Abstract
New antibiotics are needed as bacterial infections continue to be a leading cause of death, but efforts to develop compounds with promising antibacterial activity are hindered by a poor understanding of – and limited strategies for elucidating – their modes of action. We recently discovered a novel lasso peptide, ubonodin, that is active against opportunistic human lung pathogens from the Burkholderia cepacia complex (Bcc). Ubonodin inhibits RNA polymerase, but only select strains were susceptible, indicating that having a conserved cellular target does not guarantee activity. Given the cytoplasmic target, we hypothesized that cellular uptake of ubonodin determines susceptibility. Although Bcc strains harbor numerous nutrient uptake systems, these organisms lack close homologs of the single known lasso peptide membrane receptor FhuA. Thus, a straightforward homology-driven approach failed to uncover the identity of the ubonodin transporter(s). Here, we used phenotype-guided comparative genomics to identify genes uniquely associated with ubonodin-susceptible Bcc strains, leading to the identification of PupB as the ubonodin outer membrane (OM) receptor in Burkholderia. Loss of PupB renders B. cepacia resistant to ubonodin, whereas expressing PupB sensitizes a resistant strain. We also examine how a conserved iron-regulated transcriptional pathway controls PupB to further tune ubonodin susceptibility. PupB is only the second lasso peptide OM receptor to be uncovered and the first outside of enterobacteria. Finally, we elucidate the full transport pathway for ubonodin by identifying its inner membrane receptor YddA in Burkholderia. Our work provides a complete picture of the mode of action of ubonodin and establishes a general framework for deciphering the transport pathways of other natural products with cytoplasmic targets.
Graphical Abstract

INTRODUCTION
Antibiotic-resistant bacterial infections pose a serious threat to human health, but in recent years few new antibiotics have been developed even though drug resistance is on the rise. Although compounds with promising antimicrobial activity are frequently discovered, limited approaches to characterize their modes of action impede further clinical development. Throughout history, humans have looked to nature as a rich source of antimicrobial compounds and among these are ribosomally synthesized and post-translationally modified peptides (RiPPs). RiPPs are synthesized as gene-encoded precursor peptides that are post-translationally tailored with additional chemical modifications to form the mature peptide scaffold1,2. While much is known about the chemistry and biosynthesis of RiPPs, their bioactivities and the modes of action underlying those activities remain largely elusive3–5.
We recently reported the discovery of a novel lasso peptide RiPP, ubonodin, that has potent antimicrobial activity against several opportunistic human pathogens from the Gram-negative Burkholderia cepacia complex (Bcc)6. Because ubonodin represents a new lead compound that targets highly-resistant Bcc bacteria which can cause serious, often fatal infections in individuals living with cystic fibrosis or other underlying pulmonary disease7–9, we became interested in understanding its mode of action. Like other lasso peptides10,11, ubonodin adopts a right-handed lariat conformation, formed by the C-terminal tail threading through a macrolactam ring and held in place by bulky steric lock residues that straddle the ring (Figure 1A). The ring is forged by connecting the N-terminus of the peptide to the acidic side chain of Glu8 via an isopeptide bond. The integrity of this lasso fold is essential to ubonodin’s RNA polymerase (RNAP)-inhibiting activity. We were intrigued to find that despite having a highly conserved molecular target, ubonodin is selectively active, inhibiting some but not all Burkholderia strains. Because ubonodin has a cytoplasmic target, we hypothesized that its ability to access the interior of target cells is key to determining the extent of activity. That is, ubonodin transport across the bacterial outer membrane (OM) followed by the inner membrane (IM) is a prerequisite for RNAP engagement. Due to its large size, cellular uptake of ubonodin likely requires membrane transporters as seen for other lasso peptides and RiPPs4,12. Identifying the membrane transporters that are central to the mode of action of ubonodin will enable accurate prediction of its spectrum of activity, an important step toward developing ubonodin as an antibiotic.
Figure 1.
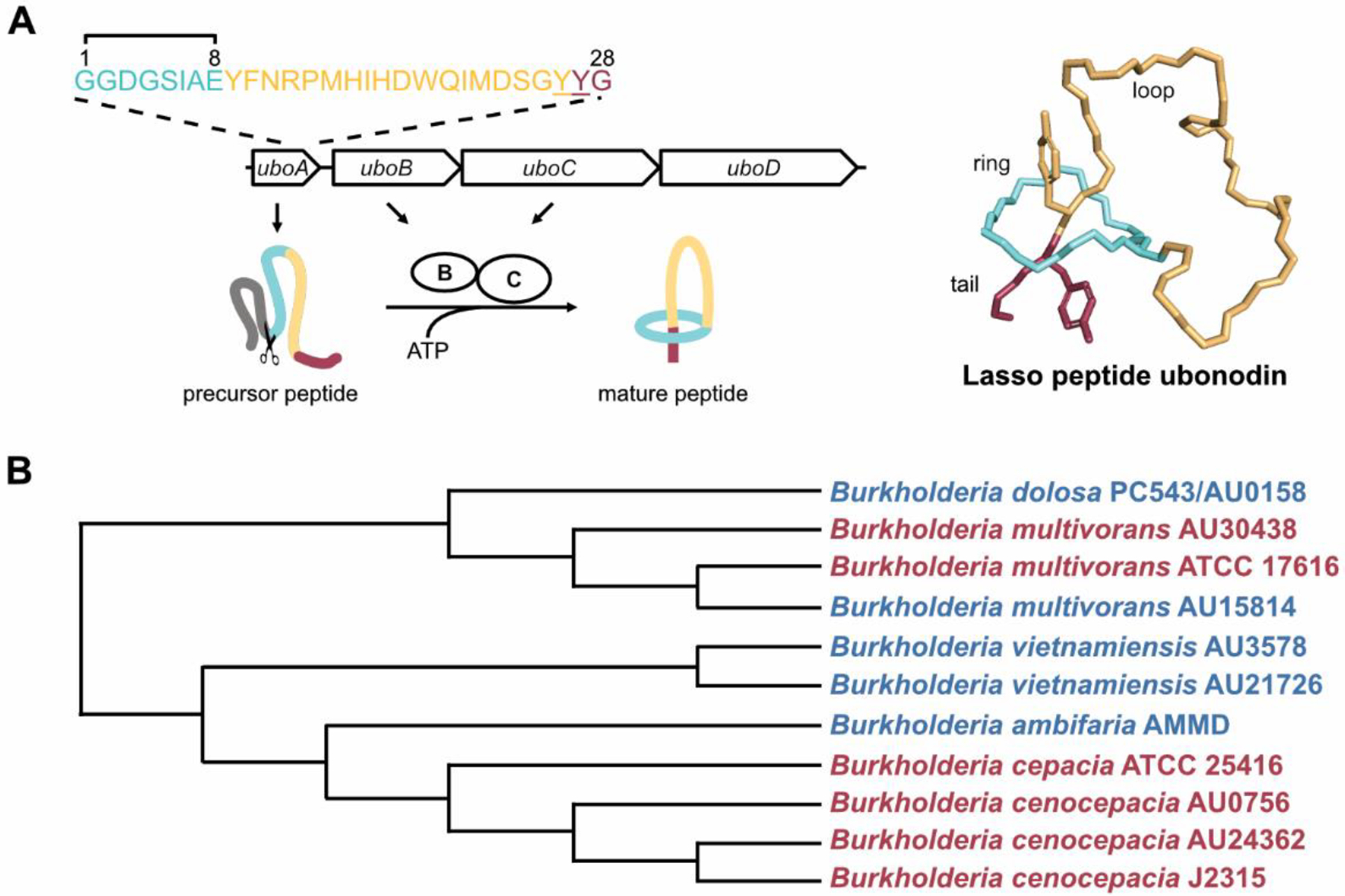
Biosynthesis and activity of ubonodin. (A) Left: Schematic showing the ubonodin biosynthetic gene cluster (BGC). Following production of the ubonodin precursor peptide specified by uboA, the B enzyme cleaves the leader peptide (gray), generating the core peptide whose amino acid sequence is outlined at the top. The C enzyme forms the final lariat structure by connecting the amino group of Gly1 to the Glu8 side chain, forging an 8-membered ring through which the tail is threaded. Residues Tyr26 and Tyr27 (underlined) act as steric locks to prevent ubonodin from unthreading. The final gene, uboD, in the BGC encodes an ABC transporter that serves as an immunity factor in the native producer B. ubonensis to export bioactive ubonodin outside the cell. Right: Ubonodin structure generated from PDB 6POR. (B) Ubonodin selectively inhibits a subset of Bcc strains. Clustal Omega alignment of 7 concatenated housekeeping gene fragments (atpD, gltB, gyrB, recA, lepA, phaC, and trpB) encoded by ubonodin-susceptible (red) and non-susceptible (blue) Bcc strains was used to generate the cladogram13–15. Shown is a Neighbor-joining tree without distance corrections.
Here, we first focus on the initial stage of cellular uptake, the OM transport step which is a key barrier to compound bioactivity. Burkholderia have unusually large, multireplicon genomes encoding dozens of possible OM transporters, consistent with the ability of Burkholderia to survive in diverse ecological niches8. The plethora of nutrient uptake systems in Burkholderia complicates the search for ubonodin transporters. To overcome this challenge, we developed a comparative genomics approach complemented with targeted mutagenesis to quickly identify the TonB-dependent transporter PupB as the ubonodin OM receptor in B. cepacia. For any Bcc strain, the presence of a close PupB homolog served as a reliable predictor of ubonodin susceptibility. We further show that PupB is subjected to iron-mediated transcriptional repression. Our finding that OM transport correlates with susceptibility provides a molecular explanation for ubonodin activity. In addition to pinpointing the OM receptor, we provide evidence that ubonodin uses the ATP-powered IM transporter YddA in the second stage of cellular uptake. Although other lasso peptide membrane transporters have been reported, our study introduces a unique, simple, and rapid approach to streamline their identification. Finally, beyond the focus on ubonodin, our studies reveal new insights into Burkholderia physiology. Our multipronged experimental and computational approach will also be useful as a general framework for other compound transport studies.
RESULTS AND DISCUSSION
Ubonodin inhibits the growth of select Bcc strains
In our original work on ubonodin, we tested for antimicrobial activity against a panel of strains, finding the most potent activity against Bcc strains6. Here, we began our search for ubonodin transporters by measuring the activity of ubonodin against an expanded panel of Bcc strains. We focused on strains from the Bcc because they are most closely related to the ubonodin producer, B. ubonensis, and RiPPs tend to have a focused spectrum of activity3,4. Moreover, we anticipated that the elusive genetic signatures differentiating ubonodin-susceptible (uboS) from non-susceptible (uboN) Bcc strains should be more apparent as Bcc members are otherwise highly similar. In total, we tested the activity of ubonodin against 12 Bcc strains representing 6 distinct Bcc species or genomovars14,16,17. These Bcc species are most associated with infections in CF patients18,19. Of the 12 Bcc strains tested, 6 were susceptible to ubonodin (Figures 1B and S1) and their levels of susceptibility were similar, ranging from 10–40 μM ubonodin in a plate-based spot assay (Table S1). Thus, ubonodin is selectively active against a subset of Bcc strains.
