Abstract
Simple Summary
Turtles are threatened all over the world. Malaysia has 24 species of turtles. This review focuses on current conservation status and some requirements for sustainability. We propose integrating concepts of ecology and molecular biology to provide almost comprehensive turtle reviews in Malaysia.
Abstract
Approximately 356 species of turtles inhabit saltwater and freshwater habitats globally, except in Antarctica. Twenty-four species of turtles have been reported in Malaysia, four of which are sea turtles. The state of Terengganu harbored the highest number of turtles, with 17 different reported species. Based on the IUCN Red List, 29% of turtle species in Malaysia are critically endangered. In comparison, another 25% are classified as endangered. Likewise, CITES reported that 67% of Malaysia’s turtles are threatened, while 25% are classified as critically endangered. This review discusses the checklists, molecular genetics work, conservation status, recent trends, and recommendations for future research. Factors contributing to their population declines and current endangered status are also discussed.
Keywords: taxonomic, sea turtles, IUCN Red List, CITES, checklist, genetic, endangered
1. Introduction
There are approximately 356 turtles living on land on every continent, except for in Antarctica, as well as in salt water and fresh water [1]. The term “turtle” is frequently used to refer to sea turtles, which only rarely leave the sea [2]. Sea turtles belong to the Cheloniidae families, except for the Leatherback turtle, which is the only genus in the Family Dermochelyidae and has a leathery carapace [3]. The seven species of sea turtles are Green turtle (Chelonia mydas), Hawksbill turtle (Eretmochelys imbricata), Leatherback turtle (Dermochelys coriacea), Loggerhead turtle (Caretta caretta), Flatback turtle (Natator depressa), Olive ridley turtle (Lepidochelys olivacea), and Kemp’s ridley turtle (Lepidochelys kempii) [4]. Malaysia is home to four sea turtle species: the Leatherback turtle, the Green turtle, the Olive ridley turtle, and the Hawksbill turtle [5]. With nearly 40% of its total body mass made up of bone, the turtle is possibly the most organized form of animal armor ever to appear [6]. As a result, this great armor is most likely why turtles appeared on the scene over 200 million years ago and miraculously survived the extinction of the dinosaurs and other devastating events [6].
In Testudines’ order, turtles are any reptile with a hard shell around its body, including tortoises [7]. They have anatomical characteristics that set them apart from other turtles. In the Chelonia order, a turtle, a tortoise, and a terrapin are all names for hard-shelled egg-laying reptiles [8]. However, the specific expression used for a particular turtle can vary depending on its natural surroundings. For example, the term “turtle” usually refers to turtles that have spent their entire lives in or near water [9]. The term “tortoise” is commonly used to refer to turtles that spend most of their time ashore, eating bushes, grass, and fruit [10]. Unlike other turtle family members, tortoises do not have webbed feet because they do not spend much time in the water [11]. Terrapins are turtles that invest energy in fresh and brackish water [12]. “Terrapin” is derived from an Algonquian Indian word that means “a small turtle” [13]. Malaysia is home to 20 different kinds of turtles. Two of them, Pelodiscus sinensis and Trachemys scripta, were brought there from other places [14]. In total, Malaysia has 24 different species of turtles [15].
The most established bodies in conservation biodiversity are the IUCN (International Union for Conservation of Nature) and CITES (Convention on International Trade in Endangered Species of Wild Fauna and Flora). The IUCN Red List is a rich compendium of information on threats, ecological requirements, habitats of species, and conservation actions that can be taken to prevent extinctions [16,17]. It is a target framework for evaluating the extinction risk of species depending on past, present, and extended threats [18]. The assessments are carried out by following a standardized procedure that employs the rigorous IUCN Red List Categories and Criteria, ensuring the highest scientific documentation standards, information management, expert review, and justification [16,19]. More assessments will aid in developing the IUCN Red List as a completely comprehensive “Barometer of Life” [20]. Threatened species include those listed as critically endangered, endangered, or vulnerable [21].
In contrast, CITES is an international agreement among governments [22]; it ensures that international trade in wild animals and plant specimens does not threaten their numbers [23]. Due to the international nature of the trade in wildlife, its regulation requires international cooperation to protect certain species from over-exploitation [24].
