Abstract
The pairing of analytical chemistry with genomic techniques represents a new wave in natural product chemistry. With an increase in the availability of sequencing and assembly of microbial genomes, interrogation into the biosynthetic capability of producers with valuable secondary metabolites is possible. However, without the development of robust, accessible, and medium to high throughput tools, the bottleneck in pairing metabolic potential and compound isolation will continue. Several innovative approaches have proven useful in the nascent stages of microbial genome-informed drug discovery. Here, we consider a number of these approaches which have led to prioritization of strain targets and have mitigated rediscovery rates. Likewise, we discuss integration of principles of comparative evolutionary studies and retrobiosynthetic predictions to better understand biosynthetic mechanistic details and link genome sequence to structure. Lastly, we discuss advances in engineering, chemistry, and molecular networking and other computational approaches that are accelerating progress in the field of omic-informed natural product drug discovery. Together, these strategies enhance the synergy between cutting edge omics, chemical characterization, and computational technologies that pitch the discovery of natural products with pharmaceutical and other potential applications to the crest of the wave where progress is ripe for rapid advances.
Genomic approaches to drug discovery have not only focused on the study of human genomes to better understand protein targets, cellular cascades, drug resistance, epigenetics, and their implications on human disease to further refine drug discovery efforts,1,2 but genomic studies have revealed a vast repertoire of diversity in microbial metabolic innovation which can also be paired with metabolomics for the study of microbially produced secondary metabolites. Secondary metabolites have garnered great interest as potential pharmaceuticals as the diverse chemical scaffolds are well suited for biological targets.3 Over 50% of FDA-approved medications have been sourced directly from or inspired by nature.4
Historically, methods of natural product isolation and characterization relied heavily on extraction of secondary metabolites from both microorganisms and macroorganisms requiring time-intensive analytical procedures for isolation and compound characterization. Over time, several barriers to natural product discovery have been identified, such as high rediscovery rates and the potential ecological impact of mass field collections, with implications on the sustainability of the source. Technological advances have now ameliorated most of these concerns. For example, omics techniques have been harnessed to prioritize samples, quickly identify metabolites of interest, and utilize genomic information to inform natural product discovery.5−7 Technological advances also enhance the potential for a more sustainable exploration of nature’s chemical wealth and for creation of an ongoing supply of compounds through biotechnology.
One of the primary ways that genomics is integrated with drug discovery of natural products is by the identification of biosynthetic gene clusters (BGCs). BGCs encode for the enzymatic machinery, ranging in organization from single iteratively acting enzymes to multidomain megaenzymes with numerous catalytic sites, that are responsible for the biosynthesis of secondary metabolites.8 These discrete genomic elements are similar to, but evolutionarily divergent from, genes involved in primary metabolism (i.e., polyketide synthases are likely derive from fatty acid synthases). The genes have been evolutionarily repurposed to produce an array of architecturally diverse compounds under tight stereochemical controls with a strong affinity toward biological targets. BGCs can be horizontally transferred from one organism to another, a phenomenon that can be identified through phylogenetic analysis, as the evolutionary history of clustered elements within a BGC can be quite divergent from the remainder of the genome.9,10
Today, there is a wealth of publicly accessible databases tailored to the fields of genomics and natural products, some linking the two disciplines, and several of which are community-curated with ongoing contributions that serve as data resources (Table 1). Data sourced from these repositories are often used in comparative analyses with a multitude of software tools resulting in powerful analyses that are becoming more integrated due to improvements in cross-communication between the fields and resources available (Table 1). There were significant barriers in the past with databases having little crosstalk, particularly between different disciplines and technologies (e.g., biosynthetic gene cluster data with metabolomic profiles). This divide demonstrates the importance of interdisciplinary collaborations and community curation in natural product chemistry and is consistent with the movement toward open-source software that evolves and incorporates new tools and strategies.
Table 1. List of Resources and Accompanying Website for Each of the Approaches Presented.
The power of analytical chemistry in natural product drug discovery remains a critical element of the field. Traditional methods for isolation and characterization of natural products, notably the use of chromatography, mass spectrometry (MS), and nuclear magnetic resonance (NMR), are a mainstay for structure isolation and characterization. When combined with genome and metagenome-informed approaches, analytical techniques are being used to advance the technological frontier of drug discovery. Examples of these approaches are discussed below, and new approaches are proposed that will continue to merge the fields of genomics and metabolomics in a synergistic way to enhance natural product discovery efforts.
Context and Scope: Integration of Interdisciplinary Omics Approaches to Advance Natural Product Discovery Efforts
In this perspective, we explore emerging technologies and computational tools that pair genomic and metabolomic data for natural product drug discovery (Figure 1). What follows is a compilation of approaches (or workflows; Table 1) that highlight the synergy between genomics and analytical chemistry which will likely continue to enhance and refine microbial natural product discovery efforts. This is not meant to serve as a comprehensive overview of all available tools and strategies as there are several reviews that discuss mass spectral databases and genomic databases as well as tools aimed at mining the big data resulting from both. We refer the reader to those reviews for further information on the tools discussed here.5−7,11−13 The tools discussed here were published before January 2022.
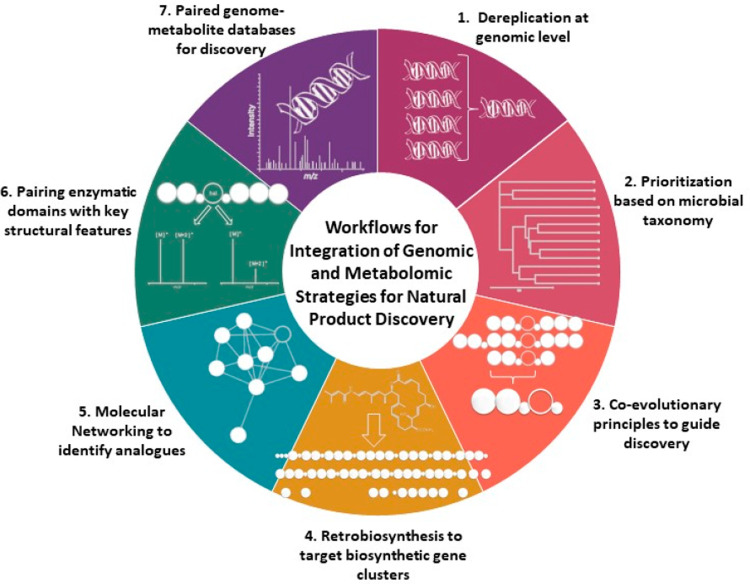
Figure 1.
Workflows for integration of genomic and metabolomic strategies for natural product discovery.
