Abstract
We recently identified a pilin-like competence factor, ComP, which is essential for natural transformation of the gram-negative soil bacterium Acinetobacter sp. strain BD413. Here we demonstrate that transcription and synthesis of the pilin-like competence factor ComP are maximal in the late stationary growth phase, whereas competence is induced immediately after inoculation of a stationary-phase culture into fresh medium. Western blot analyses revealed three forms of ComP, one with an apparent molecular mass of 15 kDa, which correlates with the molecular mass deduced from the DNA sequence, one 20-kDa form, which was found to be glycosylated, and one 23-kDa form. The glycosylation of ComP was not required for its function in DNA binding and uptake. The 20-kDa form was present in the cytoplasmic membrane, the periplasm, and the outer membrane, whereas the 23-kDa form was located in the outer membrane and might be due to a further modification. Immunological data suggest that ComP is not a subunit of the pilus structures. Possible functions of ComP in the DNA transformation machinery of Acinetobacter sp. strain BD413 are discussed.
Bacterial genetic competence, a physiological state allowing binding, processing, and internalizing of exogenous DNA, is widespread in nature. Despite the broad distribution of genetic competence among different taxonomic and trophic groups, knowledge about the DNA uptake machinery is restricted to some model organisms such as the gram-positive bacteria Bacillus subtilis and Streptococcus pneumoniae and gram-negative pathogenic bacteria, such as Neisseria gonorrhoeae and Haemophilus influenzae (22). Whereas extensive work on the DNA transformation machineries of gram-positive model organisms and gram-negative pathogenic bacteria has given many new insights into the molecular basics, information about the DNA transformation machineries of gram-negative soil bacteria is very scarce.
To obtain insights into the molecular basis of natural transformation machinery and the mechanism of DNA uptake in gram-negative soil bacteria, we studied the nutritionally versatile organism Acinetobacter sp. strain BD413. This strain, formerly designated Acinetobacter calcoaceticus BD413, is known for its high frequency of natural transformation (15). To identify components involved in the natural transformation of strain BD413, we generated transformation-deficient mutants. The analysis of these mutant strains has contributed a great deal to our understanding of the process of DNA transformation in Acinetobacter sp. strain BD413 at the molecular level. Our studies led to the identification of five novel competence factors, ComB, ComE, ComF, ComP, and ComC; the last two have been shown to be essential for binding and uptake of DNA in Acinetobacter sp. strain ADP239, a pobA (p-hydroxybenzoate hydroxylase) mutant strain of BD413 (3, 11, 21, 33). The deduced amino acid sequences of ComB, ComE, ComF, and ComP are similar to those of prepilins of type IV pili and to pilin-like components of protein translocation machineries, whereas ComC is similar to various type IV pilus biogenesis factors.
The capability for natural transformation is encoded by numerous genes. Genetic and biochemical studies indicate that homologues of type IV pilus subunits are involved in natural transformation in a variety of bacteria (4, 5, 7, 39). However, it is still an open question whether the type IV pili or a putative pilus-like structure is implicated in DNA uptake (7, 44). The deduced protein sequence of the Acinetobacter sp. strain BD413 competence gene comP is similar to those of prepilins from various microorganisms. A deletion of comP completely abolished natural transformation but had no effect on piliation, as revealed by electron microscopy (33). The results from electron microscopic studies including immunogold labeling studies together with Western blot analysis presented in this report strongly suggest that ComP is associated with neither the thin nor the thick pili of Acinetobacter sp. strain BD413. We provide evidence that ComP undergoes glycosylation and is located in the cytoplasmic and outer membranes. For a better understanding of the role of ComP in natural transformation, we have also studied the regulation of comP expression and ComP synthesis in the course of growth phase-dependent competence development.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids are listed in Table 1. Acinetobacter wild-type and mutant strains were grown in mineral medium (29). Growth conditions used were as described previously (33). DNA manipulations were done in Escherichia coli DH5α cultured in Luria-Bertani medium (36). Antibiotics were added when appropriate (kanamycin and tetracycline at 20 μg/ml for both E. coli and Acinetobacter strains and ampicillin at 100 μg/ml).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Acinetobacter sp. strains | ||
| BD413 | Wild type | 15 |
| ADP239 | Spontaneous pobA mutant of BD413 | 10 |
| Transformation-affected mutants of Acinetobacter sp. strain ADP239 | ||
| T205 | pobA ΔcomP::nptII Kmr | 33 |
| T206 | pobA comP::nptII Kmr | 33 |
| E. coli strain | ||
| DH5α | F−lacZΔM15 recA1 endA1 hsdR17 supE44 (lacZYA argF) | 8 |
| Plasmids | ||
| pRK415 | Tcrlacp/o | 16 |
| pMAL-c2 | AprlacPOZ′ oriR malE-lacZα rrB lacIq | 25 |
| pKOK6.1 | Apr Kmr Cmr | 17 |
| pRT14 | 14-kb XbaI fragment of pRK415; comP Tcr | 33 |
| pRT1 | 1.6-kb PstI fragment of pRT14 in pRK415; comP Tcr | 33 |
| pCP1 | 0.7-kb BglII-ScaI fragment of pRT14 in pRK415; comP Tcr | 33 |
| pRP1 | 1.4-kb BglII-SstI fragment of pRT14 in pRK415; comP orfC Tcr | This work |
| pRT1-4 | pRT1 with lacZ/Kmr cassette of pKOK6.1; Tcr | This work |
| pMP1 | 0.49-kb XbaI-HindIII fragment carrying comP in pMALc-2;Apr | This work |
| pMP2 | 0.24-kb EcoRI-HindIII fragment carrying 5′-truncated comP in pMALc-2; Apr | This work |
Apr, ampicillin resistance; Tcr, tetracycline resistance; Kmr, kanamycin resistance; Cmr, chloramphenicol resistance.
