Abstract
The ability of skeletal muscle to regenerate from injury is crucial for locomotion, metabolic health, and quality of life. Peroxisome proliferator-activated receptor-γ coactivator-1α (PGC1A) is a transcriptional coactivator required for mitochondrial biogenesis. Increased mitochondrial biogenesis is associated with improved muscle cell differentiation, however PGC1A's role in skeletal muscle regeneration following damage requires further investigation. The purpose of this study was to investigate the role of skeletal muscle-specific PGC1A overexpression during regeneration following damage. 22 C57BL/6J (WT) and 26 PGC1A muscle transgenic (A1) mice were injected with either phosphate-buffered saline (PBS, uninjured control) or Bupivacaine (MAR, injured) into their tibialis anterior (TA) muscle to induce skeletal muscle damage. TA muscles were extracted 3- or 28-days post-injury and analyzed for markers of regenerative myogenesis and protein turnover. Pgc1a mRNA was ∼10–20 fold greater in A1 mice. Markers of protein synthesis, AKT and 4EBP1, displayed decreases in A1 mice compared to WT at both timepoints indicating a decreased protein synthetic response. Myod mRNA was ∼75% lower compared to WT 3 days post-injection. WT mice exhibited decreased cross-sectional area of the TA muscle at 28 days post-injection with bupivacaine compared to all other groups. PGC1A overexpression modifies the myogenic response during regeneration.
Keywords: PGC-1α, Skeletal muscle, Muscle regeneration, Satellite cells, Protein turnover, p38 MAPK
Introduction
Skeletal muscle comprises ∼40% of total body mass1 and is critical for locomotion and skeletal muscle health.2 Skeletal muscle possesses an innate ability to regenerate from damage.3 Skeletal muscle regeneration is a tightly regulated process that involves the coordination of several processes including degeneration/necrosis, inflammation, regeneration, remodeling, and maturation. The initial degeneration/inflammatory response—which occurs in the first 1–2 days following muscle damage—results in neutrophil and macrophage infiltration to remove damaged and necrotic tissue.4, 5, 6 Following this inflammatory response is a period of accelerated activity for many cell types including satellite cells and myoblasts.7,8 Muscle stem cells, also known as satellite cells, begin the process of myogenic proliferation from their quiescent state, differentiate into myoblasts, and ultimately fuse with the damaged myofibers. Myoblasts reach a peak in numbers approximately 2–3 days post-injury7,8 which coincides with increased gene expression of the myogenic regulatory factor Myod. Skeletal muscle regeneration is resolved 3–4 weeks following damage. Improper regeneration of skeletal muscle—such as in muscular dystrophy—leads to decreases in muscle mass and quality and is associated with increased rates of morbidity and mortality.9,10 Recapitulation of the myogenic program during skeletal muscle regeneration requires the coordinated regulation of energy metabolism. In fact, several genes related to energy production are altered in myogenic precursor cells and during skeletal muscle regeneration.11, 12, 13, 14, 15 Proliferating cells' energy requirement increases must be met. Due to the importance of skeletal muscle to overall health and activities of daily living, understanding and evaluating potential biological avenues for improved regeneration and maintenance of skeletal muscle have received significant interest in recent literature.16,17
Mitochondrial biogenesis is stimulated during skeletal muscle regeneration and is associated with the differentiation of myoblasts into myotubes.18 In fact, when mitochondrial protein synthesis is inhibited during the regenerative response, skeletal muscle exhibits smaller muscle fibers and increased connective tissue deposition indicating an improper regenerative response.19 Peroxisome proliferator-activated receptor-γ coactivator-1α (PGC1A) is a transcriptional coactivator and the master regulator of mitochondrial biogenesis and results in marked improvements in mitochondrial content and oxidative capacity of skeletal muscle.20 PGC1A promotes increases in mitochondrial content, respiratory capacity, oxidative phosphorylation, conversion of low oxidative fibers to high oxidative fibers in mouse skeletal muscle, and induction of mitochondrial quality control regulators.21 Genetic deletion of PGC1A from skeletal muscle results in reduced oxidative capacity and decreased mitochondrial content.22,23 Following treadmill running these mice exhibited increased inflammation and increases incidence of damaged muscle fibers suggesting that PGC1A plays a role in fiber integrity.24 Prior work from Dinulovic et al.25 suggests PGC1A overexpression may accelerate the resolution of inflammation and necrosis following cardiotoxin-induced injury. However, the role of PGC1A in modulating protein turnover and myogenic responses of skeletal muscle during a regenerative phase is incompletely understood.
Therefore, the purpose of this study was to investigate skeletal muscle-specific overexpression of PGC1A on skeletal muscle regeneration following damage. We hypothesized that PGC1A overexpression would aid in the regeneration of skeletal muscle through a more favorable myogenic response. We demonstrate skeletal muscle-specific overexpression of PGC1A alters myogenic regulatory factors (MRFs) and exhibits a protective effect on skeletal muscle by preserving muscle fiber cross-sectional area. These data suggest PGC1A may play a pivotal role in skeletal muscle regeneration, and further studies to evaluate the therapeutic potential of PGC1A on muscle mass maintenance are required.
Methods
Animals
Forty-eight female muscle-specific PGC1A transgenic (A1) mice (n = 26) and their WT litter mates (n = 22) were used in this study. The A1 mice were bred from a C57BL/6 background and overexpressing the pgc1a gene under the control of the muscle creatine kinase promoter (MCK-PGC-1α), as we have previously utilized.26 Breeders were a generous gift of Dr. Bruce Spiegelman. A1 and WT mice were randomly assigned to either uninjured (UNINJ or PBS) or injured groups (INJ or MAR). Animals were housed in the University of Arkansas Central Laboratory Animal Facility. Animals were kept on a 12:12-h light-dark cycle with ad libitum access to normal chow and water throughout the study. All methods used in this study were approved by the University of Arkansas Institutional Animal Care and Use Committee (IACUC), Approval Form Reference #13020.
PBS and bupivacaine injection
At 12 weeks of age, mice were anesthetized with a cocktail of ketamine hydrochloride (45 mg/kg body weight), acepromazine (1 mg/kg body weight) and xylazine (3 mg/kg body weight).27 Mice were then injected with 0.03 mL of either phosphate buffered saline (UNINJ or PBS) or 0.75% bupivacaine, also known as Marcaine (INJ or MAR), into the left and right tibialis anterior (TA) muscle using a 25-gauge and 5/8 needle along the longitudinal axis of the muscle, as previously described12 to induce muscle damage. This is a well-established and extensively used model to induce skeletal muscle damage.7,12,25,28,29
Tibialis anterior and tibia extraction
Either 3 days or 28 days post-injection, mice were anesthetized, and the TA and tibias were extracted. This created four groups at both the 3 days and 28 days of damage-recovery time points for this study, respectively: WT-PBS (n = 5; n = 5), WT-MAR (n = 8; n = 6), A1-PBS (n = 7; n = 7), and A1-MAR (n = 6; n = 6). The left TA muscle was frozen in liquid nitrogen and stored at −80 °C for protein and mRNA abundance analysis. The right TA muscle was submerged in optimum cutting temperature compound (OCT) and then placed in liquid nitrogen cooled isopentane. OCT mounted tissue was then stored at −80 °C for morphological analysis. Following removal of the TA, tibias of both legs were extracted and measured via caliper. Tibia measurements were used to normalize muscle weights to estimate total body size.