Workflows for Integrated Approaches to Genome-Enabled Natural Product Biodiscovery
Approach 1: Dereplication at the Genomic Level to Reduce Rediscovery Rates of Known Natural Products and Enhance Discovery of Novel Natural Products
Dereplication of chemical structures, through detailed comparison of NMR and MS data of an isolated compound to the scientific literature and to chemical databases, is a long-standing strategy to reduce rediscovery in the early phases of natural product isolation process. A similar strategy can be employed for rapid dereplication using genomic information. BGCs identified in new (meta)genome sequences can be rapidly searched against global databases (e.g., NCBI)14 in addition to more curated tools (e.g., AntiSMASH)15 which assess homology across modules, genes, and full length BGCs. Due to the exponentially increasing volume of data, number of data repositories, and bioinformatic tools for known BGCs, we are in a renaissance of natural product biodiscovery.
Basic Local Alignment Search Tool (BLAST)16 searches have been a bioinformatic mainstay for comparing nucleotide and protein sequence information through the detection of regions of similarity between the inquiry sequence and a vast database of biological sequences with a broad range of taxonomic representation. There are innate limitations to BLAST searches for BGCs, however, given the lengthy multidomain nature of these cassettes. BLAST searches can help one gain a clearer understanding of the biosynthetic substructures; however, the tool is not well suited for rapid identification or comparison of entire BGCs or for unique and novel sequences.
The antiSMASH BGC annotation pipeline15 harbors annotations of putative BGCs based on their identifications using the antiSMASH algorithm. With over 150,000 putative BGCs, the ClusterBlast algorithm nested within antiSMASH compares submitted sequences to those in the database for analyses of fungal and bacterial BGCs.15 The Integrated Microbial Genome Atlas of Biosynthetic Gene Clusters (IMG/ABC)17 contains over 400,000 BGCs. Further, gene cluster families, which share similarities in gene structure, can be identified and visualized using BiGSCAPE.18 In concert, these integrated tools allow for rapid dereplication of genome-encoded BGC sequences that are critical in the biosynthesis of natural products and enhance the likelihood of discovery of not only new BGCs but also new secondary metabolites.
Recently, the scientific community has developed a common language and data standard to communicate the biosynthetic gene cluster data and associated chemistry. The standard, led by Kautsar and colleagues and referred to as the Minimum Information about a Biosynthetic Gene Cluster (MIBiG),19 is accompanied by a repository which houses nearly 2000 BGCs as of early 2022 (Figure 2). This resource allows for manual curation and annotation by the natural product community and the MIBiG developers. It serves as a centralized space to deposit and access valuable data about BGCs including information on enzymatic features, protein sequences, taxonomic origins, and associated chemical structures. MIBiG has also been incorporated into antiSMASH15 and is used to screen (meta)genomes submitted for BGC analysis for similarity to known BGCs in the MIBiG database. Beyond identification of identical BGCs and dereplication, the referencing of the MIBiG repository within antiSMASH provides a percent identity score for submitted BGCs affording the opportunity to utilize the similarities in gene structure to target novel metabolites. On the basis of the differences in the gene sequence, differences in chemical structure can be inferred. This allows for rapid dereplication as part of the antiSMASH genome mining pipeline. Metagenomic libraries can therefore be efficiently interrogated for the presence of BGCs and evaluated for homologies to known BGCs. This strategy can guide prioritization of BGCs of interest for further investigation with cultivation or biotechnological measures.
Figure 2.
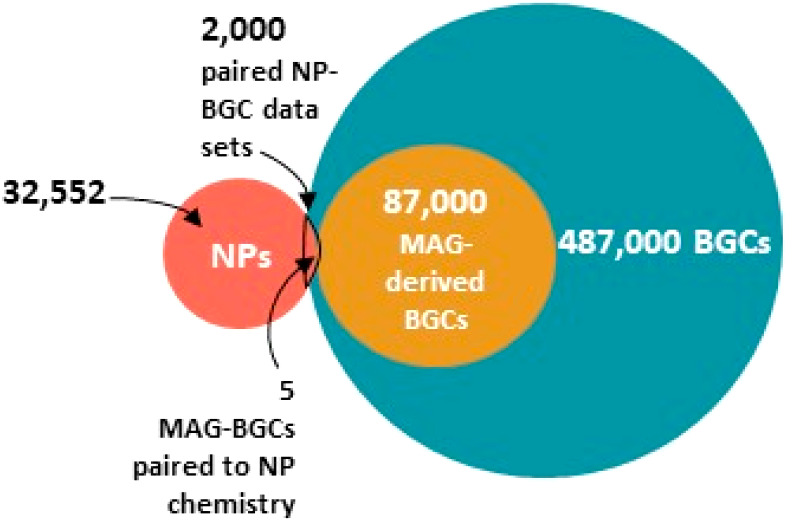
Nearly 500,000 BGCs have been identified, of which 87,000 are derived from metagenome-assembled genomes (MAGs). Of the identified natural products, only 2000 have been paired with BGCs, and only five of those are associated with metagenome-assembled genomes. Data obtained from NP Atlas,20 IMG-ABC,21 MiBIG,19 and the GEM Catalog.22
Approach 2: Prioritization of Drug Discovery Efforts Based on Biosynthetic Potential of Certain Microbial Taxa
Across the immense diversity of the bacterial domain of life, the distribution of biosynthetic gene clusters is only recently becoming understood.23 Certain bacterial taxonomic families are known to be more biosynthetically talented than others. However, biosynthetic potential, the likelihood that secondary metabolites can be produced, is understudied in many lineages. This makes the prospect for novelty high, particularly in poorly studied lineages, many of which have evaded cultivation. BGCs with low levels of similarity, in particular, those associated with poorly known phylogenetic uniqueness, can be used to mine novel BGCs and point to new compounds. Alternatively, novelty remains to be discovered even in more familiar lineages including two distinct bacterial groups that are known to be particularly rich in BGCs and known to produce bioactive compounds (filamentous cyanobacteria and Streptomyces). For example, Leão and colleagues characterized the biosynthetic potential based on BGC classes and resultant metabolites from tropical filamentous marine cyanobacteria.24 They suggested that “natural product diversity hotspots” should be prioritized, while ecosystems or niches with low beta-diversity when paired with low numbers of BGCs were deemed to be more likely to result in rediscovery of known natural products.24 Many of the FDA-approved anti-infectives are derived from actinomycetes, specifically Streptomyces spp. There are several examples applying Streptomyces-targeted investigation that are yielding novelty. Soldatou and colleagues specifically looked at the biosynthetic potential residing within Arctic and Antarctic actinomycetes and, through extensive cultivation efforts and the one strain–many compounds (OSMAC) method, were able to confirm high rates of both metabolic diversity and anti-infective bioactivity.25 The microbial diversity in Indonesian bacterial strains used a similar strategy of linking genomic and metabolomic data to leverage biosynthetic talent within their collection of Streptomyces spp. and promote the discovery of novel natural products.26 Each of these research teams utilized the Biosynthetic Gene Similarity Clustering and Prospecting engine (BiG-SCAPE)18 which groups BGCs into gene cluster families based on sequence similarity networks. This software tool is now paired with the CORe Analysis of Syntenic Orthologues to prioritize Natural Product Gene Clusters (CORASON),18 a tool that defines phylogenetic relationships within gene cluster families.