Cloning and DNA techniques.
Standard recombinant DNA techniques were performed as described by Sambrook et al. (36). Restriction enzymes (Pharmacia Biotech Europe GmbH, Freiburg, Germany) and T4 DNA ligase (Boehringer GmbH, Mannheim, Germany) were used as recommended by the manufacturers. Competent cells of E. coli were prepared and transformed (14). Conjugation, transformation, and complementation procedures were performed as described previously (33). PCR products were sequenced using the dideoxy chain termination method (37).
Construction and purification of MalE-ComP fusion proteins and production of anti-ComP antibodies.
For the construction of a MalE-ComP fusion protein, comP was amplified using the primers 5′-CAACATTGGTCTAGAAATTTTATG-3′ and 5′-TCTTTTAAAGCTTTATAGTAC-3′ containing XbaI and HindIII restriction sites (underlined), respectively. For the generation of a polyclonal antiserum directed against ComP with the N terminus deleted (designated ComP*), comP with the 5′ end deleted (encoding amino acid residues 39 to 141 of mature ComP) was amplified by primers 5′-TCTGAAGAATTCACAGCAGCATCA-3′ and 5′-TCTTTTAAAGCTTTATAGTAC-3′ containing EcoRI and HindIII restriction sites (underlined), respectively. Plasmid pRP1 (Table 1) was used as the template. The PCR was performed with 0.5 μg of template DNA, Vent polymerase, and an annealing temperature of 37°C. The PCR products were cloned in pMALc-2 (New England Biolabs GmbH, Schwalbach, Germany) with XbaI and HindIII or EcoRI and HindIII, respectively. The resulting pMALc-2 recombinant plasmids were designated pMP1 and pMP2, respectively. Correct amplification was verified by DNA sequencing. Expression of the malE-comP fusions in E. coli was induced at an optical density at 600 nm (OD600) of 0.6 by addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 0.1 mM. The MalE-ComP and MalE-ComP* fusion proteins were purified from E. coli DH5α grown at 37°C in MI (maximal induction) medium (28) by affinity chromatography on an amylose column (New England Biolabs GmbH) according to the manufacturer's instructions. The MalE-ComP* fusion protein was used directly to immunize rabbits because there is no MalE in Acinetobacter sp. strain BD413. For the production of anti-MalE-ComP* antibodies 410 μg of purified MalE-ComP* was used to immunize a rabbit (BioScience, Göttingen, Germany).
SDS-PAGE and immunoblotting.
Samples were boiled for 5 to 10 min in a water bath prior to analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 14% polyacrylamide gel as described previously (19). The separated proteins were electrotransferred onto a nitrocellulose membrane (41), which was blocked by incubation for 1 h at room temperature in TBST buffer (0.1 M Tris-HCl, 2.5 M NaCl, 0.05% Tween 20, pH 7.5) containing 3% bovine serum albumin. Subsequently, the membrane was incubated for 1 h with anti-MalE-ComP* antibodies (1:5,000). Antigen detection was carried out using alkaline phosphatase-coupled goat anti-rabbit immunoglobulin G and an alkaline phosphatase color assay (ProtoBlot Western blot AP system; Promega/Sera, Heidelberg, Germany) as recommended by the manufacturer. Silver staining was performed as described previously (2).
Cellular fractionation.
The cells were grown overnight in mineral medium. After removal of extracellular proteins, the cells were fractionated into cytoplasmic membrane, periplasm, and outer membrane. Both membrane fractions were separated as described by Hancock and Nikaido (9), and the periplasmic contents were released as described by Hoshino and Kageyama (13). The fractionation of different bacterial compartments was followed by analysis of the different protein patterns of the distinct cell fractions. Furthermore NADH dehydrogenase activity was used as a cytoplasmic protein marker. The NADH dehydrogenase activity in membrane fractions was assayed by monitoring the decrease in extinction at 340 nm due to NADH oxidation. The assays were performed in Tris-HCl buffer (20 mM, pH 7.5) with 25 μg of protein in the presence of 0.1 mM menadione as the terminal electron acceptor. The reaction was started by the addition of 0.2 mM NADH. The shear fraction was prepared as described previously (1).
Separation of thin and thick pili by sucrose density gradient.
A 4-liter culture of Acinetobacter sp. strain BD413 was grown overnight in mineral medium and harvested by centrifugation at 5,000 × g. The cells were resuspended in 20 ml of buffer (30 mM Tris-HCL, 0.9% NaCl, pH 7.5). To shear the pili, the cells were vortexed for 3 min at 4°C. The cells were separated from the shear fraction by centrifugation twice for 5 min each at 6,000 × g. After dialysis of the supernatant against high-salt buffer (30 mM Tris-HCl, 0.9% NaCl, 0.1 M MgCl2, pH 7.5) the shear fraction was pelleted by centrifugation for 1 h at 180,000 × g. The pellet was resuspended in 0.3 ml of high-salt buffer and loaded on top of a sucrose gradient (20 to 70%) in high-salt buffer. After centrifugation for 20 h at 96,000 × g, the gradient was separated into 1.3-ml fractions. The pellet was resuspended in 0.1 ml of buffer (30 mM Tris-HCl, 0.1 M NaCl, pH 7.5).
Deglycosylation and glycosylation assay.
