Abstract
Background:
MicroRNAs (miRNAs) are small noncoding RNAs of ~22 nucleotides that play a crucial role in post-transcriptional regulation of gene expression. Dysregulation of miRNA expression has been shown during microbial infections. We sought to identify miRNAs that distinguish invasive aspergillosis (IA) from non-IA in lung transplant recipients (LTRs).
Methods:
We used Nanostring nCounter Human miRNA v3 panel to measure miRNAs in bronchoalveolar lavage (BAL) samples from LTRs with Aspergillus colonization (ASP) (n=10), Aspergillus colonization and CLAD (ASPCLAD group) (n=7), IA without CLAD (IA) (n=10), IA with CLAD (IACLAD) (n=9), and control patients (Controls) (n=9). The miRNA profile was compared using the permutation test of 100000 trials for each of the comparisons. We used mirDip to obtain their gene targets and pathDIP to determine the pathway enrichment.
Results:
We performed pairwise comparisons between patient groups to identify differentially expressed miRNAs. Five miRNAs were found to be specific to IA, including four (miR-145-5p, miR-424-5p, miR-99b-5p and miR-4488) that were upregulated and the pair (miR-4454+miR-7975) that was downregulated in IA group vs. Controls. The expression change for these miRNAs was specific to IA patients; they were not significantly differentiated between IACLAD and IA group. Signaling pathways associated with an immunological response to IA were found to be significantly enriched.
Conclusion:
We report a set of five differentially-expressed miRNAs in the BAL of LTRs with IA that might help in the development of diagnostic and prognostic tools for IA in LTRs. However, further investigation is needed in a larger cohort to validate the findings.
Introduction
Lung transplantation has become an established mode of treatment for patients with end stage pulmonary disease. As the number of lung transplants continues to grow steadily every year, the two main impediments to survival remain chronic lung allograft dysfunction (CLAD) and non-CMV infections. Among lung transplant recipients (LTRs) surviving 10 years, CLAD is present in 65% of patients [1], and non-cytomegalovirus (CMV) infections, including Aspergillus infection, account for 16.7% of deaths[2].
Aspergillus spp are the most common cause of fungal infections in LTRs[3]. Aspergillus infection can present as colonization or as invasive disease (invasive aspergillosis, IA). The primary barrier to early diagnosis of Aspergillus infection is the lack of sensitivity of available diagnostic tests. Currently, diagnosis is based on clinical symptoms, and radiological and laboratory findings [4]. Diagnostic tests are predominantly dependent on direct detection of Aspergillus by culturing techniques or PCR, or by detection of pathogen-associated biomarkers, such as galactomannan and beta-D-glucan, without incorporating host immunological markers [4].
Recognition of Aspergillus conidia and hyphae by the host immune system is imperative for controlling the progression of Aspergillus colonization to invasive disease. Conidia and hyphae are identified by key players in the innate immune system, such as pathogen recognition receptors (PPRs), and are killed by alveolar macrophages and neutrophils. After the initial recognition, differentiation of naïve CD4+ T cells to Th1 or Th17 cells activates the adaptive immune response, which further assists in fungal killing [5].
Changes in the host immune response genes have been documented during Aspergillus colonization and infection in LTRs. During Aspergillus colonization, enriched expression of genes involved in host defense and inflammatory response in the bronchoalveolar lavage (BAL) cells were recently reported [6]. LTRs with IA or suspected IA also have higher expression levels of inflammatory cytokines, such as IL-12, IL-1RA and MIP-1 beta, in their BALs compared to patients with Aspergillus colonization [7].
Over the last few decades, there has been growing interest in understanding the role miRNA play in regulating the immune response [8, 9]. MicroRNAs represent a class of highly conserved small non-coding RNAs that play a critical role in post-transcriptional regulation of RNA. Recent studies have suggested that miRNAs regulate host immune response to infections by regulating innate and adaptive immune response pathways [10–12]. Furthermore, the dysregulation of miRNA expression has also been shown during bacterial, viral and fungal infections[13–15]. Due to the stability of their structures, miRNAs have been studied for their clinical application as diagnostic markers and as targets for treatment against infections[16, 17]. However, miRNA expression during Aspergillus infection in lung transplant recipients has not previously been investigated. Identification of miRNAs associated with IA may result in biomarkers of early infection and lead to the development of novel therapies.
In this study, we aimed to identify miRNA expression profiles associated with Aspergillus colonization and with IA in LTRs. We also attempted to predict genes and signaling pathways targeted by these miRNAs responsible for IA. Further validation of these results might not only provide insight into the pathogenesis of IA but might also help identify novel biomarkers for early diagnosis of IA.
Materials and Methods
Patients
This study was approved by the Research Ethics Board of University Health Network, University of Toronto. BAL samples were collected from LTRs at University Health Network between January 2012 and October 2016 for clinical purposes (REB 13-7218). Surveillance bronchoscopies are performed at 2 and 6 weeks and 3, 6, 9, 12, and 18 months after lung transplantation. In addition, diagnostic bronchoscopies were performed when infection or rejection was suspected in LTRs. Samples were stored at −80°C until analyzed. The BAL samples – illustrated in Figure 1 - were obtained from: 10 patients with only Aspergillus colonization (ASP group), 7 patient who had Aspergillus colonization and CLAD (ASPCLAD group), 10 patients who developed IA without CLAD (IA group), 9 patients who had CLAD and also developed IA (IACLAD), and 9 control patients (CONTROLS) who did not develop CLAD, IA, nor Aspergillus colonization. The BAL samples from Aspergillus colonization and IA patients only had fungal growth and the samples from control patients has no microbial growth. Patient demographics are listed in Table 1.
Figure 1.
