) |
| SIRT2 | Warburg effect | (Gao et al., 2021) | |
| SIRT1 and SIRT6 | Glycolysis | (Yang et al., 2019) | |
| p53 | P300 | lycogen metabolism | (Chen et al., 2019) |
| Glycolytic rewiring | (Di Leo et al., 2019) | ||
| FASN | ACAT1 | Lipid metabolism | (Gu et al., 2020) |
Increasing evidence suggests that hyperactive lipogenesis contribute to the establishment and maintenance of the tumorigenic state. Fatty acid synthase (FASN) is a key enzyme for the synthesis of long-chain fatty acids from malonyl-CoA, and FASN overexpression has been identified in many cancer types (Kuhajda et al., 1994). Stabilization of FASN by ACAT1-mediated GNPAT acetylation promotes lipid metabolism and HCC progression (Gu et al., 2020). Acetyl-CoA is an important metabolic intermediate that act the substrate of histone acetyltransferases regulating gene expression. It’s reported that liver mitochondrial fatty acid-derived acetyl-CoA would, like glucose-derived acetyl-CoA, be used for lipid anabolism and fuel nuclear acetylation events in citrin-deficient liver (Mention et al., 2021). Similarly, eicosapentaenoic acid (EPA), a fatty acid with anti-cancer properties, inhibited HDAC1 and DNMT expression and activity, thus promoting tumor suppressor gene expression in HCC (Ceccarelli et al., 2020).
Metabolic reprogramming plays an important role in supporting liver tumor growth. However, little is known about the histone modifications that cause HCC metabolic alterations; and whether metabolic intermediate influence the HCC progression by epigenetic manner. Those information will be helpful for better understanding the mechanisms by which oncogenic metabolites regulate the malignant phenotypes of cancer.
2.3 Histone acetylation is implicated in hepatocellular carcinoma apoptosis
Apoptosis is a precise process of programmed cell death that is crucial for progression of certain cancers including HCC. Accumulating evidence indicates that apoptotic genes can be regulated by epigenetic mechanisms (Zhou et al., 2019). Some HDAC inhibitors such as panobinostat (Choi et al., 2021), SAHA analogues (Srinivas et al., 2016), and traditional Chinese medicine galangin (Li et al., 2016) have been recently reported to regulate apoptosis in HCC through controlling the expression of pro- and anti-apoptotic genes (Buurman et al., 2016; Li and Seto, 2016). In general, administrations of HDACI can either directly prompt apoptosis through the extrinsic (death receptor)/intrinsic (mitochondria) pathway, or induce the susceptibility of tumor cells to apoptosis (Li and Seto, 2016). SIRT5 and SIRT6 were considered as the crucial lysine deacetylases that promotes HCC progression by regulating mitochondrial apoptosis (Tao et al., 2017; Zhang et al., 2019). HDAC inhibitor droxinostat could induce apoptosis in HCC cells via activation of the mitochondrial apoptotic pathway (Liu J. et al., 2016). Histone acetyltransferase PCAF also accelerates apoptosis by repressing pro-apoptotic gene BCL2-Associated X (Bax) axis or acetylating histone H4 and inactivating AKT signaling in HCC (Zheng et al., 2013; Gai et al., 2015).
One of the several biological functions of p53 is the ability to prompt apoptotic cell suicide. It’s reported that intracellular hepatitis B e antigen (HBeAg) and its precore precursors could inhibit the acetylation and translocation of p53 from cytosol to the nucleus, resulting in degradation of p53 and suppression of p53-dependent apoptosis (Liu D. et al., 2016). Long non-coding RNA (lncRNA) LOC100294145 also impedes p53 acetylation by interacting with HDAC1 and p300 to prevent HDAC1 degradation and attenuate p300 activity, leading to abrogation of p53 activity and subsequent cell proliferation and apoptosis resistance (Zhang L. Z. et al., 2020). In addition, the p53 deacetylase, SIRT1, was phosphorylated and inactivated by AMPK, resulting in p53 acetylation and apoptosis of HCC cells (Lee et al., 2012). Intriguingly, histone acetylation may regulate HCC apoptotic processes not only via p53-dependent way, but also through p53-independent pathways (Figure 1) (Lou et al., 2015; Liu D. et al., 2016; Lin et al., 2019). Treatment of pan-deacetylase inhibitor panobinostat or inducing p53 protein acetylation provide a novel therapeutic strategy for HCC by inducing apoptosis and inhibiting hepatoma cell growth (Zhu et al., 2009; Park et al., 2012; Song et al., 2013; De Matteis et al., 2018; Lim et al., 2020).
2.4 Histone acetylation is implicated in hepatocellular carcinoma immune homeostasis
Insufficient T cell infiltration in HCC limits the effectiveness of immune-checkpoint blockade (ICB) for a subset of patients. Epigenetic therapy provides further opportunities to activate cancer-associated transcriptional programs through immune regulation. It has been demonstrated that a selective HDAC8 inhibitor potentiates antitumor immunity and efficacy of immune checkpoint blockade in HCC (Yang et al., 2021). Similarly, disruption of SIRT7 increases the efficacy of checkpoint inhibitor via MEF2D regulation of programmed cell death 1 ligand 1 in HCC cells (Xiang et al., 2020). The information regarding acetylation modulation of immune in HCC is increasing, but the mechanism of selective epigenetic inhibition counteracts the immune-excluded phenotype is still unclear. Understanding the epigenomes of HCC may improve the response rate of the combination of ICB with HDACI.
2.5 Histone acetylation is implicated in cancer signaling pathway of hepatocellular carcinoma
The alterations of intracellular and extracellular cancer-associated signaling pathway have profound effects on gene transcription, cellular differentiation, and tumor microenvironment, all of which participate in the establishment and maintenance of the tumorigenic state. It has been confirmed that many cancer signaling pathways are linked with the modifications of acetylation (Table 2). An active area of research is to understand HATs/HDACs mediated-histone modifications affect key protein function and how they do so. In many cases, HDACs reverse chromatin acetylation and alter transcription of oncogenes and tumor suppressor genes by removing acetyl groups. HDACs also deacetylate nonhistone cellular substrates that govern a wide array of biological processes in liver cancer initiation and progression. HATs and HDACs activity antagonize each other to balance intracellular acetylation status. The cellular levels and biological activities of these enzymes provide a direct link between epigenetic modifications and the control of cancer signaling, transcription, and cell growth. Furthermore, acetylation of histone variant H2A.Z is also implicated in the transcriptional misregulation in cancer signaling pathway in HCC (Yuan et al., 2021). Aberrant regulation by acetylation on these signaling pathways and biological processes resulted in carcinogenesis and progression of HCC. Therefore, acetylation may function as a promising target of anti-HCC treatment.
TABLE 2.
The target cancer signaling pathways by acetylation modifications in HCC.