Additional tools for rapid evaluation of biosynthetic potential based on phylogeny and taxonomy include NaPDos and AutoMLST. The Natural Product Domain Seeker (NaPDoS) can be used to identify biosynthetic enzymes and therefore biosynthetic wealth, with a focus on PKS and NRPS genes, in genomic and metagenomic data.27 This tool phylogenetically classifies condensation domains and ketosynthase domains resulting in a phylogenetic tree of these domains with those of known BGCs to help determine similarities with known biosynthetic pathways. Another phylogenetic tool that can aid in determining biosynthetic potential is the Automated Multi-Locus Species Tree (AutoMLST)28 which builds phylogenetic trees from a simplified user interface to infer relationships of bacteria from the users’ collections. The output includes taxonomic clade information, and data points can then be used to make inferences about the biosynthetic potential of the associated species.
For laboratory groups with extensive bacterial culture collections, prioritization of strains based on their phylogenetic placement can be key in streamlining drug discovery efforts. These strategies of prioritization can be paired with targeted large-scale cultivation efforts and subsequent natural product isolation and characterization as well as biotechnological means to express specific BGCs for drug discovery.
Approach 3: Use of Coevolutionary Principles to Guide Genome-Mining
Another valuable approach has been introduced by Ziemert and colleagues through the Antibiotic Resistant Target Seeker (ARTS).29 The software tool pairs algorithms to determine phylogenic discrepancies which may indicate horizontal gene transfer, with additional features such as gene duplication and proximity to resistance markers that can be used to highlight those clusters that may have an increased likelihood of producing anti-infective bioactive molecules.29 Although the initial version was focused primarily on actinobacterial genomes, the release of the second ARTS version in 2020 expanded the tool to allow for analysis of genomes from all bacterial taxa as well as from metagenomic data. Since bacteria have been found to harbor resistance genes integral to the prevention of self-toxicity in genomic proximity to the secondary metabolites they produce,30 this strategy is a way to streamline genomic-based antibiotic discovery. These mechanisms include, but are not limited to, pro-drug formation, other chemical modification, use of efflux pumps, and self-resistant protein variants.30 Manual interrogation for specific markers not yet included in ARTS 2.0 can be used to complement and expand the search for relevant resistance genes. For example, glycosylation via glycosyl transferases can be used for pro-drug formation and self-protection.31,32
Additional tools utilize a targeted genome mining approach through analyzing genomic data sets for proteins or enzymatic domains that are believed to be a key part of biosynthesis; however, they are not readily detected in the current algorithms for BGC identification. For example, EvoMining33 utilizes coevolutionary principles for comparative genome mining based on “expansion-and-recruitment events”, gene recruitment associated with signatures of rapid evolution found as enzyme families. This tool has expanded BGC identification and annotation substantially, particularly for those biosynthetic enzymes that are not associated with PKS, NRPS, or hybrid PKS–NRPS systems, and are therefore more difficult to analyze than their more modular counterparts.33,34
Co-occurrence of tandem enzyme domains has been used to identify previously poorly annotated BGCs that are responsible for biosynthesis of oxazolone-containing natural products.35 The aptly named Co-occurrence of Enzymatic Domains (CO-ED) workflow developed by de Rond and colleagues is based on a genome mining approach that focuses on the presence of a series of catalytic domains within a protein that together perform a specific biochemical transformation.35 The workflow was successfully used to interrogate a library of genomes to guide the functional annotation of a new oxazolone synthetase, and subsequently, a suite of new oxazolone natural products was characterized through MS and spectroscopic methods after heterologous expression of the corresponding genes.35
Approach 4: Retrobiosynthesis for Targeted BGC Identification of Known Natural Products
Retrobiosynthetic analysis is the process of determining likely biosynthetic steps based on biosynthetic subunit precursors that comprise the molecule of interest. This analysis can be used to identify if the BGC responsible for a compound of interest is reasonable, particularly when colinear arrangement of genes and enzymatic activity are suspected in the biosynthesis of a given product. This type of modular organization is often seen in Type I PKS, NRPS, and hybrid PKS–NRPS systems. The secondary metabolites that result from these classes of BGCs generally demonstrate colinearity, meaning that the genomic code, enzymatic domains, and resultant chemical structures are closely linked, conferring a degree of predictability.36−38 This colinearity can be harnessed to enhance drug discovery efforts. Retrobiosynthetic strategies can predict the specific enzymatic domains that would be responsible for creating a particular structural feature or to match a known structure to the series of enzymatic domains within a BGC. Modular organization of PKS and NRP systems makes them conducive to biotechnological engineering and combinatorial biosynthesis. This, combined with the tight stereospecific control promoted by the biosynthetic enzymes for the architecturally complex natural products,39 makes identification and expression of BGCs particularly appealing.
As demonstrated by our group with palmerolide biosynthesis,40 a retrobiosynthetic strategy can be utilized to identify the BGC implicated in the biosynthesis of a specific secondary metabolite out of a diverse metagenome represented by many bacterial genomes. In the case of palmerolide A, key enzymatic features were used to interrogate the metagenome from environmental samples, and the modular arrangement of the biosynthetic pathway derived from the retrobiosynthetic analysis was used to identify the putative BGC. Identification of the BGC40 and host-associated microbial producer41 now paves the way for drug development efforts through biotechnological means (i.e., through heterologous expression or targeted cultivation efforts). Other examples of BGC identification using similar strategies within genomes of previously characterized polyketide compounds with potent bioactivity include those for calyculin (cytotoxicity),42,43 corallopyronins (broad spectrum antibiotic activity),44,45 and bryostatins (anticancer and neuroprotective activity).46−50 The implications for these strategies cross into the various compound classes and can help propel compounds with pharmaceutical promise through the drug discovery pipeline.
Approach 5: Use of Molecular Networking to Identify and Target New Analogues Arising from the Same or Highly Similar BGCs
Tandem mass spectral data from bacterial cultures or environmental samples can be analyzed in molecular networks via Global Natural Products Social Molecular Networking (GNPS).51 Useful for library searches for small molecules and peptides, for initial dereplication, and to evaluate the tandem MS data in chemical space, this increasingly robust tool is a powerful companion for drug discovery efforts. Molecular networking can be paired with genomic workflows for analogue molecule identification resulting from BGC expression, whether in the native host or in a heterologous host. Due to the sequential head-to-tail elongation steps that occur in a modular fashion in both modular Type I PKS and NRPS systems, and the diversity introduced by post-translational modifications to the established core sequence within RiPPs, these biosynthetic systems are amenable to combinatorial biosynthetic methods37,38 in which molecular networking can be an efficient way of analyzing outcomes.
Molecular networking can be used as a screening tool in novel bacterial cultivation efforts to identify analogues of known natural products as well as to highlight the presence of new metabolites arising from a single BGC. Using m/z differences between interconnected nodes within a cluster, which represent distinct compounds based on a consensus spectrum, structural differences among analogues can be inferred. Alternately, clusters of related ions with no match to known metabolites are indicative of new chemistry. In either case, MS-guided isolation using mass-selective fractionation can be pursued to focus purification strategies on the unknown masses.
