Abstract
Helicobacter pylori possesses a homolog of the luxS gene, initially identified by its role in autoinducer production for the quorum-sensing system 2 in Vibrio harveyi. The genomes of several other species of bacteria, notably Escherichia coli, Salmonella enterica serovar Typhimurium, and Vibrio cholerae, also include luxS homologs. All of these bacteria have been shown to produce active autoinducers capable of stimulating the expression of the luciferase operon in V. harveyi. In this report, we demonstrate that H. pylori also synthesizes a functional autoinducer (AI-2) that can specifically activate signaling system 2 in V. harveyi. Maximal activity is produced during early log phase, and the activity is diminished when cells enter stationary phase. We show that AI-2 is not involved in modulating any of the known or putative virulence factors in H. pylori and that a luxS null mutant has a two-dimensional protein profile identical to that of its isogenic parent strain. We discuss the implications of having an AI-2-like quorum-sensing system in H. pylori and suggest possible roles that it may play in H. pylori infection.
Since its discovery in 1982, Helicobacter pylori has attracted a great deal of attention, both because it occupies the unusual and inhospitable niche of the gastric lumen and because of the diversity of clinical outcomes associated with this infection. Research efforts have focused on identifying genes and the functions of their protein products that allow this organism to colonize and cause disease in humans. In addition, much effort has been dedicated to investigating how expression of these genes is regulated in the host.
Only a handful of the genes characterized in H. pylori have been definitively shown to be transcriptionally regulated. Studies of the H. pylori flagellar loci indicate that these genes are transcriptionally controlled by a regulatory hierarchy including sigma 54 and sigma 28 homologs and FlgR, an NtrC homolog (33). The pattern of flagellar transcription shows similarities to those of both the family Enterobacteriaceae and Caulobacter (30, 33). There have also been reports suggesting that environmental conditions, such as pH, temperature, metal concentrations, and iron availability (7, 8, 26, 37), can influence H. pylori transcription. One surprising feature of the genetic makeup of H. pylori revealed by the full genome sequence is the absence of most of the regulatory genes present in other gastrointestinal bacteria (38). Since the genome of H. pylori differs significantly from those of other organisms occupying the human gastrointestinal tract, we were interested in identifying and studying mechanisms utilized by H. pylori that contribute to its unique ability to survive and persist in the gastric lumen.
Despite the relative lack of regulatory-gene homologs, the H. pylori genome does contain putative homologs of some sensor kinase and response regulator proteins belonging to the family of two-component systems. These proteins are widespread in other organisms and are central to the transcriptional regulation of a variety of processes that respond to changing environmental conditions (22).
The H. pylori genome contains a homolog of luxS, a gene involved in autoinducer (AI) production in one of the two quorum-sensing systems of Vibrio harveyi (36). Quorum sensing involves the production and detection of membrane-permeating signaling molecules, or AIs, that function to communicate information to a population of cells about their external environment and cell density (19). The interaction of an AI with its cognate sensor protein results in activation of a signal transduction cascade that ultimately leads to changes in gene expression (19). In V. harveyi, quorum sensing controls the expression of bioluminescence.
Quorum sensing was first characterized in the regulation of bioluminescence in the marine bacterium Vibrio fischeri (reviewed by Dunlap and Greenberg [13]). In V. fischeri, quorum sensing depends on the action of two regulatory proteins, LuxI and LuxR. The product of the luxI gene directs the synthesis of the AI, an acylated homoserine lactone derivative. As cell density increases, the amount of AI also increases. At a critical concentration, the accumulated AI interacts with LuxR, the AI-binding protein. When bound to AI, LuxR acts as a transcriptional activator of the luciferase structural operon, and the bacteria emit light.
Since its discovery in V. fischeri, quorum sensing has been recognized as a widespread mechanism utilized by many gram-negative bacteria for the control of gene expression. Many elegant studies of cell-cell signaling in bacteria have recently been published (2, 9, 10, 12, 15, 16, 21, 23, 34, 36). The results of these studies underscore the importance of microbial cell-cell signaling in a variety of processes, including the development of disease in Pseudomonas aeruginosa- and Staphylococcus aureus-associated infections (1, 18, 29, 39, 40), the formation of microbial biofilms (27), and the production of light in luminous bacteria (3, 14, 24).