Proteins were deglycosylated by treatment with trifluoromethanesulfonic acid (6). Freeze-dried cell lysate of Acinetobacter sp. strain BD413 (1.5 mg) was dissolved in 150 μl of 5% SDS and freeze-dried again. Under ice-cold conditions the samples were incubated for 3 h in 500 μl of anisole-trifluoromethanesulfonic acid (1:2). The proteins were precipitated with 5 ml of 1 M sodium carbonate buffer (pH 9.2) and 22 ml of ethanol (96%). The pellets were washed in H2O and redissolved in 200 μl of H2O or Tris-buffered saline.
For labeling and detection of glycoproteins a DIG glycan detection kit (Boehringer GmbH) was used. Therefore, proteins of the cytoplasmic membrane fractions were separated by SDS-PAGE and electrotransferred onto a nitrocellulose membrane. Digoxigenin labeling and detection were performed as recommended by the manufacturer. Specificity of labeling was determined using the glycoprotein transferrin and the nonglycoprotein creatinase as controls.
Construction of comP::lacZ transcriptional fusion and assay of β-galactosidase activity.
For construction of a comP::lacZ transcriptional fusion, the 4.7-kb BamHI fragment of pKOK 6.1 containing a promoter-free lacZ gene was digested with BamHI and blunt ended using Klenow enzyme. pRT1 was digested with XbaI and KpnI and also blunt ended. The BamHI fragment was then cloned into pRT1 giving rise to pRT1-4 (Table 1), which was transformed into mutant T205. The β-galactosidase assays were performed as described previously (26), and the activities are expressed as Miller units, which are proportional to the increase in the absorbancy of free o-nitrophenol per minute and cell density.
Electron microscopy and immunogold labeling.
Negative staining and electron microscopy were performed as described previously (33). For immunogold labeling the cells and the sheared pili were attached to Formvar-coated nickel grids and incubated for 1 h with anti-MalE-ComP* antibodies. After being washed three times with PBST buffer (phosphate-buffered saline [PBS] buffer containing 0.05% Tween 20) and with 1× PBS buffer (50 mM K-PO4, 0.9% NaCl, 0.075% KCl, pH 7.2) the grids were incubated for 30 min with protein A-gold (6 nm; DAKO Diagnostika, Hamburg, Germany). After being washed with PBST and PBS buffer, the cells and the pili were subjected to negative staining.
RESULTS
Growth phase-dependent transcription of comP in Acinetobacter sp. strain BD413 and competence development.
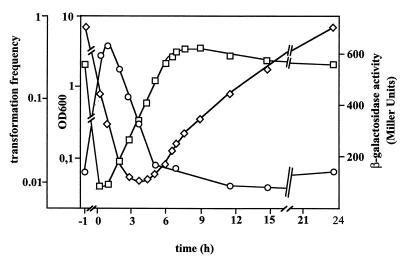
Acinetobacter sp. strain BD413 develops competence for natural transformation immediately after dilution of a stationary culture into fresh medium. During prolonged exponential growth competence is gradually lost until minimum transformation frequencies are detected in stationary phase (30). Our batch culture studies of competence induction confirmed these findings: competence was induced after dilution of a stationary culture into fresh medium and gradually decreased during exponential growth (Fig. 1). In order to gain insights into the regulation of comP expression, we constructed an in-frame comP::lacZ fusion. A 1.7-kb fragment containing the putative ς54 promoter, the start site for comP translation, and the first 210 bp of comP was fused to lacZ and integrated in the broad-host-range plasmid pRK415 giving plasmid pRT1-4 (Table 1), which was introduced into Acinetobacter sp. strain BD413. The expression of comP decreased gradually immediately after dilution into fresh medium, and minimal comP expression levels were detected in the middle of the logarithmic growth phase, but levels increased thereafter (Fig. 1). Maximal comP expression was found in the late stationary growth phase. Control experiments with pRK415 carrying a promoter-free lacZ gene in the opposite orientation to the lac promoter resulted in β-galactosidase activities ranging between 5 and 20 Miller units. Taken together these results clearly show that the transcription of comP is growth phase dependent but does not correlate with competence development. The transcription of comP does not appear to be autoregulated, since the comP null mutation in mutant T205 has no detectable effect on the expression of the comP::lacZ fusion (data not shown).
FIG. 1.
Growth phase-dependent expression of comP in Acinetobacter sp. strain BD413. comP induction was monitored using a comP::lacZ reporter fusion located on the low-copy-number plasmid pRK415. The β-galactosidase activity was monitored as described by Miller (26). Transformation frequencies were determined as described previously (33) with saturating amounts of DNA containing the marker gene pobA. OD (□), comP expression (◊), and transformation frequencies (○) in the preculture were determined just before inoculation (time point −1), immediately after inoculation (time point 0), and during growth at the time points indicated.
Heterologous expression and detection of ComP.
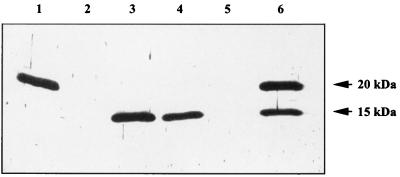
Expression of ComP in E. coli from plasmids pMP1 and pMP2 resulted in malE::comP fusion products of 58 and 53 kDa, respectively. Factor Xa treatment of the preparations revealed proteins of 15 and 10 kDa, which match exactly with the deduced masses of ComP and ComP*, respectively. When cell lysates of the BD413 wild-type strain were probed with the anti-MalE-ComP* antibodies, a single protein was detected (Fig. 2, lane 1). The apparent molecular mass of 20 kDa of this protein differs considerably from the deduced molecular mass of ComP of 15 kDa. However, since no reaction of the antiserum was observed with cell lysates prepared from the comP mutants T205 (Fig. 2, lane 2) and T206 (Fig. 2, lane 5), it is evident that the protein with a molecular mass of 20 kDa is ComP, probably in a posttranslationally modified form.