We next wanted to understand how the uboS Bcc strains differ from the uboN strains. One possible explanation would be if in the uboN strains, the molecular target RNAP has specific mutations that abolish ubonodin recognition and binding. Structural work on the transcription-inhibiting lasso peptides MccJ2520–22 and capistruin23,24 confirmed that both peptides bind within the secondary channel of RNAP blocking the path to the catalytic center25. Both MccJ25 and capistruin interact with residues belonging to the RNAP β and β’ subunits. Given the structural and functional similarities between ubonodin and these other lasso peptides6, we reasoned that their modes of binding to RNAP would also be analogous and ubonodin would contact β and β’ subunit residues. Therefore, we compared the amino acid sequence of the β and β’ subunits for the 12 Bcc strains tested for ubonodin susceptibility to determine if uboN strains encode distinctly unique β/β’ variants from uboS strains. By multiple sequence alignment, we found that the β/β’ subunits across all Bcc strains tested were >98.0% identical (Figure S2). Importantly, β/β’ variants from uboS strains are not more similar than their uboN counterparts. Ubonodin therefore likely binds RNAP of all the Bcc strains tested in a similar fashion, meaning that other cellular properties are instead responsible for the observed difference in ubonodin susceptibility.
Iron-mediated inhibition of ubonodin activity reveals a specific OM transport pathway for ubonodin uptake
The bioactivity of ubonodin, and indeed any compound with an intracellular target, is a product of its ability to first access and then bind its target. A single TonB-dependent transporter (TBDT), FhuA, has been implicated in cellular uptake of the RNAP-inhibiting lasso peptides microcin J25, klebsidin, and microcin Y in enterobacteria26–28. TBDTs are a ubiquitous class of bacterial OM proteins that import essential trace nutrients from the environment (Figure 2A)29,30. The natural substrates of many TBDTs are siderophore compounds that chelate insoluble, environmental ferric iron (Fe3+) for passage into the cell31. In Burkholderia, there are no close homologs of FhuA, so we needed another approach to identify the ubonodin OM transporter(s). Another challenge was that even if a TBDT is involved, B. cepacia encodes many more TBDTs than the enterobacteria previously examined. A protein BLAST search using E. coli FhuA and 34 known and predicted TBDTs from Pseudomonas aeruginosa32, a close relative of Burkholderia occupying similar environmental niches, revealed 29 B. cepacia TBDT homologs ranging from ~13% to ~37% sequence identity to E. coli FhuA (Table S2). As such, we first had to determine if ubonodin even uses a TBDT pathway for transport.
Figure 2.
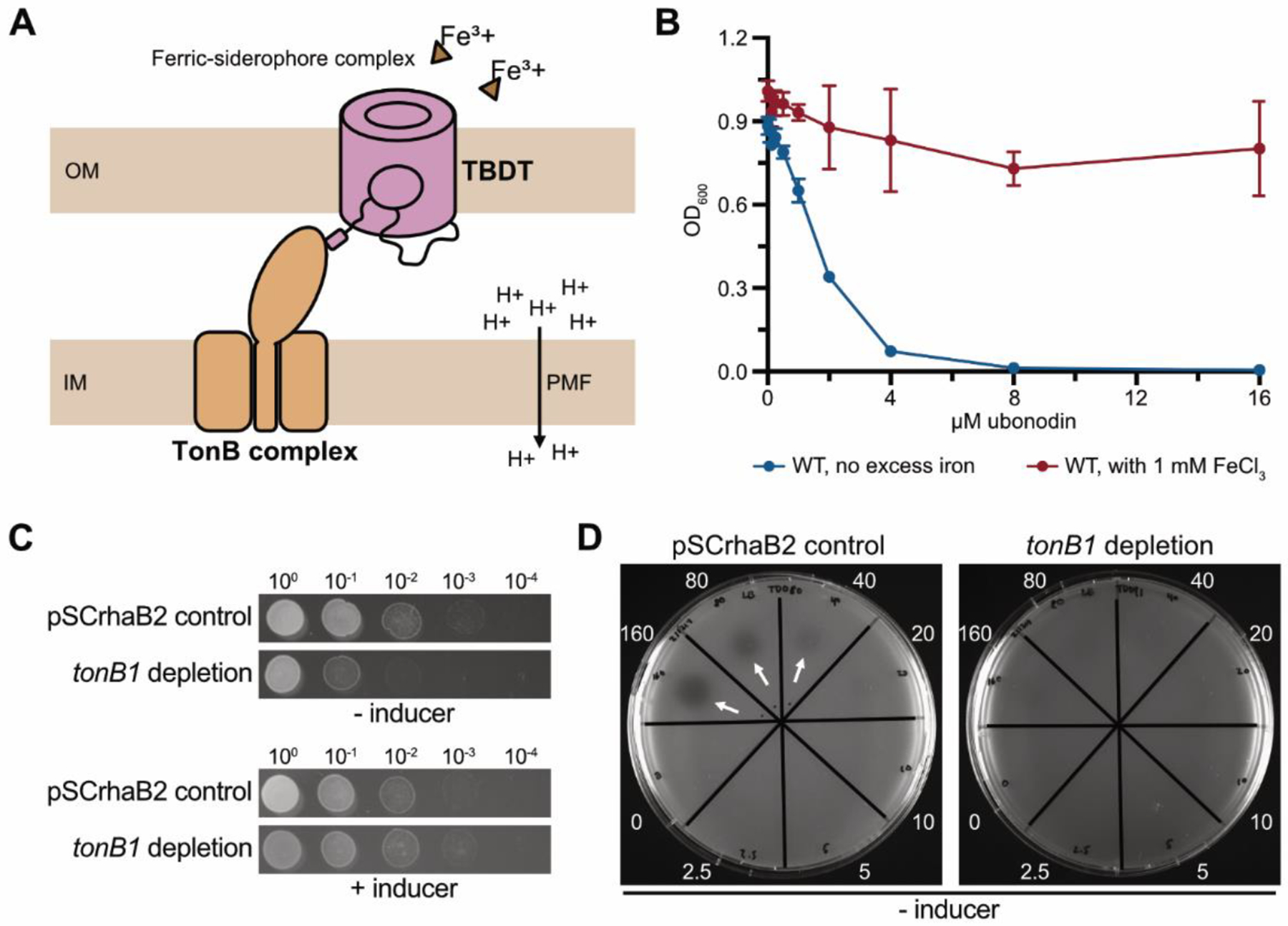
Ubonodin repurposes a specific iron transport pathway for cellular entry. (A) Cartoon describing a general TonB-dependent iron transport pathway. The TonB-dependent transporter (TBDT) localized to the bacterial outer membrane (OM) is activated by a partner TonB complex at the inner membrane (IM). TonB channels the energy from the proton motive force (PMF) to drive OM transport of siderophore-chelated ferric iron. (B) Excess iron inhibits ubonodin activity. B. cepacia was grown in plain LB or supplemented with excess iron, and the final cell density was measured after overnight growth. The mean ± standard deviation for independent biological triplicates is graphed; error bars are not apparent for points with a small standard deviation. (C) Depletion of tonB1 reduces B. cepacia cell growth but tonB1 is not required for viability. Ten-fold serial dilutions of B. cepacia propagating an empty vector (pSCrhaB2) and the tonB1 depletion mutant were spotted onto LB agar with and without the inducer (0.2% L-rhamnose). (D) The B. cepacia tonB1 depletion mutant is less susceptible to ubonodin. Cultures were plated on LB agar and 0–160 μM of ubonodin were spotted in the respective sectors (arrows: zones of inhibition).
To broadly probe whether ubonodin uses an iron receptor for passage across the B. cepacia OM, we examined how excess iron affects ubonodin activity. Bacterial iron transport systems are highly regulated to prevent toxic accumulation of intracellular iron which can lead to oxidative damage29,33–35. In iron-depleted environments, TBDT and siderophore biosynthesis genes are upregulated to maximize the ability to scavenge iron; conversely, these genes are downregulated in iron-rich environments. If ubonodin enters B. cepacia through iron-regulated TBDTs, we expected that excess iron would antagonize ubonodin activity by reducing transporter abundance. When B. cepacia was grown in media supplemented with excess iron, ubonodin indeed could no longer robustly inhibit cell growth, suggesting that ubonodin uptake occurs through one or more iron-regulated OM transporters (Figure 2B).
We next probed the specific role of TBDTs in ubonodin transport. Any number or none of the 29 predicted TBDTs in B. cepacia might be involved in ubonodin import, but all should need TonB to function. Knocking out TonB would avoid redundancy of the TBDTs. As the identity of TonB in B. cepacia is unknown, we first searched for homologs to known TonB sequences from B. cenocepacia K56–236 and B. mallei ATCC 2334437. Protein BLAST search yielded 4 hits with the top hit encoded by the B. cepacia gene GGFLHMPP_02544 (tonB1) (Table S3). When we attempted to knock out tonB1 using one-step allelic replacement38, we could only isolate single-crossover mutants that retained the wild-type allele, likely because TonB is important for Burkholderia cellular fitness36,37,39. Instead, we constructed a B. cepacia tonB1 depletion mutant in which the chromosomal copy of tonB1 is deleted while rhamnose-inducible extrachromosomal copies of tonB1 are provided on a replicative plasmid40. As expected, when the inducer was withheld to deplete tonB1, the mutant was viable but grew poorly compared to B. cepacia WT (Figure 2C). Growth was restored to near-WT levels either in the presence of L-rhamnose or when the cultures were supplemented with ferrous iron (Fe2+) to counteract iron starvation due to the defect in iron acquisition, as previously demonstrated36,37 (Figure S3). With this mutant in hand, we found that tonB1 depletion led to ubonodin resistance (Figures 2D and S4). By contrast, deletion of each of the other 3 TonB homologs, which are non-essential (Figure S5), did not impact ubonodin susceptibility (Table S4). Taken together, these findings show that ubonodin uses one specific TonB-dependent pathway to cross the B. cepacia OM.
Comparative genomics identifies siderophore receptors unique to ubonodin-susceptible Bcc strains
The next challenge was to identify the exact TBDT(s) that TonB1 presumably activates to enable ubonodin transport. All Bcc strains encode multiple TBDTs but the set of TBDTs found in each strain is variable35. Reasoning that uboS Bcc strains encode a distinct set of TBDTs from uboN Bcc strains, we developed a comparative genomics approach to predict the TBDTs that are unique to uboS strains (Figure 3A). The absence of these TBDTs in uboN strains may explain why those strains are naturally non-susceptible. Our expanded panel of Bcc strains tested for ubonodin susceptibility provided a dataset sufficient for comparison. First, we built a protein BLAST database for each Bcc strain and queried the 29 predicted B. cepacia TBDTs against each database. For each TBDT, we calculated the percent similarity normalized by query coverage between the TBDT and its highest-scoring alignment across all the Bcc strains tested. We then manually examined the heatmap summarizing the normalized BLAST scores for B. cepacia TBDTs that show high sequence conservation only or predominantly in the other uboS strains.
Figure 3.
