Abstract
The large-scale and high-intensity application of Streptococcus thermophilus species in milk fermentation processes is associated with a persistent threat of (bacterio)phage infection. Phage infection of starter cultures may cause inconsistent, slow or even failed fermentations with consequent diminished product quality and/or output. The phage life cycle commences with the recognition of, and binding to, a specific host-encoded and surface-exposed receptor, which in the case of S. thermophilus can be the rhamnose-glucose polysaccharide (RGP; specified by the rgp gene cluster) or exopolysaccharide (EPS; specified by the eps gene cluster). The genomic diversity of 23 S . thermophilus strains isolated from unpasteurized dairy products was evaluated, including a detailed analysis of the rgp and eps loci. In the present study, five novel eps genotypes were identified while variations of currently recognized rgp gene cluster types were also observed. Furthermore, the diversity of rgp genotypes amongst retrieved isolates positively correlated with phage diversity based on phageome analysis of eight representative dairy products. Our findings therefore substantially expand our knowledge on S. thermophilus’ strain and phage diversity in (artisanal) dairy products and highlight the merit of phageome analysis of artisanal and traditional fermented foods as a sensitive marker of dominant microbiota involved in the fermentation.
Keywords: rhamnose-glucose polysaccharides, RGP, exopolysaccharides, EPS, bacteriophage, phageome, Streptococcus thermophilus
Data Summary
Impact Statement.
Streptococcus thermophilus is one of the most important LAB species in dairy fermentations. This species, however, is subject to a persistent threat of phage infection, which may negatively affect the output and quality of the final fermentation product. It is known that S. thermophilus strains possess distinct saccharidic phage receptors that are linked to their phage sensitivity profiles. In the current study S. thermophilus strains were isolated from various unpasteurized dairy products, and the diversity of gene clusters involved in rhamnose glucose polysaccharide and exopolysaccharide biosynthesis was assessed. The distribution of dairy streptococcal phages in fermented dairy products was also assessed. The outcome of the present body of work provides insights for the dairy industry to assist in developing optimal and defined starter culture strain mixtures possessing distinct phage receptors/sensitivities, which in turn will minimize the risk of fermentation failure so as to ensure yield, consistency and quality of dairy products.
Genome sequences used in this study have been deposited in National Center for Biotechnology Information (NCBI) and the accession numbers are listed in Table S1 (available in the online version of this article). The authors confirm all supporting data, code and protocols have been provided within the article or through supplementary data files. Supplementary Material can be found in Figshare: https://doi.org/10.6084/m9.figshare.17080283 [1].
Introduction
Streptococcus thermophilus is one of the most widely exploited species of lactic acid bacteria (LAB) in dairy fermentations [2, 3]. S. thermophilus has been granted the generally regarded as safe (GRAS) status by the U.S. Food and Drug Administration [4, 5] and the ‘Qualified Presumption of Safety’ (QPS) status in the European Union [6]. Bacteriophage (or phage) infection poses a constant threat to industrial fermentations and can result in inconsistent, delayed or even failed fermentations and reduced product quality and output [7] with a considerable associated economic impact on the agri-food sector. Phages infecting S. thermophilus are currently differentiated into five groups termed the cos (or Moineauvirus) and pac (or Brussowvirus) [8], 5093 [9], 987 [10] and P738 [11] groups. Limited studies on the biodiversity of S. thermophilus phages have been undertaken [12–14], though phageome studies of cheese and cheese-associated (by)products have recently garnered increasing attention [15, 16]. To elucidate the phage–host interactome, it is essential to understand the biodiversity of both phages and their hosts that are applied in an industrial context, and of the phage- and host-encoded components that contribute to these interactions. Recently, protocols have been optimized for the extraction and analysis of phageomes relating to cheese and cheese wheys [15, 16]. Phageome analysis of fermented foods and their by-products provides useful insights into the prevalence, diversity and abundance of phages present in these foods, particularly where undefined starter culture mixtures are applied. While limited studies of the phageome of fermented dairy products have been reported to date, they are expected to provide a highly useful and complementary accompaniment to traditional phage-screening studies.
Phage infection commences with the recognition of, and binding to, a suitable host-encoded and surface-exposed receptor moiety. In many cases, the receptor moieties in Gram-positive ovococcoid bacteria are saccharidic and include the rhamnose-glucose polysaccharide (RGP, whose biosynthetic machinery is encoded by the rgp cluster) [17] and exopolysaccharides (EPS, specified by the eps gene cluster) in S. thermophilus [18–20]. Currently, five rgp (types A to E) and six eps genotypes (types A to F) are known to exist based on comparative sequence analysis [20]. The chemical structure and/or composition of individual EPS and RGP molecules isolated from a small number of strains have been defined [18, 21, 22], findings that have highlighted the chemical diversity and complexity of these structures. Furthermore, a multiplex PCR system has been devised to reliably classify S. thermophilus strains based on their rgp genotypes which can, in principle, be applied to establish robust strain blends and/or rotations by incorporating strains with distinct RGPs [19, 20, 22]. Therefore, while it is widely accepted that S. thermophilus as a species lacks genetic diversity [23], the rgp and eps clusters appear to represent localized regions of genomic diversity, which affect its sensitivity to phage predation. In this context it should be noted that the host range of most dairy streptococcal phages is very narrow, an observation that may, at least partially, be linked to the diversity of phage receptors and the corresponding genetic diversity of their biosynthetic machinery.
Bacteria have evolved a variety of mechanisms to defend themselves against phage infection, thus bacteria and their associated phages are constantly engaged in an antagonistic co-evolutionary process [24]. S. thermophilus incorporates defence mechanisms to limit phage proliferation, through chromosomally- or plasmid-encoded R-M (restriction and modification) systems [25] and also CRISPR-Cas (clustered regularly interspaced short palindromic repeats and CRISPR-associated genes) systems [26]. Strains of this bacterial species are known to harbour up to four CRISPR-Cas systems (CR1 to CR4), with CR1 and CR3 being the most active in spacer acquisition [27, 28]. These systems have the capacity to evolve, adapt and acquire spacers in response to phage (or plasmid) exposure, which provides increased resistance to specific phages [29, 30]. The CRISPR spacer array profiles can be used as unique identifiers of distinct strains [31, 32]. A number of studies have highlighted the presence of dairy streptococci in artisanal fermented foods from a range of geographical locations through metagenomic and/or microbiological approaches [33–37]. Isolates have been characterized largely based on technologically relevant properties including EPS production and structural characteristics [37], antibiotic sensitivity, milk acidification ability and/or gelling capabilities [34]. Within an individual product, it can be challenging to assess if more than one strain of S. thermophilus is present due to high levels of sequence conservation across the genomes of strains of this species. CRISPR spacer array profiling is a useful tool to do so but this approach is time consuming and sequence analysis of the CRISPR arrays can be difficult due to the presence of repeats. The establishment of a multiplex PCR system for the rapid differentiation and classification of dairy streptococcal strains based on variable sequences within the rgp gene cluster of strains of this species facilitates the identification of strains with distinct phage-sensitivity characteristics. Since the role of RGPs in the initial binding stage of certain S. thermophilus phages is a relatively recent observation, it is perhaps unsurprising that only a limited number of studies have focused on rgp gene cluster diversity of S. thermophilus strains, particularly when isolated from artisanal dairy products [20, 22].
In the present study, the diversity of S. thermophilus strains isolated from 27 dairy products derived from unpasteurized milk was ascertained through CRISPR spacer array analysis and rgp genotyping. Twenty three individual strains representing isolates from different food sources and possessing distinct rgp genotypes were selected for whole-genome sequencing facilitating a comparative analysis of the rgp and the eps clusters of the strains. S. thermophilus strains were classified based on defining criteria including the size of the genome, presence or absence of the cell-wall associated proteinase-encoding gene (prtS), CR2/CR3/CR4, also prophage(s), as proposed by Alexandraki et al. [38]. Furthermore, the phageomes of eight representative food samples were assessed to identify phage prevalence, abundance and diversity, in relation to the diversity and prevalence of S. thermophilus isolates obtained from each respective dairy products. The results derived from this investigation provide insights into the plausible correlation between phage and strain diversity/distribution in dairy fermented products derived from unpasteurized milk.
