Abstract
Extensively drug-resistant tuberculosis (XDR-TB), defined as resistance to at least isoniazid (INH), rifampicin (RIF), a fluoroquinolone (FQ) and a second-line injectable drug (SLID), is difficult to treat and poses a major threat to TB control. The transmission dynamics and distribution of XDR Mycobacterium tuberculosis (Mtb) strains have not been thoroughly investigated. Using whole genome sequencing data on 461 XDR-Mtb strains, we aimed to investigate the geographical distribution of XDR-Mtb strains in the Western Cape Province of South Africa over a 10 year period (2006–2017) and assess the association between Mtb sub-lineage, age, gender, geographical patient location and membership or size of XDR-TB clusters. First, we identified transmission clusters by excluding drug resistance-conferring mutations and using the 5 SNP cutoff, followed by merging clusters based on their most recent common ancestor. We then consecutively included variants conferring resistance to INH, RIF, ethambutol (EMB), pyrazinamide (PZA), SLIDs and FQs in the cluster definition. Cluster sizes were classified as small (2–4 isolates), medium (5–20 isolates), large (21–100 isolates) or very large (>100 isolates) to reflect the success of individual strains. We found that most XDR-TB strains were clustered and that including variants conferring resistance to INH, RIF, EMB, PZA and SLIDs in the cluster definition did not significantly reduce the proportion of clustered isolates (85.5–82.2 %) but increased the number of patients belonging to small clusters (4.3–12.4 %, P=0.56). Inclusion of FQ resistance-conferring variants had the greatest effect, with 11 clustered isolates reclassified as unique while the number of clusters increased from 17 to 37. Lineage 2 strains (lineage 2.2.1 typical Beijing or lineage 2.2.2 atypical Beijing) showed the large clusters which were spread across all health districts of the Western Cape Province. We identified a significant association between residence in the Cape Town metropole and cluster membership (P=0.016) but no association between gender, age and cluster membership or cluster size (P=0.39). Our data suggest that the XDR-TB epidemic in South Africa probably has its origin in the endemic spread of MDR Mtb and pre-XDR Mtb strains followed by acquisition of FQ resistance, with more limited transmission of XDR Mtb strains. This only became apparent with the inclusion of drug resistance-conferring variants in the definition of a cluster. In addition to the prevention of amplification of resistance, rapid diagnosis of MDR, pre-XDR and XDR-TB and timely initiation of appropriate treatment is needed to reduce transmission of difficult-to-treat TB.
Keywords: tuberculosis, drug resistance, XDR-TB, whole genome sequencing, transmission
Data Summary
Impact Statement.
Since the first outbreak in 2006, extensively drug-resistant tuberculosis (XDR-TB) strains have been documented in 128 countries, making prevention of transmission critical, but very little is known about the distribution and transmission dynamics of XDR strains. Additionally, there is uncertainty regarding the impact of including or excluding drug resistance-conferring mutations in transmission analysis. We aimed to investigate the locations driving transmission in a high-burden TB area as well as the epidemiological features such as age and gender of patients in transmission clusters to identify opportunities for interventions. We also aim to shed light on the role of drug resistance-conferring mutations in phylogenetic studies of drug-resistant TB.
Previously published whole genome sequencing data of Mtb genomes are published under accession numbers PRJEB35725 (https://www.ebi.ac.uk/ena/browser/view/PRJEB35725?show=reads) and PRJEB14199 (https://www.ebi.ac.uk/ena/browser/view/PRJEB14199?show=reads) by Marissa Klopper and Keertan Dheda respectively on the European Nucleotide Archive. Newly published whole genome sequences of Mtb genomes are published under accession number PRJEB43283 on the European Nucleotide Archive.
The authors confirm all supporting data, code and protocols have been provided within the article or through supplementary data files.
Introduction
The control of tuberculosis (TB) is complicated by the spread of drug-resistant Mycobacterium tuberculosis (Mtb) strains. In 2006, the first outbreak of extensively drug-resistant tuberculosis (XDR-TB) shocked the world [1–3]. Over a period of 7 months, more than 50 patients at the Tugela Ferry hospital in KwaZulu-Natal, South Africa, were diagnosed with XDR-TB given resistance to at least rifampicin (RIF), isoniazid (INH), a fluoroquinolone (FQ) and an injectable second-line drug (SLID) [4]. All but one patient died after a median of 16 days from sputum collection [2]. Since then, XDR-TB cases have been documented in 128 countries [5], making prevention of transmission critical. In 2018, 553 confirmed cases of XDR-TB were notified in South Africa, corresponding to 5 % of all multidrug-resistant/rifampicin-resistant TB (MDR/RR-TB), and about 0.18 % of all TB cases [5]. In vitro observations that drug resistance in bacteria is associated with reduced Darwinian fitness resulted in the dogma that drug-resistant Mtb strains are less likely to transmit [6]. This fitness cost is neither absolute nor universal, with drug resistance-conferring mutations having a variable impact on bacterial fitness depending on the specific resistance-conferring mutation and the Mtb strain type [6–11]. Furthermore, the difficulty in effectively treating drug-resistant TB can result in prolonged infectiousness which may lead to MDR-TB and XDR-TB outbreaks in community and congregate settings [3, 12–16].
While major progress has been made in the diagnosis and treatment of XDR-TB [5], the transmission dynamics of XDR-TB remain poorly understood. Some studies have shown that the XDR-TB epidemic in South Africa was mainly driven by acquisition of drug resistance [17] and that acquisition of INH resistance-conferring mutations occurred first, followed by acquisition of RIF resistance mutations [18, 19]. For XDR-TB, very little is known about the order of drug resistance acquisition. Transmission dynamics can be influenced by the social and behavioural patterns that determine where and with whom people spend time. Investigating the locations where clusters of cases occur, as well as the features such as age and gender of patients in transmission clusters, can identify opportunities for further investigation and clinical interventions [20].
Whole-genome sequencing (WGS) offers high resolution and is the state-of-the-art technology to investigate Mtb transmission [21, 22] and is especially useful in settings with high incidences of TB because the majority of transmission occurs in community locations between individuals who do not know each other and would not be identified through conventional contact investigations [20]. We used WGS to define clusters of XDR-TB to assess the geographical distribution of XDR-TB strains in the Western Cape Province, and to investigate the association between age and gender and the occurrence and size of XDR-TB clusters, as well as the impact of including resistance-conferring mutations in cluster analyses.
Methods
Setting and sample selection
The Western Cape Province of South Africa has a high burden of TB with an incidence rate of 739 per 100 000 population in 2016 [23], and one of the highest burdens of XDR-TB within South Africa, with an incidence of 2.8 cases per 100 000 population [16]. The Western Cape Province can be divided in six healthcare districts: Cape Town metropole, Garden Route, Cape Winelands, Overberg, West Coast and Central Karoo; and the large Cape Town metropole district can be subclassified into eight subdistricts: Eastern, Southern, Western, Northern, Khayelitsha, Klipfontein, Mitchells Plain and Tygerberg.
In the Western Cape Province, routine drug susceptibility testing is done by the National Health Laboratory Services (NHLS). Between 2006 and 2008, selected isolates were assessed by phenotypic drug susceptibility testing (pDST). In 2008, the first-line GenoType MTBDRplus line probe assay (LPA; Hain LifeSciences) was introduced. Since 2013, the initial diagnostic assay in the Western Cape Province is Xpert MTB/RIF (Cepheid) [24], with further investigation by first-line LPA (INH and RIF) of RIF-resistant isolates. Second-line DST was done using culture-based methods. Since 2006, Mtb isolates with any resistance have been archived at Stellenbosch University. This has resulted in one of the largest clinical Mtb strain collections worldwide, with over 50 000 Mtb strains of which an estimated 40 000 are drug-resistant. From this strain bank, XDR-Mtb strains collected between 2006 (first report of XDR-TB in Kwazulu-Natal, South Africa) and 2017 were selected by identifying those strains that were classified as phenotypically resistant to RIF, INH, an FQ and SLID by the NHLS through routine DST. The first available isolate from each patient that was classified as XDR-TB by routine DST was selected for further analysis.