| HATs/HDACs | Target Signaling Pathways | Cellular Function | References |
|---|---|---|---|
| P300 | TGF-β1 signaling | Cell proliferation | Guo et al. (2021) |
| MOF | Estrogen receptor α signaling pathway | Cell growth, migration, and invasion | Wei et al. (2021) |
| N-α-acetyltransferase 20 (Naa20) | AMPK-mTOR signaling pathway | Cell proliferation, autophagy | Jung et al. (2020) |
| - | PTEN signaling | Cell proliferation and angiogenesis | Zhang et al. (2020a) |
| CBP and SIRT1 | PTEN signaling and pro-apoptotic protein caspase-3 | Cell proliferation, migration, invasion, and apoptosis | Xue et al. (2020) |
| - | p38 MAPK signaling | Cell stemness and metastasis | Luk et al. (2020) |
| HDAC3 | TRAF6/c-Myc signaling | Cell proliferation | Wu et al. (2020) |
| HDAC1 | PTEN/Akt signaling | Cell proliferation, migration and invasion | Tian et al. (2017) |
| PCAF | STAT3 signaling | Cell proliferation | Zheng et al. (2016b) |
| HDAC11 | AMPK Signaling | Cell stemness | Bi et al. (2021) |
| P300/SIRT1 | YAP signaling | Cell proliferation, apoptosis | Wang et al. (2015) |
3 Anticancer effect of histone deacetylase inhibitors in hepatocellular carcinoma
The possibility to modulate epigenetic alterations of tumor cells by HDACIs provide new treatment options for HCC that exhibit an inherent resistance to cytostatic agents (Table 3). HDACs reversibly modify the acetylated histones and nonhistones, and cause widespread alterations in genes expression without a change in DNA sequence. The disrupted acetylation homeostasis in cells might contribute to tumorigenesis, and HDACIs can counteract the abnormal acetylation status of proteins existed in liver cancer cells, and can reactivate many tumor suppressors (Lai et al., 2006). Moreover, HDAC inhibitors induced considerable cellular damage in HCC-derived cells, but did not impair cellular integrity of primary human hepatocytes (Armeanu et al., 2005). However, mechanisms of anticancer effects of HDAC inhibitors are not uniform, which may depend on the cancer type, HDAC inhibitors, doses, etc. In addition to designing inhibitors against the aberrant activity of HDAC, targeting other key molecules that regulate acetylation has also been shown to exert significant effects in anti-HCC therapy, although data are limited. For example, B029-2 (a novel p300 inhibitor) disrupts the metabolic reprogramming of HCC cells by reducing H3K18Ac and H3K27Ac levels at the promoter regions of amino acid metabolism and nucleotide synthesis enzyme genes, and thus is a potential drug for the treatment of HCC (Cai et al., 2021). Bromodomains are epigenetic "readers” of histone acetylation and bromodomain inhibitors also have exhibited promising therapeutic potential for liver cancer treatment (Cheng et al., 2021).
TABLE 3.
Anti-cancer effects of HDAC inhibitors in HCC.
| HDAC Inhibitor | Specificity | Effects in HCC | References |
|---|---|---|---|
| Panobinostat | Classes I, II, IV | inhibit HCC growth and metastasis | Song et al. (2013) |
| decreased expression of an anti-apoptotic protein | (Choi et al., 2021) | ||
| elicits effective responses to sorafenib | Lachenmayer et al. (2012) | ||
| Vorinostat (SAHA) | Classes I, II, IV | induce EMT | (Xu et al., 2018; Xiao et al., 2020) |
| sensitize HCC cells to sorafenib | Yuan et al. (2014) | ||
| sensitize HCC cells to 5-FU | Wang et al. (2021) | ||
| SAHA analogues | Classes I, II, IV | inhibits cell proliferation and induces apoptosis | Srinivas et al. (2016) |
| sodium butyrate | Classes I, II | induce EMT | (Xu et al., 2018; Xiao et al., 2020) |
| suppresses HCC growth | Yang et al. (2017) | ||
| valproate (VPA) | Classes I, II | inhibits cell proliferation and induces apoptosis | Armeanu et al. (2005) |
| eicosapentaenoic acid | HDAC1 | promotes tumor suppressor gene expression | Ceccarelli et al. (2020) |
| Santacruzamate A | HDAC2 | increasing the sensitivity of radiotherapy | Jin et al. (2021) |
| Droxinostat | HDAC3 | induces apoptosis | Liu et al. (2016b) |
| PCI-34051 | HDAC8 | elicits effective responses to ICB | Yang et al. (2021) |
| Rhamnetin | SIRT1 | enhances the antitumor effect of sorafenib | (Li et al., 2021) |
After the FDA approval of HDAC inhibitors such as vorinostat and romidepsin as anticancer agents, many novel epigenetic drugs have been investigated to reverse immune resistance and synergize with ICB treatment (Zheng H. et al., 2016). It has been confirmed that disruption of SIRT7 expression or administration of a selective HDAC8 inhibitor enhances antitumor immunity and efficacy of ICB in HCC (Xiang et al., 2020; Yang et al., 2021). In addition to the effects on the tumor cell growth, HDACI promotes the expression of MHC class I-related chain molecules A and B (MICA and MICB), resulting in an increased susceptibility of HCC cells to immune therapy (Yang et al., 2015). However, nonselective HDAC inhibition have also shown immunosuppressive effects in cutaneous T-cell lymphoma patients by reducing the activation and cytokine production of natural killer cells and dendritic cells (Kelly-Sell et al., 2012), or increasing the production and immunosuppressive functions of regulatory T (Treg) cells and myeloid-derived suppressor cells (MDSCs) (Tao et al., 2007; Rosborough et al., 2012). Given the diverse outcomes of HDAC inhibition in immunoregulation, delineating isozyme-specific HDAC control of HCC tumor microenvironment may provide insights into rational design of combination immunotherapies.
In recent years, drug combination is an effective strategy to reduce cell toxicity and improve the efficacy of therapy. Epigenetic combination therapy that comprise HDACI and demethylating agents was found to exert significant antitumor effects in HCC (Venturelli et al., 2007). A portion of HCC patients can benefit from treatments with sorafenib, adriamycin, 5-fluorouracil and platinum drugs; however, most of them eventually develop drug resistance, which partly owing to overexpression of HDACs (Ceballos et al., 2018). The combination of HDAC inhibitor such as vorinostat (SAHA) and rhamnetin (an inhibitor of SIRT1) with the antineoplastic drugs could overcome the drug resistance (especially sorafenib resistance) in HCC and notably augmented the anticancer responses (Lachenmayer et al., 2012; Yuan et al., 2014; Li et al., 2021; Wang et al., 2021). Furthermore, inhibition of HDAC2 or HDAC4 expression increases the sensitivity of liver cancer radiotherapy (Tsai et al., 2018; Jin et al., 2021). However, many epigenetic drugs of small chemical compounds are cytotoxic, and epigenetic diets are emerging as relatively safe supplementations (Lewis and Tollefsbol, 2017). Pterostilbene, a small compound isolated from plants, could serve as an novel epigenetic drug by suppressing HDAC1 activity (Qian et al., 2017; Qian et al., 2018), thus opens up new avenue for the prevention and treatment of epigenetic disorders in HCC. Intriguingly, plant flavonoid luteolin also exert therapeutic impact by restoring ethanol-depleted SIRT1 activity in pre-neoplastic liver lesion mouse model (Ganai et al., 2021). These results suggest that epigenetic diets might correct aberrant HDACs abilities to maintain acetylation homeostasis in HCC.
4 Discussion
In the last decades, epigenetic modifications has been validated to contribute to the process of various kinds of cancers (Dawson and Kouzarides, 2012). Histone deacetylation is one of the earliest discovered epigenetic mechanisms, regulating many cellular events such as differentiation, proliferation, apoptosis, metabolic changes, metastasis and immune homeostasis in HCC (Figure 2). As a key regulator of acetylation status, HATs/HDACs have been found to dysregulate and/or function incorrectly in HCC, thereby providing a crucial attractive target for HCC treatment (Table 4). Currently, there are numerous HDACIs such as vorinostat, romidepsin, belinostat, panobinostat, tazemetostat, and chidamide are approved by the United States Food and Drug Administration for clinical treatment (Li and Seto, 2016; Ganai, 2020a; Nepali and Liou, 2021). However, opposite regulatory roles of HDACs were observed in HCC. Besides, the efficacies of HDAC inhibitory compounds observed against solid tumours have been disappointing, possibly owing to the lack of specificity. There is reason to believe that maintenance of the balance of histone acetylation modifications is essential for the regulation of gene expression and the maintenance of the normal status of cells. More studies are needed to systematically dissect the role and precise mechanisms of individual HDACs in HCC, which will give United States mechanistic-based rationale for the clinical use of HDACI. In addition, the effectiveness of nonselective HDACI relies on its broad-spectrum inhibition against HDACs, long-term uses of broad spectrum nonselective HDACI are potentially cytotoxic and might induce intolerable side effects in certain patients. Moreover, the activities of HDAC are often mechanistically connected with DNA methylation, miRNAs and lncRNA in HCC (Zhang et al., 2010; Yuan et al., 2011; Ding et al., 2017). Therefore, a combination of epigenetic drugs targeting multiple epigenetic alterations might incur fewer side effects. In parallel, it is anticipated that future research developing HDACI with higher target specificity that might be more efficacious with less toxicity. Because epigenetic modifications occur in a tissue, cell, or gene-specific manner, different targeted genes of HDACI might cause its distinct influences. Thus, further identification of the key genes of acetylation modifications and understanding the underlying regulatory mechanisms might lead to clinical benefits for HCC.
