Abstract
Lithium is the drug of choice for the treatment of bipolar affective disorder. The identification of an in vivo target of lithium in fission yeast as a model organism may help in the understanding of lithium therapy. For this purpose, we have isolated genes whose overexpression improved cell growth under high LiCl concentrations. Overexpression of tol1+, one of the isolated genes, increased the tolerance of wild-type yeast cells for LiCl but not for NaCl. tol1+ encodes a member of the lithium-sensitive phosphomonoesterase protein family, and it exerts dual enzymatic activities, 3′(2′),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase. tol1+ gene-disrupted cells required high concentrations of sulfite in the medium for growth. Consistently, sulfite repressed the sulfate assimilation pathway in fission yeast. However, tol1+ gene-disrupted cells could not fully recover from their growth defect and abnormal morphology even when the medium was supplemented with sulfite, suggesting the possible implication of inositol polyphosphate 1-phosphatase activity for cell growth and morphology. Given the remarkable functional conservation of the lithium-sensitive dual-specificity phosphomonoesterase between fission yeast and higher-eukaryotic cells during evolution, it may represent a likely in vivo target of lithium action across many species.
Despite its side effects and toxicity, lithium remains the treatment of choice for bipolar disorder (manic-depressive psychosis) (25, 28). However, the mechanism by which it attenuates the dramatic swings in mood between depression and mania remains unclear.
The finding that submillimolar concentrations of lithium inhibit inositol monophosphatase led to the “inositol depletion hypothesis,” which states that lithium might be exerting its therapeutic effects in bipolar disorder through attenuation of phosphatidylinositol-linked signal transduction (4). Although this hypothesis is widely accepted, evidence that does not support the hypothesis has been accumulating. A molecular genetic study in the budding yeast Saccharomyces cerevisiae showed that budding-yeast inositol monophosphatases are not essential for growth under normal or stressed conditions and are not required for inositol biosynthesis (13). Thus, it is unlikely that inhibition of inositol monophosphatase during lithium treatment results in inositol deficiency in budding yeast. Another study using a different inhibitor of inositol monophosphatase (the bisphosphonate L-690,330) failed to duplicate the developmental defects in Xenopus that are induced by lithium (10). In addition, lithium toxicity observed at high plasma concentrations is not prevented by myo-inositol, suggesting that the toxicity is not related to the modulation of phosphatidylinositol-linked signal transduction (11).
Recent genetic studies of S. cerevisiae indicate that lithium has other in vivo targets, including the Hal2 (also called Met22) 3′(2′),5′-bisphosphate nucleotidase (21, 22) and the RNA-processing enzymes (6). Another model eukaryote, Schizosaccharomyces pombe, or fission yeast, although not as extensively studied as S. cerevisiae, also has many advantages in terms of relevance to higher systems. The salt tolerance mechanism in fission yeast has been shown to be quite different from that of budding yeast (1, 7, 9, 19, 23, 32, 36). Thus, identification of an in vivo target of lithium in fission yeast should enhance our knowledge about the molecular mechanism underlying lithium action in eukaryotes in general. In this study, we isolated fission yeast genes whose overexpression improved the growth of cells under high LiCl concentrations. tol1+, one of the isolated genes, encoded a member of the lithium-sensitive phosphomonoesterase protein family and has been shown to be an in vivo target of lithium. Similar to its higher-eukaryotic homologues, it exerts dual enzymatic activities, 3′(2′),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase.
MATERIALS AND METHODS
Strains, media, genetic techniques, and nomenclature.
The fission yeast strains used in this study are listed in Table 1. The complete medium, YPD, and minimal medium, EMM, have been described previously (35). Adenine sulfate (20 μg/ml) was added to YPD (YPAD) when adenine auxotrophs were cultured. Standard methods, according to Moreno et al. (20), for S. pombe genetics were followed.
TABLE 1.
Fission yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| HM123 | h− leu1 | Our stock |
| HM528 | h+ his2 | Our stock |
| 5A/1D | h−/h+ leu1/leu1 ura4/ura4 his2/+ ade6-M210/ade6-M216 | Our stock |
| KP767 | h−/h+ leu1/leu1 ura4/ura4 his2/+ ade6-M210/ade6-M216 tol1::ura4+/+ | This study |
| KP812 | h− leu1 ura4 ade6 tol1::ura4+ | This study |
| KP656 | h− leu1 ura4 ade6 tol1::ura4+ (pDB248[tol1+]) | This study |
| KP801 | h− leu1 ura4 ade6 tol1::ura4+ (pREP81[tol1+]) | This study |
| KP803 | h− leu1 ura4 ade6 tol1::ura4+ (pDB248[HAL2]) | This study |
Gene disruptions are denoted by lowercase letters representing the disrupted gene followed by two colons and the wild-type gene marker used for disruption (for example, tol1::ura4+). Also, gene disruptions are indicated by the gene preceded by Δ (for example, Δtol1).
Expression of Tol1 in bacteria.
In order to express Tol1 in Escherichia coli, the coding region for the tol1+ gene was amplified by PCR and subcloned into the NdeI-BamHI site of an expression vector, pET3a (31). The plasmid pET3a[tol1+] was introduced into E. coli strain BL21(DE3). The recombinant protein was induced by the addition of isopropyl-β-d-thiogalactopyranoside as previously described (17).