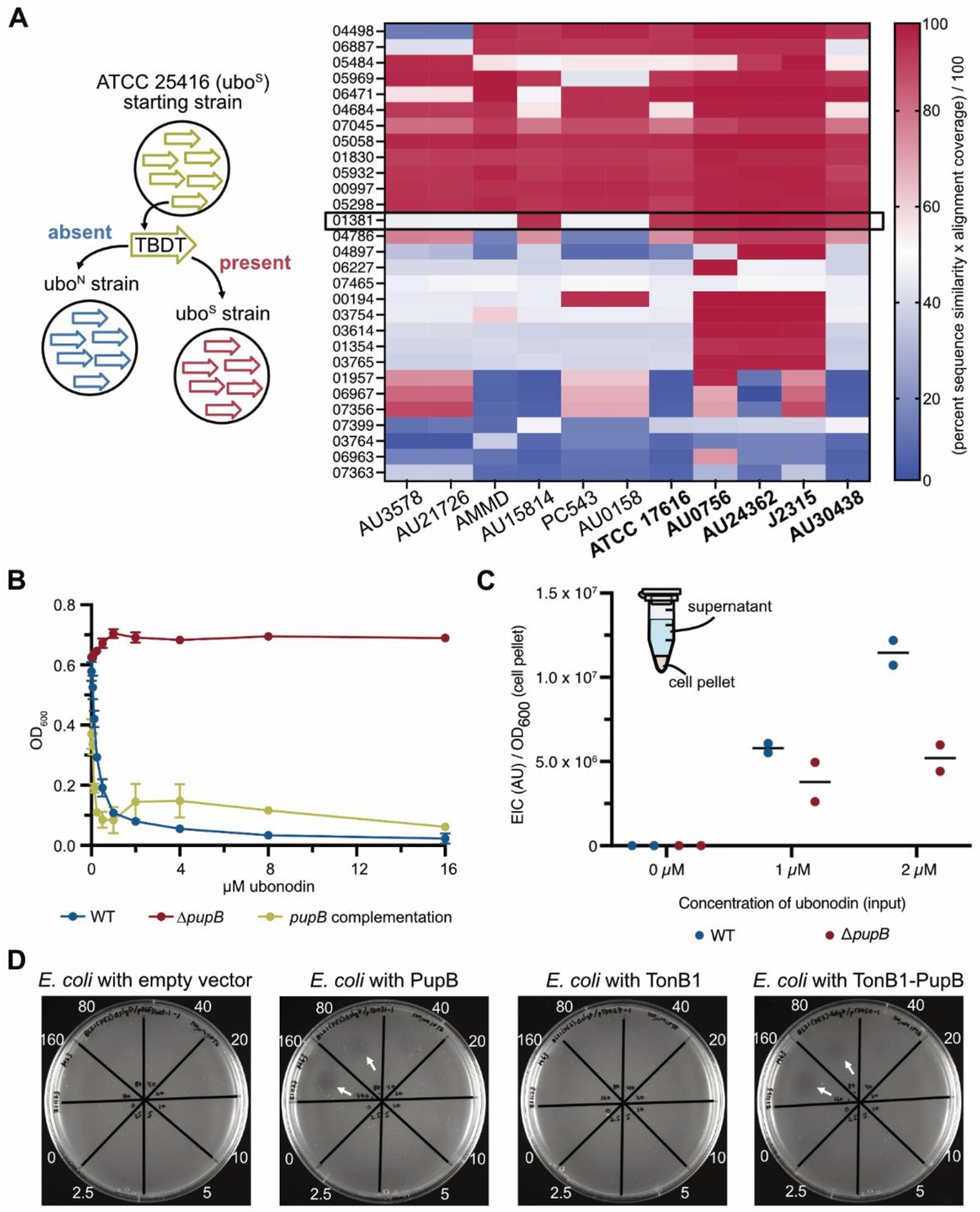
Phenotype-guided comparative genomics identifies PupB as the Burkholderia outer membrane receptor for ubonodin. (A) Comparative genomics revealed TBDT genes which are conserved in subsets of Bcc strains tested for ubonodin susceptibility. Left: Protein BLAST searches for the closest homolog of each putative B. cepacia (starting strain) TBDT in each uboS and uboN Bcc strain. Right: For each TBDT search, the percent sequence similarity of the top hit normalized to the alignment coverage was calculated. Rows of the heat map represent the 29 TBDTs assessed and columns represent the 11 Bcc strains besides B. cepacia tested for ubonodin susceptibility. The blue-to-red spectrum indicates low-to-high protein sequence conservation. Only one hit encoded by the gene GGFLHMPP_01381 was predominately conserved in uboS (bolded) but not uboN (non-bolded) strains. (B) PupB is required for ubonodin activity. Diluted cultures in cation-adjusted Mueller-Hinton II broth were exposed to ubonodin and endpoint OD600 was measured after overnight growth. Complementation of pupB was achieved by providing pupB on the pSCrhaB2 plasmid with 0.0002% L-rhamnose induction. Plots show the mean ± standard deviation for independent biological triplicates; error bars are not apparent for points with a small standard deviation. (C) Loss of PupB reduces ubonodin uptake. LC-MS analysis of cell lysates prepared from sub-MIC (minimum inhibitory concentration) ubonodin-treated B. cepacia WT and ΔpupB endpoint cultures measured the amount of ubonodin internalized by the cells. The sum of the extracted ion count (EIC) for various ubonodin species observed was normalized to the final cell density. Non-specific adsorption to the cell surface might account for some background ubonodin signal. Two biological replicates were independently measured. (D) OM transport determines ubonodin susceptibility. Growth inhibition (arrows) was observed for spot-on-lawn M63 plates of E. coli BL21(DE3) ΔslyD expressing B. cepacia PupB. Protein overexpression was achieved by the addition of 100 μM IPTG to the soft agar.
B. cepacia PupB is the ubonodin OM transporter
One B. cepacia TBDT homolog encoded by the gene GGFLHMPP_01381 (pupB) stood out because it was predicted to be highly conserved in all uboS strains and only one uboN strain (Figure 3A). PupB is named for its resemblance to TBDTs that transport pseudobactin-type siderophores in closely-related Pseudomonas41. Overall, PupB satisfies the conservation pattern expected for an ubonodin transporter that would define the spectrum of activity. No other TBDT homolog showed a similar conservation pattern. To test if PupB is involved in ubonodin transport, we deleted pupB in B. cepacia and found that the mutant was completely resistant to ubonodin, as expected with loss of cellular uptake (Figure 3B). Ubonodin susceptibility was restored when pupB was provided back in trans on a plasmid. These results strongly implicate PupB in ubonodin OM import. We also carried out cellular uptake assays comparing B. cepacia WT to the ΔpupB mutant. B. cepacia was incubated with ubonodin and intracellular levels of ubonodin were measured by LC-MS analysis of cell lysates, normalized to the endpoint cell density for each sample. In summary, we found that ubonodin accumulated to higher levels in WT cells and cellular uptake of ubonodin was reduced in the absence of PupB (Figure 3C). Our genetic and biochemical results conclusively demonstrate that ubonodin uses PupB as an OM receptor for initial cellular entry.
Next, we wondered if PupB-mediated transport is not only necessary but also sufficient for ubonodin bioactivity provided that RNAP is conserved. We previously reported that while ubonodin inhibits E. coli RNAP activity in vitro, it was not bioactive against E. coli6. We reasoned that the lack of bioactivity was due to the absence of cellular uptake and hypothesized that we could sensitize E. coli to ubonodin by heterologously expressing B. cepacia PupB. Indeed, when PupB was overexpressed in E. coli, the strain became susceptible to ubonodin at concentrations comparable to the levels of susceptibility observed for Burkholderia strains (Figure 3D). Interestingly, B. cepacia TonB1 was not needed to achieve susceptibility, as expression of PupB alone made E. coli susceptible to ubonodin and co-expression with TonB1 did not further reduce the MIC. These results suggest that E. coli TonB can efficiently bind B. cepacia PupB to relay the energy needed for ubonodin transport. OM transport is therefore a key determinant of bacterial susceptibility to ubonodin and PupB clearly plays a central role in transport which cannot be readily substituted by other transporters.
We also examined the PupB sequences in more detail to understand how the homologs belonging to the uboS strains are unique. Alignment of the closest PupB homologs for the 12 Bcc strains revealed a distinct N-terminal motif found almost exclusively in the uboS strains (Figure S6). In addition to the β-barrel domain and the periplasmic plug domain that sterically blocks the β-barrel lumen, a subset of TBDTs harbor an additional domain at the N-terminus29. This N-terminal extension is involved in signaling through a two-component FecI/FecR system to regulate the expression of the TBDT and related transport genes42–44. As intracellular iron concentrations must be precisely controlled, this added level of regulation provides a useful feedback mechanism. Besides the N-terminal extension, the PupB hits were otherwise highly similar across all strains. That PupB homologs harbor an additional N-terminal signaling domain is especially interesting in the context of the genomic location of the pupB gene, which we will further discuss below.
RNA-sequencing of B. cepacia profiles the global cellular response to excess iron
Having identified the ubonodin OM receptor, we revisited the observation that excess iron inhibits ubonodin activity to understand if excess iron does so by repressing pupB expression, thereby impeding cellular uptake. For this purpose, we performed RNA-seq to compare the B. cepacia transcriptome between 2 growth conditions: standard LB with baseline iron and supplemented with 1 mM FeCl3 to match the conditions of the ubonodin inhibition assay. An advantage of RNA-seq is that even if excess iron does not directly impact pupB expression, we might still understand how PupB transport could be affected via possible changes in the expression of other transport pathway components, e.g., TonB1.
As expected, growth in excess iron led to the repression of 183 genes, among them iron transport and utilization genes and siderophore biosynthesis genes (Figure 4A and Table S5). Several of these iron pathway-related genes were accordingly found to be induced under low-iron growth conditions in prior transcriptomics studies45–47. By contrast, 114 genes were significantly upregulated in the presence of excess iron, with none annotated as encoding for iron transporters (Table S6). Consistent with the observed gene expression changes, we noticed that B. cepacia cultures grown in excess iron were paler than the yellow-green cultures grown in standard rich media (Figure 4A, inset). As the yellow-green color is a property of fluorescent iron-scavenging siderophores34, the color change supports that siderophore production and transport were downregulated in response to excess iron.
Figure 4.
