Abstract
The T pilus, primarily composed of cyclic T-pilin subunits, is essential for the transmission of the Ti-plasmid T-DNA from Agrobacterium tumefaciens to plant cells. Although the virB2 gene of the 11-gene virB operon was previously demonstrated to encode the full-length propilin, and other genes of this operon have been implicated as members of a conserved transmembrane transport apparatus, the role of each virB gene in T-pilin synthesis and transport and T-pilus biogenesis remained undefined. In the present study, each virB gene was examined and was found to be unessential for T-pilin biosynthesis, except virB2, but was determined to be essential for the export of the T-pilin subunits and for T-pilus formation. We also find that the genes of the virD operon are neither involved in T-pilin export nor T-pilus formation. Critical analysis of three different virD4 mutants also showed that they are not involved in T-pilus biogenesis irrespective of the A. tumefaciens strains used. With respect to the environmental effects on T-pilus biogenesis, we find that T pili are produced both on agar and in liquid culture and are produced at one end of the A. tumefaciens rod-shaped cell in a polar manner. We also report a novel phenomenon whereby flagellum production is shut down under conditions which turn on T-pilus formation. These conditions are the usual induction with acetosyringone at pH 5.5 of Ti-plasmid vir genes. A search of the vir genes involved in controlling this biphasic reaction in induced A. tumefaciens cells revealed that virA on the Ti plasmid is involved and that neither virB nor virD genes are needed for this reaction. The biphasic reaction therefore appears to be mediated through a two-component signal transducing system likely involving an unidentified vir gene in A. tumefaciens.
Agrobacterium tumefaciens is uniquely adapted for the horizontal transmission of DNA. The DNA transfer system employed by this rhizoplane inhabitant is encoded by vir genes located on a resident Ti plasmid. The DNA transfer system is highly promiscuous, as evidenced by its transfer activity of the T-DNA into plants, fungi (8, 9, 14), and actinomycetes (23).
The products of the vir genes have been extensively studied in order to gain insights into the DNA transfer mechanism. Comparative studies of the nucleotide sequences of the vir genes with those of broad-host-range plasmids have revealed significant homologies between genes involved in DNA processing as well as plasmid transfer via bacterial conjugation, suggesting that the mechanism for T-DNA transfer could be a conjugation process (24, 29, 30, 35, 41, 43, 48). Using the F plasmid as a classic example, bacterial conjugation requires a conjugative pilus, whose propilin subunit is encoded by traA (17). The TraA propilin is processed from a 12.7-kDa propilin into a 7.2-kDa pilin with an acetylated N terminus (17). VirB2, encoded by the virB operon of the Ti plasmid, is a homolog of TraA and is processed like TraA by generating a 7.2-kDa protein from a 12.3-kDa full-length holoprotein (22). Unlike TraA pilin, however, the VirB2-processed pilin is not acetylated but is ligated by a peptide bond in a head-to-tail manner generating a cyclic pilin subunit (15). The processing of VirB2 occurs rapidly (22), and the processed product is assembled into an extracellular filament with a diameter of 10 nm that specifically appears upon induction of the vir genes with the inducer acetosyringone (AS) (28). We have called this filament the T pilus, since its presence is believed to be essential for virulence and for the transfer of the T-DNA (28).
T-pilus biogenesis is influenced by environmental factors. Early studies on the effects of temperature on A. tumefaciens-mediated tumorigenesis demonstrated that crown gall tumor size increased with a concomitant decrease in temperature, whereas tumorigenesis was inhibited at 31°C or above (7, 36). That crown gall tumor formation is thermosensitive appeared to be correlated with the conjugative transfer of the Ti plasmid between A. tumefaciens donor and recipient (45). Conjugative transfer of RSF1010 between A. tumefaciens donor and recipient strains was also found to lead to deficiency above 28°C, an environmental condition attributed to a nonfunctional DNA transfer machinery (18, 19). Banta et al. (3) found that steady-state levels of VirB10 are sensitive at 28°C with the protein destabilizing effect exacerbated in the absence of a chromosomally encoded product, ChvB. Recently, the formation of a 3.8-nm-diameter pilus on A. tumefaciens was correlated with virulence and the requirement of virB and virD4 genes as well as 19°C culture conditions for promoting pilus biogenesis (18, 19). Other environmental factors that promote the production of T pili have not been identified. In the present paper, we demonstrate that all the virB genes, and not the virD genes, including virD4 gene, are required for T-pilin synthesis and T-pilin export leading to T-pilus assembly. We also found that conditions inducing T-pilus biogenesis concomitantly turn off production of flagella in a strain-dependent manner, a phenomenon that could be similar to biphasic regulation in some animal pathogens (1).
MATERIALS AND METHODS
Bacteria, plasmids, and growth conditions.
The strains and plasmids used are described in Table 1. Nonpolar virB deletion mutants PC1001 to PC1011 were generously provided by Peter Christie, nonpolar virB insertion mutants were obtained from Andrew Binns, and virD4 mutant At12506 was obtained from Eugene Nester and Karla Fullner. All of these gift strains are derivatives of A. tumefaciens octopine strain A348. A. tumefaciens strains were grown on medium 523 (25), AB (pH 7.0) (11), and I-medium (pH 5.5) (28) as required. Antibiotic resistance markers were rifampin and erythromycin at 50 μg/ml each, carbenicillin at 30 μg/ml, and kanamycin at 20 μg/ml. The induction of vir genes in I-medium was performed according to the method of Lai and Kado (28) with minor modifications. Cells grown overnight in medium 523 with appropriate antibiotics were harvested by centrifugation (5,000 × g, 10 min) and resuspended in fresh I-medium (1:10 dilution). After growth at 28°C to mid-log phase (4 to 6 h), 200 μM AS (Aldrich Chemical Company) was added, and the culture was further incubated with vigorous shaking at 19°C for 2 to 3 days. For induction on agar medium, 500 μl of the mid-log-phase culture was spread on 1.5% agar I-medium (150- by 15-mm plate) containing 200 μM AS and incubated for 3 days at 19°C. Antibiotics were not added in the I-medium for all strains, since antibiotic selection is not required for the maintenance of Ti plasmids.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| Strains | ||
| C58 | Wild-type virulent strain containing nopaline Ti plasmid pTiC58 | 31 |
| NT1 | C58 cured of its Ti plasmid pTiC58 | 46 |
| NT1RE | Rifr Emr mutant of NT1 | 46 |
| NT1REB | Flagellum-free mutant of NT1RE | 10 |
| A136 | Rifr, C58 cured of its Ti plasmid pTiC58 | 46 |
| Ach5 | Wild-type virulent strain containing octopine Ti plasmid pTiAch5 | 31 |
| LBA4011 | Rifr, Ach5 cured of its Ti plasmid pTiAch5 | 27 |
| A348 | Rifr, A136 containing octopine Ti plasmid pTiA6NC | 20 |
| PC1001 to PC1011 | Rifr, ?virB1 to ?virB11 in pTiA6NC of A348, 11 virB nonpolar mutants | 5 |
| Ax56 | Kanr, virB10::Tn5virB in pTiA6NC of A348, virB10 nonpolar mutant | 6 |
| Ax42 | Kanr, virB9::Tn5virB in pTiA6NC of A348, virB9 nonpolar mutant | 13 |
| Ax46 | Kanr, virB8::Tn5virB in pTiA6NC of A348, virB8 nonpolar mutant | 13 |
| Ax68 | Kanr, virB6::Tn5virB in pTiA6NC of A348, virB6 nonpolar mutant | 6 |
| Ax59 | Kanr, virB5::Tn5virB in pTiA6NC of A348, virB5 nonpolar mutant | 6 |
| Ax79 | Kanr, virB4::Tn5virB in pTiA6NC of A348, virB4 nonpolar mutant | 6 |
| 368mx | Carbr, virB11::Tn3HoHo1 in pTiA6NC of A348, virB11 mutant | 42 |
| At12506 | Carbr, virD4::Tn3HoHo1 in pTiA6NC of A348, virD4 mutant | 42 |
| Plasmids | ||
| pJK270 | Kanr, T-DNA::Tn5 in pTiC58Trac | 26 |
| pJK107 | Kanr, virA::Tn5 in pTiC58Trac, virA mutant | 33 |
| pJK104 | Kanr, virB5::Tn5 in pTiC58Trac, virB5 polar mutant | 33 |
| pJK502 | Kanr, virB3::Tn5 in pTiC58Trac, virB3 polar mutant | 33 |
| pJK195 | Kanr, virD1::Tn5 in pTiC58Trac, virD1 polar mutant | 26 |
| pJK504 | Kanr, virD4::Tn5 in pTiC58Trac, virD4 mutant | 21 |
| pJK130 | Kanr, virD2::nptII in pTiC58Trac, virD2 nonpolar mutant | 32 |
| pJK734 | Kanr, virD4::nptII in pTiC58Trac, virD4 nonpolar mutant | 32 |
| pUCD4606 | Kanr, virB2 nonpolar deletion mutant in pJK270 | 22 |
Whole-cell lysate and supernatant fractionation.