Analogues can be formed with heterologous expression of BGCs, as seen with verticilactams,52 as well as after deletion or alteration of enzymatic domains within BGCs and subsequent expression, as seen with the BGCs for complestatin and lobophorin.53,54 Therefore, after synthetic biology experiments, a molecular network can be used to visualize the biocombinatorial chemical space through uploading the tandem mass spectral data of the metabolites produced by the wild type producer and those that arise from the synthetic biology experiments. A direct comparison can be performed that includes confirmation of contribution to nodes that represent the compound(s) of interest and an evaluation of new nodes that may appear within a compound cluster, indicating new analogues. The MS2 fingerprints for nodes representing these additional products can assist in structure elucidation and evaluation of the underlying mechanisms for their biosynthesis. A similar strategy was used following the expression of the alterchromide BGC55 and the cosmomycin BGC56 to identify previously undescribed analogues. More recently, molecular networking aided in identification of the production of several new herbicidin analogues following overexpression of the herbicidin BGC57 and in a new acylhomoserine lactone (AHL) after expression of an AHL synthase.58
Approach 6: Use of Enzymatic Domains within BGCs to Identify Key Structural Features of Unknown Products That Can Be Paired with Analytical Tools
Following annotation of enzymatic domains within a BGC, the presence of a number of domains can be used to identify a previously unknown product or feature within a product, as some metabolomic signatures on MS can be paired with functional enzymatic annotations within BGCs. For example, if a halogenase is present in a BGC, a halogenation signature on MS could be used for the isolation of a halogenated natural product. In a similar manner, the metabolomic signatures for sulfate, phosphate, and carbamate groups can be associated with enzymatic domains such as sulfatases, phosphatases, and carbamoyl transferases. These strategies have potential implications for discovery of natural products and may also play a role in linking an orphan BGC (BGC with no known product) within a bacterial genome to a previously characterized compound containing a specific functional group or groups.
Harnessing the ability to identify isotopic patterns using NMR and MS, isotopically labeled precursors as informed by BGCs, can be used for natural product discovery. In the case of the orfamides, the cultures of Pseudomonas fluorescens Pf-5 were fed isotopically labeled amino acids that were selected based upon adenylation domain specificity from the genomic information on an orphan gene cluster. Isotope-guided fractionation using NMR (in parallel with bioassay-guided fractionation) was utilized, and the structure of orfamide A was determined through NMR experiments, GC-MS, and Marfey’s analysis.59 Building upon the genomisotopic approach, Gerwick and colleagues later enriched the media of cyanobacterial cultures with 15N-nitrate and performed repeated MALDI experiments on single-filaments of Moorena producens JHB that allowed for the identification and subsequent isolation of a new natural product, cryptomaldamide, through an MS-guided fractionation approach.60 Full characterization was performed through spectroscopic methods, and the compound’s structure was linked to a putative BGC based on genomic analysis. The 28.7 kbp BGC for cryptomaldamide was subsequently heterologously expressed in a genetically tractable host, Anabaena PCC, giving further significance to this method of natural product discovery.61 Recently developed by Linington and colleagues, the IsoAnalyst platform uses isotopic labeling of biosynthetic precursors in parallel feeding experiments paired with MS to link BGCs to their natural products.62 The platform utilizes biosynthetic relatedness based on the isotopic patterns rather than deriving structural information on fragments from the tandem mass spectral data. Validated with erythromycin and its analogues, IsoAnalyst was also used to discover a new lobosamide analogue and a new desferrioxiamine compound from Micromonospora sp.62
Approach 7: Harnessing Paired Genome–Metabolite Data Sets and Tools for Natural Product Discovery
Tools to link BGCs and MS spectra have been developed, in particular, for NRPs. Released by Behsaz and colleagues, NRPminer63 pairs tandem mass spectral data with NRPS BGCs and builds upon previously developed tools, such as NRP2 Path64 and NRPquest.65 Through a series of steps which integrate antiSMASH BGC identification from a sequenced genome,15 GNPS molecular networking of the associated sample,51 and VarQuest66 searches, a list of potential enzymatic assembly lines is populated, filtered, overlaid with potential modifications, and then used to predict possible backbone structures of NRPs. These predicted structures are used to search the mass spectral data for matches that are scored based upon similarity. NRPminer is different from other tools for tandem MS and NRPS BGC pairing as it builds upon the principles of collinearity, allows for broad adenylation domain specificity (including duplication of open reading frames), and is flexible with respect to diverse post-assembly modifications.
Recently, another resource for pairing genomic and metabolomic data sets was released. The Paired Omics Data Platform (PoDP)67 is a community-curated platform that links genomic or metagenomic data, metabolomic data, and metadata regarding experimental details on culture, extraction, and instrumentation methods. The purpose of the platform is not to host the big data, but rather link the data that are deposited in various public repositories (many of which have been described above) in order to promote easy access to these carefully curated paired data sets for large computationally driven projects or for smaller scale individual analysis. This tool expands upon the efforts put forth from other teams who have developed concepts designed to link omics databases, including peptiogenomics,68 metabologenomics,69 and MetaMiner.70
Considerations and Challenges
In principle, there are no specific limitations to broad adoption of the approaches and technologies introduced here. Interdisciplinary training across biological and chemical sciences is essential, as are strong computational skills. Likewise, the value of cross-disciplinary collaboration can add paramount value to these studies. However, there are a number of considerations to the selection of an approach including the type of genomic data available and the research objective. Many of the tools and strategies discussed above are amenable to input from various sequence types, including whole genomes, partial genomes, metagenomic assemblies, and metagenome-assembled genomes or single amplified genomes. When considering “big data” and integrating different data types, the source, type, and completeness must be considered in any analyses. Completeness can be problematic in cases of phylogenetic novelty that can influence annotation accuracy and upon analysis of host-associated microbes which may have undergone genomic streamlining. Another barrier to analysis occurs during genome assembly, in which repetitive modules that are often seen in biosynthetic gene clusters can make assembling the sequences challenging. Long-read technologies and specialized assembly pipelines can help overcome this obstacle.41
Transcriptional regulation of cryptic BGCs presents a challenge to linking sequence to product and is a currently active area of research.71−73 Identification of an interesting BGC does not necessarily indicate that the cluster is translated and the product biosynthesized under laboratory culture conditions. At times, unsilencing of cryptic BGCs can be promoted through the one strain–many compounds (OSMAC) approach74 or coculture experiments; however, at other times, this is an issue best solved at the genome level through genetic engineering of promoters to control expression in addition to screening at the levels of gene and protein expression as well.
As mentioned in brief above, some BGCs, such as terpene synthases and Type II PKSs, are inherently more challenging to link to their products due to a lack of colinearity which limits structural prediction. Likewise, nascent understanding of enzymatic subtypes that catalyze specific reactions during biosynthesis limit retrobiosynthetic predictions.