Today, it protects more than 37,000 plant and animal species in different ways, depending on whether they are traded either as dried herbs, live specimens, or fur coats [25] CITES protects approximately 5950 animals and 32,800 plant species from over-exploitation through international trade [26]. They are listed in the three CITES Appendices [27]. The species are organized in the appendices based on their vulnerability to the global trade.
Therefore, the turtle checklist in Malaysia was presented previously, but the current conservation status needs to be updated. Thus, we provide the species checklist of turtles in Malaysia, document the current status of turtles in Malaysia, and compile the turtle research trends nowadays. We also review the causes of current precipitous declines in turtle populations by examining the threatened status of turtles in Malaysia and speculate on the potential reasons for extinction in the coming century. In doing so, we make a list of common threats and describe the research that needs to be performed in order to ensure long-term survival.
2. Turtle Types in Malaysia
The “hard-shelled” Cheloniidae advanced around 60 million years ago, and the “delicate-shelled” Dermochelyidae developed approximately 90 million years ago [28,29]. The Cheloniidae contain six surviving species in five genera: the Flatback turtle (Natator depressus), the Green turtle (Chelonia mydas), the Hawksbill turtle (Eretmochelys imbricata), the Loggerhead turtle (Caretta caretta), the Kemp’s ridley turtle (Lepidochelys kempii), and the Olive ridley turtle (Lepidochelys olivacea). The Leatherback turtle (Dermochelys coriacea) is the only extant species in the Dermochelyidae family [30].
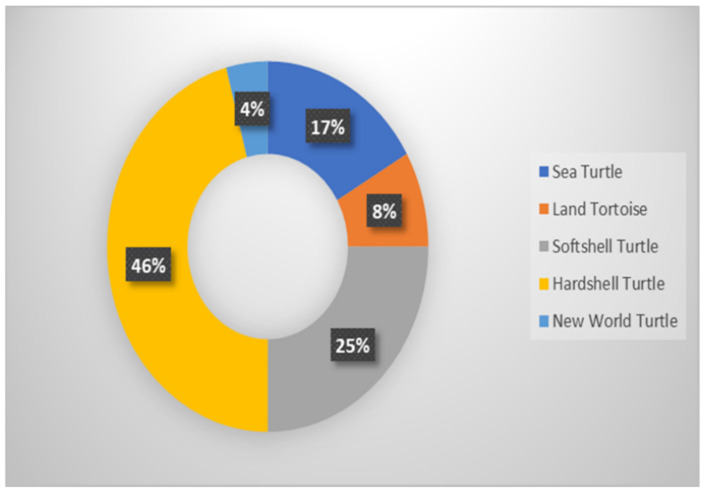
There are seven sea turtle species (Cheloniidae and Dermochelyidae) worldwide, with five nesting in Southeast Asia. Some of these species are shown in Supplementary Table S1, and Figure 1 shows that 17% of the studied sea turtles can be found locally. The other two species are the Kemp’s ridley turtle, which only lives in the Western Atlantic Ocean, and the Flatback turtle, which only lives in Australia (though its feeding grounds extend into Eastern Indonesia) [31].
Figure 1.
The diversity of turtles in Malaysia.
Testudinidae (Land Tortoises): 60 land tortoise species are recognized worldwide, accounting for 8% of the 24 turtle species in Malaysia (Figure 1). Most of them live in semi-arid, dry open habitats, including grasslands and deserts [32]. Only five Southeast Asian species have adapted to different environments, including humid, forested habitats, and cooler temperatures in a lower montane forest [31,33]. Supplementary Table S1 shows some examples.
Asian hard-shell turtles (Geoemydidae) have the most turtle family species, with approximately 40 species of 14 genera found in Southeast Asia and 70 species found worldwide. Malaysia has the most of this turtle type (46%) (Figure 1). DNA sequencing has recently revealed hidden diversity in this group; for example, the Cyclemys leaf turtles are now classified as six distinct species [31]. Supplementary Table S1 shows some examples.