FIGURE 2.
Roles of HATs/HDACs in HCC. Aberrant HATs/HDACs-mediated histone acetylation trigger oncogenes activation, and loss in tumor suppressor gene expression to lead the HCC establishment. p53 was the representative non-histone protein that shown to be acetylated/deacetylated by HATs/HDACs, and this type of modification is essential for p53 activity in HCC. In turn, the metabolic product from HCC also influence the acetylation modifications.
TABLE 4.
Expression and target genes of HATs/HDACs in HCC.
| HATs/HDACs | Differential expression in HCC | Target genes | Biological processes/cellular functions | References |
|---|---|---|---|---|
| HDAC1 | ↑ | E-cadherin | EMT | Hu et al. (2019) |
| CCAAT/enhancer binding protein β (C/EBPβ) | EMT | Huo et al. (2021) | ||
| hypoxia-inducible factor 1α (HIF-1α) | EMT | Yoo et al. (2008); Liu et al. (2013) | ||
| FBP1 | gluconeogenesis | Yang et al. (2017) | ||
| P53 | Apoptosis | Zhang et al. (2020b) | ||
| HDAC2 | ↑ | E-cadherin | EMT | Hu et al. (2019) |
| integrin αV subunit gene | cell migration | Cai et al. (2019) | ||
| HDAC3 | - | E-cadherin | EMT | Hu et al. (2019) |
| ANCR | HCC metastasis | Wen et al. (2020) | ||
| HDAC6 | ↓ | HIF-1α and VEGFA | angiogenesis | Lv et al. (2016) |
| HDAC8 | ↓ | PKM2 | Glycolysis | Zhang et al. (2020c) |
| SIRT1 | ↑ | hnRNP A1 | Glycolysis | Yang et al. (2019) |
| LC3 | Autophagy | Li et al. (2016) | ||
| P53 | Apoptosis | Lee et al. (2012); Lin et al. (2019) | ||
| SIRT2 | ↑ | protein kinase B | EMT | Chen et al. (2013) |
| SIRT5 | ↑ | cytochrome c | mitochondrial apoptosis | Zhang et al. (2019) |
| SIRT5 | ↓ | Vimentin | EMT | Guo et al. (2018) |
| SIRT6 | ↑ | FOXO3a; Beclin-1 | EMT | Han et al. (2019) |
| hnRNP A1 | Glycolysis | Yang et al. (2019) | ||
| Ku70 | Apoptosis | Tao et al. (2017) | ||
| SIRT7 | ↑ | PGK1 | cell proliferation | Hu et al. (2017) |
| MEF2D | Immunity | Xiang et al. (2020) | ||
| P300 | ↑ | HMGA2 | HCC metastasis | Liang et al. (2021) |
| DDK1 | HCC metastasis | Niu et al. (2020) | ||
| P53 | glycogen metabolism | Chen et al. (2019); Di Leo et al. (2019) | ||
| P53 | apoptosis | Zhang et al. (2020b) | ||
| hMOF | ↑ | AXL;LGALS1 | cell migration | Pote et al. (2020) |
| PCAF | ↓ | PGK1 | cell proliferation | Hu et al. (2017) |
| GLI1 | apoptosis | Gai et al. (2015) |
Acknowledgments
The authors would like to acknowledge the technical assistance provided by the staff of the Department of Hepatobiliary Surgery, The Affiliated Drum Tower Hospital of Nanjing University Medical School, Nanjing, China.
Author contributions
J-KX conceived the idea and designed the review, X-QQ, LZ, S-JL draw the figures and tables. H-ZR and X-LS analyzed data and wrote the paper.
Funding
This work was funded by the National Natural Science Foundation of China (81872359), the Fundamental Research Funds for the Central Universities (0214–14380510), the Nanjing health science and technology development project for Distinguished Young Scholars (JQX19002), the Chen Xiao-ping Foundation for the Development of Science and Technology of Hubei Province, China (CXPJJH121001-2021073).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
HCC, hepatocellular carcinoma; HBV, Hepatitis B virus; HCV, hepatitis C virus; NAFLD, non-alcoholic fatty liver disease; HATs, histone acetyltransferases; HDACs, histone deacetylases; Sir2, silent information regulator 2; HBx, hepatitis B virus X protein; EMT, epithelial-mesenchymal transition; E-cadherin; E-cadherin, epithelial-cadherin; α-SMA, α-smooth muscle actin; PKB, protein kinase B; HDACI, histone deacetylase inhibitor; HIFs, hypoxia-inducible factors; PKM2, pyruvate kinase M2 isoform; FBP1, fructose-1,6-bisphosphatase; FASN, fatty acid synthase; EPA, eicosapentaenoic acid; Bax, BCL2-Associated X; HBeAg, hepatitis B e antigen; lncRNA, long non-coding RNA; ICB, immune-checkpoint blockade; MICA and MICB, MHC class I-related chain molecules A and B; MDSCs, myeloid-derived suppressor cells.
References
- Afaloniati H., Angelopoulou K., Giakoustidis A., Hardas A., Pseftogas A., Makedou K., et al. (2020). HDAC1/2 inhibitor romidepsin suppresses DEN-induced hepatocellular carcinogenesis in mice. Onco. Targets. Ther. 13, 5575–5588. 10.2147/OTT.S250233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albadari N., Deng S., Li W. (2019). The transcriptional factors HIF-1 and HIF-2 and their novel inhibitors in cancer therapy. Expert Opin. Drug Discov. 14, 667–682. 10.1080/17460441.2019.1613370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armeanu S., Pathil A., Venturelli S., Mascagni P., Weiss T. S., Gottlicher M., et al. (2005). Apoptosis on hepatoma cells but not on primary hepatocytes by histone deacetylase inhibitors valproate and ITF2357. J. Hepatol. 42, 210–217. 10.1016/j.jhep.2004.10.020 [DOI] [PubMed] [Google Scholar]
- Bayo J., Fiore E. J., Dominguez L. M., Real A., Malvicini M., Rizzo M., et al. (2019). A comprehensive study of epigenetic alterations in hepatocellular carcinoma identifies potential therapeutic targets. J. Hepatol. 71, 78–90. 10.1016/j.jhep.2019.03.007 [DOI] [PubMed] [Google Scholar]
- Bhat K. P., Umit Kaniskan H., Jin J., Gozani O. (2021). Epigenetics and beyond: Targeting writers of protein lysine methylation to treat disease. Nat. Rev. Drug Discov. 20, 265–286. 10.1038/s41573-020-00108-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bheda P., Jing H., Wolberger C., Lin H. (2016). The substrate specificity of sirtuins. Annu. Rev. Biochem. 85, 405–429. 10.1146/annurev-biochem-060815-014537 [DOI] [PubMed] [Google Scholar]
- Bi L., Ren Y., Feng M., Meng P., Wang Q., Chen W., et al. (2021). HDAC11 regulates glycolysis through the LKB1/AMPK signaling pathway to maintain hepatocellular carcinoma stemness. Cancer Res. 81, 2015–2028. 10.1158/0008-5472.CAN-20-3044 [DOI] [PubMed] [Google Scholar]
- Biswas S., Rao C. M. (2017). Epigenetics in cancer: Fundamentals and beyond. Pharmacol. Ther. 173, 118–134. 10.1016/j.pharmthera.2017.02.011 [DOI] [PubMed] [Google Scholar]
- Buurman R., Sandbothe M., Schlegelberger B., Skawran B. (2016). HDAC inhibition activates the apoptosome via Apaf1 upregulation in hepatocellular carcinoma. Eur. J. Med. Res. 21, 26. 10.1186/s40001-016-0217-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L. Y., Chen S. J., Xiao S. H., Sun Q. J., Ding C. H., Zheng B. N., et al. (2021). Targeting p300/CBP attenuates hepatocellular carcinoma progression through epigenetic regulation of metabolism. Cancer Res. 81, 860–872. 10.1158/0008-5472.CAN-20-1323 [DOI] [PubMed] [Google Scholar]
- Cai Q., Liu Y., Zhu P., Kang C., Xu H., Qi B., et al. (2019). SIN3B promotes integrin αV subunit gene transcription and cell migration of hepatocellular carcinoma. J. Mol. Cell Biol. 11, 421–432. 10.1093/jmcb/mjy050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q. Q., Dong Y. W., Qi B., Shao X. T., Wang R., Chen Z. Y., et al. (2018). BRD1-Mediated acetylation promotes integrin αV gene expression via interaction with Sulfatide. Mol. Cancer Res. 16, 610–622. 10.1158/1541-7786.MCR-17-0527 [DOI] [PubMed] [Google Scholar]