Methods
Isolation of presumptive S. thermophilus
Twenty seven fermented dairy products including raw cow’s milk and soft, semi-soft, hard cheeses made with unpasteurized cow’s, buffalo’s, ewe’s and goat’s milk (Table 1) were screened for the presence of S. thermophilus strains. In total, 5 g of each product was transferred into 45 ml of PBS (Sigma Aldrich, MO, USA) and pummelling for 2 mins at 300 r.p.m. in a stomacher (Stomacher Circular 400; Seward, UK). Serial dilutions of each sample were prepared and plated on LM17 agar [M17 agar (Oxoid, Hampshire, UK) supplemented with 0.5% lactose (Sigma Aldrich)], incubated overnight at 42 °C anaerobically (Anaerocult A – Merck, NJ, USA). A total of 1253 individual colonies exhibiting a creamy-white colour were isolated. The isolates were maintained and stored at −80 °C in LM17 broth supplemented with 30% (v/v) glycerol (Thermo Fisher, MA, USA).
Table 1.
Number of isolates of presumptive streptococcal isolates identified by rgp typing multiplex PCR (n=325). Shaded areas represent a negative result
|
Products |
Product type |
Origin |
No. of streptococcal isolates* |
RGP† |
||
|---|---|---|---|---|---|---|
|
A |
B |
C/E |
||||
|
Raw milk A to D |
Raw milk |
Cow |
||||
|
Mozzarella A |
Soft cheese |
Cow |
25 |
1 |
2 |
22 |
|
Mozzarella B |
Buffalo |
45 |
11 |
1 |
33 |
|
|
Soft cheese A |
Goat |
1 |
1 |
|||
|
Soft cheese B |
Goat |
8 |
8 |
|||
|
Soft cheese C |
Goat |
4 |
4 |
|||
|
Brie |
Cow |
18 |
3 |
15 |
||
|
Camembert |
Cow |
|||||
|
Vacherin |
Cow |
3 |
3 |
|||
|
Reblochon A |
Cow |
|||||
|
Reblochon B |
Cow |
12 |
11 |
1 |
||
|
Stracciatella |
Cow |
5 |
4 |
1 |
||
|
Ricotta |
Buffalo |
32 |
1 |
1 |
30 |
|
|
Scamorza |
Semi-soft cheese |
Cow |
92 |
92 |
||
|
Cheddar |
Cow |
25 |
25 |
|||
|
Halloumi |
Cow |
|||||
|
Blue cheese |
Sheep |
10 |
10 |
|||
|
Semi-soft cheese A |
Cow |
1 |
1 |
|||
|
Semi-soft cheese B |
Cow |
8 |
8 |
|||
|
Semi-soft cheese C |
Cow |
4 |
4 |
|||
|
Semi-soft cheese D |
Cow |
32 |
32 |
|||
|
Pecorino |
Hard cheese |
Ewe |
||||
|
Caciocavallo |
Cow |
|||||
|
Hard cheese A |
Cow |
|||||
*Approximately 48 to 96 creamy, white colonies were isolated from every dairy products, except for raw milk A to D, vacherin, stracciatella and halloumi due to low yield of c.f.u. in the samples.
†No RGP type D isolates were retrieved in this study.
RGP genotyping by multiplex PCR
Presumptive S. thermophilus isolates were typed using an established multiplex PCR system based on the rgp gene cluster and using five primer pairs (Table S2). PCRs were performed using Phusion Green Hot Start II High-Fidelity PCR Master Mix (Thermo Fisher, Gloucester, UK) employing the following conditions: 98 °C for 10 min followed by 30 cycles of 98 °C for 15 s, 55 °C for 30 s, and 72 °C for 1 min, followed by a final extension step at 72 °C for 10 min. All PCRs were performed using an Applied Biosystems 2720 Thermal Cycler (Thermo Fisher) instrument. Amplicons were visualized on a 1% agarose gel followed by UV transillumination.
Species confirmation using 16S rRNA amplicon sequence analysis
To confirm the identity of presumptive streptococcal isolates identified by the rgp genotyping approach, the 16S rRNA gene of presumptive S. thermophilus isolates was amplified using the LucFw and LucRv primers (Table S2) using Taq DNA polymerase mastermix (Qiagen, Manchester, UK) under the following conditions: initial denaturation at 94 °C for 10 min, 30 cycles of 94 °C for 30 s, 40 °C for 30 s, 72 °C for 1 min and 30 s, followed by a final extension at 72 °C for 10 min. The amplicons were purified using the GenElute PCR Clean-Up Kit (Sigma Aldrich) according to the manufacturer’s instruction and subjected to Sanger sequencing (executed by Eurofins MWG, Waterford, Ireland). The generated sequences were analysed using blastn analysis against available sequence data on the National Center for Biotechnology Information (NCBI) database located at the following URL: https://blast.ncbi.nlm.nih.gov/Blast.cgi.
DNA extraction, genome sequencing, assembly and annotations
Bacterial DNA was extracted from a fresh 10 ml overnight culture using the Invitrogen PureLink Genomic DNA Mini Kit (Thermo Fisher) according to the manufacturer’s instruction with some modifications. The cell pellet was resuspended and incubated in TE buffer containing 25% sucrose (Thermo Fisher) and 30 mg ml−1 lysozyme (Sigma Aldrich). Chromosomal DNA was extracted from each strain and sequenced by Probiogenomics (Parma, Italy) using an Illumina MiSeq platform. Genome libraries were constructed using the TruSeq DNA PCR-Free LT Kit (Illumina) and 2.5 µg of genomic DNA, which was fragmented with a Bioruptor NGS ultrasonicator (Diagenode, USA) followed by size evaluation using Tape Station 2200 (Agilent Technologies, Santa Clara, CA, USA). Library samples were loaded into a Flow Cell V3 600 cycles (Illumina) and sequencing was performed on a MiSeq genomic platform (Illumina, Cambridge, UK) at GenProbio srl (Parma, Italy). Fastq files of the paired-end reads obtained from genome sequencing were used as input for genome assemblies through the MEGAnnotator pipeline in default mode [39]. Open reading frames prediction was performed with Prodigal v2.6 [40]. The MIRA programme v4.0.2 was used for de novo assembly of genome sequence data [41]. Following final genome assembly, putative protein-encoding genes were identified using the prediction software Prodigal v2.0 [40]. Protein-encoding genes were automatically annotated using a blastp v2.2.26 (cut-off E-value of 0.0001) sequence alignments against the non-redundant protein (nr) database curated by NCBI (ftp://ftp.ncbi.nih.gov/blast/db/). The sequences of the S. thermophilus strains sequenced in this study were deposited in the GenBank database and their associated GenBank accession numbers are presented in Table S1.
Genome and comparative analysis
The quality or completeness of genome assemblies was evaluated with the microbial genomes atlas (MiGA) webserver [42]. The genome sequences of each strain was analysed using CRISPRFinder (https://crispr.i2bc.paris-saclay.fr/Server/) to retrieve and identify CRISPR repeats and spacer sequences and to establish if each strain was distinct based on unique spacer content within the identified CRISPR arrays. Further analysis was performed to compare the spacer sequences of each strain with phageome sequences of the associated dairy product using blastn analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The presence of prtS was identified in the genomes of the sequenced strains using blastn analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi). PHAge search tool enhanced release (PHASTER) [43, 44] was used to screen for prophage-specifying DNA regions within the genome of the S. thermophilus strains sequenced in this study. Intact prophages were manually checked to confirm the presence of genes required to produce a functional phage particle including genes encoding proteins associated with replication functions, packaging, morphogenesis and lysis.
The rgp and eps gene clusters of S. thermophilus strains were identified. The rgp cluster was located between two conserved genes, encoding the 30S ribosomal protein and Rex protein, whereas the eps cluster was located between two conserved genes, encoding for chloride channel/purine nucleoside phosphorylase and VanZ. To determine and classify the rgp and eps genotype, the nucleotide sequence of the respective gene clusters of the isolates were compared using blastn analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi), with a cut-off of 50% nucleotide identity and query coverage. blastp (https://blast.ncbi.nlm.nih.gov/Blast.cgi) alignment of the rgp- and eps-biosynthetic gene clusters were performed to identify homologous gene products/regions between the analysed strains. blastp and Pfam v.33.1 [45] were used to assign functions to the individual protein-encoding regions. To identify the closest relatives of the sequenced EPS-specifying genomic regions and to establish their phylogeny based on their overall deduced proteomic content (based on the presence/absence of identified protein families with at least 50% identity over 50% of the protein), the Markov clustering (MCL) algorithm was executed via the mclblastline pipeline v12-0678 as described by Enright et al. [46], followed by hierarchical clustering (HCL) analysis performed and viewed in the multi-experiment viewer (MeV) [47]. rgp and eps genotype groupings were assigned based on similarity to previously defined rgp and eps type strains defined by Szymczak et al. [20] and/or through the identification of distinct groups observed in the heatmap generated through the HCL analysis.