The expression of bioluminescence in V. harveyi is similarly under the control of a quorum-sensing mechanism. However, in this case, the regulated expression of luciferase is not mediated by homologs of the V. fischeri LuxI and LuxR proteins. Rather, V. harveyi accomplishes quorum sensing through the integration of two parallel two-component regulatory systems (system 1 and system 2). Each system is composed of a sensor protein that contains both kinase and response regulatory domains (LuxN and LuxQ) (5, 6) and its cognate AI (AI-1 and AI-2). Interaction of either AI-1 or AI-2 with its cognate sensor activates a phosphorelay cascade that ultimately converges on the response regulator protein, LuxO (5, 6). Expression of bioluminescence is determined by the phosphorylation state of the LuxO protein. At low cell density, neither AI-1 nor AI-2 is present at high concentration and LuxO is phosphorylated and acts to repress luminescence (5, 6, 16, 17). The accumulation of AI-1 or AI-2 at high cell density leads to dephosphorylation of LuxO and derepression of the lux operon. A positive regulator, LuxR (the V. harveyi LuxR is not a homolog of V. fischeri LuxR), is also necessary to activate transcription of the lux operon.
The recently described V. harveyi luxS gene is required for the production of the AI (AI-2) in signaling system 2 (36), and luxS homologs exist in over 25 other bacterial species, including Vibrio cholerae, Salmonella enterica serovar Typhimurium, Escherichia coli, and H. pylori. Surette et al. (36) assayed several different genera possessing the luxS homolog and found that AI-2 activity was produced by many of these organisms. In this study, we present data indicating that H. pylori possesses AI-2 activity and that this activity can specifically activate lux in V. harveyi through signaling system 2.
MATERIALS AND METHODS
Bacterial strains and recombinant DNA techniques.
The bacterial strains used were the clinical isolate H. pylori Alston, a generous gift from David Cave (St. Elizabeth's Hospital, Boston, Mass.), V. harveyi BB170 (14), and E. coli DH5α (Bio-Rad Laboratories). EJ103 is a luxS deletion derivative of H. pylori Alston constructed by the following procedure. One kilobase of sequence directly flanking both the upstream and downstream regions of luxS in H. pylori was PCR amplified with Taq polymerase (Gibco BRL, Grand Island, N.Y.) using the following primers: upstream region, A (5′ TCTAGAGGTGTTTTCATGTTTTTAACTCC) and (5′ GGTACCTCATCGCTGATTTAGAAGG); downstream region, C (5′ GCGGCCGCCCTTTGCCAGATAAG) and D (5′ TCTAGAGTGGTCTGAAGTGGGG). A directed ligation containing the resulting two PCR fragments, a kanamycin (Tn903) cassette, and pBluescript II SK vector resulted in the plasmid pEJ32, in which the kanamycin cassette is flanked by the two PCR fragments such that it replaces the complete luxS sequence. pEJ32 was electroporated into H. pylori Alston, resulting in the luxS mutant strain EJ103. PCR analysis with primers B and C gave rise to a 1,300-bp product, confirming that luxS had been replaced by the kanamycin cassette. To express LuxS, the H. pylori luxS gene was amplified with the primers XP1 (5′ ATGAAAACACCAAAAATGAATGTAG) and XP2 (5′ AACCCCCACTTCAGACCAC), and the 465-bp DNA fragment was cloned into the pBAD TopoIV expression vector (Invitrogen, Carlsbad, Calif.), resulting in pEJ30, which was expressed in the luxS mutant E. coli strain DH5α (34).
Growth of bacterial cell cultures and HEp-2 tissue cultures.
H. pylori strains were grown on campylobacter agar (Difco, Detroit, Mich.) supplemented with 10 μg of vancomycin (Sigma, St. Louis, Mo.)/ml, 2.5 U of polymyxin B (Sigma)/ml, 5 μg of trimethoprimsulfamethoxazole (Elkins Sinn, Cherry Hill, N.J.)/ml, and 20 μg of kanamycin/ml (where appropriate) plus 5% defibrinated sheep blood (Binax-NEL, Waterville, Maine). H. pylori liquid cultures were grown in brucella broth (BBL; Becton Dickinson Labware, Lincoln, N.J.; and Difco) supplemented with 5% horse serum (Gibco BRL). All H. pylori cultures were incubated in GasPak jars (BBL) with Campypaks (BBL) to generate a microaerobic environment. V. harveyi was cultured in heart infusion medium (25 g of heart infusion broth [Difco], 20 g of NaCl/liter), and E. coli DH5α was grown in Luria broth (LB). The AI bioassay (AB) medium has been previously described (37). The laryngeal carcinoma cell line HEp-2 was maintained in RPMI 1640 medium containing 5% fetal calf serum and glutamine (Gibco BRL). The cells were grown at 37°C in an atmosphere of 5% CO2.
Microscopic examination, growth, and urease activity of H. pylori strains.