Another hurdle to drug development directly from microbial sources is that cultivation efforts are not always successful, or if isolated in pure culture, growth rates can be suboptimal for scale-up. Along the same lines, heterologous host suitability is an additional consideration. Finding a genetically tractable host can be difficult—particularly for novel lineages. The repertoire of heterologous hosts is dominated by Streptomyces strains, although is expanding and currently includes strains of E. coli, Saccharomonspora sp., Salinospora sp., Pseudoaltermonas sp., Anabaena sp., Synechococcus sp., among others.75
Lastly, there are also analytical challenges for compound isolation and structure elucidation. For example, low natural abundance chemical analogues identified in a molecular network may preclude subsequent characterization of the proposed analogue.
Perspectives and Possible Research Directions
The rise of genomic-based workflows for microbial natural product drug discovery is advancing the field. Genomic approaches for prioritization of bacteria enriched with biosynthetic pathways and deprioritization of BGCs known to produce previously identified compounds can help overcome the hurdle of high rediscovery rates of natural products. Taxonomy, phylogenetics, and the use of coevolutionary principles can aid in prioritization of BGCs. Additionally, in order to address orphan gene clusters or compounds that have not yet been linked to BGCs, retrobiosynthetic principles can be applied. Analytical metabolomic tools remain a mainstay in the natural product drug discovery workflows. Molecular networking is proving to be a powerful tool in identification of analogues produced by wild-type bacteria as well as from those engineered to express BGCs of high interest. The linking of paired genomic and metabolomic data sets will be conducive to large-scale and small-scale comparative and integrative analyses as natural product drug discovery efforts continue.
The field of natural product drug discovery continues to uncover novel molecules. The synergy between genomic and metabolomic approaches realized over the past decade and many of the genomic tools that have been developed have served to set the stage for a new phase of discovery. The creation of community-curated repositories for large-scale natural product data is a powerful and unique contribution to drug discovery that will continue to promote scientific advancement. As genome sequencing continues to become more accessible and more affordable, large-scale sequencing efforts will expand. With this expansion comes opportunity to pair analytical techniques for natural product discovery with genome-based approaches. Many of the approaches are currently microbially based; however, there is need to not only harness the current approaches with prokaryotes but also to extend into eukaryotic systems.
One of the primary bottlenecks for drug development of promising natural products is the issue of supply. The convergence of bacterial genomics, specifically biosynthetic gene clusters, in the drug discovery pipeline allows this issue to be addressed by cultivation or heterologous expression. Barriers beyond limited compound supply include high rediscovery rates and potential ecological impacts of large sample collections. These hurdles can be circumnavigated and the deleterious effect ameliorated by use of these genomic approaches to drug discovery.
Acknowledgments
This work was supported in part by NIH award CA205932 (to A.E.M. and B.J.B.). The development of our genomics-based workflow was informed by significant interactions with Patrick S. G. Chain, Chien-Chi Lo, Hajnalka E. Daligault and Karen W. Davenport from the Los Alamos National Laboratory, Los Alamos, New Mexico, United States.
Author Present Address
Nicole E. Avalon: Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California San Diego, La Jolla, California 92093, United States
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
References
- Ricke D. O.; Wang S.; Cai R.; Cohen D. Genomic approaches to drug discovery. Curr. Opinion Chem. Biol. 2006, 10, 303–308. 10.1016/j.cbpa.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Xia X. Bioinformatics and Drug Discovery. Curr. Top. Med. Chem. 2017, 17, 1709–1726. 10.2174/1568026617666161116143440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clardy J.; Walsh C. Lessons from natural molecules. Nature 2004, 432, 829–37. 10.1038/nature03194. [DOI] [PubMed] [Google Scholar]
- Newman D. J.; Cragg G. M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- Louwen J. J. R.; van der Hooft J. J. J. Comprehensive large-scale integrative analysis of omics data to accelerate specialized metabolite discovery. mSystems 2021, 6, e0072621–e0072621. 10.1128/mSystems.00726-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrette M. G.; Gavrilidou A.; Mantri S.; Selem-Mojica N.; Ziemert N.; Barona-Gómez F. The confluence of big data and evolutionary genome mining for the discovery of natural products. Nat. Prod. Rep. 2021, 38, 2024–2040. 10.1039/D1NP00013F. [DOI] [PubMed] [Google Scholar]
- van Santen J. A.; Kautsar S. A.; Medema M. H.; Linington R. G. Microbial natural product databases: moving forward in the multi-omics era. Nat. Prod. Rep. 2021, 38, 264–278. 10.1039/D0NP00053A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott T. A.; Piel J. The hidden enzymology of bacterial natural product biosynthesis. Nat. Rev. Chem. 2019, 3, 404–425. 10.1038/s41570-019-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt I.; Lumbsch H. T. Ancient horizontal gene transfer from bacteria enhances biosynthetic capabilities of fungi. PLoS One 2009, 4, e4437. 10.1371/journal.pone.0004437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenhall M.; Škunca N.; Lassalle F.; Dessimoz C. Inferring Horizontal Gene Transfer. PLOS Comp. Biol. 2015, 11, e1004095. 10.1371/journal.pcbi.1004095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.-Y.; Colby S. M.; Du X.; Gomez J. D.; Helf M. J.; Kechris K.; Kirkpatrick C. R.; Li S.; Patti G. J.; Renslow R. S.; Subramaniam S.; Verma M.; Xia J.; Young J. D. A practical guide to metabolomics software development. Anal. Chem. 2021, 93, 1912–1923. 10.1021/acs.analchem.0c03581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamek M.; Alanjary M.; Ziemert N. Applied evolution: phylogeny-based approaches in natural products research. Nat. Prod. Rep. 2019, 36, 1295–1312. 10.1039/C9NP00027E. [DOI] [PubMed] [Google Scholar]
