Abstract
Saccharomyces cerevisiae medium-chain acyl elongase (ELO1) mutants have previously been isolated in screens for fatty acid synthetase (FAS) mutants that fail to grow on myristic acid (C14:0)-supplemented media. Here we report that wild-type cells cultivated in myristoleic acid (C14:1Δ9)-supplemented media synthesized a novel unsaturated fatty acid that was identified as C16:1Δ11 fatty acid by gas chromatography-mass spectroscopy. Synthesis of C16:1Δ11 was dependent on a functional ELO1 gene, indicating that Elo1p catalyzes carboxy-terminal elongation of unsaturated fatty acids (α-elongation). In wild-type cells, the C16:1Δ11 elongation product accounted for approximately 12% of the total fatty acids. This increased to 18% in cells that lacked a functional acyl chain desaturase (ole1Δ mutants) and hence were fully dependent on uptake and elongation of C14:1. The observation that ole1Δ mutant cells grew almost like wild type on medium supplemented with C14:1 indicated that uptake and elongation of unsaturated fatty acids were efficient. Interestingly, wild-type cells supplemented with either C14:1 or C16:1 fatty acids displayed dramatic alterations in their phospholipid composition, suggesting that the availability of acyl chains is a dominant determinant of the phospholipid class composition of cellular membranes. In particular, the relative content of the two major phospholipid classes, phosphatidylethanolamine and phosphatidylcholine, was strongly dependent on the chain length of the supplemented fatty acid. Moreover, analysis of the acyl chain composition of individual phospholipid classes in cells supplemented with C14:1 revealed that the relative degree of acyl chain saturation characteristic for each phospholipid class appeared to be conserved, despite the gross alteration in the cellular acyl chain pool. Comparison of the distribution of fatty acids that were taken up and elongated (C16:1Δ11) to those that were endogenously synthesized by fatty acid synthetase and then desaturated by Ole1p (C16:1Δ9) in individual phospholipid classes finally suggested the presence of two different pools of diacylglycerol species. These results will be discussed in terms of biosynthesis of different phospholipid classes via either the de novo or the Kennedy pathway.
The fatty acid composition of phospholipids is an important determinant of the biophysical properties of cellular membranes and as such must be subject to dynamic exchange to ensure membrane homeostasis under varying environmental conditions (10, 15). The biochemical control of fatty acid composition of different lipid classes, i.e., the regulation of the synthesis of defined lipid molecular species, however, is not well understood. Saccharomyces cerevisiae appears to be particularly amenable to study of this process, as both lipid and fatty acid biosynthesis are well characterized, mutants blocked in most steps of the lipid biosynthetic pathways are readily available (6, 12, 21), and the acyl chain composition of the major lipid species has recently been determined by mass spectroscopy (25).
Long-chain saturated fatty acids of 16 to 18 carbon atoms are the product of de novo synthesis by the fatty acid synthetase (FAS) (26, 35). These are desaturated between the carbon atoms in positions 9 and 10 by the single essential desaturase, Ole1p, to generate cis-monounsaturated C16:1 and C18:1 long-chain fatty acids (29, 30). Unsaturated fatty acids typically comprise 70 to 80% of the total fatty acids of this organism (9). Their relative composition, however, may vary considerably, depending on the strain background (13) and the carbon source (33). Very-long-chain C24 and C26 fatty acids, on the other hand, are synthesized by a membrane-bound elongase of which two components, Elo2p and Elo3p, have recently been characterized (14, 24). Even though very-long-chain fatty acids comprise only ∼1% of the cellular fatty acid pool (36), their synthesis is essential as they form part of the yeast ceramide, found in sphingolipids and in the lipid moiety of glycosylphosphatidylinositol (glycosyl-PtdIns)-anchored membrane proteins (12).
A third elongase, Elo1p, has been identified in a screen for fasΔ mutant cells that do not grow on myristic acid-supplemented media. elo1Δ mutant cells hence fail to elongate saturated fatty acids of 14 carbon atoms to long-chain fatty acids of 16 to 18 carbon atoms (17, 32). The substrate of Elo1p, however, is only poorly defined, and the mechanism of elongation has not yet been characterized. In the present study, we thus addressed the question whether Elo1p elongates unsaturated fatty acids by carboxy-terminal elongation (α-elongation), as known from the microsomal elongation systems of mammals and plants, which catalyze the synthesis of polyunsaturated fatty acids (5, 8).
The fatty acid supplementation experiments performed in the course of this work revealed important new information about the regulation of the synthesis of lipid molecular species in yeast. Wild-type cells supplemented with either C14:1Δ9 or C16:1Δ9 fatty acids displayed dramatic alterations in their phospholipid composition, suggesting that the availability of acyl chains is a dominant determinant of the phospholipid class composition of cellular membranes. The degree of acyl chain saturation characteristic for the different lipid classes (25, 34), however, appeared to be conserved, despite the gross alteration of the cellular acyl chain pool brought about by the fatty acid supplementation. These results will be discussed in terms of biosynthesis of different phospholipid classes via the de novo or the Kennedy pathway. In the de novo pathway, decarboxylation of phosphatidylserine (PtdSer) yields phosphatidylethanolamine (PtdEtn), which is further methylated to phosphatidylcholine (PtdCho), whereas, in the Kennedy pathway, PtdEtn and PtdCho can both be synthesized from diacylglycerol (DAG) and either CDP-ethanolamine or CDP-choline (12, 21).