RNA isolation, cDNA synthesis, and quantitative Real-Time PCR
RNA was extracted from the TA muscle using a Trizol reagent (Thermo Fisher Scientific, Waltham, MA, USA) as previously described. 12 TA muscle was submerged in Trizol and homogenized. RNA was then isolated, treated with DNase and the total RNA concentration was measured using 260/280 UV Spec ratio via Bio-Tek Power Wave XS plate reader (Winoski, VT) with Take3 microvolume plate and Gen5 software. cDNA was reversed transcribed from 1 μg of total RNA using the Superscript Vilo cDNA synthesis kit (Thermo Fisher Scientific #11756050, Waltham, MA, USA). Real-time PCR from cDNA was performed, and results were analyzed by StepOne Real-Time PCR system (Life Technologies, Applied Biosystems, Grand Island, NY). cDNA was amplified in a 25 μL reaction containing desired probes and Taqman Universal Mastermix (Applied Biosystems). Samples were incubated at 95 °C for 4 min, followed by 40 cycles of denaturation, annealing and extension at 95 °C, 55 °C and 72 °C, respectively. Fluorescence was measured at the end of the extension step of each cycle. Fluorescent probes for 18S (Mm03928990_g1), Pgc1a (Mm00447183_m1), Ldha (Mm01612132_g1), Ldhb (Mm01267402_m1), Tnfa (Mm00443258_m1), Cyclin D1 (Mm00432359_m1), Myod (Mm01203489_g1), Myogenin (Mm00446194_m1), Atrogin1 (Mm00499523_m1), Murf1 (Mm01185221_m1), Chop (Mm00492097_m1), and Trb3 (Mm00454879_m1) were purchased from Applied Biosystems. RT-qPCR measured cycle threshold (Ct) and the ΔCt value was calculated as the difference between the Ct value and the 18s Ct value. Final quantification of mRNA abundance was calculated using the ΔΔCT method Ct = [ΔCt(calibrator) – ΔCt(sample)]. Relative quantifications were then calculated as 2−ΔΔCt, and 18S control genes displayed no differences across all treatment groups.
Western blotting
The left TA muscle was homogenized as previously described,12,30,31 with subsequent protein concentration measured using the RC/DC assay (500-0119, BioRad, Hercules, CA) and samples diluted to yield 40 μg total protein. Homogenate was extracted and fractioned into 6%, 10%, or a gradient gel of 6%, 10%, and 15% SDS-polyacrylamide gels. Equal loading of the gels was verified by Ponceau staining on Polyvinylidene fluoride (PVDF) membranes. Membranes were blocked using either 5% dry milk or 5% BSA solution, depending on the antibody used, containing Tris-buffered saline (TBS) with 0.1% Tween®20 (TBST) for 2 h. Primary antibodies for p-p38 (Thr180/Tyr182) (#9211), p38 (pan) (#9212), NFκB p65 (pan) (#8242), p-Akt (Ser473) (#9271), Akt (pan) (#4691), p-4EBP-1 (Thr37/46) (#9459), and 4EBP-1 (pan) (#9644) were purchased from Cell Signaling (MA, US) and were diluted 1:1 000 in 5% milk, in TBST and incubated at 4 °C overnight. Anti-mouse or anti-rabbit secondary antibody (Cell Signaling) was diluted 1:2 000 in 5% milk, in TBST and incubated at room temperature for 1 h. Enhanced Chemiluminescence (ECL) was performed using Fluorchem M Imager (Protein Simple, Santa Clara, California) to observe antibody-antigen interaction. Quantification of blotting images was performed through densitometry analysis using Alphaview software (Protein Simple). Ponceau stains were digitally scanned, and 45 kDa actin bands were quantified by densitometry to be used as a protein loading correction factor for each lane. To account for differences across multiple gels, a sample was run on all gels so that a correction factor could be made to account for gel to gel differences.
Histology
The right TA muscle was sectioned at 10 μm thickness using a Leica CM1859 clinical cryostat (Leica Biosystems, Buffalo Grove, IL). Each TA section was stained with Hematoxylin and Eosin, imaged with a Nikon camera (Sight DS-Vi1) mounted on an Olympus CKX41 inverted microscope at 20× magnification (Olympus, Pennsylvania, USA), and analyzed with Nikon NIS Elements BR software package (Nikon, Melville, NY). Fiber distribution and cross-sectional area (CSA) were performed as previously described.31 Each fiber was manually traced, and the number of pixels was calibrated to obtain CSA of the muscle. Approximately 150 fibers were traced per sample.
Statistical analyses
All data were calculated using Statistical Package for the Social Sciences (SPSS version 23.0, Armonk, NY). Results are reported as mean ± standard deviation (SD). Normality of data was established via a Shapiro Wilk test prior to any parametric analysis. A two-way ANOVA with factors of genotype (WT vs. A1) and muscle damage (PBS vs. MAR) was performed following data to analyze the main effects of genotype and injury as well as any interactions within the 3 and 28 day timepoints. Where significance was detected, Fisher's LSD post-hoc analysis was conducted. Chi-squared analysis was used to determine frequency differences between groups for cross-sectional areas. Statistical significance was determined if p ≤ 0.05.
Results
Animal weights
Descriptive measurements for body weight, tibialis anterior weight, tibia length, and tibialis anterior weight normalized to tibia length are in Table 1. For both time-points, there were no differences in body weight, tibialis anterior wet weight, or tibialis anterior weight normalized to tibia length among any of the measurements across genotype or injection group.
Table 1.