- Albarano L.; Esposito R.; Ruocco N.; Costantini M. Genome mining as new challenge in natural products discovery. Mar. Drugs 2020, 18, 199. 10.3390/md18040199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwala R.; Barrett T.; Beck J.; Benson D. A; Bollin C.; Bolton E.; Bourexis D.; Brister J R.; Bryant S. H; Canese K.; Cavanaugh M.; Charowhas C.; Clark K.; Dondoshansky I.; Feolo M.; Fitzpatrick L.; Funk K.; Geer L. Y; Gorelenkov V.; Graeff A.; Hlavina W.; Holmes B.; Johnson M.; Kattman B.; Khotomlianski V.; Kimchi A.; Kimelman M.; Kimura M.; Kitts P.; Klimke W.; Kotliarov A.; Krasnov S.; Kuznetsov A.; Landrum M. J; Landsman D.; Lathrop S.; Lee J. M; Leubsdorf C.; Lu Z.; Madden T. L; Marchler-Bauer A.; Malheiro A.; Meric P.; Karsch-Mizrachi I.; Mnev A.; Murphy T.; Orris R.; Ostell J.; O'Sullivan C.; Palanigobu V.; Panchenko A. R; Phan L.; Pierov B.; Pruitt K. D; Rodarmer K.; Sayers E. W; Schneider V.; Schoch C. L; Schuler G. D; Sherry S. T; Siyan K.; Soboleva A.; Soussov V.; Starchenko G.; Tatusova T. A; Thibaud-Nissen F.; Todorov K.; Trawick B. W; Vakatov D.; Ward M.; Yaschenko E.; Zasypkin A.; Zbicz K. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K.; Shaw S.; Steinke K.; Villebro R.; Ziemert N.; Lee S. Y.; Medema M. H.; Weber T. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F.; Gish W.; Miller W.; Myers E. W.; Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Markowitz V. M.; Chen I.-M. A.; Palaniappan K.; Chu K.; Szeto E.; Grechkin Y.; Ratner A.; Jacob B.; Huang J.; Williams P.; Huntemann M.; Anderson I.; Mavromatis K.; Ivanova N. N.; Kyrpides N. C. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012, 40, D115–D122. 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Muñoz J. C.; Selem-Mojica N.; Mullowney M. W.; Kautsar S. A.; Tryon J. H.; Parkinson E. I.; De Los Santos E. L. C.; Yeong M.; Cruz-Morales P.; Abubucker S.; Roeters A.; Lokhorst W.; Fernandez-Guerra A.; Cappelini L. T. D.; Goering A. W.; Thomson R. J.; Metcalf W. W.; Kelleher N. L.; Barona-Gomez F.; Medema M. H. A computational framework to explore large-scale biosynthetic diversity. Nature Chem. Biol. 2020, 16, 60–68. 10.1038/s41589-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautsar S. A.; Blin K.; Shaw S.; Navarro-Muñoz J. C.; Terlouw B. R.; van der Hooft J. J. J.; van Santen J. A.; Tracanna V.; Suarez Duran H. G.; Pascal Andreu V.; et al. MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 2020, 48, D454–D458. 10.1093/nar/gkz882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Santen J. A.; Jacob G.; Singh A. L.; Aniebok V.; Balunas M. J.; Bunsko D.; Neto F. C.; Castaño-Espriu L.; Chang C.; Clark T. N.; et al. The Natural Products Atlas: An open access knowledge base for microbial natural products discovery. ACS Cent. Sci. 2019, 5, 1824–1833. 10.1021/acscentsci.9b00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniappan K.; Chen I.-M. A.; Chu K.; Ratner A.; Seshadri R.; Kyrpides N. C.; Ivanova N. N.; Mouncey N. J. IMG-ABC v.5.0: an update to the IMG/Atlas of Biosynthetic Gene Clusters Knowledgebase. Nucleic Acids Res. 2020, 48, D422–D430. 10.1093/nar/gkz932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayfach S.; Roux S.; Seshadri R.; Udwary D.; Varghese N.; Schulz F.; Wu D.; Paez-Espino D.; Chen I. M.; Huntemann M.; et al. A genomic catalog of Earth’s microbiomes. Nat. Biotechnol. 2021, 39, 499–509. 10.1038/s41587-020-0718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. E.; Avalon N. E.; Bishop L.; Davenport K. W.; Delage E.; Dichosa A. E. K.; Eveillard D.; Higham M. L.; Kokkaliari S.; Lo C.-C.; Riesenfeld C. S.; Young R. M.; Chain P. S. G.; Baker B. J. Uncovering the core microbiome and distribution of palmerolide in Synoicum adareanum across the Anvers Island Archipelago, Antarctica. Mar. Drugs 2020, 18, 298. 10.3390/md18060298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leão T.; Wang M.; Moss N.; da Silva R.; Sanders J.; Nurk S.; Gurevich A.; Humphrey G.; Reher R.; Zhu Q.; et al. A multi-omics characterization of the natural product potential of tropical filamentous marine cyanobacteria. Mar. Drugs 2021, 19, 20. 10.3390/md19010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatou S.; Eldjárn G. H.; Ramsay A.; van der Hooft J. J. J.; Hughes A. H.; Rogers S.; Duncan K. R. Comparative metabologenomics analysis of polar actinomycetes. Mar. Drugs 2021, 19, 103. 10.3390/md19020103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handayani I.; Saad H.; Ratnakomala S.; Lisdiyanti P.; Kusharyoto W.; Krause J.; Kulik A.; Wohlleben W.; Aziz S.; Gross H.; Gavriilidou A.; Ziemert N.; Mast Y. Mining Indonesian microbial biodiversity for novel natural compounds by a combined genome mining and molecular networking approach. Mar. Drugs 2021, 19, 316. 10.3390/md19060316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N.; Podell S.; Penn K.; Badger J. H.; Allen E.; Jensen P. R. The Natural Product Domain Seeker NaPDoS: A phylogeny based bioinformatic Ttool to classify secondary metabolite gene diversity. PLoS One 2012, 7, e34064. 10.1371/journal.pone.0034064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alanjary M.; Steinke K.; Ziemert N. AutoMLST: an automated web server for generating multi-locus species trees highlighting natural product potential. Nucleic Acids Res. 2019, 47, W276–W282. 10.1093/nar/gkz282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungan M. D.; Alanjary M.; Blin K.; Weber T.; Medema M. H; Ziemert N. ARTS 2.0: feature updates and expansion of the Antibiotic Resistant Target Seeker for comparative genome mining. Nucleic Acids Res. 2020, 48, W546–W552. 10.1093/nar/gkaa374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almabruk K. H.; Dinh L. K.; Philmus B. Self-resistance of natural product producers: Past, present, and future focusing on self-resistant protein variants. ACS Chem. Biol. 2018, 13, 1426–1437. 10.1021/acschembio.8b00173. [DOI] [PubMed] [Google Scholar]
- Quirós L. M.; Aguirrezabalaga I.; Olano C.; Méndez C.; Salas J. A. Two glycosyltransferases and a glycosidase are involved in oleandomycin modification during its biosynthesis by Streptomyces antibioticus. Mol. Microbiol. 1998, 28, 1177–1185. 10.1046/j.1365-2958.1998.00880.x. [DOI] [PubMed] [Google Scholar]