Whole-cell lysates and supernatants were prepared according to the method of Lai and Kado (28) with minor modifications. Cells from 100 ml of liquid culture were harvested at 13,000 × g at 4°C for 20 min. The resulting supernatant, referred to as S1, contains secreted proteins. The cell pellet was resuspended in 4 ml of 10 mM sodium phosphate buffer, pH 5.3 (buffer A). Intact cells were passed through a hypodermic needle (18-gauge) 10 times to collect flagella, pili, and surface proteins. The sheared bacterial cells were collected at 13,000 × g at 4°C for 20 min, resuspended in buffer A, and adjusted to A600 of 20 (referred to as P). The supernatant containing flagella and pili is referred to as S2. Cells grown on agar were gently scraped off with a glass L-rod in the presence of 2 ml of buffer A. Only S2 and P were prepared from agar-grown cells. Bacterial cells were eliminated from S1 and S2 fractions by filtration through a 0.22-μm-pore-size membrane. Intact cells grown either on agar or in liquid I-medium were examined for pilus and flagella structures by electron microscopy (described below). Four volumes of ice-cold acetone was added to fraction S2, and the precipitate was collected by centrifugation (10,000 × g, 10 min, 4°C). The pellet was resuspended in a suitable volume (1/10 to 1/50 of original volume) of 1× sodium dodecyl sulfate (SDS)-gel loading buffer for protein analysis (see below). Proteins in fraction S1 were precipitated by adding 100 μl of 100% trichloroacetic acid (TCA) and 15 μl of 1% sodium deoxycholate in 1 ml of S1 fraction (4). The pelleted proteins were resuspended in an appropriate volume (see below) of 1 M Tris base (original pH) and incubated for at least 30 min at room temperature before adding an equal amount of 2× SDS-gel loading buffer. The volume of the collected samples from each strain was standardized according to the original cell density, so that the proteins are derived accordingly from the same number of cells. The concentrated proteins from S1 and S2 are referred to as CS1 and CS2, respectively.
Protein analysis.
Tricine-SDS-polyacrylamide gel electrophoresis (PAGE) (16.5%T, 3%C; 16.5% acrylamide–bisacrylamide, 3% bisacrylamide) (38) or 12% glycine–SDS-PAGE (37) was used for protein fractionation and analysis. Each protein sample, derived from an equivalent number of cells, was mixed with an equal volume of 2× SDS-gel loading buffer (0.1 M Tris-Cl [pH 6.8], 4% SDS, 0.1% bromophenol blue, 20% glycerol, 200 mM dithiothreitol) and incubated at 100°C for 5 min before loading. After electrophoresis, the fractionated proteins were visualized by Coomassie blue staining or by silver staining (37). For immunoblots, the proteins within the gel were electrotransferred onto nitrocellulose membranes (Hybond-C; Amersham, Arlington Heights, Ill.) and analyzed according to the method of Lai and Kado (28). Polyclonal antisera VirB2-B23 (against N-terminal region of processed VirB2 T-pilin encoded by pTiC58), VirB2-B24 (against C-terminal region of processed VirB2 T-pilin encoded by pTiC58) (40), anti-VirB9 (40), and anti-Ros (J. Archdeacon, unpublished data) were used. VirB2-B23 can be used for detecting T pilin encoded by either pTiC58 or pTiA6NC, but VirB2-B24 only recognizes T pilin encoded by pTiC58. Antibody interactions with antigen were visualized with a chemiluminescence system (ECL kit; Amersham). Molecular weight marker proteins were obtained from a commercial supplier (Amersham).
Electron microscopy.
The prepared bacterial cells (above) were adjusted to an A600 of 2 in 10 mM Tris-Cl (pH 7.5) and deposited on carbon-Formvar films on 300 mesh, 3-mm copper grids (Electron Microscopy Sciences, Fort Washington, Pa.). Usually 10 μl of sample (either bacterial cells or supernatant) was placed on each grid for 1 min, and then the grids were rinsed with sterile triple-distilled water for a few seconds and stained with 2% uranyl acetate for 1 min. The samples were examined in a Phillips EM410 electron microscope at 80 kV.
Motility assay.
Motility assays were performed in 0.4% soft agar medium as described previously (10). The motility results were examined after 3 or 7 days of incubation at 19°C. At least three independent motility assays were carried out for each strain and condition.
RESULTS
virB genes are required for T-pilin export.