- Cao M. Q., You A. B., Cui W., Zhang S., Guo Z. G., Chen L., et al. (2020). Cross talk between oxidative stress and hypoxia via thioredoxin and HIF-2α drives metastasis of hepatocellular carcinoma. FASEB J. 34, 5892–5905. 10.1096/fj.202000082R [DOI] [PubMed] [Google Scholar]
- Cavalli G., Heard E. (2019). Advances in epigenetics link genetics to the environment and disease. Nature 571, 489–499. 10.1038/s41586-019-1411-0 [DOI] [PubMed] [Google Scholar]
- Ceballos M. P., Decandido G., Quiroga A. D., Comanzo C. G., Livore V. I., Lorenzetti F., et al. (2018). Inhibition of sirtuins 1 and 2 impairs cell survival and migration and modulates the expression of P-glycoprotein and MRP3 in hepatocellular carcinoma cell lines. Toxicol. Lett. 289, 63–74. 10.1016/j.toxlet.2018.03.011 [DOI] [PubMed] [Google Scholar]
- Ceccarelli V., Ronchetti S., Marchetti M. C., Calvitti M., Riccardi C., Grignani F., et al. (2020). Molecular mechanisms underlying eicosapentaenoic acid inhibition of HDAC1 and DNMT expression and activity in carcinoma cells. Biochim. Biophys. Acta. Gene Regul. Mech. 1863, 194481. 10.1016/j.bbagrm.2020.194481 [DOI] [PubMed] [Google Scholar]
- Chai X., Guo J., Dong R., Yang X., Deng C., Wei C., et al. (2021). Quantitative acetylome analysis reveals histone modifications that may predict prognosis in hepatitis B-related hepatocellular carcinoma. Clin. Transl. Med. 11, e313. 10.1002/ctm2.313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Chan A. W., To K. F., Chen W., Zhang Z., Ren J., et al. (2013). SIRT2 overexpression in hepatocellular carcinoma mediates epithelial to mesenchymal transition by protein kinase B/glycogen synthase kinase-3β/β-catenin signaling. Hepatology 57, 2287–2298. 10.1002/hep.26278 [DOI] [PubMed] [Google Scholar]
- Chen S. L., Zhang C. Z., Liu L. L., Lu S. X., Pan Y. H., Wang C. H., et al. (2019). A GYS2/p53 negative Feedback Loop Restricts tumor growth in HBV-related hepatocellular carcinoma. Cancer Res. 79, 534–545. 10.1158/0008-5472.CAN-18-2357 [DOI] [PubMed] [Google Scholar]
- Cheng C. L., Tsang F. H., Wei L., Chen M., Chin D. W., Shen J., et al. (2021). Bromodomain-containing protein BRPF1 is a therapeutic target for liver cancer. Commun. Biol. 4, 888. 10.1038/s42003-021-02405-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C., Lee G. H., Son A., Yoo G. S., Yu J. I., Park H. C. (2021). Downregulation of Mcl-1 by panobinostat potentiates Proton Beam therapy in hepatocellular carcinoma cells. Cells 10, 554. 10.3390/cells10030554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M. A., Kouzarides T. (2012). Cancer epigenetics: From mechanism to therapy. Cell 150, 12–27. 10.1016/j.cell.2012.06.013 [DOI] [PubMed] [Google Scholar]
- De Conti A., Dreval K., Tryndyak V., Orisakwe O. E., Ross S. A., Beland F. A., et al. (2017). Inhibition of the cell death pathway in nonalcoholic steatohepatitis (NASH)-Related hepatocarcinogenesis is associated with histone H4 lysine 16 deacetylation. Mol. Cancer Res. 15, 1163–1172. 10.1158/1541-7786.MCR-17-0109 [DOI] [PubMed] [Google Scholar]
- De Matteis S., Ragusa A., Marisi G., De Domenico S., Casadei Gardini A., Bonafe M., et al. (2018). Aberrant metabolism in hepatocellular carcinoma provides Diagnostic and therapeutic opportunities. Oxid. Med. Cell. Longev. 2018, 7512159. 10.1155/2018/7512159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leo L., Vegliante R., Ciccarone F., Salvatori I., Scimeca M., Bonanno E., et al. (2019). Forcing ATGL expression in hepatocarcinoma cells imposes glycolytic rewiring through PPAR-α/p300-mediated acetylation of p53. Oncogene 38, 1860–1875. 10.1038/s41388-018-0545-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G., Li W., Liu J., Zeng Y., Mao C., Kang Y., et al. (2017). LncRNA GHET1 activated by H3K27 acetylation promotes cell tumorigenesis through regulating ATF1 in hepatocellular carcinoma. Biomed. Pharmacother. 94, 326–331. 10.1016/j.biopha.2017.07.046 [DOI] [PubMed] [Google Scholar]
- Dong Y., Zheng Q., Wang Z., Lin X., You Y., Wu S., et al. (2019). Higher matrix stiffness as an independent initiator triggers epithelial-mesenchymal transition and facilitates HCC metastasis. J. Hematol. Oncol. 12, 112. 10.1186/s13045-019-0795-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C., Huang D., Peng Y., Yao Y., Zhao Y., Yang Y., et al. (2017). 5-Fluorouracil targets histone acetyltransferases p300/CBP in the treatment of colorectal cancer. Cancer Lett. 400, 183–193. 10.1016/j.canlet.2017.04.033 [DOI] [PubMed] [Google Scholar]
- Erstad D. J., Razavi A. A., Li S., Tanabe K. K., Fuchs B. C. (2019). “Prevention strategies for hepatocellular carcinoma,” in Hepatocellular carcinoma: Translational precision medicine approaches. Editors Hoshida Y., Cham, 255–289. (CH)). [Google Scholar]
- European Association for the Study of The L., European Organisation For R., Treatment Of C. (2012). EASL-EORTC clinical practice guidelines: Management of hepatocellular carcinoma. J. Hepatol. 56, 908–943. 10.1016/j.jhep.2011.12.001 [DOI] [PubMed] [Google Scholar]
- Falkenberg K. J., Johnstone R. W. (2014). Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 13, 673–691. 10.1038/nrd4360 [DOI] [PubMed] [Google Scholar]
- Gai X., Tu K., Li C., Lu Z., Roberts L. R., Zheng X. (2015). Histone acetyltransferase PCAF accelerates apoptosis by repressing a GLI1/BCL2/BAX axis in hepatocellular carcinoma. Cell Death Dis. 6, e1712. 10.1038/cddis.2015.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajer J. M., Furdas S. D., Grunder A., Gothwal M., Heinicke U., Keller K., et al. (2015). Histone acetyltransferase inhibitors block neuroblastoma cell growth in vivo . Oncogenesis 4, e137. 10.1038/oncsis.2014.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganai S. A. (2020a). Conspectus of structurally distinct groups of histone deacetylase inhibitors of Classical histone deacetylases and sirtuins. Histone Deacetylase Inhibitors in Combinatorial Anticancer Therapy. [Google Scholar]
- Ganai S. A. (2019). HDACs and their distinct classes. Histone Deacetylase Inhibitors — Epidrugs for Neurological Disorders. [Google Scholar]
- Ganai S. A. (2020b). Histone deacetylase inhibitors in Combinatorial anticancer therapy. Histone Deacetylase Inhibitors in Combinatorial Anticancer Therapy. [Google Scholar]
- Ganai S. A. (2020c). Recap of distinct molecular signalling mechanisms modulated by histone deacetylases for cancer genesis and progression. Histone Deacetylase Inhibitors in Combinatorial Anticancer Therapy. [Google Scholar]
- Ganai S. A., Sheikh F. A., Baba Z. A., Mir M. A., Mantoo M. A., Yatoo M. A. (2021). Anticancer activity of the plant flavonoid luteolin against preclinical models of various cancers and insights on different signalling mechanisms modulated. Phytother. Res. 35, 3509–3532. 10.1002/ptr.7044 [DOI] [PubMed] [Google Scholar]