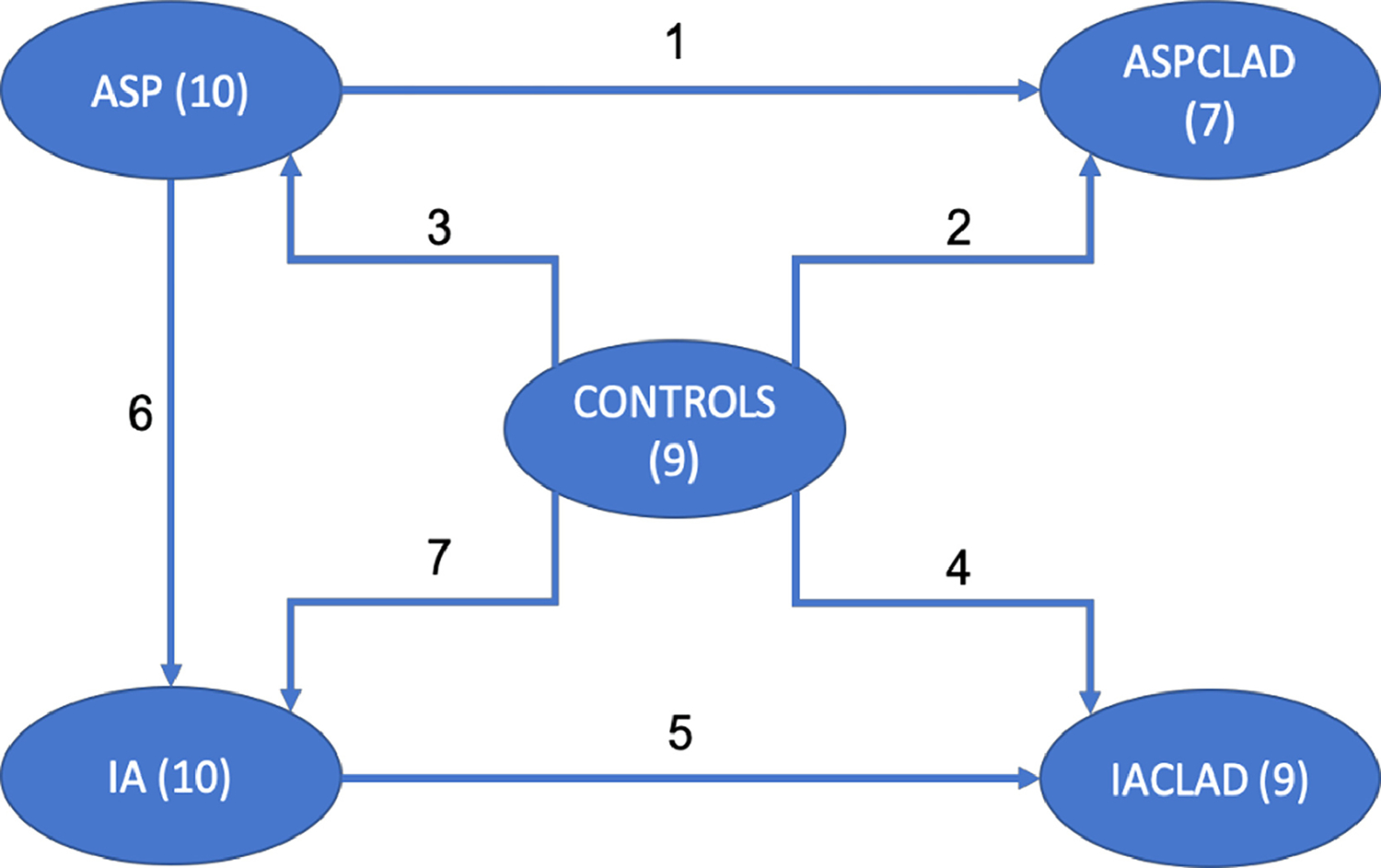
Patient sample groups (nodes) include; ASP = 10 Aspergillus Colonization, ASPCLAD = 7 Aspergillus Colonization and CLAD, IA 10 Invasive Aspergillosis, IACLAD =9 Invasive Aspergillosis and CLAD, and in the center node, 9 control (no IA and no CLAD samples).
Table 1:
Baseline characteristics of patients within the different groups.
| Controls | IA | IA CLAD | Asp | Asp CLAD | Total | |
|---|---|---|---|---|---|---|
| N | 9 | 10 | 9 | 10 | 7 | 45 |
| Sex | ||||||
| Female | 2 | 4 | 5 | 4 | 4 | 19 |
| Male | 7 | 6 | 4 | 6 | 3 | 26 |
| Type of transplant | ||||||
| Double lung | 7 | 8 | 8 | 9 | 7 | 39 |
| Single lung | 2 | 2 | 1 | 1 | 0 | 6 |
| Underlying disease | ||||||
| Pulmonary fibrosis | 3 | 2 | 1 | 1 | 1 | 8 |
| COPD | 1 | 4 | 4 | 3 | 2 | 14 |
| Cystic fibrosis | 0 | 1 | 1 | 1 | 1 | 4 |
| Interstitial Lung Disease | 3 | 1 | 0 | 3 | 1 | 8 |
| Others | 2 | 2 | 3 | 2 | 2 | 11 |
| CLAD | 0 | 0 | 9 | 0 | 7 | 16 |
| BAL fungal culture | ||||||
| Aspergillus fumigatus | 0 | 5 | 5 | 3 | 2 | 15 |
| Aspergillus niger | 0 | 0 | 0 | 0 | 3 | 3 |
| Aspergillus flavus | 0 | 0 | 1 | 3 | 1 | 5 |
| Aspergillus versicolor | 0 | 0 | 0 | 0 | 1 | 1 |
| Other | 0 | 0 | 2 | 4 | 0 | 6 |
| No growth | 9 | 0 | 1 | 0 | 0 | 10 |
| Number of patients on immunosuppressive drugs | ||||||
| Tacrolimus | 2 | 2 | 4 | 2 | 3 | |
| Cyclosporine | 8 | 8 | 6 | 8 | 3 | |
| Mycophenolate mofetil | 6 | 8 | 8 | 5 | 5 | |
| Azathioprine | 1 | 1 | 2 | 2 | 1 | |
| Prednisone | 6 | 8 | 8 | 5 | 5 | |
| Median time (days) from transplant to colonization | n/a | 97 | 193 | 238 | 272 | |
| Median time (days) from transplant to CLAD | n/a | n/a | 614 | n/a | 914 |
Definitions
The International Society of Heart and Lung Transplantation (ISHLT) guidelines for the diagnosis of fungal tracheobronchitis, bronchial anastomotic infection, and colonization were used to define invasive fungal infection [4].
Aspergillus colonization was defined as single positive culture for Aspergillus or positive galactomannan test (with GM value > 0.5) for bronchoscopy specimen, accompanied by absence of fever, no new-onset or worsening cough, normal appearing respiratory mucosa, absence of endobronchial lesions and no new or progressive infiltrate, consolidation or nodules.
Probable IA requires the presence of host factors, one clinical criterion (fever or hypothermia, leukopenia or leukocytosis, new onset of purulent sputum or change in characteristics, new-onset or worsening cough, dyspnea, tachypnea, or pleural rub, worsening gas exchange or pleural effusion), chest imaging with new or progressive infiltrate, consolidation, cavitation or nodules and one positive culture in BAL/blood or PCR or positive GM in BAL.
Proven IA requires histopathologic demonstration of fungus in a tissue/biopsy, or host factors along with radiological and laboratory evidence.