Bacterial growth was monitored by optical density of broth cultures and viable plate counts. Motility was determined by microscopic examination. The urease status of the strains was determined by osmotically shocking 24-h cultures of H. pylori strains in ice water and adding 1/10 of the supernatant to a solution of 300 mM urea, 85 mM NaCl, 10 mM KH2PO4, and 0.00045% phenol red (pH 6.4). Hydrolysis of urea results in a qualitative color change of the buffer from orange to pink.
Preparation of cell-free culture fluids for AI-2 assays.
Confluent growth from 24-h H. pylori plate cultures was collected by flooding each plate with 5 ml of brucella broth supplemented with 5% horse serum (BBH) followed by swabbing the agar surface with a sterile cotton swab. Cell suspensions were used for broth inoculations. BBH was inoculated with the appropriate culture to a starting optical density at 600 nm of 0.1, and the culture was incubated at 37°C under microaerobic conditions with vigorous shaking (220 rpm). Cell-free culture fluids were prepared by centrifugation in an Eppendorf microcentrifuge (1 min at 15,000 rpm) and filtration of the supernatant through Nalgene 0.2-μm-pore-size filters (Nalge Co., Rochester, N.Y.). The supernatants were stored on ice before being assayed for vacuolating cytotoxin and AI-2 activity (34).
AI-2 assay.
Cell-free culture fluids were prepared from the H. pylori parental and luxS mutant strains, from E. coli DH5α, and from E. coli DH5α expressing the H. pylori luxS gene as described above. Ten microliters of each preparation was added to wells of 96-well microtiter dishes and assayed for AI-2 activity with the V. harveyi BB170 AI-2 bioassay as described previously (34, 35). For each preparation, 10 μl of the corresponding sterile medium was added to the wells as a negative control, and 10 μl of serovar Typhimurium 14028 cell-free culture fluid prepared from a culture grown to mid-exponential phase in LB containing 0.5% glucose at 37°C with aeration was used as a positive control for AI-2 activity. These conditions are known to promote maximal AI-2 production in serovar Typhimurium 14028 (34). V. harveyi BB170 was grown overnight with aeration at 30°C in AB medium and then diluted 1:5,000 in fresh AB medium, and 90 μl of the diluted culture was added to the wells containing the cell-free culture fluids or medium controls. The microtiter dishes were shaken at 200 rpm in a rotary shaker at 30°C. Light production was quantified every 30 min with a Wallac Model 1450 Microbeta Plus liquid scintillation counter. The data are reported as the fold stimulation of light emission by V. harveyi BB170 over that obtained for the corresponding growth medium alone.
Vacuolating cytotoxin assay.
Cell-free supernatants were assayed for the ability to produce vacuoles in a HEp-2 epithelial monolayer using a method adapted from Cover et al. (11). Cell supernatant volumes from H. pylori grown under conditions that result in AI-2 activity in BBH were concentrated 30- to 40-fold using Centriprep30 and Centricon30 concentrators (Amicon, Beverly, Mass.). HEp-2 cells were seeded in 24-well tissue culture plates to a density of 103 cells/ml and incubated overnight in a 5% CO2 atmosphere at 37°C. After incubation, the media were aspirated, and the cells were washed with phosphate-buffered saline (PBS). Fresh medium was added, and concentrated H. pylori supernatants were applied to the cells in 5-, 10-, and 100-μl aliquots and allowed to incubate in a 5% CO2 atmosphere at 37°C overnight. The presence of vacuoles was determined by microscopic examination.
Assay for IL-8 induction by HEp-2 cells.
The assay for interleukin 8 (IL-8) was carried out as described by Sharma et al. (31). Briefly, HEp-2 cells were seeded at a density of 105/well. After a 24-h incubation period, the cells were washed twice with PBS and infected with 108 CFU of either the parent H. pylori strain or EJ103 harvested from a fresh overnight plate/ml in RPMI. The bacteria were pelleted onto the epithelial monolayer by centrifugation in an Eppendorf microcentrifuge for 5 min at 1,000 rpm and incubated for 8 h in a 5% CO2 atmosphere at 37°C. Following incubation, the supernatants were removed, centrifuged at 15,000 rpm for 5 min, and filtered as described above. The resulting cell-free culture fluid was assayed for IL-8 by enzyme-linked immunosorbent assay (catalog no. EH2-IL8-10; Endogen, Woburn, Mass.) following the manufacturer's instructions.
Protein preparation and 2-D gel analysis.
