Abstract
The redox state of nitrogenase Fe protein is shown to affect regulation of ADP-ribosylation in Klebsiella pneumoniae strains transformed by plasmids carrying dra genes from Rhodospirillum rubrum. The dra operon encodes dinitrogenase reductase ADP-ribosyltransferase and dinitrogenase reductase-activating glycohydrolase, enzymes responsible for the reversible inactivation, via ADP-ribosylation, of nitrogenase Fe protein in R. rubrum. In bacteria containing the dra operon in their chromosomes, inactivation occurs in response to energy limitation or nitrogen sufficiency. The dra gene products, expressed at a low level in K. pneumoniae, enable transformants to reversibly ADP-ribosylate nitrogenase Fe protein in response to the presence of fixed nitrogen. The activities of both regulatory enzymes are regulated in vivo as described in R. rubrum. Genetic perturbations of the nitrogenase electron transport chain were found to affect the rate of inactivation of Fe protein. Strains lacking the electron donors to Fe protein (NifF or NifJ) were found to inactivate Fe protein more quickly than a strain with wild-type background. Deletion of nifD, which encodes a subunit of nitrogenase MoFe protein, was found to result in a slower inactivation response. No variation was found in the reactivation responses of these strains. It is concluded that the redox state of the Fe protein contributes to the regulation of the ADP-ribosylation of Fe protein.
Biological nitrogen fixation is a capability found in diverse genera of bacteria, including plant root-associated symbionts, free-living bacteria, and cyanobacteria. The conversion of N2 to NH3 is catalyzed by the nitrogenase enzyme complex, which comprises two separable components. Nitrogenase MoFe protein is an α2β2 tetramer containing the cofactor FeMoco, believed to be the site of nitrogen reduction. Nitrogenase Fe protein is the obligate electron donor to MoFe protein. Fe protein is a homodimer, containing a [Fe4S4] cluster bound by cysteinyl ligands at the dimer interface (reviewed in reference 9). Nitrogen fixation is extremely expensive in terms of the biological energy requirement. No fewer than 16 high-energy phosphoanhydride bond cleavages (ATP energy equivalents) are required for the reduction of a single dinitrogen molecule (21). In response to this requirement in the unstable microbial environment, some nitrogen-fixing bacteria have adopted a posttranslational regulatory system in which the activity of the electron carrier, Fe protein, is regulated. Inhibition of Fe protein activity is achieved by ADP-ribosylation of a specific arginine residue located on the surface of Fe protein that interacts with MoFe protein (6, 26).
The ADP-ribosylation system has been best characterized in the purple nonsulfur bacterium Rhodospirillum rubrum. The NAD+-dependent ADP-ribosylation of R101 of the R. rubrum Fe protein is catalyzed by dinitrogenase reductase ADP-ribosyltransferase (DRAT) in response to energy limitation or nitrogen sufficiency (16). Upon relief of these negative stimuli, dinitrogenase reductase-activating glycohydrolase (DRAG) removes the ADP-ribose moiety, restoring the original, fully active Fe protein (17, 29). DRAT and DRAG are encoded by the dra operon, located 0.5 kb upstream from nifH (encoding nitrogenase Fe protein) and transcribed in the opposite direction (3). Also included in the dra operon is draB, located downstream of draG and encoding a protein of unknown function. A deletion in draB exhibits a weak phenotype, inactivating Fe protein more quickly and reactivating Fe protein more slowly than a wild-type strain (13). The entire draTGB operon has been identified in Azospirillum spp. (10), but in Rhodobacter capsulatus the dra operon lacks draB (20).
Regulation of in vivo DRAT and DRAG activities has been demonstrated in R. rubrum, but the means by which these enzymes are regulated has not been determined. Regulation of DRAT has been demonstrated in an R. rubrum draG mutant. Under nitrogen-fixing conditions, this mutant retains full nitrogenase activity and ADP-ribosylation is not observed (13), meaning that DRAT must be inactive under these conditions. Regulation of DRAG has been demonstrated with a 32P pulse-chase experiment (11). The [32P]ADP-ribose label on inactive Fe protein does not turn over under conditions of continual negative stimulus (nitrogen sufficiency). Therefore, DRAG must be inactive during this phase of the ADP-ribosylation cycle. Regulation of DRAT and DRAG may be, in part, in response to the nucleotide bound to Fe protein. DRAT and DRAG have opposite specificities for the ATP- and ADP-bound forms of Fe protein (15, 28). However, fluctuations in the overall cellular ATP/ADP ratio during Fe protein inactivation-reactivation cycles are insufficient to fully explain the regulation of ADP-ribosylation (25). Cellular fluctuations in NAD+ concentrations have also been suggested as a potential regulator of DRAT (23, 24).