Phageome extraction & data analysis
Phageome extractions were performed on eight representative dairy products screened (brie, mozzarella B, stracciatella, vacherin, semi-soft cheese A, semi-soft cheese D and blue cheese) in this study to determine the presence of phage in the sample. The extraction method followed a combination of elements of previous studies by Dugat-Bony et al. [15], Muhammed et al. [16] and Milani et al. [48] with some modifications. In summary, a given cheese sample (5 g) was homogenized in 30 ml sodium citrate (20 g l−1) solution and pummelling for 2 mins at 300 r.p.m. in a stomacher. Samples were centrifuged at 300 g for 10 mins to remove large aggregates, and subsequently the supernatant was centrifuged for 5000 g for 45 mins at 4 °C. The supernatant was collected and treated with 1 M NaCl (Sigma Aldrich) for 1 h at 4 °C. The supernatant was diluted (1 : 2) in cold SM buffer (10 mM CaCl2, 100 mM NaCl, 10 mM MgSO4, 50 mM Tris-HCl at pH 7.5) and the pH was adjusted to ~4.6. Samples were centrifuged at 28000 g for 15 mins, then subjected to double filtration (first with 0.45 µm, followed by 0.2 µm filter). Next, 10% (w/v) PEG 6000 was added to filtered supernatant, then incubated at 4 °C overnight and centrifuged at 15000 g for 15 mins. The pellets were collected and resuspended in 1 ml SM buffer. Samples were treated with DNAse I (20 units ml−1, Sigma Aldrich) for 15 mins at room temperature, and then inactivated by incubation at 75 °C for 10 mins. The phage DNA was extracted using Norgen Biotek Phage DNA isolation kit (Norgen Biotek, Thorold, Canada) according to the manufacturer’s instructions.
DNA was fragmented to 550–650 bp using a BioRuptor machine (Diagenode, Belgium). Samples were prepared following the TruSeq Nano DNA Sample Preparation Guide (Part no. 15041110Rev.D). Sequencing was performed using an Illumina NextSeq 500 sequencer with NextSeq Mid Output v2 Kit chemicals (Illumina, San Diego, CA 92122, USA). Read mapping of overall raw reads of the eight representative dairy products were performed using the METAnnotatorX bioinformatic platform as described by Milani et al. [48] against the NCBI database. Furthermore, read mapping of targeted phage genomes (of 489 streptococcal and lactococcal phages) was performed using MobaXterm server (https://mobaxterm.mobatek.net/). Raw reads were quality filtered using IlluQC.pl from the NGS QC Toolkit (v2.3) [49] in order to remove low-quality reads. The overall filtering process yielded 1.05 to 4.75 million reads per sample (Table S3). Quality-filtered reads were mapped for each sample against a panel of in-house database containing published and unpublished streptococcal/lactococcal phage (489 phages in total), using Bowtie2 aligner with default values [50]. Accession numbers for all samples analysed are listed in Table S1.
Phage screening and phage–host range determination
Eight of the cheese samples (brie, mozzarella B, ricotta, stracciatella, vacherin, semi-soft cheese A, semi-soft cheese D and blue cheese) were screened for the presence of phages against isolates emanating from the same product. Briefly, individual isolates of S. thermophilus obtained from a given sample were grown in 96-well microtitre plates containing 100 µl LM17 broth, incubated at 42 °C anaerobically for 24 h. The resulting culture was used to inoculate [1% (v/v)] two 96-well plates (one plate served as an untreated control) containing 100 µl LM17 broth. Plates were then incubated at 42 °C anaerobically for 1 h. Then, 10 µl of 1M CaCl2 was added to all isolates in both 96-well plates, after which 50 µl of a filter-sterilized cheese homogenate from the same food sample was added to each well in a 96-well plate. Both microtitre plates were incubated at 42 °C anaerobically overnight.
Individual phages from a streptococcal phage collection (30 phages) (Table S4) were propagated to a high titre (>107 p.f.u. ml−1) on suitable sensitive hosts, and were applied in a host-range survey against the streptococcal strains whose genomes were sequenced as part of this study. Briefly, 10 µl of each phage lysate was spotted against isolates collected in this study via the double layer LM17 agar method, as previously described [51], then the plates were incubated anaerobically for 24 h at 42 °C.
Results and discussion
Artisanal dairy products contain multiple S. thermophilus strains
From a genomic perspective S. thermophilus has long been regarded as a relatively homogenous species with limited genetic variation [38]. However, certain genomic loci allow discernment between distinct strains and permit the identification of specific genotypes within the species. Among these are the loci encoding the biosynthetic apparatus responsible for RGP and EPS production. The present study aimed to screen for and isolate S. thermophilus , utilizing the widely accepted approach of using LM17 agar incubated at 42 °C anaerobically. A total of 1253 putative S. thermophilus isolates were selected, in order to (i) identify S. thermophilus strains associated with 27 unpasteurized dairy products, and (ii) determine the extent of diversity of the rgp and eps gene clusters (where eps clusters can be retrieved from the genome sequence) of the identified S. thermophilus strains as an indicator of strain diversity (Fig. S1). No putative S. thermophilus isolates were retrieved from 10 of the 27 analysed food samples including four raw milk samples and six cheese samples. From a further eight samples less than ten putative S. thermophilus isolates per sample were obtained, while the remaining nine cheese samples possessed more than 10 viable presumptive dairy streptococci (Table 1). Although the viability of S. thermophilus was generally observed to be low, the highest counts were observed among the soft and semi-soft cheeses that had not been matured (Table 1).
All isolates were first evaluated by multiplex PCR and for consistency with current literature, the rgp genotype classification (based on the PCR system of Kouwen et al., 2019 [22]) was harmonized with that of Szymczak and colleagues [20] throughout this manuscript (where RGP 1=type B; RGP 2=type A; RGP 3=type D; RGP 4=type C or E, according to Kouwen et al. [22], and Szymczak et al. [20], respectively). Based on this genotyping approach, among the 1253 putative S. thermophilus isolates only 325 isolates were confirmed to belong to this species based on the RGP multiplex PCR (Fig. S1). Fifty six representative isolates among the 325 streptococcal isolates were further validated as S. thermophilus based on 16S rRNA gene sequence analysis. Of these 325 confirmed streptococcal isolates, the vast majority (83.1% or 270/325) were classified as RGP type C/E, 12% (39/325) as RGP type B, and 4.9% (16/325) as RGP type A (Table 1). This is consistent with a previous study in which type C strains were observed to be the most prevalent (34.8% or 8/23) [20]. The remaining 928 isolates did not produce any dairy streptococcal RGP-related amplicons suggesting that they are not dairy streptococci. In total, 171 isolates were eliminated from further investigation as they were identified as enterococcal species based on their ability to form brown precipitate around the colonies using bile aesculin agar [52]. Forty seven random isolates out of the remaining 757 isolates were evaluated by 16S rRNA gene sequencing and were identified as non-dairy streptococci, lactococci or pediococci (Fig. S1), thus the remaining isolates were discarded as unlikely S. thermophilus isolates and were not further characterized. Therefore, while the isolation procedure was selective for lactic acid bacteria it was not specifically selective for S. thermophilus .
The rgp genotype of strains within each food sample was evaluated to determine if single or multiple strains were likely present in the samples. Through this approach, the scamorza, cheddar, vacherin, blue cheese, soft cheese A to C, and semi-soft cheese A to D samples were observed to harbour isolates of a single dominant rgp genotype (Table 1). In contrast, the brie, reblochon B, stracciatella, ricotta and mozzarella A and B samples were observed to harbour strains possessing distinct rgp genotypes, highlighting the likely presence of multiple S. thermophilus strains in these cheeses. The presence of multiple dairy streptococcal strains in a given dairy product is indicative of thermophilic production regimes where S. thermophilus isolates would thrive. Among the analysed samples, there was a prevalence of RGP type C/ E S. thermophilus strains (being found in 11 dairy products), while type B strains were prevalent in six cheese products (soft cheese A, brie, vacherin, reblochon B, stracciatella, semi-soft cheese A). The mammalian origin of the milk (i.e. cow, goat, buffalo and ewe) did not appear to correlate with the presence of strains of a specific rgp genotype(s) (Table 1).