MATERIALS AND METHODS
Yeast strains and culture media.
The wild-type S. cerevisiae strain used was W303a (MATa ade2-1 his3-11,15 leu2-3, 112 trp1-1 ura3-1). The elo1Δ ole1Δ double mutant strain (MATa elo1::HIS3 ole1::LEU2 ade2 his3 leu2 ura3) was obtained from crossing elo1Δ (MATα elo1::HIS3 ade2-1 can1-100 his3-11,15 leu2-2,112 ura3-1) with ole1Δ (MATa ole1::LEU2 ade2 his3 leu2-2 ura3), both kindly provided by C. Martin (Rutgers University, Piscataway, N.J.). Standard yeast genetics methods were used for mating, sporulation, and construction of the double mutant strain (27). Cells were grown either in complete (yeast extract-peptone-dextrose) medium containing 1% yeast extract (Difco), 1% Bacto Peptone (Difco), and 2% glucose or in minimal (synthetic dextrose) medium containing 0.67% yeast nitrogen base without amino acids (Difco) and 2% glucose. Media supplemented with fatty acids contained 1% Brij 58 and 0.5 mM (either) C14:1 or C16:1 fatty acids (Sigma). For growth rate determination, optical density at 600 nm was monitored.
Lipid and fatty acid analysis.
Cells were harvested in the late exponential growth phase, washed two times with 0.2% bovine serum albumin, and used immediately or stored at −70°C before extraction of lipids. Lipids were extracted from cell homogenates by the procedure of Folch et al. (18). Phospholipid classes were separated by two-dimensional thin-layer chromatography on silica gel 60 plates (Merck, Darmstadt, Germany), using chloroform–methanol–25% ammonia (65:35:5 [vol/vol/vol]) as the first and chloroform-acetone-methanol-acetic acid-water (50:20:10:10:5 [vol/vol/vol/vol/vol]) as the second developing solvent. Spots detected after exposure to iodine vapor were scraped off, and lipids were extracted with chloroform-methanol (1:4 [vol/vol]). Total and individual phospholipids were quantified as described by Broekhuyse (4).
Fatty acids were converted to methyl esters by BF3-catalyzed methanolysis (23) and separated by gas-liquid chromatography (GLC) using a Hewlett-Packard HP 6890 Series GC, equipped with an HP Innovax column (15 m by 0.25 mm by 0.50 μm in film thickness), with a temperature gradient (20 min at 200°C, 10°C/min to 280°C, and 15 min at 280°C). Fatty acids were identified by comparison to commercially available methyl ester standards (NuCheck, Inc., Elysian, Minn.).
The double bond position of monounsaturated fatty acids was determined by GLC-mass spectrometry (GLC-MS) of dimethyl disulfide adducts of these derivatives as described previously (37), on an HP 5890 Series II Plus GC equipped with electronic pressure control, the HP Chemstation software package, and an HP 5972 mass selective detector. Injector and interface were kept at 250 and 300°C, respectively. GLC-MS analysis was performed on a capillary column, HP-5MS, 30 m by 0.25 mm by 0.25 μm in film thickness, programmed from 150 to 320°C at 20°C/min after a 2-min hold at 150°C. Finally, the column was kept at 320°C for 10 min. All analyses were carried out in the constant-flow mode. Helium was used as carrier gas with a linear velocity of 34.1 cm/s. Aliquots of 1 μl of the samples were injected with an HP 7673 autosampler in splitless mode. Electron impact ionization with 70-eV ionization energy was used for mass spectroscopy. Data were collected by scanning from 150 to 600 atomic mass units at 1.6 scans/s.
RESULTS AND DISCUSSION
Growth of ole1Δ mutant cells on C14:1-supplemented media requires a functional elongase.
In our analysis of the fatty acid requirements of an ole1Δ mutant strain, we observed that cells lacking the desaturase displayed significant growth on C14:1-supplemented media (data not shown). Since pathways for the modification of supplemented C14:1 fatty acid have not yet been described, growth of ole1Δ mutant cells on C14:1 would indicate that these cells survived by incorporating C14:1 as sole unsaturated fatty acid in their membrane lipids, a possibility that we considered unlikely. We thus investigated whether the exogenously supplemented C14:1 was metabolically converted to a longer-chain unsaturated fatty acid that, when esterified to lipid, permitted growth of an ole1Δ mutant strain on myristoleic acid.
ELO1 has previously been shown to be required for the elongation of saturated medium-chain fatty acids of 14 carbon atoms in length to saturated long-chain fatty acids of 16 to 18 carbon atoms (17, 32). We thus investigated whether Elo1p is required for ole1Δ mutant cells to grow on myristoleic acid-supplemented media. To address this question, an elo1Δ ole1Δ double mutant strain was generated and tested for growth on media supplemented with various unsaturated fatty acids. In contrast to the two parental strains, the elo1Δ ole1Δ double mutant did not grow on C14:1-supplemented media, suggesting that in an ole1Δ mutant strain the supplemented C14:1 fatty acid is elongated to longer-chain unsaturated fatty acids and that this elongation is dependent on Elo1p. On C16:1-supplemented media, on the other hand, Elo1p function appeared to be dispensable for growth of the ole1Δ mutant strain, as indicated by the vigorous growth of the elo1Δ ole1Δ double mutant strain on palmitoleic acid-supplemented media (data not shown).