FIG. 2.
Immunological detection of ComP in Acinetobacter sp. strain BD413 and in complemented mutants. Cell lysates of Acinetobacter sp. strain BD413 (lane 1), T205 (lane 2), T205(pCP1) (lane 3), T205(pRT14) (lane 4), T206 (lane 5), and T206(pCP1) (lane 6) were separated by SDS-PAGE, transferred onto nitrocellulose filters, and probed with anti-MalE-ComP∗ antibodies.
Mutant T205 carries a nondefined big deletion including comP. Complementation of this mutant with pCP1 (which harbors comP only) or pRT14 (which harbors 13.5 kb of DNA in addition to comP) revealed restored transformation phenotypes (33). To our surprise, not the modified 20-kDa ComP protein but rather a protein of 15 kDa, corresponding to the apparent molecular mass of the deduced ComP protein, was detected in the Western blots (Fig. 2, lanes 3 and 4). Since the deletion in mutant T205 is undefined, we repeated the experiment using the defined comP mutant T206. When this mutant was complemented with pCP1, both forms of ComP, the 15- and 20-kDa forms, were observed (Fig. 2, lane 6). The finding that the modification of ComP was abolished in mutant T205 suggests that the undefined deletion in mutant T205 includes the ComP modifying system. The defined comP mutant T206 still harbors the ComP modifying system, but, due to the gene dosage effect, modification might be incomplete. Recent studies have demonstrated that pilus proteins of gram-negative pathogenic bacteria are posttranslationally modified (38). One of the two reported modifications is glycosylation (27, 42). Further studies clearly showed that the 20-kDa ComP undergoes glycosylation (data not shown).
Growth phase-dependent synthesis of ComP.
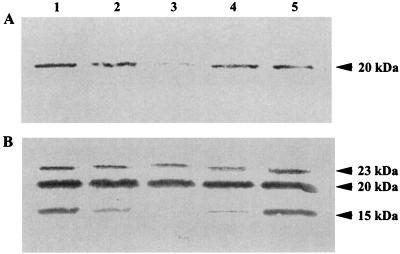
To monitor the time course of ComP synthesis, Acinetobacter sp. strain BD413 was grown in mineral medium with succinate as the carbon source. Samples were taken at time points indicated in Fig. 3 and subjected to Western blot analysis. The glycosylated 20-kDa ComP was present in the smallest amounts in the middle of the exponential growth phase (Fig. 3A, lane 3). Prolonged growth resulted in an increase in the 20-kDa form, with the largest amounts being detected in stationary phase (Fig. 3A, lanes 1 and 5). Taken together, the growth phase-dependent production of the 20-kDa ComP form correlates with the growth phase-dependent transcription of comP. The unmodified 15-kDa form was not detected under these conditions, which was attributed to concentrations of this form below the detection limit. To achieve detection of the 15-kDa form, we increased the protein concentration and repeated the Western blot analyses (Fig. 3B). These studies resulted in the detection of the unmodified 15-kDa ComP form, which was present in the largest amounts in stationary growth phase (Fig. 3B). Interestingly, a 23-kDa protein also reacted with the antibody. Since the protein was never found in comP mutants, the 23-kDa form probably represents another modification of ComP. The level of this protein did not vary significantly during growth, which might be due to an increased stability.
FIG. 3.
Growth phase-dependent cellular levels of ComP. Cell lysates of Acinetobacter sp. strain BD413 samples were taken during growth in batch culture, and 0.25 (A) and 1 (B) μg of protein were separated by SDS-PAGE, transferred onto nitrocellulose membranes, and probed with anti-MalE-ComP∗ antibodies. Lane 1, late-stationary-phase preculture; lane 2, sample taken immediately after inoculation into fresh medium; lane 3, middle of the exponential growth phase; lane 4, onset of stationary growth phase; lane 5, late stationary growth phase. Molecular masses are indicated.
Cellular localization of ComP.
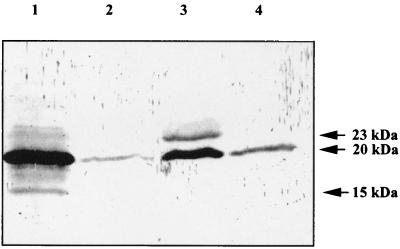
In order to determine the cellular localization of ComP, an overnight culture of Acinetobacter sp. strain BD413 was harvested and separated into cytoplasmic membrane, periplasm, outer membrane, and shear fractions. Equivalent amounts of these cell fractions were subjected to SDS-PAGE. After transfer onto nitrocellulose membranes the cell fractions were probed with anti-MalE-ComP* antibodies. The 20-kDa glycosylated form of ComP was detected in every cell compartment, although at different concentrations (Fig. 4). The largest amounts were present in the cytoplasmic membrane and the outer membrane. Minor amounts of the unmodified 15-kDa form were detectable in the cytoplasmic membrane (Fig. 4, lane 1). The 23-kDa form was mainly present in the outer membrane (Fig. 4, lane 3). Taken together these experiments show that ComP is exported and indicate that the final modification of ComP to the 23-kDa form takes place in the outer membrane. In the cytoplasmic membrane some minor polypeptides with molecular masses between 15 and 23 kDa reacted with anti-MalE-ComP* antibodies. These proteins were never observed in the comP mutants. Probably they represent ComP but at different stages of modification or processing.
FIG. 4.