Bioinformatics, phylogenetic, epidemiological and statistical analysis
Raw sequencing reads obtained by Illumina sequencing [which can be obtained at the European Nucleotide Archive (ENA) under accession numbers PRJEB14199 (previously published), PRJEB35725 (previously published) and PRJEB43283] of purified DNA from a subculture isolate were analysed using the CompleX Bacterial Samples (XBS) pipeline, built for analysis of Mtb WGS data. In brief, this pipeline maps sequencing reads to the H37Rv reference genome (version NC_000962.3), joint-calls majority variants and filters these using machine learning techniques [25]. Isolates with a mapped percentage of reads <60 % (the percentage of reads mapped against the reference genome) were considered contaminated and excluded from the study. Samples with mixed infections, indicated by the TBProfiler mutation database (20 August 2019, version 2.8.12), were also excluded from the analysis. The resulting SNP positions that were represented by ≥95 % of samples were exported to a multiple sample FASTA file that was used for phylogenetic inference, and to determine genetic distances, drug conferring mutations were initially excluded. IQ-TREE v1.6.12 was used to infer the optimal substitution model (i.e TVM+F+ASC+R) and maximum likelihood phylogeny with 10 000 ultrafast bootstraps [26].
The distribution of the Mtb (sub-)lineages and drug resistance profiles was assessed by using the TBProfiler mutation database (20 August 2019, version 2.8.12) [27, 28], which is integrated in the XBS pipeline. All variants are reported in the HGVS (Human Genome Variation Society) format as annotated by TBProfiler. We annotated the phylogenetic tree using the Interactive Tree Of Life (iTOL v6) software, an online tool for the display, annotation and management of phylogenetic trees [29]. We first annotated the phylogenetic tree with the Mtb lineage TBProfiler called for each isolate. To assess the distribution by gender, we annotated the phylogenetic tree with the patients’ gender (male/female). To assess the distribution by age, we annotated the phylogenetic tree with the patients’ age in five age-groups (0–19, 20–29, 30–39, 40–49, 50–69 years). To assess the geographical distribution of XDR-TB patients, we annotated the phylogenetic tree with the assigned (sub-)district of the clinic where the sample was collected as a surrogate for the patients’ residential address.
To identify transmission clusters, pairwise genetic distances of isolates were estimated by calculating the number of SNP differences between isolates. Resistance-causing variants were excluded as these are under strong positive selection and can mutate at vastly different rates; exclusion of these variants is therefore a commonly applied approach for phylogenetic analysis [30]. An SNP distance of ≤5 was used to identify isolates that belong to a cluster, signifying recent transmission [31]. In a sensitivity analysis, we explored the use of different SNP distances. Next, clusters were extended to include all other samples (terminal taxa) descendant from their most recent common ancestor, thereby including isolates that were above the 5 SNP cut-off but that are part of the same transmission cluster as they share a common cluster ancestor [30]. We then identified transmission subclusters with specific shared drug resistance-related profiles in the 5 SNP cut-off clusters by consecutively including variants (SNPs and indels) associated with INH, RIF, ethambutol (EMB), pyrazinamide (PZA), FQs and SLIDs resistance into the cluster definition as determined by TBProfiler. For example, a cluster defined under a 5 SNP cut-off may be further delineated into a subcluster of isolates that all share the same RIF and INH resistance mutations (an MDR-TB transmission cluster). Based on the distribution of cluster sizes observed in the data (Fig. S1, available in the online version of this article), cluster size was classified as small (2–4 isolates), medium (5–20 isolates), large (21–100 isolates) and very large (>100 isolates). To assess the effect of inclusion of resistance-conferring variants on the proportion of isolates clustered and the distribution of cluster sizes, we used a chi-square test and Fisher-exact test.
To investigate the spatial distribution of XDR-TB, clinics were grouped into the five districts in the Western Cape Province (https://municipalities.co.za/provinces/view/9/western-cape) and the Cape Town metropole district was subclassified into its eight subdistricts (https://www.westerncape.gov.za/image/2012/10/ct-sub-districts.jpg). For the subdistrict analysis, we excluded isolates for which subdistrict information of the corresponding patient was missing (n=59) or could not be allocated (n=43) because the sample was collected at a centralized facility (Brooklyn Chest MDR-TB hospital or Pollsmoor maximum security prison).
To investigate the association between clustering and gender, age and health district (Cape Town metropole vs. other, non-metropolitan, districts), we compared the distribution of these variables between XDR-Mtb isolates belonging to a cluster and XDR-Mtb isolates with a unique WGS genotype after including drug resistance-conferring variants (SNPs and indels) for INH, RIF, EMB, PZA, FQs and SLIDs. A chi-square test or Fisher-exact test was used to test for significant differences in distribution. Similarly, we assessed the association of gender, age and health district with cluster size. A P-value <0.05 was considered significant.
Results
Characteristics of XDR-TB patients and their Mtb isolates
WGS data were available for 748 isolates (including serial isolates) collected from patients diagnosed with XDR-TB. Of these, three were excluded because the patient resided outside of the Western Cape Province and 197 were excluded because they were MDR or pre-XDR strains. In addition, sequences of 54 XDR-Mtb isolates were excluded as they were sequences from duplicate isolates (n=24) or they were serial XDR isolates from the same patient (n=30); in the latter only the first sample was included. Of the remaining 494 XDR-Mtb strains, nine were excluded because of low sequencing depth or coverage, 21 because of multiple infection and three because of severe bacterial contamination. The remaining 461 XDR-Mtb isolates collected between 2006 and 2017 from individual patients residing in the Western Cape Province were included in the analysis.
Of these 461 patients, 252 (54.7 %) were male, 207 (44.9 %) were female and the mean age was 33 years (Table 1). Age was missing for 18 (3.9 %) patients. The majority of the patients (n=314, 68.1 %) sought care in a City of Cape Town metropole district facility, 31 (6.7 %) in the Garden Route district, 33 (7.2 %) in Cape Winelands, 14 (3.0 %) in the West Coast and nine (2.0 %) in Overberg district. The district was unknown for the remaining 60 isolates (13.0 %). There were no patients originating from Central Karoo, which was therefore excluded for further analyses. Of the 314 patients diagnosed in the Cape Town metropole health district, 28 (8.9 %) originated from the Eastern subdistrict, 26 (8.3 %) from the Southern subdistrict, 38 (12.1 %) from the Western subdistrict, 11 (3.5 %) from the Northern subdistrict, 54 (17.2 %) from Khayelitsha, 32 (10.2 %) from Klipfontein, 38 (12.1 %) from Mitchells Plain and 44 (14.0 %) from the Tygerberg subdistrict; 43 (13.7 %) were excluded from the analyses as they were diagnosed in centralized facilities.
Table 1.