Acute cellular rejection was classified as present and graded according to ISHLT criteria.
Sample Collection and processing
Stored respiratory samples collected by the Toronto Lung Transplant Biobank were used for this study. BAL samples were collected post-bronchoscopy for all patients included in this study. The collected BAL samples were centrifuged at 4000 rpm for 20 mins at 4°C. The supernatant was removed; the sedimented cell pellet was reconstituted in TRIzol (Thermo Fisher Scientific, CA, USA), and stored at −80°C until assessment.
RNA extraction
Samples were removed from storage, thawed and 200uL of chloroform was added. The solution was thoroughly mixed and incubated at room temperature for 10 min. Samples were centrifuged for 15 mins at 12, 000 × g at 4 °C. The aqueous phase of the sample was removed into a new tube and 500 uL of isopropanol was added. The samples were thoroughly mixed and incubated for 20 mins at room temperature, and centrifuged at 12,000 × g for 10 min at 4 °C. The supernatant was removed, and the resulting pellet was washed twice with 75% ethanol. RNA was reconstituted in 30 uL ultrapure water. RNA was quantified using NanoDrop ND-100 UV-Vis Spectrophotometer (Nanodrop Technologies, Rockland, USA) with the NanoDrop 2000/2000c software (Thermo Fischer Scientific, Canada). The A260/A280 and A260/A230 ratio for all samples were >2.0 and >1.5, respectively. Samples were also analyzed using Agilent 2100 Bioanalyzer, the RNA integrity number for all samples was > 9. All RNA samples were stored at −80°C until required.
Nanostring nCounter Analysis of miRNA Expression
Digital quantification of the differentially expressed miRNAs amongst the diagnostic groups was performed using the nCounter System® (NanoString Technologies Inc., Seattle, WA) at the UHN Microarray Center as per the manufacturer’s protocol. Briefly, the NanoString nCounter platform involves mixing total RNA with pairs of capture and reporter probes tailored to each miRNA, hybridizing, washing away excess probes, immobilizing probe bound miRNAs on a surface and then scanning color-coded bar tags on the reporter probes. The microarray dataset was normalized using Quantile normalization methods developed for NanoString technology; these include performing quality control checks, background corrections and signal normalization using negative samples, positive controls, and housekeeping genes.
Computational Data Analysis
Our objective is to study the role of miRNA in IA. We identified differential miRNAs in patient samples based on comparing miRNA expressions of their patient groups. The connecting edges between nodes shown in Figure 1 illustrate the pairwise miRNA expression comparisons that were carried out between these patient groups including: IA vs. CONTROLS, IA vs. ASP, IACLAD vs. IA, IACLAD vs. CONTROLS, ASP vs. CONTROLS, ASPCLAD vs. CONTROLS, and ASPCLAD vs. ASP.
To compare miRNA expressions, we used the permutation test to identify miRNAs whose expression was differential by more than 2 folds and with p-value less than 0.05 in each pairwise comparison as described above. We apply the test iteratively to various patient groups in pairwise comparisons to select those significantly differential miRNAs that are specific to IA. For the selected miRNAs, we obtained their predicted gene targets using the mirDIP database which integrates results of commonly available miRNA target prediction tools and databases [18, 19]. Then, the resulting top gene targets are used to construct an extended Protein-Protein Interaction (PPI) to enable us to perform a pathway enrichment analysis to identify signaling pathways invoked by these targets. The enrichment analysis are performed using two tools; (i) the pathDIP system [15] whose results offer a comprehensive integration of all miRNA predictions available by most of the tools available in the literature, and (ii) to demonstrate the utility of results in a detailed discussion later in the paper[20].
Results
miRNA Expression Analysis
For this study, only five patient groups were available and are shown in Figure 1. We compared these patient groups in a pairwise pattern (also in Figure 1) to examine the differentiation of miRNA expressions in the corresponding groups. To identify the miRNAs that were differential to IA, we needed to compare IA group to the Control group. However, it is expected that the presence of CLAD could have an impact on miRNA expression, and therefore, we compared CLAD groups against controls and against those patients who had IA. Contrasting the results of the results of these comparisons allows for the identification of miRNAs whose expressions are differential in IA and resist the influence of CLAD. In total, we performed seven pairwise comparisons including IA vs. CONTROLS, IA vs. ASP, IACLAD vs. IA, IACLAD vs. CONTROLS, ASP vs. CONTROLS, ASPCLAD vs. CONTROLS, and ASPCLAD vs. ASP. Among the seven pairwise comparisons above, a total of 143 miRNAs were significantly differential in any one comparison. In other words, 143 miRNAs had at least 2-fold change and p-value less than 0.05 in at least one of the seven pairwise comparisons. However, only 43 miRNAs were found significantly differential in IA vs. CONTROLS, these are listed in Table 2.
Table 2:
Significantly differential miRNA expressions resulting from pairwise comparisons of patient groups illustrated in Figure 1. Using the permutation test, we iteratively measure random differences between groups by repeating random permutations without exceeding the maximum number of permutations due to the limited sample size. The groups compared were: Aspergillus colonization (ASP group), Aspergillus colonization and CLAD (ASPCLAD group), IA without CLAD (IA group), IA with CLAD (IACLAD), and (CONTROLS) who did not develop CLAD, IA, nor Aspergillus colonization. The number of miRNAs whose expression was significantly differential is shown in the last column.
| Comparison | Group1 (K) | Group2 | N | NchooseK (max. iterations) | Iteration Used | Significantly Differential miRNAs |
|---|---|---|---|---|---|---|
| ASPCLAD vs ASP | 7 | 10 | 17 | 19448 | 19000 | 48 |
| ASPCLAD vs CTL | 7 | 9 | 16 | 11440 | 11000 | 41 |
| ASP vs CTL | 10 | 9 | 19 | 92378 | 92000 | 2 |
| IACLAD vs CTL | 9 | 9 | 18 | 48620 | 48000 | 114 |
| IACLAD vs IA | 9 | 10 | 19 | 92378 | 92000 | 41 |
| IA vs ASP | 10 | 10 | 20 | 184756 | 184000 | 38 |
| IA vs CTL | 10 | 9 | 19 | 92378 | 92000 | 33 |
| Total = 143 unique miRNAs |
With the high potential of gene targets for all the miRNAs we have identified, we selected five miRNAs that are differentially expressed during IA. While four out of these five miRNAs were upregulated during IA (miR-145-5p, miR-424-5p, miR-99b-5p, and miR-4488), one miRNA pair (miR-4454+miR-7975) was downregulated during IA. The boxplots in Figure 2 show the log expression of these differential miRNAs, and their fold change between patients groups are highlighted in yellow in Table 3.