Recently, another potential regulatory mechanism has been suggested by the finding that DRAT and DRAG have opposite specificities for the redox state of Fe protein. DRAT ADP-ribosylates only the oxidized form of Fe protein, while DRAG reacts only with the reduced form of ADP-ribosylated Fe protein (8). Accordingly, the rate of inactivation of Fe protein in an R. rubrum nifD mutant was found to be much slower than that in wild type. Fe protein unable to donate electrons to MoFe protein (as in the nifD mutant) is expected to exist primarily in the reduced form. Due to the physiology of R. rubrum, the effect of promoting the oxidation of Fe protein could not be studied. The in vivo electron donor(s) to nitrogenase Fe protein in R. rubrum is unknown, and therefore it is not possible to prepare mutants unable to reduce Fe protein in vivo. In contrast, the nitrogen-fixing enteric bacterium Klebsiella pneumoniae contains a single, obligate electron donor to Fe protein, the flavodoxin NifF (22, 27). Pyruvate-flavodoxin oxidoreductase (NifJ), in turn, is the obligate electron donor to NifF (27, 31). In this report, we demonstrate the functional expression of the R. rubrum dra operon in K. pneumoniae and the regulation of DRAT and DRAG activities in the K. pneumoniae transformants. The effects of in vivo perturbation of the nitrogen fixation electron transport chain on the modification of Fe protein were examined.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains used in this study are listed in Table 1. Escherichia coli DH5α (30) was used to maintain plasmids.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype and description | Source or reference |
|---|---|---|
| K. pneumoniae | ||
| UN | Wild type, strain M5a1 | P. W. Wilson |
| UN1072 | nifD4396 | 19 |
| UN1487 | nifJ4617 | 19 |
| UN3409 | nifF5520 | 18 |
| UN5482 | Transformant of UN with pCH1, Smr | This study |
| UN5483 | Transformant of UN with pCH3, Smr | This study |
| UN5484 | Transformant of UN1072 with pCH1, Smr | This study |
| UN5485 | Transformant of UN1487 with pCH1, Smr | This study |
| UN5486 | Transformant of UN3409 with pCH1, Smr | This study |
| UN5491 | Transformant of UN with pEXT21, Smr | This study |
| Plasmids | ||
| pEXT21 | Ptac expression vector, LacIq, IncW ori, Smr | 2 |
| pYPZ148 | R. rubrum nifH′draTGB (3.3-kb PstI fragment) in pUC19, Apr | Unpublished data |
| pCH1 | R. rubrum draTGB (2.7-kb BamHI-PstI fragment) in pEXT21, Smr | This study |
| pCH3 | R. rubrum draTG′ (1.7-kb BamHI-HindIII fragment) in pEXT21, Smr | This study |
Construction of plasmids for the expression of the dra operon.
To create a system for low-level expression of the dra gene products in K. pneumoniae, the draTGB operon was cloned into the low-copy-number plasmid pEXT21 under the control of the Ptac promoter. The draTGB operon from R. rubrum, including a portion of nifH located upstream and in the opposite orientation, has been previously cloned into pUC19, yielding plasmid pYPZ148 (Y. Zhang, unpublished results). This plasmid was digested with BamHI and PstI, yielding two BamHI-PstI DNA fragments of 2.7 kb (the draTGB operon and a pUC19 fragment), as well as other, smaller DNA fragments. The 2.7-kb DNA fragments of pYPZ148 were ligated into pEXT21 that had been digested with BamHI and PstI at unique sites in the multicloning region. E. coli DH5α cells were transformed by the ligated plasmids using a standard heat shock method (30). The resulting draTGB-bearing clones were resolved from those bearing the pUC19 fragment by replicate plating; bacteria bearing the pUC19 fragment were Apr, while draTGB-bearing clones were Aps. The plasmid containing the draTGB operon under the control the Ptac promoter of pEXT21 was designated pCH1. Note that, in this plasmid, the translation of DRAT is dependent on the native ribosome binding site. A plasmid expressing only draT was produced by excision of a 1.0-kb HindIII fragment from pCH1. The excised fragment includes the last 70 bp of draG, the entire draB gene, and a portion of the pEXT21 multicloning site. This deletion in draG has been shown to be sufficient to abrogate DRAG activity (7). The plasmid resulting from the HindIII fragment excision was designated pCH3.
K. pneumoniae transformation and growth conditions.
Recipient K. pneumoniae strains (see Table 1) were transformed with plasmids pCH1 and pCH3 using a freeze-thaw procedure (32). K. pneumoniae strains were isolated and grown in LC media (1% tryptone, 0.5% yeast extract, 0.5% NaCl) at 30°C. Stocks of each strain were stored in 10% dimethyl sulfoxide at −80°C. For nif derepression of K. pneumoniae, frozen stock was inoculated into minimal medium with 0.2% ammonium acetate (25 mM) as the nitrogen source (22) and was grown aerobically at 30°C for 24 h. The overnight cultures were inoculated (1:50) into the same minimal medium and again grown aerobically for 24 h at 30°C, reaching a maximal optical density at 600 nm (OD600) of ∼6. The cells were collected by centrifugation (4,000 × g, 10 min at 4°C) and resuspended in 5 volumes of minimal medium, in the absence of ammonium but in the presence of 0.015% serine. When appropriate, the expression of draTGB was induced by the addition of 50 μM isopropyl-β-d-thiogalactopyranoside (IPTG). Cultures were degassed and flushed with argon three times and were then incubated anaerobically for 5 h in anaerobic 60-ml glass vials.
In vivo nitrogenase assay.
Nitrogenase activity was measured by the acetylene reduction method (1). To measure whole-cell activity, 1 ml of K. pneumoniae cells was transferred anaerobically into a 9-ml anaerobic vial containing 10% acetylene (in argon) headspace. The assays were incubated 2 min at 30°C with shaking and then were terminated by the addition of 0.4 N NaOH. The ethylene produced was detected by flame-ionization-monitored gas chromatography, using an Alltech Porapak N 80/100 column on a Shimadzu GCF gas chromatograph. Nitrogenase activities are expressed as nanomoles of ethylene formed per milliliter of cell culture per hour, normalized to a cell culture OD600 of 1.0.
Determination of [32P]ADP-ribose turnover on Fe protein.