Socio-demographic, clinical and phylogenetic characteristics of 461 patients diagnosed with XDR-TB in the Western Cape Province of South Africa, 2006–2017
|
N (%) |
||
|---|---|---|
|
Gendera |
Male |
252 (54.7) |
|
Female |
207 (44.9) |
|
|
Ageb |
0–19 years |
32 (6.9) |
|
20–29 years |
128 (27.8) |
|
|
30–39 years |
147 (31.9) |
|
|
40–49 years |
82 (17.8) |
|
|
50–69 years |
54 (11.7) |
|
|
Health (sub)districtc |
Cape Winelands |
33 (7.2) |
|
Garden Route |
31 (6.7) |
|
|
Overberg |
9 (2.0) |
|
|
West Coast |
14 (3.0) |
|
|
Cape Town |
314 (68.1) |
|
|
Eastern |
28 (8.9) |
|
|
Southern |
26 (8.3) |
|
|
Western |
38 (12.1) |
|
|
Northern |
11 (3.5) |
|
|
Khayelitsha |
54 (17.2) |
|
|
Klipfontein |
32 (10.2) |
|
|
Mitchells Plain |
38 (12.1) |
|
|
Tygerberg |
44 (14.0) |
|
|
Centralized hospital or prison |
43 (13.7) |
|
|
Molecular drug resistance profile |
Resistant to INH, RIF, FQ and AMK or KM +ETO +EMB+ETO +PZA+ETO +EMB+SM +EMB+PZA +EMB+PZA+ETO +EMB+PZA+SM +EMB+SM+ETO +PZA+SM+ETO +EMB+PZA+SM+ETO +EMB+PZA+SM+ ETO+LZD |
461 (100.0) 8 (1.7) 57 (12.4) 1 (0.2) 1 (0.2) 2 (0.4) 30 (6.5) 19 (4.1) 4 (0.9) 1 (0.2) 330 (71.6) 8 (1.7) |
|
Year diagnosedd |
2006–2009 |
84 (18.2) |
|
2010–2013 |
198 (43.0) |
|
|
2014–2017 |
178 (38.6) |
|
|
Lineage |
Lineage 2 |
429 (93.1) |
|
Lineage 2.2.1 |
124 (28.9) |
|
|
Lineage 2.2.2 |
305 (71.1) |
|
|
Lineage 4 |
32 (6.9) |
|
|
Lineage 4.1 |
19 (59.4) |
|
|
Lineage 4.3 |
9 (28.1) |
|
|
Lineage 4.4 |
3 (9.4) |
|
|
Lineage 4.8 |
1 (3.1) |
|
Missing data on agender (n=2), bage (n=18), chealth (sub)district (n=60), dyear diagnosed (n=1).
ETO, ethionamide; EMB, ethambutol; PZA, pyrazinamide; SM, streptomycin; LZD, linezolid.
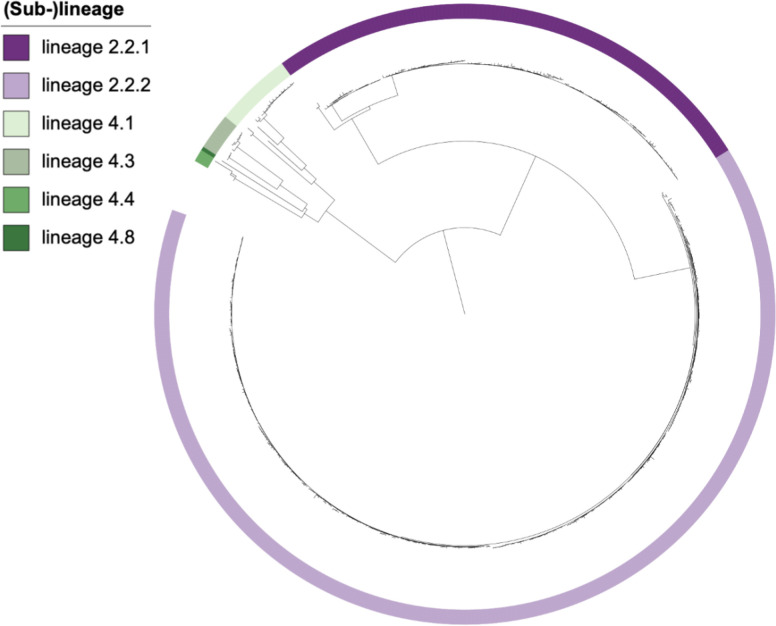
Almost all XDR-Mtb isolates belonged to lineage 2 (n=429, 93.1 %) and the remainder to lineage 4 (n=32, 6.9 %). The 429 lineage 2 isolates could be grouped into sub-lineage 2.2, of which 124 (28.9 %) belonged to the typical Beijing lineage (lineage 2.2.1) and 305 (71.1 %) belonged to the atypical Beijing lineage (lineage 2.2.2). Of the 32 lineage 4 isolates, 19 (59.4 %) belonged to sub-lineage 4.1, nine (28.1 %) belonged to sub-lineage 4.3, three (9.4 %) belonged to sub-lineage 4.4 and one (3.1 %) belonged to sub-lineage 4.8 (Table 1, Fig. 1).
Fig. 1.
Distribution of XDR-Mtb (sub-)lineages across the phylogenetic tree of individual XDR-Mtb isolates collected from 461 patients in the Western Cape Province of South Africa, 2006–2017.
Of the 461 individual patient XDR-Mtb isolates, all were confirmed to be genotypically resistant to the four drug classes defining XDR-TB: INH, RIF, an FQ and an SLID (Table 1). Resistance to additional drugs occurred in all isolates. Almost all (97.8 %) were resistant to EMB, 95.2 % were resistant to ethionamide (ETO), 84.8 % to PZA and 78.8 % to streptomycin (SM). Eight isolates (1.7 %) were additionally resistant to linezolid (LZD). INH resistance was caused by mutations in the inhA promotor, katG, inhA and ahpC. Mutations in rpoB and rpoC resulted in RIF resistance. The majority of EMB resistance was due to embB mutations and more limited due to embA mutations. All PZA resistance was caused by mutations in the pncA gene. Mutations in rrs and eis promotor resulted in SLID resistance. The vast majority of FQ resistance was caused by gyrA and more limited by gyrB mutations (Tables S1 and S2).
XDR-TB clusters
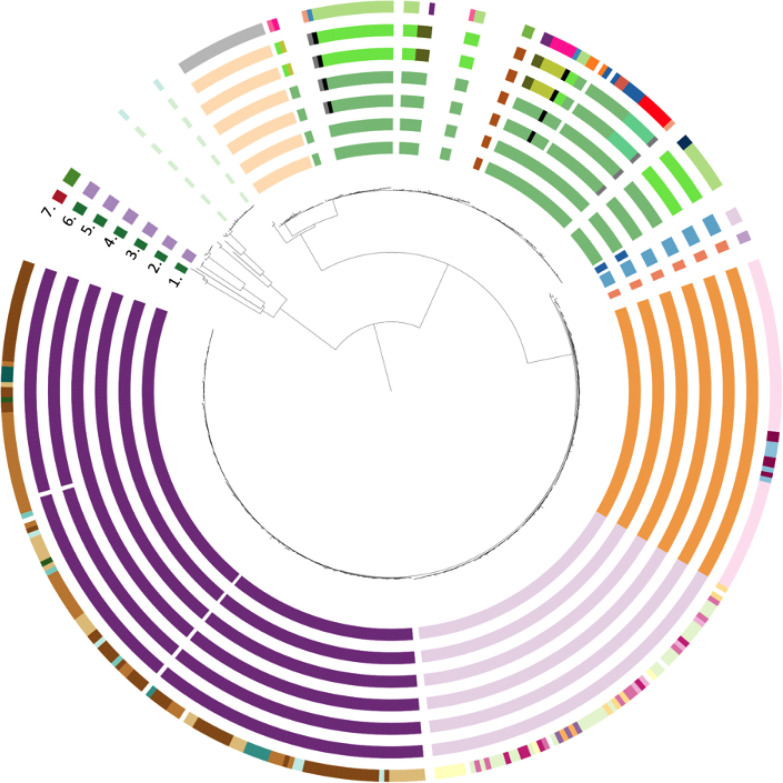
Using the accepted 5 SNP cut-off and excluding drug resistance-conferring variants, we identified 183 (39.7 %) unique isolates and 278 (60.3 %) isolates divided over 38 clusters. Taking common ancestors into account resulted in 116 unique isolates being integrated within a cluster and 32 smaller clusters being collapsed within larger clusters. This led to 394 (85.5 %) isolates belonging to one of 12 clusters and 67 isolates (14.5 %) not being associated with any cluster (Fig. 2). The size of XDR-TB clusters ranged from two to 150 patients: seven clusters were small (2–4 isolates), one medium (19 isolates), three large (66–72 isolates) and one very large (150 isolates) (Table 2). Raising the SNP cut-off from 5 to 8, 10, 12 or 15 had limited impact on the number of clusters identified (Table S3)
Fig. 2.
Transmission clusters with drug resistance-conferring mutations excluded and clusters merged on common ancestors (circle 1) and subsequent inclusion of INH (circle 2), RIF (circle 3), EMB (circle 4), PZA (circle 5), SLIDs (circle 6) and FQs (circle 7) drug resistance-conferring mutations.