Figure 2.
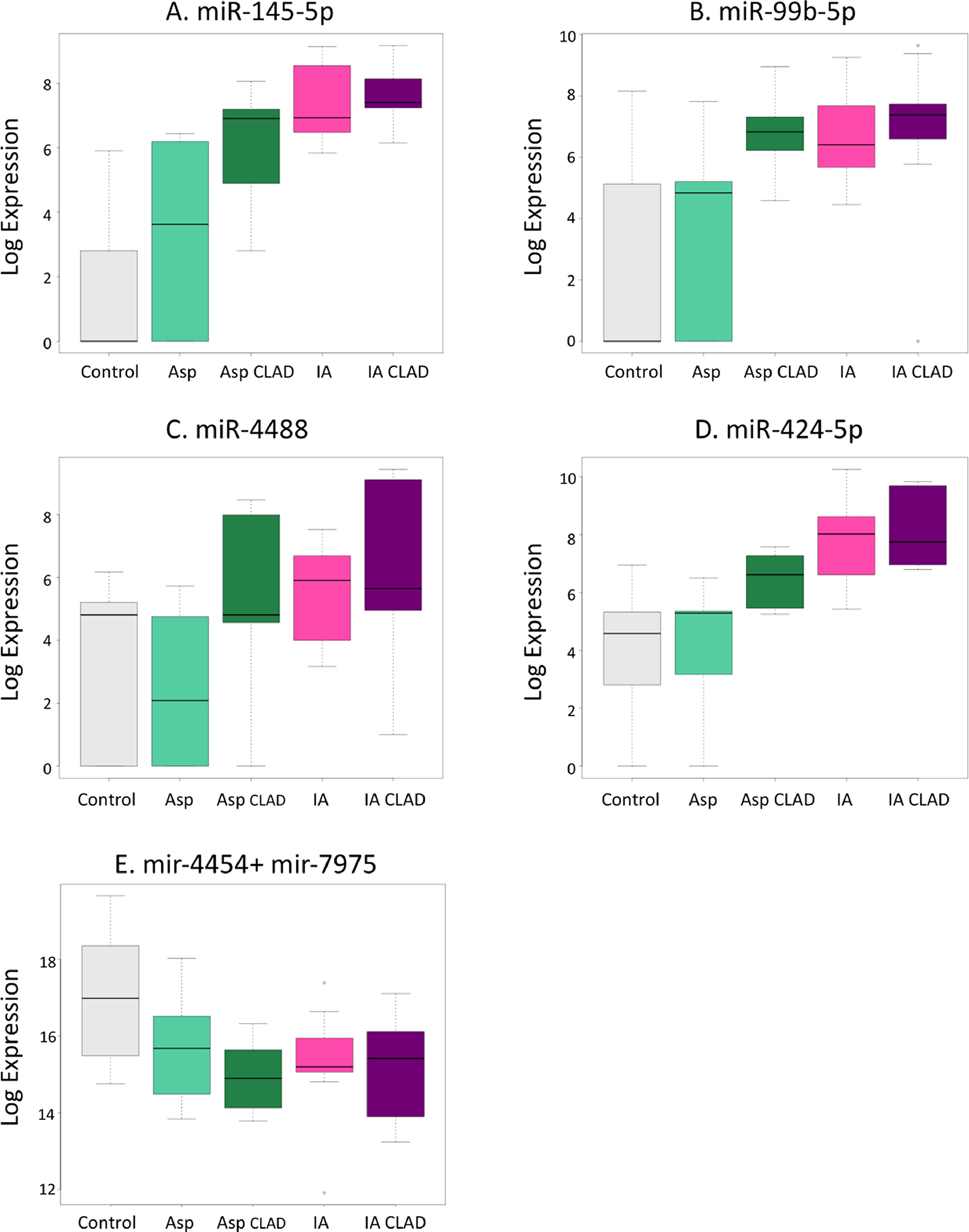
Boxplots of candidate miRNA expression that are differentially expressed (more than 2 fold change and p value < 0.05) in patient group comparisons illustrated in Figure 1 and listed in Table 2. Plots (A) througt (D) show high miRNA expressions are for IA regardless of the presence of CLAD and low expression for CONTROLS and ASP only. Plot (E) slows a candidate miRNA that is down regulated for non-CONTROL samples. This represents a candidate marker to combine with the above four markers.
Table 3:
Fold change and p-values of significantly differential miRNAs in IA vs. CONTROL. Most of the miRNAs below are also significantly differential in IACLAD vs CONTROL but are not in IA vs. IACLAD. While miRNAs highlighted in yellow are specific to IA and resistant to the effect of CLAD, those highlighted in blue are also significantly differential in IA vs. IACLAD suggesting that they are CLAD specific miRNAs and are included for illustration purposes only. Bolded and yellow miRNAs are candidates for IA signature.