K. pneumoniae cells (strain UN5482) were grown, derepressed for nitrogen fixation, and assayed for nitrogenase activity, as described above, except that the cultures were grown in the presence of 2 μCi of H332PO4 ml−1 (3 mM total Pi) and the cells were buffered with 0.3 M MOPS (morpholinepropanesulfonic acid; pH 8.0). After derepression for 5 h, 25 mM ammonium acetate was added to the cells to effect inactivation of nitrogenase Fe protein. Upon complete inactivation of Fe protein, the 32P label was chased by the addition of 0.3 M unlabeled Pi (pH 8.0). At 1-h intervals, 5-ml samples of cell cultures were precipitated by the addition of 10% trichloroacetic acid. After centrifugation, the precipitated pellets were washed successively with 1:1 and 1:3 ethanol-ether solutions. The dried pellets were resuspended in 1 ml of 50 mM Tris (pH 7.5)–150 mM NaCl–0.1% Igepal CA-630 (Sigma)–1 mM EDTA–0.02% sodium azide. After removal of cell debris by centrifugation, the supernatant was incubated with 0.2 mg of anti-Fe protein immunoglobulin G (IgG) purified from a crude rabbit antiserum raised against Azotobacter vinelandii and R. rubrum Fe protein. The IgG-Fe protein complex was coprecipitated with protein A affixed to Sepharose beads. Proteins were solubilized from the Sepharose beads in Laemmli sodium dodecyl sulfate (SDS) sample buffer (12). The proteins were resolved in a 10% polyacrylamide gel (0.6% cross-linked) by SDS-polyacrylamide gel electrophoresis (PAGE). The [32P]ADP-ribose label on nitrogenase Fe protein was quantitated from a Cyclone Storage Phosphor screen (Packard) exposed to the gel for 72 h.
At each sampling time, the OD600 of each culture was obtained. Additionally, 0.5 ml of culture was collected by centrifugation (5,000 × g, 1 min) and was resuspended in 0.5 ml of distilled, deionized H2O. The total cellular 32P content was determined by liquid scintillation counting of 50 μl of resuspended culture.
Quantitation of nitrogenase Fe protein by immunoblotting.
Samples of K. pneumoniae cultures derepressed for nitrogen fixation were precipitated by the addition of 10% trichloroacetic acid. After centrifugation, the precipitated pellet was resuspended in Laemmli SDS sample buffer. Nitrogenase Fe protein composition was quantitated by enhanced chemiluminescence (ECL; Amersham Pharmacia Biotech)-developed immunoblots of samples electrophoresed in a 10% polyacrylamide gel (0.6% bisacrylamide). Primary and secondary immunoblotting antibodies were anti-A. vinelandii Fe protein raised in rabbit and horseradish peroxidase-linked anti-rabbit IgG raised in goat. The intensities of the ADP-ribosylated subunit (slower migrating) and the unmodified subunit (faster migrating) of nitrogenase Fe protein were determined by scanning the ECL (Amersham)-developed X-ray film with a Personal Densitometer SI (Molecular Dynamics). The percentage of subunits modified was calculated from the proportion of nitrogenase Fe protein in the slower-migrating form.
RESULTS
Expression of draTGB in K. pneumoniae.
Plasmids were constructed that expressed the dra operon in low quantities, similar to the levels observed in wild-type R. rubrum. In plasmids pCH1 (draTGB) and pCH3 (draTG′), the dra genes were cloned into the pEXT21 low-copy-number expression vector (2). In these constructs, the transcription of dra was placed under the control of the Ptac promoter. The strong transcriptional terminator rrnBT2 was located downstream of the dra genes. The lacIq gene was present in the construct, reducing IPTG-independent expression of draTGB. The cloned region of the dra operon included ∼40 bp upstream of the TTG draT start codon. This region included the ribosome binding site required for DRAT translation.
K. pneumoniae strains transformed with pCH1 retained the original nif phenotype. Cultures of K. pneumoniae UN (wild type) transformed with pCH1 (strain UN5482) exhibited the same initial nitrogenase activity (∼1,000 nmol/ml/h) as UN transformed with pEXT1 (strain UN5491) (Fig. 1). Transformants derived from Nif− strains retained the Nif− phenotype. UN5484 (nifD) and UN5485 (nifJ) exhibited no acetylene reduction activity (data not shown). UN5486 (nifF) had a trace of activity (3 nmol ml−1 h−1), reflecting the slightly leaky phenotype previously described for nifF mutants of K. pneumoniae (27).
FIG. 1.
Response of K. pneumoniae strains to exogenous ammonium. Strains UN5482 (wild type transformed with pCH1, containing draTGB) (■), UN5483 (draTG′) (▴), and UN5491 (vector-only control) (□) were derepressed for nitrogenase activity. After 5 h of derepression, 0.5 mM ammonium acetate was added to each culture (time = 0). The nitrogenase activities of each culture were monitored by the acetylene reduction method at the time points indicated, after the addition of ammonium acetate. Results are expressed as nanomoles of ethylene produced per milliliter of culture per hour.