Cellular localization of ComP. Acinetobacter sp. strain BD413 cells were grown overnight and separated into cytoplasmic membrane (lane 1), periplasm (lane 2), outer membrane (lane 3), and extracellular shear (lane 4) fractions. Equivalent amounts of the different cell fractions were separated by SDS-PAGE, transferred onto nitrocellulose membranes, and probed with anti-MalE-ComP∗ antibodies. Molecular masses are indicated.
To investigate whether the recently identified Acinetobacter sp. strain BD413 competence factors ComE, ComF, and ComC influence the cellular location of ComP, its cellular location in comE, comF, and comC mutants was determined. Mutant cells showed the same cellular distribution of ComP as wild-type cells, indicating that ComE, ComF, and ComC are not involved in export, cellular targeting, or glycosylation of ComP (data not shown).
Immunocytochemical and Western blot analyses of Acinetobacter sp. strain BD413 pilus structures.
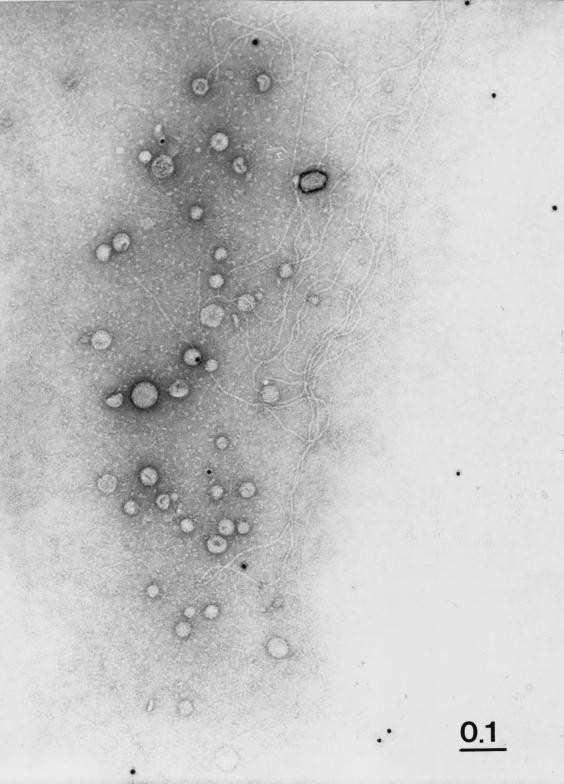
Recently, we have shown that Acinetobacter sp. strain BD413 possesses two types of pili, bundle-forming thin pili and single thick pili (21). To determine whether ComP is part of the pilus structures, immunogold labeling of cells carrying thin and thick pilus structures was performed with polyclonal antiserum raised against purified MalE-ComP*. We did not observe binding of gold-labeled antibodies to either the BD413 cells or to the pilus structures at the cell surface (data not shown). We repeated the immunogold labeling studies with sheared pili of Acinetobacter sp. strain BD413 but never observed binding of the immunogold-labeled antibodies to the pilus structures (Fig. 5). These results suggest that ComP is not part of the pili seen on electron micrographs of Acinetobacter sp. strain BD413.
FIG. 5.
Immunocytochemical analysis of thin and thick pili from Acinetobacter sp. strain BD413 for the presence of ComP. Immunogold labeling was performed with sheared Acinetobacter sp. strain BD413 pili using anti-MalE-ComP∗ antibodies. The representative sample shows thick pili and thin pili. Bar, 0.1 μm.
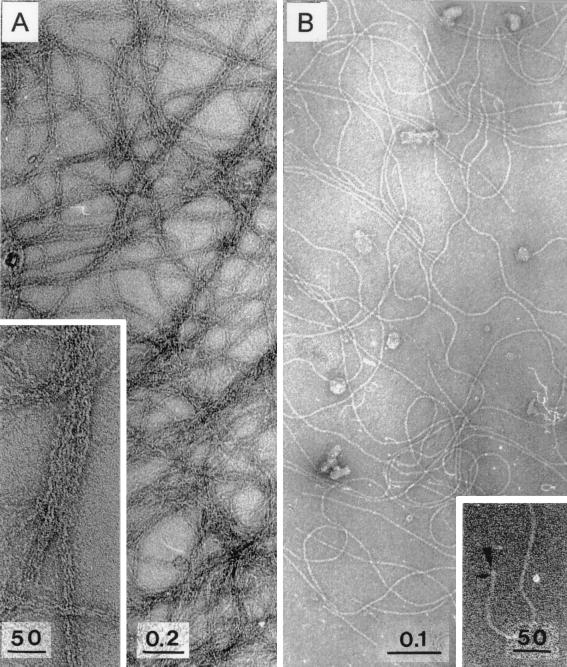
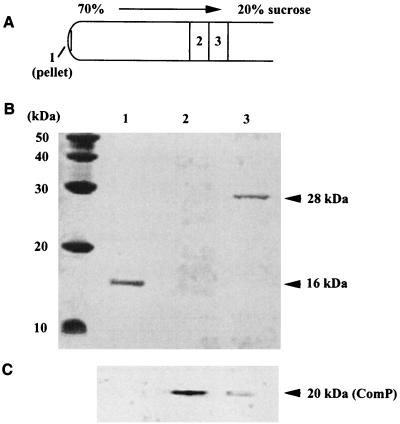
To exclude the possibility that ComP is part of the pilus structures but cannot be detected by immunogold labeling due to inaccessibility in the native pilus structures, we separated the shear fractions by centrifugation in a sucrose density gradient. The gradient was fractionated and analyzed for pili by electron microscopy. The thick pili were enriched in fraction 3 (Fig. 6B), whereas the pellet contained highly aggregated thin pili (Fig. 6A). In all other fractions only very few single pili were found. Analysis of the protein pattern of the pilus fractions revealed a distinct 16-kDa protein in the pellet (Fig. 7B, lane 1) and a 28-kDa protein in fraction 3 (Fig. 7B, lane 3). From these findings we conclude that the 16-kDa protein is the major subunit of the thin pili, whereas the 28-kDa protein is the major subunit of the thick pili.