Phosphatase assay.
The enzyme activity was determined by quantifying the inorganic phosphate released from substrate using a colorimetric method as previously described (17). A standard assay was conducted in a 0.3-ml reaction mixture containing 50 mM Tris-HCl (pH 7.8), 1.5 mM 3′-phosphoadenosine 5′-phosphate (PAP; Sigma), 1 mM MgCl2, and 1 μg of the purified protein. The mixture was incubated at 30°C for 30 min. For determination of the Km values, the mixture containing different concentrations of substrates was incubated for 20 min and the data were analyzed by a Lineweaver-Burk plot. The Li+ was removed from 3′-phosphoadenosine 5′-phosphosulfate (PAPS; Sigma) by passing it over P11 cation-exchange columns at 4°C just before use, as described by Peng and Verma (26).
Disruption of tol1+ gene.
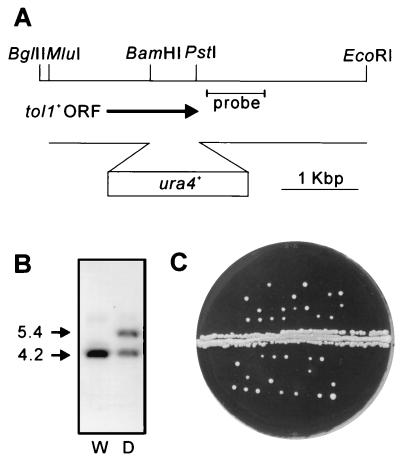
A one-step gene disruption by homologous recombination (29) was performed. The tol1::ura4+ disruption was constructed as follows. The 4.4-kb BglII-EcoRI fragment containing the tol1+ gene was subcloned into the BamHI-EcoRI site of pGEM-7Zf (Promega). Then, a BamHI-PstI fragment containing the ura4+ gene was inserted into the BamHI-PstI site of the previous construct (see Fig. 5A). The MluI-EcoRI fragment containing the disrupted tol1+ gene was transformed into diploid cells (5A/1D). Stable integrants were selected on medium lacking uracil, and disruption of the gene was checked by genomic Southern hybridization (see Fig. 5B).
FIG. 5.
Disruption of tol1+ gene. (A) Restriction map of the genomic tol1+ locus. The arrow indicates the extent and direction of the tol1+ open reading frame (ORF), which encodes 353 amino acids. A disruption construct used to replace the chromosomal tol1+ gene is shown schematically below the arrow. (B) Southern blot analysis showing proper disruption of the chromosomal gene. Genomic DNAs from a wild-type tol1+/tol1+ diploid (W) and the heterozygous tol1+/tol1::ura4+ diploid (D) were digested with BglII and EcoRI and resolved on a 0.8% agarose gel. The DNAs were blotted onto Hybond-N+ (Amersham) and hybridized with the labeled probe indicated in panel A. The deletion results in the production of a new band of 5.4 kb in addition to the 4.2-kb parental band. (C) Tetrad analysis of a heterozygous diploid. The diploid (KP767), which consisted of one copy of wild-type tol1+ and its complete deletion allele, was manipulated and incubated at 33°C on a YPAD plate for 5 days.
To express Tol1 under the control of a thiamine-repressible promoter in a haploid strain with tol1+ deleted, the coding region for the tol1+ gene was amplified by PCR and subcloned into the pREP81 vector (2, 18). A diploid strain, KP767 (Δtol1/tol1+), was transformed with the resultant plasmid (pREP81[tol1+]), and Ura+ Leu+ haploid progeny (KP801) were recovered after sporulation. Haploid strains with tol1+ deleted and carrying other multicopy plasmids (KP656 and KP803) were prepared similarly.
Nucleotide sequence accession number.
The nucleotide sequence of the 2,088-bp MluI-EcoRI fragment containing the entire tol1+ gene reported in this study will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases under the accession number D86083.
RESULTS
Isolation of tol1+ gene.
To identify genes whose products may be in vivo targets of lithium, we isolated genes that when overexpressed improved the growth of cells under high LiCl concentrations. The wild-type cells were transformed with an S. pombe genomic library constructed in the multicopy plasmid pDB248 (3). Transformants were obtained and subsequently screened for the ability to grow on a YPD plate containing 10 mM LiCl at 33°C. The plasmids fell into two classes by restriction enzyme analyses. One class contained the sod2+ gene (9), and cells carrying the plasmid were resistant to high concentrations of NaCl (data not shown). Another class contained a gene whose restriction map was different from that of the sod2+ gene, which was designated tol1+ (target of lithium).
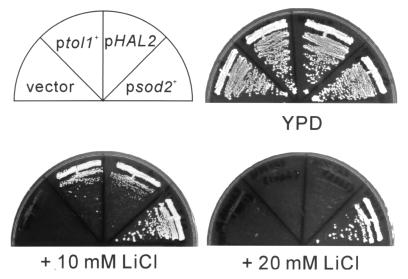
In rich YPD medium, wild-type cells carrying a multicopy plasmid containing sod2+ or tol1+ grew normally. In the YPD containing 10 mM LiCl, wild-type cells carrying sod2+ or tol1+ could grow while wild-type cells carrying a vector failed to grow. In this medium, cells carrying tol1+ exhibited smaller colony size than cells carrying sod2+ (Fig. 1). In the YPD containing 20 mM LiCl, cells carrying tol1+ or vector alone could not grow while cells carrying sod2+ could grow (Fig. 1).