The big-headed turtle (Platysternon megacephalum) is mainly the only individual from this family, Platysternidae; it is remembered for the superfamily Testudinoidea, which likewise incorporates the Testudinidae (land turtles), Geoemydidae (Asian hard-shelled turtles), and Emydidae (new world reptiles). This turtle has a large head that it cannot retract into its shell [34].
Softshell turtles (Trionychidae) have 30 species worldwide [35], with 15 species in Southeast Asia and six found in Malaysia (Supplementary Table S1). They also have a flexible, rugged carapace [36], and Southeast Asia is home to approximately half of the world’s softshell turtles [31,33].
The snake-necked turtles of the Chelodina family are an ancient group of expert fish-eaters whose long necks must be turned sideways to reach beneath the carapace [37]. There are 16 species in the world under this family [31,38].
The pig-nosed turtle (Carettochelys insculpta) is the only species in the Carettochelyidae family [39]. It is found in just three nations—Indonesia, Papua, New Guinea, and Australia [40]. These species are kept as pets in Malaysia. Their flippers resemble those of sea turtles, and their carapace is rough, but their most unique feature is their pig-like nose [31,41].
Turtles from the Emydidae family originated in the Americas and are widely available in the world as the most popular pet [42]. For instance, consider the Red-Eared Slider [31].
The turtles in the family Emydidae belong to the order Testudines and the suborder Cryptodira. There are about 52 species in this family, which is divided into 12 genera: Actinemys, Chrysemys, Clemmys, Deirochelys, Emydoidea, Emys, Glyptemys, Graptemys, Malaclemys, Pseudemys, Terrapene, and Trachemys [43]. Except for Trachemys, which is found in South America and the West Indies, and Emys, which inhabits Southern Europe, Northern Africa, and Western Asia, all of these species are restricted to North America [44]. The relationships between the 12 genera and the species that make up the family are yet unknown [45].
3. Molecular Research Trends
3.1. Mating System
In turtles, molecular markers have been used to describe the mating system in the existing species, with multiple paternity and sperm storage being distinguishing features [46,47,48,49]. In contrast, the IUCN lists the Hawksbill turtle (Eretmochelys imbricata) as critically endangered [50,51]. Eretmochelys imbricata was shown to be primarily monogamous in two studies [52,53] so that sperm could be stored [53]. Nonetheless, one of these examinations was conducted in an isolated area of Malaysia [52]. Moreover, the other one was found in a remote part of the Republic of Seychelles [53]. It has been suggested that different sea turtles may have different levels of paternity depending on where they live and how abundant there are [47,48,54].
3.2. Population Genetics Analysis
Table 1 briefly describes genetic markers and how they were used in turtle studies. Prior studies have also used multilocus minisatellites (“DNA fingerprinting”) [55]. Microsatellites, along with arbitrary intensified polymorphic DNA (RAPDs) [56], allozymes [57,58,59,60], and the anonymous single-copy nuclear DNA (scnDNA) [61,62,63,64,65], are the most commonly used markers for paternity analysis. Other studies have used microsatellites to analyze population genetics [46,64,66,67,68,69,70,71,72] and molecular evolution [73,74]. A few were reported to be the development of new markers [75,76] or developed attributes for species identification [77]. Microsatellite markers were compared to mtDNA haplotype markers, which are passed down from the mother and have been used for a long time to study the genetic structure and phylogeography of turtle populations [48,67,68].
3.3. Genetic Variability
Due to molecular markers in various fields of knowledge, there has been a significant advancement in molecular genetics techniques in recent years. These markers are practical and risk-free tools that can facilitate an accurate diagnosis. Comparative studies of the genetic variation described by molecular markers and regional morphometric patterns have centered on several wild turtle species [78]. The following molecular markers (Table 1) stand out among those used to detect genetic variability in DNA sequences, such as restriction fragment length polymorphism (RFLP), random amplified polymorphic DNA (RAPD), single nucleotide polymorphism (SNP), mitochondrial DNA (mtDNA), and microsatellites or short tandem repeats (STR) [79]. They stand out because they provide ecological and morphological data that can be used to make different plans for protecting most genetic resources [48].