Bodyweight, tibialis anterior (TA) weight, tibia length, and tibialis anterior (TA) weight normalized to tibia length 3 days and 28 days post-injection of phosphate buffered saline (PBS) or Marcaine (MAR) in wildtype (WT) or muscle specific PGC-1α transgenic (A1) mice. Values are means ± SD. Statistical significance we set at an alpha of p ≤ 0.05.
| Group | Body Weight (g) | Tibialis Anterior (mg) | Tibia Length (mm) | TA mass/TL (mg∙mm−1) | |
|---|---|---|---|---|---|
| 3 Day | |||||
| WT | |||||
| PBS | 20.76 ± 0.93 | 38.42 ± 1.17 | 17.02 ± 0.85 | 2.26 ± 0.06 | |
| MAR | 20.61 ± 0.67 | 37.79 ± 2.90 | 17.13 ± 0.82 | 2.21 ± 0.20 | |
| A1 | |||||
| PBS | 20.96 ± 0.72 | 40.34 ± 1.20 | 17.14 ± 0.55 | 2.37 ± 0.11 | |
| MAR | 20.28 ± 1.18 | 36.73 ± 2.21 | 15.87 ± 0.22 | 2.32 ± 0.13 | |
| 28 Day | |||||
| WT | |||||
| PBS | 21.42 ± 0.73 | 40.72 ± 0.75 | 16.32 ± 0.55 | 2.53 ± 0.04 | |
| MAR | 22.02 ± 1.61 | 42.34 ± 3.52 | 16.94 ± 0.51 | 2.51 ± 0.42 | |
| A1 | |||||
| PBS | 21.98 ± 2.26 | 42.54 ± 2.53 | 16.74 ± 0.27 | 2.54 ± 0.13 | |
| MAR | 22.75 ± 1.51 | 41.86 ± 3.63 | 16.70 ± 0.57 | 2.51 ± 0.20 | |
PGC-1α and metabolic markers
There was a main effect for A1 mice to have ∼7 to 18-fold higher Pgc1a mRNA abundance compared to their WT counterpart (Fig. 1A & 1B) confirming overexpression of PGC1A. A1-INJ mice displayed a ∼30%–35% lower Pgc1a mRNA abundance compared to A1-UNINJ mice (Fig. 1A, p ≤ 0.05). There were no differences in Pgc1a mRNA abundance between UNINJ and INJ A1 mice at 28 days. There was a main effect of genotype for Lactate Dehydrogenase A (Ldha) mRNA abundance to be ∼50% lower in A1 mice compared to WT (Fig. 1C and D, p ≤ 0.05). There were no differences in Ldha mRNA abundance in WT mice at either 3 days or 28 days post-injection. There was a main effect of genotype for A1 mice to exhibit ∼4-fold and ∼6-fold greater mRNA abundance of Lactate Dehydrogenase B (Ldhb) at 3 and 28 days post-injection, respectively, compared to their WT counterparts (Fig. 1E and F, p ≤ 0.05). There were no differences in Ldhb mRNA abundance in WT mice at 3- and 28-days post-injection (Fig. 1E and F, p ≤ 0.05). A1-UNINJ mice exhibited ∼2-fold greater Ldhb mRNA abundance compared to their A1 mice compared to their A1-INJ counterpart (Fig. 1F, p ≤ 0.05). There was a main effect of genotype for a 70%–90% lower ratio of Ldha:Ldhb at both 3 and 28 days post-injection (Fig. 1G and H). There was a main effect of genotype for A1 mice to exhibit ∼50% greater abundance of Cd147 mRNA compared to WT mice at 3 days and 28 days post-injection, regardless of injection type (Fig. 1I and J, p ≤ 0.05).
Fig. 1.
mRNA abundance of transgenic identifier PGC-1α and associated phenotypic, metabolic markers at 3 days (left) and 28 days (right) post-injection in wildtype (WT) or muscle specific PGC-1α transgenic (A1) mice. (A–B) PGC-1α mRNA abundance. (C–D) LDHA mRNA abundance. (E–F) LDHB mRNA abundance. (G–H) LDHA:LDHB ratio. (I–J) CD-147 mRNA abundance. The uninjured phosphate buffered saline (PBS) group is denoted UNINJ. The injured Marcaine (MAR) group is denoted INJ. Main effects (ME) of genotype and/or injury appear on each graph when relevant, # signifies difference from genetic counterpart, ∗ signifies difference between injections within the same genotype following an interaction. Values are means ± SE. Statistical significance we set at an alpha of p ≤ 0.05.
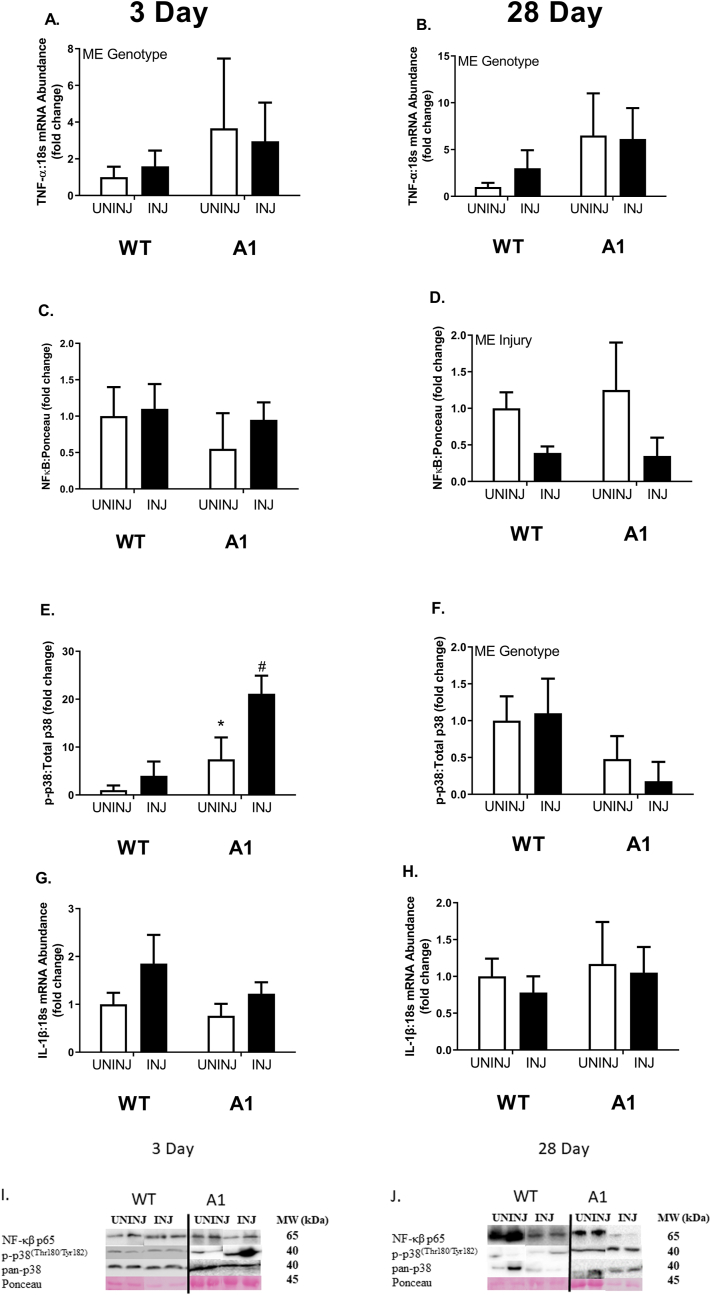
p38 and inflammatory markers
At 3 days and 28 days, there was a main effect of genotype for A1 mice to display ∼2-4-fold greater mRNA abundance of tumor necrosis factor-α (Tnfa) regardless of injection type compared to their WT counterparts (Fig. 2A & 2B, p ≤ 0.05). There were no differences between any groups for total nuclear factor kappa-β (NFκB) protein at 3 days post-injection (Fig. 2C). At 28 days post-injection, there was a main effect of MAR injection to have less total NFκB protein content by ≥ 50% (Fig. 2D, p ≤ 0.05). There was a significant interaction between genotype and injury on relative p38 phosphorylation 3 days post-bupivacaine-injection (Fig. 2E, p ≤ 0.05). At 3 days post-injection, A1-UNINJ mice exhibited ∼5-fold greater phosphorylation of p38 protein to total p38 protein than WT-UNINJ mice (Fig. 2E, p ≤ 0.05). At 3 days post-injection, A1-INJ mice exhibited ∼20-fold greater phosphorylation of p38 protein to total p38 protein than WT-UNINJ mice (Fig. 2E, p ≤ 0.05). At 28 days post-injection there was a main effect of genotype for A1 mice to have ∼50% less phosphorylation of p38 to total p38 protein than their WT counterparts (Fig. 2F, p ≤ 0.05). There was no difference in interleukin-1β (Il1b) mRNA abundance between any groups at both 3 day and 28 days post-injection (Fig. 2G and H).