FIG. 1.
The LiCl sensitivity of wild-type cells transformed with the multicopy plasmid pDB248 (vector), pDB248[tol1+] (ptol1+), pDB248[HAL2] (pHAL2), or pDB248[sod2+] (psod2+). Transformed cells were streaked onto each plate containing YPD, YPD plus 10 mM LiCl, or YPD plus 20 mM and incubated at 33°C for 2 days (for YPD) or 3 days (for YPD plus 10 mM LiCl or YPD plus 20 mM LiCl).
Nucleotide sequence analysis showed that tol1+ encodes a protein of 353 amino acid residues which has significant sequence similarity to Hal2 (21, 22) and Sal1 (27), 3′(2′),5′-bisphosphate nucleotidases of budding yeast and Arabidopsis thaliana, respectively (Fig. 2).
FIG. 2.
Amino acid sequence alignment of Tol1, Hal2, and Sal1. Identical amino acid residues are indicated by solid boxes.
3′(2′),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activity of Tol1.
HAL2 was identified by functional assay as supporting the growth of cells under high-salinity stress and is identical to MET22, a methionine auxotrophy mutation that results in the inability to use sulfate, sulfite, sulfide, or cysteine as sulfur sources (33, 34). Hal2 is a 3′(2′),5′-bisphosphate nucleotidase, which is required for the turnover of PAP, a toxic side product of sulfate assimilation (21, 22). In addition to its 3′(2′),5′-bisphosphate nucleotidase activity, Sal1 has been reported to exhibit the typical activity of an inositol polyphosphate 1-phosphatase, which plays a role in the phosphoinositide signaling pathway (14, 27). The high homology of the translated amino acid sequences of tol1+ with those of Hal2 and Sal1 suggested that these proteins have similar enzyme activities.
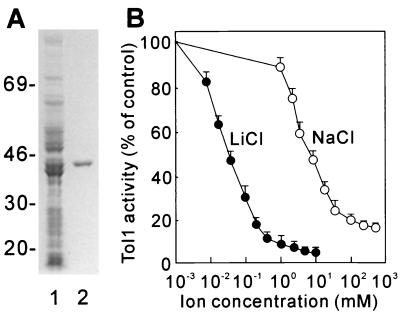
In order to examine the enzyme activity of the protein encoded by tol1+, the open reading frame was cloned into a bacterial expression vector, which was introduced into E. coli. The expressed recombinant protein accumulated only in the soluble fraction of the host cells. The enzyme was purified to homogeneity by a combination of conventional chromatographic steps, as described previously (17) (Fig. 3A). The size of the purified protein is approximately 40 kDa, the size expected from the translation of the Tol1 open reading frame. As shown in Table 2, the Tol1 protein efficiently hydrolyzed PAP, 2′-PAP, and PAPS, while low or no enzymatic activity was seen with inositol 1,4,5-triphosphate, myo-inositol 1-phosphate, fructose-1,6-bisphosphate, AMP, or ATP. Tol1 shared high sequence homology with Sal1, which reportedly hydrolyzes inositol 1,4-bisphosphate and inositol 1,3,4-triphosphate (27). Consistently, inositol 1,4-bisphosphate and inositol 1,3,4-triphosphate also served as substrates for the enzyme (Table 2). The Km value for PAP hydrolysis was estimated to be lower than 10 μM but was too low to be accurately calculated by our assay method, and the Km value for inositol 1,4-bisphosphate was 77 μM. These enzymatic characteristics are very similar to those of Sal1 (27). Tol1 activity was inhibited by submillimolar concentrations of LiCl (Fig. 3B), and its nucleotidase activity was strictly Mg2+ dependent (with an optimal concentration of 1 mM [data not shown]), common properties of the metal-dependent, Li+-inhibited phosphomonoesterase protein family (37). The enzyme was also inhibited by submolar concentrations of NaCl (Fig. 3B) and was significantly activated by K+ (data not shown), properties that have been observed for Hal2-related 3′(2′),5′-bisphosphate nucleotidases (21, 26, 27).
FIG. 3.
Purification and characterization of bacterially expressed Tol1. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of Tol1. Lane 1, crude extract from E. coli transformed with the Tol1 expression plasmid; lane 2, the purified Tol1 protein. The numbers on the left indicate the molecular masses of the markers in kilodaltons. (B) Inhibition of the 3′(2′),5′-bisphosphate nucleotidase activity of Tol1 by LiCl (●) and NaCl (○). Assays were performed under standard conditions as described in Materials and Methods, and cations were added to the mixture as indicated. The data represent an average of three independent experiments, each sample done in duplicate. The bars indicate standard errors of the mean.
TABLE 2.
The substrate specificity of Tol1 enzymea
| Substrate (2 mM) | Relative activity (% of PAP) |
|---|---|
| PAP | 100 |
| 2′-PAP | 120 |
| PAPS | Activeb |
| Inositol 1,4-bisphosphate | 105 |
| Inositol 1,3,4-trisphosphate | 100 |
| Inositol 1,4,5-trisphosphate | <5 |
| myo-Inositol 1-phosphate | <5 |
| Fructose 1,6-bisphosphate | <5 |
| AMP | <5 |
| ATP | <5 |
The activities (initial velocities) obtained with different substrates (1.5 mM) are expressed as percentages, with PAP as 100%. The results are the means of three independent experiments.