- Gao F., Zhang X., Wang S., Zheng L., Sun Y., Wang G., et al. (2021). TSP50 promotes the Warburg effect and hepatocyte proliferation via regulating PKM2 acetylation. Cell Death Dis. 12, 517. 10.1038/s41419-021-03782-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes A. S., Ramos H., Soares J., Saraiva L. (2018). p53 and glucose metabolism: an orchestra to be directed in cancer therapy. Pharmacol. Res. 131, 75–86. 10.1016/j.phrs.2018.03.015 [DOI] [PubMed] [Google Scholar]
- Gray S. G., Dangond F. (2006). Rationale for the use of histone deacetylase inhibitors as a dual therapeutic modality in multiple sclerosis. Epigenetics 1, 67–75. 10.4161/epi.1.2.2678 [DOI] [PubMed] [Google Scholar]
- Gu L., Zhu Y., Lin X., Tan X., Lu B., Li Y. (2020). Stabilization of FASN by ACAT1-mediated GNPAT acetylation promotes lipid metabolism and hepatocarcinogenesis. Oncogene 39, 2437–2449. 10.1038/s41388-020-1156-0 [DOI] [PubMed] [Google Scholar]
- Guo D., Song X., Guo T., Gu S., Chang X., Su T., et al. (2018). Vimentin acetylation is involved in SIRT5-mediated hepatocellular carcinoma migration. Am. J. Cancer Res. 8, 2453–2466. [PMC free article] [PubMed] [Google Scholar]
- Guo L., Li H., Fan T., Ma Y., Wang L. (2021). Synergistic efficacy of curcumin and anti-programmed cell death-1 in hepatocellular carcinoma. Life Sci. 279, 119359. 10.1016/j.lfs.2021.119359 [DOI] [PubMed] [Google Scholar]
- Han L. L., Jia L., Wu F., Huang C. (2019). Sirtuin6 (SIRT6) promotes the EMT of hepatocellular carcinoma by Stimulating autophagic degradation of E-cadherin. Mol. Cancer Res. 17, 2267–2280. 10.1158/1541-7786.MCR-19-0321 [DOI] [PubMed] [Google Scholar]
- Han Q., Chen C. A., Yang W., Liang D., Lv H. W., Lv G. S., et al. (2021). ATP-citrate lyase regulates stemness and metastasis in hepatocellular carcinoma via the Wnt/β-catenin signaling pathway. Hepatobiliary Pancreat. Dis. Int. 20, 251–261. 10.1016/j.hbpd.2020.05.010 [DOI] [PubMed] [Google Scholar]
- Han T. S., Ban H. S., Hur K., Cho H. S. (2018). The epigenetic regulation of HCC metastasis. Int. J. Mol. Sci. 19, E3978. 10.3390/ijms19123978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Zhu W., Qin J., Chen M., Gong L., Li L., et al. (2017). Acetylation of PGK1 promotes liver cancer cell proliferation and tumorigenesis. Hepatology 65, 515–528. 10.1002/hep.28887 [DOI] [PubMed] [Google Scholar]
- Hu Y., Zheng Y., Dai M., Wang X., Wu J., Yu B., et al. (2019). G9a and histone deacetylases are crucial for Snail2-mediated E-cadherin repression and metastasis in hepatocellular carcinoma. Cancer Sci. 110, 3442–3452. 10.1111/cas.14173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo T. X., Wang X. P., Yu Z., Kong B., He Y., Guo Q. L., et al. (2021). Oroxylin A inhibits the migration of hepatocellular carcinoma cells by inducing NAG-1 expression. Acta Pharmacol. Sin. 43, 724–734. 10.1038/s41401-021-00695-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q., Hu H., Yan S., Jin L., Pan Y., Li X., et al. (2021). lncRNA mir22hg-derived miR-22-5p enhances the Radiosensitivity of hepatocellular carcinoma by increasing histone acetylation through the inhibition of HDAC2 activity. Front. Oncol. 11, 572585. 10.3389/fonc.2021.572585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T. Y., Ryu J. E., Jang M. M., Lee S. Y., Jin G. R., Kim C. W., et al. (2020). Naa20, the catalytic subunit of NatB complex, contributes to hepatocellular carcinoma by regulating the LKB1-AMPK-mTOR axis. Exp. Mol. Med. 52, 1831–1844. 10.1038/s12276-020-00525-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly-Sell M. J., Kim Y. H., Straus S., Benoit B., Harrison C., Sutherland K., et al. (2012). The histone deacetylase inhibitor, romidepsin, suppresses cellular immune functions of cutaneous T-cell lymphoma patients. Am. J. Hematol. 87, 354–360. 10.1002/ajh.23112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhajda F. P., Jenner K., Wood F. D., Hennigar R. A., Jacobs L. B., Dick J. D., et al. (1994). Fatty acid synthesis: A potential selective target for antineoplastic therapy. Proc. Natl. Acad. Sci. U. S. A. 91, 6379–6383. 10.1073/pnas.91.14.6379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachenmayer A., Toffanin S., Cabellos L., Alsinet C., Hoshida Y., Villanueva A., et al. (2012). Combination therapy for hepatocellular carcinoma: Additive preclinical efficacy of the HDAC inhibitor panobinostat with sorafenib. J. Hepatol. 56, 1343–1350. 10.1016/j.jhep.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J. P., Yu C., Moser C. D., Aderca I., Han T., Garvey T. D., et al. (2006). SULF1 inhibits tumor growth and potentiates the effects of histone deacetylase inhibitors in hepatocellular carcinoma. Gastroenterology 130, 2130–2144. 10.1053/j.gastro.2006.02.056 [DOI] [PubMed] [Google Scholar]
- Lasko L. M., Jakob C. G., Edalji R. P., Qiu W., Montgomery D., Digiammarino E. L., et al. (2017). Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550, 128–132. 10.1038/nature24028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M., Daujat S., Schneider R. (2016). Lateral thinking: How histone modifications regulate gene expression. Trends Genet. 32, 42–56. 10.1016/j.tig.2015.10.007 [DOI] [PubMed] [Google Scholar]
- Lee C. W., Wong L. L., Tse E. Y., Liu H. F., Leong V. Y., Lee J. M., et al. (2012). AMPK promotes p53 acetylation via phosphorylation and inactivation of SIRT1 in liver cancer cells. Cancer Res. 72, 4394–4404. 10.1158/0008-5472.CAN-12-0429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. D., Kim W., Jeong J. W., Park J. W., Kim J. E. (2016). AK-1, a SIRT2 inhibitor, destabilizes HIF-1α and diminishes its transcriptional activity during hypoxia. Cancer Lett. 373, 138–145. 10.1016/j.canlet.2016.01.031 [DOI] [PubMed] [Google Scholar]
- Lewis K. A., Tollefsbol T. O. (2017). The influence of an epigenetics diet on the cancer epigenome. Epigenomics 9, 1153–1155. 10.2217/epi-2017-0077 [DOI] [PubMed] [Google Scholar]
- Li B., Feng F., Jia H., Jiang Q., Cao S., Wei L., et al. (2021). Rhamnetin decelerates the elimination and enhances the antitumor effect of the molecular-targeting agent sorafenib in hepatocellular carcinoma cells via the miR-148a/PXR axis. Food Funct. 12, 2404–2417. 10.1039/d0fo02270e [DOI] [PubMed] [Google Scholar]
- Li X., Wang Y., Xiong Y., Wu J., Ding H., Chen X., et al. (2016). Galangin induces autophagy via deacetylation of LC3 by SIRT1 in HepG2 cells. Sci. Rep. 6, 30496. 10.1038/srep30496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Seto E. (2016). HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb. Perspect. Med. 6, a026831. 10.1101/cshperspect.a026831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Niu J., Wang X., Zhang Z. S., Yang R. H., Yao X., et al. (2021). P300-dependent acetylation of histone H3 is required for epidermal growth factor receptor-mediated high-mobility group protein A2 transcription in hepatocellular carcinoma. Cancer Sci. 112, 679–690. 10.1111/cas.14729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J. Y., Liu C., Hu K. Q., Smith D. E., Wu D., Lamon-Fava S., et al. (2020). Xanthophyll beta-Cryptoxanthin inhibits highly Refined Carbohydrate diet-Promoted hepatocellular carcinoma progression in mice. Mol. Nutr. Food Res. 64, e1900949. 10.1002/mnfr.201900949 [DOI] [PubMed] [Google Scholar]