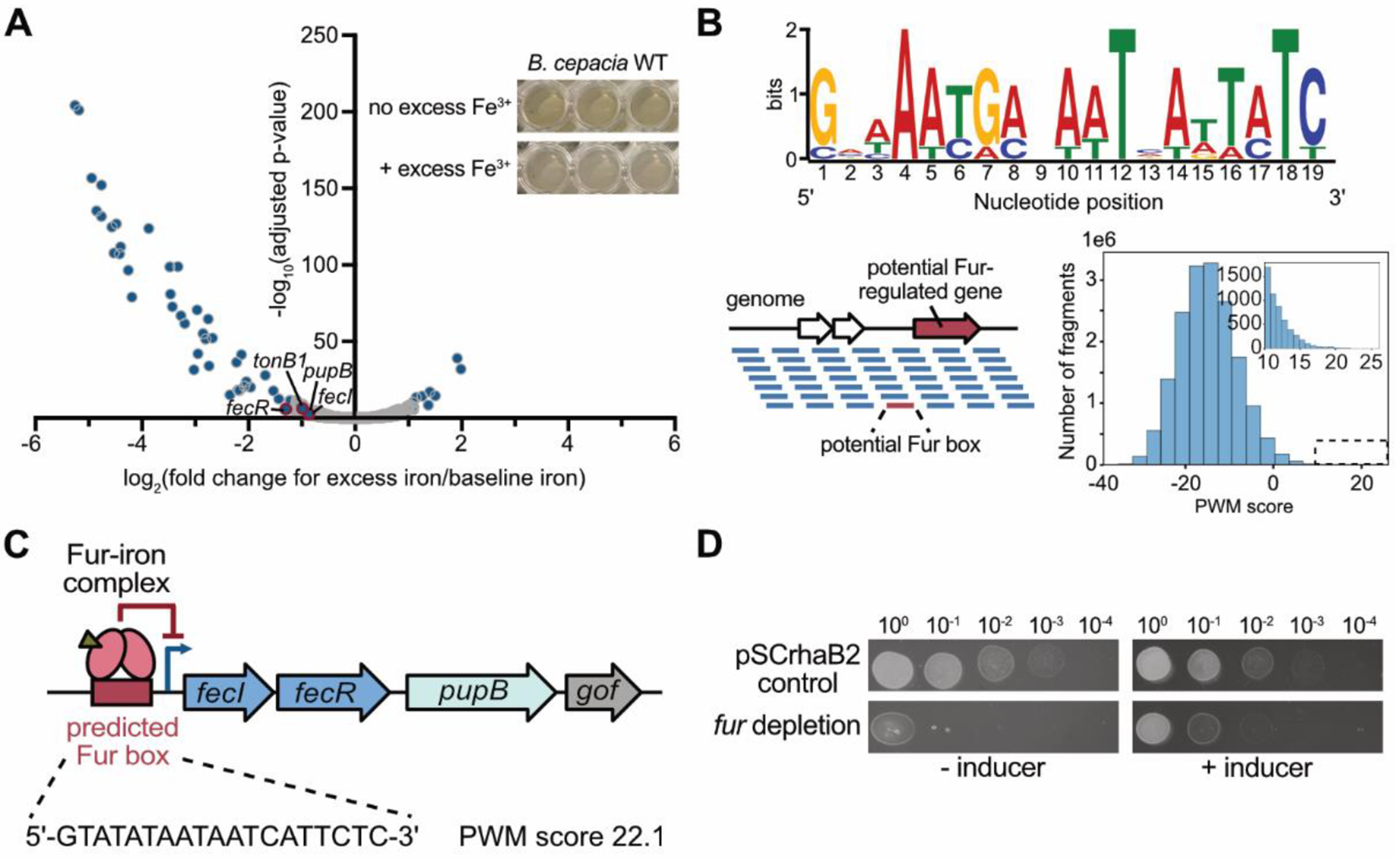
PupB is a member of the Burkholderia iron regulon. (A) Volcano plot showing iron-repressed and upregulated genes in B. cepacia ATCC 25416. Total RNA was extracted from independent biological triplicate cultures of B. cepacia WT grown in LB ± 1 mM FeCl3. Illumina RNA-seq libraries were prepared and sequenced. Genes which exhibited significant changes (adjusted p-value < 0.01) in read count between the 2 growth conditions are shown as blue dots superimposed onto all detectable genes in gray. Select genes of interest in the PupB transport pathway are denoted with a red outline and labeled. Inset: Excess iron triggers a change in the color of B. cepacia cultures grown in LB ± 1 mM FeCl3. (B) Prediction of the B. cepacia Fur regulon. Top: Seven experimentally determined P. aeruginosa Fur DNA-binding sites48–51 were aligned using the MEME (Multiple EM for Motif Elicitation) tool to generate the Fur consensus sequence52. Bottom left: A position-weight matrix (PWM) built using the Fur DNA-binding sites was used to scan the B. cepacia genome for similar sequences indicating a potential Fur box. Bottom right: Histogram of PWM scores for all scanned genome fragments with the inset showing the distribution of scores >10. (C) A potential Fur box is present upstream of the 4-gene operon predicted to encode pupB. The sequence and PWM score of the identified Fur box are denoted. The Operon-mapper web server was used for operon prediction53. The last gene in the predicted operon is a gene of unknown function (gof). (D) Fur is essential in B. cepacia. Ten-fold serial dilutions of cultures were spotted onto LB agar ± 0.2% L-rhamnose.
We examined the list of iron-repressed genes in more detail to identify any overlap with known components of the ubonodin transport pathway. Strikingly, both pupB (alternate gene designation, APZ15_10615) and tonB1 (APZ15_16310) were significantly repressed when B. cepacia was grown in the presence of excess iron. Of note, iron-mediated gene repression was not a general attribute for all TBDT genes because only 8 of the 29 predicted B. cepacia TBDT genes showed any significant expression changes (Table S7). The most highly repressed TBDT gene is associated with the biosynthetic operon of ornibactin, a major siderophore produced by Burkholderia34,54–56. That both PupB and its TonB activator are downregulated likely explains why excess iron inhibits ubonodin activity. Excess iron triggers gene expression changes that reduce the abundance of the ubonodin OM transport machinery, in turn likely reducing cellular uptake of the peptide and therefore activity.
A strong Fur DNA-binding site is predicted upstream of the pupB operon
Our finding that excess iron negatively regulates pupB and tonB1 expression points to an underlying iron-responsive signaling pathway in B. cepacia. As previously discussed, bacteria have sophisticated mechanisms to maintain cellular iron homeostasis. Central to bacterial iron homeostasis is a highly conserved transcription factor called the ferric uptake regulator (Fur) protein57,58. Fur represses the transcription of genes within its regulon when bound to Fe2+, but in the absence of the Fe2+ cofactor when iron is depleted, Fur regulon genes are derepressed59. Likewise, excess iron might repress pupB and tonB expression through Fur, but overall, there is limited information on Fur-regulated genes in Burkholderia60,61.
As an initial approach to determine if pupB and/or tonB1 gene expression is Fur-regulated, we applied bioinformatics to predict the Fur regulon in B. cepacia. We leveraged the fact that the 19-bp Fur binding site sequence (i.e., Fur box)62 is conserved and well-established to scan the B. cepacia genome for candidate Fur-regulated genes (Figure 4B and Table S8). From this analysis, a high-scoring sequence that closely matches the Fur consensus sequence was identified immediately upstream of a predicted 4-gene operon53 harboring pupB, suggesting that pupB is a member of the Fur regulon (Figure 4C and Supplementary file 2). As expected with co-regulated genes, excess iron also repressed the expression of the two genes residing upstream of pupB in the same operon, fecI (APZ15_10605) and fecR (APZ15_10610) (Figure 4A and Supplementary file 3). In contrast, no probable Fur box was predicted directly upstream of tonB1.
Because the ubonodin OM receptor PupB appears to be transcriptionally regulated by the Fur repressor, we decided to delete fur (GGFLHMPP_00610) and test if the mutant becomes hyper-susceptible to ubonodin. Our attempts to delete fur in B. cepacia were unsuccessful via one-step allelic replacement but we were able to construct a fur depletion mutant. When the inducer was withheld, growth of the fur depletion mutant was severely attenuated and only partially restored when the inducer was reintroduced, showing that Fur is essential in B. cepacia (Figure 4D). As the mutant was already very sick, we could not clearly determine if it is hypersensitive to ubonodin. Additional studies are needed to pinpoint the role of Fur in regulating pupB expression.
The putative ABC transporter YddA is the ubonodin IM transporter
The ability to sensitize otherwise resistant E. coli to ubonodin by enabling OM uptake without interfering with IM uptake suggests that there is an E. coli IM transporter that can substitute for the one found natively in B. cepacia. The E. coli polytopic IM protein SbmA is required for cellular uptake of several antimicrobial peptides including the RNAP-inhibiting lasso peptides microcin J25 and citrocin63,64. Thus, we wondered if there exists a homolog of SbmA in B. cepacia and whether such a homolog is involved in ubonodin transport. Through protein BLAST search, we found that the closest SbmA homolog in B. cepacia is a predicted ATP-binding cassette (ABC) transporter encoded by the gene GGFLHMPP_00373 (yddA). While the YddA protein sequence is only ~25% identical to that of E. coli SbmA and YddA contains a cytoplasmic nucleotide binding domain not present in SbmA, YddA is highly conserved (>80% sequence identity) across all the Bcc strains tested for ubonodin susceptibility (Figure 5A). E. coli encodes an ABC transporter also called YddA65, but it is only ~32% identical to B. cepacia YddA. Consistent with YddA acting as an ubonodin IM transporter, the PSORTb bacterial protein subcellular localization prediction tool66 localized YddA to the IM and the Phobius topology prediction tool67,68 predicted 6 transmembrane helices. When we deleted yddA in B. cepacia, the bacterium was no longer susceptible to ubonodin like how E. coli SbmA loss-of-function mutants are resistant to microcin J25 and citrocin (Figure 5B)63,64. These results strongly implicate B. cepacia YddA in ubonodin IM transport.
Figure 5.
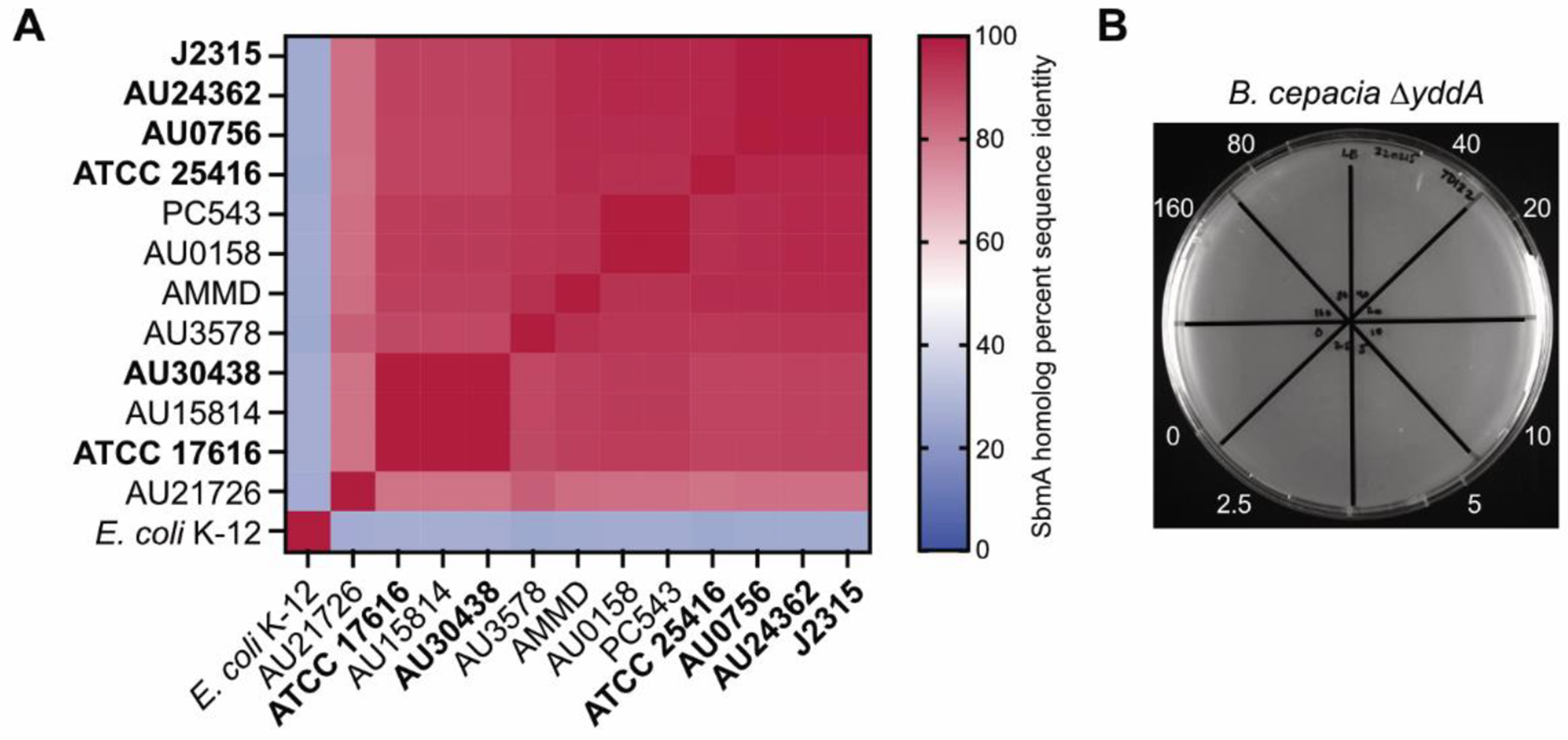
Ubonodin requires B. cepacia YddA for bioactivity. (A) Conservation of the inner membrane protein SbmA across the Bcc strains tested for ubonodin susceptibility. The closest homolog in strain ATCC 25416 to E. coli SbmA was identified using protein BLAST. This homolog, B. cepacia YddA, was used to identify the closest homolog in each of the other Bcc strains. Heat map shows percent sequence identity from Clustal Omega alignment of the SbmA homologs. A darker red shade represents more identical SbmA sequences for a pair of strains along the x- and y-axes. Ubonodin-susceptible strains are bolded. (B) Loss of YddA makes B. cepacia resistant to ubonodin. Spot-on-lawn LB plate showing no growth inhibition for 0–160 μM ubonodin.