- Wencewicz T. A. Crossroads of antibiotic resistance and biosynthesis. J. Mol. Biol. 2019, 431, 3370–3399. 10.1016/j.jmb.2019.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sélem-Mojica N.; Aguilar C.; Gutiérrez-García K.; Martínez-Guerrero C. E.; Barona-Gómez F. EvoMining reveals the origin and fate of natural product biosynthetic enzymes. Microb. Genom. 2019, 5, e000260. 10.1099/mgen.0.000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Morales P.; Kopp J. F.; Martínez-Guerrero C. E.; Yáñez-Guerra L. A.; Sélem-Mojica N.; Ramos-Aboites H. E.; Feldmann J.; Barona-Gómez F. Phylogenomic analysis of natural products biosynthetic gene clusters allows discovery of arseno-organic metabolites in model Streptomycetes. Genom. Biol. Evol. 2016, 8, 1906–1916. 10.1093/gbe/evw125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rond T.; Asay J. E.; Moore B. S. Co-occurrence of enzyme domains guides the discovery of an oxazolone synthetase. Nature Chem. Biol. 2021, 17, 794–799. 10.1038/s41589-021-00808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich E. J.; Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 2016, 33, 231–316. 10.1039/C5NP00125K. [DOI] [PubMed] [Google Scholar]
- Cane D. E.; Walsh C. T.; Khosla C. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science 1998, 282, 63–8. 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- Sardar D.; Schmidt E. W. Combinatorial biosynthesis of RiPPs: docking with marine life. Curr. Opinion Chem. Biol. 2016, 31, 15–21. 10.1016/j.cbpa.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaser R.; Luesch H. Marine natural products: a new wave of drugs?. Future Med. Chem. 2011, 3, 1475–1489. 10.4155/fmc.11.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avalon N. E.; Murray A. E.; Daligault H. E.; Lo C.-C.; Davenport K. W.; Dichosa A. E. K.; Chain P. S. G.; Baker B. J. Bioinformatic and mechanistic analysis of the palmerolide PKS-NRPS biosynthetic pathway from the microbiome of an Antarctic ascidian. Front. Chem. 2021, 9, 802574. 10.3389/fchem.2021.802574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. E.; Lo C.-C.; Daligault H. E.; Avalon N. E.; Read R. W.; Davenport K. W.; Higham M. L.; Kunde Y.; Dichosa A. E. K.; Baker B. J.; Chain P. S. G. Discovery of an Antarctic ascidian-associated uncultivated Verrucomicrobia with antimelanoma palmerolide biosynthetic potential. mSphere 2021, 6, e0075921. 10.1128/mSphere.00759-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma M.; Fujiki H.; Furuya-Suguri H.; Yoshizawa S.; Yasumoto S.; Kato Y.; Fusetani N.; Sugimura T. Calyculin A, an inhibitor of protein phosphatases, a potent tumor promoter on CD-1 mouse skin. Cancer Res. 1990, 50, 3521–3525. [PubMed] [Google Scholar]
- Wakimoto T.; Egami Y.; Nakashima Y.; Wakimoto Y.; Mori T.; Awakawa T.; Ito T.; Kenmoku H.; Asakawa Y.; Piel J.; Abe I. Calyculin biogenesis from a pyrophosphate protoxin produced by a sponge symbiont. Nature Chem. Biol. 2014, 10, 648–655. 10.1038/nchembio.1573. [DOI] [PubMed] [Google Scholar]
- Jansen R.; Höfle G.; Irschik H.; Reichenbach H. Antibiotika aus Gleitenden Bakterien, XXIV. Corallopyronin A, B und C – drei neue Antibiotika aus Corallococcus coralloides Cc c127 (Myxobacterales). Liebigs Ann. Chem. 1985, 1985, 822–836. 10.1002/jlac.198519850418. [DOI] [Google Scholar]
- Erol Ö.; Schäberle T. F.; Schmitz A.; Rachid S.; Gurgui C.; El Omari M.; Lohr F.; Kehraus S.; Piel J.; Müller R.; König G. M. Biosynthesis of the myxobacterial antibiotic corallopyronin A. ChemBioChem. 2010, 11, 1253–1265. 10.1002/cbic.201000085. [DOI] [PubMed] [Google Scholar]
- Sudek S.; Lopanik N. B.; Waggoner L. E.; Hildebrand M.; Anderson C.; Liu H.; Patel A.; Sherman D. H.; Haygood M. G. Identification of the putative bryostatin polyketide synthase gene cluster from “Candidatus Endobugula sertula”, the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J. Nat. Prod. 2007, 70, 67–74. 10.1021/np060361d. [DOI] [PubMed] [Google Scholar]
- Buchholz T. J.; Rath C. M.; Lopanik N. B.; Gardner N. P.; Håkansson K.; Sherman D. H. Polyketide β-branching in bryostatin biosynthesis: Identification of surrogate acetyl-ACP donors for BryR, an HMG-ACP synthase. Chem. Biol. 2010, 17, 1092–1100. 10.1016/j.chembiol.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S.; Wang Z.; Mohammad M.; Sarkar F. H.; Mohammad R. M. Efficacy of selected natural products as therapeutic agents against cancer. J. Nat. Prod. 2008, 71, 492–496. 10.1021/np0705716. [DOI] [PubMed] [Google Scholar]
- Nelson T. J.; Alkon D. L. Neuroprotective versus tumorigenic protein kinase C activators. Trend. Biochem. Sci. 2009, 34, 136–145. 10.1016/j.tibs.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Sun M.-K.; Alkon D. L. Dual effects of bryostatin-1 on spatial memory and depression. Eur. J. Pharmacol. 2005, 512, 43–51. 10.1016/j.ejphar.2005.02.028. [DOI] [PubMed] [Google Scholar]
- Wang M. X.; Carver J. J.; Phelan V. V.; Sanchez L. M.; Garg N.; Peng Y.; Nguyen D. D.; Watrous J.; Kapono C. A.; Luzzatto-Knaan T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogawa T.; Terai A.; Amagai K.; Hashimoto J.; Futamura Y.; Okano A.; Fujie M.; Satoh N.; Ikeda H.; Shin-ya K.; Osada H.; Takahashi S. Heterologous expression of the biosynthetic gene cluster for verticilactam and identification of analogues. J. Nat. Prod. 2020, 83, 3598–3605. 10.1021/acs.jnatprod.0c00755. [DOI] [PubMed] [Google Scholar]
- Park O.-K.; Choi H.-Y.; Kim G.-W.; Kim W.-G. Generation of new complestatin analogues by heterologous expression of the complestatin biosynthetic gene cluster from Streptomyces chartreusis AN1542. ChemBioChem. 2016, 17, 1725–1731. 10.1002/cbic.201600241. [DOI] [PubMed] [Google Scholar]
- Tan B.; Chen S.; Zhang Q.; Chen Y.; Zhu Y.; Khan I.; Zhang W.; Zhang C. Heterologous expression leads to discovery of diversified lobophorin analogues and a fexible glycosyltransferase. Org. Lett. 2020, 22, 1062–1066. 10.1021/acs.orglett.9b04597. [DOI] [PubMed] [Google Scholar]