Identification of an Elo1p-dependent elongation product of unsaturated fatty acids in wild-type cells.
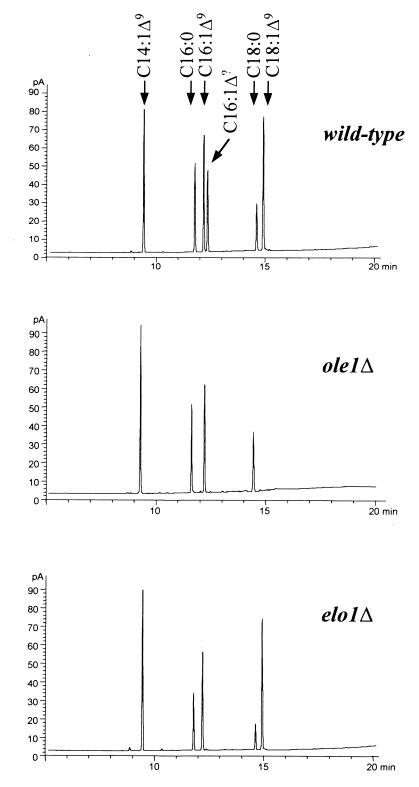
To determine the chemical nature and relative abundance of the predicted Elo1p-dependent product of medium-chain unsaturated fatty acid elongation, wild-type and ole1Δ and elo1Δ mutant strains were cultivated in C14:1-supplemented media and their fatty acid profiles were analyzed. As shown in Fig. 1, a novel peak, eluting in the range of C16 fatty acid methyl esters, was detected in wild-type and ole1Δ mutant cells. This peak was absent from the fatty acid profile of the elo1Δ mutant, indicating that synthesis of this compound was directly or indirectly dependent on Elo1p. In wild-type cells, this elongation product comprised 12.3% of all the fatty acids. In ole1Δ mutant cells, uptake and elongation of C14:1 were only slightly increased, with the elongation product accounting for 18.1% of the total fatty acids (Table 1). Under these conditions, elo1Δ mutant cells had approximately 4.5-fold-more unsaturated than saturated fatty acids and a roughly equal ratio of acyl chains with 16 carbon atoms to those with 18 carbon atoms. In comparison, the ole1Δ mutant strain displayed a ratio of unsaturated to saturated fatty acids that was intermediate between those observed in wild-type cells and those observed for elo1Δ mutant cells. The ratio of C16 to C18 fatty acids, however, was grossly perturbed in the desaturase mutant (Table 1).
FIG. 1.
Fatty acid profiles of wild-type and elo1Δ and ole1Δ mutant cells cultivated in C14:1Δ9-supplemented media. Wild-type and ole1Δ and elo1Δ mutant cells were cultivated in C14:1Δ9-supplemented minimal media to late exponential growth phase, cells were harvested, lipids were extracted, and fatty acid methyl esters were analyzed by gas chromatography as described in Materials and Methods. Positions of saturated and unsaturated fatty acids in the chromatograms are indicated. C16:1Δ? indicates the ELO1-dependent product.
TABLE 1.
Comparison of fatty acid composition of total lipids from wild-type and ole1Δ and elo1Δ mutant cells cultivated in C14:1Δ9- and C16:1Δ9-supplemented mediaa
| Fatty acid | Composition (mol%) in medium
|
|||||
|---|---|---|---|---|---|---|
| C14:1Δ9 supplemented
|
C16:1Δ9 supplemented
|
|||||
| Wild type | ole1Δ | elo1Δ | Wild type | ole1Δ | elo1Δ | |
| C14:0 | ND | ND | 0.9 ± 0.1 | 0.5 ± 0.1 | 1.3 ± 0.1 | 3.1 ± 0.3 |
| C14:1 | 21.4 ± 1.6 | 58.1 ± 4.3 | 38.7 ± 2.9 | ND | ND | ND |
| C16:0 | 13.5 ± 0.9 | 14.3 ± 2.1 | 10.4 ± 0.8 | 14.3 ± 1.2 | 22.8 ± 1.9 | 22.7 ± 2.1 |
| C16:1Δ9 | 17.5 ± 1.2 | ND | 18.6 ± 2.1 | 62.0 ± 4.6 | 69.7 ± 5.3 | 62.0 ± 4.4 |
| C16:1Δ11 | 12.3 ± 1.1 | 18.1 ± 1.3 | ND | ND | ND | ND |
| C18:0 | 7.6 ± 0.6 | 6.2 ± 0.4 | 4.9 ± 0.3 | 5.8 ± 0.4 | 4.6 ± 0.6 | 7.1 ± 0.6 |
| C18:1Δ9 | 20.1 ± 1.4 | ND | 24.3 ± 1.8 | 10.8 ± 0.9 | ND | ND |
| C18:1Δ11 | ND | ND | ND | 1.0 ± 0.9 | ND | ND |
| C18:1Δ13 | 0.5 ± 0.1 | 0.5 ± 0.1 | ND | ND | ND | ND |
| C26:0 | 5.9 ± 0.4 | 2.7 ± 0.3 | 1.8 ± 0.2 | 5.1 ± 0.4 | 1.5 ± 0.2 | 5.2 ± 0.4 |
| Ratio | ||||||
| Unsat./sat. | 2.7 | 3.3 | 4.5 | 2.9 | 2.3 | 1.9 |
| C16:x/C18:x | 1.5 | 4.8 | 1.0 | 4.3 | 20.1 | 11.9 |
Cells were grown in C14:1Δ9- or C16:1Δ9-supplemented media to late exponential growth phase. Lipids were extracted and subjected to alkaline hydrolysis, and fatty acids were converted to methyl esters by BF3-catalyzed esterification. Fatty acid methyl esters were separated by gas chromatography as described in Materials and Methods. Ratios of unsaturated to saturated fatty acids (Unsat./sat.) and that of 16-carbon-atom-chain-length to 18-carbon-atom-chain-length fatty acids are indicated. Values represent means ± standard deviations of three independent determinations. ND, not detected.
