Abstract
We have previously identified and characterized the alkyl hydroperoxide reductase of Streptococcus mutans, which consists of two components, Nox-1 and AhpC. Deletion of both nox-1 and ahpC had no effect on the sensitivity of S. mutans to cumene hydroperoxide or H2O2, implying that the existence of another antioxidant system(s) independent of the Nox-1–AhpC system compensates for the deficiency. Here, a new antioxidant gene (dpr for Dps-like peroxide resistance gene) was isolated from the S. mutans chromosome by its ability to complement an ahpCF deletion mutant of Escherichia coli with a tert-butyl hydroperoxide-hypersensitive phenotype. The dpr gene complemented the defect in peroxidase activity caused by the deletion of nox-1 and ahpC in S. mutans. Under aerobic conditions, the dpr disruption mutant carrying a spectinomycin resistance gene (dpr::Spcr mutant) grew as well as wild-type S. mutans in liquid medium. However, the dpr::Spcr mutant could not form colonies on an agar plate under air. In addition, neither the dpr::Spcr ahpC::Emr::nox-1 triple mutant nor the dpr::Spcr sod::Emr double mutant was able to grow aerobically in liquid medium. The 20-kDa dpr gene product Dpr is an iron-binding protein. Synthesis of Dpr was induced by exposure of S. mutans cells to air. We propose a mechanism by which Dpr confers aerotolerance on S. mutans.
Bacteria contain certain enzymes capable of reacting with oxygen, and they cannot avoid confronting harmful reactive oxygen species, including superoxide anion (O2−), hydrogen peroxide (H2O2), and organic hydroperoxide, if they are exposed to air. To live in the presence of oxygen, they have to convert these reactive oxygen species to nontoxic molecules. Therefore, enzymes such as superoxide dismutases (SOD), catalases, and peroxidases are ubiquitously distributed in aerotolerant bacteria.
Lactic acid bacteria, including Streptococcus mutans, lack cytochromes and other heme-containing proteins. Most lactic acid bacteria, except several lactobacilli which acquire catalase activity if a source of heme is added to their growth medium (42), also lack catalase. However, they can grow in the presence of air. In view of the defense against oxygen toxicity, the lack of catalase in lactic acid bacteria is inconsistent with their aerotolerance. Mechanisms by which lactic acid bacteria cope with peroxide-mediated stress are therefore an area under active investigation. We previously identified H2O2-forming NADH oxidase (Nox-1) in S. mutans and found that Nox-1 is homologous with a flavoprotein component, AhpF, of Salmonella typhimurium alkyl hydroperoxide reductase (AhpR), consisting of AhpF and AhpC (16, 17, 30). We also identified ahpC, which is homologous with ahpC of S. typhimurium, upstream of nox-1 in S. mutans (30). Analyses of purified AhpC together with Nox-1 have verified that these proteins act as a bicomponent peroxidase system in S. mutans, catalyzing the NADH-dependent reduction of organic hydroperoxides or H2O2 to their respective alcohols and/or H2O (31). Furthermore, the bicomponent system restored the resistance of Escherichia coli TA4315 (ahpCF-defective mutant) (36) to cumene hydroperoxide (18). However, the nox-1 ahpC double deletion mutant of S. mutans still showed the same level of peroxide tolerance as did the wild-type strain (18). These results suggested the existence of another antioxidant system(s) in addition to Nox-1 and AhpC in S. mutans. Here we identified a new gene, dpr (for Dps-like peroxide resistance gene); its product, Dpr, as an iron-binding protein responsible for peroxide tolerance in S. mutans. In this study, we also propose a mechanism by which Dpr confers aerotolerance on S. mutans.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and cell growth.
The bacterial strains and plasmids used in this study are described in Table 1. S. mutans was grown aerobically or anaerobically at 37°C in Todd-Hewitt broth (THB; Difco Laboratories, Detroit, Mich.), TY medium containing 1% glucose (TYG) (15), or THB supplemented with 5% horse serum for generation of component cells. Overnight cultures of wild-type and mutant S. mutans strains were incubated in an anaerobic glove box (Hirasawa Works, Tokyo, Japan) under an atmosphere of 80% nitrogen, 10% hydrogen, and 10% carbon dioxide. For aerobic growth, overnight cultures were diluted 50-fold and incubated to late exponential phase (an A660 of 1.0) under aerobic conditions without shaking. The cultures were then diluted again to an A660 of 0.020 in a 100-ml flask containing 5 ml of TYG medium and incubated with vigorous shaking under aerobic conditions (150 cycles/min). Fifty units of bovine liver catalase (Sigma) per ml, with or without pretreatment at 80°C for 10 min, or 100 nmol of deferoxamine mesylate (deferoxamine) (ICN Pharmaceuticals, K. K. Tokyo, Japan) per ml was added to the growth medium when indicated. Cell density was monitored by determination of A660 during growth. For measurement of viable cells, aliquots of cultures were diluted and spread onto TYG plates during growth. After 48 h of incubation in air at 37°C, the CFU on plates were counted. For anaerobic growth, 10 ml of fresh medium was inoculated with 0.1 ml of the late exponential phase (A660 of 1.0) anaerobic subculture and incubated without shaking in an anaerobic glove box. E. coli was grown aerobically at 37°C in LB broth (25). Solid media were supplemented with 1.5% agar. When present in selective plates, antibiotics were used at the following concentrations: for S. mutans, erythromycin at 10 μg/ml and/or spectinomycin at 250 μg/ml; for E. coli, ampicillin at 100 μg/ml, erythromycin at 300 μg/ml, chloramphenicol at 25 μg/ml, and spectinomycin at 50 μg/ml. Plasmids pUC118 (TaKaRa Biochemicals, Tokyo, Japan) and pACYC184 (Nippon Gene Co., Ltd., Tokyo, Japan) were used as vectors for gene cloning, and pSPC1 (23) was the source of a spectinomycin resistance (Spcr) determinant.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference |
|---|---|---|
| Strains | ||
| S. mutans | ||
| GS-5 | Wild type, serotype c | 41 |
| BEE | ahpC::Emr::nox-1 | 18 |
| DES | dpr::Spcr | This study |
| KD251 | sod::Emr | This study |
| BEE-DES | dpr::SpcrahpC::Emr::nox-1 | This study |
| KD251-DES | dpr::Spcrsod::Emr | This study |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 14 |
| TA4315 | ΔahpC ΔahpF | 36 |
| Plasmids | ||
| pUC118 | AprlacPOZ′ | 44 |
| pACYC184 | Tetr Cmr | 6 |
| pSPC1 | Spcr 1.1-kb EcoRI fragment | 23 |
| pS23 | 5.3-kb fragment containing dpr in pUC118 | This study |
| pS23B1 | 2.6-kb BamHI-BamHI fragment containing dpr from pS23 in pUC118 | This study |
| pS23E12 | 1.8-kb EcoRI-EcoRI fragment containing dpr from pS23 in pUC118 | This study |
| pS23E121 | 0.94-kb EcoRI-EcoRI fragment from pS23E12 | This study |
| pS23E122 | 0.86-kb EcoRI-EcoRI fragment from pS23E12 | This study |
| pD13184 | dpr in pACYC184 | This study |
| pS23B1ES | dpr::Spcr from pS23B1 | This study |
| pS23E12ES | dpr::Spcr from pS23E12 | This study |
| pKD251 | sod::Emr | 27 |
Construction of a chromosomal gene library from the nox-1 and ahpC double deletion mutant (AhpR deletion mutant) of S. mutans.