- Ross A. C.; Gulland L. E. S.; Dorrestein P. C.; Moore B. S. Targeted capture and heterologous expression of the Pseudoalteromonas alterochromide gene cluster in Escherichia coli represents a promising natural product exploratory platform. ACS Synth. Biol. 2015, 4, 414–420. 10.1021/sb500280q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson C. B.; Crüsemann M.; Moore B. S. PCR-independent method of transformation-associated recombination reveals the cosmomycin biosynthetic gene cluster in an ocean Streptomycete. J. Nat. Prod. 2017, 80, 1200–1204. 10.1021/acs.jnatprod.6b01121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y.; Gu R.; Li Y.; Wang X.; Ren W.; Li X.; Wang L.; Xie Y.; Hong B. Exploring novel herbicidin analogues by transcriptional regulator overexpression and MS/MS molecular networking. Microb. Cell Factor. 2019, 18, 175. 10.1186/s12934-019-1225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albataineh H.; Duke M.; Misra S. K.; Sharp J. S.; Stevens D. C. Identification of a solo acylhomoserine lactone synthase from the myxobacterium Archangium gephyra. Sci. Rep. 2021, 11, 3018. 10.1038/s41598-021-82480-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross H.; Stockwell V. O.; Henkels M. D.; Nowak-Thompson B.; Loper J. E.; Gerwick W. H. The genomisotopic approach: A systematic method to isolate products of orphan biosynthetic gene clusters. Chem. Biol. 2007, 14, 53–63. 10.1016/j.chembiol.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Kinnel R. B.; Esquenazi E.; Leao T.; Moss N.; Mevers E.; Pereira A. R.; Monroe E. A.; Korobeynikov A.; Murray T. F.; Sherman D.; Gerwick L.; Dorrestein P. C.; Gerwick W. H. A maldiisotopic approach to discover natural products: Cryptomaldamide, a hybrid tripeptide from the marine cyanobacterium Moorea producens. J. Nat. Prod. 2017, 80, 1514–1521. 10.1021/acs.jnatprod.7b00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taton A.; Ecker A.; Diaz B.; Moss N. A.; Anderson B.; Reher R.; Leão T. F.; Simkovsky R.; Dorrestein P. C.; Gerwick L.; Gerwick W. H.; Golden J. W. Heterologous expression of cryptomaldamide in a cyanobacterial host. ACS Synth. Biol. 2020, 9, 3364–3376. 10.1021/acssynbio.0c00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughey C. S.; van Santen J. A.; van der Hooft J. J. J.; Medema M. H.; Linington R. G. An isotopic labeling approach linking natural products with biosynthetic gene clusters. Nature Chem. Biol. 2022, 18, 295–304. 10.1038/s41589-021-00949-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behsaz B.; Bode E.; Gurevich A.; Shi Y.-N.; Grundmann F.; Acharya D.; Caraballo-Rodríguez A. M.; Bouslimani A.; Panitchpakdi M.; Linck A.; Guan C.; Oh J.; Dorrestein P. C.; Bode H. B.; Pevzner P. A.; Mohimani H. Integrating genomics and metabolomics for scalable non-ribosomal peptide discovery. Nature Commun. 2021, 12, 3225. 10.1038/s41467-021-23502-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema M. H.; Paalvast Y.; Nguyen D. D.; Melnik A.; Dorrestein P. C.; Takano E.; Breitling R. Pep2Path: Automated mass spectrometry-guided genome mining of peptidic natural products. PLOS Comp. Biol. 2014, 10, e1003822. 10.1371/journal.pcbi.1003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohimani H.; Liu W.-T.; Kersten R. D.; Moore B. S.; Dorrestein P. C.; Pevzner P. A. NRPquest: Coupling mass spectrometry and genome mining for nonribosomal peptide discovery. J. Nat. Prod. 2014, 77, 1902–1909. 10.1021/np500370c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A.; Mikheenko A.; Shlemov A.; Korobeynikov A.; Mohimani H.; Pevzner P. A. Increased diversity of peptidic natural products revealed by modification-tolerant database search of mass spectra. Nature Microbiol. 2018, 3, 319–327. 10.1038/s41564-017-0094-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorn M. A.; Verhoeven S.; Ridder L.; Huber F.; Acharya D. D.; Aksenov A. A.; Aleti G.; Moghaddam J. A.; Aron A. T.; Aziz S.; et al. A community resource for paired genomic and metabolomic data mining. Nature Chem. Biol. 2021, 17, 363–368. 10.1038/s41589-020-00724-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten R. D.; Yang Y.-L.; Xu Y.; Cimermancic P.; Nam S.-J.; Fenical W.; Fischbach M. A.; Moore B. S.; Dorrestein P. C. A mass spectrometry–guided genome mining approach for natural product peptidogenomics. Nature Chem. Biol. 2011, 7, 794–802. 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goering A. W.; McClure R. A.; Doroghazi J. R.; Albright J. C.; Haverland N. A.; Zhang Y.; Ju K.-S.; Thomson R. J.; Metcalf W. W.; Kelleher N. L. Metabologenomics: Correlation of microbial gene clusters with metabolites drives discovery of a nonribosomal peptide with an unusual amino acid monomer. ACS Cent. Sci. 2016, 2, 99–108. 10.1021/acscentsci.5b00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L.; Gurevich A.; Alexander K. L.; Naman C. B.; Leao T.; Glukhov E.; Luzzatto-Knaan T.; Vargas F.; Quinn R.; Bouslimani A.; Nothias L. F.; Singh N. K.; Sanders J. G.; Benitez R. A.S.; Thompson L. R.; Hamid M.-N.; Morton J. T.; Mikheenko A.; Shlemov A.; Korobeynikov A.; Friedberg I.; Knight R.; Venkateswaran K.; Gerwick W. H.; Gerwick L.; Dorrestein P. C.; Pevzner P. A.; Mohimani H. MetaMiner: A scalable peptidogenomics approach for discovery of ribosomal peptide natural products with blind modifications from microbial communities. Cell Systems 2019, 9, 600–608. 10.1016/j.cels.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington B. C.; Xu F.; Seyedsayamdost M. R. A natural product chemist’s guide to unlocking silent biosynthetic gene clusters. Annu. Rev. Biochem. 2021, 90, 763–788. 10.1146/annurev-biochem-081420-102432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y.; Huang H.; Liang J.; Wang M.; Lu L.; Shao Z.; Cobb R. E.; Zhao H. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nature Commun. 2013, 4, 2894. 10.1038/ncomms3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushchuk O.; Ostash I.; Mösker E.; Vlasiuk I.; Deneka M.; Rückert C.; Busche T.; Fedorenko V.; Kalinowski J.; Süssmuth R. D.; Ostash B. Eliciting the silent lucensomycin biosynthetic pathway in Streptomyces cyanogenus S136 via manipulation of the global regulatory gene adpA. Sci. Rep. 2021, 11, 3507. 10.1038/s41598-021-82934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode H. B.; Bethe B.; Hofs R.; Zeeck A. Big effects from small changes: possible ways to explore nature’s chemical diversity. Chembiochem 2002, 3, 619–627. . [DOI] [PubMed] [Google Scholar]
- Wang G.; Zhao Z.; Ke J.; Engel Y.; Shi Y.-M.; Robinson D.; Bingol K.; Zhang Z.; Bowen B.; Louie K.; et al. CRAGE enables rapid activation of biosynthetic gene clusters in undomesticated bacteria. Nature Microbiol. 2019, 4, 2498–2510. 10.1038/s41564-019-0573-8. [DOI] [PubMed] [Google Scholar]