Comparative genome analysis of S. thermophilus
One to two representative isolates with distinct RGP profiles derived from each of 17 products that yielded S. thermophilus isolates shown in Table 1 (32 isolates in total) were selected for genome sequencing. Preliminary analysis of the whole genome and CRISPR spacer arrays confirmed that 23 of the 32 strains were distinct (Table 2) and the remaining nine isolates were eliminated from downstream analyses. Since the genome sequence data was typically assembled into multiple contigs, the genomes were checked for completeness based on the presence of 106 essential genes using the webserver MiGA and all strains were predicted to be (100%) complete and of ‘excellent’ quality (>95%). The genome sizes ranged from 1.78 to 2.05 mega base-pairs (Mbp), with average mol % GC content of 38.63–39.04%, in keeping with the genomes of previously analysed strains of the species.
Table 2.
General genome features of S. thermophilus strains sequenced in this study
|
Strain |
Average coverage |
No. of bases |
Contigs |
No. of predicted ORFs |
Average GC % |
Presence of prtS (Yes/No) |
No. of prophage(s)* |
Genome completeness (%)† |
|---|---|---|---|---|---|---|---|---|
|
Moz109 |
180.8 |
1 807 378 |
33 |
1903 |
38.87 |
Yes |
four inc. |
100 |
|
Brie1 |
242 |
1 799 678 |
42 |
1904 |
38.93 |
No |
one inc. |
100 |
|
Vach57 |
252 |
1 815 322 |
41 |
1931 |
38.81 |
No |
three inc. |
100 |
|
Vach60 |
129.8 |
1 771 565 |
57 |
1877 |
38.91 |
No |
two inc. |
100 |
|
Rico65 |
438.2 |
1 779 950 |
39 |
1867 |
38.95 |
Yes |
two inc. |
100 |
|
Strac48 |
468.1 |
1 853 447 |
41 |
1954 |
38.81 |
No |
one inc. |
100 |
|
Moz111 |
137.8 |
1 820 649 |
22 |
1910 |
38.95 |
Yes |
two inc. |
100 |
|
Brie16 |
154 |
1 798 726 |
25 |
1891 |
38.87 |
No |
two inc. |
100 |
|
Moz83 |
164.5 |
1 753 946 |
22 |
1844 |
39.04 |
Yes |
three inc. |
100 |
|
Moz76 |
126.3 |
1 807 399 |
33 |
1910 |
38.91 |
Yes |
two inc. |
100 |
|
Rico66 |
342.5 |
1 801 536 |
34 |
1871 |
38.93 |
Yes |
three inc. |
100 |
|
Brie28 |
82.6 |
1 769 315 |
26 |
1870 |
38.90 |
No |
two inc. |
100 |
|
FDL19 |
247 |
1 805 448 |
29 |
1873 |
38.92 |
Yes |
two inc. |
100 |
|
Nect1 |
325 |
1 799 567 |
36 |
1896 |
38.93 |
Yes |
three inc. |
100 |
|
Scam27 |
259 |
1 787 154 |
27 |
1903 |
38.85 |
No |
four inc. |
100 |
|
FDL17 |
274 |
1 790 125 |
30 |
1895 |
38.84 |
No |
four inc. |
100 |
|
Strac42 |
251.2 |
1 796 889 |
50 |
1898 |
38.86 |
Yes |
three inc. |
100 |
|
Racle124 |
458.17 |
1 933 220 |
67 |
2048 |
38.78 |
No |
one intact, two inc., one q. |
100 |
|
Nect13 |
222 |
1 801 872 |
34 |
1892 |
38.92 |
Yes |
two inc. |
100 |
|
Moz77 |
191.4 |
1 786 575 |
27 |
1857 |
38.89 |
Yes |
two inc. |
100 |
|
Moz74 |
534.97 |
1 805 715 |
26 |
1898 |
38.91 |
Yes |
four inc. |
100 |
|
Roque89 |
425.4 |
1 969 840 |
72 |
2071 |
38.63 |
No |
three inc. |
100 |
|
Douc24 |
187.1 |
2 050 171 |
75 |
2167 |
38.81 |
No |
three inc., two q. |
100 |
*No. of prophage(s) predicted, inc., incomplete, and q., questionable.
†Genome completeness as calculated by MiGA webserver considering 106 essential genes.
The presence of prtS is associated with rapid growth in milk and is typically prevalent in industrial strains, which indicates lateral gene transfer in S. thermophilus population [38, 53] and was identified in 12/23 (or 52.2%) of the genomes analysed in this study. Two major clusters of S. thermophilus genotypes were proposed by Alexandraki et al. [38] in which the genomes of cluster A strains were observed to possess larger genomes (>1.83 Mbp), harbour prtS, CRISPR systems CR2, CR3 and CR4 and in which cluster B genomes are typically smaller (<1.83 Mbp), do not harbour prtS and often do not harbour CR2, CR3 and CR4. Based on these criteria seven strains (Brie1, Vach57, Vach60, Brie16, Brie28, Scam27, FDL17) analysed in the present study belong to cluster B, while none of the other genomes fulfil all criteria of the proposed cluster A genotype strains. In this study, all strains possessing the larger genomes (Strac48, Racle124, Roque89, Douc24) do not possess prtS (Table 2). Two strains (Brie28 and Douc24) harbour only CR1 in the genome with absence of prtS, nonetheless only Brie28 fits the requirement for cluster B as Douc24 has a relatively large genome (2.05 Mbp) (Tables 2 and 3). Based on our findings, we suggest a genome size cut-off of >1.80 Mbp for cluster A and <1.80 Mbp for cluster B. With this proposed cut-off, the genomes of seven strains could be characterized as cluster A and six as cluster B genotypes. The presence of intact prophage(s) is considerably more rare amongst S. thermophilus [38] compared to other species associated with dairy products such as Lactococcus lactis [54]. Therefore, the finding that only one (of 23 or 4.3%) genome (Table 2) was predicted to harbour an intact prophage is consistent with the previous literature [55]. This prophage (in the genome of Racle124) has a predicted size of 50.8 kb and harbours genes that encode proteins that are associated with tail and capsid morphogenesis and DNA replication functions. blastn analysis of the predicted prophage of Racle124 revealed 93.66% sequence identity over 57 % of the genome of the Brussowvirus CHPC1248, which was isolated from a French cheese sample [20]. Remarkably, 19 of the 23 analysed genomes harbour an identical prophage remnant of 10.7 kb, while four genomes harbour an identical prophage remnant region of 14.1 kb.
Table 3.