Chromosomal DNA prepared from bacterial cells as described previously (17) was partially digested with Sau3AI and separated by agarose gel electrophoresis. DNA fragments comprising 4 to 6 kb were eluted from the gel using EASYTRAP Ver.2 (TaKaRa) and ligated into the BamHI site of plasmid vector pUC118 DNA. E. coli strain TA4315 (ΔahpC ΔahpF), which is hypersensitive to tert-butyl hydroperoxide (tBHP; Katayama Chemical Co., Ltd., Osaka, Japan), was transformed with the S. mutans-derived library and subjected to selection by the method of Mongkolsuk et al. (26) on LB agar plates containing ampicillin and 0.5 mM tBHP to identify transformants in which the organic hydroperoxide-hypersensitive phenotype was suppressed.
Primers used for amplification of dpr and sod by PCR.
Five primers, dpr1 (5′-CAAAGAAAGAGCCAAGAGAAGC-3′), dpr2 (5′-ACTGGAATTCTAATCCAACGTCTGGGA-3′), dpr3 (5′-TTTCTATTCTGTGCTCCCTGCG-3′), sod1 (5′-TCCCCCGGGATGATTTCTGT CAAAGC-3′), and sod2 (5′-TTGAATTCTAGCGTATAGCGACTTACGG-3′), were used to amplify dpr and sod by PCR.
Construction of the plasmid for knockout of dpr.
An Spcr determinant was obtained by digestion of plasmid pSPC1 with EcoRI, and its cohesive ends were blunted. Knockout plasmid pS23B1ES was constructed by introducing the Spcr-encoding gene into the EcoRV site of pS23B1. The plasmid had a dpr::Spcr fusion gene and did not confer tBHP tolerance on E. coli TA4315.
Transformation of S. mutans and homologous recombination.
Genetic transformation of S. mutans with DNA fragments was performed as described previously (18).
Measurement of plating efficiency of S. mutans.
Overnight anaerobic cultures of S. mutans were diluted 100-fold and incubated while standing under air. At the exponential phase (A660 of 0.6), the cultures were spread onto TYG plates and incubated at 37°C anaerobically (A) and aerobically (B). After 48 h of incubation, the CFU on both plates were counted. Plating efficiency under air was defined as the ratio of CFU in B relative to those in A. Five hundreds units of catalase was spread on top of the plate immediately before the cells were spread when indicated.
Preparation of cell extracts.
Cells grown aerobically or anaerobically were harvested by centrifugation at 12,000 × g for 10 min, washed twice with 50 mM potassium phosphate buffer containing 0.2 mM EDTA (pH 7.0), and disrupted by sonication. After unbroken cells and cell debris were removed by centrifugation at 25,000 × g for 30 min, the clear lysate was collected.
Assay for SOD.
SOD activity was measured by the xanthine oxidase-cytochrome c method (39). One unit of SOD activity was defined as the amount of protein that gave a 50% decrease in the rate of reduction of cytochrome c.
Analytical procedures.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was done as described by Laemmli (22). Protein was measured by the method of Bradford (4). The N-terminal amino acid sequence was determined by a gas-phase protein sequencer (model ABI 491; Applied Biosystems) equipped with an on-line amino acid analyzer (model 610A; Applied Biosystems). For staining of iron-binding proteins, Fe(NH4)2(SO4) · 6H2O was added to the crude extract at a final concentration of 1 mM and the mixture was incubated on ice for 30 min. The crude extract was then resolved by nondenaturing PAGE (native PAGE) (16). The gel was stained with 3-(2-pyridyl)-5,6-bis(2-[5-furyl sulfonic acid])-1,2,4-triazine (Ferene S; Sigma) by the method of Chung (8).
Nucleotide sequencing.
The cloned DNA was fragmented by digestion with various restriction enzymes, subcloned into plasmid pUC118, and sequenced. The cycle sequencing reaction was done with a Thermo Sequenase cycle sequencing kit (Amersham) with M13 forward and reverse IRD-41-labeled dye primers (Aloka, Ltd., Tokyo, Japan). A DNA sequencing system (model 4000; Li-Cor, Lincoln, Nebr.) was used for sequencing.
Homology search, ORF identification, and alignment of multiple nucleotide and amino acid sequences.
Protein and nucleotide sequences were compared with database sequences using the BLAST (version 1.49) programs implemented at the EMBL, GenBank, and DDBJ nucleotide sequence databases and the SWISSPROT and NBRF-PIR protein sequence databases. Open reading frame (ORF) identification and multiple-sequence alignment were performed using the GENETYX program (Software Development Co., Tokyo, Japan) and multiple-sequence alignment with hierarchical clustering (Laboratoire de Genetique Cellulaire, Toulouse, France). Secondary-structure prediction was performed by the method of Rost and Sander (32).