Fig. 2.
mRNA abundance and protein content of inflammatory signaling factors, TNF-α, NFκB, p38 MAPK, and IL-1β at 3 days (left) and 28 days (right) post-injection in wildtype (WT) or muscle specific PGC-1α transgenic (A1) mice. (A–B) TNF-α mRNA abundance. (C–D) NFκB protein content. (E–F) p38 MAPK protein content. (G–H) IL-1β mRNA abundance. The uninjured phosphate buffered saline (PBS) group is denoted UNINJ. The injured Marcaine (MAR) group is denoted INJ. Main effects (ME) of genotype and/or injury appear on each graph when relevant, # signifies difference from genetic counterpart, ∗ signifies difference between injections within the same genotype following an interaction. Values are means ± SE. Statistical significance we set at an alpha of p ≤ 0.05.
Protein synthetic and atrophic markers
There were no differences in phosphorylated AKT to total AKT protein content between WT and A1 mice (Fig. 3A & 3B). At 3 days post-injection, phosphorylation of AKT protein to total AKT protein content was ∼3-fold greater in the WT-INJ mice compared to all other groups (Fig. 3A, p ≤ 0.05). There were no significant differences in phosphorylation of AKT 28 days post-injection regardless of genotype or injury (Fig. 3B). Phosphorylation of 4EBP1 protein to total 4EBP1 protein content was ∼50%–67% lower and in the A1-INJ group compared to both WT injection groups, respectively, as well as ∼50% lower than its A1-UNINJ counterpart at 3 days post-injection (Fig. 3C, p ≤ 0.05). At 28 days post-injection, there was a main effect of genotype for A1 mice to have ∼1-fold lowered phosphorylation of 4EBP1 protein to total 4EBP1 protein content than WT mice (Fig. 3D, p ≤ 0.05). There were no differences in mRNA abundance of Atrogin1 or Murf1 at either the 3-day or 28-day time point (Fig. 3E–H).
Fig. 3.
mRNA abundance and protein content of protein synthetic and catabolic factors, Akt, 4E-BP1, and the E3 ligases Atrogin-1 and MuRF-1 at 3 days (left) and 28 days (right) post-injection in wildtype (WT) or muscle specific PGC-1α transgenic (A1) mice. (A–B) Akt protein content. (C–D) 4E-BP1 protein content. (E–F) Atrogin-1 mRNA abundance. (G–H) MuRF-1 mRNA abundance. The uninjured phosphate buffered saline (PBS) group is denoted UNINJ. The injured Marcaine (MAR) group is denoted INJ. Main effects (ME) of genotype and/or injury appear on each graph when relevant, # signifies difference from genetic counterpart, ∗ signifies difference between injections within the same genotype following an interaction. Values are means ± SE. Statistical significance we set at an alpha of p ≤ 0.05.
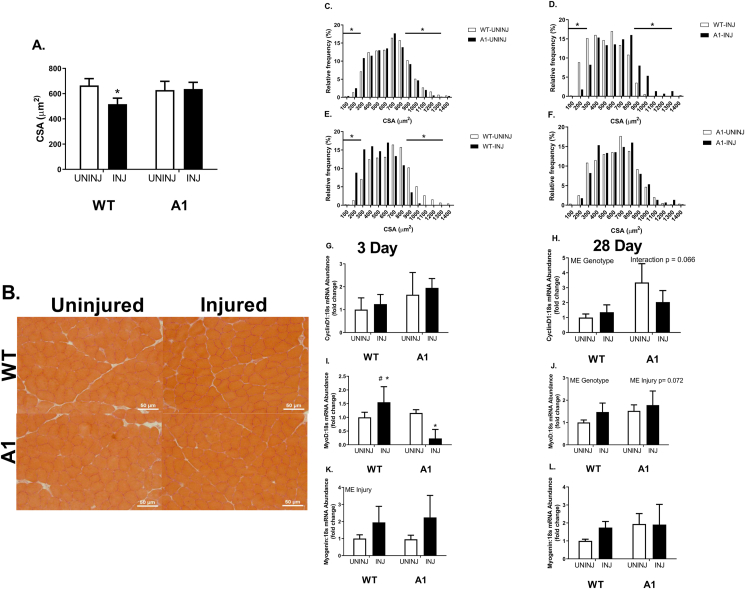
CSA, fiber size distribution, and myogenic markers
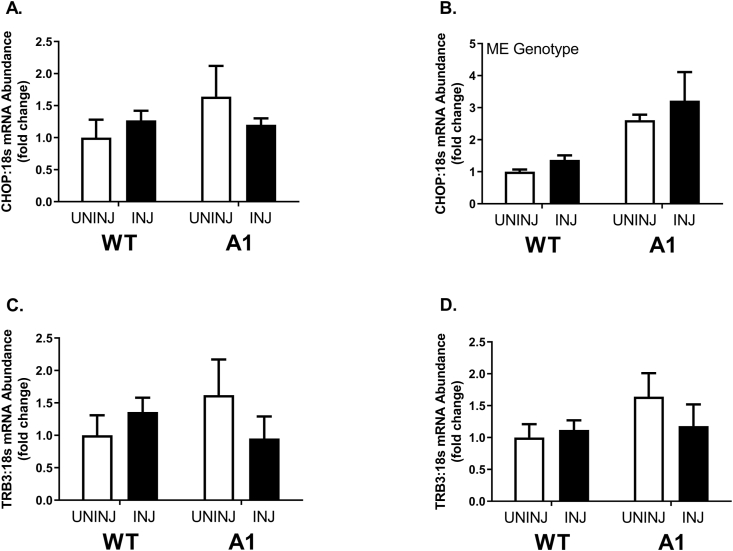
At 28 days post-injection, the WT-INJ group had a 20% lower muscle fiber CSA compared to all other groups, who shared no differences (Fig. 4A & 4B, p ≤ 0.05). The lowered CSA observed in the WT-INJ group was largely due to the higher volume of smaller muscle fibers highlighted in the representative images provided (Fig. 4A and B). WT-UNINJ mice displayed a higher frequency of larger fibers compared to the A1-UNINJ group (Fig. 4C, p ≤ 0.05). There were no differences in fiber size distribution between A1-UNINJ and A1-INJ mice (Fig. 4F). Fiber size distribution of muscle fibers reflected a higher frequency of small fibers (≤ 300 μm) and decreased frequency of larger fibers (≥ 900 μm) in the WT-INJ mice compared to the WT-UNINJ and A1-INJ groups at 28 days post-injection (Fig. 4D and E, p ≤ 0.05). There was no difference in Cyclin D1 mRNA abundance between WT-UNINJ and A1-UNINJ mice (Fig. 4G and H). There were no differences in mRNA abundance of Cyclin D1 between any groups at 3 days post-injection (Fig. 4G). There was a main effect of genotype in the A1 mice to have higher Cyclin D1 mRNA abundance ∼2-fold compared to WT mice at 28 days post-injection (Fig. 4H). There was no difference in Myod mRNA abundance between the WT-UNINJ and A1-UNINJ mice (Fig. 4I and J). Myod mRNA abundance was ∼50% higher 3 days post MAR injection in WT mice, however A1 mice demonstrated ∼75% less mRNA abundance compared to the WT and A1 uninjured mice (Fig. 4I and J, p ≤ 0.05). There was no difference in Myogenin mRNA abundance between WT-PBS and A1-PBS mice (Fig. 4K and L). There was a main effect of injury following injection with MAR for Myogenin mRNA abundance to be ∼ 2-fold higher in both WT and A1 mice 3 days post-injection (Fig. 4K, p ≤ 0.05). There were no significant differences in Myogenin mRNA abundance 28 days post-injection (Fig 4L). Finally, there was a main effect of genotype for greater Chop mRNA in A1 mice at 28 d with no other significant effects on Chop or Trb3 (Fig. 5).