Table 2.
Distribution of the proportion of unique and clustered XDR-TB isolates and distribution of cluster size by cluster definition among 461 XDR-TB patients diagnosed with XDR-TB in the Western Cape Province of South Africa, 2006–2017
|
Type of DR-TB variants included in cluster definition |
None |
INH resistance-conferring mutations |
INH+RIF resistance-conferring mutations |
INH+RIF+ EMB resistance-conferring mutations |
INH+RIF+ EMB+PZA resistance-conferring mutations |
INH+RIF+ EMB+PZA+SLIDs resistance-conferring variants |
INH+RIF+ EMB+PZA+SLIDs+FQs resistance-conferring variants |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Cluster size |
Clusters n (%) |
Patients n (%) |
Clusters n (%) |
Patients n (%) |
Clusters n (%) |
Patients n (%) |
Clusters n (%) |
Patients n (%) |
Clusters n (%) |
Patients n (%) |
Clusters n (%) |
Patients n (%) |
Clusters n (%) |
Patients n (%) |
|
Unique |
na |
67 (14.5) |
na |
67 (14.5) |
na |
70 (15.2) |
na |
70 (15.2) |
na |
70 (15.2) |
na |
71 (15.4) |
na |
82 (17.8) |
|
Clustered (any size) |
12 |
394 (85.5) |
12 |
394 (85.5) |
13 |
391 (84.8) |
13 |
391 (84.8) |
17 |
391 (84.8) |
17 |
390 (84.6) |
37 |
379 (82.2) |
|
Small (n=2–4) |
7 (58.3) |
17 (4.3) |
7 (58.3) |
17 (4.3) |
8 (61.5) |
19 (4.9) |
8 (61.5) |
19 (4.9) |
8 (47.1) |
19 (4.9) |
8 (47.1) |
19 (4.9) |
19 (51.4) |
47 (12.4) |
|
Medium (n=5–20) |
1 (8.3) |
19 (4.8) |
1 (8.3) |
19 (4.8) |
1 (7.7) |
19 (4.9) |
1 (7.7) |
19 (4.9) |
5 (29.4) |
49 (12.5) |
5 (29.4) |
48 (12.3) |
12 (32.4) |
86 (22.7) |
|
Large (n=21–100) |
3 (25.0) |
208 (52.8) |
3 (25.0) |
208 (52.8) |
3 (23.1) |
203 (51.9) |
3 (23.1) |
203 (51.9) |
3 (17.6) |
173 (44.2) |
3 (17.6) |
173 (44.4) |
6 (16.2) |
246 (64.9) |
|
Very large (n>100) |
1 (8.3) |
150 (38.1) |
1 (8.3) |
150 (38.1) |
1 (7.7) |
150 (38.4) |
1 (7.7) |
150 (38.4) |
1 (5.9) |
150 (38.4) |
1 (5.9) |
150 (38.5) |
0 |
0 |
na, Not applicable; INH, isoniazid; RIF, rifampicin; EMB, ethambutol; PZA, pyrazinamide; SLIDs, second line injectable drugs; FQs, fluoroquinolones.
By including the mutations that confer resistance to INH, we did not observe a change in the proportion of clustering (P=1) nor cluster size (P=1). Next, additionally including mutations conferring resistance to RIF increased the number of clusters from 12 to 13 but had no impact on the number of clustered isolates as only three were re-classified as unique (P=0.85). Correspondingly, it did not significantly change the proportion of isolates belonging to a transmission cluster (85.5 vs. 84.8 %, P=1). Similarly, the inclusion of variants conferring resistance to INH and RIF, and subsequently to EMB, did not change the proportion of clustering nor cluster size (P=1 and P=1, respectively). By including PZA resistance-conferring mutations, the number of clusters increased from 13 to 17 and resulted in four additional medium-sized clusters, thereby re-classifying 30 patients from large clusters to medium-sized clusters. This led to a redistribution of patients over the cluster sizes (P=0.001) but not to a change in the proportion of clustered isolates (P=1). The addition of SLIDs drug resistance-conferring mutations had no impact on cluster distribution (size or proportion, P=1) and only resulted in reclassifying one isolate as unique.
Inclusion of variants conferring resistance to FQs in addition to those conferring resistance to INH, RIF, EMB, PZA and SLIDs reclassified 11 clustered isolates as unique and increased the number of clusters from 17 to 37. This had no significant impact on the proportion of isolates belonging to a cluster (84.6 vs. 82.2 %, P=0.38), or the proportion of unique isolates (15.4 vs. 17.8 %, P=0.38). We did observe a significant decrease in the number of patients in large clusters (P<0.001), with the largest cluster containing 65 isolates occurring in all districts and sub-districts in Cape Town (Table 2).
Phylogenetic and epidemiological findings
All isolates that formed part of large (n=3) and very large clusters (n=1) belonged to lineage 2. All belonged to lineage 2.2.2 (atypical Beijing strain), except for one large cluster (72 isolates), which belonged to lineage 2.2.1 (typical Beijing strain) (Fig. S2).
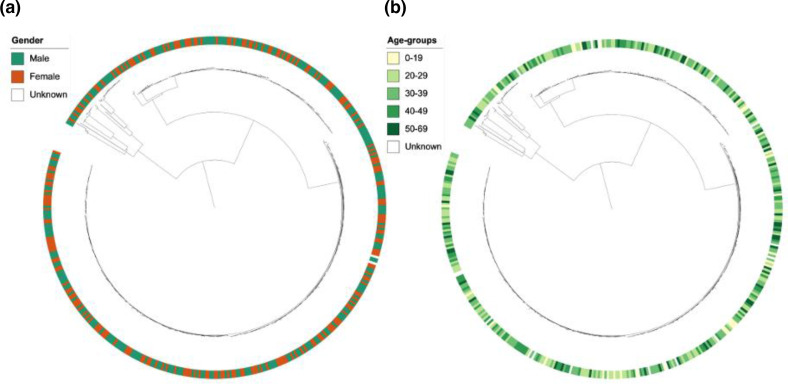
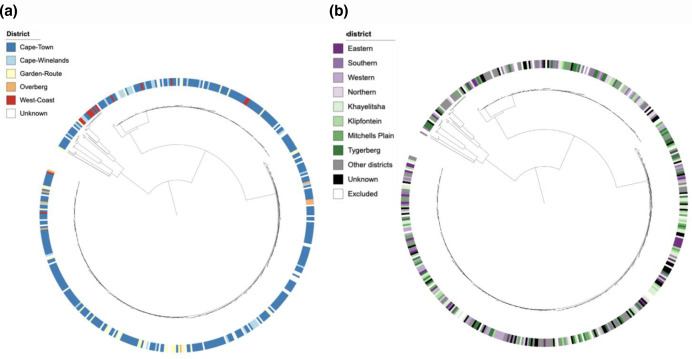
Visual inspection of annotation of the phylogenetic tree with patient gender did not suggest an association between XDR-TB clusters and gender (Fig. 3a). The proportion of unique and clustered isolates was similar between male and female patients (P=0.39) and there was no association between gender and distribution of cluster size among clustered isolates (P=0.86,Fig. S3a). Similarly, annotation of the phylogenetic tree with age did not suggest a correlation between XDR-TB clusters and age (Fig. 3b). The age distribution of patients with a unique WGS genotype did not differ from patients whose isolates belonged to a cluster (P=0.37) nor did we find an association between age and cluster size (P=0.61) (Fig. S3b). Annotation of the phylogenetic tree by geographical origin of the patients showed that XDR-TB strains were distributed over the entire Western Cape Province, with no obvious clustering based on district (Fig. 4a) or subdistricts of the Cape Town metropole (Fig. 4b). We identified a significant difference in the distribution of clustered and unique isolates between the urban Cape Town metropole and other districts, with 80.4 % of isolates in the Cape Town metropole being clustered versus 68.6 % in more rural districts (P=0.016). There was no significant association between geographical location and cluster size (P=0.39) (Fig. S3c) (Table 3).
Fig. 3.