Active, detectable activity with undetermined efficiency.
Overexpression of tol1+ gene improved LiCl tolerance but did not affect NaCl tolerance in S. pombe.
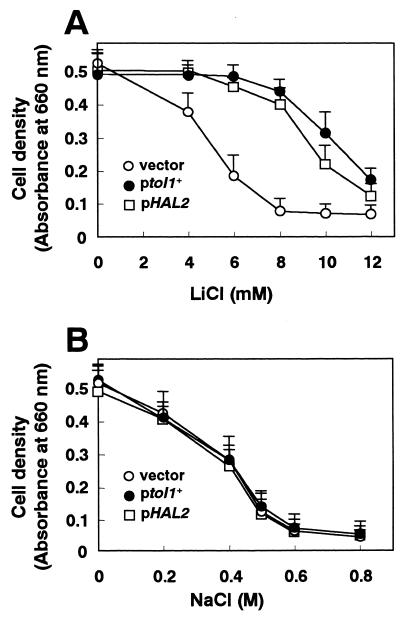
Transformation with a multicopy plasmid carrying the tol1+ gene dramatically increased the LiCl tolerance of the wild-type cells (Fig. 1 and 4A). However, overexpression of tol1+ did not affect NaCl tolerance (Fig. 4B). At the same time, a budding-yeast genomic fragment containing the coding region and 450 bp of upstream sequence of the HAL2 gene was amplified by PCR and cloned into the multicopy plasmid pDB248. Overexpression of HAL2 also increased the LiCl tolerance of the wild-type cells, but it did not affect the NaCl tolerance of S. pombe (Fig. 1 and 4). On the other hand, overexpression of tol1+ or HAL2 conferred lithium tolerance as well as sodium tolerance on S. cerevisiae (data not shown). Thus, NaCl toxicity seems to be caused by intracellular sodium toxicity and not by the osmotic effect of the solute, because high concentrations of KCl (0.6 to 0.8 M) had only slight inhibitory effects on cell growth and tol1+ did not confer sodium tolerance upon overexpression in an S. pombe sod2 mutant (data not shown).
FIG. 4.
Overexpression of tol1+ or HAL2 gene improved LiCl tolerance (A) but did not affect NaCl tolerance (B) in S. pombe. Saturated cultures of wild-type cells transformed with the multicopy plasmid pDB248 (○; vector), pDB248[tol1+] (●; ptol1+), or pDB248[HAL2] (□; pHAL2) were diluted in fresh minimal medium supplemented with various concentrations of LiCl (A) or NaCl (B) as indicated. Growth was recorded by measurement of the absorbance at 660 nm after 18 h. Data were averaged from three independent experiments, each sample done in duplicate. The bars indicate standard errors of the mean.
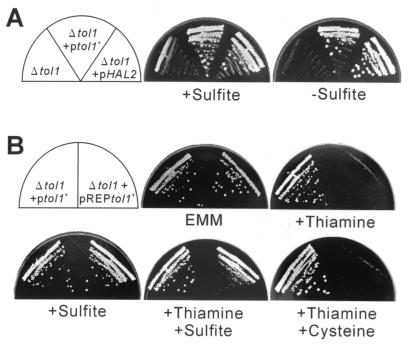
Deletion of tol1+ gene leads to sulfite auxotrophy.
To construct tol1+ gene-disrupted cells, a one-step gene replacement method (29) was used. One copy of tol1+ was replaced with tol1::ura4+ in a diploid strain (Fig. 5A and B). Tetrad analysis on the YPAD plate showed that only two of four spores formed a colony and that all of the viable haploid cells derived from this diploid were uracil auxotrophs, indicating that the tol1::ura4+ mutation caused lethality (Fig. 5C). This was also true when spores were germinated in the medium supplemented with methionine or cysteine. However, when sulfite was added to the medium, three to four spores were viable (data not shown). Thus, Ura+ haploid progeny were recovered as a Δtol1 strain (KP812). Δtol1 cells grew very slowly in the medium supplemented with sulfite, and after transfer to unsupplemented medium, the cells ceased to grow (Fig. 6A). Δtol1 cells carrying pDB248[tol1+] or pDB248[HAL2] grew normally in the unsupplemented medium (Fig. 6A). The sulfite auxotrophy of Δtol1 cells was confirmed by placing the tol1+ gene under the control of a thiamine-repressible promoter (Fig. 6B). Δtol1 cells carrying pREP81[tol1+] grew normally in the absence of thiamine. The growth of Δtol1 cells carrying pREP81[tol1+] on thiamine-containing medium was markedly inhibited, while Δtol1 cells carrying pDB248[tol1+] grew normally both in the absence and in the presence of thiamine (Fig. 6B). Addition of sulfite to the thiamine-containing medium partially restored growth of the disruptant carrying pREP81[tol1+]. On the other hand, cysteine supplementation had no effect (Fig. 6B).
FIG. 6.