CONCLUSIONS
In this work, we have developed a comparative genomics approach to dissect the membrane transport pathway of compounds with an intracellular target. Our approach predicts the genes that differentiate two phenotypically distinct groups of strains, with respect to their susceptibility to a compound, to hone in on the molecular basis for the different phenotypes. We applied this approach to uncover how the anti-Burkholderia lasso peptide ubonodin crosses the cell membranes that surround target bacteria as essential steps in exerting antimicrobial activity. By identifying the PupB OM transporter and demonstrating that it is a key determinant of ubonodin susceptibility, we have provided a molecular explanation for ubonodin’s focused spectrum of activity (Figure 6A). Our finding also provides the first example of a lasso peptide that uses a different receptor from FhuA to breach the notoriously impenetrable bacterial OM. Finally, having this more complete understanding of ubonodin’s mode of action will enable us to anticipate steps at which resistance can arise and extrapolate which other strains may be susceptible to ubonodin.
Figure 6.
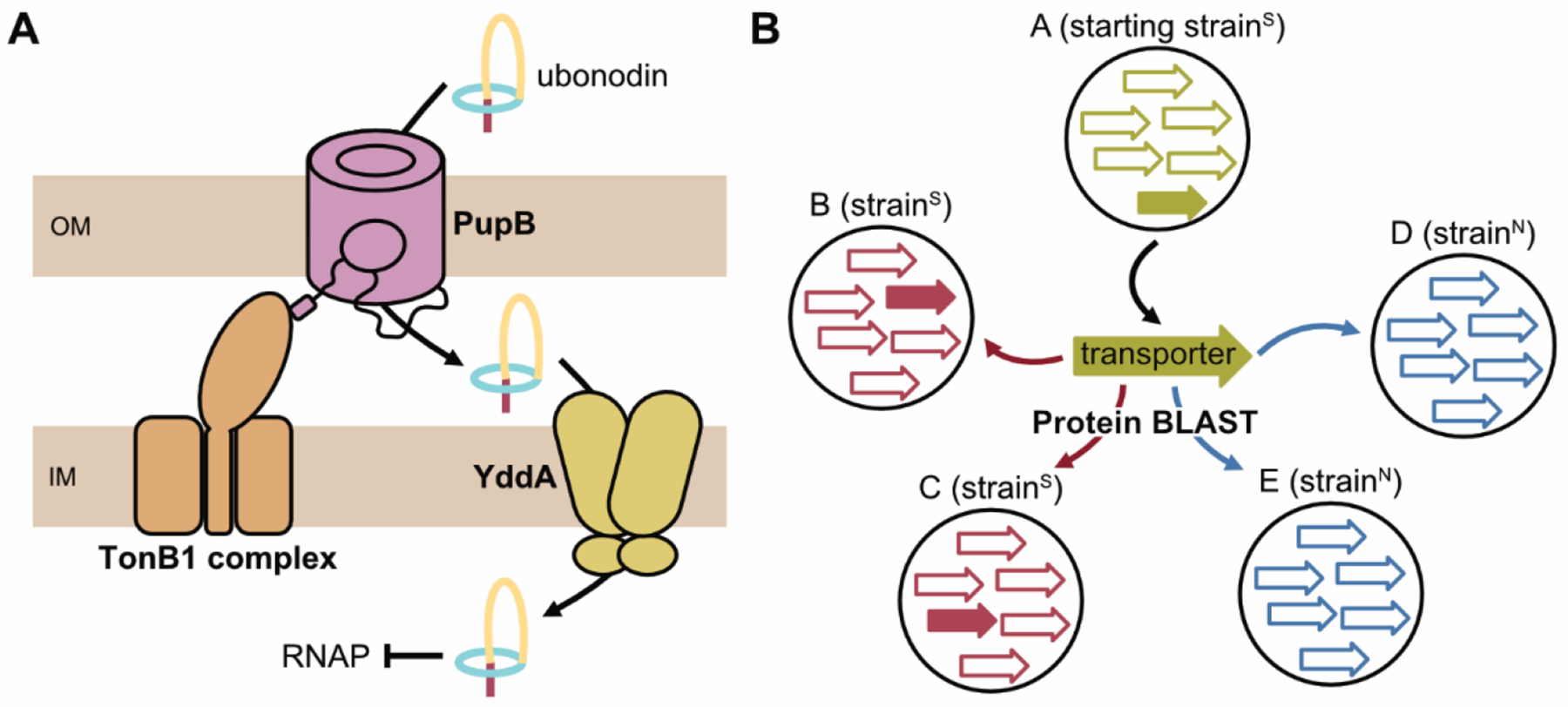
Model for the membrane transport pathway of ubonodin. (A) To cross the bacterial OM, ubonodin hijacks the siderophore receptor PupB native to certain Bcc strains. PupB contacts TonB1 through the TonB box. The IM transporter of ubonodin is the putative ABC transporter YddA. (B) Comparative genomics was key to honing in on PupB and this approach should be broadly useful for investigating the transport pathways of other antimicrobial RiPPs with a defined susceptibility profile. First, antimicrobial activity is measured against a panel of strains to sort susceptible from non-susceptible strains. All transporters from a starting susceptible strain are then individually assessed for conservation in the other susceptible strains versus lack of homology in non-susceptible strains using protein BLAST search.
Lasso peptides that act as RNAP inhibitors comprise a major lasso peptide subclass and an emerging theme is that these peptides use TBDTs localized in the OM of target bacterial cells for initial cellular entry. When the microcin J2526,69, klebsidin27, and microcin Y28 OM transporters were identified, the ability to readily isolate resistant mutants that have a mutation in the OM transporter or to predict a close homolog to E. coli FhuA simplified their identification. With ubonodin, we have observed a very low rate of bacterial resistance. Moreover, multireplicon Burkholderia bacteria generally encode multiple homologs for a protein that is otherwise found only once in other bacteria8, such as the 4 TonB homologs found in B. cepacia. This unprecedented level of redundancy created an additional challenge when we initially searched for TBDT homologs in B. cepacia, as there were 29 predicted TBDTs and the closest homolog was only ~37% identical to E. coli FhuA. Without a clear candidate to assess, knocking out each TBDT one-by-one would be prohibitively time-consuming because genetic tools to manipulate Burkholderia are limited38,70–73. The comparative genomics strategy we developed bypassed these limitations, enabling us to rapidly prioritize a single B. cepacia TBDT for further genetic studies to confirm its role in ubonodin transport. The PupB OM receptor eventually linked to ubonodin transport is only ~25% identical to E. coli FhuA and would otherwise have not been an obvious initial candidate. We anticipate that our comparative genomics pipeline will be broadly useful in deciphering the transport pathways of other lasso peptides and RiPPs with a similarly focused spectrum of activity (Figure 6B). We do note that a limitation of this strategy is that it cannot resolve whether strains encoding a susceptibility-defining gene actually express those genes, which might explain why B. multivorans AU15814 encodes pupB but is uboN. For such exceptions, comparative transcriptomics on a panel of differentially-susceptible strains might be more appropriate.
Another key insight that associated PupB with ubonodin transport was our finding that excess iron inhibits ubonodin activity. RNA-seq profiling of B. cepacia indicated that excess iron represses pupB expression; fewer transporters would mean reduced cellular uptake of ubonodin and therefore bioactivity. Initial bioinformatic analysis suggested a signaling pathway involving the iron-sensing transcriptional repressor Fur. We had hoped to test that Fur directly controls pupB expression by deleting the fur gene in B. cepacia and showing that the mutant is hypersensitive to ubonodin, but we found that fur is essential. Although iron regulation in Burkholderia has been an area of significant interest, the specific role of Burkholderia Fur is poorly understood. A fur deletion mutant of B. multivorans has been reported, but this mutant has severe growth defects60,74. While we could not delete fur, this line of investigation led us to another interesting observation. The operon that is predicted to contain pupB and to be under Fur regulation also encodes FecI and FecR, proteins that resemble an extracytoplasmic σ factor and an IM σ regulator pair. FecIR-type protein pairs cooperate with a special class of TBDTs that are structurally distinct from conventional TBDTs42–44. PupB harbors the N-terminal extension domain found uniquely in this TBDT subclass unlike the FhuA receptors for microcin J25, klebsidin, and microcin Y, which all lack this extra domain. An intriguing question for future studies is whether PupB and FecIR form an iron-regulated signaling cascade in the native cellular context.
In addition to transcriptional regulation, as a TBDT PupB is also regulated through direct interaction with its TonB partner. Although B. cepacia encodes 4 TonB homologs, only TonB1 is required for ubonodin activity. Either the PupB-TonB1 interaction is highly specific or the other TonBs are not expressed under the experimental conditions, meaning that the ΔtonB2, ΔtonB3, and ΔtonB4 mutants would effectively mimic WT. Given that expressing PupB alone in E. coli is sufficient for ubonodin susceptibility, there may be some degree of promiscuity in the interaction of PupB with TonB. E. coli has one TonB and it is only ~28% identical to B. cepacia TonB1. Unless PupB is independently active in E. coli, it likely cooperates with E. coli TonB; expressing PupB in an E. coli ΔtonB background may provide an answer. By contrast, the interaction of PupB with ubonodin appears quite specific as no other B. cepacia or E. coli TBDTs can apparently stand in for its ubonodin transport function. Mapping the PupB-ubonodin binding interface will expose how the transporter engages its substrate, a subject for future studies.
While not the main focus of this study, we additionally show that ubonodin’s ability to cross the IM using the putative ABC transporter YddA is also required for bioactivity as its intended cellular target is cytoplasmic. The native biological function of YddA in B. cepacia is not known but YddA is highly conserved in the Bcc strains examined herein. As both uboN and uboS Bcc strains encode near-identical YddA homologs, they all appear to be inherently capable of ubonodin IM transport. IM translocation is necessary but clearly not sufficient for ubonodin bioactivity, as further evidenced by the requirement for PupB to observe activity against E. coli. Similar to our reconstitution of PupB function in E. coli, other groups have shown that expressing the native microcin J2575 and klebsidin27 OM receptors in resistant strains without providing their native IM transporters is sufficient to achieve bioactivity. Thus, IM transporters capable of recognizing ubonodin and lasso peptides in general may be highly prevalent. Although SbmA homologs have been associated with the transport of lasso peptides and other antimicrobial peptides, how they interact with their cargo is poorly understood. In the case of E. coli SbmA-mediated transport of microcin J25, it is thought that SbmA makes specific contacts with the peptide and is powered by a proton gradient76. Unlike SbmA, which possesses the transmembrane but not nucleotide-binding domain of ABC transporters76,77, B. cepacia YddA is predicted to have a C-terminal ATP-binding domain. As such, YddA may use a transport mechanism distinct from SbmA, another intriguing question for future studies.