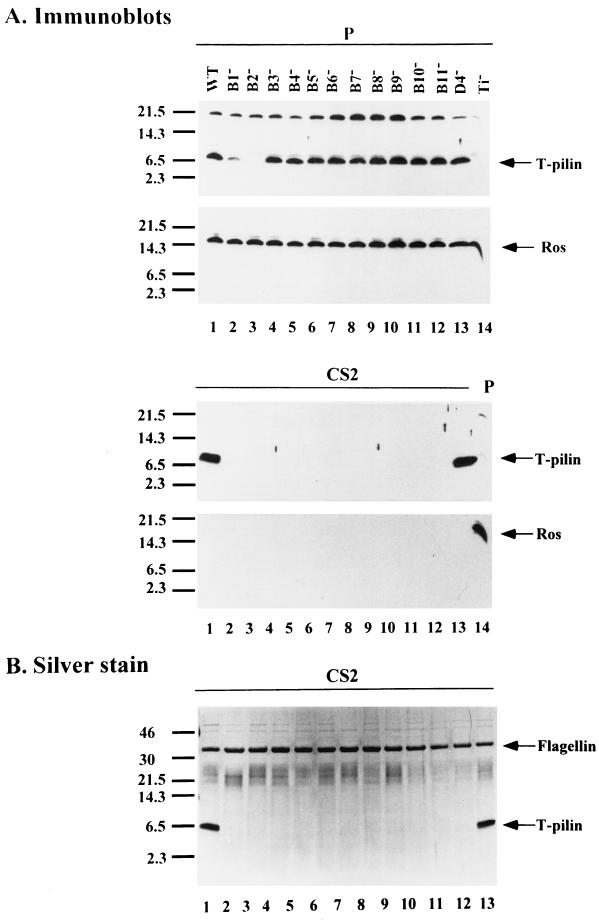
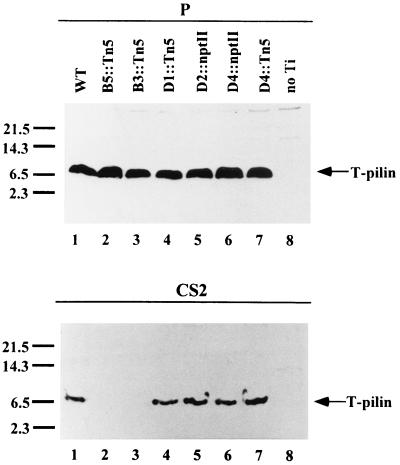
We examined virB nonpolar mutants to determine whether virB genes are required for the export of T pilin to the cell surface. Although it has been shown that nonpolar mutations in each of virB1 to virB11 and virD4 genes are unable to form the virB- and virD4-dependent pilus with a width of 3.8 nm (18), no biochemical data showed whether VirB2 propilin is expressed, processed, and exported to the bacterial exterior for pilus assembly in each mutant. Here, we examined the expression and export of T pilin in each virB nonpolar mutant, derived from octopine Ti plasmid pTiA6NC (both insertion and deletion mutants). The sheared bacterial cell lysate (P) and the concentrated proteins in supernatant S2 (CS2) collected from each AS-induced mutant strain were fractionated by SDS-PAGE to determine the fate of VirB2 by immunoblotting. As shown in Fig. 1A, except for the virB2 mutant, where no T pilin is produced, the 7.2-kDa T pilin (processed VirB2) accumulates inside the cells of wild-type strain A348 as well as in the virB mutants with nonpolar deletion mutations in each of the 11 virB genes (PC1001 to -1011) and a virD mutant with a Tn3HoHo1 insertion in the virD4 gene (At12506). T pilin accumulates in the cells of all mutants except for the virB1 mutant, where the T-pilin consistently accumulates at a lower level than that in the wild-type strain, suggesting that VirB1 might play a role in stabilizing the T pilin. As an internal control, the transcriptional regulator Ros was detected at the same level in all strains, showing that the small amount of T-pilin in the virB1 mutant is not due to an uneven protein loading or to a low level of protein translation in this strain. These results indicate that full-length VirB2 propilin is processed into T pilin independent of any other virB genes, supporting and extending previous observations (15, 28, 39). As shown in Fig. 1, T pilin was exported exocellularly in the wild-type strain A348, but was absent in each CS2 preparation from each of the 11 virB nonpolar mutants. Each virB gene other than virB2 is therefore required for the export of T pilin to the bacterial exterior. T pilin also accumulates inside the cells, but is not exported to the cell exterior in the virB nonpolar insertion virB4(Ax79), virB5(Ax59), virB6(Ax68), virB8(Ax46), virB9(Ax42), virB10(Ax56), and virB11(368mx) (data not shown). In contrast to that found with virB mutants, T pilin is still exported exocellularly in virD4 mutant strain At12506 to levels equivalent to that of the wild-type strain (Fig. 1). As a control, Ros protein, which is not secreted, is only detected in the bacterial whole-cell lysate and is not detected in the concentrated supernatant, indicating that the T pilin detected in the supernatant derived from strain At12506 did not originate from lysed cells (Fig. 1A). The proteins present in fraction S2, concentrated by TCA precipitation, also gave the same results (data not shown). T pilin is only detected in wild-type strain A348 and in virD4 mutant At12506, while the 33-kDa flagellin protein (10) is clearly present in all tested strains shown in Fig. 1B.
FIG. 1.
T-pilin excocellular export leading to T-pilus assembly requires virB genes. Sheared whole-cell lysate (P) and concentrated supernatant S2 (CS2) collected from AS-induced Agrobacterium grown on I-medium agar for 3 days at 19°C were fractionated by tricine-SDS-PAGE followed by immunoblotting (A) or silver staining (B). The strains tested were A348 (wild type) (lane 1), PC1001 to PC1011 (virB1 to virB11 nonpolar mutants, respectively) (lanes 2 to 12), At12506 (virD4 mutant) (lane 13), and A136(no Ti) (lane 14). Nonspecific cross-reactive protein appears above the processed T pilin. The relevant characteristics of each sample are indicated as the top of the gel. The molecular-mass markers are indicated on the left in kilodaltons and the positions of T-pilin, Ros, and flagellin proteins are indicated on the right.
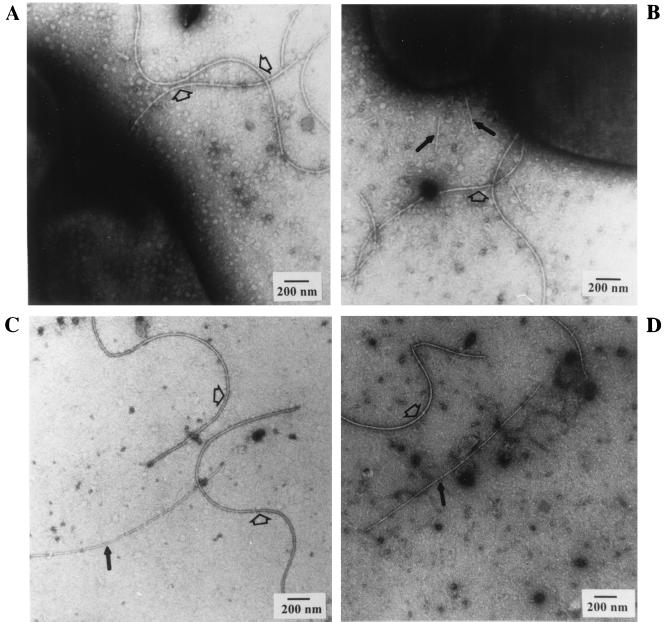
To determine whether the presence of the exocellular T-pilin is correlated with the presence of the T pilus, we examined all of the virB nonpolar mutants by electron microscopy. As reported by Fullner et al. (18), we also found that each virB gene (virB1 to -11) is essential for pilus formation, since we observed the absence of T pili in any of the virB mutants examined (see Fig. 3A). However, we found that the virD4 mutant (At12506) still produced T pili, as shown in Fig. 3B. The latter result is in contrast to a previous report that the same virD4 mutant is defective in vir-specific pilus formation (18). The T pili that we examined in both strains A348 and At12506 are approximately 10 nm in diameter (Fig. 3 and 6) rather than 3.8 nm as reported by Fullner et al. (18). The larger diameter of T pilus has been independently observed by several laboratories (15, 28, 39), including ours, as a flexuous 10-nm-wide filament (Fig. 3, 5, and 6).
FIG. 3.
Electron microscopy of virB and virD mutants. Intact bacterial whole cells (A and B) or S2 fractions (C and D) derived from cells grown on agar media at 19°C for 3 days were negatively stained with uranyl acetate and visualized by transmission electron microscopy. The strains are PC1003 (virB3 nonpolar mutant of pTiA6NC) (A), At12506 (virD4 mutant of pTiA6NC) (B), NT1RE(pJK130) (virD2 nonpolar mutant of pTiC58) (C), and NT1RE(pJK734) (virD4 nonpolar mutant of pTiC58) (D). The flagella are indicated by open arrows, and T pili are shown as solid black arrows. Scale bar, 200 nm.
