Abstract
Oral actinomycetes produce fructosyltransferase (FTF) enzymes which convert sucrose into polymers of d-fructose, known as levans, and these polymers are thought to contribute to the persistence and virulence of the organisms. A gene encoding FTF was isolated from Actinomyces naeslundii WVU45; the deduced amino acid sequence showed significant similarity to known levansucrases of gram-negative environmental isolates but was less similar to FTFs from gram-positive bacteria. A transcriptional start site was mapped by primer extension 70 bp 5′ from the putative start codon. Promoter fusions to a chloramphenicol acetyltransferase gene were used to confirm that there was a functional promoter driving ftf expression and to show that sequences located 86 to 218 bp upstream of the transcription initiation site were required for optimal ftf expression. Quantitative slot blot analysis against total RNA from cells grown on different sugars or from different growth phases revealed that ftf was constitutively transcribed. Thus, the A. naeslundii FTF is more similar in primary sequence and the regulation of expression to levansucrases of gram-negative bacteria than gram-positive bacteria.
Actinomyces naeslundii is a gram-positive, facultative anaerobe that is one of the primary colonizers of mucosal and tooth surfaces (45). It has been suggested that A. naeslundii may be involved in the pathogenesis of root caries (40) and periodontal diseases (25, 38). A. naeslundii isolated from human root caries has been demonstrated to produce root caries and alveolar bone loss in germ-free rats fed a high-sucrose diet (38). In a similar study, gnotobiotic rats which were infected with Actinomyces and fed a high-carbohydrate diet developed root caries and periodontal lesions (25). More recently, though, epidemiological studies which enumerated the bacteria isolated from human root caries and noncarious surfaces indicated that the presence of A. naeslundii is not positively correlated with caries (6, 42). Considering the abundance of Actinomyces in plaque and the likelihood that these organisms play key roles in oral health and disease, comparatively little is known about the pathogenic potential of these bacteria or their role in plaque ecology.
Actinomyces spp. and many oral streptococci produce fructosyltransferases (FTFs), which use dietary sucrose to produce extracellular homopolymers of fructose (fructans), which are predominantly β2,6 linked (levans) or primarily β2,1 linked (inulin), depending on the source of the FTF (4). FTFs that produce levans are often called levansucrases. Organisms that synthesize fructans have the ability to hydrolyze them via fructanases or levanases (9). Fructans accumulate rapidly in human dental plaque following the ingestion of sucrose, and the levels of fructans decrease steadily for about an hour after exogenous carbohydrates are exhausted (22, 24). Based on this observation and experiments conducted with strains of Streptococcus mutans with defects in fructan metabolism (7), fructans appear to serve principally as storage polysaccharides, which can be hydrolyzed by fructanases when other carbohydrate sources are depleted. It is hypothesized that fructan metabolism contributes to the development of dental caries by allowing plaque bacteria to utilize a greater proportion of dietary sucrose over a longer period of time, thus enhancing acid production. The β2,6-linked fructans, such as those produced by Actinomyces, are effective T-cell-independent antigens, can trigger inflammation, and are mitogenic for B cells (14, 18). Thus, it is possible that an immune response to levans may contribute to the inflammation seen in periodontal diseases.
Only a few investigations of FTF of Actinomyces have been conducted, and those were performed more than 25 years ago. In those early studies, the biochemical properties of FTF were explored in Actinomyces viscosus (33, 41), a strain related to A. naeslundii. An analysis of the fructan product from A. viscosus revealed that the enzyme synthesizes primarily levan-type polymers, rich in β2,6 linkages (34, 41). The A. viscosus FTF was characterized as having both cell-associated and extracellular forms. Since that time, no additional insights into the genetics or biochemistry of Actinomyces FTFs have been realized, in spite of the potential contribution of fructans to the persistence and pathogenesis of Actinomyces, nor has the role of these polymers in modulation of oral biofilm composition and virulence been elucidated. The purpose of this study was to begin a detailed molecular characterization of the FTF of A. naeslundii.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and reagents.
A. naeslundii WVU45 (ATCC 12104) was grown in brain heart infusion broth. For gene regulation experiments, WVU45 and promoter fusion strains were grown in Actinomyces defined media (ADM) (12) supplemented with 1% desired carbohydrate source. Escherichia coli DH10B was grown in Luria broth, and Streptococcus salivarius was grown in brain heart infusion broth. Kanamycin (50 μg/ml), streptomycin (50 μg/ml), and ampicillin (100 μg/ml) were added to media when necessary. All chemical reagents were obtained from Sigma Chemical Co. (St. Louis, Mo.).
DNA manipulations.
Chromosomal DNA was isolated from A. naeslundii by the method of Donkersloot et al. (19). E. coli plasmid DNA was isolated by a rapid boiling method or by using a QiaPrep spin plasmid kit (Qiagen, Inc., Chatsworth, Calif.). Restriction and DNA-modifying enzymes were obtained from Life Technologies (Bethesda, Md.), MBI Fermentas (Amherst, N.Y.), New England Biolabs (Beverly, Mass.), or U.S. Biochemicals (Cleveland, Ohio). A genomic library of A. naeslundii WVU45 was constructed in λGEM-12, and subclones were generated as previously detailed (31).