- Lin C. Y., Islam A., Su C. J., Tikhomirov A. S., Shchekotikhin A. E., Chuang S. M., et al. (2019). Histone Demethylase KDM4C stimulates the proliferation of prostate cancer cells via activation of AKT and c-Myc. Cancers (Basel) 11, E1785. 10.3390/cancers11111785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Cui L., Wang Y., Yang G., He J., Hao R., et al. (2016a). Hepatitis B e antigen and its precursors promote the progress of hepatocellular carcinoma by interacting with NUMB and decreasing p53 activity. Hepatology 64, 390–404. 10.1002/hep.28594 [DOI] [PubMed] [Google Scholar]
- Liu G. M., Zhang Y. M. (2018). Targeting FBPase is an emerging novel approach for cancer therapy. Cancer Cell Int. 18, 36. 10.1186/s12935-018-0533-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Li G., Wang X., Wang L., Zhao R., Wang J., et al. (2016b). Droxinostat, a histone deacetylase inhibitor, induces apoptosis in hepatocellular carcinoma cell lines via activation of the mitochondrial pathway and Downregulation of FLIP. Transl. Oncol. 9, 70–78. 10.1016/j.tranon.2016.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Y., Tang S. H., Wu S. L., Luo Y. H., Cao M. R., Zhou H. K., et al. (2015). Epigenetic modulation of insulin-like growth factor-II overexpression by Hepatitis B virus X protein in hepatocellular carcinoma. Am. J. Cancer Res. 5, 956–978. [PMC free article] [PubMed] [Google Scholar]
- Liu Y. R., Wang J. Q., Huang Z. G., Chen R. N., Cao X., Zhu D. C., et al. (2021). Histone deacetylase2: A potential regulator and therapeutic target in liver disease (review). Int. J. Mol. Med. 48, 131. 10.3892/ijmm.2021.4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang J. B., Qin Y., Wang W., Wei L., Teng Y., et al. (2013). PROX1 promotes hepatocellular carcinoma metastasis by way of up-regulating hypoxia-inducible factor 1α expression and protein stability. Hepatology 58, 692–705. 10.1002/hep.26398 [DOI] [PubMed] [Google Scholar]
- Llovet J. M., Kelley R. K., Villanueva A., Singal A. G., Pikarsky E., Roayaie S., et al. (2021). Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 7, 6. 10.1038/s41572-020-00240-3 [DOI] [PubMed] [Google Scholar]
- Lou G., Liu Y., Wu S., Xue J., Yang F., Fu H., et al. (2015). The p53/miR-34a/SIRT1 positive Feedback Loop in quercetin-induced apoptosis. Cell. Physiol. biochem. 35, 2192–2202. 10.1159/000374024 [DOI] [PubMed] [Google Scholar]
- Lu M., Zhu W. W., Wang X., Tang J. J., Zhang K. L., Yu G. Y., et al. (2019). ACOT12-Dependent alteration of acetyl-CoA drives hepatocellular carcinoma metastasis by epigenetic induction of epithelial-mesenchymal transition. Cell Metab. 29, 886–900.e5. 10.1016/j.cmet.2018.12.019 [DOI] [PubMed] [Google Scholar]
- Luk S. T., Ng K. Y., Zhou L., Tong M., Wong T. L., Yu H., et al. (2020). Deficiency in embryonic Stem cell marker reduced expression 1 activates Mitogen-activated protein kinase kinase 6-dependent p38 Mitogen-activated protein kinase signaling to drive hepatocarcinogenesis. Hepatology 72, 183–197. 10.1002/hep.31020 [DOI] [PubMed] [Google Scholar]
- Lv Z., Weng X., Du C., Zhang C., Xiao H., Cai X., et al. (2016). Downregulation of HDAC6 promotes angiogenesis in hepatocellular carcinoma cells and predicts poor prognosis in liver transplantation patients. Mol. Carcinog. 55, 1024–1033. 10.1002/mc.22345 [DOI] [PubMed] [Google Scholar]
- Martin M., Kettmann R., Dequiedt F. (2007). Class IIa histone deacetylases: Regulating the regulators. Oncogene 26, 5450–5467. 10.1038/sj.onc.1210613 [DOI] [PubMed] [Google Scholar]
- Mcclure J. J., Li X., Chou C. J. (2018). Advances and Challenges of HDAC inhibitors in cancer therapeutics. Adv. Cancer Res. 138, 183–211. 10.1016/bs.acr.2018.02.006 [DOI] [PubMed] [Google Scholar]
- Mcglynn K. A., Petrick J. L., El-Serag H. B. (2020). Epidemiology of hepatocellular carcinoma. Hepatology 73, 4–13. 10.1002/hep.31288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mention K., Joncquel Chevalier Curt M., Dessein A. F., Douillard C., Dobbelaere D., Vamecq J. (2021). Citrin deficiency: Does the reactivation of liver aralar-1 come into play and promote HCC development? Biochimie 190, 20–23. 10.1016/j.biochi.2021.06.018 [DOI] [PubMed] [Google Scholar]
- Morse M. A., Sun W., Kim R., He A. R., Abada P. B., Mynderse M., et al. (2019). The role of angiogenesis in hepatocellular carcinoma. Clin. Cancer Res. 25, 912–920. 10.1158/1078-0432.CCR-18-1254 [DOI] [PubMed] [Google Scholar]
- Nepali K., Liou J. P. (2021). Recent developments in epigenetic cancer therapeutics: Clinical advancement and emerging trends. J. Biomed. Sci. 28, 27. 10.1186/s12929-021-00721-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J., Li W., Liang C., Wang X., Yao X., Yang R. H., et al. (2020). EGF promotes DKK1 transcription in hepatocellular carcinoma by enhancing the phosphorylation and acetylation of histone H3. Sci. Signal. 13, eabb5727. 10.1126/scisignal.abb5727 [DOI] [PubMed] [Google Scholar]
- Park S. Y., Lee K. B., Lee M. J., Bae S. C., Jang J. J. (2012). Nicotinamide inhibits the early stage of carcinogen-induced hepatocarcinogenesis in mice and suppresses human hepatocellular carcinoma cell growth. J. Cell. Physiol. 227, 899–908. 10.1002/jcp.22799 [DOI] [PubMed] [Google Scholar]
- Pavlova N. N., Thompson C. B. (2016). The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47. 10.1016/j.cmet.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pote N., Cros J., Laouirem S., Raffenne J., Negrao M., Albuquerque M., et al. (2020). The histone acetyltransferase hMOF promotes vascular invasion in hepatocellular carcinoma. Liver Int. 40, 956–967. 10.1111/liv.14381 [DOI] [PubMed] [Google Scholar]
- Pugh C. W., Ratcliffe P. J. (2003). Regulation of angiogenesis by hypoxia: Role of the HIF system. Nat. Med. 9, 677–684. 10.1038/nm0603-677 [DOI] [PubMed] [Google Scholar]
- Qian Y. Y., Liu Z. S., Pan D. Y., Li K. (2017). Tumoricidal activities of pterostilbene depend upon destabilizing the MTA1-NuRD complex and enhancing P53 acetylation in hepatocellular carcinoma. Exp. Ther. Med. 14, 3098–3104. 10.3892/etm.2017.4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y. Y., Liu Z. S., Yan H. J., Yuan Y. F., Levenson A. S., Li K. (2018). Pterostilbene inhibits MTA1/HDAC1 complex leading to PTEN acetylation in hepatocellular carcinoma. Biomed. Pharmacother. 101, 852–859. 10.1016/j.biopha.2018.03.022 [DOI] [PubMed] [Google Scholar]