Deletion of tol1+ gene leads to sulfite auxotrophy. (A) Δtol1 cells (Δtol1) and Δtol1 cells carrying pDB248[tol1+] (Δtol1+ptol1+) or pDB248[HAL2] (Δtol1+pHAL2) were streaked onto each plate containing YPAD plus 16 mM sodium sulfite (+Sulfite) or YPAD (−Sulfite) and incubated at 33°C for 4 days. (B) Δtol1 cells carrying pDB248[tol1+] (Δtol1+ptol1+) or pREP81[tol1+] (Δtol1+pREPtol1+) were streaked onto each plate containing EMM (EMM), EMM plus 4 μM thiamine (+Thiamine), EMM plus 16 mM sodium sulfite (+Sulfite), EMM plus 4 μM thiamine and 16 mM sodium sulfite (+Thiamine +Sulfite), or EMM plus 4 μM thiamine and cysteine (30 μg/ml) (+Thiamine +Cysteine). The plates were incubated at 33°C for 4 days. Each EMM plate was supplemented with adenine sulfate (20 μg/ml).
Repression of the sulfate assimilation pathway by sulfur compounds in S. pombe.
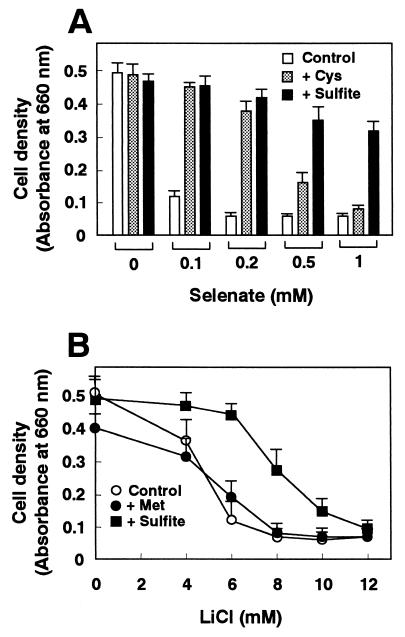
The sulfite auxotrophy of Δtol1 cells suggests that the sulfate assimilation pathway is suppressed by sulfite and thus may prevent the accumulation of PAP in strains lacking Tol1 activity. We tested the effects of various sulfur compounds on the repression of the sulfate assimilation pathway by using selenate, a toxic analogue of sulfate, as reported by Brzywczy and Paszewski (5). Cells become resistant to selenate if the sulfate assimilation pathway, through which selenate is assimilated, is repressed. As shown in Fig. 7A, the addition of selenate inhibited the growth of S. pombe cells. Consistent with sulfite auxotrophy of Δtol1 cells, sulfite counteracted the inhibitory effect of selenate. Sulfite supplementation consistently showed improved changes in cell viability for a wide range of selenate concentrations. Cysteine failed to counteract the inhibitory effect of high selenate concentrations (Fig. 7A). Methionine supplementation failed to improve cell growth in the presence of selenate (data not shown). These results indicate that sulfite represses the sulfate assimilation pathway in S. pombe.
FIG. 7.
Repression of the sulfate assimilation pathway (A) and improvement of LiCl tolerance (B) by sulfite in S. pombe. (A) Repression of the sulfate assimilation pathway was examined using selenate, a toxic analogue of sulfate. Saturated cultures of wild-type cells were diluted in fresh minimal medium (Control) or in the medium supplemented with cysteine (30 μg/ml; + Cys) or 16 mM sodium sulfite (+ Sulfite) in the presence of various concentrations of sodium selenate as indicated. (B) Saturated cultures of wild-type cells carrying an empty vector, pDB248, were diluted in fresh minimal medium (Control; ○), or in the medium supplemented with methionine (10 μg/ml) (+ Met; ●) or 4 mM sodium sulfite (+ Sulfite; ■) and tested for tolerance to various concentrations of LiCl as indicated. Growth was determined as for Fig. 4. The data were averaged from three independent experiments, each sample done in duplicate. The bars indicate standard errors of the mean.
Sulfite supplementation improved LiCl tolerance in S. pombe.
The growth of wild-type cells in medium containing LiCl was improved by sulfite (Fig. 7B). Consistent with the observation that methionine failed to repress sulfate assimilation (5), methionine supplementation did not improve the LiCl tolerance of cells (Fig. 7B). Overexpression of the tol1+ gene had only a slight additive effect on LiCl tolerance compared to that of sulfite supplementation, and tolerance to NaCl was not affected by the addition of sulfite to the medium (data not shown).
Terminal morphological phenotypes caused by loss of tol1+ expression and by toxic concentration of LiCl.
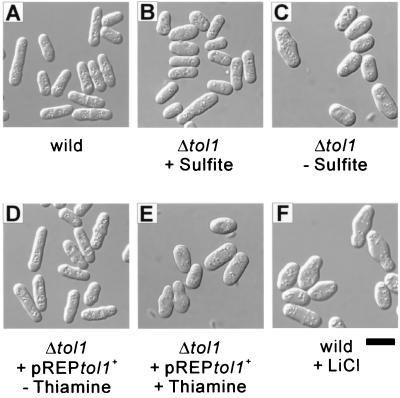
Terminal morphological phenotypes caused by loss of tol1+ expression and by toxic concentration of LiCl were compared. In the medium supplemented with sulfite, cells with tol1+ deleted grew slowly and showed a slightly swollen spherical appearance compared to wild-type cells (Fig. 8A and B); after transfer to unsupplemented medium, the cells ceased to grow and became more swollen (Fig. 8C). Δtol1 cells carrying pREP81[tol1+] began to cease growth after the addition of thiamine and displayed similar swollen spherical cell shapes (Fig. 8D and E). Wild-type cells ceased to grow within 3 h after the addition of 10 mM LiCl. Approximately 10 h after the addition of LiCl, swollen spherical cells appeared, and the percentage of swollen cells increased with the duration of LiCl treatment (Fig. 8F). Sulfite supplementation markedly attenuated the effect of 10 mM LiCl treatment on growth and cell morphology (data not shown).