For set preservation systems, molecular markers help us to better understand the genetic variety of faunal species and segment relations. The low genetic variety and star-shaped haplotype network of sea turtles in the Eastern Pacific suggest that the species most likely originated in the Indo-Pacific on a generally late transformative timescale [80,81]. Most sea turtles can be found all over the world, meaning that natural damage and environmental loss will probably affect their movements [82,83], assuming that these effects reduce availability and population size. If that happens, genetic diversity will be lost, and this will increase the risk of extinction [84] caused by potential inbreeding depression, which makes animals less able to change and reduces fitness [85].
The rate at which genetic variability is lost in specific animal varieties is a component of the successful population size. This boundary is impacted by an organic entity’s characteristic history and segment history [85]. An assessment of crucial boundaries that shape compelling population size is expected to measure the danger of hereditary disintegration and inbreeding depression [86]. The principal is information on both quality streams via an animal type’s geographic appropriation and the mating framework. The mating framework fact is that, when the successful population’s size is less than the registered size, the regenerative slant is high [87].
Table 1.
Genetic markers for molecular turtle research.
| Allozymes | Alleles of a catalyst can be identified by various electrophoretic versatility (usually starch or cellulose acetic acid derivation gels). Since mtDNA haplotypes and microsatellites have become more popular as markers for population genetics and paternity analysis [48,57,88], a well-developed method has been used only a few times in turtle concentrates. |
| Anonymous scnDNA |
Various PCR preliminaries have been created for sea turtles to intensify mysterious single-copy nuclear DNA loci. Variety in intensified items is inspected by RFLP or sequencing and has not been utilized much since microsatellites became the more mainstream nuclear DNA marker [48,61,62,63,64,65,89]. |
| Microsatellites | Pair rehashes of a 1–6 bp “core” grouping. The evaluation strategy gives single-locus data. Bespoke introductions for PCR intensification are intended for the microsatellite’s flanking areas. The hypervariable idea of a microsatellite is several rehashes of the left change effectively. Moreover, single-locus information implies that more impressive scientific strategies are conceivable than multilocus fingerprinting. It has become a mainstream marker for the population’s genetic qualities. It is a technique for determining paternity in turtles [48,90,91,92]. |
| Minisatellites | This is the first DNA fingerprinting technique. Dispersed across the nuclear genome are families of tandemly repetitive “minisatellite” regions sharing a 10–15 bp “core” sequence. Variation in the recurrent number is acquired. Moreover, minisatellites are exceptionally polymorphic (hypervariable) as genetic markers at the individual and population levels. It has been used only once for sea turtles since microsatellites became the more common marker [55,93]. |
| MtDNA Haplotypes | For sea turtles, it is normal to utilize arrangement variety in the control district of mitochondrial DNA (mtDNA). These are named “haplotypes” because the mitochondrial genome exits as a single copy (haploid). An approximately 400 bp piece is enhanced with a standard arrangement of groundworks. In current examinations, the variety is composed of the improved items by sequencing. More seasoned investigations may utilize Restriction Fragment Length Polymorphism (RFLP) or other fast screening methods. The haplotypes at the Archie Carr Center for Sea Turtle Research are put in order by a normalized classification [94,95]. |
| RAPDs | Random Amplified Polymorphic DNA (RAPD). A PCR-based procedure utilizes short (10 bp) oligonucleotide primers in random arrangement to create different PCR results of contrasting sizes isolated on an agarose gel. The multilocus technique was momentarily famous in molecular ecology because of its modest and straightforward convention. However, it tumbled from favor when reproducibility turned into an issue. I am mindful of just one turtle study that utilized it [56,96,97]. |
| RFLP | Restriction Fragment Length Polymorphism (RFLP). Limitation compounds have the potential to cut DNA at explicit acknowledgement groupings. In order to create parts of reproducible size from any substrate DNA particle (nuclear or mitochondrial DNA), they should be be isolated by the electrophoresis process. The variation between individuals in the sizes of DNA fragments is caused by mutations that create or eliminate restriction enzyme recognition sequences and is used as an indication of genetic variation. It was previously popular for assessing mtDNA haplotype variation in turtles, but direct sequencing has largely replaced RFLP. In addition, it was briefly used in turtle studies for anonymous scnDNA [48,61,62,63,64,65,98,99]. |
4. Conservation Status
4.1. The Status of the IUCN Red List
The IUCN Red List categorizes species into nine groups (Table 2), which Reference [100] defined based on population size, rate of decline, geographic distribution area, fragmentation distribution, and population degree. The importance of applying any measures without extensive information, including suspicion and potential future threats, is emphasized “so long as these can reasonably be supported” [19]. The “Threatened” category includes “Critically Endangered”, “Endangered”, and “Vulnerable” [21] on its Red List.