Exogenous ammonium has previously been shown to cause inactivation of nitrogenase in K. pneumoniae strains overexpressing DRAT and DRAG (4). To determine the minimum level of DRAT expression required to cause this inactivation, expression of draT was induced by varying amounts of IPTG in nitrogen-fixing UN5483 (draTG′, UN background) cultures. In low concentrations of IPTG (≤10 μM), a gradual reduction of nitrogenase activity (50% loss of activity over 30 min) was observed in response to the addition of 0.5 mM ammonium acetate. This inactivation appears to be due to background expression from the strong Ptac promoter. Rapid inactivation (100% loss of activity in 10 min) was achieved by dra induction with 50 μM IPTG. Expression of dra genes was induced by 50 μM IPTG in all further experiments. At this level of induction, neither DRAT nor DRAG could be detected in K. pneumoniae strains by ECL-developed immunoblots (data not shown). From the known detection limits of this method, DRAT and DRAG were present at <0.5 μg per ml of cell culture (OD = 1.0). This places DRAT and DRAG, at most, at levels similar to those described in R. rubrum (0.1 to 0.3 μg per ml of cell culture at an OD of 1.0) (14, 29). Note that DRAT and DRAG are also undetectable in R. rubrum UR2 (wild type) crude extracts by immunoblotting; the proteins are only detected after partial purification or in crude extracts of overexpressing strains (7).
Reversible inactivation of nitrogenase Fe protein.
In the K. pneumoniae system using the parameters described above, functional activities of both DRAT and DRAG in vivo in response to exogenous ammonium were demonstrated. In response to the addition of 0.5 mM ammonium acetate, UN5482 (draTGB) exhibited a complete loss of nitrogenase activity in 15 min (Fig. 1). After approximately 15 min (presumably upon exhaustion of ammonium), a rapid, complete restoration of nitrogenase activity was observed. In UN5483 (draTG′), inactivation was observed, but nitrogenase activity was not recovered even after a long time period (2 h). A K. pneumoniae strain carrying only the expression vector (pEXT21) in a wild-type background did not lose nitrogenase activity in response to added ammonium (Fig. 1).
Regulation of DRAT and DRAG in K. pneumoniae transformants.
Regulation of DRAT and DRAG activities in K. pneumoniae was demonstrated by experiments similar to those conducted by Liang et al. (13) and Kanemoto and Ludden (11), which demonstrated that DRAT and DRAG activities are regulated in R. rubrum. A K. pneumoniae strain expressing only draT (UN5483), analogous to a draG mutant in R. rubrum, has full nitrogenase activity, relative to a draTGB transformant (Fig. 1). Thus, DRAT becomes active only upon signaling initiated by ammonium.
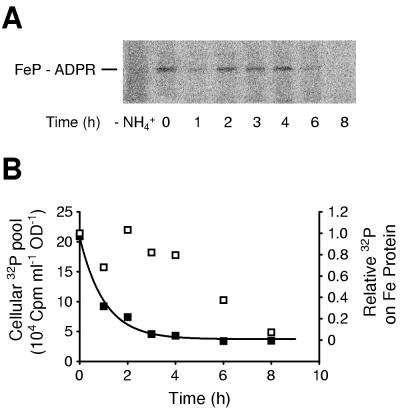
Regulation of DRAG in K. pneumoniae transformants was demonstrated by showing that the ADP-ribose moiety on Fe protein turns over more slowly than the overall 32P pool under conditions of constant negative stimulus. UN5482 (draTGB) cells grown on 2 μCi of 32P ml−1 did not contain any radiolabeled material immunoreactive to anti-Fe protein antibody (Fig. 2A, lane −NH4+). Upon treatment with 25 mM ammonium acetate for 30 min, radiolabel appeared on Fe protein (Fig. 2A, lane 0), and this label persisted for at least 4 h before disappearing. In contrast, the overall 32P pool turned over much more rapidly, with a t1/2 of 42 min (Fig. 2B). Because the rate of exchange of the ADP-ribose label did not reflect the exchange rate of the overall cellular phosphorus pool, futile cycling of ADP-ribose must not be occurring and DRAG must be inactive under these conditions.
FIG. 2.
Regulation of DRAG in K. pneumoniae. UN5482 (draTGB transformant of wild type) was grown on depleted phosphate medium containing 2 μCi of [32P]orthophosphate ml−1. After derepression for 5 h, the culture was treated with 25 mM ammonium acetate (30 min) and then 0.3 M Pi (pH 8.0). (A) Phosphor storage screen image of SDS-PAGE gel of anti-Fe protein immunoprecipitated samples. The migration position of ADP-ribosylated Fe protein is indicated (FeP-ADPR). The culture was sampled immediately before the addition of ammonium acetate (−NH4+), immediately after the addition of unlabeled phosphate (time = 0), and at 1-h intervals thereafter. (B) Comparison of the turnover times of overall cellular 32P pool (■) and 32P label on Fe protein (□). Cellular 32P was determined from liquid scintillation counting of cells collected by centrifugation, normalized to cell content as estimated by the OD600. 32P label on Fe protein was calculated from the intensities of bands on the phosphor storage screen, normalized to the background intensity of the lane of the phosphor storage screen.
Correlation of immunoblots with nitrogenase activity.