Nucleotide sequence analysis was performed with a Ladderman sequencing kit (Takara Shuzo Co., Otsu, Japan) or by using the Taq Trak sequencing reagents (Promega, Madison, Wis.). Nucleotide sequencing was completed by generating subclones and by primer walking with oligonucleotides that were complementary to the ftf open reading frame. Sequencing reactions were radiolabeled using [α-35S]dATP (New England Nuclear, Boston, Mass.). Southern blotting experiments were performed as described by Sambrook et al. (36) under conditions of high stringency.
Primer extension analysis.
A. naeslundii WVU45 was grown in ADM, containing 1% sucrose or glucose as the carbohydrate source, to mid-exponential phase, and total RNA was isolated as described by Chen et al. (11). Primer extensions were carried out with the oligonucleotide PE3 (5′-GACAGGCTCGGAACGAAGTGTG-3′). The protocol followed was that of McKnight and Kingsbury (29) as described elsewhere (3), with reverse transcription and annealing at 42°C and using annealing buffer containing 1.5 M KCl, 0.1 M Tris (pH 8.0), and 10 mM EDTA.
Promoter fusion construction.
DNA fragments containing the promoter region and ribosome binding sites of the ftf gene of A. naeslundii WVU45 were amplified by PCR using two sets of primers, engineered to contain a BamHI site 5′ and a KpnI site on the 3′ end of the product. The first set was PF1 (5′-GACCACTGGTACCGACAGGATGGGAGACCAG-3′) plus PF3 (5′-GGTCATGGATCCTACTGTCCTTACTCATGAGGG-3′), yielding a 300-bp product. The second primer pair, PF2 (5′GCAGGGGGTACCTCTTTGGTAGGAGACGAC3′) and PF3, produced a 160-bp fragment. The correct nucleotide sequence of the PCR products was confirmed, and a BamHI/KpnI fragment was subcloned into pCW24 containing an E. coli chloramphenicol acetyltransferase (cat) gene (11). The cat gene in pCW24 lacks a ribosome binding site and was engineered such that a promoter and cognate translational initiation sites of the gene of interest could be juxtaposed immediately 5′ to cat. A fragment containing the promoter region fused to cat was generated by digestion with KpnI and HindIII and subcloned into pJRD215 (16). Plasmid pJRD215, containing the ftf promoter fused to the cat reporter gene, was introduced into A. naeslundii WVU45 by the protocol of Yeung and Kozelsky (47), using a Gene Pulser (Bio-Rad Laboratories, Richmond, Calif.) connected to a pulse controller. Plasmid pJ218CAT contained the full-length promoter including 218 bp 5′ to the transcriptional initiation site. Plasmid pJ86CAT was the 5′ deletion derivative containing the promoter and additional sequences located 86 bp upstream of the transcription start site, and pJCAT contained only the promoterless cat. Plasmid DNA was extracted from Actinomyces as described by Anderson and McKay (1).
Enzyme assays.
Cell extracts were prepared from E. coli FTFLB4 by collecting cells from a 50-ml culture and washing in 10 mM potassium phosphate buffer (pH 6.0). Cells were resuspended in 1 ml of 50 mM phosphate buffer (pH 6.0) and added to 600 μl of glass beads in a screw-cap microcentrifuge tube. Cells were homogenized in a Bead Beater (Biospec, Bartlesville, Okla.) for 20 s, placed on ice for 1 min, and homogenized again. For measuring sucrose hydrolysis, cleared lysates generated by centrifugation at 14,000 × g for 15 min at 4°C were incubated with 25 mM sucrose in 25 mM phosphate buffer (pH 6.0) at 37°C. Total reducing sugar released during the reaction was measured by the method of Luchsinger and Cornesky (27). Alternatively, incorporation of fructose into polymer was assayed as previously described (35) by incubating the cleared cell lysates with [fructose-1-3H]sucrose at 37°C in 25 mM phosphate buffer (pH 6.0) for 4 h; 5 ml of ice-cold methanol was added, and the samples were applied to glass fiber filters on a vacuum manifold. Filters were washed twice with 5 ml of ice-cold methanol, dried, and counted in a scintillation counter. Activity was expressed in units, defined as the amount of enzyme required to incorporate 1 μmol of fructose into methanol-insoluble material per milligram of protein per hour.
CAT assays.
A. naeslundii strains carrying cat gene fusions were grown to exponential phase in ADM containing 1% glucose. One hour before the cells were harvested, sucrose was added to a final concentration of 1%. Cells were collected by centrifugation and washed with 10 mM Tris-HCl (pH 7.8). The pellets were resuspended in 750 μl of 10 mM Tris-HCl (pH 7.8), homogenized with 500 μl of glass beads for 20 s, and placed on ice for 2 min; then this process was repeated. The cleared lysates were used for determining CAT activity by the method of Shaw (37). One unit of CAT activity was defined as the amount of enzyme needed to acetylate 1 nmol of chloramphenicol per minute. Activities were normalized to protein concentrations, which were determined by the method of Bradford (5) with a commercially available reagent (Bio-Rad, Hercules, Calif.), using bovine serum albumin as the standard.