Nucleotide sequence accession number.
The sequence of the cloned DNA described here has been deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession number AB036428.
RESULTS
Identification of a peroxide resistance gene from S. mutans.
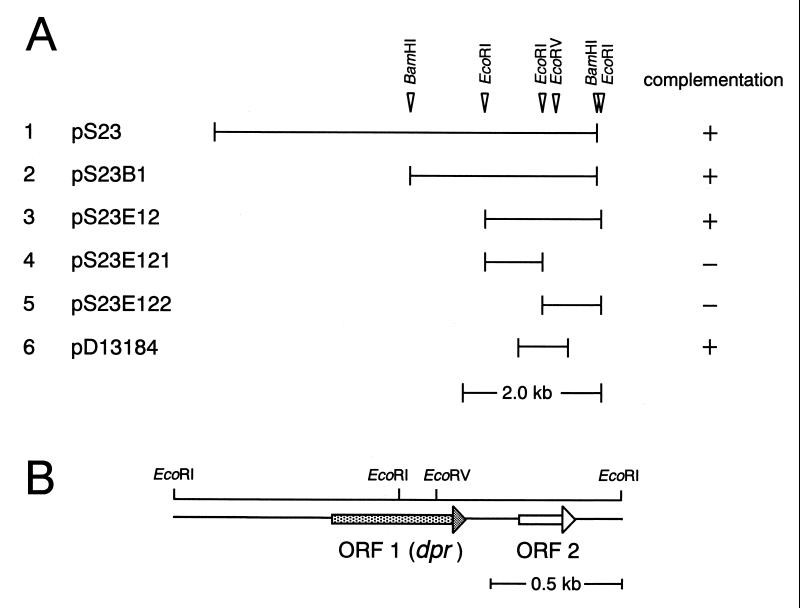
We started with the cloning of a potential peroxide tolerance gene(s) from a Sau3AI genomic library of S. mutans strain BEE (ahpC::Emr::nox-1) ligated into the BamHI site of pUC118 and transformed into E. coli TA4315 (ΔahpC ΔahpF) with a tBHP-hypersensitive phenotype. One positive transformant was obtained from approximately 10,000 colonies plated on LB broth plates containing ampicillin (100 μg/ml) and 0.5 mM tBHP. The positive clone contained a DNA insert of about 5.3 kbp. The plasmid carrying the 5.3-kbp fragment was designated pS23 (Fig. 1A, lane 1). pS23 DNA was digested with BamHI or EcoRI, and the fragments were ligated into the BamHI and EcoRI sites of pUC118, respectively, and transformed into E. coli TA4315. After selection for colony growth on LB broth-ampicillin plates containing tBHP, positive clones with 2.6-kbp BamHI and 1.8-kbp EcoRI fragments were obtained (pS23B1 and pS23E12, respectively; Fig. 1A). Sequence analysis revealed that the 1.8-kbp fragment of pS23E12 included two ORFs (ORFs 1 and 2 in Fig. 1B) and that an EcoRI site was present within ORF 1.
FIG. 1.
DNA segments from S. mutans which suppressed the tBHP-hypersensitive phenotype of E. coli TA4315 (ΔahpC ΔahpF) (A) and physical map of the 1.8-kbp segment of pS23E12 (B). The solid lines in panel A indicate the regions of the chromosomal DNA of S. mutans (lanes 1 to 5) and the DNA amplified by PCR (lane 6). In panel B, the arrows indicate the locations of ORFs.
To examine whether ORF 1 and/or 2 is responsible for peroxide tolerance in plasmid pS23E12, the 1.8-kbp fragment was digested with EcoRI to obtain 0.94- and 0.86-kbp fragments. The 0.94- and 0.86-kbp fragments were subcloned into the EcoRI site of pUC118 to obtain plasmids pS23E121 and pS23E122, respectively (Fig. 1A). Neither plasmid could confer peroxide resistance on E. coli TA4315, suggesting that ORF 1 is responsible for peroxide tolerance in E. coli TA4315. ORF 1, including its putative promoter region, was then amplified by PCR using primers dpr1 and dpr2, ligated into the EcoRV site of pACYC184 to obtain plasmid pD13184 (Fig. 1A), and then transformed into E. coli TA4315. The recombinant E. coli generated in this manner exhibited a peroxide resistance phenotype. In addition, ORF 1 was disrupted by introducing an Spcr-encoding gene into the EcoRV site in ORF 1 in pS23E12 to obtain plasmid pS23E12ES containing a dpr::Spcr fusion gene and the intact ORF 2 gene. E. coli TA4315(pS23E12ES) was sensitive to tBHP (data not shown). Thus, we concluded that ORF 1 is the tBHP resistance gene.
ORF 1 consisted of 525 bp encoding a protein of 19,617 Da (20-kDa protein) with 175 amino acid residues. This ORF had putative −35 (5′-TAGAAT-3′) and −10 (5′-TATAAA-3′) promoter regions upstream of the translation start codon and palindromic sequences downstream from the translation stop codon. A putative Shine-Dalgarno sequence (5′-AGGAG-3′) was also found 13 bp upstream of the start codon. The deduced amino acid sequence of the 20-kDa protein showed low homology with that of the Dps (DNA-binding protein from starved cells) (1) family of proteins (Table 2). The secondary structure of the 20-kDa protein was also predicted to be similar to that of Dps (as described later). Accordingly, the gene encoding ORF 1 was designated dpr (dps-like peroxide resistance gene). On the other hand, ORF 2, 204 nucleotides long, encoded a protein of 8,332 Da comprising 68 amino acid residues. The deduced amino acid sequence of the putative 8.3-kDa protein showed 52% identity with that of a hypothetical protein from Bacillus megaterium (35).
TABLE 2.