RGP and EPS type classification, number of CRISPR spacers, number of acquired CRISPR spacers compared to the corresponding phageome sequences and source of origin of 23 strains of S. thermophilus . Absence of spacers in a given CRISPR locus are shaded grey
|
Strain |
RGP type* |
EPS type† |
No. of spacers in CRISPR locus |
No. of CRISPR spacers that matched phageome sequence‡ |
Source of origin |
|||
|---|---|---|---|---|---|---|---|---|
|
CR1 |
CR2 |
CR3 |
CR4 |
|||||
|
Moz109 |
B |
D |
17 |
3 |
23 |
7 |
Mozzarella B_buffalo |
|
|
Brie1 |
B |
H |
49 |
1 |
18 |
0 |
Brie_cow |
|
|
Vach57 |
B |
D |
16 |
1 |
19 |
0 |
Vacherin_cow |
|
|
Vach60 |
B |
H |
20 |
7 |
19 |
0 |
Vacherin _cow |
|
|
Rico65 |
B |
A |
24 |
1 |
5 |
Ricotta_buffalo |
||
|
Strac48 |
B |
J |
15 |
1 |
11 |
15 |
Straciatella_cow |
|
|
Moz111 |
A |
A |
26 |
4 |
18 |
6 |
Mozzarella B_buffalo |
|
|
Brie16 |
A |
C2 |
41 |
1 |
34 |
1 |
Brie_cow |
|
|
Moz83 |
A |
C2 |
27 |
1 |
28 |
9 |
Mozzarella B_buffalo |
|
|
Moz76 |
A |
I |
18 |
3 |
9 |
4 |
Mozzarella B_buffalo |
|
|
Rico66 |
A |
C1 |
62 |
3 |
20 |
20 |
Ricotta_buffalo |
|
|
Brie28 |
A |
F |
30 |
1 |
Brie_cow |
|||
|
FDL19 |
A |
C1 |
45 |
3 |
20 |
n/a |
Mozzarella A_cow |
|
|
Nect1 |
C |
J |
35 |
1 |
26 |
28 |
Semi-soft cheese D_cow |
|
|
Scam27 |
C |
C1 |
19 |
1 |
18 |
n/a |
Scamorza_cow |
|
|
FDL17 |
C |
C1 |
18 |
1 |
28 |
n/a |
Mozzarella A_cow |
|
|
Strac42 |
C |
G |
24 |
11 |
25 |
3 |
Straciatella_cow |
|
|
Racle124 |
C |
A |
18 |
3 |
0 |
Semi-soft cheese A_cow |
||
|
Nect13 |
C |
D |
42 |
3 |
19 |
12 |
11 |
Semi-soft cheese D_cow |
|
Moz77 |
E |
C1 |
56 |
3 |
29 |
9 |
Mozzarella B_buffalo |
|
|
Moz74 |
E |
C2 |
12 |
1 |
6 |
3 |
Mozzarella B_buffalo |
|
|
Roque89 |
E |
C1 |
44 |
5 |
36 |
1 |
Blue cheese_sheep |
|
|
Douc24 |
E |
C1 |
63 |
n/a |
Reblochon B_cow |
|||
*RGP typing based on multiplex PCR, following classification by Kouwen et al. [22].
†Predicted RGP and EPS type followed classification by Szymczak et al. [20].
‡Number of spacers with a hit in phageome sequences. N/A means the food sample was not sequenced and analysed for presence of phage genome.
RGP- and EPS-encoding cluster diversity analysis
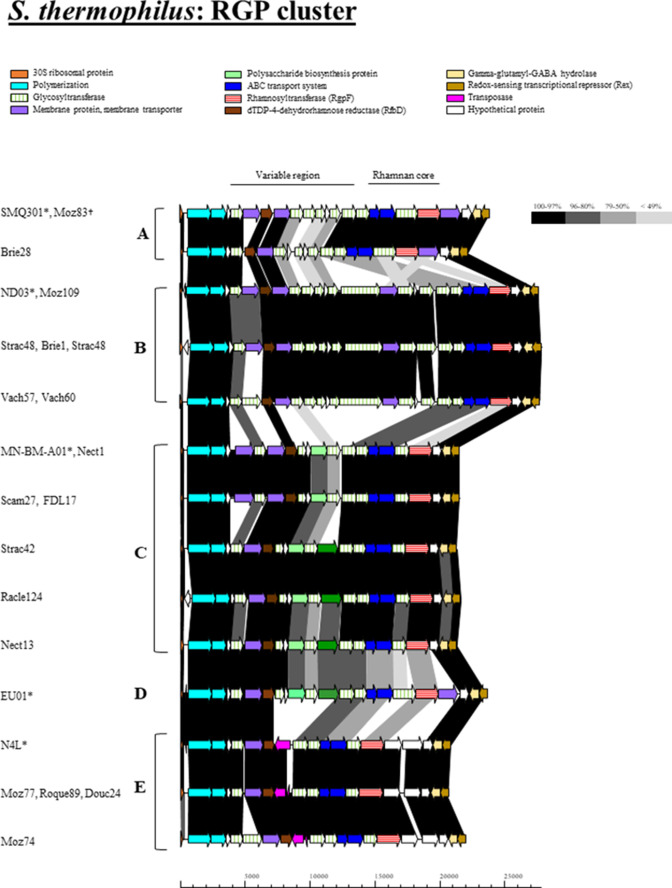
The rgp and eps clusters of each S. thermophilus strain’s genome were identified and compared to establish the diversity of these loci among the isolates. The rgp cluster is located between two conserved genes: at the 5′ end it is flanked by a gene encoding the 30S ribosomal subunit, while a gene encoding the Rex protein borders the 3′ end of the rgp gene cluster (Fig. 1). RGP-specifying gene clusters are comprised of two regions: (i) the 3′ portion, which is associated with biosynthesis of the rhamnan backbone component and which is believed to be covalently linked to and embedded in the peptidoglycan, and (ii) the 5′ portion, which is involved in the biosynthesis of the variable side chain, which is attached to the rhamnan backbone and being exposed at the cell surface (Fig. 1). Comparative analysis of rgp gene clusters identified in the sequenced genomes highlighted a high level of intra-group conservation of the clusters (Fig. 1). However, minor modifications were observed among selected isolates. For example, the RGP A strain Brie28 harbours several glycosyltransferase-encoding genes with limited sequence homology to other A type isolates (Fig. 1). Similarly, the RGP type B and E strains Vach57/60 and Moz74, respectively, each harbour a unique glycosyltransferase-encoding gene, suggesting that these strains possess unique glycosidic components in their respective RGP glycan structures relative to members of their genotypic groups. Despite the overall conservation and the lack of apparently novel rgp genotypes observed amongst the isolated strains, these clusters exhibit minor modifications, which may have occurred through insertion/deletion events.
Fig. 1.
Schematic overview of the organization and sequence relatedness the RGP-specifying gene clusters of S. thermophilus strains (A–E), including five reference strains of S. thermophilus (*) for each respective RGP type. Regions of homology (% amino acid identity) are joined by blocks of different shades of grey to black as indicated in the figure. The proposed functions of the individual protein-encoding regions are colour coded and indicated within the figure. Scale bar was measured in base pair (bp). † indicates where representative examples of identical gene clusters are presented; the RGP-encoding operons of Moz111, Moz76, Brie16, FDL19 and Rico66 are identical to SMQ-301 and Moz83.
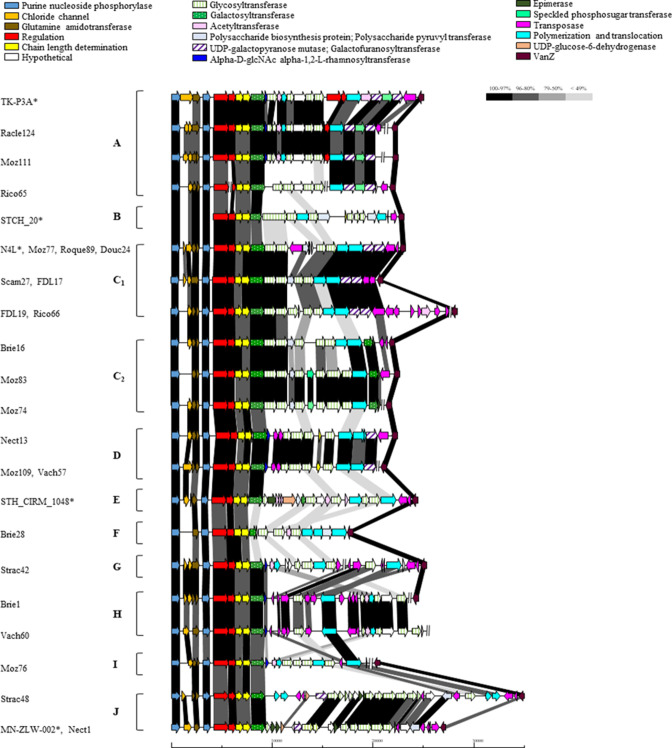
The eps cluster is located between conserved genes encoding chloride channel/purine nucleoside phosphorylase (on the 5′ border) and VanZ (on the 3′ flank) (Fig. 2). The eps cluster is present in each of the 23 assessed genomes and is often interjected by transposons likely contributing to the diversification of these clusters through recombination events (Fig. 2). Eleven distinct eps genotypes (Table 3, Fig. 2) were assigned based on HCL analysis, including type A to F based on a previous study by Szymczak et al. [20] (Fig. S2). Among the 23 identified eps clusters, seven strains (Moz77, Roque89, Douc 24, Scam27, FDL17, FDL19) were classified as type C. Furthermore, based on the observation that certain C-type strains possess a unique gene content while maintaining certain genes that are specific to C-type strains, two C sub-types were arbitrarily defined in this study (C1 and C2; Fig. 2). The eps loci of Racle124, Moz111 and Rico65 cluster among EPS type A strains; Nect13, Moz109 and Vach57 to D type; and Brie28 to F type (Table 3, Fig. 2). Additional eps genotypes were identified in this study in extension to those previously defined by Szymczak et al. [20]: Strac42 is proposed as a type G; Brie 1 and Vach60 as type H; Moz76 as type I; and Strac48 and Nect1 as type J (Table 3, Fig. 2). While rgp gene clusters show high levels of conservation, the identified eps clusters appear highly variable in terms of their genetic composition and locus size (Figs 1 and 2), a finding that is consistent with a previous report [18]. Intact eps gene clusters were aligned, revealing that overall the glycosyltransferase-encoding genes are significantly different between groups (<49% identity) and vary moderately between strains of the same groups (for instance, between Brie16 and Moz83, glycosyltransferase-encoding genes share 50–96% sequence identity, with the Moz83 cluster encoding one unique glycosyltransferase).