Fig. 4.
Cross-sectional area and muscle fiber size distribution at 28 days post-injection, followed by mRNA abundance of satellite cell dynamics-associated factors at 3 days (left) and 28 days (right) post-injection in wildtype (WT) or muscle specific PGC-1α transgenic (A1) mice. (A) Cross-sectional area (CSA). (B) Representative images of H&E staining of tibialis anterior cross-sections 28 days post-injection. (C–F) Relative frequency of the muscle fiber sizes across each possible pairwise comparison. (G–H) mRNA abundance of Cyclin D1. (I–J) mRNA abundance of MyoD. (K–L) mRNA abundance of Myogenin. The uninjured phosphate buffered saline (PBS) group is denoted UNINJ. The injured Marcaine (MAR) group is denoted INJ. Main effects (ME) of genotype and/or injury appear on each graph when relevant, # signifies difference from genetic counterpart, ∗ signifies difference between injections within the same genotype following an interaction. Values are means ± SE. Statistical significance we set at an alpha of p ≤ 0.05.
Fig. 5.
mRNA abundance of CHOP and TRB3 at 3 days (left) and 28 days (right) post-injection in wildtype (WT) or muscle specific PGC-1α transgenic (A1) mice. (A–B) CHOP mRNA content. (C–D) TRB3 mRNA content. The uninjured phosphate buffered saline (PBS) group is denoted UNINJ. The injured Marcaine (MAR) group is denoted INJ. Values are means ± SE. Statistical significance we set at an alpha of p ≤ 0.05.
Discussion
The process of muscle regeneration is critical for skeletal muscle maintenance and allows for proper recovery and adaptations following damage. This regenerative capacity is largely controlled by a complex genetic response, and the capacity of skeletal muscle to regenerate is a critical homeostatic process.32 PGC1A has long been studied due to its wide range of transcriptional and regulatory effects on muscle. A recent observation of PGC1A altering muscle regeneration by Dinulovic et al.,25 revealed that muscles with higher levels of PGC1A exhibited faster resolution of inflammation and prevention of fibrosis following injury. However, how PGC1A regulates molecular signaling events during muscle regeneration such as regenerative myogenesis and mTOR signaling remains unclear. Herein, we provide evidence for PGC1A overexpression altering the myogenic response of damaged skeletal muscle through myogenic regulatory factor Myod. The faster resolution of damage observed in PGC1A transgenic mice noted by Dinulovic et al.25 combined with the altered Myod response observed in the current study suggests a potentially advantageous environment for skeletal muscle regeneration.
Successful muscle-specific overexpression of PGC1A in the A1 mice was strongly supported by increased Pgc1a mRNA abundance along with increased Ldhb mRNA abundance and decreased Ldha mRNA abundance. This increase in Pgc1a mRNA is consistent with prior work from our group where a similar increase was matched by 2-fold increases in PGC1A protein.26 Our observations are congruent with the findings of Summermatter et al.,33 where PGC1A itself was found to fuel this isoform switching through cooperation with estrogen-related receptor-α (ERRA) increasing Ldhb mRNA abundance, while simultaneously inhibiting the transcription of Ldha through inactivation of its transcription factor myelocytomatosis oncogene (c-Myc). Functionally, a shift in LDH isoforms toward LDH-B is congruent with PGC1A's role in enhanced oxidative metabolism.
Skeletal muscle regeneration is associated with alterations in the metabolic profile of the muscle. It has been demonstrated by us and others that 3 days post-injury, citrate synthase activity, mitochondrial protein yield, and mitochondrial content are decreased.12,18,19 This would likely shift the muscle to a more glycolytic profile to deal with the increased energy demands associated with the early stages of skeletal muscle regeneration. Due to a decrease in mitochondrial function and content it would be expected that during the early stages of skeletal muscle regeneration mitochondrial biogenesis would be a priority for the muscle. It has been well established that skeletal muscle regeneration is associated with increased mitochondrial biogenesis.18,19 Interestingly, PGC1A has been shown not to increase during skeletal muscle regeneration.18,19 This is consistent with our data. However, markers of mitochondrial protein synthesis, mitochondrial respiration, and other markers of mitochondrial biogenesis are upregulated during skeletal muscle regeneration implying that mitochondrial biogenesis is critical during skeletal muscle regeneration.18,19 The decrease in Pgc1a mRNA abundance in A1 mice at 3 days post-injection could directly contribute to the observed downregulation of Ldhb mRNA abundance—through Pgc1a and ERRα regulating Ldhb transcription—which may imply a more glycolytic profile in the early regenerative phase in A1 mice. A1 mice also displayed higher mRNA abundance of Cd147, an extracellular matrix metalloproteinase inducer used in skeletal muscle primarily for targeting monocarboxylate transporters (MCT) 1 and 4 to the plasma membrane during lactate shuttling.34 Where, upregulation of Cd147 mRNA abundance could indicate preferential increases in lactate transport into the muscle for further substrate utilization. The alterations in metabolic markers shown here can be best described as a possible optimization of metabolism for the oxidative environment created in skeletal muscle by the chronic overexpression of PGC1A. However, as metabolism was not directly measured further study is needed to validate this claim.