Pairwise identities of amino acid sequences between S. mutans Dpr and Dps family proteins
| Organism and protein | % Sequence identity with:
|
||||||
|---|---|---|---|---|---|---|---|
| S. mutans Dpr | L. innocua Fera | B. subtilis MrgAb | E. coli Dpsc | B. subtilis Dpsd | H. pylori NapAe | H. ducreyi pilinf | |
| L. innocua Fera | 40.0 | ||||||
| B. subtilis MrgAb | 30.9 | 36.0 | |||||
| E. coli Dpsc | 27.6 | 25.6 | 22.4 | ||||
| B. subtilis Dpsd | 26.3 | 43.0 | 45.5 | 20.0 | |||
| H. pylori NapAe | 26.1 | 32.1 | 29.5 | 22.4 | 34.2 | ||
| H. ducreyi pilinf | 23.0 | 21.5 | 21.2 | 35.1 | 21.6 | 23.6 | |
| Synechococcus DpsAg | 21.5 | 26.3 | 26.8 | 22.6 | 22.7 | 20.5 | 21.9 |
Nonheme ferritin of L. innocua (accession no. P80725).
MrgA of B. subtilis (accession no. P37960).
Dps of E. coli (accession no. P27430).
Dps of B. subtilis (accession no. AF008220).
Neutrophil-activating protein of H. pylori (accession no. AE000543).
Fine-tangled pilus major subunit of H. ducreyi (accession no. Q47953).
DpsA of Synechococcus sp. strain PCC7492 (accession no. Q55024).
Abortive colony formation of dpr disruption mutants of S. mutans under aerobic conditions.
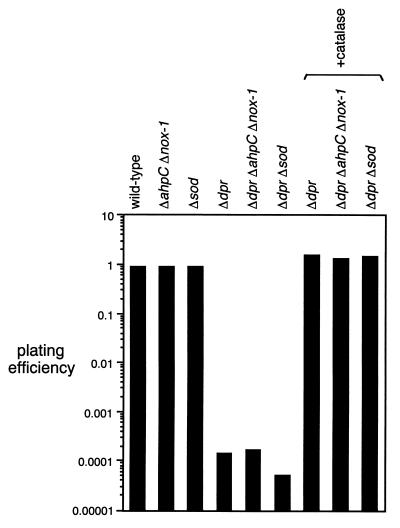
Three dpr disruption mutants of S. mutans, dpr::Spcr, dpr::Spcr ahpC::Emr::nox-1, and dpr::Spcr sod::Emr, were constructed, and their ability to form colonies was examined under both anaerobic and aerobic conditions. For disruption of dpr on the S. mutans chromosome, homologous recombination was performed between a linearized DNA fragment carrying a mutated dpr gene and the functional genomic gene. Plasmid pS23B1ES (Table 1), containing dpr::Spcr, was digested with both HindIII and SacI to obtain a 3.8-kbp linearized DNA fragment. The linearized DNA fragment was transformed into S. mutans GS-5 (wild type), BEE (ahpC::Emr::nox-1 mutant), and KD251 (sod disruption mutant). Transformants were selected on a THB-agar plate containing spectinomycin (250 μg/ml) under anaerobic conditions in an anaerobic glove box. The insertion of the Spcr-encoding gene into the dpr gene on the chromosomal DNA of the transformants was confirmed by direct PCR using a pair of primers, dpr1 and dpr3. S. mutans KD251 was constructed as described above, using plasmid pKD251 (27) containing sod interrupted by an Emr-encoding gene, and also confirmed by direct PCR using primers sod1 and sod2 (data not shown). Absence of SOD activity in crude extracts of strain KD251 cells was confirmed (data not shown). From the results obtained for the colony-forming ability of a series of dpr disruption mutants of S. mutans, the following findings became evident. (i) All dpr disruption mutants, including those generated in the ahpC::Emr::nox-1, sod::Emr, and wild-type backgrounds, were equally capable of forming colonies of normal numbers and size under anaerobic conditions (data not shown). (ii) As shown previously (18, 27), the ahpC::Emr::nox-1 double mutant and the sod::Emr mutant with an intact dpr gene showed the same level of aerobic plating efficiency as did the wild-type strain (Fig. 2), except that the colony size of the sod mutant is significantly smaller than that of the wild type in the presence of air. (iii) In contrast, all dpr disruption mutants showed a decrease of about 104-fold in the number of colonies formed compared to the wild-type strain in the presence of air (Fig. 2). Thus, we concluded that dpr is an essential gene for colony formation by S. mutans in the presence of air. The defective colony formation of the dpr mutants in the presence of air was completely reversed by addition of catalase to the plate (Fig. 2).
FIG. 2.
Aerobic plating efficiency of wild-type and mutant S. mutans. Wild-type and mutant cells of S. mutans were grown without shaking in TYG at 37°C. At exponential phase, cells were spread onto TYG plates and incubated at 37°C in an anaerobic glove box or under air. After 48 h of incubation, plating efficiency was measured as described in Materials and Methods. The data shown are the means of triplicate determinations.
Growth of dpr disruption mutants in liquid medium.