FIG. 6.
Electron microscopy of pili separated in a sucrose density gradient. Pili were sheared off and separated as described in Materials and Methods. Each fraction was analyzed by electron microscopy. The pellet and fraction 3 of the sucrose density gradient were found to contain enriched intact thin or thick pili. All other fractions were found to contain nearly no pilus structures. (A) Enriched thin pili (inset shows distinct bundles of thin pili). (B) Enriched thick pili in fraction 3. (Inset) View of the end of a thick pilus structure, located perpendicular to the support film (arrowhead); small arrow, penetration of the negative stain into the pilus structure, suggesting an axial pilus region of lower protein density. Bars, 50 nm, 0.2 μm, and 0.1 μm, as indicated.
FIG. 7.
Western blot of pili separated by the sucrose density gradient. The pellet, fraction 2, and fraction 3 of the sucrose density gradient (A) were separated by SDS-PAGE (B), transferred onto nitrocellulose membranes, and probed with anti-MalE-ComP∗ antibodies (C). Molecular masses are indicated.
Of special interest was the question of whether the pilus-containing fractions reacted with the anti-MalE-ComP* antibodies. As evident from Fig. 7C (lane 1), the proteins in the pellet did not react with anti-MalE-ComP* antibodies. Western blot analysis of all fractions of the sucrose density gradient revealed major amounts of ComP in fraction 2. This fraction was found in electron micrographs to contain nearly no pilus structures. In fraction 3 only small amounts of ComP were detected (Fig. 7C, lane 3). Taken together these findings strongly suggest that ComP is not a subunit of the thin and thick pilus structures of Acinetobacter sp. strain BD413.
DISCUSSION
In this study we show that the expression of comP is not correlated with competence induction; comP expression increased during prolonged exponential growth and in stationary phase, whereas the transformation frequencies did not increase. These findings strongly suggest that components of the DNA uptake apparatus are already synthesized in the stationary growth phase, prior to maximal competence induction after dilution into fresh medium. If so, this would suggest that competence induction is not due to induction of protein expression. This view is supported by the finding that during incubation of BD413 cells with DNA the formation of transformants is not inhibited by the addition of chloramphenicol, indicating that no new proteins of the DNA transformation machinery are synthesized during DNA uptake (31).
Competence induction in Acinetobacter sp. strain BD413 is strictly regulated, depending on growth phase (30). Results of detailed physiological studies of competence regulation gave rise to the assumption that regulation involves promoters which react to nutrient upshifts or which seem to be affected by the free-energy load of the cell (30). It was suggested that promoters involved in competence might have similarities to the E. coli fis promoter (30). The results from our transcriptional studies do not support the hypothesis of the involvement of a fis-like promoter in competence development. Our data rather suggest that the comP promoter responds to nutrient limitations.
Apparently, activation of the DNA uptake apparatus after inoculation into fresh medium is very fast; it does not involve protein synthesis (31). Since DNA uptake is an energy-dependent process (32), the low competence observed in the stationary phase could be due to the low energy charge of the cells. Starvation-induced promoters have been subjected to intensive investigations (for a review of stationary-phase promoters see reference 18). Examples are the glucose and amino acid transport systems in several marine Vibrio spp. Analogous to what is found for these transport systems, the different components of the DNA uptake machinery could be synthesized long before their activity is needed, resulting in cells primed for DNA uptake. Inoculation into fresh medium would then provide new energy and a carbon source, the prerequisites for the observed drastic increase in transformation frequencies.
The nonglycosylated ComP was found to completely restore the transformation phenotype when provided in trans in T205 (pCP1) (33). This shows that glycosylation of ComP is not required for its essential function within the DNA binding and uptake process. The occurrence of a 23-kDa form of ComP specifically in the outer membrane might be attributed to a further modification. This has been shown for the meningococcal pilin; it contains an α-glycerophosphate substituent attached to Ser93 by a phosphodiester linkage (42). It has been suggested that glycerol residues might serve as a substrate for fatty acylation, thereby being involved in membrane anchoring of the pilin (42). Since ComP is similar to the meningococcal pilin and also contains several central serine residues (33), it is tempting to speculate that ComP might also contain an α-glycerophosphate substituent.
The similarities between several proteins involved in DNA uptake, protein secretion, or type IV pilus systems indicate that they might belong to evolutionarily related systems comprising cell envelope-spanning structures with a general architecture and general components (12, 34). It is known that PilA, the type IV pilus subunit in Pseudomonas aeruginosa, not only constitutes the main structural pilus protein but is also involved in protein secretion (24). Hence one might anticipate that the respective components from Acinetobacter sp. strain BD413 might also have dual functions. Due to the conserved peptidase cleavage site in ComP it is highly likely that a single peptidase might process both the pilin-like competence factor ComP and the subunits of the BD413 pilus structures, as described for peptidase PilD in P. aeruginosa, which is involved in processing PilA and pilin-like proteins (23). Since pilus mutants of strain BD413 have not been isolated up to now, studies addressing the question of whether components of the Acinetobacter sp. strain BD413 pili or the pilus structures are involved in DNA transformation have not been possible. Components with dual function in DNA uptake and pilus biogenesis in Acinetobacter sp. strain BD413 have not been identified.