Table 3 shows that Malaysia has 24 turtle species, four of which are sea turtles, and the other 20 are freshwater turtles (two of which are introduced species) [14,101]. According to the IUCN Red List, a sea turtle (Eretmochelys imbricata) and six freshwater turtle populations (Manouria emys, Batagur affinis, Orlitia borneensis, Batagur borneoensis, Indotestudo elongata, and Chitra chitra) are critically endangered in Malaysia (Figure 2). In contrast, a sea turtle (Chelonia mydas) and five freshwater turtles (Heosemys annandalii, Cuora amboinensis, Heosemys spinosa, Chitra indica, and Pelochelys cantorii) were endangered in Malaysia. Two sea turtles (Dermochelys coriacea and Lepidochelys olivacea) and six freshwater turtles were vulnerable (Malayemys macrocephala, Notochelys platynotan, Siebenrockiella crassicollis, Amyda cartilaginea, Manouria iimpressa, and Pelodiscus sinensis). However, two sea turtles were reported by Reference [102] in The ASEAN Post; a source from the World Wildlife Foundation (WWF) Malaysia shows that the Leatherback turtle is critically endangered, and the Olive Ridley turtle is endangered in the Malaysian ocean. Moreover, one species, Cyclemys dentata, is near threatened, and two species, Dogania subplana and Trachemys scoundaripta, are less concerned.
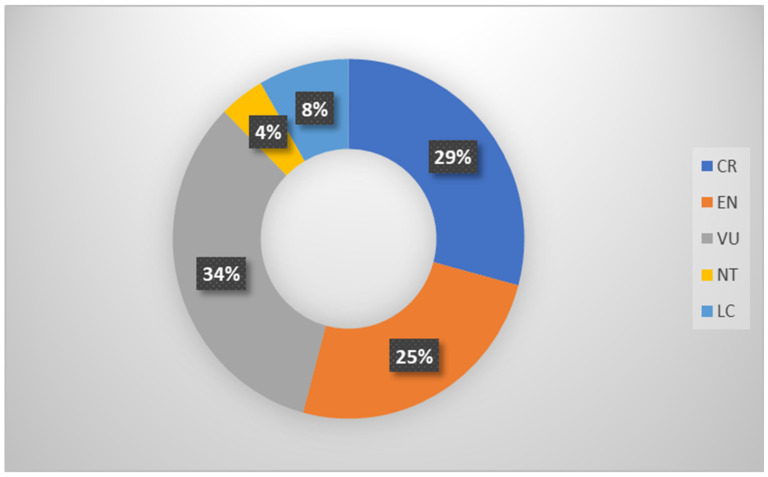
All the turtle species are distributed all over Malaysia. However, Terengganu is home to 17 species, including four species of sea turtles (Chelonia mydas, Dermochelys coriacea, Lepidochelys olivacea, and Eretmochelys imbricata) and 13 species of freshwater turtles (Trachemys scripta, Batagur affinis, Batagur borneonsis, Coura amboinensis, Siebenrockiella crassicollis, Manouria emys, Amyda cartilaginea, Dogania subplana, and Pelochelys cantorii) [14]. In addition, referring to Figure 3, the IUCN Red List analysis shows that 29 percent of Malaysia’s turtle species are critically endangered and 25 percent are endangered.