| IA vs Controls | IACLAD vs Controls | IA vs IACLAD | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| miRNA | Fold Change | pValue | Fold Change | pValue | Fold Change | pValue |
|
| ||||||
| hsa-miR-145-5p | 6.0533 | 1.0870E-05 | 6.3739 | 4.1667E-05 | 0.3206 | 0.5232 |
| hsa-miR-199a-3p+hsa-miR-199b-3p | 4.6122 | 0.0001 | 6.2736 | 0.0001 | 1.6614 | 0.1334 |
| hsa-miR-99b-5p | 4.2809 | 0.0019 | 4.4817 | 0.0086 | 0.2008 | 0.8651 |
| hsa-miR-424-5p | 3.7435 | 0.0005 | 4.0088 | 4.1667E-05 | 0.2653 | 0.7022 |
| hsa-miR-450a-5p | 3.6766 | 0.0050 | 4.4318 | 0.0028 | 0.7552 | 0.5656 |
| hsa-miR-132-3p | 3.5454 | 0.0020 | 3.3363 | 0.0271 | −0.2090 | 0.8583 |
| hsa-miR-195-5p | 3.5252 | 0.0075 | 5.4320 | 0.0002 | 1.9068 | 0.1882 |
| hsa-miR-4284 | 3.4999 | 0.0243 | 4.5749 | 0.0160 | 1.0750 | 0.5678 |
| hsa-miR-199b-5p | 3.4256 | 0.0030 | 6.2736 | 0.0001 | 1.8162 | 0.1891 |
| hsa-miR-2116-5p | 3.0650 | 0.0177 | 5.3854 | 0.0014 | 2.3204 | 0.0635 |
| hsa-miR-503-5p | 3.0500 | 0.0155 | 2.3849 | 0.0464 | −0.6651 | 0.6037 |
| hsa-miR-1290 | 2.9679 | 0.0149 | 5.5064 | 0.0012 | 2.5385 | 0.0595 |
| hsa-miR-548d-3p | 2.9633 | 0.0267 | 5.8144 | 0.0012 | 2.8512 | 0.0460 |
| hsa-miR-542-5p | 2.8258 | 0.0096 | 2.9901 | 0.0123 | 0.1643 | 0.8929 |
| hsa-miR-7-5p | 2.7860 | 0.0052 | 3.8222 | 0.0045 | 1.0363 | 0.4043 |
| hsa-miR-548d-5p | 2.7477 | 0.0294 | 5.8144 | 0.0012 | 2.8512 | 0.0460 |
| hsa-miR-548n | 2.7428 | 0.0437 | 5.8587 | 0.0006 | 3.1159 | 0.0400 |
| hsa-miR-1285-5p | 2.6835 | 0.0255 | 4.8189 | 0.0012 | 2.1354 | 0.1251 |
| hsa-miR-101-3p | 2.6689 | 0.0395 | 1.7690 | 0.1995 | −0.8999 | 0.3981 |
| hsa-miR-2117 | 2.5884 | 0.0183 | 6.1587 | 0.0001 | 3.5703 | 0.0359 |
| hsa-miR-378f | 2.5205 | 0.0194 | 3.4736 | 0.0256 | 3.0899 | 0.0704 |
| hsa-miR-514b-5p | 2.4222 | 0.0210 | 4.3024 | 0.0031 | 1.8802 | 0.0868 |
| hsa-miR-532-3p | 2.4045 | 0.0423 | 1.9728 | 0.1305 | −0.4318 | 0.7189 |
| hsa-miR-217 | 2.4041 | 0.0030 | 1.9702 | 0.0287 | −0.4339 | 0.6403 |
| hsa-miR-125a-3p | 2.3875 | 0.0317 | 3.6409 | 0.0022 | 1.2534 | 0.2945 |
| hsa-miR-4488 | 2.2880 | 0.0351 | 2.9029 | 0.0446 | 0.6149 | 0.5554 |
| hsa-miR-196a-5p | 2.2388 | 0.0045 | 4.9516 | 0.0004 | 2.7127 | 0.0197 |
| hsa-miR-1257 | 2.2140 | 0.0437 | 4.0344 | 0.0060 | 1.8205 | 0.1468 |
| hsa-miR-210-3p | 2.2116 | 0.0224 | 3.3812 | 0.0279 | 1.1696 | 0.4099 |
| hsa-miR-4532 | 2.0959 | 0.0317 | 4.8586 | 0.0025 | 2.7627 | 0.0764 |
| hsa-miR-1249-3p | 2.0465 | 0.0294 | 1.1148 | 0.1875 | −0.9317 | 0.3680 |
| hsa-miR-1305 | 1.9852 | 0.0352 | 4.9723 | 0.0161 | 2.9871 | 0.1099 |
| hsa-miR-574-5p | 1.9384 | 0.0418 | 4.1222 | 0.0032 | 2.1838 | 0.1398 |
| hsa-miR-4516 | 1.9208 | 0.0153 | 3.6549 | 0.0013 | 1.7341 | 0.1798 |
| hsa-miR-877-5p | 1.8178 | 0.0358 | 2.9057 | 0.0086 | 1.0879 | 0.3154 |
| hsa-miR-152-3p | 1.7710 | 0.0384 | 2.4456 | 0.0222 | 0.6746 | 0.5214 |
| hsa-miR-1915-3p | 1.7187 | 0.0358 | 2.5902 | 0.0132 | 0.8715 | 0.2630 |
| hsa-miR-187-3p | −1.3462 | 0.0441 | −2.8924 | 0.0297 | −1.5462 | 0.2263 |
| hsa-miR-4454+hsa-miR-7975 | −1.7458 | 0.0284 | −1.9256 | 0.0219 | −0.1798 | 0.7878 |
| hsa-miR-218-5p | −2.3920 | 0.0083 | −1.1567 | 0.2284 | 1.2354 | 0.2981 |
| hsa-miR-708-5p | −2.8788 | 0.0281 | −1.5909 | 0.1892 | 1.2879 | 0.2931 |
We found that the expression of miR-145-5p is increased by 2-folds in the ASP group but is not statistically significant difference (p=0.07), and it is increased further by 6-folds during IA (p<0.01). miR-145-5p expression is also increased by 6.3-folds during IACLAD (p<0.01). The negligible fold change difference between IA and IACLAD of miR-145-5p (p=0.523) expression demonstrates that it is altered during IA and is not impacted by the presence of CLAD. Similarly, the fold changes of miR-424-5p, miR-99b-5p, and miR-4488 are not impacted by the presence of CLAD but maintain higher expression levels when compared to Controls in IA and in IACLAD groups as shown by the boxplots of Figure 2. The fold changes when comparing ASP group to Controls or ASP to IA were not statistically significant for these miRNAs.
In contrast, the expression of the pair miR-4454+miR-7975 is decreased during IA and IACLAD compared to that of controls by nearly two folds. However, the differentiation of their expression between IACLAD and IA shows a negligible fold change (−0.18) and p=0.78, signifying that the downregulation of this miRNA pair is correlated to IA and is not further influenced by CLAD. We include them in our analysis due to the change in regulation direction compared to the other miRNAs identified above.
More importantly, the expressions of all the above five miRNAs (highlighted in Table 3), appear unaffected by the presence of CLAD, their expressions are not differential when comparing IA vs. IACLAD. In the absence of CLAD only patient samples, this suggests that these miRNAs are specific to IA only.
It is important to note that miR-132-3p was more than 3-fold higher in patients with IA as compared to Controls (p=0.002) and ASP (p=0.0003). However, no difference was noted between IA CLAD and IA patients (p=0.8) suggesting a lack of specificity in the presence of CLAD in lung transplant recipients. Other miRNAs (miR-129-5p, miR-132-5p, miR-212-3p, miR-212-5p, miR-146, miR-155p, and miR-21) were not significantly different between IA and Controls.
miRNA Gene Target Predictions
After identifying the miRNAs that are significantly differentiated in patient groups (the five miRNAs highlighted in Table 3), we obtained their predicted gene targets to enable enrichment analysis to examine the enriched signaling pathways.
We used The mirDIP v4.1 to predict gene targets for the given miRNA [18]. The number of the resulting target genes of the five miRNAs highlighted in Table 3, and after accounting for their overlap, the total number of gene targets we obtained was 6748.