FIG. 8.
Terminal morphological phenotypes caused by loss of tol1+ expression and caused by toxic concentration of LiCl. Exponentially growing wild-type cells (A), Δtol1 cells (B), and Δtol1 cells carrying pREP81[tol1+] (D) cultured in minimal medium at 33°C are shown. Δtol1 cells were cultured in the medium supplemented with 16 mM sodium sulfite. The terminal morphological phenotype of Δtol1 cells was observed 24 h after their transfer to the medium without sulfite supplementation (C), and that of Δtol1 cells carrying pREP81[tol1+] was observed 48 h after the addition of 4 μM thiamine (E). The terminal phenotype of wild-type cells caused by toxic concentration of lithium was observed 20 h after the addition of 10 mM LiCl to the medium (F). The cells were observed under Nomarski differential interference contrast microscopy. Bar, 10 μm.
DISCUSSION
In this study, we isolated an S. pombe gene, tol1+, that upon overexpression improved the LiCl tolerance of cells. Notably, improvement in cell growth under high LiCl or NaCl concentrations has been previously identified by overexpression of the Hal2 3′(2′),5′-bisphosphate nucleotidase in budding yeast, suggesting that Hal2 is an in vivo target of LiCl or NaCl (21, 22). Further characterization showed that Tol1 is a 3′(2′),5′-bisphosphate nucleotidase with high sequence homology with Hal2. Tol1 and Hal2 dephosphorylate PAP, and their nucleotidase activities in vitro were inhibited by submillimolar concentrations of LiCl (21). In addition, the lethality of the tol1+ deletion was complemented by HAL2 in a multicopy plasmid. These results suggest that Tol1 is an in vivo target of LiCl in fission yeast, that is, LiCl stress would produce a PAP accumulation due to inhibition of Tol1 activity in vivo, and overexpression of tol1+ would decrease the PAP accumulation during the stress.
Mammalian and plant 3′(2′),5′-bisphosphate nucleotidases have been shown to have inositol polyphosphate 1-phosphatase activity, suggesting an involvement of these enzymes in the phosphoinositide signaling pathway as well as in the sulfur assimilation pathway (12, 27, 30). Tol1 also exhibited significant inositol polyphosphate 1-phosphatase activity, as shown in this study. Thus, the identification of a fission yeast homologue of these dual-specificity phosphomonoesterase proteins suggests a remarkable functional conservation of sulfur and inositol metabolisms between fission yeast and higher eukaryotes. Cells with tol1+ deleted grew very slowly and had a slightly swollen spherical appearance, suggesting that repression of sulfate assimilation by sulfite was still partial. This also suggests that defects in inositol metabolism caused by the absence of the inositol polyphosphate 1-phosphatase activity of Tol1 may result in abnormal cell growth and morphology. However, the catalytic efficiency of Tol1 toward nucleotide substrates is highly favored, suggesting that the biologically relevant activity is the 3′(2′),5′-bisphosphate nucleotidase. Further studies are needed to elucidate the physiological significance of the inositol polyphosphate 1-phosphatase activity of Tol1.
Interestingly, while the overexpression of HAL2 improved NaCl tolerance (22), overexpression of tol1+ as well as HAL2 could not improve the growth of fission yeast under high NaCl concentrations. Since the NaCl sensitivity of Tol1 in vitro is almost identical to that of Hal2, a putative system necessary for S. pombe growth but not for S. cerevisiae growth may have higher sensitivity to sodium ion than does the Tol1 system. In fact, wild-type S. pombe cells were more sensitive to growth in high NaCl concentrations than wild-type S. cerevisiae cells (23, 32). Moreover, it is possible that the inability of Tol1 and Hal2 to confer sodium tolerance on S. pombe may indicate some difference between budding and fission yeasts that could be pertinent to the understanding of the physiological role of bisphosphate nucleotidases.
Another notable difference between the two yeast strains involves sulfur metabolism. Methionine auxotrophy (MET22) has been identified as a mutation in HAL2 (16). Methionine, but not sulfite or cysteine, represses the sulfate assimilation pathway in S. cerevisiae (5); thus, methionine may prevent the accumulation of PAP in strains lacking Hal2 activity. In S. pombe, on the other hand, it has been reported that no repression of sulfate assimilation is observed when methionine is added to the growth medium (5). In this study, we have shown that sulfite supplementation represses the sulfate assimilation pathway in S. pombe. Thus, it appears that the molecular mechanisms underlying sulfur control in these two yeast strains differ significantly. In addition, sulfite supplementation improved the growth of cells under high LiCl concentrations. Also, overexpression of the tol1+ gene had only a slight additive effect on the LiCl tolerance of cells compared to that of sulfite supplementation. These results again suggest that sulfite supplementation, as well as overexpression of the tol1+ gene, improved the LiCl tolerance of cells by eliminating PAP, the toxic metabolite in the sulfate assimilation pathway.