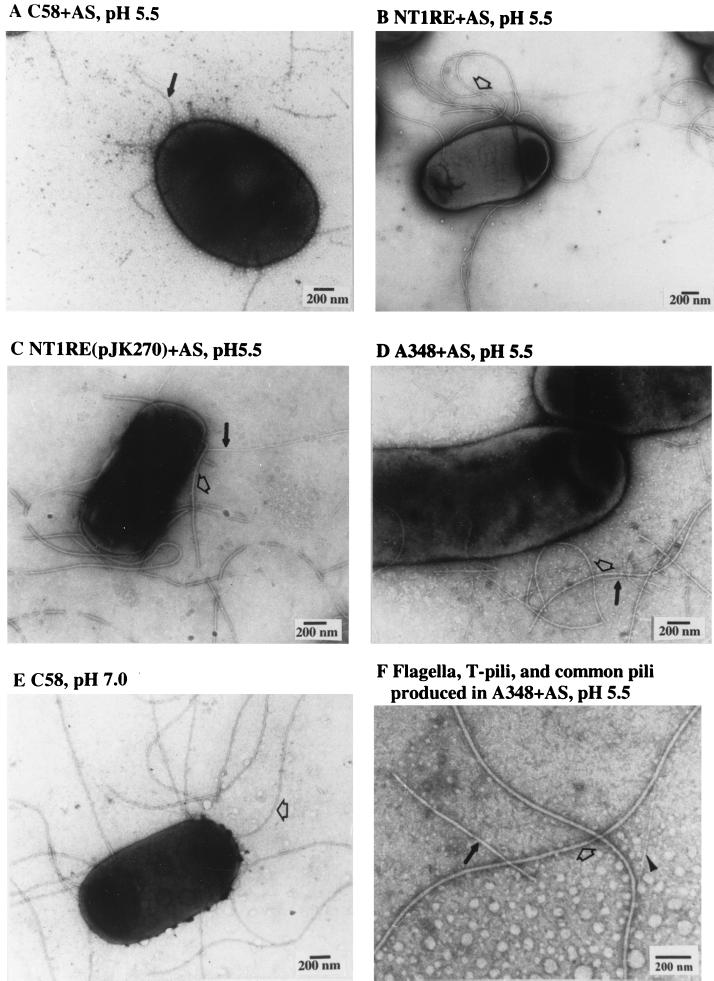
FIG. 6.
Electron microscopy of expression of T pili and flagella in different strains and under various growth conditions. Intact bacterial whole cells grown on agar media at 19°C for 3 days were negatively stained with uranyl acetate and visualized by transmission electron microscopy. The strains and culture conditions are as follows: C58 (A), NT1RE (B), NT1RE(pJK270) (C), and A348 (D and F) were grown on agar I-medium with AS. (E) C58 was grown on agar AB medium (pH 7.0) without AS. The flagella are indicated by open arrows, T pili are shown as solid arrows, and common pilus is indicated by an arrowhead. Scale bar, 200 nm.
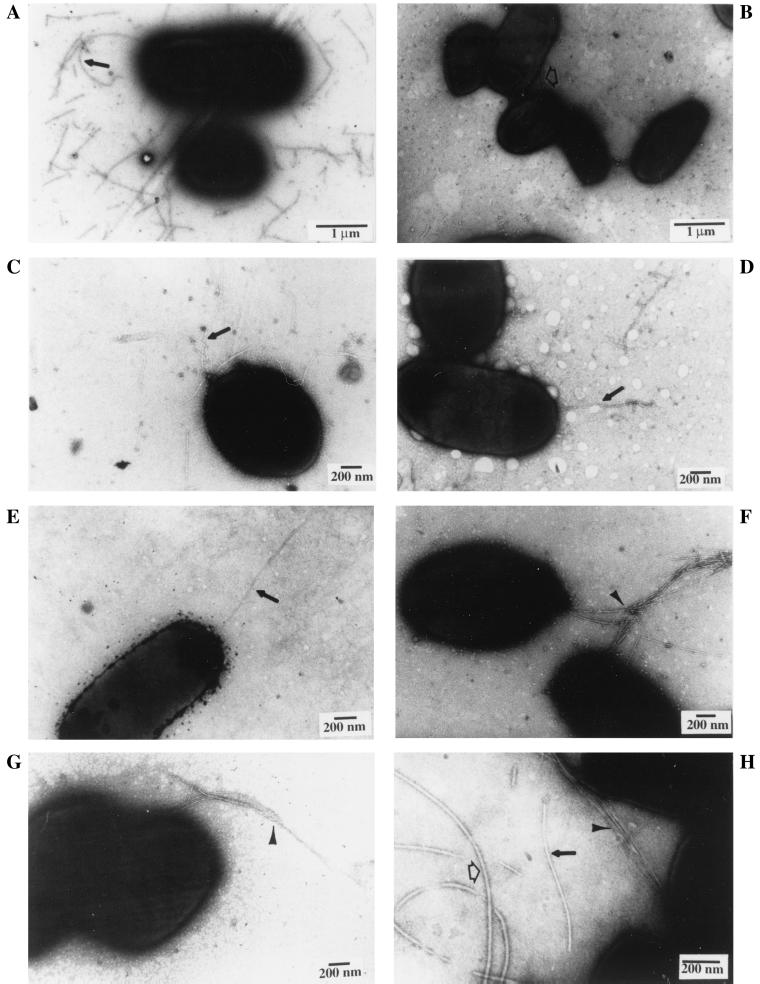
FIG. 5.
Electron microscopy of polar T-pili, common pili, and flagella. Intact bacterial whole cells collected from agar or liquid media at 19°C were negatively stained with uranyl acetate and visualized by transmission electron microscopy. The strains and culture conditions were as follows: C58 on agar I-medium with AS for 3 days (A and D), C58 on agar I-medium without AS for 3 days (B and G), C58 in liquid I-medium with AS for 2 days (C), NT1REB(pJK270) on agar I-medium with AS for 3 days (E), NT1REB (no Ti plasmid) on agar I-medium with AS for 3 days (F), and NT1RE(pJK270) on agar I-medium with AS for 3 days (H). The scale bar, corresponding to 1 μm (A and B) or 200 nm (C to H), is shown in each panel. T-pili (10 nm in width) are indicated by solid arrows as shown in panels A, C, D, E, and H; thin common pili (ca. 3 nm in width) of unknown identity are indicated by arrowheads in panels F, G, and H; and flagella (ca. 15 nm in width) are indicated by open arrows as observed in panels B and H.
virD4 gene is not required for T-pilin exocellular export and T-pilus biogenesis.
To determine whether or not virD4 is required for T-pilin export and its composite assembly into the T pilus, we examined several virD insertion mutants on nopaline Ti plasmid pTiC58. As expected, T-pilin subunits accumulated to the same level inside the cells of all tested strains, except for the Ti plasmid-less strain NT1RE (Fig. 2). T pilin was detected at wild-type levels in CS2 collected from virD1, virD2, and virD4 mutants, whereas T pilin was absent in the two virB polar mutants used as controls (Fig. 2). All of the tested virD mutants also produced T pili, as evidenced by electron microscopy (Fig. 3C and D). The presence of T pili in virD1 and virD2 mutants is expected, since several studies have demonstrated that T-DNA processing by VirD1 and VirD2 and the formation of a functional T-DNA transport system by VirB and VirD4 machinery are independent events (12). Taken together, the three different virD4 mutants—one polar mutation on pTiA6NC (At12506), one polar mutation on pTiC58 (pJK504), and one nonpolar mutation on pTiC58 (pJK734)—are all proficient in T-pilin export and T-pilus formation. These results indicate that virD4 is not involved in T-pilus formation.
FIG. 2.
Immunoblot analysis for T-pilin expression in virD mutants. Sheared whole-cell lysate (P) and concentrated supernatant S2 (CS2) were prepared as described in the legend to Fig. 1, and the proteins were analyzed by immunoblotting with anti-VirB2 antibody. Strains NT1RE containing wild-type or mutant Ti plasmids are shown as follows: pJK270 (wild type), lane 1; pJK104 (virB5::Tn5), lane 2; pJK502 (virB3::Tn5), lane 3; pJK195 (virD1::Tn5), lane 4; pJK130 (virD2::Tn5), lane 5; pJK734 (virD4::nptII), lane 6; pJK504 (virD4::Tn5), lane 7; and no Ti plasmid, lane 8. The molecular mass markers are indicated on the left in kilodaltons, and the position of T pilin is indicated on the right.
T-pilus biogenesis is not dependent on semisolid media.