Slot blot analysis.
Total RNA was isolated from A. naeslundii WVU45 grown in ADM containing 1% desired carbohydrate. RNA concentrations were determined spectrophotometrically, and equivalent amounts of RNA were loaded onto the slot blot apparatus as described by Sambrook et al. (36). Blots were probed with internal fragments of the ftf gene. Hybridization and wash conditions were as described by Church and Gilbert (13).
RESULTS
Identification of an ftf gene from A. naeslundii WVU45.
As part of a separate project in the laboratory, a genomic library of A. naeslundii WVU45 was constructed in the bacteriophage λGEM-12 and screened for urease genes (31). Nucleotide sequence analysis of a plasmid subclone derived from λLM9 (31) revealed similarity to known levansucrases. A 9-kbp BamHI fragment containing the entire ftf gene was subcloned into pGEM7 (Promega) to yield pWC1. Southern hybridization using an internal fragment of pWC1 to A. naeslundii WVU45 chromosomal DNA under high-stringency conditions confirmed that the clone isolated was indeed derived from A. naeslundii (data not shown).
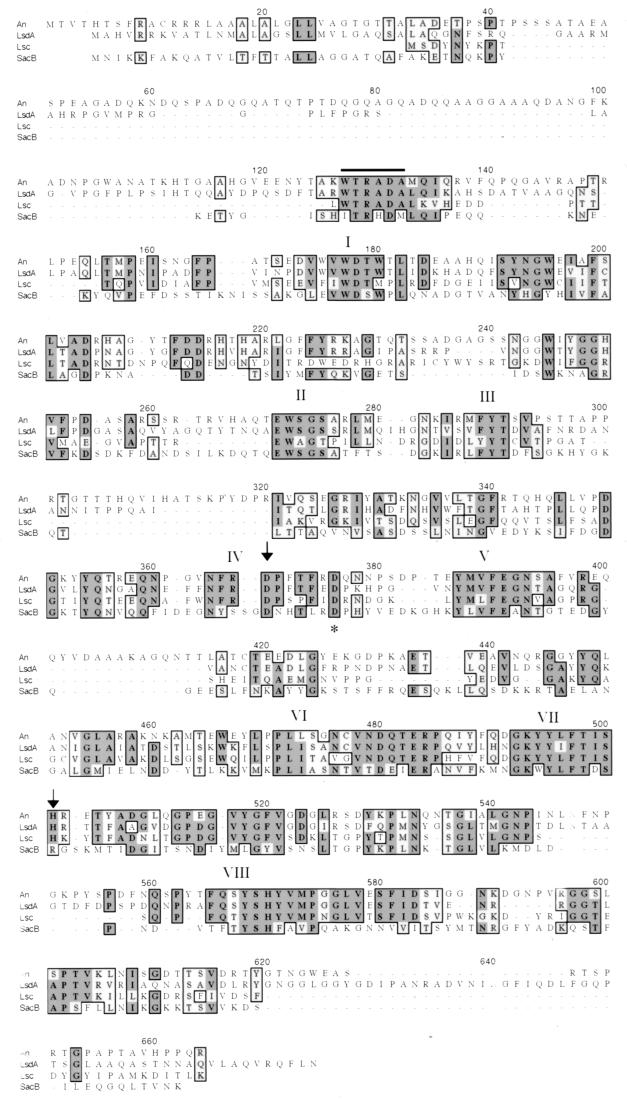
DNA sequence of the ftf gene was obtained by generating subclones and by primer walking. An open reading frame of 1,889 bp was identified. The deduced amino acid sequence had the highest degrees of similarity (57%) and identity (51%) to the levansucrase of the gram-negative soil and plant bacterium Acetobacter diazotrophicus. The protein contained a sequence characteristic of signal sequences of gram-positive bacteria. The calculated molecular mass of 68,251 Da was similar to those of many but not all levansucrases. In addition, eight regions that have been shown to be conserved in all levansucrases (2), including an RDP sequence containing the putative catalytic aspartic acid residue located at position 357, were conserved in the A. naeslundii FTF (Fig. 1). Overall the A. naeslundii FTF shared the strongest similarities with FTFs of gram-negative bacteria; lesser similarities were observed with the FTFs of gram-positive bacteria, including S. mutans, S. salivarius, and Bacillus subtilis. Also, the sequence WTRADA (positions 127 to 132), which has been observed only in levansucrases of gram-negative bacteria, including A. diazotrophicus, Erwinia amylovora, Zymomonas mobilis, and Pseudomonas syringae, was present in the A. naeslundii FTF.
FIG. 1.