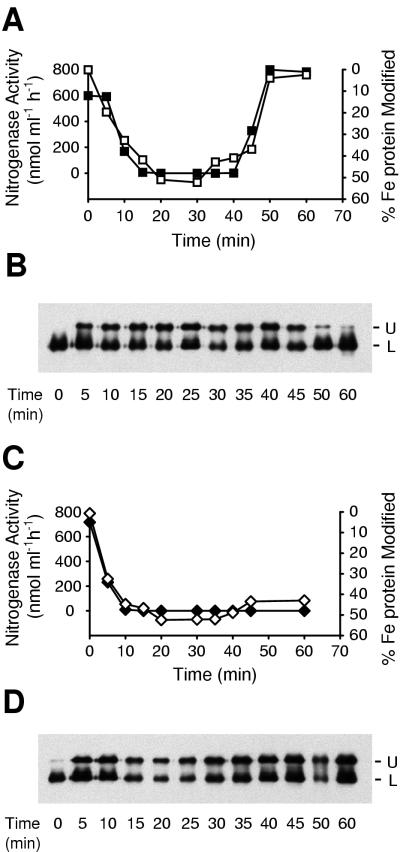
The fraction of nitrogenase Fe protein in the ADP-ribosylated (inactive) state was correlated to the decrease in cellular nitrogenase activity. Nitrogen-fixing cultures of UN5482 (draTGB) and UN5483 (draTG′) were treated with 0.5 mM ammonium acetate, and the state of the Fe protein was monitored by acetylene reduction activity and by immunoblotting. The Fe protein of UN5482 was initially unmodified (Fig. 3B), and the fraction of Fe protein dimers modified was correlated to the nitrogenase activity (Fig. 3A and C) (note that when 50% of the subunits are modified, one subunit of each dimer is modified, yielding inactive protein). Fe protein in UN5483 remained fully modified after long time periods (Fig. 3D). These results demonstrated that the immunoblotting protocol could be used as a valid estimation of the ADP-ribosylation state of nitrogenase Fe protein.
FIG. 3.
Correlation of nitrogenase activity with modification of Fe protein. K. pneumoniae cultures were derepressed for nitrogenase activity. Inactivation of nitrogenase was initiated by addition of 0.5 mM ammonium acetate. (A) Traces of nitrogenase activity (■) and modification of Fe protein (□) in UN5482 (draTGB transformant, wild-type background) are recorded. (B) Corresponding immunoblot of nitrogenase Fe protein from UN5482. The upper band (U) corresponds to the ADP-ribosylated form of Fe protein. The lower band (L) represents unmodified Fe protein. (C) Traces of nitrogenase activity (⧫) and modification of Fe protein (◊) in UN5483 (draTG′ transformant, wild-type background). (D) Corresponding immunoblot of nitrogenase Fe protein from UN5483. The positions of the upper (U) and lower (L) bands are indicated.
Inactivation response in nif mutants.
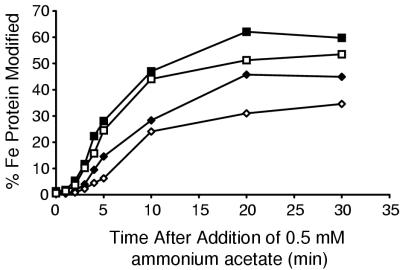
K. pneumoniae strains containing mutations in nifD, nifF, or nifJ were transformed with plasmid pCH1 (draTGB) in order to examine the importance of the redox state of nitrogenase Fe protein on the rate of ADP-ribosylation. Cultures of these transformants were grown under nitrogen-fixing conditions. Inactivation of Fe protein was initiated by the addition of 0.5 mM ammonium acetate to the cultures. The extent of modification of Fe protein was followed by immunoblotting (Fig. 4). In a nifD mutant (UN5484), in which Fe protein is thought to be predominantly reduced, the rate of modification was just over half that in the wild-type background (UN5482). Therefore, DRAT activity was inhibited when Fe protein was predominantly reduced. In nifF and nifJ mutants (UN5485 and UN5486), the rate of ADP-ribosylation was nearly twice that in UN5482, indicating that DRAT activity was enhanced when Fe protein could not be reduced. Note that in UN5485, modification of >50% of Fe protein subunits was observed consistently.
FIG. 4.
Effect of Fe protein redox state on DRAT-dependent inactivation rate. K. pneumoniae cultures were derepressed for nitrogen fixation. Inactivation of nitrogenase was initiated by the addition of 0.5 mM ammonium acetate. Modification of Fe protein was monitored by anti-Fe protein immunoblotting. Traces represent data from UN5482 (draTGB, wild-type background) (⧫), UN5484 (nifD background) (◊), UN5485 (nifJ background) (■), and UN5486 (nifF background) (□). Data represent the averages from four experiments. The standard deviation for each datum point is <30%.
Reactivation response in nif mutants.
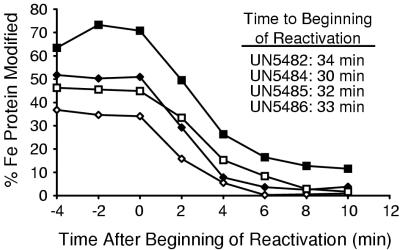
Strains UN5482 (wild-type), UN5484 (nifD), UN5485 (nifF), and UN5486 (nifJ) were further monitored for the demodification of nitrogenase Fe protein. Demodification of Fe protein was observed in all four strains after a similar period of maximal modification (Fig. 5, inset). Note that Fe protein from the nifD strain (UN5484) was never fully modified (<50% subunits ADP-ribosylated) and that Fe protein from the nifJ strain (UN5485) was initially >50% modified. Reactivation of Fe protein was generally faster than inactivation (t1/2 = ∼3 min), and rates of reactivation were similar for all four strains. Fe protein from the nifJ mutant (UN5485) was observed to retain a residual level of modification (∼10% of subunits) that decreased very slowly over an extended period of time (∼5% modified after 1 h).
FIG. 5.
Effect of Fe protein redox state on reaction on DRAG-dependent reactivation. K. pneumoniae cultures were derepressed for nitrogen fixation and inactivated by addition of 0.5 mM ammonium acetate as in Fig. 4. The extent of modification of Fe protein was monitored by anti-Fe protein immunoblotting. Traces represent data from ⧫ UN5482 (draTGB, wild-type background) (⧫), UN5484 (nifD background) (◊), UN5485 (nifJ background) (■), and UN5486 (nifF background) (□). The data represent averages from four experiments. The standard deviation for each datum point is <30%. The beginning of reaction (inset) was defined as the last datum point at which modification was maximal for each experiment.