Fig. 2.
Schematic overview of the organization and sequence relatedness the EPS-specifying gene clusters of S. thermophilus strains (type A to J), including five reference strains of S. thermophilus (*). Regions of homology (% amino acid identity) are joined by blocks of different shades of grey to black as indicated in the schematic. The proposed functions of the individual protein-encoding regions are colour coded and indicated above the figure. Scale bar is measured in base pair (bp). In the case where the eps cluster was retrieved from different contigs, break symbol (//) was used.
In agreement with Szymczak et al. [20], this study demonstrates that the RGP and EPS-specifying gene clusters harboured by S. thermophilus strains are not correlated highlighting the range of combinations of cell-wall polysaccharide biosynthetic gene clusters that may be present among strains of the species (Table 2). However, the one apparent exception to this was the observation that all RGP type E strains, including the reference strain N4L, possess EPS type C (Table 3), which was also observed in a study by Szymczak et al. [20] where all four strains (CHPC1042, CHPC1246, CHPC1247, CHPC1248) identified as rgp type E possessed eps type C. Understanding the diversity (and combinations) of host cell-wall (RGP and EPS) polysaccharide structures and their encoding gene clusters may facilitate predictions of phage sensitivity [56]. Furthermore, it was previously suggested that S. thermophilus genomes are homogenous with limited genetic variation [38], but the present study highlights the presence of specific loci that exhibit significant diversity [20]. Significant diversity was observed in the eps loci despite a relatively small number (n=23) of sequenced/analysed strains (Fig. 2, Table 3). This may represent a biological response to phage (or other external) pressure as a previous study demonstrated that mutations in the eps locus resulted in increased resistance to phages [18].
Phageome composition correlation to S. thermophilus strains’ diversity
To evaluate if phages were present in the food samples that could infect the retrieved isolates, filtered cheese suspensions were tested against the isolates emanating from the respective cheese samples to ascertain if phage–host combinations could be identified. However, using a liquid testing system in 96-well microtitre plates, no such interactions were observed. The screened dairy products did not appear to contain phages that targeted the retrieved dairy streptococcal strains isolated from the corresponding dairy samples. Nonetheless, since it is widely accepted that phages are ubiquitous in food fermentations, it seems unlikely that the foods analysed in the study were devoid of phages. A panel of 30 phages from our in-house collection (listed in Table S4) was used to evaluate the phage robustness of the isolates. Among the 23 strains tested, three of the seven RGP A type strains were sensitive to phages in our collection. Phages SW11 (cos), SW6 (cos) and SW24 (5093) were able to form plaques on strains Brie28, Moz83 and Rico66, respectively (Table 4). The region encoding the rgp gene cluster in the genomes of strains Moz83 and Rico66 displays almost 100% sequence identity across the entire cluster, which suggests that the cos phage SW6 and the 5093 phage SW24 require distinct moieties since the RGP composition and structure is likely to be highly similar between these strains. Perhaps Moz83 (EPS C1 type) and Rico66 (EPS C2 type) displayed different sensitivity to phages as they possess different EPS genotypes, as it has previously been suggested that EPS represents the receptor for the 5093 phages [10]. The RGP B-type strain Moz109 was shown to be sensitive to infection by the cos phages STP1 and SW9, which is consistent with the finding that the deduced Bpp’s of these two phages share 96% aa identity across their entire protein length. The RGP C-type strain Nect13 strain was shown to be sensitive to cos phage SW41 (Tables 3 and 4). This analysis confirms the narrow host range that is typical of dairy streptococcal phages and highlights that the majority (18 of 23 strains) of tested strains was insensitive to any of the assessed phages.
Table 4.
Phage–host relationships identified among the S. thermophilus strains. Formation of plaques was shown as ‘+’, whereas absence of plaques was shown as ‘–’
|
Strain (RGP type) |
SW11 (cos) |
SW6 (cos) |
SW24 (5093) |
SW9 (cos) |
STP1 (cos) |
SW41 (cos) |
|---|---|---|---|---|---|---|
|
Brie28 (RGP A) |
+ |
− |
− |
− |
− |
− |
|
Moz83 (RGP A) |
− |
+ |
− |
− |
− |
− |
|
Rico66 (RGP A) |
− |
− |
+ |
− |
− |
− |
|
Moz109 (RGP B) |
− |
− |
− |
+ |
+ |
− |
|
Nect13 (RGP C) |
− |
− |
− |
− |
− |
+ |
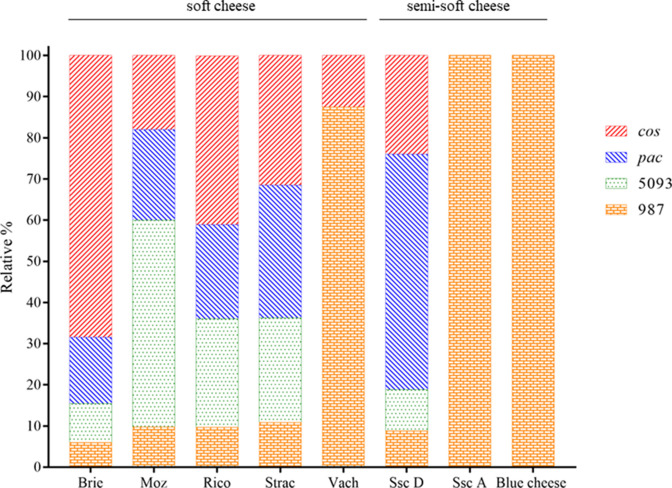
Given the observed diversity among the rgp and eps loci and their reported role as phage receptors, it was hypothesized that samples containing dairy streptococcal isolates would likely harbour streptococcal phages. To evaluate the presence of phages in the foods and to establish if any correlations could be made between the isolates and the harboured phages, eight representative dairy products (brie, mozzarella B, ricotta, stracciatella, vacherin, semi-soft cheese D, semi-soft cheese A and blue cheese) containing strains with distinct S. thermophilus RGP profiles were selected for phageome sequencing and analysis. Microbiological analysis of vacherin, semi-soft cheese A and blue cheese yielded strains with limited rgp genotype diversity (Table 3). This correlates to a narrow streptococcal phage diversity in the corresponding phageome analysis with only 987 group phages present in semi-soft cheese A and blue cheese, and 987 and cos group phages present in vacherin (Fig. 3). Conversely, in brie, mozzarella B, ricotta, stracciatella and semi-soft cheese D, members of four streptococcal phage groups were present, while at least two to four distinct RGP types of S. thermophilus are present in these products (Table 3). Therefore, in general, samples possessing streptococci with diverse rgp genotypes are correlated with a higher number of phage groups being present in the food. Vacherin, stracciatella, blue and semi-soft cheese A yielded low numbers of viable dairy streptococci (n≤10 isolates), which may be due to prevalence of other species such as L. lactis . Analysis of the overall distribution of phages in the eight selected dairy products revealed a prevalence of lactococcal phages in four of the eight analysed foods (Table S5), i.e. vacherin, stracciatella, blue and semi-soft cheese A (Fig. S3).
Fig. 3.
Relative % of read mapping to streptococcal phage (cos, pac, 5093, 987) distribution in eight cheese samples – brie, mozzarella B (moz), ricotta (rico), stracciatella (strac), vacherin (vach), semi-soft cheese D (ssc D), semi-soft cheese A (ssc A) and blue cheese, based on phageome analysis.