Multiple sequence alignments of levansucrase from A. naeslundii (An), Acetobacter diazotrophicus (LsdA), Erwinia amylovora (Lsc), and Bacillus subtilis (SacB). Alignment was done by ClustalW. The WTRADA sequence is overlined. Arrows indicate the aspartic residue of the RDP motif and the histidine residue thought to be involved in enzyme catalysis. An asterisk marks the aspartic residue of the RDP motif of SacB. The eight regions conserved in all levansucrases are numbered I to VIII.
Enzyme characterization.
A. naeslundii produces more than one enzyme capable of hydrolyzing sucrose (30, 32). A variety of chromatographic purification steps were implemented to purify FTF from A. naeslundii. However, all of these failed because of rapid loss of enzymatic activity, possibly due to proteolytic activity or aggregation of the enzyme. Because of this, initial characterization of the enzyme was performed using enzyme preparations from E. coli, which is normally devoid of sucrase activity. Plasmid pWC1 was digested with SmaI and ClaI, and a 6-kbp fragment was subcloned into EcoRV- and ClaI-digested pBCIISk (Stratagene, La Jolla, Calif.) in E. coli DH10B. The resulting strain, FTFLB4, expressed a sucrase activity not detectable in DH10B and exhibited levan synthesis as measured by incorporation of 2.6 μmol of radiolabeled fructose mg of protein−1 h−1 into a methanol-insoluble polymer. To provide a frame of reference, S. salivarius 57.1, a known levan-synthesizing bacterium, produced 1.08 μmol mg of protein−1 h−1 in culture supernatants. The ftf gene in pFTFLB4 was in the same orientation as the lacZ promoter in the vector, but the transcriptional start site (see below) was located 2.5 kb from the lacZ promoter. Therefore, it is likely that E. coli transcriptional machinery recognized promoter-like sequences upstream of ftf.
Mapping of the transcriptional initiation site.
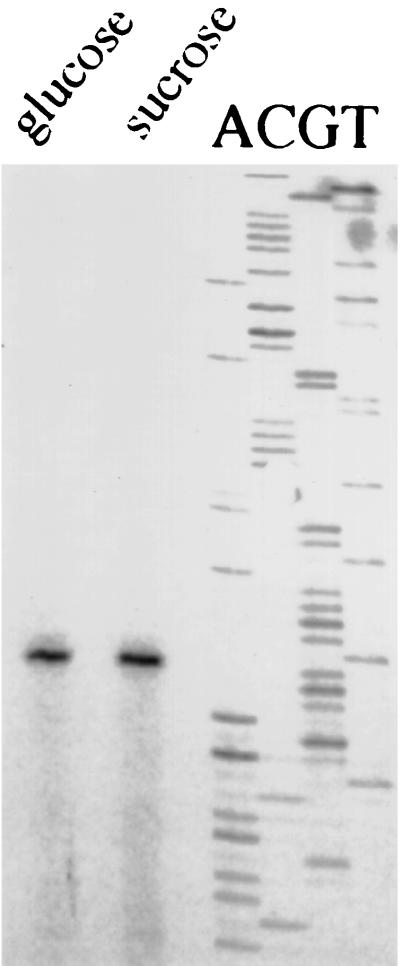
The primer PE3, complementary to the region located 13 to 34 nucleotides 3′ to the ATG start codon, was used in the primer extension analysis. A single transcription initiation site was mapped to a thymidine residue located 70 bases 5′ of the start site, regardless of whether the cells were grown in sucrose or glucose (Fig. 2). Not surprisingly, no similarities to canonical promoters were identified within 200 bp 5′ to the start site, nor did a comparison of the putative promoter with other known Actinomyces promoters reveal any obvious consensus sequences. Of note though, the DNA found within 100 bases 5′ to the transcriptional initiation site was 57% G+C, contrasted with a 68% G+C content for the ftf open reading frame and the 65 to 68% G+C content estimated for the A. naeslundii chromosome. A relatively low G+C content has been noted for other Actinomyces promoter regions (31).
FIG. 2.
Mapping of the ftf promoter by primer extension. Lanes labeled sucrose and glucose contain the primer extension product obtained with the primer PE3 from cells grown to mid-log phase in ADM with 1% indicated carbohydrate. The lanes marked ACGT are sequence reactions obtained from the same primer, using plasmid pFTFLB4 DNA.
Expression of ftf is constitutive.
Expression of FTFs by gram-positive bacteria is generally regulated by environmental factors, including carbohydrate source. To determine if the ftf of A. naeslundii could be regulated by growth domain, slot blot analysis was performed with total RNA from early-, mid-, and late-exponential-phase cells of A. naeslundii WVU45 growing with sucrose or glucose as the sole carbohydrate source. The results indicated that ftf was expressed at the same level regardless of the growth phase or carbohydrate source (data not shown). To determine if ftf was regulated by other carbohydrate sources, RNA was isolated from A. naeslundii grown in the presence of 1% sucrose, glucose, fructose, or galactose. The results of the slot blot indicated that the transcription of ftf was independent of carbohydrate source under the conditions tested (data not shown). The ability of sucrose to induce ftf expression was also examined in cells growing on glucose because A. naeslundii clumps in the presence of sucrose. For these experiments, cells were grown to an optical density at 600 nm of 0.5 or 1.0, and then sucrose was added to a final concentration of 1% 1 h before cells were harvested. No significant differences in the amounts of ftf mRNA were noted between controls, i.e., cells incubated for an additional hour in glucose alone, and the sucrose-treated cells (data not shown).