- Rosborough B. R., Castellaneta A., Natarajan S., Thomson A. W., Turnquist H. R. (2012). Histone deacetylase inhibition facilitates GM-CSF-mediated expansion of myeloid-derived suppressor cells in vitro and in vivo . J. Leukoc. Biol. 91, 701–709. 10.1189/jlb.0311119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagnelli E., Macera M., Russo A., Coppola N., Sagnelli C. (2020). Epidemiological and etiological variations in hepatocellular carcinoma. Infection 48, 7–17. 10.1007/s15010-019-01345-y [DOI] [PubMed] [Google Scholar]
- Song X., Wang J., Zheng T., Song R., Liang Y., Bhatta N., et al. (2013). LBH589 Inhibits proliferation and metastasis of hepatocellular carcinoma via inhibition of gankyrin/STAT3/Akt pathway. Mol. Cancer 12, 114. 10.1186/1476-4598-12-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas C., Swathi V., Priyanka C., Anjana Devi T., Subba Reddy B. V., Janaki Ramaiah M., et al. (2016). Novel SAHA analogues inhibit HDACs, induce apoptosis and modulate the expression of microRNAs in hepatocellular carcinoma. Apoptosis. 21, 1249–1264. 10.1007/s10495-016-1278-6 [DOI] [PubMed] [Google Scholar]
- Sun L., Kong Y., Cao M., Zhou H., Li H., Cui Y., et al. (2017). Decreased expression of acetyl-CoA synthase 2 promotes metastasis and predicts poor prognosis in hepatocellular carcinoma. Cancer Sci. 108, 1338–1346. 10.1111/cas.13252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao N. N., Ren J. H., Tang H., Ran L. K., Zhou H. Z., Liu B., et al. (2017). Deacetylation of Ku70 by SIRT6 attenuates Bax-mediated apoptosis in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 485, 713–719. 10.1016/j.bbrc.2017.02.111 [DOI] [PubMed] [Google Scholar]
- Tao R., De Zoeten E. F., Ozkaynak E., Chen C., Wang L., Porrett P. M., et al. (2007). Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 13, 1299–1307. 10.1038/nm1652 [DOI] [PubMed] [Google Scholar]
- Tapadar S., Fathi S., Wu B., Sun C. Q., Raji I., Moore S. G., et al. (2020). Liver-targeting class I selective histone deacetylase inhibitors potently suppress hepatocellular tumor growth as Standalone agents. Cancers (Basel) 12, E3095. 10.3390/cancers12113095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., Ge C., Zhao F., Zhu M., Zhang L., Huo Q., et al. (2017). Downregulation of AZGP1 by Ikaros and histone deacetylase promotes tumor progression through the PTEN/Akt and CD44s pathways in hepatocellular carcinoma. Carcinogenesis 38, 207–217. 10.1093/carcin/bgw125 [DOI] [PubMed] [Google Scholar]
- Tomaselli D., Lucidi A., Rotili D., Mai A. (2020). Epigenetic polypharmacology: A new frontier for epi-drug discovery. Med. Res. Rev. 40, 190–244. 10.1002/med.21600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. L., Liu W. L., Hsu F. M., Yang P. S., Yen R. F., Tzen K. Y., et al. (2018). Targeting histone deacetylase 4/ubiquitin-conjugating enzyme 9 impairs DNA repair for radiosensitization of hepatocellular carcinoma cells in mice. Hepatology 67, 586–599. 10.1002/hep.29328 [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K. (2012). Role of oxidative stress in hepatocarcinogenesis induced by hepatitis C virus. Int. J. Mol. Sci. 13, 15271–15278. 10.3390/ijms131115271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturelli S., Armeanu S., Pathil A., Hsieh C. J., Weiss T. S., Vonthein R., et al. (2007). Epigenetic combination therapy as a tumor-selective treatment approach for hepatocellular carcinoma. Cancer 109, 2132–2141. 10.1002/cncr.22652 [DOI] [PubMed] [Google Scholar]
- Verza F. A., Das U., Fachin A. L., Dimmock J. R., Marins M. (2020). Roles of Histone Deacetylases and Inhibitors in Anticancer Therapy, 12.Cancers (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Liu T., Wu J. C., Luo S. Z., Chen R., Lu L. G., et al. (2018). STAT3 aggravates TGF-β1-induced hepatic epithelial-to-mesenchymal transition and migration. Biomed. Pharmacother. 98, 214–221. 10.1016/j.biopha.2017.12.035 [DOI] [PubMed] [Google Scholar]
- Wang J., Tang X., Weng W., Qiao Y., Lin J., Liu W., et al. (2015). The membrane protein melanoma cell adhesion molecule (MCAM) is a novel tumor marker that stimulates tumorigenesis in hepatocellular carcinoma. Oncogene 34, 5781–5795. 10.1038/onc.2015.36 [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhu Q., Hu H., Zhu H., Yang B., He Q., et al. (2021). Upregulation of histone acetylation reverses organic anion transporter 2 repression and enhances 5-fluorouracil sensitivity in hepatocellular carcinoma. Biochem. Pharmacol. 188, 114546. 10.1016/j.bcp.2021.114546 [DOI] [PubMed] [Google Scholar]
- Wei S., Liu W., Sun N., Wu Y., Song H., Wang C., et al. (2021). MOF upregulates the estrogen receptor alpha signaling pathway by its acetylase activity in hepatocellular carcinoma. Cancer Sci. 112, 1865–1877. 10.1111/cas.14836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z., Lian L., Ding H., Hu Y., Xiao Z., Xiong K., et al. (2020). LncRNA ANCR promotes hepatocellular carcinoma metastasis through upregulating HNRNPA1 expression. RNA Biol. 17, 381–394. 10.1080/15476286.2019.1708547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Yang T. Y., Li Y., Ye W. L., Liu F., He X. S., et al. (2020). Tumor necrosis factor receptor-associated factor 6 promotes hepatocarcinogenesis by interacting with histone deacetylase 3 to enhance c-Myc gene expression and protein stability. Hepatology 71, 148–163. 10.1002/hep.30801 [DOI] [PubMed] [Google Scholar]
- Xiang J., Zhang N., Sun H., Su L., Zhang C., Xu H., et al. (2020). Disruption of SIRT7 increases the efficacy of checkpoint inhibitor via MEF2D regulation of programmed cell death 1 ligand 1 in hepatocellular carcinoma cells. Gastroenterology 158, 664–678.e24. 10.1053/j.gastro.2019.10.025 [DOI] [PubMed] [Google Scholar]
- Xiao Q., Liu H., Wang H. S., Cao M. T., Meng X. J., Xiang Y. L., et al. (2020). Histone deacetylase inhibitors promote epithelial-mesenchymal transition in Hepatocellular Carcinoma via AMPK-FOXO1-ULK1 signaling axis-mediated autophagy. Theranostics 10, 10245–10261. 10.7150/thno.47045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Liu H., Liu Z. G., Wang H. S., Zhang F., Wang H., et al. (2018). Histone deacetylase inhibitors upregulate Snail via Smad2/3 phosphorylation and stabilization of Snail to promote metastasis of hepatoma cells. Cancer Lett. 420, 1–13. 10.1016/j.canlet.2018.01.068 [DOI] [PubMed] [Google Scholar]
- Xue J., Cao Z., Cheng Y., Wang J., Liu Y., Yang R., et al. (2020). Acetylation of alpha-fetoprotein promotes hepatocellular carcinoma progression. Cancer Lett. 471, 12–26. 10.1016/j.canlet.2019.11.043 [DOI] [PubMed] [Google Scholar]
- Xue X., Gao W., Sun B., Xu Y., Han B., Wang F., et al. (2013). Vasohibin 2 is transcriptionally activated and promotes angiogenesis in hepatocellular carcinoma. Oncogene 32, 1724–1734. 10.1038/onc.2012.177 [DOI] [PubMed] [Google Scholar]