PPI Network and Signaling Pathway Enrichment
Using the 6748 gene targets for our miRNAs, we performed an enrichment analysis on them to identify significantly enriched signaling pathways. For this purpose, we use the pathDIP database [15], which annotates signaling cascades that combines core signaling pathways curated in various existing databases with pathways predicted based on protein-protein interactions (PPIs). For this purpose, we search for enriched pathways by combining core pathways (curated) with extended pathways based on experimentally detected and computationally predicted PPIs with 99% minimum confidence level of association.
As a result, pathDIP revealed that 2367 signaling pathways are significantly enriched at p < 0.05 with false discovery rate FDR <0.05 as calculated by Benjamini-Hochberg Procedure [20].
We divide the resulting pathways into groups based on the organization of immune system related signaling pathways provided by the REACTOM database (https://reactome.org)- -- this is one of the many pathway database sources integrated by the pathDIP database [15].
Many of the major innate immune system signaling pathways have been found to be highly and significantly enriched by the gene targets set obtained for the miRNAs we identified as significantly differential and specific for IA (Toll Like Receptor Cascades, FC gamma receptor (FCGR), FC Epsilon receptor (FCERI), and C-Type Lectin receptors (CLRS) consisting of various sub signaling pathways like TNF signaling, TNGF (1 and 2), Dectin-1, NF-KB signaling). Table 4 lists the pathways names and significance statistics for enriched pathways we obtained from our analysis. Downstream pathways involved in cytokine signaling were also enriched (IL-4, IL-6, IL-17, IL-12, IL-23, TNFa, IFN gamma) along with upregulation of transcription factors, such as STAT3, STAT5 and NF-KB.
Table 4:
Summary of host immune system signaling pathways detected by enrichment analysis of the five significantly differentiated miRNAs specific to IA.
| Pathway Name | p-value | q-value (FDR: BH-method) | q-value (Bonferroni) | |
|---|---|---|---|---|
| Pathogen Recognition Receptors | CLEC7A (Dectin-1) signaling | 5.16E-09 | 2.15E-08 | 2.72E-05 |
| Toll Like Receptor 2 (TLR2) Cascade | 1.90E-16 | 1.37E-15 | 1.01E-12 | |
| Toll Like Receptor 3 (TLR3) Cascade | 1.56E-19 | 1.42E-18 | 8.25E-16 | |
| Toll Like Receptor 4 (TLR4) Cascade | 2.91E-17 | 2.22E-16 | 1.54E-13 | |
| Toll Like Receptor 5 (TLR5) Cascade | 1.05E-18 | 9.04E-18 | 5.56E-15 | |
| Toll Like Receptor 9 (TLR9) Cascade | 9.98E-18 | 7.91E-17 | 5.27E-14 | |
| Transcription Factors | Stat3 Signaling | 2.25E-12 | 1.16E-11 | 1.19E-08 |
| STAT5A_mediated_PC | 1.97E-19 | 1.78E-18 | 1.04E-15 | |
| JAK STAT pathway and regulation | 7.59E-24 | 9.48E-23 | 4.01E-20 | |
| NF-kappa B signaling | 1.06E-09 | 4.57E-09 | 5.58E-06 | |
| Cytokines | IFN-gamma | 4.92E-29 | 9.91E-28 | 2.60E-25 |
| IL12 signaling mediated by STAT4 | 2.54E-05 | 7.92E-05 | 1.34E-01 | |
| Interleukin-12 signaling | 3.45E-03 | 8.53E-03 | 1.00E+00 | |
| Interleukin-17 signaling | 4.78E-18 | 3.90E-17 | 2.52E-14 | |
| Interleukin-6 signaling | 3.66E-08 | 1.44E-07 | 1.93E-04 | |
| IL23-mediated signaling events | 1.45E-04 | 4.25E-04 | 7.65E-01 | |
| TNF alpha Signaling | 3.90E-26 | 5.89E-25 | 2.06E-22 | |
| Interleukin 4 (IL-4) Pathway | 7.64E-19 | 6.64E-18 | 4.04E-15 | |
| Interleukin-17 signaling | 4.78E-18 | 3.90E-17 | 2.52E-14 | |
Discussion
Invasive aspergillosis continues to be a significant cause of morbidity and mortality in LTRs [21]. Although mortality in LTRs has remarkably decreased, in LTRs with IA the mortality rate is still 20% [22]. Several clinical risk factors, such as single lung transplantation, pre or post-transplant colonization with Aspergillus species, or CMV infection have been assessed. However, the role of regulators of the immune response to IA has not been well defined. MiRNAs have been shown to play a critical role in maintaining various biological processes, such as cellular development, differentiation and proliferation, metabolism, apoptosis, autophagy, and immune responses[23–29]. It has been suggested that miRNAs are key players in regulating maturation and differentiation of immune cells and thus influencing both innate and adaptive immune systems. The profile of host miRNAs during IA has not previously been studied in LTRs. In this study, we identified five miRNAs (miR-145-5p, miR-424-5p, miR-99b-5p, miR-4488, miR-4454+miR-7975) specific to IA in LTRs. Pathway analysis linked these miRNAs to be involved in regulation of PRR, transcription factors, and cytokines associated with IA.
The coordinated interactions between the innate and adaptive immunity protect individuals from developing IA. To our understanding, initial fungal recognition occurs through PRR receptors such as dectin-1 and TLR 2/TLR4, and it leads to complement activation, phagocytosis, and killing of ingested fungi. Adaptive immunity develops when dendritic cells present fungal peptides to Aspergillus-specific CD4+–naive T cells and induce their differentiation into T-helper (Th) 1 and Th17 via cytokine-induced activation of signal transducer and activator of transcription (STAT) 4 and STAT3, respectively[5]. We have previously shown that individuals with IA had a defective expression of dectin-1. Fungal recognition via dectin-1 favors T helper 17 responses. Th17 responses are highly dependent on the activation of the signal transducer and activator of transcription (STAT) 3. We showed while IFN-γ/STAT1 signaling was similar between groups, naïve T cells from patients with IA, but not those with mucormycosis, exhibited reduced responsiveness to IL-6 as measured by STAT3 phosphorylation [30].
There is a paucity of data regarding the regulation of miRNA on pulmonary and systemic responses to fungal exposures and their role in the immunological mechanisms associated with them in humans, particularly in lung transplantation. Das Gupta et al. have shown that in vitro exposure of monocytes and dendritic cells to A. fumigatus upregulates expression of miR-132. This response was specific to A. fumigatus and could not be elicited by LPS, suggesting miR-132 may regulate immune response against A. fumigatus [31].