CAT activity of promoter fusions in A. naeslundii.
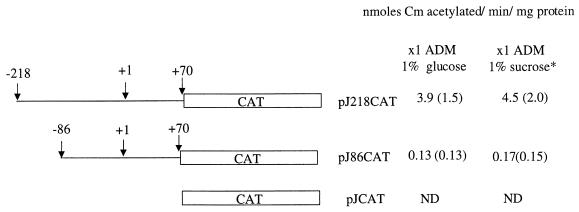
Expression of cat driven from the A. naeslundii promoter and a 5′ deletion derivative was assessed in cells grown in glucose and in cells induced with sucrose as described above (Fig. 3). Cells carrying the full-length promoter fusion, which extends to position −218 from the transcriptional initiation site, expressed the highest level of CAT activity. The smaller fusion, containing only 86 bp upstream from the transcriptional initiation site, had significantly less activity. A. naeslundii that harbored the construct that contained the promoterless CAT had no detectable activity.
FIG. 3.
Schematic representation and functional characterization of the A. naeslundii ftf promoter CAT fusions. CAT activity was measured in cell extracts of A. naeslundii grown in ADM and 1% glucose. *, cells were grown in 1% glucose; 1 h before they were harvested, 1% sucrose was added. Values are the average of four separate experiments. Levels are expressed as units of CAT per milligram of protein, where 1 U is defined as the amount of enzyme required to acetylate 1 nmol of chloramphenicol (Cm) per minute. Standard deviations are indicated in parentheses. ND, none detected.
DISCUSSION
The major fructan-producing organisms in plaque are streptococci and Actinomyces spp. S. mutans, a known cariogenic bacterium, synthesizes an inulin-type polymer, whereas Actinomyces spp. synthesize the levan-type polymer (4). Interestingly, the S. mutans fructan hydrolase (FruA) was found to have a two- to threefold-higher rate of attack on levans compared to inulins (8). This suggests that levan-type fructans may be more abundant than inulins in human dental plaque, and that there may be an ecological advantage afforded by levan catabolism. Although the soft tissue colonizer S. salivarius can make a levan-type polymer, dental plaque streptococci generally do not (20). Therefore, levan production by Actinomyces in supra- and subgingival dental plaque may be a major factor in the compositional balance of oral biofilms and a contributor to the initiation of oral diseases. This study has provided the first insights into the primary sequence, activity, and regulation of expression of the A. naeslundii FTF.
The resemblance of the A. naeslundii FTF to gram-negative levansucrases extends beyond the overall percentage of sequence identity and similarity. In particular, when Arg331 of the B. subtilis levansucrase is changed to a histidine residue, the efficiency of polymerization decreases significantly (10). Arg331 is conserved in both the S. mutans and B. subtilis FTFs and is thought to be part of the active site of the enzyme (10, 28). In levansucrases from gram-negative bacteria and in the A. naeslundii FTF, a histidine residue occupies this position. It has been postulated that this amino acid substitution is responsible for the high amounts of 1-kestose (glucose-β2,1-fructose-β2,1-fructose) and lower levels of high-molecular-weight levans synthesized by A. diazotrophicus (23) and Z. mobilis (15). Based on these observations, the A. naeslundii FTF may synthesize products more similar to the gram-negative levansucrases.
Interestingly, the A. naeslundii urease operon is located 6 kbp downstream of ftf, but its gene products share strongest similarities with ureases of gram-positive bacteria (31). The similarity of the A. naeslundii FTF to gram-negative FTFs may be due to the fact that the ftf gene was acquired by horizontal gene transfer from a gram-negative organism. Although the G+C content of the ftf gene does not differ from that estimated for the A. naeslundii chromosome (45), there are high-G+C gram-negative organisms that produce levansucrase. For example, the A. diazotrophicus levansucrase, with which the A. naeslundii enzyme shares the highest similarity, is encoded by a gene with a high-G+C content, 63%, which is in good agreement with its genomic DNA G+C content (2). At this time, though, there are no additional data to indicate that the gene could have been acquired by gene transfer, and a search of 500 bp 5′ and 300 bp 3′ of the ftf open reading frame did not reveal the presence of mobile genetic elements.
Attempts to align the ftf promoter with the relatively few other Actinomyces promoters that have been mapped, or with known Streptomyces promoters, did not yield any consensus sequences. These results are similar to the few other studies of Actinomyces promoters, which have led to the conclusion that Actinomyces spp. may possess a distinct class of promoter sequences (31, 45). The only common feature discovered thus far in Actinomyces promoters is that the region within 100 bp of the transcriptional initiation site is unusually AT rich, which is also the case for the ftf gene.