Comparison of growth rates of wild-type and elo1Δ and ole1Δ mutant cells in media supplemented with either C14:1 or C16:1 indicated that elongation of C14:1 was not growth limiting for ole1Δ mutant cells, suggesting that uptake and elongation of the supplemented C14:1 are rapid and efficient. ole1Δ mutant cells required 207 min for doubling in C14:1-supplemented media, compared to 198 min in C16:1-supplemented media (data not shown).
Elo1p is required for carboxy-terminal elongation of unsaturated fatty acids.
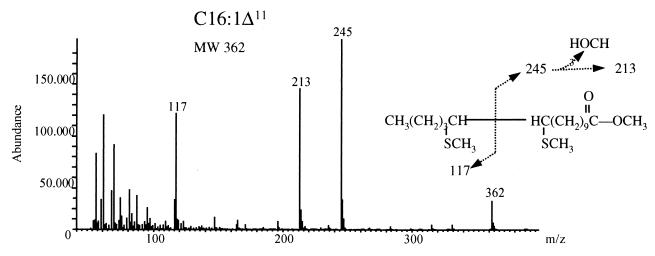
The position of the double bond in the elongation product was determined by GLC-MS analysis of fatty acid dimethyl disulfide adducts, as previously described (37). Fragmentation of C16:1Δ? into two main products of m/z 117 and 245 allowed unequivocal identification of the Elo1p-dependent products as being a C16:1Δ11 fatty acid, indicating that Elo1p is required for α-elongation of C14:1Δ9 to C16:1Δ11 (Fig. 2). Furthermore, fragmentation of the Elo1p-dependent product was identical to that of a chemically synthesized C16:1Δ11 fatty acid. In contrast, GLC-MS analysis of an authentic C16:1Δ9 fatty acid methyl ester yielded major products at m/z 217 and 145, consistent with the predicted fragmentation properties of C16:1Δ9 fatty acid (data not shown).
FIG. 2.
Characterization of the double bond position present in the ELO1-dependent product of C14:1Δ9, C16:1Δ11. Dimethyl disulfide adducts of fatty acid methyl esters were prepared and analyzed as described in Materials and Methods.
To determine the substrate specificity of the elongation process that is operating on unsaturated fatty acids, wild-type and ole1Δ and elo1Δ mutant cells were cultivated in C16:1Δ9-supplemented media and their fatty acid profiles were analyzed. A novel peak in the C18:1 region of the chromatogram was apparent in lipid extracts from wild-type cells. This peak was not detected in lipid extracts from elo1Δ mutant cells, suggesting that its synthesis depends on the presence of a functional elongase. Determination of the double bond position in this elongation product by GLC-MS analysis of fatty acid dimethyl disulfide adducts allowed unequivocal identification of the product as C18:1Δ11 (data not shown). As shown in Table 1, elongation efficiency of C16:1 in vivo is approximately 12-fold lower than that of C14:1. Only 1% of C18:1Δ11 was detected in wild-type cells grown in the presence of C16:1Δ9, compared to 12.3% of C16:1Δ11 that was synthesized by the elongation of C14:1 (Table 1). The notion that C16:1 is a poorer substrate for Elo1p-dependent elongation is also apparent from the fact that only 0.5% of C18:1Δ13 was detected in cells supplemented with C14:1, in which case elongation of C16:1Δ11 by a second cycle generates a C18:1Δ13 fatty acid.
The fatty acid profile of cells supplemented with C16:1Δ9 (Table 1) furthermore revealed that all three strains appeared to maintain a ratio of unsaturated to saturated fatty acids of between 2 and 3. The ratio of C16 to C18 fatty acids, however, was grossly disturbed in the two mutants. Under these conditions, the endogenous synthesis of C18:1Δ9 appeared to be strongly repressed in the elo1Δ mutant but not in the wild type, suggesting that Ole1p activity is tightly downregulated by C16:1Δ9 (3), but not by C14:1Δ9, and that this repression is somewhat relieved by the presence of functional Elo1p.
Effects of altered fatty acid profile on the phospholipid composition.