The repression mechanism of sulfate assimilation has been studied in other organisms. In S. cerevisiae, S-adenosylmethionine acts as the metabolite responsible for sulfur repression (34). In E. coli, mutations in the cysQ gene encoding a 3′(2′),5′-bisphosphate nucleotidase, which is homologous to Tol1, resulted in a requirement for sulfite or cysteine (24). In the case of Neurospora crassa, cysteine or a compound closely related to or derived from cysteine may represent the true repressing sulfur metabolite (8, 15). Thus, in S. pombe, the repression mechanism of the sulfate assimilation pathway seems to be more similar to that of E. coli or N. crassa than to that of S. cerevisiae. However, it is also suggested that the mechanism is distinct from that of E. coli or N. crassa, because cysteine could not restore the growth of cells with tol1+ deleted and only partially repressed the sulfate assimilation pathway. Although the nature of the repression mechanism triggered by sulfite remains to be discovered, suppression of lithium toxicity in fission yeast may have important implications in the management of the toxic side effects of lithium therapy. In summary, our results identified an in vivo target of lithium in fission yeast that may be involved in sulfur and inositol metabolisms. These results may help us to explain the molecular basis for the beneficial and toxic effects of lithium treatment.
ACKNOWLEDGMENTS
We thank M. Yamamoto and R. Kudo for technical assistance.
This work was supported in part by research grants from the Ministry of Education, Science and Culture of Japan.
REFERENCES
- 1.Balcells L, Gomez N, Casamayor A, Clotet J, Arino J. Regulation of salt tolerance in fission yeast by a protein-phosphatase-Z-like Ser/Thr protein phosphatase. Eur J Biochem. 1997;250:476–483. doi: 10.1111/j.1432-1033.1997.0476a.x. [DOI] [PubMed] [Google Scholar]
- 2.Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- 3.Beach D, Piper M, Nurse P. Construction of a Schizosaccharomyces pombe gene bank in a yeast bacterial shuttle vector and its use to isolate genes by complementation. Mol Gen Genet. 1982;187:326–329. doi: 10.1007/BF00331138. [DOI] [PubMed] [Google Scholar]
- 4.Berridge M J, Downes C P, Hanley M R. Neural and developmental actions of lithium: a unifying hypothesis. Cell. 1989;59:411–419. doi: 10.1016/0092-8674(89)90026-3. [DOI] [PubMed] [Google Scholar]
- 5.Brzywczy J, Paszewski A. Sulfur amino acid metabolism in Schizosaccharomyces pombe: occurrence of two O-acetylhomoserine sulfhydrylases and the lack of the reverse transsulfuration pathway. FEMS Microbiol Lett. 1994;121:171–174. doi: 10.1111/j.1574-6968.1994.tb07095.x. [DOI] [PubMed] [Google Scholar]
- 6.Dichtl B, Stevens A, Tollervey D. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 1997;16:7184–7195. doi: 10.1093/emboj/16.23.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haro R, Garciadeblas B, Rodriguez N A. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 1991;291:189–191. doi: 10.1016/0014-5793(91)81280-l. [DOI] [PubMed] [Google Scholar]
- 8.Jacobson E S, Metzenberg R L. Control of arylsulfatase in a serine auxotroph of Neurospora. J Bacteriol. 1977;130:1397–1398. doi: 10.1128/jb.130.3.1397-1398.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia Z P, McCullough N, Martel R, Hemmingsen S, Young P G. Gene amplification at a locus encoding a putative Na+/H+ antiporter confers sodium and lithium tolerance in fission yeast. EMBO J. 1992;11:1631–1640. doi: 10.1002/j.1460-2075.1992.tb05209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein P S, Melton D A. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci USA. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kofman O, Belmaker R H. Ziskind-Somerfeld Research Award 1993. Biochemical, behavioral, and clinical studies of the role of inositol in lithium treatment and depression. Biol Psychiatry. 1993;34:839–852. doi: 10.1016/0006-3223(93)90052-f. [DOI] [PubMed] [Google Scholar]
- 12.Lopez C J, Belles J M, Lesage F, Serrano R, Rodriguez P L. A novel mammalian lithium-sensitive enzyme with a dual enzymatic activity, 3′-phosphoadenosine 5′-phosphate phosphatase and inositol-polyphosphate 1-phosphatase. J Biol Chem. 1999;274:16034–16039. doi: 10.1074/jbc.274.23.16034. [DOI] [PubMed] [Google Scholar]
- 13.Lopez F, Leube M, Gil M R, Navarro A J, Serrano R. The yeast inositol monophosphatase is a lithium- and sodium-sensitive enzyme encoded by a non-essential gene pair. Mol Microbiol. 1999;31:1255–1264. doi: 10.1046/j.1365-2958.1999.01267.x. [DOI] [PubMed] [Google Scholar]
- 14.Majerus P W. Inositol phosphate biochemistry. Annu Rev Biochem. 1992;61:225–250. doi: 10.1146/annurev.bi.61.070192.001301. [DOI] [PubMed] [Google Scholar]
- 15.Marzluf G A. Molecular genetics of sulfur assimilation in filamentous fungi and yeast. Annu Rev Microbiol. 1997;51:73–96. doi: 10.1146/annurev.micro.51.1.73. [DOI] [PubMed] [Google Scholar]