It appears that E. coli harboring pFTFLB4 could express the ftf gene, although not necessarily from the cognate Actinomyces promoter. This assumption is based on the fact that the start site of the ftf gene was located about 2.5 kbp from the lacZ promoter in the E. coli vector and on the fact that addition of isopropyl-β-d-thiogalactopyranoside to growing cultures of FTFLB4 had no influence on ftf expression (data not shown). The capacity of E. coli to express the fimbrial gene from A. naeslundii (46) has also been observed. Clearly, E. coli strains carrying the ftf gene acquire the ability to cleave sucrose and to synthesize levan-type polymers, as would be predicted, and as has been confirmed by using a mouse monoclonal antibody specific for β2,6-linked fructans (data not shown). Recently, we have insertionally inactivated the ftf gene in A. naeslundii WVU45 and have confirmed its essential role in levan production by these organisms (L. J. Bergeron and R. A. Burne, unpublished data), and studies are under way to study more closely the biochemical characteristics of the enzyme and its product.
To our knowledge, there are no published reports of the successful use of gene fusions to study the expression of any Actinomyces genes, although we have also demonstrated the utility of cat in the study of urease gene expression in this organism (E. Morou-Bermudez and R. A. Burne, unpublished data). The use of reporter fusions, coupled with mRNA analysis, has revealed that FTF expression in A. naeslundii is unaffected by carbohydrate source or growth domain, which is not the case for the FTFs of other gram-positive bacteria. For example, the use of cat fusions to the ftf promoter of S. mutans revealed that expression is constitutive but that transcription could be induced about threefold by addition of sucrose (43), mediated through cis elements 5′ to the promoter (26). Additionally, pH and growth rate influence ftf transcription in S. mutans (43). The regulation of the sacB gene, which encodes the levansucrase of B. subtilis, is mediated by an elaborately controlled antitermination mechanism (17). More similar to the A. naeslundii ftf, many FTF genes from gram-negative bacteria are constitutively transcribed (21, 23, 39). For example, E. amylovora levansucrase is produced constitutively, and its expression is modified by the levansucrase regulatory gene rlsA (48). RlsA is a member of the LysR family of transcriptional activators, and it positively regulates transcription of levansucrase independently of the carbohydrate source (48). Based on the data obtained with the cat fusion strains, it seems also that optimal expression of the A. naeslundii ftf gene could require a transcriptional activator(s). Therefore, the A. naeslundii FTF is more similar to the gram-negative levansucrases not only in terms of the primary sequence but also in terms of the regulation of expression.
To better understand the pathogenic potential of oral bacteria, substantial efforts have been directed at analysis of the metabolism by oral streptococci of sucrose, the most common carbohydrate associated with dental caries. Actinomyces species are abundant in supra- and subgingival dental plaque and on oral mucosal surfaces. However, the role of Actinomyces in oral biofilm formation, biofilm homeostasis, and oral diseases is still largely undefined. We have identified the ftf gene from A. naeslundii WVU45 in an effort to begin to dissect the role of sucrose metabolism and polymer production by this organism as it applies to oral health and diseases. Future studies will be oriented toward understanding biochemical properties of this enzyme and its role in persistence of A. naeslundii in the oral cavity.
ACKNOWLEDGMENTS
We especially acknowledge the late Maria K. Yeung (University of Texas Health Science Center at San Antonio) for bacterial strains, advice, and encouragement. We thank George H. W. Bowden (University of Manitoba) and Margaret Chen (University of Rochester) for providing bacterial strains, plasmids, and/or technical advice.
This work was supported by PHS grants DE12236, DE10362, and DE07165 from the National Institute of Dental and Craniofacial Research.