Next, we wished to determine whether an altered fatty acid composition, generated by cultivation of the yeast strains in the presence of Δ9-unsaturated fatty acids, affected the phospholipid composition of these cells. For this purpose, phospholipids of wild-type and ole1Δ and elo1Δ mutant cells cultivated in either C14:1Δ9- or C16:1Δ9-supplemented media were analyzed. As shown in Table 2, phospholipid compositions of wild-type and mutant cells deviated greatly from each other and were strongly dependent on the chain length of the unsaturated fatty acid supplied. The most notable observations were as follows. (i) In wild-type cells, the relative content of the two major phospholipid classes, PtdEtn and PtdCho, was strongly dependent on the type of unsaturated fatty acid supplemented. While PtdEtn was the most abundant phospholipid (74.5%) in wild-type cells supplemented with C14:1, PtdEtn levels were reduced to 38.9% when cells were supplemented with C16:1. Concomitantly, PtdCho increased from 17.9% in C14:1-supplemented cells to 48.7% in cells supplemented with C16:1. This tendency toward an increased content of PtdEtn at the expense of PtdCho in cells supplemented with C14:1 was also observed for the ole1Δ mutant. (ii) Furthermore, in ole1Δ mutant cells grown in C14:1-supplemented media, the relative content of phosphatidic acid (PtdOH) was greatly elevated (∼10-fold compared to the wild type), suggesting that the unfavorable fatty acid composition of these cells results in increased steady-state levels of PtdOH, possibly as a result of an altered phospholipid turnover. (iii) In all three strains, the relative content of PtdIns was lower in C14:1-supplemented cells than in C16:1-supplemented cells. This chain-length-dependent decrease of PtdIns was most pronounced (eight-fold) in the elo1Δ mutant. (iv) PtdSer levels appeared to be least affected by chain length alterations or mutations in ELO1 and OLE1.
TABLE 2.
Comparison of the phospholipid composition of wild-type and ole1Δ and elo1Δ mutant cells cultivated in C14:1Δ9- and C16:1Δ9-supplemented mediaa
| Phospholipid | Composition (mol%) of cell type in medium
|
|||||
|---|---|---|---|---|---|---|
| Wild type
|
ole1Δ
|
elo1Δ
|
||||
| C14:1Δ9 | C16:1Δ9 | C14:1Δ9 | C16:1Δ9 | C14:1Δ9 | C16:1Δ9 | |
| PtdSer | 1.3 ± 0.1 | 3.1 ± 0.4 | 5.9 ± 0.9 | 1.6 ± 0.1 | 2.1 ± 0.2 | 1.3 ± 0.4 |
| PtdEtn | 74.5 ± 4.6 | 38.9 ± 2.2 | 36.7 ± 1.8 | 29.3 ± 3.7 | 42.6 ± 0.3 | 21.8 ± 4.2 |
| PtdCho | 17.9 ± 3.6 | 48.7 ± 1.8 | 27.0 ± 2.7 | 51.3 ± 0.8 | 50.4 ± 0.9 | 45.9 ± 6.2 |
| PtdIns | 3.7 ± 0.6 | 6.0 ± 0.4 | 8.2 ± 3.9 | 11.5 ± 0.7 | 2.6 ± 0.6 | 20.2 ± 2.3 |
| PtdOH | 1.3 ± 0.3 | 0.5 ± 0.1 | 14.2 ± 1.0 | 3.8 ± 2.9 | 0.6 ± 0.1 | 5.3 ± 1.4 |
| CLb | 1.4 ± 0.3 | 2.9 ± 0.6 | 7.9 ± 2.0 | 2.6 ± 0.8 | 0.6 ± 0.2 | 5.5 ± 2.9 |
Cells were grown in either C14:1Δ9- or C16:1Δ9-supplemented media to late exponential growth phase. Lipids were extracted, and individual phospholipid classes were separated by two-dimensional thin-layer chromatography and quantified as described in Materials and Methods. Values represent means ± standard deviations of three independent determinations.
CL, cardiolipin.
On one hand, this analysis illustrates the flexibility of the cellular phospholipid composition; on the other hand, it demonstrates that the phospholipid composition is greatly affected by the availability of acyl chains. These data suggest, in fact, that acyl chain supply is a dominant determinant of the phospholipid composition of a cellular membrane, suggesting that the lipid head group composition is more modulatory. This notion is supported by biophysical data showing that the introduction of a single double bond decreases the melting temperature of dipalmitoyl-PtdCho by 46°C. Head group exchange, on the other hand, appears to affect the phase behavior in a degree that is more comparable to that exerted by a two-carbon increase in acyl chain length (11). While lipid phase properties are certainly not the sole parameters in vivo, additional variables such as charge, size, and polarity of the lipid head group; the intrinsic curvature of a lipid species; and even superlattice formation may all significantly affect the head group composition of cellular lipid bilayers (2, 28, 31).
Analysis of the distribution of the elongation product in individual phospholipid classes.
Having analyzed the dependency of the phospholipid class composition on the acyl chain length of the supplemented unsaturated fatty acid, we tested whether incorporation of the C16:1Δ11 elongation product displayed a preference for certain phospholipid classes. Therefore, the fatty acid composition of individual phospholipid classes, isolated from wild-type and ole1Δ and elo1Δ mutant cells cultivated in media supplemented with C14:1Δ9, was determined. As shown in Table 3, this analysis revealed that, in ole1Δ mutant cells, the C16:1Δ11 elongation product was incorporated in all of the phospholipid classes, with the highest relative content in PtdCho (18.8%) and PtdEtn (15.8%), compared to PtdSer (8.7%) and PdtIns (6.1%). This enrichment of unsaturated C16:1Δ11 in order from PtdIns and PtdSer via PtdEtn to PtdCho parallels the general increase in the content of unsaturated acyl chains from PtdIns via PtdSer and PtdEtn to PtdCho observed both in C14:1-supplemented cells and in wild-type cells grown in rich medium (25, 34), an observation which suggests that the relative degree of acyl chain saturation of the different phospholipid classes is maintained even under conditions where the endogenous acyl coenzyme A pool is grossly altered.