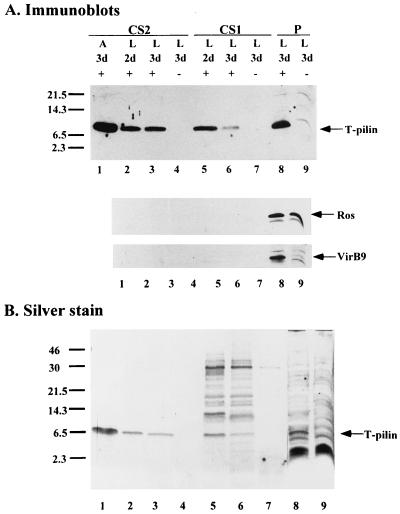
Semisolid agar medium was previously used for the detection and purification of the T pili (15, 18, 28, 39). However, it was unclear whether A. tumefaciens cells required a semisolid surface such as agar to generate T pili, presumably through some type of contact-mediated pilus expression system. Therefore, A. tumefaciens strain C58 cells were cultured in liquid and on agar media at 19°C for 2 to 3 days, with and without AS induction. Both whole-cell lysates and concentrated supernatants were analyzed for the presence of T pilin and T pili (Fig. 4 and 5). The results showed that T pilin was present exocellularly irrespective of the liquid or agar environment, although the exocellular T pilin in CS2 is consistently detected in smaller amounts when grown in liquid culture than that on agar medium (Fig. 4). CS1, the supernatant collected from liquid culture and concentrated by TCA precipitation, also contains significant amounts of T pilin, as demonstrated by Western blotting (Fig. 4A). To verify that the T pilin present in both CS1 and CS2 did not originate from lysed cells, Ros and VirB9 proteins were used as internal controls. The results show that both Ros and VirB9 remained inside the cells, while T pilin was found in both CS1 and CS2 in addition to the confines of the cells. We also examined the protein profiles of both CS1 and CS2 by silver staining. As shown in Fig. 4, T pilins are the major protein bands detected in CS2 derived from either induced liquid culture or agar medium. Several protein bands including T pilin are present in CS1 derived from culture medium with AS induction, but are absent from a parallel CS1 sample without AS induction. These results suggest that several proteins are also secreted into the medium when A. tumefaciens is induced in liquid culture. The nature of these proteins remains to be determined.
FIG. 4.
Protein analysis for T-pilin export on agar and in liquid media. Sheared whole-cell lysate (P) and concentrated supernatant (CS1 and CS2) were collected from C58 grown on agar (A) or in liquid (L), with (+) or without (−) AS induction, for 2 or 3 days (d) at 19°C. The proteins derived from equivalent numbers of cells were fractionated by Tricine-SDS-PAGE followed by immunoblotting (A) or silver staining (B). The molecular mass markers are indicated on the left in kilodaltons, and the positions of T pilin, Ros, and VirB9 proteins are indicated on the right.
When intact whole cells were examined by electron microscopy, T-pili were observed on cells cultured either in liquid or on agar (Fig. 5). More T pili were consistently observed on cells grown on agar medium than in liquid medium (data not shown). As shown in Fig. 5A, T pili are detached easily by handling, because they were scattered and not attached to the Agrobacterium cells. However, T pili were also observed extending from the bacterial cell mainly in a polar or subpolar fashion with one or several pili in clusters (Fig. 5C, D, and E). These data suggest that T pili are likely produced at one end of the A. tumefaciens cell and mechanically detached easily from the bacterial cell.
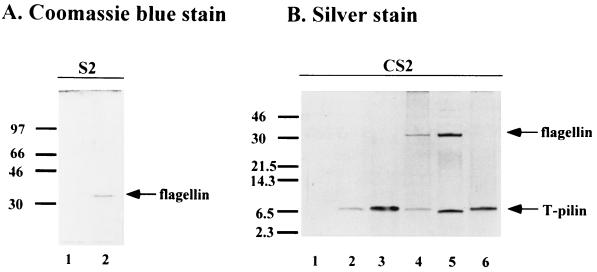
Common pili (or fimbriae) of a width of ca. 3 nm are observed in all A. tumefaciens strains tested, including wild-type, virB/virD mutant, and Ti plasmid-less strains. The Ti plasmidless bald mutant, NT1REB, and the Ti-plasmid-containing strains with or without AS induction both produce common pili (Fig. 5F to H and 6F). Because of its size compared with that of the larger 10-nm-diameter T pilus (Fig. 5 and 6), common pili are easily distinguishable and tend to aggregate or bundle together (Fig. 5F and G). The nature of these common pili is unknown, and they are often observed tightly associated with bacterial cells and are rarely detected in the S2 fraction where T pili are present (data not shown). No consistently significant protein bands other than T pilin and flagellin are present in CS2 derived from different strains when examined by silver staining (Fig. 1, 4, and 7). Perhaps the shearing method used for isolation of T pili is not harsh enough to detach common pili from the bacterial cells. It is clear, however, their formation is independent of the Ti plasmid and AS induction and likely represents fimbriae commonly found in many gram-negative bacteria.
FIG. 7.
SDS-PAGE analysis of supernatant fractions collected from cells grown in liquid culture or on agar media at 19°C for 3 days. The left gel was supernatant S2 (without concentration) analyzed by 12% glycine–SDS-PAGE followed by Coomassie blue staining to visualize flagellin proteins. The right gel was CS2 fractionated by 16.5% T, 3% C Tricine–SDS-PAGE followed by silver staining to visualize both flagellins and T pilins. The analyzed strains and growth conditions are as follows: A, lane 1, C58 from agar I-medium without AS; lane 2, C58 from agar AB medium without AS; B, lane 1, C58 from liquid I-medium without AS; lane 2, C58 from liquid I-medium with AS; lane 3, C58 from agar I-medium with AS; lane 4, NT1RE(pJK270) from agar I-medium with AS; lane 5, A348 from agar I-medium with AS; lane 6, NT1REB(pJK270) from agar I-medium with AS. The molecular mass markers are indicated on the left in kilodaltons, and the positions of flagellin and T pilin are indicated on the right.
Flagellum production is inhibited under virulence induction conditions and at low pH.