The question of whether the type IV pilus structures themselves are involved in DNA transformation has not yet been answered unequivocally. The DNA transformation systems of B. subtilis and H. influenzae, microorganisms which have never been found to exhibit type IV pilus structures, contain components similar to proteins implicated in type IV pilus biogenesis in other organisms (5, 20, 40). In N. gonorrhoeae, which exhibits a type IV piliation phenotype, the structural subunits of type IV pili (PilE) and the type IV pilus factor PilC are essential for DNA transformation (7, 35). Furthermore it was demonstrated that gonococcal PilT mutants exhibit simultaneous defects in competence for natural transformation and twitching motility (43). Nevertheless, these mutants exhibit structurally and morphologically intact pili (43). Recently a pilin-like gonococcal competence factor, ComP, has been identified and has been found to be dispensable for pilus biogenesis in N. gonorrhoeae (44). Very recently Legionella pneumophila mutants defective in DNA transformation were generated, and the molecular analysis of one mutant mapped the mutation within pilEL, the gene encoding the structural subunit of type IV pili (39). Taken together these results suggest that type IV pili and DNA transformation machineries are somehow linked; further studies are needed.
Our finding that transformation-affected Acinetobacter mutants carrying mutations within one or more of the pilin-like competence genes comB, comE, comF, and comP exhibit morphologically and functionally intact pili (3, 11, 33) led to the proposal that the pilus structure itself is not involved in DNA transformation. The results presented here strongly suggest that ComP is at least not a major subunit of the two fimbrial structures detected by electron microscopy. However, we cannot exclude the possibility that minor amounts of ComP, not detectable by Western blotting, are present in the thin and thick pili. Furthermore an association of ComP with the subsurface base of these pilus structures cannot be absolutely excluded. Nevertheless, our data are not in favor of the idea that the thin and thick pili are part of DNA binding or uptake machinery in Acinetobacter sp. strain BD413. Rather, ComP, which is preferentially located in the outer membrane, is part of a specific DNA binding and/or transporting structure. Attempts to isolate such a complex from Acinetobacter sp. strain BD413 have just begun.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (grant Av 9/4-2 and Graduiertenkolleg “Chemische Aktivitäten von Mikroorganismen”).
We are grateful to G. Gottschalk, Göttingen, Germany, for generous support. The contribution of C. Rosenplänter, Göttingen, Germany, to the studies concerning the cellular localization of ComP in comE and comF mutants is gratefully acknowledged. Many thanks are due to V. Müller, München, Germany, for critical reading of the manuscript.
REFERENCES
- 1.Alm R A, Mattick J S. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol Microbiol. 1995;16:485–496. doi: 10.1111/j.1365-2958.1995.tb02413.x. [DOI] [PubMed] [Google Scholar]
- 2.Blum H, Beier H, Gross H J. Improved silver staining method of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 3.Busch S, Rosenplänter C, Averhoff B. Identification and characterization of comE and comF, two novel pilin-like competence factors involved in natural transformation of Acinetobacter sp. strain BD413. Appl Environ Microbiol. 1999;65:4568–4574. doi: 10.1128/aem.65.10.4568-4574.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dougherty B A, Hamilton O S. Identification of Haemophilus influenzae Rd transformation genes using cassette mutagenesis. Microbiology. 1999;145:401–409. doi: 10.1099/13500872-145-2-401. [DOI] [PubMed] [Google Scholar]
- 5.Dubnau D. Binding and transport of transforming DNA by Bacillus subtilis: the role of type IV pilin-like proteins. Gene. 1997;192:191–198. doi: 10.1016/s0378-1119(96)00804-9. [DOI] [PubMed] [Google Scholar]
- 6.Edge A S, Faltynek C R, Hof L, Reichert L E, Weber P. Deglycosylation of glycoproteins by trifluoromethanesulfonic acid. Anal Biochem. 1981;118:131–137. doi: 10.1016/0003-2697(81)90168-8. [DOI] [PubMed] [Google Scholar]
- 7.Fussenegger M, Rudel T, Barten R, Ryll R, Meyer T F. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae. Gene. 1997;192:125–134. doi: 10.1016/s0378-1119(97)00038-3. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 9.Hancock R E W, Nikaido H. Outer membranes of gram-negative bacteria. XIX. Isolation from Pseudomonas aeruginosa PAO1 and use in reconstitution and definition of the permeability barrier. J Bacteriol. 1978;136:381–390. doi: 10.1128/jb.136.1.381-390.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartnett G B, Averhoff B, Ornston L N. Selection of Acinetobacter calcoaceticus mutants deficient in the p-hydroxybenzoate hydroxylase gene (pobA), a member of a supraoperonic cluster. J Bacteriol. 1990;172:6160–6161. doi: 10.1128/jb.172.10.6160-6161.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herzberg C, Friedrich A, Averhoff B. comB, a novel competence gene required for natural transformation of Acinetobacter sp. BD413: identification, characterization, and analysis of growth-phase-dependent regulation. Arch Microbiol. 2000;173:220–228. doi: 10.1007/s002039900134. [DOI] [PubMed] [Google Scholar]