REFERENCES
- 1.Anderson D G, McKay L L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrieta J, Hernandez L, Coego A, Suarez V, Balmori E, Menendez C, Petit-Glatron M F, Chambert R, Selman-Housein G. Molecular characterization of the levansucrase gene from the endophytic sugarcane bacterium Acetobacter diazotrophicus SRT4. Microbiol. 1996;142:1077–1085. doi: 10.1099/13500872-142-5-1077. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1989. [Google Scholar]
- 4.Birkhed D, Rosell K G, Granath K. Structure of extracellular water-soluble polysaccharides synthesized by oral strains of Streptococcus mutans, Streptococcus salivarius, Streptococcus sanguis and Actinomyces viscosus. Arch Oral Biol. 1979;24:53–61. doi: 10.1016/0003-9969(79)90175-4. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Brown L R, Billings R J, Kaster A G. Quantitative comparisons of potentially cariogenic microorganisms cultured form noncarious and carious root and coronal tooth surfaces. Infect Immun. 1986;51:765–770. doi: 10.1128/iai.51.3.765-770.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burne R A, Chen Y Y, Wexler D L, Kuramitsu H, Bowen W H. Cariogenicity of Streptococcus mutans strains with defects in fructan metabolism assessed in a program-fed specific-pathogen-free rat model. J Dent Res. 1996;75:1572–1577. doi: 10.1177/00220345960750080801. [DOI] [PubMed] [Google Scholar]
- 8.Burne R A, Schilling K, Bowen W H, Yasbin R E. Cloning and partial purification of a β-d-fructosidase of Streptococcus mutans GS-5. In: Ferretti J J, Curtiss III R, editors. Streptococcal genetics. Washington, D.C.: American Society for Microbiology; 1987. pp. 220–224. [Google Scholar]
- 9.Burne R A, Schilling K, Bowen W H, Yasbin R E. Expression, purification, and characterization of an exo-β-d-fructosidase of Streptococcus mutans. J Bacteriol. 1987;169:4507–4517. doi: 10.1128/jb.169.10.4507-4517.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambert R, Petit-Glatron M F. Polymerase and hydrolase activities of Bacillus subtilis levansucrase can be separately modulated by site-directed mutagenesis. Biochem J. 1991;279:35–41. doi: 10.1042/bj2790035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y M, Weaver C A, Mendelsohn D R, Burne R A. Transcriptional regulation of the Streptococcus salivarius 57.1 urease operon. J Bacteriol. 1998;180:5769–5775. doi: 10.1128/jb.180.21.5769-5775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christie A O, Porteus J W. The cultivation of a single strain of Actinomyces israelii in a simplified and chemically defined medium. J Gen Microbiol. 1962;28:443–454. doi: 10.1099/00221287-28-3-443. [DOI] [PubMed] [Google Scholar]
- 13.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Couthino A, Moller G. B cell mitogenic properties of thymus independent antigens. Nat New Biol. 1973;245:12. doi: 10.1038/newbio245012a0. [DOI] [PubMed] [Google Scholar]
- 15.Crittenden R G, Doelle H W. Structural identification of oligosaccharides produced by Zymomonas mobilis levansucrase. Biotechnol Lett. 1993;15:1055–1060. [Google Scholar]
- 16.Davison J, Heusterspreute M, Chevalier N, Ha-Thi V, Brunel F. Vectors with restriction site banks. V. pJRD215, a wide host range cosmid vector with multiple cloning sites. Gene. 1987;51:275–280. doi: 10.1016/0378-1119(87)90316-7. [DOI] [PubMed] [Google Scholar]
- 17.Debarbouille M, Martin-Verstraete I, Arnaud M, Klier A, Rapoport G. Positive and negative regulation controlling the expression of the sac genes in Bacillus subtilis. Res Microbiol. 1991;142:757–764. doi: 10.1016/0923-2508(91)90052-c. [DOI] [PubMed] [Google Scholar]
- 18.Desaymard C, Ivanyl L. Comparison of in vitro immunogenicity, tolergenicity and mitogenicity of dinitrophenyl-levan conjugates with varying epitope density. Immunology. 1976;30:647. [PMC free article] [PubMed] [Google Scholar]
- 19.Donkersloot J A, Cisar J O, Wax M E, Harr R J, Chassy B M. Expression of Actinomyces viscosus antigens in Escherichia coli: Cloning of a structural gene (fimA) for type 2 fimbriae. J Bacteriol. 1985;162:1075–1078. doi: 10.1128/jb.162.3.1075-1078.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebisu S, Keijiro K, Kotani S, Misaka A. Structural differences in the fructans elaborated by Streptococcus mutans and Strep. salivarius. J Biochem. 1975;78:879–887. doi: 10.1093/oxfordjournals.jbchem.a130993. [DOI] [PubMed] [Google Scholar]
- 21.Geier G, Geider K. Characterization and influence on virulence of the levansucrase gene from the fireblight pathogen Erwinia amylovora. Physiol Mol Plant Pathol. 1993;42:387–404. [Google Scholar]
- 22.Gold W, Preston F B, Lache M C, Blechman H. Production of levan and dextran in plaque in vivo. J Dent Res. 1974;53:442–446. doi: 10.1177/00220345740530024401. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez L, Arrieta J, Menendez C, Vazquez R, Coego A, Suarez V, Selman G, Petit-Glatron M F, Chambert R. Isolation and enzymic properties of levansucrase secreted by Acetobacter diazotrophicus SRT4, a bacterium associated with sugar cane. Biochem J. 1995;309:113–118. doi: 10.1042/bj3090113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higuchi M, Iwami Y, Yamada T, Araya S. Levan synthesis and accumulation by human dental plaque. Arch Oral Biol. 1970;15:563–567. doi: 10.1016/0003-9969(70)90111-1. [DOI] [PubMed] [Google Scholar]