DISCUSSION
A system of heterologous expression, in K. pneumoniae, of genes encoding the R. rubrum ADP-ribosylation regulatory system for nitrogenase Fe protein has been used to study the regulation of DRAT and DRAG. At low levels of expression, the activities and regulation of DRAT and DRAG are similar to those observed in R. rubrum. This heterologous expression system lends itself to a number of useful applications. Since K. pneumoniae is more genetically tractable than R. rubrum, it may be possible to analyze the signal transduction pathway giving rise to ammonium-induced inactivation of Fe protein. Although energy limitation signals have been described in the ADP-ribosylation systems of R. rubrum and Azospirillum brasilense, no corresponding response has been tested in the K. pneumoniae heterologous system.
The heterologous expression system functioned in a manner strikingly similar to that of R. rubrum, suggesting that the regulation of DRAT and DRAG occurs in response to compounds found in K. pneumoniae, which does not contain draTGB genes in the wild-type strain. Expression of the draT gene was required for the inactivation of Fe protein, and draG was required for reactivation. As in R. rubrum, the rate of inactivation appears to depend on the presence of the draG and draB gene products (13), as a slightly faster inactivation rate was observed in UN5483 (draTG′) than in UN5482 (draTGB). Unlike the native R. rubrum system, the K. pneumoniae expression system is capable of complete inactivation of nitrogenase (Fig. 1). Also, the rate of inactivation in response to exogenous ammonium is faster in K. pneumoniae (t1/2 ≈ 5 min at 0.5 mM NH4+) than in R. rubrum (t1/2 ≈ 20 min at 2 mM NH4+). These two phenomena may be interpreted as representing a stronger activating signal for DRAT, resulting in a longer transient activity. Because K. pneumoniae Fe protein is a better substrate for DRAT than R. rubrum Fe protein in vitro (14), it is possible that DRAT is able to modify K. pneumoniae Fe protein in vivo under less-favorable conditions, resulting in a longer period of DRAT activity.
Previous studies have described the functional expression of draT and draG in K. pneumoniae (4, 5). However, a number of drawbacks in this previously described system have prompted the developments described in this study. Previously, the dra genes were expressed at high levels (Ptac promoter in pKK223-3 expression vector, 1 mM IPTG) (4, 5). At this level of expression, expression of draT alone resulted in a Nif− phenotype, due to constitutive ADP-ribosylation of Fe protein (5). While reversible ADP-ribosylation was observed upon coexpression of draT and draG, it is probable that DRAT and DRAG were improperly regulated, resulting in futile cycling of ADP-ribose. These results are analogous to those observed by Grunwald et al. (7). Overexpression of draT alone in R. rubrum resulted in drastically decreased nitrogenase activity (although not due to constitutive ADP-ribosylation), while coexpression of draT and draG restored apparently regulated activity, although with a lowered initial nitrogenase activity. The previously described K. pneumoniae system also failed to express draB, which has been shown to have a role in ADP-ribosylation activities (13). The expression system described here produces DRAT, DRAG, and DRAB at the low levels observed in R. rubrum, resulting in an initial nitrogenase activity as high as that of wild type (1,000 nmol ml−1 h−1).
Expression of a regulated ADP-ribosylation system in K. pneumoniae allowed examination of the role of the redox state of Fe protein in posttranslational regulation. Previously, we have shown that in an R. rubrum nifD mutant, in which Fe protein is presumed to be predominantly in the reduced state, inactivation of Fe protein occurs at a much slower rate than in the wild type (8). This result was also observed in the K. pneumoniae system, but the difference in inactivation rates was less than in R. rubrum. The lessened difference may be due to the fact that K. pneumoniae Fe protein is a better substrate for DRAT. As observed for in vitro results with A. vinelandii Fe protein (8), some DRAT activity may be possible with reduced Fe protein. In K. pneumoniae nifJ and nifF mutants, inactivation was faster than in the wild-type background, but the inability to effectively reduce Fe protein was not, in itself, sufficient to cause inactivation. This indicates that the redox state of Fe protein must not be the sole regulatory component for the activities of DRAT and DRAG. In UN5485 (nifJ, draTGB) and UN5486 (nifF, draTGB), Fe protein remains unmodified until initiation of an additional signal, caused by the ammonium stimulus. Nevertheless, the redox state of Fe protein upon initiation of negative stimulus is demonstrated to have an effect on the activity of DRAT in vivo.
Surprisingly, there was very little difference in the rates of reactivation of Fe protein in wild-type or nif mutant strains of K. pneumoniae. The similarity of the times to initiation of reactivation indicates that all strains perceive similarly the signal produced by the exhaustion of fixed nitrogen. In this experiment, the state of nitrogenase Fe protein is less well understood than in the inactivation period. It is unknown if ADP-ribosylated Fe protein can receive electrons from (or donate electrons to) the flavodoxin NifF or other electron carriers. It is possible that, in all strains, reduced Fe protein accumulates in the 30-min period during which Fe protein is ADP-ribosylated. Certainly in strain UN5486 (nifF), the leaky phenotype observed could lead to accumulation of reduced Fe protein when electron donation to MoFe protein is blocked by ADP-ribosylation. However, these results do not discount the possibility of regulation of DRAG activity in response to oxidation of Fe protein. None of the experiments in this study were conducted under growth conditions that would be expected to cause limitations in cellular reducing power. Rather, electron transfer was inhibited by deletion of electron carriers. Under growth conditions that give rise to a limitation in available reducing power, Fe protein might indeed accumulate in the oxidized state, inhibiting DRAG activity.