(a) Distribution of gender across the phylogenetic tree of XDR-Mtb isolates collected from 461 patients in the Western Cape Province of South Africa, 2006–2017. (b) Distribution of age across the phylogenetic tree of XDR-Mtb isolates collected from 461 patients in the Western Cape Province of South Africa, 2006–2017.
Fig. 4.
(a) Distribution of geographicl origin (district level) across the phylogenetic tree of individual XDR-Mtb isolates collected from 461 patients in the Western Cape Province of South Africa, 2006–2017. (b) Distribution of geographical origin (local district level) across the phylogenetic tree of individual XDR-Mtb isolates collected from 461 patients in the Western Cape Province of South Africa, 2006–2017.
Table 3.
Association of gender, age and geographical location with cluster and cluster size among 461 XDR-TB patients in the Western Cape province of South Africa, 2006–2017
|
Unique isolates N (%) |
Clustered isolates N (%) |
P-value |
Small clusters N (%) |
Medium cluster N (%) |
Large clusters N (%) |
P-value |
||
|---|---|---|---|---|---|---|---|---|
|
Gender |
Male |
49 (59.8) |
203 (53.6) |
0.39* |
27 (7.1) |
45 (11.9) |
131 (34.6) |
0.86* |
|
Female |
33 (40.2) |
174 (46.4) |
20 (5.3) |
41 (10.8) |
113 (29.8) |
|||
|
Age, years |
0–19 |
3 (3.7) |
29 (7.6) |
0.37* |
4 (1.1) |
4 (1.1) |
21 (5.5) |
0.61** |
|
20–29 |
20 (24.4) |
108 (28.5) |
14 (3.7) |
22 (5.8) |
72 (19.0) |
|||
|
30–39 |
30 (36.6) |
117 (30.9) |
18 (4.7) |
28 (7.4) |
71 (18.7) |
|||
|
40–49 |
19 (23.2) |
63 (16.6) |
4 (1.1) |
13 (3.4) |
46 (12.1) |
|||
|
50–69 |
8 (9.8) |
46 (12.1) |
6 (1.6) |
13 (3.4) |
27 (7.1) |
|||
|
Location |
Urban |
49 (59.8) |
265 (69.9) |
0.016* |
32 (8.4) |
65 (17.2) |
168 (44.3) |
0.39* |
|
Rural |
24 (29.3) |
63 (16.6) |
5 (1.3) |
12 (3.2) |
46 (12.1) |
*P value for chi square test, **P value for Fisher’s exact test.
Discussion
Transmission dynamics
In this study, 461 patients diagnosed with XDR-TB in the Western Cape Province of South Africa between 2006 and 2017 were investigated. The vast majority of XDR-Mtb strains (85.5 %) were clustered and only 14.5 % patients had a unique XDR-Mtb strain, when identifying clusters based on a 5 SNP distance cut-off, merging clusters based on common ancestors, and excluding drug resistance-conferring mutations [30, 32]. Including variants that confer resistance had minimal impact on the proportion of isolates that were clustered (from 84.8 % when including INH and RIF resistance-conferring variants to 82.2 % when also including EMB, PZA, SLIDs and FQs). The number of clusters increased when consecutively including variants conferring resistance (from 12 to 13 for INH and RIF resistance, to 17 for INH, RIF, EMB and PZA resistance, and to 37 when adding INH, RIF, EMB, PZA, SLIDs and FQs resistance). These results demonstrate that clustering using the 5 SNP cut-off with subsequent inclusion of resistance-conferring variants is required to evaluate transmission of (pre-)XDR strains. Excluding SNPs in resistance-associated genes from the clustering definition of (pre-)XDR results in neglecting an important proportion of genetic variation in (pre-)XDR-TB strains and can lead to misinterpretation of XDR-TB transmission dynamics. The observation that the inclusion of the FQ resistance-conferring mutations had the greatest impact raises the hypothesis that, in this setting, transmission may mainly occur at the stage of MDR-TB or pre-XDR-TB, with subsequent acquisition of FQ resistance and probably less transmission thereafter. These findings support results from a mathematical model that showed that more than 80 % of MDR-TB is transmitted, and extends these findings to pre-XDR and XDR-TB [33]. However, it is difficult to compare the proportion of XDR-TB patients who are clustered in our study to the reports of other studies as details on the inclusion or exclusion of drug-resistant variants in phylogenetic analyses are not provided [19, 34]. Our findings do support that emergence and spread of MDR- and XDR-TB is a gradual process starting with acquisition of resistance-conferring mutations followed by selection of mutations with low fitness cost (e.g. rpoB Ser450Leu) [19] or co-occurrence with compensatory mutations (e.g. ahpC mutations as compensatory mutations associated with katG Ser315Thr) [35] and successful transmission and clonal expansion of these drug-resistant strains [36]. Our findings are in line with the acquisition of drug resistance-conferring mutations in KZN [19] where the introduction of INH in the 1950s [37] and ineffective single drug treatments allowed clonal expansion of INH-resistant strains [36]. The introduction of RIF without systematic screening for drug resistance contributed to the MDR-TB epidemic and the introduction of SLIDs and FQs [38] for treatment of MDR-TB in the absence of an efficient DST programme gave rise to the first XDR-TB outbreak in 2006. The more recent introduction of FQs may explain the limited clonality observed when including FQ resistance-conferring variants and the significant impact thereof on the distribution of cluster sizes.
Cluster sizes
The majority (64.9 %) of the clustered XDR-TB isolates belonged to large (21–100 isolates) clusters and were spread across the province. The largest cluster of 65 XDR-Mtb strains occurred in all districts of the Western Cape Province and all subdistricts of Cape Town. Within the Western Cape Province, 68.1 % of XDR-TB cases occurred in the densely populated Cape Town metropole district and we identified a significant association between clustered and unique isolates in the Cape Town metropole and rural districts, with 80.4 % of isolates originating from the Cape Town metropole being clustered versus 68.6 % of the isolates originating from more rural districts. This underlines the role that Cape Town city plays as an urban hotspot of Mtb transmission. The decade-long spread of XDR-TB highlights that XDR-TB strains are now endemic in the Western Cape Province, which is alarming as the province is home to millions of migrant labourers and the Western Cape is one of the world’s most popular tourist destinations [2]. These observations also confirm findings from the KwaZulu Natal province in South Africa where transmission of XDR-TB strains was shown not only to occur in congregate settings but also within and between communities, indicating the threat and rapid spread of XDR-TB strains [15].
Almost all (93.1 %) XDR-Mtb strains belonged to lineage 2, either typical Beijing (lineage 2.2.1) or atypical Beijing (lineage 2.2.2), a few (6.9 %) belonged to lineage 4 and none to lineages 1, 3, 5, 6, 7, 8 or 9. The low proportion of lineage 4 is somewhat surprising as lineage 4 is widespread in South Africa and has shown enhanced virulence in animal models, although with varied transmission success in molecular epidemiological studies [39]. The high proportion of lineage 2 isolates among XDR-TB strains was expected as Beijing strains have been associated with an increased likelihood of acquisition of drug resistance [40–42] and have caused MDR-TB outbreaks in countries such as Russia [43], South Africa [44] and Colombia [45]. The majority (71.1 %) of isolates in this study belonged to lineage 2.2.2, a subgroup of atypical Beijing strains (Asia Ancestral 1, AA1). With the exception of Japan, Vietnam and Taiwan, this genotype is usually present at low frequency [11]. In South Africa, the atypical AA1 Beijing strains originated from an AA1-progenitor strain that was introduced in the Eastern Cape Province and then spread across the country [11]. Some deleterious SNPs defining the AA1 strains include variants in genes that play a role in transport of drugs across the membrane, macrotetrolide resistance, pathogenesis and entry of hydrophilic molecules [11]. These variants may contribute to a phenotype that is better adapted to gain drug resistance-conferring mutations and survive the fitness cost thereof. Studies have also reported a higher fitness of lineage 2 strains [46–48] and the presence of polymorphisms in genes involved in DNA replication, recombination and repair. This suggests that Beijing strains are more mutable than other Mtb strains [49] and more frequently acquire compensatory mutations compared to other lineages [50]. We therefore hypothesize that the higher fitness of lineage 2 strains may explain our finding that XDR-TB strains are spread over the entire province, with the largest cluster occurring in all districts and subdistricts.