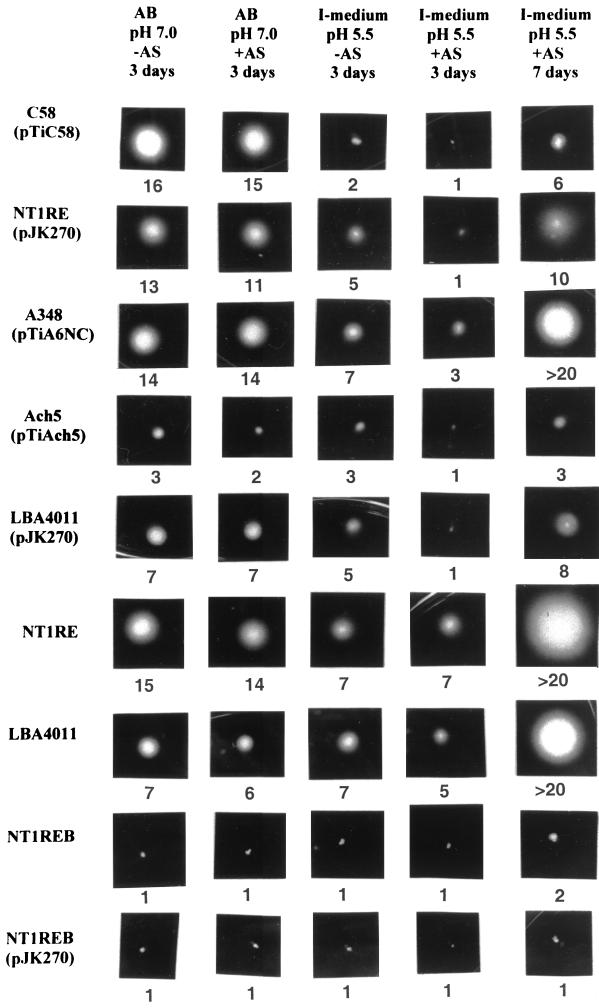
When strain C58 cells were examined for the production of T pili, we commonly observed that flagella were absent when the cells were grown in I-medium (pH 5.5) containing AS (Fig. 5A and 6A). Flagella were rarely produced when AS was omitted from the culture (Fig. 5B). Since the Ti plasmid-less strain NT1RE, NT1RE containing pJK270 (a nopaline-type pTiC58 derivative), and A136 containing pTiA6NC (an octopine-type Ti plasmid) (A348) are proficient for flagellum production (Fig. 6B, C, and D, respectively), we investigated whether our particular C58 strain is defective in flagellum synthesis. However, we found that strain C58 produced abundant flagella when cultured on medium at neutral pH, as shown in Fig. 6E. This result is supported by the presence of the 33-kDa flagellin encoded by flaA, which was detected in the supernatant only when strain C58 was grown on medium at neutral pH (Fig. 7). On the other hand, flagellin was absent in the supernatant of strain C58 when it was grown on I-medium (pH 5.5), with or without AS. Flagellin is detected, however, in the supernatant derived from strains NT1RE(pJK270) and A348(pTiA6NC) when grown under the same conditions (Fig. 7). These results are supported by the motility studies shown in Fig. 8. The motility of C58 was greatly reduced in I-medium, and it was almost nonmotile in the presence of AS for 3 days at 19°C (Fig. 8). In contrast, NT1RE(pJK270) and A348 still showed detectable motility when grown on I-medium with or without AS induction. However, in all Ti-plasmid-bearing strains, the motility zone was largest at neutral pH and was decreased at pH 5.5 and inhibited under induction conditions. At neutral pH with or without AS, strain C58 was fully motile (Fig. 8). The ingredient MES (morpholineethanesulfonic acid), which is present in I-medium and absent in AB medium, is not responsible for inhibiting motility, since I-medium when adjusted to pH 7.0 showed motility results similar to that seen with cells in AB medium (pH 7.0) (data not shown). Moreover, the addition of AS at neutral pH did not affect motility. The results of the motility assays with C58, NT1RE(pJK270), and A348 correlate well with flagellum and flagellin production, as evidenced by electron microscopy and SDS-PAGE (Fig. 6 and 7). Furthermore, we also found that A. tumefaciens strains Ach5 and LBA4011(pJK270) also exhibited an inhibition of motility under AS induction. In strains C58, Ach5, and LBA4011(pJK270), the motility was inhibited under AS induction conditions. As controls, Ti plasmid-less strains NT1RE and LBA4011 were motile and their motility zones were not significantly decreased under any of the conditions tested. As negative controls, the flagellum-deficient mutant NT1REB alone or containing pJK270 remained nonmotile under all conditions. Upon prolonged incubation, the inhibition of motility in Ti-plasmid-containing strains was partially overcome, while the bald mutant NT1REB remained nonmotile (Fig. 8).
FIG. 8.
Motility assay. The representative motility assay was photographed showing the bacterial cells forming the swimming zone. The strain name is indicated on the left, and the conditions used for each assay are indicated on the top. The number below each photograph represents the average diameter (in millimeters) of the motility zone of three independent assays (standard deviation of ±0.5). The motility assay was carried out at 19°C for 3 or 7 days.
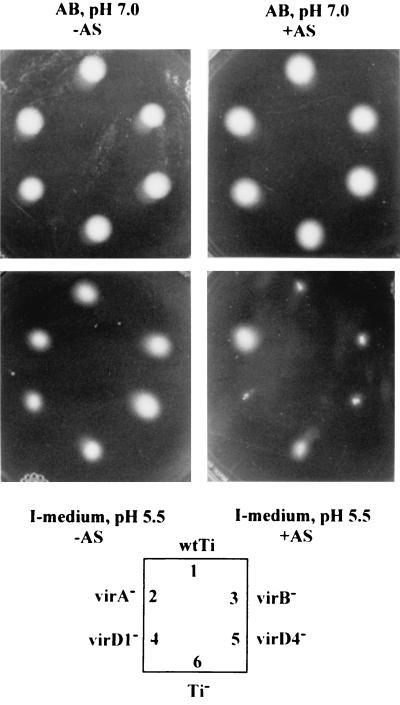
Motility was consistently inhibited under AS induction conditions in strains containing Ti plasmids. Since AS is the inducer of Ti plasmid virulence genes by the VirA VirG two-component signal transduction system, the identity of the vir gene responsible for inhibiting motility would be of significance. We found that mutations in virB and virD genes did not affect the motility inhibition effect at low pH with AS induction. On the other hand, a mutation in virA indeed overcame the suppression effect of low pH with AS, resulting in the same motility ability as that with Ti plasmid-less strain NT1RE. A representative motility assay is shown in Fig. 9. It therefore is apparent that in strain C58, flagellum production and motility are repressed when grown under vir gene induction conditions. Although the size of the motility zone could be affected by the rate of bacterial growth and by different strains and growth media, examination of strain C58 for flagellation by electron microscopy and SDS-PAGE clearly showed that the inhibition of motility is due to a deficiency in flagellation. The restoration of motility in the virA mutant suggests that the repression of A. tumefaciens flagellation might be controlled by the same two-component system involved in controlling the expression of vir genes (47). Low temperature is not required to induce this phenotype, since motility assays performed at 23 or 28°C for 2 days also showed similar results (data not shown).
FIG. 9.
Motility analyses of vir mutants. The representative motility assay was photographed after incubation at 19°C for 3 days. Inoculation positions of bacterial strains are denoted in the key. The strains shown here are as follows: 1, NT1RE(pJK270); 2, NT1RE(pJK107); 3, NT1RE(pUCD4606); 4, NT1RE(pJK195); 5, NT1RE(pJK504); and 6, NT1RE (no pTi).
DISCUSSION
Our present studies have shown that all 11 virB genes are essential for T-pilin synthesis and export leading to the assembly of the T pilus. We also found that the virD genes, including virD4, although essential for T-DNA transfer, are not required for the biogenesis of the T pilus. The T pilus appears to be mainly produced at one end of the A. tumefaciens cell either on agar medium or in liquid culture. During the initial stages of T-pilin biosynthesis, which involves AS induction at low pH, flagellar biogenesis is down regulated; this observation represents a novel phenomenon that needs to be explored further and strongly suggests that an unidentified vir gene or genes could be involved. One explanation for this phenomenon could be that the chemotactic response to a target host may no longer be needed by A. tumefaciens and that the T pilus would become the critical element to facilitate plant transformation.
T-pilin biosynthesis occurs in the absence of other Ti-plasmid genes (15), but requires the transmembrane virB-specific transport machinery for the biogenesis of the T pilus. Analyses of nonpolar virB mutants demonstrated that each of the 10 virB genes, other than virB2, is required for the export of the T pilin (Fig. 1). Likewise analyses of virD mutants showed that the exocellular appearance of T pilin remains unaffected irrespective of the A. tumefaciens strain tested (Fig. 1 and 2). We found that the presence of T pilin directly correlates with T-pilus formation (Fig. 3).
Although we have shown that the virB operon is essential for T-pilus biogenesis, the kinetics of T-pilin export from the bacterial cytoplasmic (inner) membrane to the cell surface culminating in the formation of the T pilus remains to be determined. T-pilin processing occurs rapidly (22), resulting in a 7.2-kDa cyclic peptide subunit (15). How the cyclized T pilin is shuttled through the VirB transport apparatus is unknown. VirB5 has been reported to be associated with T pili, possibly as a minor component (39), but we predict that VirB5 might be a plausible candidate as a chaperone or usher, aiding the T-pilin transport process. The cyclic feature of T pilin appears to be an evolved component to ensure that the T pilus can tolerate and resist denaturing agents in the exocellular environment, particularly on the rhizoplane and in plant tissues.