TABLE 3.
Fatty acid composition of individual phospholipid classes from wild-type and ole1Δ and elo1Δ mutant cells cultivated in C14:1Δ9-supplemented mediaa
| Fatty acid | Composition (mol%) of phospholipid from cell type
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PtdSer
|
PtdEtn
|
PtdCho
|
PtdIns
|
|||||||||
| Wild type | ole1Δ | elo1Δ | Wild type | ole1Δ | elo1Δ | Wild type | ole1Δ | elo1Δ | Wild type | ole1Δ | elo1Δ | |
| 14:1Δ9 | ND | 14.6 | ND | 8.0 | 36.6 | 20.1 | 24.5 | 42.8 | 37.2 | 11.1 | 27.8 | 7.4 |
| 16:0 | 42.3 | 72.2 | 50.8 | 18.5 | 44.4 | 19.6 | 17.0 | 22.7 | 15.3 | 27.3 | 37.5 | 31.3 |
| 16:1Δ9 | 12.2 | ND | 4.9 | 30.6 | ND | 27.8 | 17.6 | ND | 21.3 | 6.9 | ND | 6.1 |
| 16:1Δ11 | 2.7 | 8.7 | ND | 7.9 | 15.8 | ND | 15.8 | 18.8 | ND | 5.8 | 6.1 | ND |
| 18:0 | 3.2 | 4.5 | 4.2 | 1.7 | 2.5 | 1.3 | 9.0 | 15.8 | 7.8 | 26.4 | 28.0 | 32.7 |
| 18:1Δ9 | 39.6 | ND | 40.0 | 33.3 | ND | 31.2 | 16.1 | ND | 18.4 | 21.9 | ND | 22.1 |
| Acyl chain saturation | 45.5 | 76.7 | 55.0 | 21.0 | 46.9 | 20.9 | 26.0 | 38.5 | 23.1 | 53.7 | 65.5 | 64.0 |
| 16:x | 57.2 | 80.9 | 55.7 | 57.0 | 60.2 | 47.4 | 50.4 | 41.5 | 36.6 | 40.0 | 43.6 | 37.4 |
Cells were grown in C14:1Δ9-supplemented media to late exponential growth phase. Lipids were extracted, individual phospholipid classes were separated by two-dimensional thin-layer chromatography and subjected to alkaline hydrolysis, and fatty acids were converted to methyl esters by BF3-catalyzed methanolysis. Fatty acid methyl esters were separated by gas chromatography as described in Materials and Methods. Values represent means of two determinations with standard deviations between experiments of less than 10%. The degree of acyl chain saturation and that of acyl chains 16 carbon atoms in length (C16:x) are indicated. ND, not detected.
The ratio of C16:1Δ9 to C16:1Δ11 in the different phospholipid classes, however, varied markedly. In wild-type cells, PtdSer and PtdEtn contained approximately fourfold-more C16:1Δ9 than C16:1Δ11. In PtdCho and PtdIns, on the other hand, the ratio of C16:1Δ9 to C16:1Δ11 was close to 1. This apparent acyl chain preference of different phospholipid classes is particularly noteworthy in light of the fact that both PtdSer and PtdIns are synthesized from a common precursor, CDP-DAG. The observation that PtdEtn has a C16:1Δ9-to-C16:1Δ11 ratio similar to that of PtdSer suggests that the bulk of PtdEtn is synthesized by decarboxylation of PtdSer (i.e., the de novo pathway). In contrast, the similarity in the C16:1Δ9-to-C16:1Δ11 ratio of PtdCho and PtdIns suggests that, in this case, the bulk of PtdCho is synthesized through the Kennedy (CDP-choline) pathway, possibly by reutilizing molecular species of DAG that are produced from the turnover of PtdIns. Turnover of PtdIns is required for the maturation of sphingolipids, in which the synthesis of inositolphosphorylceramide from ceramide and that of mannosyl-diinositolphosphorylceramide from mannosylinositol-phosphorylceramide each consumes PtdIns and yields DAG (1). Interestingly, a recent study of the molecular species of PtdCho synthesized in hepatocytes either through the de novo methylation pathway or through the Kennedy pathway revealed that different molecular species of PtdCho are produced by the two pathways, with saturated species predominating in the Kennedy pathway and arachidonic-acid-containing species predominating in the methylation pathway (16). Remarkably, these two classes of PtdCho species appear to be functionally distinct, since overexpression of the PtdEtn N-methyltransferase-2 in conditional mutants of the CDP-choline pathway does not rescue the temperature-sensitive growth defect of these cells (19). A functional distinction of PtdCho synthesized by the de novo pathway from that formed by the Kennedy pathway is also evident in yeast, where attenuation of PtdCho synthesis via the Kennedy pathway, but not via the methylation pathway, can rescue the temperature-sensitive growth phenotype of sec14 mutants (7, 20, 22).