Cell pellets from both the parent H. pylori strain and EJ103 grown under conditions resulting in AI-2 activity were resuspended in an osmotic lysis buffer containing 10 mM Tris (pH 7.4) and 0.3% sodium dodecyl sulfate, plus 1× (final concentration) of the following 100× protease inhibitor stock: pepstatin (0.34 mg/ml), leupeptin (1 mg/ml), E64 (0.36 mg/ml), benzamidine (5.6 mg/ml), and phenylmethylsulfonyl fluoride (0.2 mg/ml). RNase (500 mg/ml) (Worthington Biochemical, Lakewood, N.J.) and 350 U of DNase (Worthington Biochemical) were added, and the suspension was incubated on ice for 2 min. Aliquots were removed for protein concentration determination with the Bio-Rad (Hercules, Calif.) protein assay (catalog no. 500-0006), and the remaining sample was solubilized in an equal volume of 5% sodium dodecyl sulfate–5% β-mercaptoethanol boiling buffer. Samples were boiled for 5 min, frozen in dry ice, and stored at −70°C. Two-dimensional (2-D) gel electrophoresis services were performed by Kendrick Laboratories (Madison, Wis.).
RESULTS
Identification of AI-2 activity in H. pylori.
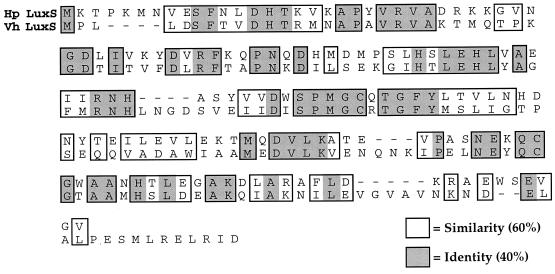
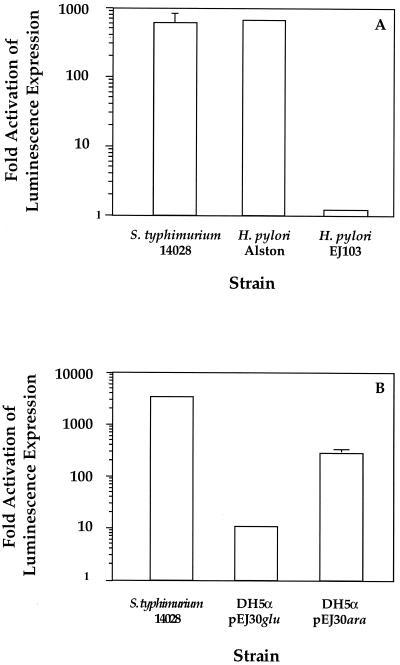
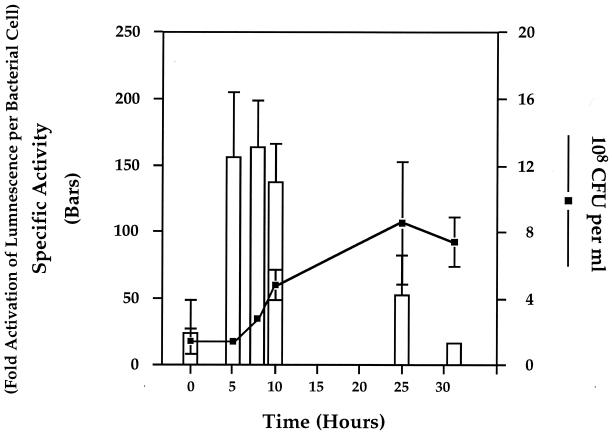
Analysis of the H. pylori genome database revealed a hypothetical protein with 40% identity and 60% homology to LuxS of V. harveyi (Fig. 1). To determine if H. pylori possessed an AI-2-like activity, we tested H. pylori Alston cell-free supernatants derived from late-log-phase growth for the ability to induce luminescence in the V. harveyi reporter strain BB170 (sensor 1− sensor 2+) (4). The results shown in Fig. 2A demonstrate that the addition of 10% H. pylori cell-free supernatant stimulates luminescence in the V. harveyi reporter strain BB170 equally to addition of the positive control AI-2 from serovar Typhimurium. Specifically, cell-free supernatants from both H. pylori and serovar Typhimurium induced signaling system 2 in the V. harveyi reporter strain approximately 500-fold. To determine when H. pylori produced this activity, cell-free supernatants obtained from early-, middle-, and late-log-phase cultures were assayed with BB170. These results, shown in Fig. 3, suggest that AI-2 activity per cell reaches a maximum during early log phase and is greatly diminished in late-log-phase cultures. Activation of luminescence was not observed when H. pylori supernatants were tested in the V. harveyi reporter strain BB886 (sensor 1+ sensor 2−), which does not have the AI-2 sensor LuxQ (data not shown). Taken together, these data indicate that H. pylori produces an AI-2-like activity at early log phase that activates luminescence specifically through LuxQ and system 2 in V. harveyi.
FIG. 1.
Alignment of the V. harveyi and putative H. pylori LuxS proteins.
FIG. 2.