One prominent result in this study was the consistent modification of >50% of the subunits of nitrogenase Fe protein in UN5485 (nifJ) in response to exogenous ammonium. The consensus opinion has long been that only one subunit of each Fe protein dimer can be ADP-ribosylated by DRAT. However, there are now several reports of in vivo overmodification of Fe protein. In the study of Fu et al. (5), draT was expressed at a high level in a K. pneumoniae nifF mutant. Even without the addition of an inactivating stimulus, an immunoblot of the extract of this mutant shows, clearly, more Fe protein-immunoreactive material in the slower-migrating position than in the unmodified position. Also, in an R. rubrum mutant bearing an “always active” form of DRAT (K103E), >50% of Fe protein subunits may be modified (Zhang, unpublished). Overmodification has also been demonstrated in A. brasilense cells in which protein synthesis has been terminated by chloramphenicol or tetracycline (Zhang, unpublished). It remains unclear if these examples are related. However, it is possible that DRAT is able to modify a previously uncharacterized form of Fe protein that is present only under oxidizing conditions (e.g., a monomeric Fe protein subunit) or that Fe dimers can undergo subunit exchange to form the doubly-modified species after formation of the mono-ADP-ribosylated dimer.
We have demonstrated that DRAT and DRAG activities, in K. pneumoniae, can be regulated in a manner consistent with the regulation observed in R. rubrum. This demonstration casts doubt upon models of the regulation of DRAT and DRAG that rely on a specific signal transduction pathway for regulation. DRAT and DRAG might be regulated in response to the proportion of Fe protein in the ATP- or ADP-bound state and the proportion in the reduced or oxidized state. Given at least these two parameters available for regulation, considerable fine-tuning of DRAT and DRAG activities could be possible. Thus, the point of convergence of signal transduction pathways from different stimuli could be nitrogenase Fe protein itself, with some inactivation signals giving rise to oxidation of Fe protein and others giving rise to a decrease in the ATP/ADP ratio. Many aspects of the dra regulatory system remain to be incorporated in this model, including the role of the draB gene product and the signal transduction pathway from the ammonium inactivation signal.
ACKNOWLEDGMENTS
We thank E. L. Pohlman for assistance with growth of bacterial cultures and R. L. Kerby and L. M. Rubio for helpful advice. We gratefully acknowledge D. M. Peters for advice on immunoprecipitation.
This work was supported by National Institute of General Medical Science grant GM54910 to P. W. Ludden. C. M. Halbleib is a trainee of the NIH Biotechnology Training Program (grant NIH 5 T32 GM08349).
REFERENCES
- 1.Burris R H. Nitrogen fixation—assay methods and techniques. Methods Enzymol. 1972;24:415–431. doi: 10.1016/0076-6879(72)24088-5. [DOI] [PubMed] [Google Scholar]
- 2.Dykxhoorn D M, St. Pierre R, Linn T. A set of compatible tac promoter expression vectors. Gene. 1996;177:133–136. doi: 10.1016/0378-1119(96)00289-2. [DOI] [PubMed] [Google Scholar]
- 3.Fitzmaurice W P, Saari L L, Lowery R G, Ludden P W, Roberts G P. Genes coding for the reversible ADP-ribosylation system of dinitrogenase reductase from Rhodospirillum rubrum. Mol Gen Genet. 1989;218:340–347. doi: 10.1007/BF00331287. [DOI] [PubMed] [Google Scholar]
- 4.Fu H, Burris R H, Roberts G P. Reversible ADP-ribosylation is demonstrated to be a regulatory mechanism in prokaryotes by heterologous expression. Proc Natl Acad Sci USA. 1990;87:1720–1724. doi: 10.1073/pnas.87.5.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu H A, Wirt H J, Burris R H, Roberts G P. Functional expression of a Rhodospirillum rubrum gene encoding dinitrogenase reductase ADP-ribosyltransferase in enteric bacteria. Gene. 1989;85:153–160. doi: 10.1016/0378-1119(89)90475-7. [DOI] [PubMed] [Google Scholar]
- 6.Georgiadis M M, Komiya H, Chakrabarti P, Woo D, Kornuc J J, Rees D C. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science. 1992;257:1653–1659. doi: 10.1126/science.1529353. [DOI] [PubMed] [Google Scholar]
- 7.Grunwald S K, Lies D P, Roberts G P, Ludden P W. Posttranslational regulation of nitrogenase in Rhodospirillum rubrum strains overexpressing the regulatory enzymes dinitrogenase reductase ADP-ribosyltransferase and dinitrogenase reductase activating glycohydrolase. J Bacteriol. 1995;177:628–635. doi: 10.1128/jb.177.3.628-635.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halbleib C M, Zhang Y, Ludden P W. Regulation of dinitrogenase reductase ADP-ribosyltransferase and dinitrogenase reductase-activating glycohydrolase by a redox-dependent conformational change of nitrogenase Fe protein. J Biol Chem. 2000;275:3493–3500. doi: 10.1074/jbc.275.5.3493. [DOI] [PubMed] [Google Scholar]
- 9.Howard J B, Rees D C. Nitrogenase: a nucleotide-dependent molecular switch. Annu Rev Biochem. 1994;63:235–264. doi: 10.1146/annurev.bi.63.070194.001315. [DOI] [PubMed] [Google Scholar]