Table 2.
| Classification | Describtion |
|---|---|
| Not evaluated (NE) | Not yet assessed by the IUCN, they indicate species that have not been reviewed enough to be assigned to a category. |
| Data deficiency (DD) | Offering insufficient information for a proper assessment of conservation status to be made. |
| Least concern (LC) | It is unlikely to become extinct soon. |
| Near threatened (NT) | Close to being at an increased risk of extinction soon. |
| Vulnerable (VU) | It is considered at an increased risk of unnatural (human-caused) extinction without further human intervention. |
| Endangered (EN) | A very high risk of extinction in the wild. |
| Critically endangered (CR) | Points in a particular and extremely critical state. |
| Extinct in the wild (EW) | Point only lives on in zoos, farms, and places outside of its native range, as surveys have shown. |
| Extinct (EX) | Beyond a reasonable doubt, the species is no longer extant. |
Table 3.
| Common Name | Scientific Name | GenBank Accession |
IUCN Red List Status | CITES Appendix |
Reference |
|---|---|---|---|---|---|
| Asian Narrow Headed Softshell Turtle | Chitra chitra | HQ329770 | CR | I | [105] |
| Hawksbill Turtle | Eretmochelys imbricata | GQ152887 | CR | I | [71] |
| Southern River Terrapin | Batagur affinis | MN069310 | CR | I | [106] |
| Asian Giant Tortoise | Manouria emys | KP268838 | CR | II | [107] |
| Elongated Tortoise | Indotestudo elongata | KP268857 | CR | II | [108] |
| Malaysian Giant Turtle | Orlitia borneensis | HQ329693 | CR | II | [105] |
| Painted Terrapin | Batagur borneoensis | HQ329672 | CR | II | [105] |
| Green Turtle | Chelonia mydas | MN124278 | EN | I | [109] |
| Asian Giant Softshell Turtle | Pelochelys cantorii | HQ329785 | EN | II | [105] |
| Indian Narrow-headed Softshell Turtle | Chitra indica | HQ329771 | EN | II | [105] |
| Malaysian Box Turtle | Cuora amboinensis | JN860217 | EN | II | [108] |
| Spiny Turtle | Heosemys spinosa | HQ329684 | EN | II | [105] |
| Yellow-headed Temple Turtle | Heosemys annandalii | HQ329681 | EN | II | [105] |
| Leatherback Turtle | Dermochelys coriacea | KU883273 | VU | I | [110] |
| Olive Ridley Turtle | Lepidochelys olivacea | KF894766 | VU | I | [111] |
| Asiatic Softshell Turtle | Amyda cartilaginea | HQ329768 | VU | II | [105] |
| Black Marsh Turtle | Siebenrockiella crassicollis | HQ329704 | VU | II | [105] |
| Impressed Tortoise | Manouria impressa | GQ867670 | VU | II | [112] |
| Malayan Flat-shelled Turtle | Notochelys platynota | HQ329692 | VU | II | [105] |
| Malayan Snail-eating Turtle | Malayemys macrocephala | HQ329686 | VU | II | [105] |
| Chinese Softshell Turtle | Pelodiscus sinensis | JQ844545 | VU | None | [113] |
| Asian Leaf Turtle | Cyclemys dentata | HQ329676 | NT | II | [105] |
| Malayan Softshell Turtle | Dogania subplana | NC_002780 | LC | II | [114] |
| Yellow-bellied Slider Turtle | Trachemys scripta | JF700194 | LC | None | [115] |
Figure 2.
The critically endangered turtles in Malaysia. Top left to right: Chitra chitra [116], Manouria emys [117], Eretmochelys imbricata [118], Batagur borneensis [119], Indotestudo elongata [120], Orlitia borneensis [121], and Batagur affinis [122].
Figure 3.
Chart of IUCN Red List status on turtles.
4.2. The CITES Appendices
The Convention’s Appendices I, II, and III are lists of species with different levels of protection from over-exploitation [123]. Appendix I lists the most endangered plants and animals on the CITES list. They are almost extinct, but CITES allows international trade in specimens of these species as long as the import is not for commercial use (i.e., a scientific research study) [124].
In Appendix II, there is a list of species that are not threatened with extinction right now, but if the trade is not controlled, there is a high chance that they will be in the future. It also includes supposed “similar species”, such as species whose standards in exchange resemble species recorded for conservation purposes. Trade-in specimens of Appendix-II species may be authorized by issuing an export permit or re-export permit certificate. No import permit is necessary for these species under CITES (although a permit is needed in some countries with stricter measures than CITES requires) [125].