PGC-1A is activated by phosphorylation of its negative regulatory domain via p38 MAPK, a known kinase activated by inflammatory cytokines or other environmental stressors.35,36 Addition of TNFA, a major inflammatory cytokine, to human mesenchymal stem cells and adipose progenitor cells results in significant increases in VEGF, HGF, and IGF1 production via a p38 MAPK-dependent pathway.37 Furthermore, we have recently demonstrated locally induced TNFA in atrophic oxidative muscles (soleus) further suggesting the interplay between the oxidative muscle phenotype elicited by PGC1A and TNFA.38 TNFA has also been shown to induce a greater abundance of Atrogin1 and Murf1 mRNA through a p38-mediated pathway as well.39,40 This relationship between TNFA, p38, and ubiquitin ligases Atrogin1 and Murf1 was not directly reflected in the data presented here, with Atrogin1 and Murf1 exhibiting no changes in mRNA abundance at the conclusion of regeneration with altered phosphorylation of p38, even with the elevated Tnfa mRNA abundance in A1 mice. This indicates PGC1A overexpression increases basal levels of Tnfa transcription but does not result in an atrophic phenotype, as further supported by the lack of change in NFκB protein content in A1 mice.
The reduction in phosphorylation of the protein synthetic markers AKT and 4EBP1 in mice overexpressing PGC1A presented here is also indicative of a lowered protein synthetic response from injury induced by bupivacaine. Activation of AKT via phosphorylation results in activation of mTOR, where mTOR will then phosphorylate and inactivate the downstream target 4EBP1, ultimately leading to an upregulation in protein synthesis.41 The reduction in phosphorylation of AKT and 4EBP1 in A1 mice shown here suggests A1 mice did not induce classic protein synthetic signaling via mTOR to undergo successful regeneration of skeletal muscle at the measured timepoints. WT mice however, showed a clear increase in phosphorylation of AKT at 3 days post-injection with bupivacaine and a non-significant mean increase in phosphorylation of 4EBP1, indicating a present protein synthetic response in WT mice following the injury that was not observed in the A1 mice. The lack of an increase in markers of protein synthesis observed in the A1 mice could be due to a lowered requirement for protein synthesis to accomplish proper muscle regeneration. Where alterations to satellite cell dynamics could be sufficient for healing of damaged muscle tissue. This observation is speculative, however, due to a lack of a robust and direct measure of protein synthesis beyond the protein content of these markers. These interpretations, however, are limited by the measurement timeframe and primary consideration of phosphorylation signaling.
As presented here, the mean cross-sectional area of wild-type mice was lower 28 days following damage typically seen via bupivacaine injection,42 but this reduction was not observed in A1 mice. In conjunction, wild-type mice had a greater frequency of smaller muscle fibers compared to the transgenic mice following damage. In this study, we demonstrated a decrease in Myod mRNA abundance at 3 days post-injury in A1 mice whereas wild-type mice exhibited classically elevated Myod. This could be indicative of lowered myoblast proliferation. There was also a trend for A1 mice to exhibit elevated Myogenin mRNA abundance when compared to WT mice at 28 days post-injury. This, in conjunction with lowered MyoD mRNA abundance, could indicate an earlier switch in transcriptional signaling of satellite cells from proliferation to differentiation following PGC1A overexpression. The trend towards a decrease in Cyclin D1 in A1 mice at 28 days post-bupivacaine injection further supports this finding. Where a decrease in Cyclin D1 could indicate cell cycle exiting for the onset of myotube differentiation and preservation of myofibers.43 Where MyoD has been shown to induce the termination of the cell cycle through upregulation of p21 which has been shown to inhibit Cyclin-dependent kinase activity during terminal myocyte differentiation.44,45 p38 MAPK has been demonstrated to promote myoblast differentiation while simultaneously restricting excess proliferation during postnatal growth in mice.46 Myoblasts lacking the p38α MAPK isoform have also been found unable to differentiate and form myotubes.47 Therefore, it appears that increased phosphorylation of p38 MAPK in A1 mice may contribute to satellite cell differentiation over proliferation, providing a differential effect on skeletal muscle following the damage when compared to wild-type counterparts in the early stages of regeneration.
Finally, considering our large induction of p38 phosphorylation in 3 days post-bupivacaine injected A1 mice, we considered further mechanisms which may impact downstream signaling on repressed MyoD in this timeframe. p38 MAPK is known to activate CHOP48 which in turn can repress MyoD49 and activate TRB350 where TRB3, in turn, inhibits AKT.51 To provide evidence of whether such a mechanism may be in effect in our A1 mice we assessed Chop and Trb3 mRNA contents identifying only elevated Chop mRNA in A1 mice regardless of injury. However, we should note p38 activation of CHOP occurs primarily through direct phosphorylation and thus we may be missing the best opportunity to assess this system. This CHOP/TRB3 axis deserves further scrutiny in future studies of muscle regeneration especially considering TRB3 may exert specific effects on muscle protein turnover.52
In conclusion, we provide evidence for skeletal muscle with chronic overexpression of PGC1A to exhibit no morphological changes following the damage via bupivacaine injection. Although it is important to note that although there was an observed decrease in mean CSA of WT mice following injection of bupivacaine, by day 28 muscle wet weight and mean CSA had returned to control levels and thus damage was resolved as reported in similar studies.53 The lack of morphological changes in A1 mice could most likely be a result of modulation of satellite cell proliferation/differentiation dynamics, and of note, in the presence of reduced markers of protein synthesis. However, these results could be due to either a lesser extent of or the earlier resolution of damage in the A1 mice. While both are possible, the distinction concerning which is the greatest contributor to earlier resolution of damage in A1 mice cannot be elucidated at this time. Overall, this study contributes to the continuously growing repertoire of literature concerning the effects of muscle-specific overexpression of PGC1A on conditions and pathologies of muscle damage or wasting. This study adds to the evidence that conditions which increase mitochondrial content (e.g., endurance training) could be beneficial for the ability of the skeletal muscle to recover from injury.
Ethical approval statement
All procedures are approved by the University of Arkansas Institutional Animal Care and Use Committee (Protocol 13020). All animals were housed at the University of Arkansas Central Laboratory Animal Facility according to the guidelines set forth by the Institutional Animal Care and Use Committee.
Funding
The Arkansas Biological Institute, the major research component of the Arkansas Tobacco Settlement Proceeds Act of 2000, funded this study.
Authors' contributions
TAW conceived the study and designed the experiment. WSH, ERS, RAP, LAB, MER, DEL, and JLB handled data collection. TAW, WSH, and ERS conducted final data analysis and wrote the original draft of the manuscript. TAW, WSH, ERS, RAP, LAB, MER, DEL, and JLB edited the original manuscript.