Further analysis was conducted to investigate whether the strains had acquired CRISPR spacers that matched the phageome sequence of the respective dairy products they were isolated from. CRISPR has been described as an effective adaptive immunity acquired by bacteria through provision of sequence-specific interference against phages, within the case of S. thermophilus novel spacer acquisition occurring mostly in CR1 and CR3, and only rarely in CR4 [57]. Such patterns of CR1 and CR3 being the most active were also observed in this study and most strains, which had acquired spacers that matched respective phageome sequences, had the spacers located mainly in CR1 and CR3 (Table 3). Spacers of short DNA fragments are obtained from infecting phages, thereby providing immunity against subsequent exposure to the same phage [56]. However, Vach57, Vach60 (sourced from vacherin) and Racle124 (sourced from semi-soft cheese A) strains, do not appear to have spacer sequences that correlate to the respective phageome sequences, hence they do not appear to be recently evolved CRISPR-mediated bacteriophage insensitive mutants (BIMs) (Table 3). Their survival in the dairy products may be due to relative low abundance and less variation of streptococcal phage with only 987 being the dominant phage group in vacherin and semi-soft cheese A (Figs 3 and S3).
Conclusions
In the present study, we observed a prevalence of S. thermophilus strains among soft and semi-soft cheeses. Genome analysis based on RGP and EPS clusters, the presence of prophages and the component CRISPR arrays provided novel insights into strain diversity and the composition of dairy streptococci in dairy foods. Strains isolated in this study exhibited significant diversity within the EPS loci with four novel EPS genotypes identified (G to J) and a subgrouping of the C type EPS cluster genotype (C1 and C2) while subtle modifications were observed among the RGP clusters contributing to their diversification (type A to E). Microbiological isolation of surviving strains of S. thermophilus has permitted an evaluation of the diversity of strains that may be present in artisanal dairy products. Data generated in this study has provided insights for the dairy industry to follow nature’s recipe in developing optimal and mixed defined starter cultures of strains with diverse RGP and EPS types thereby enhancing the production consistency and organoleptic properties of the product.
Supplementary Data
Funding information
This publication has emanated from research conducted with the financial support of/supported in part by a grant from Science Foundation Ireland under Starting Investigator Research Grant (SIRG) (Ref. No. 15/SIRG/3430) awarded to J.M, and Principal Investigator award (Ref. No. 13/IA/1953) awarded to DvS. For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Author contributions
Conceptualization, J.M.; data curation, E.P.; formal analysis, E.P., J.M., G.A.L.; funding acquisition, J.M, DvS.; investigation, E.P.; methodology, E.P., B.M.D.; project administration, J.M., DvS.; resources, J.M., DvS., M.V.; software, G.A.L.; supervision J.M., DvS., M.V; validation, E.P., G.A.L.; visualization, E.P.; writing – original draft, E.P.; writing – review and editing, J.M., DvS.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CRISPR-Cas, clustered regularly interspaced short palindromic repeats and CRISPR-associated genes; EPS, exopolysaccharide; GRAS, generally regarded as safe; LAB, lactic acid bacteria; PHASTER, PHAge search tool enhanced release; QPS, qualified presumption of safety; RGP, rhamnose-glucose polysaccharide.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Three supplementary figures and five supplementary tables are available with the online version of this article.
References
- 1.Parlindungan E, McDonnell B, Lugli GA, Ventura M, van Sinderen D, et al. Dairy streptococcal cell wall and exopolysaccharide genome diversity. Figshare. 2022 doi: 10.6084/m9.figshare.17080283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hols P, Hancy F, Fontaine L, Grossiord B, Prozzi D, et al. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol Rev. 2005;29:435–463. doi: 10.1016/j.femsre.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Yamamoto E, Watanabe R, Koizumi A, Ishida T, Kimura K. Isolation and characterization of Streptococcus thermophilus possessing prts gene from raw milk in Japan. Biosci Microbiota Food Health. 2020;39:169–174. doi: 10.12938/bmfh.2019-052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song A-L, In LLA, Lim SHE, Rahim RA. A review on Lactococcus lactis: from food to factory. Microb Cell Fact. 2017;16:55. doi: 10.1186/s12934-017-0669-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iyer R, Tomar SK, Uma Maheswari T, Singh R. Streptococcus thermophilus strains: Multifunctional lactic acid bacteria. Int Dairy J. 2010;20:133–141. doi: 10.1016/j.idairyj.2009.10.005. [DOI] [Google Scholar]
- 6.EFSA Panel on Biological Hazards (BIOHAZ) Koutsoumanis K, Allende A, Álvarez-Ordóñez A, Bolton D, et al. Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 9: suitability of taxonomic units notified to EFSA until September 2018. EFSA J. 2019;17:e05555. doi: 10.2903/j.efsa.2019.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahony J, van Sinderen D. Novel strategies to prevent or exploit phages in fermentations, insights from phage-host interactions. Curr Opin Biotechnol. 2015;32:8–13. doi: 10.1016/j.copbio.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Le Marrec C, van Sinderen D, Walsh L, Stanley E, Vlegels E, et al. Two groups of bacteriophages infecting Streptococcus thermophilus can be distinguished on the basis of mode of packaging and genetic determinants for major structural proteins. Appl Environ Microbiol. 1997;63:3246–3253. doi: 10.1128/aem.63.8.3246-3253.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mills S, Griffin C, O’Sullivan O, Coffey A, McAuliffe OE, et al. A new phage on the ‘Mozzarella’ block: Bacteriophage 5093 shares A low level of homology with other Streptococcus thermophilus phages. Int Dairy J. 2011;21:963–969. doi: 10.1016/j.idairyj.2011.06.003. [DOI] [Google Scholar]
- 10.McDonnell B, Mahony J, Neve H, Hanemaaijer L, Noben J-P, et al. Identification and analysis of a novel group of bacteriophages infecting the lactic acid bacterium Streptococcus thermophilus . Appl Environ Microbiol. 2016;82:5153–5165. doi: 10.1128/AEM.00835-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philippe C, Levesque S, Dion MB, Tremblay DM, Horvath P, et al. Novel genus of phages infecting Streptococcus thermophilus: genomic and morphological characterization. Appl Environ Microbiol. 2020;86:e00227-20. doi: 10.1128/AEM.00227-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavelle K, Martinez I, Neve H, Lugli GA, Franz CMAP, et al. Biodiversity of Streptococcus thermophilus phages in global dairy fermentations. Viruses. 2018;10:E577. doi: 10.3390/v10100577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szymczak P, Janzen T, Neves AR, Kot W, Hansen LH, et al. Novel variants of Streptococcus thermophilus bacteriophages are indicative of genetic recombination among phages from different bacterial species. Appl Environ Microbiol. 2017;83:e02748-16. doi: 10.1128/AEM.02748-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quiberoni A, Tremblay D, Ackermann HW, Moineau S, Reinheimer JA. Diversity of Streptococcus thermophilus phages in a large-production cheese factory in Argentina. J Dairy Sci. 2006;89:3791–3799. doi: 10.3168/jds.S0022-0302(06)72420-1. [DOI] [PubMed] [Google Scholar]
- 15.Dugat-Bony E, Lossouarn J, De Paepe M, Sarthou A-S, Fedala Y, et al. Viral metagenomic analysis of the cheese surface: A comparative study of rapid procedures for extracting viral particles. Food Microbiol. 2020;85:103278. doi: 10.1016/j.fm.2019.103278. [DOI] [PubMed] [Google Scholar]
- 16.Muhammed MK, Kot W, Neve H, Mahony J, Castro-Mejía JL, et al. Metagenomic analysis of dairy bacteriophages: extraction method and pilot study on whey samples derived from using undefined and defined mesophilic starter cultures. Appl Environ Microbiol. 2017;83:e00888-17. doi: 10.1128/AEM.00888-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romero DA, Magill D, Millen A, Horvath P, Fremaux C. Dairy lactococcal and streptococcal phage-host interactions: an industrial perspective in an evolving phage landscape. FEMS Microbiol Rev. 2020;44:909–932. doi: 10.1093/femsre/fuaa048. [DOI] [PubMed] [Google Scholar]