Association of gender, age and geographical location
In our study, 54.7 % of XDR-TB patients were male but we did not identify an association between gender and cluster membership or cluster size. The majority of the patients in our study (62.1 %) were between 20 and 39 years old, the age group with the highest TB burden in Cape Town [51]. Work-related travel among this economically active group and social movements could have contributed to the spread of XDR-TB across the province. In settings with a high TB burden such as sub-Saharan Africa, Mtb transmission has been shown to occur often in schools, public transportation, workplaces, healthcare facilities, mines, prisons and public places in addition to households [52]. Additionally, patients travelling farther than their nearest facilities to seek care because of the stigma that is associated with TB [53] may have contributed to the spread of XDR-TB beyond small communities as travel to more distant healthcare facilities is mostly done using public transport [52].
Limitations
This study of a large XDR-Mtb WGS dataset collected from a defined geographical region over a 10-year period created a unique opportunity to study the phylo-epidemiology of XDR-TB. To date, most WGS studies have focused on transmission of MDR [54, 55], or drug-susceptible Mtb [56] and the few studies that focused on XDR-TB transmission were smaller (one to 404 strains) [14, 20, 57–62]. Our study had some limitations. First, the isolates included in the analyses do not represent all XDR-TB cases in the Western Cape Province over the 10-year study period as some cases may not have been diagnosed, culturing of isolates for DST may have failed or cultures may have been contaminated. The exact case detection rate of XDR-TB in the area is, however, unknown. Second, we focused on the high-confidence mutations included in TBProfiler and did not assess for heteroresistance. This may have resulted in misclassification of some strains that were phenotypically XDR as genotypically pre-XDR and may have underestimated the proportion of clustered isolates. Third, because the study included isolates collected between 2006 and 2017, we focused on INH, RIF, EMB, PZA, SLIDs and FQs drug resistance-conferring variants and cannot extrapolate on the need to include variants conferring resistance to BDQ and LZD to accurately describe the transmission of difficult-to-treat TB under the novel XDR-TB definition [62]. Fourth, because of homoplasy, where different isolates independently acquire the same resistance-conferring mutations, it is possible that we have over-estimated transmission events. Fifth, the absence of HIV (human immunodeficiency virus) status data for most patients prohibited assessment of the role of HIV in XDR-TB transmission, and an absence of contact and social network data prohibited confirmation of transmission events between patients whose isolates belonged to the same cluster. Furthermore, the location where the XDR-TB diagnosis was made may not represent the site where transmission occurred due to mobility of people between infection and development of symptoms. Sixth, we used a 5 SNP cut-off to classify a strain belonging to a cluster even though this cut-off is inferred from a study performed in a country with low TB burden [31]. Raising the SNP cut-off had limited impact, suggesting that a SNP cut-of of 5 can also be applied in high-burden settings [30].
Finally, we assessed clustering by age, gender and health district without taking time into account. A three-dimensional analysis could allow a more refined investigation of the movement of clusters over time and space.
Conclusion
The XDR-TB epidemic in the Western Cape Province of South Africa probably has its origin in the endemic spread of MDR-Mtb and pre-XDR-Mtb strains followed by acquisition of FQ resistance, with more limited transmission of XDR-Mtb strains. In recent decades, XDR-TB has become endemic with large XDR-TB clusters spread across the entire province and especially high clustering observed in the urban Cape Town metropole. Our results highlight the importance of including drug resistance-conferring variants (especially FQ resistance-conferring variants) in transmission analyses of (pre-)XDR-TB as their exclusion eliminates a substantial proportion of genetic information, which may lead to misinterpretation of transmission dynamics. To further improve our insights into the transmission dynamics of XDR-TB, future studies should perform analyses of the movement of clusters over time and space to identify the determinants of transmission and locate common transmission hotspots. Efforts to combat the XDR-TB epidemic will require a multifaceted response aimed at timely diagnosing MDR, pre-XDR and XDR-TB and administering effective treatment to reduce the duration of infectiousness, prevent the acquisition of drug resistance, as well as addressing social determinants of health.
Supplementary Data
Funding information
This work was funded by the Research Foundation Flanders (FWO), under grant No. G0F8316N (FWO Odysseus) and No. 11M5422N. K.D. acknowledges funding from the SA MRC (RFA-EMU-02-2017), EDCTP (TMA-2015SF-1043, TMA-1051-TESAII, TMA-CDF2015), UK Medical Research Council (MR/S03563X/1) and the Wellcome Trust (MR/S027777/1). This work was supported by the National Research Foundation (NRF), the South African Medical Research Council (SAMRC), and the Stellenbosch University Faculty of Medicine Health Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the FWO, NRF or SAMRC. The funders had no role in study design, data collection or interpretation, or the decision to submit the work for publication
Acknowledgements
We would also like to thank the members of the international TORCH consortium for helpful discussions. We also thank Dr James Posey and Scott Burns at the Centers for Disease Control and Prevention, Atlanta, GA, USA, for next-generation sequencing analyses for the sequencing of a subset of the Mtb genomes.
Author contributions
S.O.: Conceptualization, Methodology, Formal Analysis, Investigation, Writing – Original Draft Preparation, Visualization. S.D.L.: Conceptualization, Data Curation, Writing – Review and Editing. T.H.H.: Methodology, Software, Writing – Review and Editing, Visualization. A.D.: Methodology, Data Curation, Supervision, Writing – Review and Editing. E.M.S.: Data Curation, Resources, Writing – Review and Editing. E.D.V.: Writing – Review and Editing. C.J.M.: Software, Writing – Review and Editing. K.D.: Resources, Funding, Writing – Review and Editing. R.W.: Conceptualization, Resources, Methodology, Writing – Review and Editing, Supervision, Funding. A.V.R.: Conceptualization, Methodology, Writing – Original Draft Preparation, Writing – Review and Editing, Visualization, Supervision, Project Administration, Funding.
Conflicts of interest
The authors declare that there are no conflicts of interest
Ethical statement
All isolates and sequences used in this project were collected, stored and analysed under the ethical clearance obtained from Stellenbosch University Health Research Ethics Committee (N09/11/269). The ethics approval has a waiver of consent to link routine clinical data to the laboratory data prior to de-identification of patient identifiers.
Footnotes
Abbreviations: EMB, ethambutol; ENA, European Nucleotide Archive; ETO, ethionamide; FQ, fluoroquinolone; HGVS, Human Genome Variation Society; HIV, human immunodeficiency virus; INH, isoniazid; LPA, line probe assay; LZD, linezolid; MDR, multidrug-resistant; Mtb, Mycobacterium tuberculosis; NHLS, National Health Laboratory Services; pDST, phenotypic drug susceptibility testing; PZA, pyrazinamide; RIF, rifampicin; RR, rifampicin-resistant; SLID, second-line injectable drug; SM, streptomycin; SNP, single nucleotide polymorphism; TB, tuberculosis; WGS, whole genome sequencing; XBS, Complex Bacterial Samples; XDR-TB, extensively drug-resistant tuberculosis.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Three supplementary figures and three supplementary tables are available with the online version of this article.
References
- 1.Cohen J. Infectious disease. Extensively drug-resistant TB gets foothold in South Africa. Science. 2006;313:1554. doi: 10.1126/science.313.5793.1554a. [DOI] [PubMed] [Google Scholar]
- 2.Singh JA, Upshur R, Padayatchi N. XDR-TB in South Africa: no time for denial or complacency. PLoS Med. 2007;4:e50. doi: 10.1371/journal.pmed.0040050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gandhi NR, Moll A, Sturm AW, Pawinski R, Govender T, et al. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet. 2006;368:1575–1580. doi: 10.1016/S0140-6736(06)69573-1. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization The global MDR-TB and XDR-TB response plan 2007-2008. 2007.