- 12.Hobbs M, Mattick J S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 13.Hoshino T, Kageyama M. Purification and properties of a binding protein for branched-chain amino acids in Pseudomonas aeruginosa. J Bacteriol. 1980;141:1055–1063. doi: 10.1128/jb.141.3.1055-1063.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue H, Nojima H, Okayama H. High efficiency transformation of E. coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 15.Juni E, Janik A. Transformation of Acinetobacter calcoaceticus (Bacterium anitratum) J Bacteriol. 1969;98:281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 17.Kokotek W, Lotz W. Construction of a lacZ-kanamycin-resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene. 1989;84:467–471. doi: 10.1016/0378-1119(89)90522-2. [DOI] [PubMed] [Google Scholar]
- 18.Kolter R. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Larson T G, Goodgal S H. Sequence and transcriptional regulation of com101A, a locus required for genetic transformation in Haemophilus influenzae. J Bacteriol. 1991;173:4683–4691. doi: 10.1128/jb.173.15.4683-4691.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Link C, Eickernjäger S, Porstendörfer D, Averhoff B. Identification and characterization of a novel competence gene, comC, required for DNA binding and uptake in Acinetobacter sp. strain BD413. J Bacteriol. 1998;180:1592–1595. doi: 10.1128/jb.180.6.1592-1595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorenz M G, Wackernagel W. Bacterial gene transfer by genetic transformation in the environment. Microbiol Rev. 1994;58:563–603. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lory S, Strom M S. Structure-function relationship of type-IV prepilin peptidase of Pseudomonas aeruginosa. Gene. 1997;192:117–121. doi: 10.1016/s0378-1119(96)00830-x. [DOI] [PubMed] [Google Scholar]
- 24.Lu H M, Motley S T, Lory S. Interactions of the components of the general secretion pathway: role of Pseudomonas aeruginosa type IV pilin subunits in complex formation and protein secretion. Mol Microbiol. 1997;25:247–259. doi: 10.1046/j.1365-2958.1997.4561818.x. [DOI] [PubMed] [Google Scholar]
- 25.Maina C V, Riggs P D, Grandea III A G I, Slatko B E, Moran L S, Tagliamonte J A, McReynolds L A, Guan C D. An Escherichia coli vector to express and purify foreign proteins by fusion to and separation from maltose-binding protein. Gene. 1988;74:365–373. doi: 10.1016/0378-1119(88)90170-9. [DOI] [PubMed] [Google Scholar]
- 26.Miller J H. Assay of β-galactosidase. In: Platt T, Müller-Hill B, Miller J H, editors. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. pp. 351–355. [Google Scholar]
- 27.Moens S, Vanderleyden J. Glycoproteins in procaryotes. Arch Microbiol. 1997;168:169–175. doi: 10.1007/s002030050484. [DOI] [PubMed] [Google Scholar]
- 28.Mott J E, Grant R A, Ho J, Platt T. Maximizing gene expression from plasmid vectors containing the lambda P1 promoter: strategies for overproducing transcription factor. Proc Natl Acad Sci USA. 1985;82:88–92. doi: 10.1073/pnas.82.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ornston L N, Stanier R Y. The conversion of catechol and protocatechuate to β-ketoadipate by Pseudomonas putida. 1. Biochemistry. J Biol Chem. 1966;241:3776–3786. [PubMed] [Google Scholar]
- 30.Palmen R, Buijsman P, Hellingwerf K J. Physiological regulation of competence induction for natural transformation in Acinetobacter calcoaceticus. Arch Microbiol. 1994;162:344–351. [Google Scholar]
- 31.Palmen R, Hellingwerf K J. Uptake and processing of DNA by Acinetobacter calcoaceticus. Gene. 1997;192:179–190. doi: 10.1016/s0378-1119(97)00042-5. [DOI] [PubMed] [Google Scholar]
- 32.Palmen R, Vosman B, Buijsman P, Breek C K D, Hellingwerf K J. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J Gen Microbiol. 1993;139:295–305. doi: 10.1099/00221287-139-2-295. [DOI] [PubMed] [Google Scholar]
- 33.Porstendörfer D, Drotschmann U, Averhoff B. A novel competence gene, comP, is essential for natural transformation of Acinetobacter sp. strain BD413. Appl Environ Microbiol. 1997;63:4150–4157. doi: 10.1128/aem.63.11.4150-4157.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudel T, Facius D, Barten R, Scheuerpflug I, Nonnenmacher E, Meyer T F. Role of pili and the phase-variable PilC protein in natural competence for transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1995;92:7986–7990. doi: 10.1073/pnas.92.17.7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-termination inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stinson E, Virij M, Makepeace K, Dell A, Morris H R, Payne G, Saunders J R, Jennings M P, Barker S, Panico M, Blench I, Moxon E R. Meningococcal pilin: a glycoprotein substituted with digalactosyl 2,4-diacetamido-2,4,6-trideoxyhexose. Mol Microbiol. 1995;17:1201–1214. doi: 10.1111/j.1365-2958.1995.mmi_17061201.x. [DOI] [PubMed] [Google Scholar]
- 39.Stone B J, Abu Kwaik Y. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J Bacteriol. 1999;181:1395–1402. doi: 10.1128/jb.181.5.1395-1402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomb J F, El-Hajj H, Smith H O. Nucleotide sequence of a cluster of genes involved in the transformation of Haemophilus influenzae Rd. Gene. 1991;104:1–10. doi: 10.1016/0378-1119(91)90457-m. [DOI] [PubMed] [Google Scholar]
- 41.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Virji M. Post-translational modifications of meningococcal pili. Identification of common substituents: glycans and α-glycerophosphate. Gene. 1997;192:141–147. doi: 10.1016/s0378-1119(97)00082-6. [DOI] [PubMed] [Google Scholar]
- 43.Wolfgang M, Lauer P, Park H, Brossay L, Hebert J, Koomey M. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol. 1998;29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- 44.Wolfgang M, van Putten J P M, Hayes S F, Koomey M. The comP locus of Neisseria gonnorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol Microbiol. 1999;31:1345–1357. doi: 10.1046/j.1365-2958.1999.01269.x. [DOI] [PubMed] [Google Scholar]