- 18.McDonnell B, Hanemaaijer L, Bottacini F, Kelleher P, Lavelle K, et al. A cell wall-associated polysaccharide is required for bacteriophage adsorption to the Streptococcus thermophilus cell surface. Mol Microbiol. 2020;114:31–45. doi: 10.1111/mmi.14494. [DOI] [PubMed] [Google Scholar]
- 19.Szymczak P, Filipe SR, Covas G, Vogensen FK, Neves AR, et al. Cell wall glycans mediate recognition of the dairy bacterium Streptococcus thermophilus by bacteriophages. Appl Environ Microbiol. 2018;84:e01847-18. doi: 10.1128/AEM.01847-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szymczak P, Rau MH, Monteiro JM, Pinho MG, Filipe SR, et al. A comparative genomics approach for identifying host-range determinants in Streptococcus thermophilus bacteriophages. Sci Rep. 2019;9:7991. doi: 10.1038/s41598-019-44481-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Q, Tun HM, Leung F-C, Shah NP. Genomic insights into high exopolysaccharide-producing dairy starter bacterium Streptococcus thermophilus ASCC 1275. Sci Rep. 2014;4:4974. doi: 10.1038/srep04974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kouwen RHM, Van Sinderen D, McDonnell B, Ver Loren Van Themaat P, Emiel Mahony J, inventors Streptococcus thermophilus starter cultures. Netherlands Patent. 2019;20190367866 [Google Scholar]
- 23.Delorme C, Legravet N, Jamet E, Hoarau C, Alexandre B, et al. Study of Streptococcus thermophilus population on a world-wide and historical collection by a new MLST scheme. Int J Food Microbiol. 2017;242:70–81. doi: 10.1016/j.ijfoodmicro.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 24.Stern A, Sorek R. The phage-host arms race: shaping the evolution of microbes. Bioessays. 2011;33:43–51. doi: 10.1002/bies.201000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burrus V, Bontemps C, Decaris B, Guédon G. Characterization of a novel type II restriction-modification system, Sth368I, encoded by the integrative element ICESt1 of Streptococcus thermophilus CNRZ368. Appl Environ Microbiol. 2001;67:1522–1528. doi: 10.1128/AEM.67.4.1522-1528.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Common J, Morley D, Westra ER, van Houte S. CRISPR-Cas immunity leads to a coevolutionary arms race between Streptococcus thermophilus and lytic phage. Philos Trans R Soc Lond B Biol Sci. 2019;374:1772. doi: 10.1098/rstb.2018.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Achigar R, Scarrone M, Rousseau GM, Philippe C, Machado F, et al. Ectopic spacer acquisition in Streptococcus thermophilus CRISPR3 Array. Microorganisms. 2021;9:512. doi: 10.3390/microorganisms9030512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli . Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horvath P, Romero DA, Coûté-Monvoisin A-C, Richards M, Deveau H, et al. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus . J Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Achigar R, Magadán AH, Tremblay DM, Julia Pianzzola M, Moineau S. Phage-host interactions in Streptococcus thermophilus: Genome analysis of phages isolated in Uruguay and ectopic spacer acquisition in CRISPR array. Sci Rep. 2017;7:43438. doi: 10.1038/srep43438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu T, Cui Y, Qu X. Characterization and comparison of CRISPR Loci in Streptococcus thermophilus . Arch Microbiol. 2020;202:695–710. doi: 10.1007/s00203-019-01780-3. [DOI] [PubMed] [Google Scholar]
- 32.Dion MB, Labrie SJ, Shah SA, Moineau S. CRISPRStudio: A User-Friendly Software for Rapid CRISPR Array Visualization. Viruses. 2018;10:E602. doi: 10.3390/v10110602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moh LG, Etienne PT, Jules-Roger K. Seasonal diversity of lactic acid bacteria in artisanal yoghurt and their antibiotic susceptibility pattern. Int J Food Sci. 2021;2021:6674644. doi: 10.1155/2021/6674644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fagbemigun O, Cho G-S, Rösch N, Brinks E, Schrader K, et al. Isolation and characterization of potential starter cultures from the nigerian fermented milk product nono . Microorganisms. 2021;9:640. doi: 10.3390/microorganisms9030640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng C, Sun Z, Sun Y, Ma T, Li W, et al. Characterization and association of bacterial communities and nonvolatile components in spontaneously fermented cow milk at different geographical distances. J Dairy Sci. 2021;104:2594–2605. doi: 10.3168/jds.2020-19303. [DOI] [PubMed] [Google Scholar]
- 36.Zago M, Bardelli T, Rossetti L, Nazzicari N, Carminati D, et al. Evaluation of bacterial communities of Grana Padano cheese by DNA metabarcoding and DNA fingerprinting analysis. Food Microbiol. 2021;93:103613. doi: 10.1016/j.fm.2020.103613. [DOI] [PubMed] [Google Scholar]
- 37.Hu T, Cui Y, Zhang Y, Qu X, Zhao C. Genome analysis and physiological characterization of four Streptococcus thermophilus strains isolated from Chinese traditional fermented milk. Front Microbiol. 2020;11:184. doi: 10.3389/fmicb.2020.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexandraki V, Kazou M, Blom J, Pot B, Papadimitriou K, et al. Comparative genomics of Streptococcus thermophilus support important traits concerning the evolution, biology and technological properties of the species. Front Microbiol. 2019;10:2916. doi: 10.3389/fmicb.2019.02916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lugli GA, Milani C, Mancabelli L, van Sinderen D, Ventura M. MEGAnnotator: A user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol Lett. 2016;363:fnw049. doi: 10.1093/femsle/fnw049. [DOI] [PubMed] [Google Scholar]
- 40.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chevreux B, Wetter T, Suhai S., editors. Genome Sequence Assembly Using Trace Signals and Additional Sequence Information. German Conference on Bioinformatics; 1999. [Google Scholar]
- 42.Rodriguez-R LM, Gunturu S, Harvey WT, Rosselló-Mora R, Tiedje JM, et al. The Microbial Genomes Atlas (MiGA) webserver: taxonomic and gene diversity analysis of Archaea and Bacteria at the whole genome level. Nucleic Acids Res. 2018;46:W282–W288. doi: 10.1093/nar/gky467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: A fast phage search tool. Nucleic Acids Res. 2011;39:W347–52. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saeed AI, Sharov V, White J, Li J, Liang W, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 48.Milani C, Casey E, Lugli GA, Moore R, Kaczorowska J, et al. Tracing mother-infant transmission of bacteriophages by means of a novel analytical tool for shotgun metagenomic datasets: METAnnotatorX. Microbiome. 2018;6:145. doi: 10.1186/s40168-018-0527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel RK, Jain M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLOS ONE. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lillehaug D. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J Appl Microbiol. 1997;83:85–90. doi: 10.1046/j.1365-2672.1997.00193.x. [DOI] [PubMed] [Google Scholar]
- 52.Chuard C, Reller LB. Bile-esculin test for presumptive identification of enterococci and streptococci: effects of bile concentration, inoculation technique, and incubation time. J Clin Microbiol. 1998;36:1135–1136. doi: 10.1128/JCM.36.4.1135-1136.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delorme C, Bartholini C, Bolotine A, Ehrlich SD, Renault P. Emergence of a cell wall protease in the Streptococcus thermophilus population. Appl Environ Microbiol. 2010;76:451–460. doi: 10.1128/AEM.01018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kelleher P, Mahony J, Schweinlin K, Neve H, Franz CM, et al. Assessing the functionality and genetic diversity of lactococcal prophages. Int J Food Microbiol. 2018;272:29–40. doi: 10.1016/j.ijfoodmicro.2018.02.024. [DOI] [PubMed] [Google Scholar]
- 55.Arioli S, Eraclio G, Della Scala G, Neri E, Colombo S, et al. Role of temperate bacteriophage ϕ20617 on Streptococcus thermophilus DSM 20617T autolysis and biology. Front Microbiol. 2018;9:2719. doi: 10.3389/fmicb.2018.02719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahony J, Bottacini F, van Sinderen D, Fitzgerald GF. Progress in lactic acid bacterial phage research. Microb Cell Fact. 2014;13 Suppl 1:S1. doi: 10.1186/1475-2859-13-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paez-Espino D, Sharon I, Morovic W, Stahl B, Thomas BC, et al. CRISPR immunity drives rapid phage genome evolution in Streptococcus thermophilus . mBio. 2015;6:e00262-15. doi: 10.1128/mBio.00262-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.