To date, many RiPPs with promising antimicrobial activity have been discovered but for most the mode of action is still unknown, hampering efforts to develop them as drugs. Before the genomic era, new RiPPs were typically found through bioactivity-guided studies, starting from isolation of a compound with antimicrobial activity to elucidation of its structure and target. Genomics-guided discovery of RiPPs has now overtaken activity-focused approaches, but our understanding of the function of these new compounds has not kept pace in part due to the lack of tools. The pipeline we have developed – from phenotype/drug susceptibility to computational genomics to target discovery – is generalizable to other RiPPs for which mode of action data are sorely needed.
METHODS
Detailed materials and methods are provided in Supplementary file 1.
Strain and plasmid construction.
The list of strains, plasmids, and oligonucleotides used in this study can be found in Tables S9, S10, and S11, respectively.
Spot-on-lawn assay.
A standard spot-on-lawn assay to assess bacterial susceptibility to ubonodin was carried out as previously reported6 with a few changes denoted here. Overnight cultures of test strains were diluted 1:100 into fresh LB with antibiotics as needed the following day and grown at the appropriate temperature with shaking to an OD600 of ~0.4–0.6. Once the cultures reached mid-exponential phase, the volume equivalent to 108 CFUs (assuming OD600 = 1 contains 109 CFUs mL−1) was added to 10 mL of melted LB or M63 soft agar (0.65% agar). The cell and agar mixture was poured onto 10 mL of LB or M63 base agar (1.5% agar, M63 base agar lacked amino acids) and the plate was left to dry in the biosafety cabinet with the lid slightly ajar. When needed, IPTG or L-rhamnose was also added to the soft agar to induce protein expression. M63 was used for the Bcc clinical isolates and E. coli BL21(DE3) ΔslyD overexpression strains, whereas the ATCC 25416 strains were assessed on LB. The ubonodin MIC of B. cepacia WT is similar on M63 and LB. Two-fold serial dilutions of ubonodin (0–160 μM) were prepared using sterile ultrapure water and 10 μL of each dilution was spotted onto the dried lawn of cells in their respective sector. The spots were left to dry in the biosafety cabinet once again with the lid slightly ajar. The plates were incubated ~15 h at the appropriate temperature and imaged the next day using the Bio Rad ChemiDoc XRS Gel Imaging System under epi white illumination and the Quantity One 4.6.6 imaging software. Images were processed using FIJI. Spot-on-lawn assays were performed with at least 2 biological replicates (independent cultures) for each tested ATCC 25416 strain and the Bcc clinical isolates AU0158 and AU15814.
Protein BLAST-based comparative genomics.
Protein BLAST-based comparative genomics was performed using the standalone NCBI BLAST+ suite installed on a local machine78. For each strain, genome annotation files in GenBank format were downloaded from NCBI. The GenBank files were converted to FASTA format using the custom Python script gbk_to_fasta.py adapted from source code available through the University of Warwick at the following web link: https://warwick.ac.uk/fac/sci/moac/people/students/peter_cock/python/genbank2fasta/. Then, using the makeblastdb program within the BLAST+ suite, a protein BLAST database was built from each of the FASTA files.
The input sequences for the next step of analysis were retrieved by first predicting TBDT homologs in B. cepacia ATCC 25416. Using the E. coli FhuA sequence and 34 known and predicted P. aeruginosa TBDT sequences32 as queries, protein BLAST search was performed against the ATCC 25416 protein BLAST database to identify 29 B. cepacia TBDT homologs. A high e-value cutoff of 1000 was used in this search to maximize retrieval of any potential homologs. Using the BLAST+ suite blastp program, the 29 predicted TBDT homologs encoded by B. cepacia ATCC 25416 then served as queries for protein BLAST against each of the custom-built Bcc protein BLAST database generated in the first step. This process identifies the closest homolog to each predicted ATCC 25416 TBDT in each Bcc strain. Finally, to identify patterns of TBDT conservation across all strains, the custom blast_comparison_genes.py Python script was implemented. Briefly, this script calculates the percentage of positive-scoring matches (ppos; percent protein sequence similarity for the alignment of the query TBDT and subject TBDT hit) normalized by multiplying to the length of the alignment (qcovhsp; query coverage per high-scoring segment pair) for each query-top subject hit pair. A clustered 29 × 11 (TBDTs x BCC strains) heat map matrix was generated based on these normalized scores to visualize how conserved each predicted B. cepacia TBDT is across the other Bcc strains. This BLAST matrix analysis was adapted from a large-scale genomics study on Burkholderia strains79. GraphPad Prism version 9.3.1 was used to make all plots. All custom Python scripts used for this study were uploaded to the Link lab Github page at the following web link: https://github.com/ajlinklab/PupB.
Extraction, preparation, and sequencing of total RNA.
B. cepacia ATCC 25416 was freshly streaked out on LB agar. For each condition, three biological replicates derived from separate colonies were prepared. Single colonies were inoculated into LB ± 1 mM FeCl3 (ferric chloride stock was made fresh in water and filter-sterilized) and grown overnight at 30 °C with shaking. The next morning, the overnight cultures were diluted 1:100 into fresh LB ± 1 mM FeCl3 and grown at 30 °C with shaking to an OD600 of ~0.5–0.6. The volume of mid-exponential phase culture to achieve 5 × 108 CFUs was calculated (assuming OD600 of 1.0 yields 109 CFUs mL−1) and added to 2 volumes of RNAprotect Bacteria Reagent (QIAGEN, cat. no. 74124). The mixture was immediately vortexed for 5 sec, incubated at room temperature for 5 min, and then centrifuged at 5000 xg for 10 min at room temperature. The supernatant was decanted and the cell pellet was stored at −80 °C until RNA extraction.
Total RNA was extracted from RNAprotect-stabilized cell pellets using the RNeasy Protect Kit (QIAGEN, cat. no. 74124) according to manufacturer’s protocol. Briefly, the frozen cell pellets were thawed at room temperature and cell lysis was achieved using enzymatic lysis with 200 μL of 1X TE lysis buffer (30 mM Tris-HCl, 1 mM EDTA, pH 8.0) containing 1 mg mL−1 of lysozyme and supplemented with 10 μL of Proteinase K (QIAGEN, cat. no. 19131). The mixture was incubated on a shaker at room temperature for 30 min with intermittent vortexing, 700 μL of Buffer RLT was added, the mixture was vortexed, and 500 μL of 100% ethanol was added and mixed. Total RNA was then purified from the bacterial cell lysate using the RNeasy Mini Kit protocol and eluted with 2 × 35–40 μL of nuclease-free water. The miniprepped RNA was further treated with in-solution rigorous TURBO DNase treatment using the TURBO DNA-free™ Kit (Thermo Fisher Scientific, cat. no. AM1907) to remove contaminant genomic DNA according to manufacturer’s protocol. The concentration and integrity of purified RNA were estimated using a NanoDrop ND-1000 Spectrophotometer and a 2% ethidium bromide non-denaturing agarose gel observing for intact rRNA bands, respectively. RNA samples were submitted to the Princeton Genomics Core Facility for library construction, quality control check, and sequencing on an Illumina NovaSeq 6000 Illumina sequencing platform. The samples were depleted of ribosomal RNA and checked on a Bioanalyzer prior to sequencing.
Analysis of RNA-Seq results was carried out in collaboration with research-computing staff at the Princeton University Lewis-Sigler Institute for Integrative Genomics using Galaxy. The B. cepacia ATCC 25416 genome assembly (ASM141149v1; GenBank assembly accession: GCA_001411495.1) was downloaded from NCBI in fna and gtf format; the gtf file was edited to avoid tool issues on Galaxy. Forward and reverse sequences were uploaded to and demultiplexed on the Princeton HTSeq database system and transferred to the Princeton Galaxy instance. Read quality was assessed using FastQC (Galaxy Version 0.72), Read Distribution (Galaxy Version 2.6.4.1), BAM/SAM Mapping Stats (Galaxy Version 2.6.4), IdxStats (Galaxy Version 2.0.2), and Gene Body Coverage (Galaxy Version 2.6.4.3). Top over-represented sequences and ribosomal RNA content was assessed using in-house Galaxy workflows. Quality control stats were viewed using a MultiQC (Galaxy Version 1.8+galaxy0) report. Sequences were aligned using Burrows-Wheeler Alignment (BWA; Galaxy Version 0.7.17.4)80. Reads aligning to genes according to NCBI’s gene annotations were counted using featureCounts (Galaxy Version 1.6.4+galaxy1). Iron-treated and untreated samples were then compared using DESeq2 (Galaxy Version 2.11.40.6+galaxy1)81, which generated QC plots, rLog normalized counts, and differential expression data, including adjusted p-values to account for multiple testing with the Benjamini-Hochberg procedure which controls false discovery rate (FDR).
Accession Codes.
RNA-seq data (accession number PRJNA813900) can be found in the NCBI BioProject database.
Supplementary Material
Supplementary file 1 contains additional materials and methods; Figures S1–6 summarizing susceptibility data and bacterial growth assays; Tables S1–11 summarizing MIC data, RNA-seq results, and additional bioinformatics results; and supplementary references.
Supplementary file 2 summarizes the predicted Fur DNA-binding sites in the B. cepacia ATCC 25416 genome.
Supplementary file 3 summarizes the Operon-mapper predicted operons in B. cepacia ATCC 25416.
SYNOPSIS.
Members of the Burkholderia cepacia complex encode numerous outer membrane transporters for nutrient uptake. A phenotype-driven comparative genomics approach was used to discover that the lasso peptide antibiotic ubonodin co-opts the nutrient transporter PupB to enter susceptible Burkholderia.
ACKNOWLEDGEMENTS
We thank W. Wang and his staff at the Princeton Genomics Core Facility for advice on RNA isolation and for construction and sequencing of the RNA-seq libraries. We also thank M. Thomas (University of Sheffield) for sharing his pSHAFT-series, p34E-Km, and p34E-TpTer plasmids and both him and S. Cardona (University of Manitoba) for useful advice in making Burkholderia mutants. We also thank M. Seyedsayamdost for sharing the DH5α/pRK2013 helper strain for triparental mating and Z. Gitai for the S17-1(λpir) and CC118(λpir) strains. Finally, we thank J. LiPuma for sharing the Bcc clinical strains to test ubonodin susceptibility. Funding for this work was provided by the National Institutes of Health grant GM107036 to A. Link.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet.