- 16.Masselot M, De-Robichon S H. Methionine biosynthesis in Saccharomyces cerevisiae. I. Genetical analysis of auxotrophic mutants. Mol Gen Genet. 1975;139:121–132. doi: 10.1007/BF00264692. [DOI] [PubMed] [Google Scholar]
- 17.Matsuhisa A, Suzuki N, Noda T, Shiba K. Inositol monophosphatase activity from the Escherichia coli suhB gene product. J Bacteriol. 1995;177:200–205. doi: 10.1128/jb.177.1.200-205.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 19.Mendoza I, Rubio F, Rodriguez N A, Pardo J M. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J Biol Chem. 1994;269:8792–8796. [PubMed] [Google Scholar]
- 20.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 21.Murguia J R, Belles J M, Serrano R. A salt-sensitive 3′(2′),5′-bisphosphate nucleotidase involved in sulfate activation. Science. 1995;267:232–234. doi: 10.1126/science.7809627. [DOI] [PubMed] [Google Scholar]
- 22.Murguia J R, Belles J M, Serrano R. The yeast HAL2 nucleotidase is an in vivo target of salt toxicity. J Biol Chem. 1996;271:29029–29033. doi: 10.1074/jbc.271.46.29029. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura T, Liu Y, Hirata D, Namba H, Harada S, Hirokawa T, Miyakawa T. Protein phosphatase type 2B (calcineurin)-mediated, FK506-sensitive regulation of intracellular ions in yeast is an important determinant for adaptation to high salt stress conditions. EMBO J. 1993;12:4063–4071. doi: 10.1002/j.1460-2075.1993.tb06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neuwald A F, Krishnan B R, Brikun I, Kulakauskas S, Suziedelis K, Tomcsanyi T, Leyh T S, Berg D E. cysQ, a gene needed for cysteine synthesis in Escherichia coli K-12 only during aerobic growth. J Bacteriol. 1992;174:415–425. doi: 10.1128/jb.174.2.415-425.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peet M, Pratt J P. Lithium. Current status in psychiatric disorders. Drugs. 1993;46:7–17. doi: 10.2165/00003495-199346010-00002. [DOI] [PubMed] [Google Scholar]
- 26.Peng Z, Verma D P. A rice HAL2-like gene encodes a Ca2+-sensitive 3′(2′),5′-diphosphonucleoside 3′(2′)-phosphohydrolase and complements yeast met22 and Escherichia coli cysQ mutations. J Biol Chem. 1995;270:29105–29110. doi: 10.1074/jbc.270.49.29105. [DOI] [PubMed] [Google Scholar]
- 27.Quintero F J, Garciadeblas B, Rodriguez N A. The SAL1 gene of Arabidopsis, encoding an enzyme with 3′(2′),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell. 1996;8:529–537. doi: 10.1105/tpc.8.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenthal N E, Goodwin F K. The role of the lithium ion in medicine. Annu Rev Med. 1982;33:555–568. doi: 10.1146/annurev.me.33.020182.003011. [DOI] [PubMed] [Google Scholar]
- 29.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 30.Spiegelberg B D, Xiong J P, Smith J J, Gu R F, York J D. Cloning and characterization of a mammalian lithium-sensitive bisphosphate 3′-nucleotidase inhibited by inositol 1,4-bisphosphate. J Biol Chem. 1999;274:13619–13628. doi: 10.1074/jbc.274.19.13619. [DOI] [PubMed] [Google Scholar]
- 31.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 32.Sugiura R, Toda T, Shuntoh H, Yanagida M, Kuno T. pmp1+, a suppressor of calcineurin deficiency, encodes a novel MAP kinase phosphatase in fission yeast. EMBO J. 1998;17:140–148. doi: 10.1093/emboj/17.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas D, Barbey R, Surdin K Y. Gene-enzyme relationship in the sulfate assimilation pathway of Saccharomyces cerevisiae. Study of the 3′-phosphoadenylylsulfate reductase structural gene. J Biol Chem. 1990;265:15518–15524. [PubMed] [Google Scholar]
- 34.Thomas D, Rothstein R, Rosenberg N, Surdin K Y. SAM2 encodes the second methionine S-adenosyl transferase in Saccharomyces cerevisiae: physiology and regulation of both enzymes. Mol Cell Biol. 1988;8:5132–5139. doi: 10.1128/mcb.8.12.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toda T, Dhut S, Superti F G, Gotoh Y, Nishida E, Sugiura R, Kuno T. The fission yeast pmk1+ gene encodes a novel mitogen-activated protein kinase homolog which regulates cell integrity and functions coordinately with the protein kinase C pathway. Mol Cell Biol. 1996;16:6752–6764. doi: 10.1128/mcb.16.12.6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Withee J L, Sen R, Cyert M S. Ion tolerance of Saccharomyces cerevisiae lacking the Ca2+/CaM-dependent phosphatase (calcineurin) is improved by mutations in URE2 or PMA1. Genetics. 1998;149:865–878. doi: 10.1093/genetics/149.2.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.York J D, Ponder J W, Majerus P W. Definition of a metal-dependent/Li+-inhibited phosphomonoesterase protein family based upon a conserved three-dimensional core structure. Proc Natl Acad Sci USA. 1995;92:5149–5153. doi: 10.1073/pnas.92.11.5149. [DOI] [PMC free article] [PubMed] [Google Scholar]