- 10.Inoue A, Shigematsu T, Hidaka M, Masaki H, Uozumi T. Cloning, sequencing and transcriptional regulation of the draT and draG genes of Azospirillum lipoferum FS. Gene. 1996;170:101–106. doi: 10.1016/0378-1119(95)00852-7. [DOI] [PubMed] [Google Scholar]
- 11.Kanemoto R H, Ludden P W. Effect of ammonia, darkness, and phenazine methosulfate on whole-cell nitrogenase activity and Fe protein modification in Rhodospirillum rubrum. J Bacteriol. 1984;158:713–720. doi: 10.1128/jb.158.2.713-720.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Liang J H, Nielsen G M, Lies D P, Burris R H, Roberts G P, Ludden P W. Mutations in the draT and draG genes of Rhodospirillum rubrum result in loss of regulation of nitrogenase by reversible ADP-ribosylation. J Bacteriol. 1991;173:6903–6909. doi: 10.1128/jb.173.21.6903-6909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowery R G, Ludden P W. Purification and properties of dinitrogenase reductase ADP-ribosyltransferase from the photosynthetic bacterium Rhodospirillum rubrum. J Biol Chem. 1988;263:16714–16719. [PubMed] [Google Scholar]
- 15.Lowery R G, Ludden P W. Effect of nucleotides on the activity of dinitrogenase reductase ADP-ribosyltransferase from Rhodospirillum rubrum. Biochemistry. 1989;28:4956–4961. doi: 10.1021/bi00438a008. [DOI] [PubMed] [Google Scholar]
- 16.Lowery R G, Saari L L, Ludden P W. Reversible regulation of the nitrogenase iron protein from Rhodospirillum rubrum by ADP-ribosylation in vitro. J Bacteriol. 1986;166:513–518. doi: 10.1128/jb.166.2.513-518.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ludden P W, Burris R H. Activating factor for the iron protein of nitrogenase from Rhodospirillum rubrum. Science. 1976;194:424–426. doi: 10.1126/science.824729. [DOI] [PubMed] [Google Scholar]
- 18.MacNeil T. Complementation and deletion analysis of nitrogen fixation genes in Klebsiella pneumoniae. Ph.D. thesis. Madison: University of Wisconsin-Madison; 1978. [Google Scholar]
- 19.MacNeil T, MacNeil D, Roberts G P, Supiano M A, Brill W J. Fine-structure mapping and complementation analysis of nif (nitrogen fixation) genes in Klebsiella pneumoniae. J Bacteriol. 1978;136:253–266. doi: 10.1128/jb.136.1.253-266.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masepohl B, Krey R, Klipp W. The draTG gene region of Rhodobacter capsulatus is required for post-translational regulation of both the molybdenum and the alternative nitrogenase. J Gen Microbiol. 1993;139:2667–2675. doi: 10.1099/00221287-139-11-2667. [DOI] [PubMed] [Google Scholar]
- 21.Mortenson L E, Thorneley R N. Structure and function of nitrogenase. Annu Rev Biochem. 1979;48:387–418. doi: 10.1146/annurev.bi.48.070179.002131. [DOI] [PubMed] [Google Scholar]
- 22.Nieva-Gomez D, Roberts G P, Klevickis S, Brill W J. Electron transport to nitrogenase in Klebsiella pneumoniae. Proc Natl Acad Sci USA. 1980;77:2555–2558. doi: 10.1073/pnas.77.5.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noren A, Nordlund S. Changes in the NAD(P)H concentration caused by addition of nitrogenase ‘switch-off’ effectors in Rhodospirillum rubrum G-9, as measured by fluorescence. FEBS Lett. 1994;356:43–45. doi: 10.1016/0014-5793(94)01233-4. [DOI] [PubMed] [Google Scholar]
- 24.Noren A, Soliman A, Nordlund S. The role of NAD+ as a signal during nitrogenase switch-off in Rhodospirillum rubrum. Biochem J. 1997;322:829–832. doi: 10.1042/bj3220829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul T D, Ludden P W. Adenine nucleotide levels in Rhodospirillum rubrum during switch-off of whole-cell nitrogenase activity. Biochem J. 1984;224:961–969. doi: 10.1042/bj2240961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pope M R, Murrell S A, Ludden P W. Covalent modification of the iron protein of nitrogenase from Rhodospirillum rubrum by adenosine diphosphoribosylation of a specific arginine residue. Proc Natl Acad Sci USA. 1985;82:3173–3177. doi: 10.1073/pnas.82.10.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts G P, MacNeil T, MacNeil D, Brill W J. Regulation and characterization of protein products coded by the nif (nitrogen fixation) genes of Klebsiella pneumoniae. J Bacteriol. 1978;136:267–279. doi: 10.1128/jb.136.1.267-279.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saari L L, Pope M R, Murrell S A, Ludden P W. Studies on the activating enzyme for iron protein of nitrogenase from Rhodospirillum rubrum. J Biol Chem. 1986;261:4973–4977. [PubMed] [Google Scholar]
- 29.Saari L L, Triplett E W, Ludden P W. Purification and properties of the activating enzyme for iron protein of nitrogenase from the photosynthetic bacterium Rhodospirillum rubrum. J Biol Chem. 1984;259:15502–15508. [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Shah V K, Stacey G, Brill W J. Electron transport to nitrogenase. Purification and characterization of pyruvate:flavodoxin oxidoreductase. The nifJ gene product. J Biol Chem. 1983;258:12064–12068. [PubMed] [Google Scholar]
- 32.Shokolenko I N, Alexeyev M F. Transformation of Escherichia coli TG1 and Klebsiella oxytoca VN13 by freezing-thawing procedure. BioTechniques. 1995;18:596–598. [PubMed] [Google Scholar]