H. pylori produces a luxS-dependent AI-2 activity. Cell-free supernatants were tested for the ability to induce luminescence expression in V. harveyi BB170. Ten percent cell-free supernatants or sterile media were mixed with the reporter strain in microtiter plates and incubated at 30°C on a rotary shaker. Aliquots were taken, and both cell density and light production were determined. Activity is reported as fold activation of luminescence of BB170 over the level of luminescence when sterile media were added. Assays were repeated at least two times. (A) Wild-type H. pylori and Typhimurium 14028 supernatants contain signaling substances that induce expression of luminescence in V. harveyi BB170, while the luxS deletion strain EJ103 does not. (B) H. pylori luxS expression in E. coli DH5α restores AI-2 activity. The error bars indicate standard deviations.
FIG. 3.
Specific activity of H. pylori AI-2 is maximal in early log phase and decreases in late log phase. H. pylori Alston was grown for 31 h under microaerobic conditions in BBH. At 0, 5, 8, 10, 25, and 31 h, cell-free culture fluids were prepared. Cell density was measured by diluting and plating cell aliquots at each time point (line). AI-2 activity was assayed for the ability to stimulate expression of bioluminescence in the V. harveyi reporter strain BB170 (bars). Specific activity was determined by dividing the fold activation of luminescence values by the total number of bacterial cells present at each time point.
luxS is responsible for AI-2 activity in H. pylori.
To determine whether H. pylori luxS is involved in AI-2 production, we constructed a luxS null mutant (EJ103). Taking advantage of H. pylori's natural tendency for homologous recombination, plasmid pEJ32 was electroporated into H. pylori Alston, followed by selection for the double-crossover event, resulting in the replacement of luxS with the kanamycin marker. The chromosomal luxS deletion was confirmed by PCR analysis. Cell-free supernatants prepared from EJ103 were tested for the ability to activate expression of luminescence in V. harveyi BB170. Unlike the parent H. pylori strain, results from the supernatants prepared from the luxS deletion strain, EJ103, did not activate luminescence of BB170 above background levels, as shown in Fig. 2A.
H. pylori luxS can complement a LuxS null E. coli mutant in trans. H. pylori luxS was expressed under the control of the inducible ara promoter in pEJ30. After transforming this expression vector into the LuxS− E. coli DH5α strain, the cells were grown to mid-log phase and induction from the arabinose promoter was accomplished by pelleting the cells and resuspending them in LB plus 2% arabinose. After a 2-h incubation period, cell-free supernatants were tested for the ability to activate expression of luminescence in BB170. The supernatants prepared from the DH5α/pEJ30 culture grown under inducing conditions showed a 400-fold activation of light production in V. harveyi BB170 (Fig. 2B). In contrast, supernatants prepared from DH5α/pEJ30 culture grown under repressing conditions showed only minimal activation (less than 10-fold) of the reporter strain (Fig. 2B). These results suggest that the inability of EJ103 supernatants to activate luminescence in BB170 is due to the absence of luxS. Neither growth medium nor supernatants prepared from E. coli DH5α alone caused stimulation of bioluminescence expression in V. harveyi BB170.
AI-2 activity in H. pylori does not affect expression of known virulence factors.
Only a few genes in H. pylori have been demonstrated to play a role in virulence. Mutation of the luxS homolog in E. coli O157 results in loss of virulence, suggesting that cell-cell communication plays a role in pathogenesis (32, 36). By analogy, we wanted to test if the expression of any of the known or putative virulence factors described for H. pylori was under the regulatory control of AI-2. The parental strain, Alston, and the luxS deletion strain, EJ103, were grown under conditions that result in AI-2 activity and compared for any differences in the growth kinetics in BBH medium, motility, urease activity, Cag-mediated induction of IL-8 production in HEp-2 cells, and H. pylori-induced vacuolization of HEp-2 cells. The growth kinetics of the two strains in BBH (liquid) medium were similar, and microscopic examination of these cultures showed that both strains were motile. There were no discernible differences between Alston and EJ103 in urease activity or in the ability to induce IL-8 production. In addition, when equivalent amounts of concentrated supernatants isolated from Alston and EJ103 were used in a vacuolating cytotoxin assay (11, 28), the luxS mutant was as effective as the parent strain in causing accumulation of vacuoles in HEp-2 cells (data not shown). Taken together, these data indicate that luxS is not involved in the in vitro expression of these factors previously shown to play a role in virulence in H. pylori.
2-D gel analysis of proteins from the wild-type H. pylori strain and the ΔluxS mutant EJ103.