COMPETING INTERESTS
A. Link has applied for a patent covering the antimicrobial activity of ubonodin.
REFERENCES
- (1).Arnison PG; Bibb MJ; Bierbaum G; Bowers AA; Bugni TS; Bulaj G; Camarero JA; Campopiano DJ; Challis GL; Clardy J; et al. Ribosomally Synthesized and Post-Translationally Modified Peptide Natural Products: Overview and Recommendations for a Universal Nomenclature. Nat Prod Rep 2012, 30 (1), 108–160. 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Montalbán-López M; Scott TA; Ramesh S; Rahman IR; van Heel AJ; Viel JH; Bandarian V; Dittmann E; Genilloud O; Goto Y; et al. New Developments in RiPP Discovery, Enzymology and Engineering. Nat Prod Rep 2020, 38 (1), 130–239. 10.1039/d0np00027b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Li Y; Rebuffat S The Manifold Roles of Microbial Ribosomal Peptide–Based Natural Products in Physiology and Ecology. J Biol Chem 2020, 295 (1), 34–54. 10.1074/jbc.rev119.006545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Cao L; Do T; Link AJ Mechanisms of Action of Ribosomally Synthesized and Posttranslationally Modified Peptides (RiPPs). J Ind Microbiol Biot 2021, 48 (3–4), kuab005. 10.1093/jimb/kuab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Kodani S; Unno K How to Harness Biosynthetic Gene Clusters of Lasso Peptides. J Ind Microbiol Biot 2020, 47 (9–10), 703–714. 10.1007/s10295-020-02292-6. [DOI] [PubMed] [Google Scholar]
- (6).Cheung-Lee WL; Parry ME; Zong C; Cartagena AJ; Darst SA; Connell ND; Russo R; Link AJ Discovery of Ubonodin, an Antimicrobial Lasso Peptide Active against Members of the Burkholderia Cepacia Complex. Chembiochem 2020, 21 (9), 1335–1340. 10.1002/cbic.201900707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Leitão JH; Feliciano JR; Sousa SA; Pita T; Guerreiro SI Burkholderia Cepacia Complex Infections Among Cystic Fibrosis Patients: Perspectives and Challenges. In Progress in Understanding Cystic Fibrosis; 2017. 10.5772/67712. [DOI] [Google Scholar]
- (8).Mahenthiralingam E; Urban TA; Goldberg JB The Multifarious, Multireplicon Burkholderia Cepacia Complex. Nat Rev Microbiol 2005, 3 (2), 144–156. 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- (9).Chiarini L; Bevivino A; Dalmastri C; Tabacchioni S; Visca P Burkholderia Cepacia Complex Species: Health Hazards and Biotechnological Potential. Trends Microbiol 2006, 14 (6), 277–286. 10.1016/j.tim.2006.04.006. [DOI] [PubMed] [Google Scholar]
- (10).Hegemann JD; Zimmermann M; Xie X; Marahiel MA Lasso Peptides: An Intriguing Class of Bacterial Natural Products. Accounts Chem Res 2015, 48 (7), 1909–1919. 10.1021/acs.accounts.5b00156. [DOI] [PubMed] [Google Scholar]
- (11).Maksimov MO; Pan SJ; Link AJ Lasso Peptides : Structure, Function, Biosynthesis, and Engineering. Nat Prod Rep 2012, 29 (9), 996–1006. 10.1039/c2np20070h. [DOI] [PubMed] [Google Scholar]
- (12).Mathavan I; Beis K The Role of Bacterial Membrane Proteins in the Internalization of Microcin MccJ25 and MccB17. Biochem Soc T 2012, 40 (6), 1539–1543. 10.1042/bst20120176. [DOI] [PubMed] [Google Scholar]
- (13).Vandamme P; Peeters C Time to Revisit Polyphasic Taxonomy. Antonie Van Leeuwenhoek 2014, 106 (1), 57–65. 10.1007/s10482-014-0148-x. [DOI] [PubMed] [Google Scholar]
- (14).Depoorter E; Canck ED; Peeters C; Wieme AD; Cnockaert M; Zlosnik JEA; LiPuma JJ; Coenye T; Vandamme P Burkholderia Cepacia Complex Taxon K: Where to Split? Front Microbiol 2020, 11, 1594. 10.3389/fmicb.2020.01594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Winsor GL; Khaira B; Rossum TV; Lo R; Whiteside MD; Brinkman FSL The Burkholderia Genome Database: Facilitating Flexible Queries and Comparative Analyses. Bioinformatics 2008, 24 (23), 2803–2804. 10.1093/bioinformatics/btn524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Mahenthiralingam E; Bischof J; Byrne SK; Radomski C; Davies JE; Av-Gay Y; Vandamme P DNA-Based Diagnostic Approaches for Identification of Burkholderia Cepacia Complex, Burkholderia Vietnamiensis, Burkholderia Multivorans, Burkholderia Stabilis, and Burkholderia Cepacia Genomovars I and III. J Clin Microbiol 2000, 38 (9), 3165–3173. 10.1128/jcm.38.9.3165-3173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Coenye T; Vandamme P; Govan JRW; LiPuma JJ Taxonomy and Identification of the Burkholderia Cepacia Complex. J Clin Microbiol 2001, 39 (10), 3427–3436. 10.1128/jcm.39.10.3427-3436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).LiPuma JJ The Changing Microbial Epidemiology in Cystic Fibrosis. Clin Microbiol Rev 2010, 23 (2), 299–323. 10.1128/cmr.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zlosnik JEA; Henry DA; Hird TJ; Hickman R; Campbell M; Cabrera A; Chiavegatti GL; Chilvers MA; Sadarangani M Epidemiology of Burkholderia Infections in People with Cystic Fibrosis in Canada between 2000 and 2017. Ann Am Thorac Soc 2020, 17 (12), 1549–1557. 10.1513/annalsats.201906-443oc. [DOI] [PubMed] [Google Scholar]
- (20).Salomón RA; Farías RN Microcin 25, a Novel Antimicrobial Peptide Produced by Escherichia Coli. J Bacteriol 1992, 174 (22), 7428–7435. 10.1128/jb.174.22.7428-7435.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Delgado MA; Rintoul MR; Farías RN; Salomón RA Escherichia Coli RNA Polymerase Is the Target of the Cyclopeptide Antibiotic Microcin J25. J Bacteriol 2001, 183 (15), 4543–4550. 10.1128/jb.183.15.4543-4550.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Yuzenkova J; Delgado M; Nechaev S; Savalia D; Epshtein V; Artsimovitch I; Mooney RA; Landick R; Farias RN; Salomon R; et al. Mutations of Bacterial RNA Polymerase Leading to Resistance to Microcin J25. J Biol Chem 2002, 277 (52), 50867–50875. 10.1074/jbc.m209425200. [DOI] [PubMed] [Google Scholar]
- (23).Knappe TA; Linne U; Zirah S; Rebuffat S; Xie X; Marahiel MA Isolation and Structural Characterization of Capistruin, a Lasso Peptide Predicted from the Genome Sequence of Burkholderia Thailandensis E264. J Am Chem Soc 2008, 130 (34), 11446–11454. 10.1021/ja802966g. [DOI] [PubMed] [Google Scholar]
- (24).Kuznedelov K; Semenova E; Knappe TA; Mukhamedyarov D; Srivastava A; Chatterjee S; Ebright RH; Marahiel MA; Severinov K The Antibacterial Threaded-Lasso Peptide Capistruin Inhibits Bacterial RNA Polymerase. J Mol Biol 2011, 412 (5), 842–848. 10.1016/j.jmb.2011.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Braffman NR; Piscotta FJ; Hauver J; Campbell EA; Link AJ; Darst SA Structural Mechanism of Transcription Inhibition by Lasso Peptides Microcin J25 and Capistruin. P Natl Acad Sci Usa 2019, 116 (4), 1273–1278. 10.1073/pnas.1817352116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Salomón RA; Farías RN The FhuA Protein Is Involved in Microcin 25 Uptake. J Bacteriol 1993, 175 (23), 7741–7742. 10.1128/jb.175.23.7741-7742.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Metelev M; Arseniev A; Bushin LB; Kuznedelov K; Artamonova TO; Kondratenko R; Khodorkovskii M; Seyedsayamdost MR; Severinov K Acinetodin and Klebsidin, RNA Polymerase Targeting Lasso Peptides Produced by Human Isolates of Acinetobacter Gyllenbergii and Klebsiella Pneumoniae. Acs Chem Biol 2017, 12 (3), 814–824. 10.1021/acschembio.6b01154. [DOI] [PubMed] [Google Scholar]
- (28).Li Y; Han Y; Zeng Z; Li W; Feng S; Cao W Discovery and Bioactivity of the Novel Lasso Peptide Microcin Y. J Agr Food Chem 2021, 69 (31), 8758–8767. 10.1021/acs.jafc.1c02659. [DOI] [PubMed] [Google Scholar]
- (29).Noinaj N; Guillier M; Barnard TJ; Buchanan SK TonB-Dependent Transporters: Regulation, Structure, and Function. Nat Microbiol 2010, 64 (1), 43–60. 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Krewulak KD; Vogel HJ TonB or Not TonB: Is That the Question? Biochem Cell Biol 2011, 89 (2), 87–97. 10.1139/o10-141. [DOI] [PubMed] [Google Scholar]
- (31).Hider RC; Kong X Chemistry and Biology of Siderophores. Nat Prod Rep 2010, 27 (5), 637–657. 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- (32).Luscher A; Moynié L; Auguste PS; Bumann D; Mazza L; Pletzer D; Naismith JH; Köhler T TonB-Dependent Receptor Repertoire of Pseudomonas Aeruginosa for Uptake of Siderophore-Drug Conjugates. Antimicrob Agents Ch 2018, 62 (6), e00097–18. 10.1128/aac.00097-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Andrews SC; Robinson AK; Rodríguez-Quiñones F Bacterial Iron Homeostasis. FEMS Microbiol Rev 2003, 27 (2–3), 215–237. 10.1016/s0168-6445(03)00055-x. [DOI] [PubMed] [Google Scholar]
- (34).Thomas MS Iron Acquisition Mechanisms of the Burkholderia Cepacia Complex. Biometals 2007, 20 (3–4), 431–452. 10.1007/s10534-006-9065-4. [DOI] [PubMed] [Google Scholar]
- (35).Butt AT; Thomas MS Iron Acquisition Mechanisms and Their Role in the Virulence of Burkholderia Species. Front Cell Infect Mi 2017, 7, 460. 10.3389/fcimb.2017.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Pradenas GA; Myers JN; Torres AG Characterization of the Burkholderia Cenocepacia TonB Mutant as a Potential Live Attenuated Vaccine. Vaccines 2017, 5 (4), 33. 10.3390/vaccines5040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Mott TM; Vijayakumar S; Sbrana E; Endsley JJ; Torres AG Characterization of the Burkholderia Mallei tonB Mutant and Its Potential as a Backbone Strain for Vaccine Development. Plos Neglect Trop D 2015, 9 (6), e0003863. 10.1371/journal.pntd.0003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Shastri S; Spiewak HL; Sofoluwe A; Eidsvaag VA; Asghar AH; Pereira T; Bull EH; Butt AT; Thomas MS An Efficient System for the Generation of Marked Genetic Mutants in Members of the Genus Burkholderia. Plasmid 2017, 89, 49–56. 10.1016/j.plasmid.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Higgins S; Sanchez-Contreras M; Gualdi S; Pinto-Carbó M; Carlier A; Eberl L The Essential Genome of Burkholderia Cenocepacia H111. J Bacteriol 2017, 199 (22). 10.1128/jb.00260-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Cardona ST; Valvano MA An Expression Vector Containing a Rhamnose-Inducible Promoter Provides Tightly Regulated Gene Expression in Burkholderia Cenocepacia. Plasmid 2005, 54 (3), 219–228. 10.1016/j.plasmid.2005.03.004. [DOI] [PubMed] [Google Scholar]
- (41).Koster M; Vossenberg J; Leong J; Weisbeek PJ Identification and Characterization of the pupB Gene Encoding an Inducible Ferric-pseudobactin Receptor of Pseudomonas Putida WCS358. Mol Microbiol 1993, 8 (3), 591–601. 10.1111/j.1365-2958.1993.tb01603.x. [DOI] [PubMed] [Google Scholar]