Further confirmation of this comes from Dix et al., who infected monocyte-derived dendritic cells with Candida albicans and A. fumigatus to identify specific miRNA signatures for each fungal infection[32]. They identified five differentially expressed miRNA (miR-129-5p, miR-132-3p, miR-132-5p, miR-212-3p, miR-212-5p) that were expressed when dendritic cells were challenged with fungi, but not during challenge with LPS, including miRNA from the miR-132 family[32]. On the other hand, Wen et al. have suggested miR-21 as a possible serum diagnostic marker for invasive pulmonary aspergillosis [33]. The miRNA miR-146a was noted to regulate dectin-1 negatively induced inflammatory responses, while several other miRNAs are associated with TLR signaling [34].
In our study, the six-fold increase in miR-145-5p with insignificant fold change difference between IA and IACLAD (p=0.523) expression demonstrates that it is altered during IA and not impacted by the presence of CLAD. Similarly, the fold changes of miR-424-5p, miR-99b-5p, and miR-4488 are not affected by the presence of CLAD but maintain higher expression levels when compared to Controls in IA and IACLAD groups. On the other hand, the expression of the pair miR-4454+miR-7975 is decreased during IA and IACLAD compared to that of Controls by nearly two folds without much change in the between IA and IA CLAD group signifying strong association with IA in lung transplant recipients. It is intuitive to compare miRNA expression in IA samples to those in Control. However, the potential prevalence of CLAD in lung transplant patients renders sufficient grounds for a robust confounding signal because CLAD will affect miRNA expression in samples.
In the absence of samples of LTRs who only CLAD have, we compare miRNA expressions in IACLAD vs. ASPCLAD to identify miRNAs whose expression is not altered by CLAD but is found significantly different in IA. This helps us account for CLAD as a potential confounding variable. Therefore, to identify differentially expressed miRNAs solely due to IA, we examined expression fold changes that were statistically significant in the IA vs. CTL comparison, but their fold change in IACLAD vs. CTL comparison were not statistically significant. These miRNAs were concluded to be differentially expressed due to IA alone. We examined previously reported miR-132-3p, which was reported to be specific to IA by Das et al [31]. We also observed a significant increase in miR-132-3p in BAL of LTRs with IA. At the same time, miR-155p was not significantly different between the IA vs. non-IA groups. However, when we compared miR-132-3p between IA and IA CLAD, the fold changes were not significant, suggesting that background noise with CLAD may affect its specificity to IA. We did not find statistically significant differences between other miRNAs that have been previously reported to be associated with IA (miR-129-5p, miR-212-3p, miR-212-5p, miR-146, and miR-21). This may be due to the threshold of 2-fold change we have used in our study. It is important to note that as compared to targeted miRNA analyses reported previously, we evaluated more than 800 miRNAs and were able to detect novel miRNAs that were more specific than previously reported miRNAs in the BAL of lung transplants recipients. We, however, were not able to measure miRNAs using PCR due to the insufficient quantities of RNA left after completing the Nanostring nCounter Analysis of miRNA Expression.
We did not observe significant changes in miRNA expression due to Aspergillus colonization. The gene expression of miRNA candidates that were selected for IA were not altered in the Aspergillus colonization patients (ASP group). This suggests that colonization does not affect miRNA expression, however the smaller sample size may have prevented us from observing any significant effects of colonization. Future studies with a larger sample size would be needed to understand the effects of Aspergillus colonization on expression of miRNAs.
We also assessed the gene targets and signaling pathways for the five miRNAs. As the selected miRNAs are IA specific, we expected the resulting gene targets and consequently, the signaling pathways enriched by them, to also be associated with IA. We do realize that the task of predicting genes targeted by miRNAs is a difficult challenge for which there exist various tools and methods. These tools produce an enormous number of gene targets, and due to the sheer volume of these predictions, the process of validation becomes intractable. Many online tools search for putative targets in pre-calculated data, instead of performing real interactive calculations, making it much harder to predict targets for novel miRNA sequences. A critical shortcoming is that most of the programs primarily search only for sites conserved across several species. Although this may lead to important discoveries, it certainly will miss targets that are different between distinct species. However, pathways and gene targets reported in this study through mirDIP v4.1 [14] have previously been reported to play a role in the development of IA in various studies [5].
MiRNAs can serve as novel infection biomarkers and therapeutic targets. Host miRNAs have been shown to directly target pathogens and inhibit their replications[17]. Identification of highly specific miRNA expression profiles during certain infections have provided a new method of infection therapy. Studies have shown efficacy of miRNA targeted therapy for hepatitis C virus, however no such work has been done for Aspergillus thus far[35]. Further studies to confirm miRNA profile during IA could assist in improving IA diagnosis and provide miRNAs that can be targeted for therapeutic purposes.
Conclusion
We report a novel set of miRNAs in the BAL of LTRs specific to IA. Further validation in a larger cohort and elucidation of an exact target will enhance our understanding of IA in lung transplant recipients and aid in improving IA diagnosis. These findings may help us identify host immune markers for IA, which will aid with early identification of IA with more certainty.
Funding Sources
This study was funded by NIH CTOT-20 Ancillary Grant (Subaward No: 0255-1369-4609).
Dr. Shahid Husain has received research grants from Merck, research grants from Pfizer, research grants from Gilead, research grant from Astellas, and educational grant from Avir.