We next examined the 2-D protein profiles of both strains grown under conditions that result in AI-2 activity. This experiment was designed to identify candidate proteins that could be under AI-2 regulatory control in H. pylori. The resulting profiles showed a complex pattern of approximately 400 to 500 expressed proteins with high resolution. Under the conditions used, no apparent differences between the parent strain and EJ103 were detectable (data not shown).
DISCUSSION
The identification of luxS and examination of its critical role in signaling system 2 quorum sensing in V. harveyi led to the discovery of luxS homologs in a variety of other bacteria, including many in which quorum-sensing systems had not been previously described. One of these organisms, H. pylori, has a LuxS homolog that shares 40% identity and 60% similarity with the V. harveyi LuxS protein (Fig. 1). Homology exists over the entire length of the protein. Despite its clear role in quorum sensing in V. harveyi, an exact function in AI-2 production has not been assigned. Furthermore, there are no motifs in the protein that suggest a particular enzymatic or regulatory activity. Since H. pylori possesses an open reading frame whose predicted protein product has such high homology to the V. harveyi LuxS, we wanted to determine if H. pylori could synthesize AI-2 activity and investigate whether this activity might be associated with a potential quorum-sensing system. Our results indicate that H. pylori (luxS+) does produce an active signaling molecule (AI-2) that activates expression of bioluminescence in V. harveyi in a system-2-specific manner (Fig. 2). Activation of luminescence by supernatants harvested at various times during the growth of H. pylori revealed that AI-2-specific activity reaches a maximum early in log phase and diminishes as cells enter late log phase. This suggests that the activity of the molecule is density dependent and, specifically, that conditions of low cell density favor activity (Fig. 3). The loss of activity in stationary phase indicates that early-log-phase activity is degraded when H. pylori reaches stationary phase, an observation that mirrors the findings in serovar Typhimurium (34).
Despite the association between infection with H. pylori and gastric disease, very few gene products have been conclusively shown to play a role in H. pylori-mediated pathogenesis. Since LuxS has been implicated in pathogenesis (32), we wanted to test whether our luxS null mutation would have an effect on the expression or activities of any of the confirmed or putative virulence factors described for H. pylori. Our results indicate that none of the factors or processes that we examined, including growth, motility, urease activity, vacuolating cytotoxin activity, and induction of IL-8 response in epithelial cells, were affected in EJ103. It is possible that any impact that the H. pylori system 2 may have on regulating gene expression involves processes other than those directly involved in pathogenesis. In addition to several reports implicating a role for quorum sensing in pathogenesis (20, 25, 32, 34–36, 39), it is known to control a variety of other phenotypes not directly associated with pathogenesis, such as biofilm formation, luminescence, sporulation, and natural competence. Nevertheless, as the molecular mechanisms and factors underlying H. pylori-mediated pathogenesis become more fully understood and characterized, a necessity for cell density-dependent regulation via luxS may be revealed.
In an effort to undertake a global examination of luxS-mediated gene expression in H. pylori, we performed 2-D gel analysis of the parental strain and an isogenic luxS deletion strain, EJ103. This analysis suggested that a luxS deletion has little effect on protein complexity and abundance during steady-state analysis, since no obvious and reproducible differences in protein profiles between the two strains were detected. There are several potential explanations for this result. Several studies have reported that levels of target gene activation in response to quorum-sensing signals are as little as two- or threefold (20, 25, 27). In these cases, 2-D gel analysis may not be sensitive enough to detect modest changes in protein synthesis.
One limitation of all studies of expression of pathogenic factors in vitro is the difficulty in approximating an in vivo environment. Controlling for multiple environmental factors that may simultaneously influence gene expression is difficult, if not impossible, to accomplish. In this regard, it is also possible that sensory information from several environmental cues may be channeled through other, AI-2-independent, pathways that converge at some critical junction. Thus, a combination of transmitted signals may be required to influence downstream gene expression. If so, elimination of any single pathway may not necessarily result in an obvious phenotype. Our burgeoning understanding of the variety and combinations of molecular mechanisms employed by bacteria to establish quorum sensing as a critical mode of communicating information suggests that this is very likely to be the case. For instance, in V. harveyi, the expression of bioluminescence is only partially due to density-dependent signaling. Other environmental cues, such as the availability of iron, oxygen, and carbohydrates, also contribute to the regulation of bioluminescence in an AI-2-independent fashion.