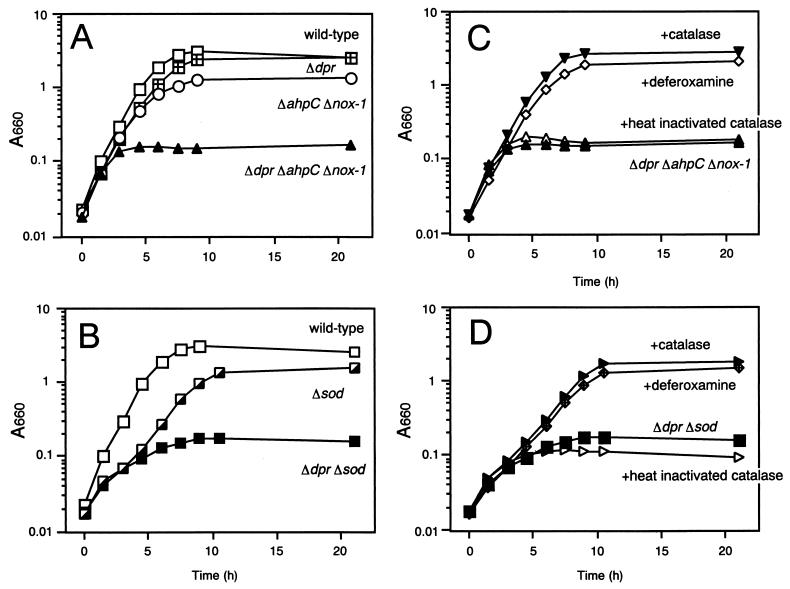
To examine dpr function in liquid cultures of S. mutans, the growth of dpr::Spcr, dpr::Spcr ahpC::Emr::nox-1, and dpr::Spcr sod::Emr mutants was measured under aerobic and anaerobic conditions and compared to that of the wild-type parent and the ahpC::Emr::nox-1 and sod::Emr mutants of S. mutans. The following findings became evident. (i) All mutants grew as well as the wild-type strain under anaerobic conditions (data not shown). (ii) Under aerobic conditions, the dpr::Spcr mutant grew as well as the wild-type strain. However, viable cells of the dpr::Spcr mutant at 21 h of incubation were reduced to approximately 40% of the wild-type level (data not shown). The growth of the ahpC::Emr::nox-1 mutant was partially inhibited (Fig. 3A). However, the dpr::Spcr ahpC::Emr::nox-1 triple mutant was severely hampered in its ability to grow aerobically (Fig. 3A). (iii) Under aerobic conditions, growth inhibition of the sod::Emr mutant was observed in the first few hours but the sod::Emr mutant grew to almost the same level as the wild type after 10 h of incubation (Fig. 3B). In contrast, the growth of the dpr::Spcr sod::Emr double mutant stopped after 10 h (Fig. 3B). (iv) Addition of catalase or deferoxamine (100 μM), which is a cell-permeating iron chelator, to the growth medium compensated for the growth defect of both the dpr::Spcr ahpC::Emr::nox-1 and dpr::Spcr sod::Emr mutants (Fig. 3C and D). Heat-inactivated catalase did not compensate for the growth defect of these mutants (Fig. 3C and D).
FIG. 3.
Aerobic growth of wild-type and mutant S. mutans in liquid medium. Wild-type and mutant cells of S. mutans were grown in TYG at 37°C without shaking. At late exponential phase, cultures were added to fresh TYG medium with or without catalase or deferoxamine and incubated aerobically at 37°C with shaking. Growth was monitored by measuring A660. Panels A and C emphasize the growth characteristics of dpr::Spcr, ahpC::Emr::nox-1, and combined mutants in the absence or presence, respectively, of catalase or deferoxamine, while panels B and D emphasize the growth characteristics of the sod::Emr and dpr::Spcr mutants in the absence or presence, respectively, of catalase or deferoxamine. The data shown are the means of triplicate determinations.
Identification and iron-binding ability of Dpr.
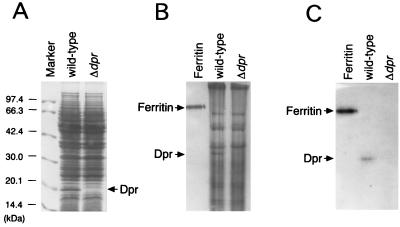
We identified the dpr gene product, Dpr, from a crude extract of wild-type S. mutans. The wild-type and dpr disruption mutant S. mutans strains were grown aerobically to early stationary phase (A660 of approximately 3.0). The crude extracts obtained from the cells by sonication were analyzed for Dpr by SDS-PAGE and native PAGE. A protein band of 20 kDa, which corresponds to the size of Dpr, was detected in wild-type cells but not in the dpr mutant (Fig. 4A). The 20-kDa protein in the SDS-polyacrylamide gel was blotted onto a polyvinylidene difluoride membrane (Millipore, Tokyo, Japan), and its amino acid sequence was determined. The N-terminal 14 residues of the protein were identical to those of the deduced amino sequence of codons 2 to 15 of Dpr. Thus, the 20-kDa protein identified by SDS-PAGE in wild-type S. mutans was found to be the dpr gene product, Dpr. Native PAGE analysis also revealed the presence of a protein band in the wild-type S. mutans crude extract whose N-terminal 11 residues were identical to those of Dpr (Fig. 4B).
FIG. 4.
Identification of Dpr and its iron-binding activity. Crude extracts from cells of wild-type and dpr::Spcr mutant S. mutans grown to early stationary phase were analyzed. (A) SDS-PAGE and staining with Coomassie brilliant blue R250. (B) Native PAGE and staining with Coomassie brilliant blue R250. (C) Native PAGE and staining with Ferene S. Forty micrograms of total protein of wild-type S. mutans or the dpr::Spcr mutant was loaded into each well. Molecular mass markers (A) and 2 μg of ferritin (B and C) were used as standards.
It is known that the Dps family of proteins form spherical complexes like ferritin and that some of them bind iron (3, 37). Therefore, the iron-binding ability of Dpr was examined. Native PAGE gels were stained with Ferene S, which is an iron-specific staining reagent. Protein bands corresponding to Dpr and ferritin (used as a positive control) were stained with Ferene S (Fig. 4C). Thus, it was demonstrated that Dpr can bind iron.
Aerobic induction of Dpr.
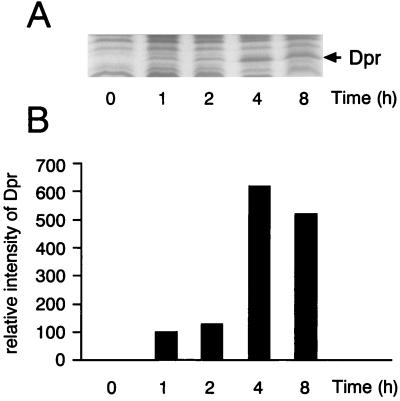
The aerobic induction of Dpr in the wild-type GS-5 strain of S. mutans was examined. At early log phase (A660 of 0.3), 50 ml of an anaerobic culture (time zero) in an anaerobic box was transferred to a 500-ml flask and incubated with shaking under air. At the times indicated in Fig. 5, samples were taken and the cells were harvested by centrifugation. After cells were disrupted, crude extracts were prepared and analyzed for the 20-kDa protein by SDS-PAGE (Fig. 5A). Dpr was not detected in cells grown anaerobically (time zero in Fig. 5B). In contrast, cells which were exposed to air started to synthesize Dpr and its relative amount per unit number of cells reached a maximum at 4 h of exposure (Fig. 5B). Thus, it was concluded that the synthesis of Dpr in GS-5 is induced by air.