- 5.World Health Organization Global tuberculosis report 2019. 2019.
- 6.de Vos M, Müller B, Borrell S, Black PA, van Helden PD, et al. Putative compensatory mutations in the rpoC gene of rifampin-resistant Mycobacterium tuberculosis are associated with ongoing transmission. Antimicrob Agents Chemother. 2013;57:827–832. doi: 10.1128/AAC.01541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Böttger EC, Springer B, Pletschette M, Sander P. Fitness of antibiotic-resistant microorganisms and compensatory mutations. Nat Med. 1998;4:1343–1344. doi: 10.1038/3906. [DOI] [PubMed] [Google Scholar]
- 8.Billington OJ, McHugh TD, Gillespie SH. Physiological cost of rifampin resistance induced in vitro in Mycobacterium tuberculosis . Antimicrob Agents Chemother. 1999;43:1866–1869. doi: 10.1128/AAC.43.8.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sander P, Springer B, Prammananan T, Sturmfels A, Kappler M, et al. Fitness cost of chromosomal drug resistance-conferring mutations. Antimicrob Agents Chemother. 2002;46:1204–1211. doi: 10.1128/AAC.46.5.1204-1211.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shcherbakov D, Akbergenov R, Matt T, Sander P, Andersson DI, et al. Directed mutagenesis of Mycobacterium smegmatis 16S rRNA to reconstruct the in vivo evolution of aminoglycoside resistance in Mycobacterium tuberculosis . Mol Microbiol. 2010;77:830–840. doi: 10.1111/j.1365-2958.2010.07218.x. [DOI] [PubMed] [Google Scholar]
- 11.Klopper M, Heupink TH, Hill-Cawthorne G, Streicher EM, Dippenaar A, et al. A landscape of genomic alterations at the root of A near-untreatable tuberculosis epidemic. BMC Med. 2020;18:24. doi: 10.1186/s12916-019-1487-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valway SE, Greifinger RB, Papania M, Kilburn JO, Woodley C, et al. Multidrug-resistant tuberculosis in the New York State prison system, 1990-1991. J Infect Dis. 1994;170:151–156. doi: 10.1093/infdis/170.1.151. [DOI] [PubMed] [Google Scholar]
- 13.van Rie A, Warren RM, Beyers N, Gie RP, Classen CN, et al. Transmission of a multidrug-resistant Mycobacterium tuberculosis strain resembling “strain W” among noninstitutionalized, human immunodeficiency virus-seronegative patients. J Infect Dis. 1999;180:1608–1615. doi: 10.1086/315054. [DOI] [PubMed] [Google Scholar]
- 14.Arnold A, Witney AA, Vergnano S, Roche A, Cosgrove CA, et al. XDR-TB transmission in London: Case management and contact tracing investigation assisted by early whole genome sequencing. J Infect. 2016;73:210–218. doi: 10.1016/j.jinf.2016.04.037. [DOI] [PubMed] [Google Scholar]
- 15.Shah NS, Auld SC, Brust JCM, Mathema B, Ismail N, et al. Transmission of extensively drug-resistant tuberculosis in South Africa. N Engl J Med. 2017;376:243–253. doi: 10.1056/NEJMoa1604544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan EA, Kreiswirth BN, Palumbo L, Kapur V, Musser JM, et al. Emergence of fluoroquinolone-resistant tuberculosis in New York City. Lancet. 1995;345:1148–1150. doi: 10.1016/s0140-6736(95)90980-x. [DOI] [PubMed] [Google Scholar]
- 17.Mlambo CK, Warren RM, Poswa X, Victor TC, Duse AG, et al. Genotypic diversity of extensively drug-resistant tuberculosis (XDR-TB) in South Africa. Int J Tuberc Lung Dis. 2008;12:99–104. [PubMed] [Google Scholar]
- 18.Dixit A, Freschi L, Vargas R, Calderon R, Sacchettini J, et al. Whole genome sequencing identifies bacterial factors affecting transmission of multidrug-resistant tuberculosis in a high-prevalence setting. Sci Rep. 2019;9:5602. doi: 10.1038/s41598-019-41967-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen KA, Abeel T, Manson McGuire A, Desjardins CA, Munsamy V, et al. Evolution of extensively drug-resistant tuberculosis over four decades: whole genome sequencing and dating analysis of Mycobacterium tuberculosis isolates from KwaZulu-Natal. PLoS Med. 2015;12:e1001880. doi: 10.1371/journal.pmed.1001880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson KN, Jenness SM, Mathema B, Lopman BA, Auld SC, et al. Social mixing and clinical features linked with transmission in a network of extensively drug-resistant tuberculosis cases in KwaZulu-Natal, South Africa. Clin Infect Dis. 2020;70:2396–2402. doi: 10.1093/cid/ciz636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meehan CJ, Goig GA, Kohl TA, Verboven L, Dippenaar A, et al. Whole genome sequencing of Mycobacterium tuberculosis: current standards and open issues. Nat Rev Microbiol. 2019;17:533–545. doi: 10.1038/s41579-019-0214-5. [DOI] [PubMed] [Google Scholar]
- 22.Jamieson FB, Teatero S, Guthrie JL, Neemuchwala A, Fittipaldi N, et al. Whole-genome sequencing of the Mycobacterium tuberculosis Manila sublineage results in less clustering and better resolution than mycobacterial interspersed repetitive-unit-variable-number tandem-repeat (MIRU-VNTR) typing and spoligotyping. J Clin Microbiol. 2014;52:3795–3798. doi: 10.1128/JCM.01726-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt B-M, Geldenhuys H, Tameris M, Luabeya A, Mulenga H, et al. Impact of Xpert MTB/RIF rollout on management of tuberculosis in a South African community. S Afr Med J. 2017;107:1078–1081. doi: 10.7196/SAMJ.2017.v107i12.12502. [DOI] [PubMed] [Google Scholar]
- 25.Heupink TH, Verboven L, Warren RM, Van Rie A. Comprehensive and accurate genetic variant identification from contaminated and low-coverage Mycobacterium tuberculosis whole genome sequencing data. Microb Genom. 2021;7 doi: 10.1099/mgen.0.000689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coll F, McNerney R, Preston MD, Guerra-Assunção JA, Warry A, et al. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med. 2015;7:51. doi: 10.1186/s13073-015-0164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phelan JE, O’Sullivan DM, Machado D, Ramos J, Oppong YEA, et al. Integrating informatics tools and portable sequencing technology for rapid detection of resistance to anti-tuberculous drugs. Genome Med. 2019;11:41. doi: 10.1186/s13073-019-0650-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meehan CJ, Moris P, Kohl TA, Pečerska J, Akter S, et al. The relationship between transmission time and clustering methods in Mycobacterium tuberculosis epidemiology. EBioMedicine. 2018;37:410–416. doi: 10.1016/j.ebiom.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker TM, Ip CLC, Harrell RH, Evans JT, Kapatai G, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaziri F, Kohl TA, Ghajavand H, Kargarpour Kamakoli M, Merker M, et al. Genetic diversity of multi- and extensively drug-resistant Mycobacterium tuberculosis isolates in the Capital of Iran, revealed by whole-genome sequencing. J Clin Microbiol. 2019;57:e01477-18. doi: 10.1128/JCM.01477-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McBryde ES, Meehan MT, Doan TN, Ragonnet R, Marais BJ, et al. The risk of global epidemic replacement with drug-resistant Mycobacterium tuberculosis strains. Int J Infect Dis. 2017;56:14–20. doi: 10.1016/j.ijid.2017.01.031. [DOI] [PubMed] [Google Scholar]
- 34.Casali N, Nikolayevskyy V, Balabanova Y, Harris SR, Ignatyeva O, et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet. 2014;46:279–286. doi: 10.1038/ng.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naidoo CC, Pillay M. Fitness-compensatory mutations facilitate the spread of drug-resistant F15/LAM4/KZN and F28 Mycobacterium tuberculosis strains in KwaZulu-Natal, South Africa. J Genet. 2017;96:599–612. doi: 10.1007/s12041-017-0805-8. [DOI] [PubMed] [Google Scholar]