Interestingly, the supplemented C14:1 was not detected in PtdSer species of wild-type and elo1Δ mutant cells but was incorporated to 14.6% in PtdSer of ole1Δ cells (Table 3). This apparent exclusion of C14:1 from PtdSer in desaturation-competent cells is remarkable and may reflect the preference and/or restricted accessibility of PtdSer synthase for/to certain molecular species of CDP-DAG (12). In ole1Δ cells, on the other hand, this species preference appears to have been overcome, possibly due to an acute requirement of the cell to increase the degree of lipid unsaturation (Table 1).
Conclusions.
Taken together, the present study provides evidence that the yeast ELO1 gene is required for carboxy-terminal elongation of unsaturated fatty acids. Since the two very- long-chain elongases, Elo2p and Elo3p, are closely related to Elo1p, with more than 52% identity and 72% homology (24), it appears reasonable to suggest that these operate by a similar mechanism. The second important observation made in this study is that the exogenous addition of unsaturated fatty acids results in a dramatic alteration in the phospholipid composition, suggesting that acyl chain supply is a dominant parameter of the lipid composition. Altering the cellular acyl chain pool, however, appears not to affect the degree of acyl chain saturation characteristic of each phospholipid class (25, 34). The third and possibly most interesting observation is that the C16:1Δ9-to-C16:1Δ11 ratio of individual phospholipid classes may be used to trace the biosynthetic origin of the corresponding lipid species. Experiments to validate this approach utilizing mutants in the de novo pathway and those defective in the Kennedy pathway are currently being performed.
ACKNOWLEDGMENTS
We thank G. Daum for liberal support and valuable comments on the manuscript, C. Martin for generously providing the elo1Δ and ole1Δ mutant strains used throughout this study, and D. Ribitsch for synthesizing the C16:1Δ11 fatty acid employed as a standard for the GLC-MS analysis.
This work was supported by grants from the Fonds zur Förderung der wissenschaftlichen Forschung in Österreich P11731 (to S.D.K.) and M00304 and 13767 (to R.S.) and the Swiss National Science Foundation (823A-046702 to R.S.).
REFERENCES
- 1.Becker G W, Lester R L. Biosynthesis of phosphoinositol-containing sphingolipids from phosphatidylinositol by a membrane preparation from Saccharomyces cerevisiae. J Bacteriol. 1980;142:747–754. doi: 10.1128/jb.142.3.747-754.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloom M, Evans E, Mouritsen O G. Physical properties of the fluid lipid-bilayer component of cell membranes: a perspective. Q Rev Biophys. 1991;24:293–397. doi: 10.1017/s0033583500003735. [DOI] [PubMed] [Google Scholar]
- 3.Bossie M A, Martin C E. Nutritional regulation of yeast delta-9 fatty acid desaturase activity. J Bacteriol. 1989;171:6409–6413. doi: 10.1128/jb.171.12.6409-6413.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broekhuyse R M. Phospholipids in tissues of the eye. Isolation, characterization and quantitative analysis by two-dimensional thin-layer chromatography of diacyl-ether phospholipids. Biochim Biophys Acta. 1968;260:449–459. doi: 10.1016/0005-2760(68)90038-6. [DOI] [PubMed] [Google Scholar]
- 5.Browse J, Somerville C. Glycerolipid synthesis: biochemistry and regulation. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:467–506. [Google Scholar]
- 6.Carman G M, Henry S A. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog Lipid Res. 1999;38:361–399. doi: 10.1016/s0163-7827(99)00010-7. [DOI] [PubMed] [Google Scholar]
- 7.Cleves A E, McGee T P, Whitters E A, Champion K M, Aitken J R, Dowhan W, Goebl M, Bankaitis V A. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 1991;64:789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook H W. Fatty acid desaturation and chain elongation in eukaryotes. New Compr Biochem. 1996;31:129–152. [Google Scholar]
- 9.Cottrell M, Viljoen B C, Kock J L F, Lategan P M. The long-chain fatty acid compositions of species representing the genera Saccharomyces, Schanniomyces and Lipomyces. J Gen Microbiol. 1986;132:2401–2403. [Google Scholar]
- 10.Cullis P R, de Kruijff B. Lipid polymorphism and the functional roles of lipids in biological membranes. Biochim Biophys Acta. 1979;559:399–420. doi: 10.1016/0304-4157(79)90012-1. [DOI] [PubMed] [Google Scholar]
- 11.Cullis P R, Fenske D B, Hope M J. Physical properties and functional roles of lipids in membranes. New Compr Biochem. 1996;31:1–33. [Google Scholar]