- (42).Visca P; Leoni L; Wilson MJ; Lamont IL Iron Transport and Regulation, Cell Signalling and Genomics: Lessons from Escherichia Coli and Pseudomonas. Mol Microbiol 2002, 45 (5), 1177–1190. 10.1046/j.1365-2958.2002.03088.x. [DOI] [PubMed] [Google Scholar]
- (43).Braun V; Mahren S; Ogierman M Regulation of the FecI-Type ECF Sigma Factor by Transmembrane Signalling. Curr Opin Microbiol 2003, 6 (2), 173–180. 10.1016/s1369-5274(03)00022-5. [DOI] [PubMed] [Google Scholar]
- (44).Braun V; Mahren S Transmembrane Transcriptional Control (Surface Signalling) of the Escherichia Coli Fec Type. FEMS Microbiol Rev 2005, 29 (4), 673–684. 10.1016/j.femsre.2004.10.001. [DOI] [PubMed] [Google Scholar]
- (45).Tuanyok A; Kim HS; Nierman WC; Yu Y; Dunbar J; Moore RA; Baker P; Tom M; Ling JML; Woods DE Genome-wide Expression Analysis of Iron Regulation in Burkholderia Pseudomallei and Burkholderia Mallei Using DNA Microarrays. FEMS Microbiol Lett 2005, 252 (2), 327–335. 10.1016/j.femsle.2005.09.043. [DOI] [PubMed] [Google Scholar]
- (46).Sass AM; Schmerk C; Agnoli K; Norville PJ; Eberl L; Valvano MA; Mahenthiralingam E The Unexpected Discovery of a Novel Low-Oxygen-Activated Locus for the Anoxic Persistence of Burkholderia Cenocepacia. Isme J 2013, 7 (8), 1568–1581. 10.1038/ismej.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Tyrrell J; Whelan N; Wright C; Sá-Correia I; McClean S; Thomas M; Callaghan M Investigation of the Multifaceted Iron Acquisition Strategies of Burkholderia Cenocepacia. Biometals 2015, 28 (2), 367–380. 10.1007/s10534-015-9840-1. [DOI] [PubMed] [Google Scholar]
- (48).Dudek C-A; Jahn D PRODORIC: State-of-the-Art Database of Prokaryotic Gene Regulation. Nucleic Acids Res 2021, 50 (D1), D295–D302. 10.1093/nar/gkab1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Ochsner UA; Vasil AI; Vasil ML Role of the Ferric Uptake Regulator of Pseudomonas Aeruginosa in the Regulation of Siderophores and Exotoxin A Expression: Purification and Activity on Iron-Regulated Promoters. J Bacteriol 1995, 177 (24), 7194–7201. 10.1128/jb.177.24.7194-7201.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Hassett DJ; Howell ML; Ochsner UA; Vasil ML; Johnson Z; Dean GE An Operon Containing fumC and sodA Encoding Fumarase C and Manganese Superoxide Dismutase Is Controlled by the Ferric Uptake Regulator in Pseudomonas Aeruginosa: Fur Mutants Produce Elevated Alginate Levels. J Bacteriol 1997, 179 (5), 1452–1459. 10.1128/jb.179.5.1452-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Ochsner UA; Johnson Z; Vasil ML Genetics and Regulation of Two Distinct Haem-Uptake Systems, Phu and Has, in Pseudomonas Aeruginosa. Microbiology+ 2000, 146 (1), 185–198. 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- (52).Bailey TL; Boden M; Buske FA; Frith M; Grant CE; Clementi L; Ren J; Li WW; Noble WS MEME Suite: Tools for Motif Discovery and Searching. Nucleic Acids Res 2009, 37 (Web Server issue), W202–W208. 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Taboada B; Estrada K; Ciria R; Merino E Operon-Mapper: A Web Server for Precise Operon Identification in Bacterial and Archaeal Genomes. Bioinformatics 2018, 34 (23), 4118–4120. 10.1093/bioinformatics/bty496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Stephan H; Freund S; Beck W; Jung G; Meyer J-M; Winkelmann G Ornibactins—a New Family of Siderophores from Pseudomonas. Biometals 1993, 6 (2), 93–100. 10.1007/bf00140109. [DOI] [PubMed] [Google Scholar]
- (55).Darling P; Chan M; Cox AD; Sokol PA Siderophore Production by Cystic Fibrosis Isolates of Burkholderia Cepacia. Infect Immun 1998, 66 (2), 874–877. 10.1128/iai.66.2.874-877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Sokol PA; Darling P; Lewenza S; Corbett CR; Kooi CD Identification of a Siderophore Receptor Required for Ferric Ornibactin Uptake in Burkholderia Cepacia. Infect Immun 2000, 68 (12), 6554–6560. 10.1128/iai.68.12.6554-6560.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Escolar L; Pérez-Martín J; de Lorenzo V. Opening the Iron Box: Transcriptional Metalloregulation by the Fur Protein. J Bacteriol 1999, 181 (20), 6223–6229. 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Schäffer S; Hantke K; Braun V Nucleotide Sequence of the Iron Regulatory Gene Fur. Mol Gen Genetics 1985, 200 (1), 110–113. 10.1007/bf00383321. [DOI] [PubMed] [Google Scholar]
- (59).Bagg A; Neilands JB Ferric Uptake Regulation Protein Acts as a Repressor, Employing Iron(II) as a Cofactor to Bind the Operator of an Iron Transport Operon in Escherichia Coli. Biochemistry-us 1987, 26 (17), 5471–5477. 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- (60).Yuhara S; Komatsu H; Goto H; Ohtsubo Y; Nagata Y; Tsuda M Pleiotropic Roles of Iron-Responsive Transcriptional Regulator Fur in Burkholderia Multivorans. Microbiology 2008, 154 (6), 1763–1774. 10.1099/mic.0.2007/015537-0. [DOI] [PubMed] [Google Scholar]
- (61).Sass AM; Coenye T Low Iron-Induced Small RNA BrrF Regulates Central Metabolism and Oxidative Stress Responses in Burkholderia Cenocepacia. PloS One 2020, 15 (7), e0236405. 10.1371/journal.pone.0236405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).de Lorenzo V; Wee S; Herrero M; Neilands JB Operator Sequences of the Aerobactin Operon of Plasmid ColV-K30 Binding the Ferric Uptake Regulation (Fur) Repressor. J Bacteriol 1987, 169 (6), 2624–2630. 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Salomón RA; Farías RN The Peptide Antibiotic Microcin 25 Is Imported through the TonB Pathway and the SbmA Protein. J Bacteriol 1995, 177 (11), 3323–3325. 10.1128/jb.177.11.3323-3325.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Cheung-Lee WL; Parry ME; Cartagena AJ; Darst SA; Link AJ Discovery and Structure of the Antimicrobial Lasso Peptide Citrocin. J Biol Chem 2019, 294 (17), 6822–6830. 10.1074/jbc.ra118.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Feng Z; Liu D; Liu Z; Liang Y; Wang Y; Liu Q; Liu Z; Zang Z; Cui Y Cloning and Functional Characterization of Putative Escherichia Coli ABC Multidrug Efflux Transporter YddA. J Microbiol Biotechn 2020, 30 (7), 982–995. 10.4014/jmb.2003.03003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Yu NY; Wagner JR; Laird MR; Melli G; Rey S; Lo R; Dao P; Sahinalp SC; Ester M; Foster LJ; et al. PSORTb 3.0: Improved Protein Subcellular Localization Prediction with Refined Localization Subcategories and Predictive Capabilities for All Prokaryotes. Bioinformatics 2010, 26 (13), 1608–1615. 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Käll L; Krogh A; Sonnhammer ELL A Combined Transmembrane Topology and Signal Peptide Prediction Method. J Mol Biol 2004, 338 (5), 1027–1036. 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- (68).Käll L; Krogh A; Sonnhammer ELL Advantages of Combined Transmembrane Topology and Signal Peptide Prediction—the Phobius Web Server. Nucleic Acids Res 2007, 35 (Web Server issue), W429–W432. 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Mathavan I; Zirah S; Mehmood S; Choudhury HG; Goulard C; Li Y; Robinson CV; Rebuffat S; Beis K Structural Basis for Hijacking Siderophore Receptors by Antimicrobial Lasso Peptides. Nat Chem Biol 2014, 10 (5), 340–342. 10.1038/nchembio.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Flannagan RS; Linn T; Valvano MA A System for the Construction of Targeted Unmarked Gene Deletions in the Genus Burkholderia. Environ Microbiol 2008, 10 (6), 1652–1660. 10.1111/j.1462-2920.2008.01576.x. [DOI] [PubMed] [Google Scholar]
- (71).Barrett AR; Kang Y; Inamasu KS; Son MS; Vukovich JM; Hoang TT Genetic Tools for Allelic Replacement in Burkholderia Species. Appl Environ Microb 2008, 74 (14), 4498–4508. 10.1128/aem.00531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Hogan AM; Rahman ASMZ; Lightly TJ; Cardona ST A Broad-Host-Range CRISPRi Toolkit for Silencing Gene Expression in Burkholderia. Acs Synth Biol 2019, 8 (10), 2372–2384. 10.1021/acssynbio.9b00232. [DOI] [PubMed] [Google Scholar]
- (73).Hogan AM; Jeffers KR; Palacios A; Cardona ST Improved Dynamic Range of a Rhamnose-Inducible Promoter for Gene Expression in Burkholderia Spp. Appl Environ Microb 2021, 87 (18), e00647–21. 10.1128/aem.00647-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Sato T; Nonoyama S; Kimura A; Nagata Y; Ohtsubo Y; Tsuda M The Small Protein HemP Is a Transcriptional Activator for the Hemin Uptake Operon in Burkholderia Multivorans ATCC 17616. Appl Environ Microb 2017, 83 (16). 10.1128/aem.00479-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Vincent PA; Delgado MA; Farías RN; Salomón RA Inhibition of Salmonella Enterica Serovars by Microcin J25. Fems Microbiol Lett 2004, 236 (1), 103–107. 10.1111/j.1574-6968.2004.tb09634.x. [DOI] [PubMed] [Google Scholar]
- (76).Ghilarov D; Inaba-Inoue S; Stepien P; Qu F; Michalczyk E; Pakosz Z; Nomura N; Ogasawara S; Walker GC; Rebuffat S; et al. Molecular Mechanism of SbmA, a Promiscuous Transporter Exploited by Antimicrobial Peptides. Sci Adv 2021, 7 (37), eabj5363. 10.1126/sciadv.abj5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Runti G; Ruiz M. d C. L.; Stoilova T; Hussain R; Jennions M; Choudhury HG; Benincasa M; Gennaro R; Beis K; Scocchi M Functional Characterization of SbmA, a Bacterial Inner Membrane Transporter Required for Importing the Antimicrobial Peptide Bac7(1–35). J Bacteriol 2013, 195 (23), 5343–5351. 10.1128/jb.00818-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Camacho C; Coulouris G; Avagyan V; Ma N; Papadopoulos J; Bealer K; Madden TL BLAST+: Architecture and Applications. BMC Bioinformatics 2009, 10 (1), 421–421. 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Ussery DW; Kiil K; Lagesen K; Sicheritz-Pontén T; Bohlin J; Wassenaar TM The Genus Burkholderia: Analysis of 56 Genomic Sequences. Genome Dyn 2009, 6, 140–157. 10.1159/000235768. [DOI] [PubMed] [Google Scholar]
- (80).Li H; Durbin R Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25 (14), 1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Love MI; Huber W; Anders S Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol 2014, 15 (12), 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file 1 contains additional materials and methods; Figures S1–6 summarizing susceptibility data and bacterial growth assays; Tables S1–11 summarizing MIC data, RNA-seq results, and additional bioinformatics results; and supplementary references.
Supplementary file 2 summarizes the predicted Fur DNA-binding sites in the B. cepacia ATCC 25416 genome.
Supplementary file 3 summarizes the Operon-mapper predicted operons in B. cepacia ATCC 25416.