In addition to the virB genes, the reported requirement of the virD4 gene for the production of a pilus with a diameter of 3.8 nm (18) raised questions with regards to pilus morphology (below) and the role of virD4 for T-pilus biogenesis. We examined the virD4 mutant encoded by pTiA6NC (18), as well as our own virD4 mutants, and found that T-pilin export and T-pilus formation remain unaffected (Fig. 1, 2, and 3). Our results are consistent with the fact that VirD4 homologues such as TraG of RP4 and TraD of F plasmids are essential for conjugative plasmid DNA transfer, but are not involved in the elaboration of their respective conjugative pili (2, 16). Firth et al. (16) proposed that TraD and its homologues might play a role for linking protein complexes required for DNA processing and for DNA translocation. Hence, virD4 encodes a protein presumably used for T-DNA translocation rather than for T-pilus biogenesis.
Although previous studies used T pili generated on agar medium (15, 28, 39), we found that T pili are also produced in liquid culture. Irrespective of the medium used to generate T pili, morphologically the T pili remain the same; they are long flexuous filaments somewhat like F pili, whereas R pili of IncPα plasmid RP4 have a more or less rigid structure (15). Despite their morphological differences, all three pili facilitate the transfer of DNA. Interestingly, our present electron microscope studies revealed the presence of another pilus, designated here as the “common” pilus, which is morphologically distinct from the T pilus. As opposed to the 10-nm-diameter T pilus, the common pilus is much thinner, with a diameter of approximately 3.0 nm, somewhat similar to the 3.8-nm-diameter pilus reported by Fullner et al. (18). This common pilus is also similar to the one observed in a previous study in which the fine filaments were observed in an uninduced strain of A. tumefaciens (44). Common pili are produced independent of AS induction and appear at one end of the A. tumefaciens cell. The Ti-plasmid-free, bald strain NT1REB also produces common pili, indicating that the common pilus and its pilin transport system are independent of Ti plasmid genes. We propose that this common pilus is encoded by either the Agrobacterium chromosomes or the 450-kb cryptic plasmid pAtC58.
The third appendage that we examined is the flagellum. The switching-off of flagellum biosynthesis and thus A. tumefaciens cell motility under conditions that promote T-pilus production proved interesting. Our genetic evidence implies that flagellum-mediated motility is controlled via the two-component signal transduction pathway initiated by the virA gene product. The down regulation of flagellum genes and the concomitant up regulation of T-pilus biosynthesis loci resemble the virulence and flagellar control system in Bordetella bronchiseptica (1). In this mammalian pathogen, the bvgAS two-component system activates and represses gene expression in response to environmental signals and mediates a biphasic phenotype transition by activating the virulence genes and repressing the flagellum regulon (1). Upon reaching the target host, shutting down motility is a reasonable and economic strategy for bacterial pathogens.
Although common among A. tumefaciens strains, variation in motility is observed between strains, such as the nopaline strain C58, which has the propensity to clump appreciably in liquid culture, while the nonclumping derivatives of strain C58, such as NT1RE and A348, are slightly motile. Thus, cell clumping could impede motility. On the other hand, growth stage might also affect the degree of motility. The growth rates of these latter two derivative strains are noticeably higher than that of the wild-type C58 strain. Interestingly, constraints on motility can be alleviated by prolonged incubation (Fig. 8), suggesting that the inhibition of motility is reversible and that the growth stage might affect its regulation.
Overall, the biogenesis of the T pilus is central for the transmission of the T-DNA from A. tumefaciens into its host cell. That the T-pilin subunits accumulate in the bacterial cell independent of all other virB genes, including all other Ti-plasmid genes, indicates that the T pilin itself is stably maintained at a certain level intracellularly, with the exception that virB1 could play an accessory role for T-pilin stability (Fig. 1A). On the other hand, T-pilus biogenesis is totally dependent on virB genes that encode the transmembrane transport apparatus. How the cyclized T-pilin subunits are assembled into the T pilus is a critical question. The machinery for T-pilus synthesis could be like that used in the synthesis of flagella, where flagellin subunits are transferred through the 2-nm lumen of the growing flagellum filament at their distal end (34).
ACKNOWLEDGMENTS
We thank Peter Christie and Andrew Binns for generously supplying the virB nonpolar mutants, Eugene Nester for kindly providing the virD4 mutant of pTiA6NC, Robert Munn for help with electron microscopy, and Jeff Hall for scientific photography. We also thank Karla Fullner for candid discussion and Petar Pujic and Laurence Leloup for critical reading and comments on the manuscript.
This work is supported by NIH grant GM45550 from the National Institute of General Medical Sciences to C.I.K., NSF grant MCB9506144 to L.M.B., and the Jastro-Shields Graduate Student Research Award of University of California, Davis, to E.M.L.
REFERENCES
- 1.Akerley B J, Miller J F. Understanding signal transduction during bacterial infection. Trends Microbiol. 1996;4:141–146. doi: 10.1016/0966-842x(96)10024-x. [DOI] [PubMed] [Google Scholar]
- 2.Balzer D, Pansegrau W, Lanka E. Essential motifs of relaxase (TraI) and TraG proteins involved in conjugative transfer of plasmid RP4. J Bacteriol. 1994;176:4285–4295. doi: 10.1128/jb.176.14.4285-4295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banta L M, Bohne J, Lovejoy S D, Dostal K. Stability of the Agrobacterium tumefaciens VirB10 protein is modulated by growth temperature and periplasmic osmoadaption. J Bacteriol. 1998;180:6597–6606. doi: 10.1128/jb.180.24.6597-6606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bensadoun A, Weinstein D. Assay of proteins in the presence of interfering materials. Anal Biochem. 1976;70:241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- 5.Berger B R, Christie P J. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binns A N, Beaupré C E, Dale E M. Inhibition of VirB-mediated transfer of diverse substrates from Agrobacterium tumefaciens by the IncQ plasmid RSF1010. J Bacteriol. 1995;177:4890–4899. doi: 10.1128/jb.177.17.4890-4899.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun A C. Thermal studies on the factors responsible for tumor initiation in crown gall. Am J Bot. 1947;34:234–240. [PubMed] [Google Scholar]
- 8.Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas P J. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 1995;14:3206–3214. doi: 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bundock P, Mróczek K, Winkler A A, Steensma H Y, Hooykaas P J. T-DNA from Agrobacterium tumefaciens as an efficient tool for gene targeting in Kluyveromyces lactis. Mol Gen Genet. 1999;261:115–121. doi: 10.1007/s004380050948. [DOI] [PubMed] [Google Scholar]
- 10.Chesnokova O, Coutinho J B, Khan I H, Mikhail M S, Kado C I. Characterization of flagella genes of Agrobacterium tumefaciens, and the effect of a bald strain on virulence. Mol Microbiol. 1997;23:579–590. doi: 10.1046/j.1365-2958.1997.d01-1875.x. [DOI] [PubMed] [Google Scholar]