Submission statement
This article has not been published previously and it is not under consideration for publication. This publication is approved by all authors and by the responsible authorities where the work was carried out. After accepted, this article will not be published elsewhere including electronically in the same form, in English or any other language, without the written consent of the copyright-holder.
Conflict of interest
The authors do not have any conflicts of interest to declare.
Acknowledgements
The authors would like to thank the Exercise Science Research Center staff at the University of Arkansas for all of their hard work and all administrative contributions towards this project.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.smhs.2022.06.005.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Frontera WR, Ochala J. Skeletal muscle: a brief review of structure and function. Calcif Tissue Int. 2015;96(3):183–195. doi: 10.1007/s00223-014-9915-y. [DOI] [PubMed] [Google Scholar]
- 2.Tieland M, Trouwborst I, Clark BC. Skeletal muscle performance and ageing. J Cachexia Sarcopenia Muscle. 2018;9(1):3–19. doi: 10.1002/jcsm.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flück M, Hoppeler H. Molecular basis of skeletal muscle plasticity--from gene to form and function. Rev Physiol Biochem Pharmacol. 2003;146:159–216. doi: 10.1007/s10254-002-0004-7. [DOI] [PubMed] [Google Scholar]
- 4.Orimo S, Hiyamuta E, Arahata K, Sugita H. Analysis of inflammatory cells and complement C3 in bupivacaine-induced myonecrosis. Muscle Nerve. 1991;14(6):515–520. doi: 10.1002/mus.880140605. [DOI] [PubMed] [Google Scholar]
- 5.Tidball JG, Berchenko E, Frenette J. Macrophage invasion does not contribute to muscle membrane injury during inflammation. J Leukoc Biol. 1999;65(4):492–498. https://www.ncbi.nlm.nih.gov/pubmed/10204578 [PubMed] [Google Scholar]
- 6.Warren GL, Summan M, Gao X, Chapman R, Hulderman T, Simeonova PP. Mechanisms of skeletal muscle injury and repair revealed by gene expression studies in mouse models. J Physiol. 2007;582(Pt 2):825–841. doi: 10.1113/jphysiol.2007.132373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan Z, Choi S, Liu X, et al. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J Biol Chem. 2003;278(10):8826–8836. doi: 10.1074/jbc.M209879200. [DOI] [PubMed] [Google Scholar]
- 8.Saito Y, Nonaka I. Initiation of satellite cell replication in bupivacaine-induced myonecrosis. Acta Neuropathol. 1994;88(3):252–257. doi: 10.1007/BF00293401. [DOI] [PubMed] [Google Scholar]
- 9.Brooke MH, Fenichel GM, Griggs RC, et al. Duchenne muscular dystrophy: patterns of clinical progression and effects of supportive therapy. Neurology. 1989;39(4):475–481. doi: 10.1212/wnl.39.4.475. [DOI] [PubMed] [Google Scholar]
- 10.Emery AE. Duchenne muscular dystrophy--Meryon’s disease. Neuromuscul Disord. 1993;3(4):263–266. doi: 10.1016/0960-8966(93)90018-f. [DOI] [PubMed] [Google Scholar]
- 11.Koopman R, Ly CH, Ryall JG. A metabolic link to skeletal muscle wasting and regeneration. Front Physiol. 2014;5:32. doi: 10.3389/fphys.2014.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Washington TA, Brown L, Smith DA, Davis G, Baum J, Bottje W. Monocarboxylate transporter expression at the onset of skeletal muscle regeneration. Phys Rep. 2013;1(4) doi: 10.1002/phy2.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pala F, Di Girolamo D, Mella S, et al. Distinct metabolic states govern skeletal muscle stem cell fates during prenatal and postnatal myogenesis. J Cell Sci. 2018;131(14):jcs212977. doi: 10.1242/jcs.212977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dell’Orso S, Juan AH, Ko KD, et al. Single cell analysis of adult mouse skeletal muscle stem cells in homeostatic and regenerative conditions. Development. 2019;146(12):dev174177. doi: 10.1242/dev.174177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKellar DW, Walter LD, Song LT, et al. Large-scale integration of single-cell transcriptomic data captures transitional progenitor states in mouse skeletal muscle regeneration. Commun Biol. 2021;4(1):1280. doi: 10.1038/s42003-021-02810-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanay N, Rabie M, Nevo Y. Impaired regeneration in dystrophic muscle-new target for therapy. Front Mol Neurosci. 2020;13:69. doi: 10.3389/fnmol.2020.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rugowska A, Starosta A, Konieczny P. Epigenetic modifications in muscle regeneration and progression of Duchenne muscular dystrophy. Clin Epigenet. 2021;13(1):13. doi: 10.1186/s13148-021-01001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duguez S, Feasson L, Denis C, Freyssenet D. Mitochondrial biogenesis during skeletal muscle regeneration. Am J Physiol Endocrinol Metab. 2002;282(4):E802–E809. doi: 10.1152/ajpendo.00343.2001. [DOI] [PubMed] [Google Scholar]
- 19.Wagatsuma A, Kotake N, Yamada S. Muscle regeneration occurs to coincide with mitochondrial biogenesis. Mol Cell Biochem. 2011;349(1–2):139–147. doi: 10.1007/s11010-010-0668-2. [DOI] [PubMed] [Google Scholar]
- 20.Kang C, Ji LL. Role of PGC-1α signaling in skeletal muscle health and disease. Ann N Y Acad Sci. 2012;1271:110–117. doi: 10.1111/j.1749-6632.2012.06738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J, Wu H, Tarr PT, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418(6899):797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 22.Hatazawa Y, Minami K, Yoshimura R, et al. Deletion of the transcriptional coactivator PGC1alpha in skeletal muscles is associated with reduced expression of genes related to oxidative muscle function. Biochem Biophys Res Commun. 2016;481(3–4):251–258. doi: 10.1016/j.bbrc.2016.10.133. [DOI] [PubMed] [Google Scholar]
- 23.Adhihetty PJ, Uguccioni G, Leick L, Hidalgo J, Pilegaard H, Hood DA. The role of PGC-1alpha on mitochondrial function and apoptotic susceptibility in muscle. Am J Physiol Cell Physiol. 2009;297(1):C217–C225. doi: 10.1152/ajpcell.00070.2009. [DOI] [PubMed] [Google Scholar]