Footnotes
Conflict of Interest
Authors have no conflicts of interest to report.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Finlen Copeland CA, Snyder LD, Zaas DW, Turbyfill WJ, Davis WA, Palmer SM: Survival after bronchiolitis obliterans syndrome among bilateral lung transplant recipients. Am J Respir Crit Care Med 2010, 182(6):784–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khush KK, Cherikh WS, Chambers DC, Harhay MO, Hayes D Jr., Hsich E, Meiser B, Potena L, Robinson A, Rossano JW et al. : The International Thoracic Organ Transplant Registry of the International Society for Heart and Lung Transplantation: Thirty-sixth adult heart transplantation report - 2019; focus theme: Donor and recipient size match. J Heart Lung Transplant 2019, 38(10):1056–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kennedy CC, Razonable RR: Fungal Infections After Lung Transplantation. Clin Chest Med 2017, 38(3):511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Husain S, Mooney ML, Danziger-Isakov L, Mattner F, Singh N, Avery R, Ison M, Humar A, Padera RF, Lawler LP et al. : A 2010 working formulation for the standardization of definitions of infections in cardiothoracic transplant recipients. J Heart Lung Transplant 2011, 30(4):361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camargo JF, Husain S: Immune correlates of protection in human invasive aspergillosis. Clin Infect Dis 2014, 59(4):569–577. [DOI] [PubMed] [Google Scholar]
- 6.Weigt SS, Wang X, Palchevskiy V, Patel N, Derhovanessian A, Shino MY, Sayah DM, Lynch JP 3rd, Saggar R, Ross DJ et al. : Gene Expression Profiling of Bronchoalveolar Lavage Cells During Aspergillus Colonization of the Lung Allograft. Transplantation 2018, 102(6):986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herrera S, Gohir W, Foroutan F, Aguilar C, Juvet S, Martinu T, Kumar D, Humar A, Rotstein C, Keshavjee S et al. : Cytokine profile in lung transplant recipients with Aspergillus spp colonization. Transpl Infect Dis 2019, 21(3):e13060. [DOI] [PubMed] [Google Scholar]
- 8.Cohen TS: Role of MicroRNA in the Lung’s Innate Immune Response. J Innate Immun 2017, 9(3):243–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cobb BS, Hertweck A, Smith J, O’Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG et al. : A role for Dicer in immune regulation. J Exp Med 2006, 203(11):2519–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang T, Ge B: miRNAs in immune responses to Mycobacterium tuberculosis infection. Cancer Lett 2018, 431:22–30. [DOI] [PubMed] [Google Scholar]
- 11.Cardin SE, Borchert GM: Viral MicroRNAs, Host MicroRNAs Regulating Viruses, and Bacterial MicroRNA-Like RNAs. Methods Mol Biol 2017, 1617:39–56. [DOI] [PubMed] [Google Scholar]
- 12.Cadamuro AC, Rossi AF, Maniezzo NM, Silva AE: Helicobacter pylori infection: host immune response, implications on gene expression and microRNAs. World J Gastroenterol 2014, 20(6):1424–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X, Li X, Wu M: miRNAs reshape immunity and inflammatory responses in bacterial infection. Signal Transduct Target Ther 2018, 3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girardi E, Lopez P, Pfeffer S: On the Importance of Host MicroRNAs During Viral Infection. Front Genet 2018, 9:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Croston TL, Lemons AR, Beezhold DH, Green BJ: MicroRNA Regulation of Host Immune Responses following Fungal Exposure. Front Immunol 2018, 9:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ojha R, Nandani R, Pandey RK, Mishra A, Prajapati VK: Emerging role of circulating microRNA in the diagnosis of human infectious diseases. J Cell Physiol 2019, 234(2):1030–1043. [DOI] [PubMed] [Google Scholar]
- 17.Drury RE, O’Connor D, Pollard AJ: The Clinical Application of MicroRNAs in Infectious Disease. Front Immunol 2017, 8:1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokar T, Pastrello C, Rossos AEM, Abovsky M, Hauschild AC, Tsay M, Lu R, Jurisica I: mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res 2018, 46(D1):D360–D370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shirdel EA, Xie W, Mak TW, Jurisica I: NAViGaTing the micronome--using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS One 2011, 6(2):e17429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahmati S, Abovsky M, Pastrello C, Jurisica I: pathDIP: an annotated resource for known and predicted human gene-pathway associations and pathway enrichment analysis. Nucleic Acids Res 2017, 45(D1):D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguilar CA, Hamandi B, Fegbeutel C, Silveira FP, Verschuuren EA, Ussetti P, Chin-Hong PV, Sole A, Holmes-Liew C, Billaud EM et al. : Clinical risk factors for invasive aspergillosis in lung transplant recipients: Results of an international cohort study. J Heart Lung Transplant 2018, 37(10):1226–1234. [DOI] [PubMed] [Google Scholar]
- 22.Steinbach WJ, Marr KA, Anaissie EJ, Azie N, Quan SP, Meier-Kriesche HU, Apewokin S, Horn DL: Clinical epidemiology of 960 patients with invasive aspergillosis from the PATH Alliance registry. J Infect 2012, 65(5):453–464. [DOI] [PubMed] [Google Scholar]
- 23.Hwang HW, Mendell JT: MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 2006, 94(6):776–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J et al. : The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 2007, 67(16):7713–7722. [DOI] [PubMed] [Google Scholar]
- 25.Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, Dangwal S, Kumarswamy R, Bang C, Holzmann A et al. : The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun 2012, 3:1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng AM, Byrom MW, Shelton J, Ford LP: Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res 2005, 33(4):1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M et al. : miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A 2005, 102(39):13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jordan SD, Kruger M, Willmes DM, Redemann N, Wunderlich FT, Bronneke HS, Merkwirth C, Kashkar H, Olkkonen VM, Bottger T et al. : Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol 2011, 13(4):434–446. [DOI] [PubMed] [Google Scholar]
- 29.Wilfred BR, Wang WX, Nelson PT: Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab 2007, 91(3):209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camargo JF, Bhimji A, Kumar D, Kaul R, Pavan R, Schuh A, Seftel M, Lipton JH, Gupta V, Humar A et al. : Impaired T cell responsiveness to interleukin-6 in hematological patients with invasive aspergillosis. PLoS One 2015, 10(4):e0123171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das Gupta M, Fliesser M, Springer J, Breitschopf T, Schlossnagel H, Schmitt AL, Kurzai O, Hunniger K, Einsele H, Loffler J: Aspergillus fumigatus induces microRNA-132 in human monocytes and dendritic cells. Int J Med Microbiol 2014, 304(5–6):592–596. [DOI] [PubMed] [Google Scholar]
- 32.Dix A, Czakai K, Leonhardt I, Schaferhoff K, Bonin M, Guthke R, Einsele H, Kurzai O, Loffler J, Linde J: Specific and Novel microRNAs Are Regulated as Response to Fungal Infection in Human Dendritic Cells. Front Microbiol 2017, 8:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen ZN, Ling ZG, Huang Y, Li X: [Expression and differential diagnostic value of serum microRNA for invasive pulmonary aspergillosis]. Zhonghua Jie He He Hu Xi Za Zhi 2017, 40(4):272–277. [DOI] [PubMed] [Google Scholar]
- 34.Du L, Chen X, Duan Z, Liu C, Zeng R, Chen Q, Li M: MiR-146a negatively regulates dectin-1-induced inflammatory responses. Oncotarget 2017, 8(23):37355–37366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Ree MH, van der Meer AJ, de Bruijne J, Maan R, van Vliet A, Welzel TM, Zeuzem S, Lawitz EJ, Rodriguez-Torres M, Kupcova V et al. : Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antiviral Res 2014, 111:53–59. [DOI] [PubMed] [Google Scholar]