FIG. 5.
Quantitative analysis of Dpr in S. mutans after exposure of cells to air. Crude extracts of S. mutans GS-5 (wild type) were analyzed before and after exposure to air for 1, 2, 4, and 8 h in liquid TYG medium. Forty micrograms of protein from the corresponding extracts was subjected to SDS-PAGE, and the gel was stained with Coomassie brilliant blue R250 (A). Bands corresponding to Dpr (20 kDa) were scanned by a Bio-Rad 620 video densitometer (B).
Comparison of amino acid sequences of Dpr, Dps, and Dps homologues.
Dps was found to be a nonspecific DNA-binding protein which accumulates in stationary-phase cells of E. coli (1). To date, three family members, including E. coli Dps, have been shown to bind to DNA for protection from oxidative stress (1, 7, 28). On the other hand, functional divergence of other Dps family proteins has also been reported, i.e., in the nonheme ferritin of Listeria innocua (3), the fine-tangled pilus major subunit of Haemophilus ducreyi (5), and the neutrophil-activating protein of Helicobacter pylori (37).
The amino acid sequence alignment of S. mutans Dpr and proteins of the Dps family which were previously reported was examined. Seven amino acid residues, H50, G55, F58, H62, E81, R82, and D144, in Dpr were conserved in Dps homologue proteins, suggesting that Dpr is a member of the Dps family. Dps family proteins commonly form multimers of a size smaller than that of ferritin (1, 3, 7, 28, 37). The multimer of E. coli Dps is a dodecamer of four-α-helix bundle monomers (12). The primary structure of Dpr showed only 27.6% identity with that of E. coli Dps, but computer prediction of the secondary structure of Dpr indicated that Dpr may also have α-helical segments similar to those of Dps, except for the presence of one additional α-helical segment (E7-S13) in the N terminus of Dpr. These results suggested that Dpr also forms spherical oligomers like ferritin. Recently, Ilari et al. solved the X-ray crystal structure of L. innocua ferritin at 2.35 Å resolution and identified five iron-binding sites, H31, H43, D47, D58, and E62 (18a). These amino acid residues were also conserved in Dpr (H50, H62, D66, D77, and E81).
DISCUSSION
Iron has toxic properties in the presence of oxygen. Iron ions stimulate the generation of highly reactive and toxic oxygen species such as hydroxyl radicals. In vitro experiments have shown that Fe2+ catalyzes nonenzymatic hydroxyl radical synthesis from H2O2 via the Fenton reaction (13), but H2O2 remained intact in the absence of iron ions at physiological pH (13). Therefore, tight regulation of the intracellular free iron ion concentration is believed to be a significant factor in organisms survival under aerobic conditions.
In this study, we have identified a new antioxidant gene, dpr, in the chromosomal DNA of S. mutans. After constructing a series of dpr::Spcr mutants which lost the ability to grow under air, we demonstrated that Dpr plays a vital role in the aerobic survival of S. mutans. Dpr was found to be a member of the Dps family of proteins, which form spherical oligomers like ferritin. In fact, Dpr was found to have iron-binding ability (Fig. 4C). Thus, a possible protective role of Dpr might be to sequester iron, thereby protecting cells from peroxides and conferring oxygen tolerance.
Dpr was induced by exposure of S. mutans to air and accumulated abundantly in S. mutans (Fig. 5). The existence of a high concentration of Dpr in S. mutans might enable intracellular free iron ion concentrations to be kept low. The ability of the ahpC::Emr::nox-1 mutant of S. mutans to grow aerobically might be due to the titration of intracellular free iron ions by Dpr (Fig. 3A), resulting in nonconversion of H2O2 to hydroxyl radicals. Neither the dpr::Spcr ahpC::Emr::nox-1 mutant nor the dpr::Spcr sod::Emr mutant could grow under air (Fig. 3A and B). In the former case, the high concentrations of intracellular H2O2 and free intracellular iron ions might prompt the conversion of H2O2 to hydroxyl radicals via the Fenton reaction and lead to cell death. Addition of catalase or deferoxamine to the growth medium complemented the defective growth of not only the former mutant but also the latter mutant (Fig. 3C and D), indicating that the defective growth of the latter mutant may also have been caused by hydroxyl radical formation. It has been reported that O2−, derived from SOD deficiency, enhances the Fenton reaction by releasing Fe2+ from iron-containing proteins (19, 24). In the dpr::Spcr sod::Emr mutant, an increase in the amount of hydroxyl radicals caused by Fe2+ would be enough to kill the cell. Wai et al. reported that a ferritin-deficient mutant of Campylobacter jejuni shows H2O2 and O2− sensitivity, indicating that sequestration of iron by ferritin contributes to oxygen tolerance (40). Touati et al. reported that nonheme ferritin, FtnA, which is overexpressed by a multicopy plasmid carrying ftnA, suppressed the iron-mediated oxygen sensitivity of E. coli (Δfur ΔrecA) (38). These findings agree well with the results we obtained with S. mutans.
One more significant role of protection by Dps and Dps family proteins was considered to be DNA-binding ability. Wolf and Hammes have shown that E. coli Dps forms an extensive crystalline lattice in the presence of DNA in vitro and in vivo, proposing that the DNA-Dps cocrystallization mode provides protection of DNA by sequestration (42). On the other hand, it was reported that two members of the Dps family of proteins, L. innocua ferritin and H. pylori HP-NAP, function as authentic ferritin and cannot bind DNA (3, 37). To elucidate the details of the mechanism of the aerotolerance conferred by Dpr, the molecular properties of Dpr, including its DNA-binding ability, should be explored.