- 24.Handschin C, Chin S, Li P, et al. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. J Biol Chem. 2007;282(41):30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- 25.Dinulovic I, Furrer R, Di Fulvio S, Ferry A, Beer M, Handschin C. PGC-1α modulates necrosis, inflammatory response, and fibrotic tissue formation in injured skeletal muscle. Skeletal Muscle. 2016;6:38. doi: 10.1186/s13395-016-0110-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greene NP, Lee DE, Brown JL, et al. Mitochondrial quality control, promoted by PGC-1α, is dysregulated by Western diet-induced obesity and partially restored by moderate physical activity in mice. Phys Rep. 2015;3(7) doi: 10.14814/phy2.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perry RA, Brown LA, Lee DE, et al. Differential effects of leucine supplementation in young and aged mice at the onset of skeletal muscle regeneration. Mech Ageing Dev. 2016;157:7–16. doi: 10.1016/j.mad.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown LA, Lee DE, Patton JF, et al. Diet-induced obesity alters anabolic signalling in mice at the onset of skeletal muscle regeneration. Acta Physiol. 2015;215(1):46–57. doi: 10.1111/apha.12537. [DOI] [PubMed] [Google Scholar]
- 29.van der Poel C, Gosselin LE, Schertzer JD, et al. Ageing prolongs inflammatory marker expression in regenerating rat skeletal muscles after injury. J Inflamm. 2011;8(1):41. doi: 10.1186/1476-9255-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greene NP, Nilsson MI, Washington TA, et al. Impaired exercise-induced mitochondrial biogenesis in the obese Zucker rat, despite PGC-1α induction, is due to compromised mitochondrial translation elongation. Am J Physiol Endocrinol Metab. 2014;306(5):E503–E511. doi: 10.1152/ajpendo.00671.2013. [DOI] [PubMed] [Google Scholar]
- 31.Washington TA, White JP, Davis JM, et al. Skeletal muscle mass recovery from atrophy in IL-6 knockout mice. Acta Physiol. 2011;202(4):657–669. doi: 10.1111/j.1748-1716.2011.02281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Musaro A. Muscle homeostasis and regeneration: from molecular mechanisms to therapeutic opportunities. Cells. 2020;9(9):2033. doi: 10.3390/cells9092033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Summermatter S, Santos G, Pérez-Schindler J, Handschin C. Skeletal muscle PGC-1α controls whole-body lactate homeostasis through estrogen-related receptor α-dependent activation of LDH B and repression of LDH A. Proc Natl Acad Sci U S A. 2013;110(21):8738–8743. doi: 10.1073/pnas.1212976110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benton CR, Yoshida Y, Lally J, Han XX, Hatta H, Bonen A. PGC-1alpha increases skeletal muscle lactate uptake by increasing the expression of MCT1 but not MCT2 or MCT4. Physiol Genom. 2008;35(1):45–54. doi: 10.1152/physiolgenomics.90217.2008. [DOI] [PubMed] [Google Scholar]
- 35.Fan M, Rhee J, St-Pierre J, et al. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1alpha: modulation by p38 MAPK. Genes Dev. 2004;18(3):278–289. doi: 10.1101/gad.1152204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298(5600):1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 37.Wang M, Crisostomo PR, Herring C, Meldrum KK, Meldrum DR. Human progenitor cells from bone marrow or adipose tissue produce VEGF, HGF, and IGF-I in response to TNF by a p38 MAPK-dependent mechanism. Am J Physiol Regul Integr Comp Physiol. 2006;291(4):R880–R884. doi: 10.1152/ajpregu.00280.2006. [DOI] [PubMed] [Google Scholar]
- 38.Lim S, Dunlap KR, Rosa-Caldwell ME, et al. Comparative plasma proteomics in muscle atrophy during cancer-cachexia and disuse: the search for atrokines. Phys Rep. 2020;8(19) doi: 10.14814/phy2.14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li YP, Chen Y, John J, et al. TNF-alpha acts via p38 MAPK to stimulate expression of the ubiquitin ligase atrogin1/MAFbx in skeletal muscle. Faseb J. 2005;19(3):362–370. doi: 10.1096/fj.04-2364com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J, Won KJ, Lee HM, et al. p38 MAPK participates in muscle-specific RING finger 1-mediated atrophy in cast-immobilized rat gastrocnemius muscle. KOREAN J PHYSIOL PHARMACOL. 2009;13(6):491–496. doi: 10.4196/kjpp.2009.13.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolster DR, Jefferson LS, Kimball SR. Regulation of protein synthesis associated with skeletal muscle hypertrophy by insulin-, amino acid- and exercise-induced signalling. Proc Nutr Soc. 2004;63(2):351–356. doi: 10.1079/PNS2004355. [DOI] [PubMed] [Google Scholar]
- 42.Plant DR, Colarossi FE, Lynch GS. Notexin causes greater myotoxic damage and slower functional repair in mouse skeletal muscles than bupivacaine. Muscle Nerve. 2006;34(5):577–585. doi: 10.1002/mus.20616. [DOI] [PubMed] [Google Scholar]
- 43.Perdiguero E, Cornelison D. Erratum to: isolation, culture, and immunostaining of skeletal muscle myofibers from wildtype and nestin-GFP mice as a means to analyze satellite cells. Methods Mol Biol. 2017;1556:C1. doi: 10.1007/978-1-4939-6771-1_21. [DOI] [PubMed] [Google Scholar]
- 44.Guo K, Wang J, Andrés V, Smith RC, Walsh K. MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Mol Cell Biol. 1995;15(7):3823–3829. doi: 10.1128/mcb.15.7.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Halevy O, Novitch BG, Spicer DB, et al. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267(5200):1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 46.Brien P, Pugazhendhi D, Woodhouse S, Oxley D, Pell JM. p38α MAPK regulates adult muscle stem cell fate by restricting progenitor proliferation during postnatal growth and repair. Stem Cell. 2013;31(8):1597–1610. doi: 10.1002/stem.1399. [DOI] [PubMed] [Google Scholar]
- 47.Perdiguero E, Ruiz-Bonilla V, Serrano AL, Muñoz-Cánoves P. Genetic deficiency of p38alpha reveals its critical role in myoblast cell cycle exit: the p38alpha-JNK connection. Cell Cycle. 2007;6(11):1298–1303. doi: 10.4161/cc.6.11.4315. [DOI] [PubMed] [Google Scholar]
- 48.Wang XZ, Ron D. Stress-induced phosphorylation and activation of the transcription factor CHOP (GADD153) by p38 MAP Kinase. Science. 1996;272(5266):1347–1349. doi: 10.1126/science.272.5266.1347. [DOI] [PubMed] [Google Scholar]
- 49.Alter J, Bengal E. Stress-induced C/EBP homology protein (CHOP) represses MyoD transcription to delay myoblast differentiation. PLoS One. 2011;6(12) doi: 10.1371/journal.pone.0029498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 2005;24(6):1243–1255. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kato S, Du K. TRB3 modulates C2C12 differentiation by interfering with Akt activation. Biochem Biophys Res Commun. 2007;353(4):933–938. doi: 10.1016/j.bbrc.2006.12.161. [DOI] [PubMed] [Google Scholar]
- 52.Choi RH, McConahay A, Jeong HW, et al. Tribbles 3 regulates protein turnover in mouse skeletal muscle. Biochem Biophys Res Commun. 2017;493(3):1236–1242. doi: 10.1016/j.bbrc.2017.09.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones EJ, Bishop PA, Woods AK, Green JM. Cross-sectional area and muscular strength: a brief review. Sports Med. 2008;38(12):987–994. doi: 10.2165/00007256-200838120-00003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.