- 12.Daum G, Lees N D, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 13.Daum G, Tuller G, Nemec T, Hrastnik C, Balliano G, Cattel L, Milla P, Rocco F, Conzelmann A, Vionnet C, Kelly D E, Kelly S, Schweizer E, Schuller H J, Hojad U, Greiner E, Finger K. Systematic analysis of yeast strains with possible defects in lipid metabolism. Yeast. 1999;15:601–614. doi: 10.1002/(SICI)1097-0061(199905)15:7<601::AID-YEA390>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 14.David D, Sundarababu S, Gerst J E. Involvement of long chain fatty acid elongation in the trafficking of secretory vesicles in yeast. J Cell Biol. 1998;143:1167–1182. doi: 10.1083/jcb.143.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Kruijff B. Lipid polymorphism and biomembrane function. Curr Opin Chem Biol. 1997;1:564–569. doi: 10.1016/s1367-5931(97)80053-1. [DOI] [PubMed] [Google Scholar]
- 16.DeLong C J, Shen Y J, Thomas M J, Cui Z. Molecular distinction of phosphatidylcholine synthesis between the CDP-choline pathway and phosphatidylethanolamine methylation pathway. J Biol Chem. 1999;274:29683–29688. doi: 10.1074/jbc.274.42.29683. [DOI] [PubMed] [Google Scholar]
- 17.Dittrich F, Zajonc D, Hühne K, Hoja U, Ekici A, Greiner E, Klein H, Hofmann J, Bessoule J J, Sperling P, Schweizer E. Fatty acid elongation in yeast—biochemical characteristics of the enzyme system and isolation of elongation-defective mutants. Eur J Biochem. 1998;252:477–485. doi: 10.1046/j.1432-1327.1998.2520477.x. [DOI] [PubMed] [Google Scholar]
- 18.Folch J, Lees M, Sloane-Stanley G H. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 19.Houweling M, Cui Z, Vance D E. Expression of phosphatidylethanolamine N-methyltransferase-2 cannot compensate for an impaired CDP-choline pathway in mutant Chinese hamster ovary cells. J Biol Chem. 1995;270:16277–16282. doi: 10.1074/jbc.270.27.16277. [DOI] [PubMed] [Google Scholar]
- 20.Kearns B G, Alb J G, Bankaitis V A. Phosphatidylinositol transfer proteins: the long and winding road to physiological function. Trends Cell Biol. 1998;8:276–282. doi: 10.1016/s0962-8924(98)01281-1. [DOI] [PubMed] [Google Scholar]
- 21.Kent C. Eukaryotic phospholipid biosynthesis. Annu Rev Biochem. 1995;64:315–343. doi: 10.1146/annurev.bi.64.070195.001531. [DOI] [PubMed] [Google Scholar]
- 22.Kent C, Carman G M. Interactions among pathways for phosphatidylcholine metabolism, CTP synthesis and secretion through the Golgi apparatus. Trends Biochem Sci. 1999;24:146–150. doi: 10.1016/s0968-0004(99)01365-1. [DOI] [PubMed] [Google Scholar]
- 23.Morrison W R, Smith L M. Preparation of fatty acid methyl esters and dimethylacetals from lipids with boron fluoride-methanol. J Lipid Res. 1964;5:600–608. [PubMed] [Google Scholar]
- 24.Oh C S, Toke D A, Mandala S, Martin C E. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- 25.Schneiter R, Brugger B, Sandhoff R, Zellnig G, Leber A, Lampl M, Athenstaedt K, Hrastnik C, Eder S, Daum G, Paltauf F, Wieland F T, Kohlwein S D. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schweizer E. Genetics of fatty acid biosynthesis in yeast. New Compr Biochem. 1984;7:59–83. [Google Scholar]
- 27.Sherman F, Fink G R, Hicks J B N. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 28.Somerharju P, Virtanen J A, Cheng K H. Lateral organisation of membrane lipids. The superlattice view. Biochim Biophys Acta. 1999;1440:32–48. doi: 10.1016/s1388-1981(99)00106-7. [DOI] [PubMed] [Google Scholar]
- 29.Stukey J E, McDonough V M, Martin C E. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J Biol Chem. 1989;264:16537–16544. [PubMed] [Google Scholar]
- 30.Stukey J E, McDonough V M, Martin C E. The OLE1 gene of Saccharomyces cerevisiae encodes the delta 9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J Biol Chem. 1990;265:20144–20149. [PubMed] [Google Scholar]
- 31.Tate M W, Eikenberry E F, Turner D C, Shyamsunder E, Gruner S M. Nonbilayer phase of membrane lipids. Chem Phys Lipids. 1991;57:147–164. doi: 10.1016/0009-3084(91)90073-k. [DOI] [PubMed] [Google Scholar]
- 32.Toke D A, Martin C E. Isolation and characterization of a gene affecting fatty acid elongation in Saccharomyces cerevisiae. J Biol Chem. 1996;271:18413–18422. doi: 10.1074/jbc.271.31.18413. [DOI] [PubMed] [Google Scholar]
- 33.Tuller G, Nemec T, Hrastnik C, Daum G. Lipid composition of subcellular membranes of an FY1679-derived haploid yeast wild-type strain grown on different carbon sources. Yeast. 1999;15:1555–1564. doi: 10.1002/(SICI)1097-0061(199910)15:14<1555::AID-YEA479>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 34.Wagner S, Paltauf F. Generation of glycerophospholipid molecular species in the yeast Saccharomyces cerevisiae. Fatty acid pattern of phospholipid classes and selective acyl turnover at sn-1 and sn-2 positions. Yeast. 1994;10:1429–1437. doi: 10.1002/yea.320101106. [DOI] [PubMed] [Google Scholar]
- 35.Wakil S J. Fatty acid synthase, a proficient multifunctional enzyme. Biochemistry. 1989;28:4523–4530. doi: 10.1021/bi00437a001. [DOI] [PubMed] [Google Scholar]
- 36.Welch J W, Burlingame A L. Very long-chain fatty acids in yeast. J Bacteriol. 1973;115:464–466. doi: 10.1128/jb.115.1.464-466.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto K, Shibahara A, Nakayama T, Kajimoto G. Determination of double-bond positions in methylene-interrupted dienoic fatty acids by GC-MS as their dimethyl disulfide adducts. Chem Phys Lipids. 1991;60:39–50. [Google Scholar]