The initial identification of luxS in V. harveyi took advantage of the observation that E. coli DH5α was phenotypically LuxS− by complementing the LuxS defect with a V. harveyi library (34). Subsequently, the E. coli DH5α luxS gene was found to have a frameshift mutation. The observation that luxS genes from many other organisms complement the defect in E. coli DH5α suggests that LuxS is a highly conserved protein. The availability of several bacterial genome sequences reveals that LuxS homologs exist in at least 25 different species. Our data from the trispecies complementation experiment shown in Fig. 2B demonstrating that expression of the H. pylori luxS gene in E. coli DH5α results in active AI-2 molecules that complement the AI-2 defect in V. harveyi underscores the high level of LuxS conservation among species. There is mounting evidence from a few organisms that have a luxS homolog for the existence of LuxS-homologous signaling systems (32). Taken together, while it is currently unknown what functions are controlled by AI-2 in H. pylori, these observations suggest that a LuxS signaling system is present in H. pylori. Understanding how this complex microbial communication process contributes to the biology of H. pylori will necessitate developing a facility for “bacterial lexicology.”
Since H. pylori is unique in its ability to colonize the gastric mucosa, it likely has developed specialized strategies allowing it to exploit, grow, and persist in the restrictive niche of the stomach. The lack of any other characterized reservoir may imply that the organism spends most of its life cycle in this unique environment, the gastric lumen of its host. This raises the possibility that H. pylori has little need to regulate gene expression to survive the low pH of the stomach. However, because H. pylori depends on a host organism for its survival, it must strike a dynamic balance between growing in the gastric niche and avoiding the host immune response in order to successfully persist. Density-dependent cell signaling could provide a mechanism for H. pylori to keep its population size in check in response to changes in its host's environment in order to avoid alerting the host immune system to the infection.
ACKNOWLEDGMENTS
We gratefully acknowledge the technical support provided by Anne Kane and her staff at the GRASP Digestive Disease Center at New England Medical Center, Boston, Mass., which is supported by a grant from the National Institutes of Health (NIDDK,P30DK34928). Special thanks are due to Bryan Hurley and Ramona Chitrakar-Walson for performing the IL-8 assays.
We also thank Dorothy Fallows, D. Scotty Merrell, Anne Kane, and Stephen Popper for critical review of the manuscript and thoughtful scientific discussions.
REFERENCES
- 1.Abdelnour A, Arvidson S, Bremell T, Ryden C, Tarkowski A. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect Immun. 1993;61:3879–3885. doi: 10.1128/iai.61.9.3879-3885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmer B M, van Reeuwijk J, Watson P R, Wallis T S, Heffron F. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol. 1999;31:971–982. doi: 10.1046/j.1365-2958.1999.01244.x. [DOI] [PubMed] [Google Scholar]
- 3.Bassler B L, Greenberg E P, Stevens A M. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassler B L, Wright M, Showalter R E, Silverman M R. Intercellular signalling in Vibrio harveyi—sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 5.Bassler B L, Wright M, Silverman M R. Multiple signaling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 6.Bassler B L, Wright M, Silverman M R. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol Microbiol. 1994;12:403–412. doi: 10.1111/j.1365-2958.1994.tb01029.x. [DOI] [PubMed] [Google Scholar]
- 7.Beier D, Spohn G, Rappuoli R, Scarlato V. Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J Bacteriol. 1997;179:4676–4683. doi: 10.1128/jb.179.15.4676-4683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bereswill S, Waidner U, Odenbreit S, Lichte F, Fassbinder F, Bode G, Kist M. Structural, functional and mutational analysis of the pfr gene encoding a ferritin from Helicobacter pylori. Microbiology. 1998;144:2505–2516. doi: 10.1099/00221287-144-9-2505. [DOI] [PubMed] [Google Scholar]
- 9.Birkey S M, Liu W, Zhang X, Duggan M F, Hulett F M. Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol Microbiol. 1998;30:943–953. doi: 10.1046/j.1365-2958.1998.01122.x. [DOI] [PubMed] [Google Scholar]
- 10.Brito B, Marenda M, Barberis P, Boucher C, Genin S. prhJ, and hrpG, two new components of the plant signal-dependent regulatory cascade controlled by PrhA in Ralstonia solanacearum. Mol Microbiol. 1999;31:237–251. doi: 10.1046/j.1365-2958.1999.01165.x. [DOI] [PubMed] [Google Scholar]
- 11.Cover T L, Cao P, Lind C D, Tham K T, Blaser M J. Correlation between vacuolating cytotoxin production by Helicobacter pylori isolates in vitro and in vivo. Infect Immun. 1993;61:5008–5012. doi: 10.1128/iai.61.12.5008-5012.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deiwick J, Nikolaus T, Erdogan S, Hensel M. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol. 1999;31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 13.Dunlap P V, Greenberg E P. Role of intracellular chemical communication in the Vibrio fischeri-monocentrid fish symbiosis. Washington, D.C.: American Society for Microbiology; 1991. [Google Scholar]