- Yang H., Lan P., Hou Z., Guan Y., Zhang J., Xu W., et al. (2015). Histone deacetylase inhibitor SAHA epigenetically regulates miR-17-92 cluster and MCM7 to upregulate MICA expression in hepatoma. Br. J. Cancer 112, 112–121. 10.1038/bjc.2014.547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Zhu R., Zhao X., Liu L., Zhou Z., Zhao L., et al. (2019). Sirtuin-mediated deacetylation of hnRNP A1 suppresses glycolysis and growth in hepatocellular carcinoma. Oncogene 38, 4915–4931. 10.1038/s41388-019-0764-z [DOI] [PubMed] [Google Scholar]
- Yang J., Jin X., Yan Y., Shao Y., Pan Y., Roberts L. R., et al. (2017). Inhibiting histone deacetylases suppresses glucose metabolism and hepatocellular carcinoma growth by restoring FBP1 expression. Sci. Rep. 7, 43864. 10.1038/srep43864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Feng Y., Zhou J., Cheung O. K., Cao J., Wang J., et al. (2021). A selective HDAC8 inhibitor potentiates antitumor immunity and efficacy of immune checkpoint blockade in hepatocellular carcinoma. Sci. Transl. Med. 13, eaaz6804. 10.1126/scitranslmed.aaz6804 [DOI] [PubMed] [Google Scholar]
- Yeo W., Chung H. C., Chan S. L., Wang L. Z., Lim R., Picus J., et al. (2012). Epigenetic therapy using belinostat for patients with unresectable hepatocellular carcinoma: A multicenter phase I/II study with biomarker and pharmacokinetic analysis of tumors from patients in the Mayo Phase II Consortium and the cancer therapeutics research group. J. Clin. Oncol. 30, 3361–3367. 10.1200/JCO.2011.41.2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo Y. G., Na T. Y., Seo H. W., Seong J. K., Park C. K., Shin Y. K., et al. (2008). Hepatitis B virus X protein induces the expression of MTA1 and HDAC1, which enhances hypoxia signaling in hepatocellular carcinoma cells. Oncogene 27, 3405–3413. 10.1038/sj.onc.1211000 [DOI] [PubMed] [Google Scholar]
- Yuan H., Li A. J., Ma S. L., Cui L. J., Wu B., Yin L., et al. (2014). Inhibition of autophagy significantly enhances combination therapy with sorafenib and HDAC inhibitors for human hepatoma cells. World J. Gastroenterol. 20, 4953–4962. 10.3748/wjg.v20.i17.4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J. H., Yang F., Chen B. F., Lu Z., Huo X. S., Zhou W. P., et al. (2011). The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology 54, 2025–2035. 10.1002/hep.24606 [DOI] [PubMed] [Google Scholar]
- Yuan K., Xie K., Lan T., Xu L., Chen X., Li X., et al. (2020). TXNDC12 promotes EMT and metastasis of hepatocellular carcinoma cells via activation of beta-catenin. Cell Death Differ. 27, 1355–1368. 10.1038/s41418-019-0421-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Cao W., Zhou H., Qian H., Wang H. (2021). H2A.Z acetylation by lincZNF337-AS1 via KAT5 implicated in the transcriptional misregulation in cancer signaling pathway in hepatocellular carcinoma. Cell Death Dis. 12, 609. 10.1038/s41419-021-03895-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Li H., Wang Y., Liu W., Zhang Q., Zhang T., et al. (2010). Epigenetic inactivation of the tumor suppressor gene RIZ1 in hepatocellular carcinoma involves both DNA methylation and histone modifications. J. Hepatol. 53, 889–895. 10.1016/j.jhep.2010.05.012 [DOI] [PubMed] [Google Scholar]
- Zhang H., Xu H. B., Kurban E., Luo H. W. (2020a). LncRNA SNHG14 promotes hepatocellular carcinoma progression via H3K27 acetylation activated PABPC1 by PTEN signaling. Cell Death Dis. 11, 646. 10.1038/s41419-020-02808-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yang F., Yuan J. H., Yuan S. X., Zhou W. P., Huo X. S., et al. (2013). Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis 34, 577–586. 10.1093/carcin/bgs381 [DOI] [PubMed] [Google Scholar]
- Zhang L. Z., Yang J. E., Luo Y. W., Liu F. T., Yuan Y. F., Zhuang S. M. (2020b). A p53/lnc-Ip53 negative Feedback Loop regulates tumor growth and Chemoresistance. Adv. Sci. 7, 2001364. 10.1002/advs.202001364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Shen M., Wu C., Chen Y., Lu J., Li J., et al. (2020c). HDAC8-dependent deacetylation of PKM2 directs nuclear localization and glycolysis to promote proliferation in hepatocellular carcinoma. Cell Death Dis. 11, 1036. 10.1038/s41419-020-03212-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang C., Tian Y., Yao Y., Mao J., Wang H., et al. (2019). SIRT5 promotes hepatocellular carcinoma progression by regulating mitochondrial apoptosis. J. Cancer 10, 3871–3882. 10.7150/jca.31266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Sun H. C., Wang W. Q., Zhang Q. B., Zhuang P. Y., Xiong Y. Q., et al. (2012). Sorafenib down-regulates expression of HTATIP2 to promote invasiveness and metastasis of orthotopic hepatocellular carcinoma tumors in mice. Gastroenterology 143, 1641–1649.e5. e1645. 10.1053/j.gastro.2012.08.032 [DOI] [PubMed] [Google Scholar]
- Zhang X., Dou P., Akhtar M. L., Liu F., Hu X., Yang L., et al. (2021). NEU4 inhibits motility of HCC cells by cleaving sialic acids on CD44. Oncogene 40, 5427–5440. 10.1038/s41388-021-01955-7 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Zhang Z., Li J., Xu F., Zhang B., Liu M., et al. (2020). Lysine acetylome study of human hepatocellular carcinoma tissues for Biomarkers and therapeutic targets discovery. Front. Genet. 11, 572663. 10.3389/fgene.2020.572663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., et al. (2010). Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004. 10.1126/science.1179689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Zhao W., Yan C., Watson C. C., Massengill M., Xie M., et al. (2016a). HDAC inhibitors enhance T-cell Chemokine expression and augment response to PD-1 immunotherapy in Lung adenocarcinoma. Clin. Cancer Res. 22, 4119–4132. 10.1158/1078-0432.CCR-15-2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Gai X., Ding F., Lu Z., Tu K., Yao Y., et al. (2013). Histone acetyltransferase PCAF up-regulated cell apoptosis in hepatocellular carcinoma via acetylating histone H4 and inactivating AKT signaling. Mol. Cancer 12, 96. 10.1186/1476-4598-12-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Xu M., Yao B., Wang C., Jia Y., Liu Q. (2016b). IL-6/STAT3 axis initiated CAFs via up-regulating TIMP-1 which was attenuated by acetylation of STAT3 induced by PCAF in HCC microenvironment. Cell. Signal. 28, 1314–1324. 10.1016/j.cellsig.2016.06.009 [DOI] [PubMed] [Google Scholar]
- Zhou H. Z., Zeng H. Q., Yuan D., Ren J. H., Cheng S. T., Yu H. B., et al. (2019). NQO1 potentiates apoptosis evasion and upregulates XIAP via inhibiting proteasome-mediated degradation SIRT6 in hepatocellular carcinoma. Cell Commun. Signal. 17, 168. 10.1186/s12964-019-0491-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Luo Z., Li Y., Ni C., Li H., Zhu M. (2009). Human inhibitor of growth 1 inhibits hepatoma cell growth and influences p53 stability in a variant-dependent manner. Hepatology 49, 504–512. 10.1002/hep.22675 [DOI] [PubMed] [Google Scholar]