All three mutants of S. mutans, dpr::Spcr, dpr::Spcr ahpC::Emr::nox-1, and dpr::Spcr sod::Emr, were unable to form colonies on solid media under air (Fig. 2). Unlike the results obtained with liquid cultures, the ability to form colonies was absolutely dependent on Dpr. Addition of catalase to agar plates compensated for the growth defect (Fig. 2), indicating that there exists a significant amount of H2O2 in the cell in spite of the presence of ahpC and nox-1. Our previous study indicated that the level of expression of AhpC, which is the peroxidatic protein of the bicomponent peroxidase system, was low during the early growth phase under air (18). At an early growth phase of S. mutans on solid medium, the cells would also contain a small amount of AhpC protein. Therefore, hydroxyl radicals could be generated from the remaining H2O2 to cause cell death in the absence of Dpr on the solid medium. In contrast, the dpr::Spcr mutant could grow aerobically in liquid medium. As H2O2 is a highly diffusible oxidant in liquid medium, endogenously synthesized H2O2 might permeate the cell membrane and be released easily from the cells until its intracellular and extracellular concentrations reached equilibrium (11). Consequently, at early exponential phase, when both cell density and AhpC levels are low, the intracellular concentration of H2O2 might be kept low enough to enable dpr::Spcr mutants to grow (Fig. 3A). From mid-log phase (A660 of 0.2), the increase in cell mass would lead to increased production of H2O2, causing both internal and external accumulation of H2O2. At this stage, the dpr::Spcr ahpC::Emr::nox-1 mutant could not grow, as mentioned above.
Dpr homologues were also present in other streptococci, including Streptococcus pyogenes (B. A. Roe, S. Clifton, M. McShan, and J. Ferretti, Streptococcal Genome Sequencing Project at the University of Oklahoma) and Streptococcus pneumoniae (29), and their deduced amino acid sequences showed 75.4 and 57.1% identity, respectively, with that of S. mutans Dpr. To date, nonheme peroxidases (30, 31), a pseudocatalase (20), and oxidases (2, 10, 16, 34) of lactic acid bacteria, including streptococci, have been reported to substitute for catalase, which is present in other aerotolerant bacteria but not lactic acid bacteria, and some of them, including SOD, were proven to play a role in aerotolerance (9, 10, 21, 27, 33). Unlike them, Dpr did not directly react with oxygen and reactive oxygen species. The iron-binding ability of Dpr might indirectly contribute oxygen tolerance in S. mutans. This mechanism of aerotolerance conferred by Dpr is rational for lactic acid bacteria that are known to lack catalase and to require only low levels of iron for growth.
ACKNOWLEDGMENTS
We are grateful to K. Nakayama of Kyushu University for the gift of plasmid pKD251 for disruption of sod in S. mutans. We are also grateful to N. Takahashi of Tohoku University and E. Sato of Osaka City University for their fruitful discussion.
REFERENCES
- 1.Almiron M, Link A J, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 2.Auzat I, Chapuy-Regaud S, Le Bras G, Dos Santos D, Ogunniyi A D, Le Thomas I, Garel J R, Paton J C, Trombe M C. The NADH oxidase of Streptococcus pneumoniae: its involvement in competence and virulence. Mol Microbiol. 1999;34:1018–1028. doi: 10.1046/j.1365-2958.1999.01663.x. [DOI] [PubMed] [Google Scholar]
- 3.Bozzi M, Mignogna G, Stefanini S, Barra D, Longhi C, Valenti P, Chiancone E. A novel non-heme iron-binding ferritin related to the DNA-binding proteins of the Dps family in Listeria innocua. J Biol Chem. 1997;272:3259–3265. doi: 10.1074/jbc.272.6.3259. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive methods for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Brentjens R J, Ketterer M, Apicella M A, Spinola S M. Fine tangled pili expressed by Haemophilus ducreyi are a novel class of pili. J Bacteriol. 1996;178:808–816. doi: 10.1128/jb.178.3.808-816.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang A C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Helmann J D. Bacillus subtilis MrgA is a Dps(PexB) homologue: evidence for metalloregulation of an oxidative-stress gene. Mol Microbiol. 1995;18:295–300. doi: 10.1111/j.1365-2958.1995.mmi_18020295.x. [DOI] [PubMed] [Google Scholar]
- 8.Chung M C. A specific iron stain for iron-binding proteins in polyacrylamide gels: application to transferrin and lactoferrin. Anal Biochem. 1985;148:498–502. doi: 10.1016/0003-2697(85)90258-1. [DOI] [PubMed] [Google Scholar]
- 9.Gibson C M, Caparon M G. Insertional inactivation of Streptococcus pyogenes sod suggests that prtF is regulated in response to a superoxide signal. J Bacteriol. 1996;178:4688–4695. doi: 10.1128/jb.178.15.4688-4695.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson C M, Mallett T C, Claiborne A, Caparon M G. Contribution of NADH oxidase to aerobic metabolism of Streptococcus pyogenes. J Bacteriol. 2000;182:448–455. doi: 10.1128/jb.182.2.448-455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Flecha B, Demple B. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J Biol Chem. 1995;270:13681–13687. doi: 10.1074/jbc.270.23.13681. [DOI] [PubMed] [Google Scholar]
- 12.Grant R A, Filman D J, Finkel S E, Kolter R, Hogle J M. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 13.Halliwell B, Gutteridge J M. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi M. Effect of oxygen on the growth and mannitol metabolism of Streptococcus mutans. J Gen Microbiol. 1984;130:1819–1826. doi: 10.1099/00221287-130-7-1819. [DOI] [PubMed] [Google Scholar]
- 16.Higuchi M, Shimada M, Yamamoto Y, Hayashi T, Kamio Y. Identification of two distinct NADH oxidases corresponding to H2O2-forming oxidase and H2O-forming oxidase induced in Streptococcus mutans. J Gen Microbiol. 1993;139:2343–2351. doi: 10.1099/00221287-139-10-2343. [DOI] [PubMed] [Google Scholar]