- 14.Egland K A, Greenberg E P. Quorum sensing in Vibrio fischeri: elements of the luxI promoter. Mol Microbiol. 1999;31:1197–1204. doi: 10.1046/j.1365-2958.1999.01261.x. [DOI] [PubMed] [Google Scholar]
- 15.Ellehauge E, Norregaard-Madsen M, Sogaard-Andersen L. The FruA signal transduction protein provides a checkpoint for the temporal co-ordination of intercellular signals in Myxococcus xanthus development. Mol Microbiol. 1998;30:807–817. doi: 10.1046/j.1365-2958.1998.01113.x. [DOI] [PubMed] [Google Scholar]
- 16.Freeman J A, Bassler B L. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol. 1999;31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- 17.Freeman J A, Bassler B L. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J Bacteriol. 1999;181:899–906. doi: 10.1128/jb.181.3.899-906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuqua C, Greenberg E P. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr Opin Microbiol. 1998;1:183–189. doi: 10.1016/s1369-5274(98)80009-x. [DOI] [PubMed] [Google Scholar]
- 19.Fuqua W C, Winans S C, Greenberg E P. Quorum sensing in bacteria: the LuxR/LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gambello M J, Kaye S, Iglewski B H. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hastings J W, Greenberg E P. Quorum sensing: the explanation of a curious phenomenon reveals a common characteristic of bacteria. J Bacteriol. 1999;181:2667–2668. doi: 10.1128/jb.181.9.2667-2668.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoch J A, Silhavy T J, editors. Two-Component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. [Google Scholar]
- 23.Jungnitz H, West N P, Walker M J, Chhatwal G S, Guzman C A. A second two-component regulatory system of Bordetella bronchiseptica required for bacterial resistance to oxidative stress, production of acid phosphatase, and in vivo persistence. Infect Immun. 1998;66:4640–4650. doi: 10.1128/iai.66.10.4640-4650.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaplan H B, Greenberg E P. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol. 1985;163:1210–1214. doi: 10.1128/jb.163.3.1210-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Latifi A, Winson M K, Foglino M, Bycroft B W, Stewart G S, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 26.McGowan C C, Necheva A, Thompson S A, Cover T L, Blaser M J. Acid-induced expression of an LPS-associated gene in Helicobacter pylori. Mol Microbiol. 1998;30:19–31. doi: 10.1046/j.1365-2958.1998.t01-1-01079.x. [DOI] [PubMed] [Google Scholar]
- 27.O'Toole G A, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 28.Papini E, Debernard M, Milia E, Bugnoli M, Zerial M, Rappuoli R, Montecucco C. Cellular vacuoles induced by Helicobacter pylori originate from late endosomal compartments. Proc Natl Acad Sci USA. 1994;91:9720–9724. doi: 10.1073/pnas.91.21.9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pesci E C, Iglewski B H. The chain of command in Pseudomonas quorum sensing. Trends Microbiol. 1997;5:132–134. doi: 10.1016/S0966-842X(97)01008-1. [DOI] [PubMed] [Google Scholar]
- 30.Schmitz A, Josenhans C, Suerbaum S. Cloning and characterization of the Helicobacter pylori flbA gene, which codes for a membrane protein involved in coordinated expression of flagellar genes. J Bacteriol. 1997;179:987–997. doi: 10.1128/jb.179.4.987-997.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma S A, Tummuru M K R, Miller G G, Blaser M J. Interleukin-8 response of gastric epithelial cell lines to Helicobacter pylori stimulation in vitro. Infect Immun. 1995;63:1681–1687. doi: 10.1128/iai.63.5.1681-1687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sperandio V, Mellies J L, Nguyen W, Shin S, Kaper J B. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc Natl Acad Sci USA. 1999;96:15196–15201. doi: 10.1073/pnas.96.26.15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spohn G, Scarlato V. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J Bacteriol. 1999;181:593–599. doi: 10.1128/jb.181.2.593-599.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Surette M G, Bassler B L. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc Natl Acad Sci USA. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Surette M G, Bassler B L. Regulation of autoinducer production in Salmonella typhimurium. Mol Microbiol. 1999;31:585–595. doi: 10.1046/j.1365-2958.1999.01199.x. [DOI] [PubMed] [Google Scholar]
- 36.Surette M G, Miller M B, Bassler B L. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szczebara F, Dhaenens L, Armand S, Husson M O. Regulation of the transcription of genes encoding different virulence factors in Helicobacter pylori by free iron. FEMS Microbiol Lett. 1999;175:165–170. doi: 10.1111/j.1574-6968.1999.tb13615.x. [DOI] [PubMed] [Google Scholar]
- 38.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzgerald L M, Lee N, Adams M D, Venter J C, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 39.Van Delden C, Iglewski B H. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis. 1998;4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whiteley M, Lee K M, Greenberg E P. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]