- 11.Chilton M D, Currier T C, Farrand S K, Bendich A J, Gordon M P, Nester E W. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc Natl Acad Sci USA. 1974;71:3672–3676. doi: 10.1073/pnas.71.9.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christie P J. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dale E M, Binns A N, Ward J E., Jr Construction and characterization of Tn5virB, a transposon that generates nonpolar mutations, and its use to define virB8 as an essential virulence gene in Agrobacterium tumefaciens. J Bacteriol. 1993;175:887–891. doi: 10.1128/jb.175.3.887-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Groot M J, Bundock P, Hooykaas P J, Beijersbergen A G. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol. 1998;16:839–842. doi: 10.1038/nbt0998-839. [DOI] [PubMed] [Google Scholar]
- 15.Eisenbrandt R, Kalkum M, Lai E M, Lurz R, Kado C I, Lanka E. Conjugative pili of IncP plasmids and the Ti plasmid T-pilus are composed of cyclic subunits. J Biol Chem. 1999;274:22548–22555. doi: 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- 16.Firth N, Ippen-Ihler K, Skurray R A. Structure and function of the F factor and mechanism of conjugation. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1996. pp. 2377–2401. [Google Scholar]
- 17.Frost L S, Paranchych W, Willetts N S. DNA sequence of the F traALE region that includes the gene for F pilin. J Bacteriol. 1984;160:395–401. doi: 10.1128/jb.160.1.395-401.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fullner K J, Lara J C, Nester E W. Pilus assembly by Agrobacterium T-DNA transfer genes. Science. 1996;273:1107–1109. doi: 10.1126/science.273.5278.1107. [DOI] [PubMed] [Google Scholar]
- 19.Fullner K J, Nester E W. Temperature affects the T-DNA transfer machinery of Agrobacterium tumefaciens. J Bacteriol. 1996;178:1498–1504. doi: 10.1128/jb.178.6.1498-1504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garfinkel D J, Simpson R B, Ream L W, White F F, Gordon M P, Nester E W. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell. 1981;27:143–153. doi: 10.1016/0092-8674(81)90368-8. [DOI] [PubMed] [Google Scholar]
- 21.Hirooka T, Kado C I. Location of the right boundary of the virulence region on Agrobacterium tumefaciens plasmid pTiC58 and a host-specifying gene next to the boundary. J Bacteriol. 1986;168:237–243. doi: 10.1128/jb.168.1.237-243.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones A L, Lai E M, Shirasu K, Kado C I. VirB2 is a processed pilin-like protein encoded by the Agrobacterium tumefaciens Ti plasmid. J Bacteriol. 1996;178:5706–5711. doi: 10.1128/jb.178.19.5706-5711.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kado C I. Agrobacterium-mediated horizontal gene transfer. Genet Eng. 1998;20:1–24. doi: 10.1007/978-1-4899-1739-3_1. [DOI] [PubMed] [Google Scholar]
- 24.Kado C I. Promiscuous DNA transfer system of Agrobacterium tumefaciens: role of the virB operon in sex pilus assembly and synthesis. Mol Microbiol. 1994;12:17–22. doi: 10.1111/j.1365-2958.1994.tb00990.x. [DOI] [PubMed] [Google Scholar]
- 25.Kado C I, Heskett M G. Selective media for isolation of Agrobacterium, Corynebacterium, Erwinia, Pseudomonas, and Xanthomonas. Phytopathology. 1970;60:969–976. doi: 10.1094/phyto-60-969. [DOI] [PubMed] [Google Scholar]
- 26.Kao J C, Perry K L, Kado C I. Indoleacetic acid complementation and its relation to host range specifying genes on the Ti plasmid of Agrobacterium tumefaciens. Mol Gen Genet. 1982;188:425–432. doi: 10.1007/BF00330044. [DOI] [PubMed] [Google Scholar]
- 27.Klapwijk P M, Scheulderman T, Schilperoort R A. Coordinated regulation of octopine degradation and conjugative transfer of Ti plasmids in Agrobacterium tumefaciens: evidence for a common regulatory gene and separate operons. J Bacteriol. 1978;136:775–785. doi: 10.1128/jb.136.2.775-785.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai E M, Kado C I. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J Bacteriol. 1998;180:2711–2717. doi: 10.1128/jb.180.10.2711-2717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lessl M, Balzer D, Pansegrau W, Lanka E. Sequence similarities between the RP4 Tra2 and the Ti VirB region strongly support the conjugation model of T-DNA transfer. J Biol Chem. 1992;267:20471–20480. [PubMed] [Google Scholar]
- 30.Lessl M, Lanka E. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell. 1994;77:321–324. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 31.Lin B C, Kado C I. Studies on Agrobacterium tumefaciens. VIII. Avirulence induced by temperature and ethidium bromide. Can J Microbiol. 1977;23:1554–1561. doi: 10.1139/m77-229. [DOI] [PubMed] [Google Scholar]
- 32.Lin T S, Kado C I. The virD4 gene is required for virulence while virD3 and orf5 are not required for virulence of Agrobacterium tumefaciens. Mol Microbiol. 1993;9:803–812. doi: 10.1111/j.1365-2958.1993.tb01739.x. [DOI] [PubMed] [Google Scholar]
- 33.Lundquist R C, Close T J, Kado C I. Genetic complementation of Agrobacterium tumefaciens Ti plasmid mutants in the virulence region. Mol Gen Genet. 1984;193:1–7. doi: 10.1007/BF00327406. [DOI] [PubMed] [Google Scholar]
- 34.Macnab R M. The bacterial flagellum: reversible rotary propellor and type III export apparatus. J Bacteriol. 1999;181:7149–7153. doi: 10.1128/jb.181.23.7149-7153.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pohlman R F, Genetti H D, Winans S C. Common ancestry between IncN conjugal transfer genes and macromolecular export systems of plant and animal pathogens. Mol Microbiol. 1994;14:655–668. doi: 10.1111/j.1365-2958.1994.tb01304.x. [DOI] [PubMed] [Google Scholar]
- 36.Riker A J. Studies on the influence of some environmental factors on the development of crown gall. J Agric Res. 1926;32:83–96. [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt-Eisenlohr H, Domke N, Angerer C, Wanner G, Zambryski P C, Baron C. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J Bacteriol. 1999;181:7485–7492. doi: 10.1128/jb.181.24.7485-7492.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirasu K, Kado C I. Membrane location of the Ti plasmid VirB proteins involved in the biosynthesis of a pilin-like conjugative structure on Agrobacterium tumefaciens. FEMS Microbiol Lett. 1993;111:287–294. doi: 10.1111/j.1574-6968.1993.tb06400.x. [DOI] [PubMed] [Google Scholar]
- 41.Shirasu K, Kado C I. The virB operon of the Agrobacterium tumefaciens virulence regulon has sequence similarities to B, C and D open reading frames downstream of the pertussis toxin-operon and to the DNA transfer-operons of broad-host-range conjugative plasmids. Nucleic Acids Res. 1993;21:353–354. doi: 10.1093/nar/21.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stachel S E, An G, Flores C, Nester E W. A Tn3 lacZ transposon for the random generation of beta-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 1985;4:891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stachel S E, Zambryski P C. Agrobacterium tumefaciens and the susceptible plant cell: a novel adaptation of extracellular recognition and DNA conjugation. Cell. 1986;47:155–157. doi: 10.1016/0092-8674(86)90437-x. [DOI] [PubMed] [Google Scholar]
- 44.Stemmer W P C, Sequeira L. Fimbriae of phytopathogenic and symbiotic bacteria. Phytopathology. 1987;77:1633–1639. [Google Scholar]
- 45.Tempé J, Petit A, Holsters M, van Montagu M, Schell J. Thermosensitive step associated with transfer of the Ti plasmid during conjugation: possible relation to transformation in crown gall. Proc Natl Acad Sci USA. 1977;74:2848–2849. doi: 10.1073/pnas.74.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watson B, Currier T C, Gordon M P, Chilton M D, Nester E W. Plasmid required for virulence of Agrobacterium tumefaciens. J Bacteriol. 1975;123:255–264. doi: 10.1128/jb.123.1.255-264.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winans S C. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol Rev. 1992;56:12–31. doi: 10.1128/mr.56.1.12-31.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zupan J R, Zambryski P. Transfer of T-DNA from Agrobacterium to the plant cell. Plant Physiol. 1995;107:1041–1047. doi: 10.1104/pp.107.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]