- 36.Welekidan LN, Skjerve E, Dejene TA, Gebremichael MW, Brynildsrud O, et al. Frequency and patterns of first- and second-line drug resistance-conferring mutations in Mycobacterium tuberculosis isolated from pulmonary tuberculosis patients in a cross-sectional study in Tigray Region, Ethiopia. J Glob Antimicrob Resist. 2021;24:6–13. doi: 10.1016/j.jgar.2020.11.017. [DOI] [PubMed] [Google Scholar]
- 37.Fernandes GFDS, Salgado HRN, Santos JLD. Isoniazid: a review of characteristics, properties and analytical methods. Crit Rev Anal Chem. 2017;47:298–308. doi: 10.1080/10408347.2017.1281098. [DOI] [PubMed] [Google Scholar]
- 38.World Health Organization Guidelines for the programmatic management of drug-resistant tuberculosis, emergency update 2008. 2008. [PubMed]
- 39.Stucki D, Brites D, Jeljeli L, Coscolla M, Liu Q, et al. Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat Genet. 2016;48:1535–1543. doi: 10.1038/ng.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buu TN, van Soolingen D, Huyen MNT, Lan NTN, Quy HT, et al. Increased transmission of Mycobacterium tuberculosis Beijing genotype strains associated with resistance to streptomycin: a population-based study. PLoS One. 2012;7:e42323. doi: 10.1371/journal.pone.0042323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Albanna AS, Reed MB, Kotar KV, Fallow A, McIntosh FA, et al. Reduced transmissibility of East African Indian strains of Mycobacterium tuberculosis . PLoS One. 2011;6:e25075. doi: 10.1371/journal.pone.0025075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Couvin D, Rastogi NT. Tuberculosis - A global emergency: Tools and methods to monitor, understand, and control the epidemic with specific example of the Beijing lineage. Tuberculosis (Edinb) 2015;95 Suppl 1:S177–89. doi: 10.1016/j.tube.2015.02.023. [DOI] [PubMed] [Google Scholar]
- 43.Lasunskaia E, Ribeiro SCM, Manicheva O, Gomes LL, Suffys PN, et al. Emerging multidrug resistant Mycobacterium tuberculosis strains of the Beijing genotype circulating in Russia express a pattern of biological properties associated with enhanced virulence. Microbes Infect. 2010;12:467–475. doi: 10.1016/j.micinf.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 44.Marais BJ, Mlambo CK, Rastogi N, Zozio T, Duse AG, et al. Epidemic spread of multidrug-resistant tuberculosis in Johannesburg, South Africa. J Clin Microbiol. 2013;51:1818–1825. doi: 10.1128/JCM.00200-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nieto Ramirez LM, Ferro BE, Diaz G, Anthony RM, de Beer J, et al. Genetic profiling of Mycobacterium tuberculosis “Modern” Beijing strains from Southwest Colombia. Microbiology. 2019 doi: 10.1101/819425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.European Concerted Action on New Generation Genetic Markers and Techniques for the Epidemiology and Control of Tuberculosis Beijing/W genotype Mycobacterium tuberculosis and drug resistance. Emerg Infect Dis. 2006;12:736–743. doi: 10.3201/eid1205.050400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Spuy GD, Kremer K, Ndabambi SL, Beyers N, Dunbar R, et al. Changing Mycobacterium tuberculosis population highlights clade-specific pathogenic characteristics. Tuberculosis (Edinb) 2009;89:120–125. doi: 10.1016/j.tube.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 48.Tuite AR, Guthrie JL, Alexander DC, Whelan MS, Lee B, et al. Epidemiological evaluation of spatiotemporal and genotypic clustering of Mycobacterium tuberculosis in Ontario, Canada. Int J Tuberc Lung Dis. 2013;17:1322–1327. doi: 10.5588/ijtld.13.0145. [DOI] [PubMed] [Google Scholar]
- 49.Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet. 2013;45:784–790. doi: 10.1038/ng.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuo TY, Liu QY, Gan MY, Gao Q. Comprehensive identification of compensatory mutations in rifampicin-resistant Mycobacterium tuberculosis strains. Zhonghua Jie He He Hu Xi Za Zhi. 2018;41:207–212. doi: 10.3760/cma.j.issn.1001-0939.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 51.Blaser N, Zahnd C, Hermans S, Salazar-Vizcaya L, Estill J, et al. Tuberculosis in Cape Town: An age-structured transmission model. Epidemics. 2016;14:54–61. doi: 10.1016/j.epidem.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Churchyard G, Kim P, Shah NS, Rustomjee R, Gandhi N, et al. What we know about tuberculosis transmission: an overview. J Infect Dis. 2017;216:S629–S635. doi: 10.1093/infdis/jix362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kapwata T, Morris N, Campbell A, Mthiyane T, Mpangase P, et al. Spatial distribution of extensively drug-resistant tuberculosis (XDR TB) patients in KwaZulu-Natal, South Africa. PLoS One. 2017;12:e0181797. doi: 10.1371/journal.pone.0181797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang X-Y, Li Y-P, Mei Y-W, Yu Y, Xiao J, et al. Time and spatial distribution of multidrug-resistant tuberculosis among Chinese people, 1981-2006: a systematic review. Int J Infect Dis. 2010;14:e828–37. doi: 10.1016/j.ijid.2010.02.2244. [DOI] [PubMed] [Google Scholar]
- 55.Lalor MK, Casali N, Walker TM, Anderson LF, Davidson JA, et al. The use of whole-genome sequencing in cluster investigation of a multidrug-resistant tuberculosis outbreak. Eur Respir J. 2018;51:1702313. doi: 10.1183/13993003.02313-2017. [DOI] [PubMed] [Google Scholar]
- 56.Chirenda J, Gwitira I, Warren RM, Sampson SL, Murwira A, et al. Spatial distribution of Mycobacterium Tuberculosis in metropolitan Harare, Zimbabwe. PLoS One. 2020;15:e0231637. doi: 10.1371/journal.pone.0231637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peterson ML, Gandhi NR, Clennon J, Nelson KN, Morris N, et al. Extensively drug-resistant tuberculosis “hotspots” and sociodemographic associations in Durban, South Africa. Int J Tuberc Lung Dis. 2019;23:720–727. doi: 10.5588/ijtld.18.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gallo JF, Pinhata JMW, Simonsen V, Galesi VMN, Ferrazoli L, et al. Prevalence, associated factors, outcomes and transmission of extensively drug-resistant tuberculosis among multidrug-resistant tuberculosis patients in São Paulo, Brazil: a cross-sectional study. Clin Microbiol Infect. 2018;24:889–895. doi: 10.1016/j.cmi.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 59.Perdigão J, Gomes P, Miranda A, Maltez F, Machado D, et al. Using genomics to understand the origin and dispersion of multidrug and extensively drug resistant tuberculosis in Portugal. Sci Rep. 2020;10:2600. doi: 10.1038/s41598-020-59558-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roycroft E, O’Toole RF, Fitzgibbon MM, Montgomery L, O’Meara M, et al. Molecular epidemiology of multi- and extensively-drug-resistant Mycobacterium tuberculosis in Ireland, 2001-2014. J Infect. 2018;76:55–67. doi: 10.1016/j.jinf.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 61.Bainomugisa A, Lavu E, Hiashiri S, Majumdar S, Honjepari A, et al. Multi-clonal evolution of multi-drug-resistant/extensively drug-resistant Mycobacterium tuberculosis in a high-prevalence setting of Papua New Guinea for over three decades. Microb Genom. 2018;4 doi: 10.1099/mgen.0.000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hadifar S, Shamkhali L, Kargarpour Kamakoli M, Mostafaei S, Khanipour S, et al. Genetic diversity of Mycobacterium tuberculosis isolates causing pulmonary and extrapulmonary tuberculosis in the capital of Iran. Mol Phylogenet Evol. 2019;132:46–52. doi: 10.1016/j.ympev.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 63.World Health Organization Meeting report of the WHO expoert consultation on the definition of extensively drug-resistant tuberculosis. 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.