- 17.Higuchi M, Shimada M, Matsumoto J, Yamamoto Y, Rhaman A, Kamio Y. Molecular cloning and sequence analysis of the gene encoding the H2O2-forming NADH oxidase from Streptococcus mutans. Biosci Biotech Biochem. 1994;58:1603–1607. doi: 10.1271/bbb.58.1603. [DOI] [PubMed] [Google Scholar]
- 18.Higuchi M, Yamamoto Y, Poole L B, Shimada M, Sato Y, Takahashi N, Kamio Y. Functions of two types of NADH oxidases in energy metabolism and oxidative stress of Streptococcus mutans. J Bacteriol. 1999;181:5940–5947. doi: 10.1128/jb.181.19.5940-5947.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Ilari A, Stefanini S, Chiancone E, Tsernoglou D. The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site. Nat Struct Biol. 2000;7:38–43. doi: 10.1038/71236. [DOI] [PubMed] [Google Scholar]
- 19.Keyer K, Imlay J A. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci USA. 1996;93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kono Y, Fridovich I. Isolation and characterization of the pseudocatalase of Lactobacillus plantarum. J Biol Chem. 1983;258:6015–6019. [PubMed] [Google Scholar]
- 21.Kono Y, Fridovich I. Functional significance of manganese catalase in Lactobacillus plantarum. J Bacteriol. 1983;155:742–746. doi: 10.1128/jb.155.2.742-746.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.LeBlanc D J, Lee L N, Inamine J M. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob Agents Chemother. 1991;35:1804–1810. doi: 10.1128/aac.35.9.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liochev S I, Fridvich I. The role of O2− in the production of OH ·: in vitro and in vivo. Free Radic Biol Med. 1994;16:29–33. doi: 10.1016/0891-5849(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 25.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Mongkolsuk S, Praituan W, Loprasert S, Fuangthong M, Chamnongpol S, Maciver I, Hansen E J. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J Bacteriol. 1998;180:2636–2643. doi: 10.1128/jb.180.10.2636-2643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakayama K. Nucleotide sequence of Streptococcus mutans superoxide dismutase gene and isolation of insertion mutants. J Bacteriol. 1992;174:4928–4934. doi: 10.1128/jb.174.15.4928-4934.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pena M M, Bullerjahn G S. The DpsA protein of Synechococcus sp. strain PCC7942 is a DNA-binding hemoprotein. Linkage of the Dps and bacterioferritin protein families. J Biol Chem. 1995;270:22478–22482. doi: 10.1074/jbc.270.38.22478. [DOI] [PubMed] [Google Scholar]
- 29.Pikis A, Donkersloot J A, Rodriguez W J, Keith J M. A conservative amino acid mutation in the chromosome-encoded dihydrofolate reductase confers trimethoprim resistance in Streptococcus pneumoniae. J Infect Dis. 1998;178:700–706. doi: 10.1086/515371. [DOI] [PubMed] [Google Scholar]
- 30.Poole L B, Higuchi M, Shimada M, Calzi M L, Kamio Y. Streptococcus mutans H2O2-forming NADH oxidase is an alkyl hydroperoxide reductase protein. Free Radic Biol Med. 2000;28:108–120. doi: 10.1016/s0891-5849(99)00218-x. [DOI] [PubMed] [Google Scholar]
- 31.Ross R P, Claiborne A. Cloning, sequence and overexpression of NADH peroxidase from Streptococcus faecalis 10C1. Structural relationship with the flavoprotein disulfide reductases. J Mol Biol. 1991;221:857–871. doi: 10.1016/0022-2836(91)80180-3. [DOI] [PubMed] [Google Scholar]
- 32.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 33.Sanders J W, Leenhouts K J, Haandrikman A J, Venema G, Kok J. Stress response in Lactococcus lactis: cloning, expression analysis, and mutation of the lactococcal superoxide dismutase gene. J Bacteriol. 1995;177:5254–5260. doi: 10.1128/jb.177.18.5254-5260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt H L, Stocklein W, Danzer J, Kirch P, Limbach B. Isolation and properties of an H2O-forming NADH oxidase from Streptococcus faecalis. Eur J Biochem. 1986;156:149–155. doi: 10.1111/j.1432-1033.1986.tb09560.x. [DOI] [PubMed] [Google Scholar]
- 35.Spath C, Kraus A, Hillen W. Contribution of glucose kinase to glucose repression of xylose utilization in Bacillus megaterium. J Bacteriol. 1997;179:7603–7605. doi: 10.1128/jb.179.23.7603-7605.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storz G, Jacobson F S, Tartaglia L A, Morgan R W, Silveira L A, Ames B N. An alkyl hydroperoxide reductase induced by oxidative stress in Salmonella typhimurium and Escherichia coli: genetic characterization and cloning of ahp. J Bacteriol. 1989;171:2049–2055. doi: 10.1128/jb.171.4.2049-2055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tonello F, Dundon W G, Satin B, Molinari M, Tognon G, Grandi G, Del Giudice G, Rappuoli R, Montecucco C. The Helicobacter pylori neutrophil-activating protein is an iron-binding protein with dodecameric structure. Mol Microbiol. 1999;34:238–246. doi: 10.1046/j.1365-2958.1999.01584.x. [DOI] [PubMed] [Google Scholar]
- 38.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vance G P, Keele B B. Superoxide dismutase from Streptococcus mutans. J Biol Chem. 1972;247:4782–4786. [PubMed] [Google Scholar]
- 40.Wai S N, Nakayama K, Umene K, Moriya T, Amako K. Construction of a ferritin-deficient mutant of Campylobacter jejuni: contribution of ferritin to iron storage and protection against oxidative stress. Mol Microbiol. 1996;20:1127–1134. doi: 10.1111/j.1365-2958.1996.tb02633.x. [DOI] [PubMed] [Google Scholar]
- 41.Wetherell J R, Jr, Bleiweis A S. Antigens of Streptococcus mutans: characterization of a polysaccharide antigen from walls of strain GS-5. Infect Immun. 1975;12:1341–1348. doi: 10.1128/iai.12.6.1341-1348.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf G, Hammes W P. Effect of hematin on the activities of nitrite reductase and catalase in lactobacilli. Arch Microbiol. 1988;149:220–224. [Google Scholar]
- 43.Wolf S G, Frenkiel D, Arad T, Finkel S E, Kolter R, Minsky A. DNA protection by stress-induced biocrystallization. Nature. 1999;400:83–85. doi: 10.1038/21918. [DOI] [PubMed] [Google Scholar]
- 44.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]