- 25.Jordan H V, Keyes P H, Bellack S. Periodontal lesions in hamsters and gnotobiotic rats infected with Actinomyces of human origin. J Periodontal Res. 1972;7:21–28. doi: 10.1111/j.1600-0765.1972.tb00627.x. [DOI] [PubMed] [Google Scholar]
- 26.Kiska D L, Macrina F L. Genetic regulation of fructosyltransferase in Streptococcus mutans. Infect Immun. 1994;62:1241–1251. doi: 10.1128/iai.62.4.1241-1251.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luchsinger W W, Cornesky R A. Reducing power by the dinitrosalicyclic acid method. Anal Biochem. 1962;4:346–347. doi: 10.1016/0003-2697(62)90098-2. [DOI] [PubMed] [Google Scholar]
- 28.Mantsala P, Puntala M. Comparison of levansucrase from Bacillus subtilis and from Bacillus amyloliquefaciens. FEMS Microbiol Lett. 1982;13:395–399. [Google Scholar]
- 29.McKnight S L, Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982;217:316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- 30.Miller C H, Somers P J B. Degradation of levan by Actinomyces viscosus. Infect Immun. 1978;22:266–274. doi: 10.1128/iai.22.1.266-274.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morou-Bermudez E, Burne R A. Genetic and physiologic characterization of urease of Actinomyces naeslundii. Infect Immun. 1999;67:504–512. doi: 10.1128/iai.67.2.504-512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norman J M, Bunny K L, Giffard P M. Characterization of levJ, a sucrase/fructanase-encoding gene from Actinomyces naeslundii T14V and comparison with other sucrose-cleaving enzymes. Gene. 1994;152:93–98. doi: 10.1016/0378-1119(94)00695-o. [DOI] [PubMed] [Google Scholar]
- 33.Pabst M J. Levan and levansucrase of Actinomyces viscosus. Infect Immun. 1977;15:518–526. doi: 10.1128/iai.15.2.518-526.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pabst M J, Cisar J O, Trummel C L. The cell wall-associated levansucrase of Actinomyces viscosus. Biochim Biophys Acta. 1979;566:274–282. doi: 10.1016/0005-2744(79)90031-7. [DOI] [PubMed] [Google Scholar]
- 35.Robrish S A, Reid W, Krichevsky M. Distribution of enzymes forming polysaccharides from sucrose and the composition of extracellular polysaccharide synthesized by Streptococcus mutans. Appl Microbiol. 1972;24:184–190. doi: 10.1128/am.24.2.184-190.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Shaw W V. Chloramphenicol acetyltransferase activity from chloramphenicol-resistant bacteria. Methods Enzymol. 1979;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- 38.Socransky S S, Hubersak C, Propas D. Induction of periodontal destruction in gnotobiotic rats by a human oral strain of Actinomyces naeslundii. Arch Oral Biol. 1970;15:993–995. doi: 10.1016/0003-9969(70)90095-6. [DOI] [PubMed] [Google Scholar]
- 39.Song K B, Joo H K, Rhee S-K. Nucleotide sequence of levansucrase gene (levU) of Zymomonas mobilis ZM1 (ATCC 10988) Biochim Biophys Acta. 1993;1173:320–324. doi: 10.1016/0167-4781(93)90130-6. [DOI] [PubMed] [Google Scholar]
- 40.Sumney D L, Jordan H V. Characterization of bacteria isolated from human root surface carious lesions. J Dent Res. 1974;53:343–351. doi: 10.1177/00220345740530022701. [DOI] [PubMed] [Google Scholar]
- 41.van der Hoeven J S, Vogels G D, Bekkers M F J. A levansucrase from Actinomyces viscosus. Caries Res. 1976;10:33–48. doi: 10.1159/000260187. [DOI] [PubMed] [Google Scholar]
- 42.Van Houte J, Jordan H V, Laraway R, Kent R, Soparkar P M, DePaola P F. Association of the microbial flora of dental plaque and saliva with human root-surface caries. J Dent Res. 1990;69:1463–1468. doi: 10.1177/00220345900690080301. [DOI] [PubMed] [Google Scholar]
- 43.Wexler D L, Hudson M C, Burne R A. Streptococcus mutans fructosyltransferase (ftf) and glucosyltransferase (gtfBC) operon fusion strains in continuous culture. Infect Immun. 1993;61:1259–1267. doi: 10.1128/iai.61.4.1259-1267.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeung M K. Complete nucleotide sequence of the Actinomyces viscosus T14V sialidase gene: presence of a conserved repeating sequence among strains of Actinomyces spp. Infect Immun. 1993;61:109–116. doi: 10.1128/iai.61.1.109-116.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeung M K. Molecular and genetic analyses of Actinomyces spp. Crit Rev Oral Biol Med. 1999;10:120–138. doi: 10.1177/10454411990100020101. [DOI] [PubMed] [Google Scholar]
- 46.Yeung M K, Cisar J O. Cloning and nucleotide sequence of a gene for Actinomyces naeslundii WVU45 type 2 fimbriae. J Bacteriol. 1988;170:3803–3809. doi: 10.1128/jb.170.9.3803-3809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeung M K, Kozelsky C S. Transformation of Actinomyces spp. by a gram-negative broad-host-range plasmid. J Bacteriol. 1994;176:4173–4176. doi: 10.1128/jb.176.13.4173-4176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Geider K. Molecular analysis of the rlsA gene regulating levan production by the fireblight pathogen Erwinia amylovora. Physiol Mol Plant Pathol. 1999;54:187–201. [Google Scholar]