Appendix III contains a list of species added at the request of a party that already regulates international trade in the species. Specimens of the species in this appendix can be traded around the world only if the proper permits or certificates are shown [123].
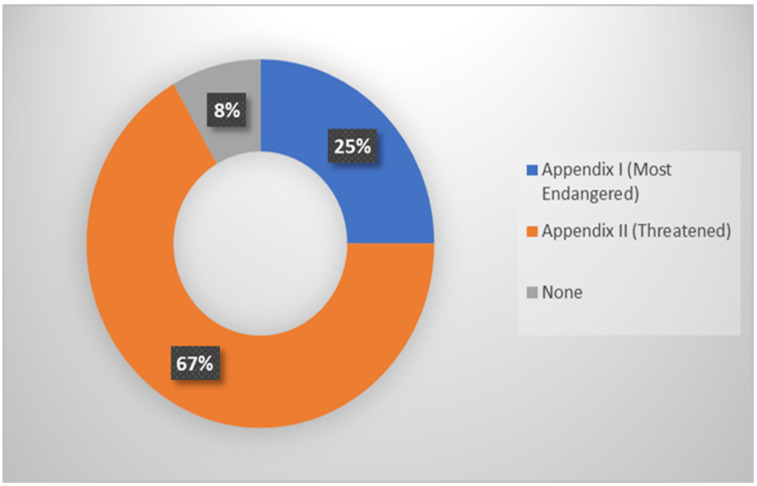
An analysis of Figure 4 reveals that CITES has classified Malaysian turtles as 67 percent threatened, including Manouria emys, Orlitia borneensis, Batagur borneoensis, Indotestudo elongate, Heosemys annandalii, Cuora amboinensis, Heosemys spinosa, Chitra indica, Pelochelys cantorii, Malayemys macrocephala, Notochelys platynotan, Siebenrockiella crassicollis, Amyda cartilaginea, Manouria impressa, Cyclemys dentata, and Dogania subplana. About 25% (Batagur affinis, Chitra chitra, Eretmochelys imbricata, Chelonia mydas, Dermochelys coriacea, and Lepidochelys olivacea) are the most endangered species.
Figure 4.
Chart of CITES’s appendices on turtles.
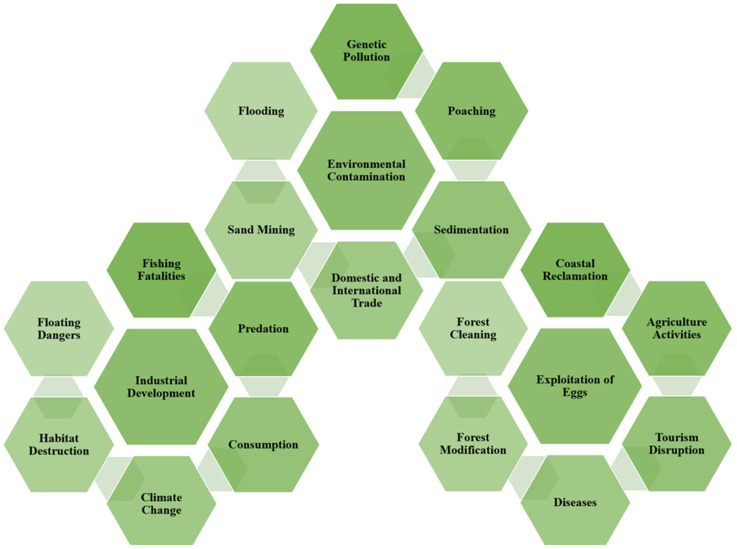
5. Threat Factors
The [21] population trends for all VU, EN, and CR turtle populations are decreasing. In conclusion, many factors contribute to threats. This review article compiles and documents the work of other Malaysian researchers and decision-makers for future reference (Figure 5). The primary causes of concern are egg consumption and trade [4,126]. The main threats to turtles are illegal and unregulated turtle poaching by Hainan (China) vessels and Vietnam [1]. Turtles are hunted for food, medicine, and ornaments [127]. In Malaysia, religious beliefs have reduced the killing of adult turtles for food [128]. On the other hand, building dams, taking turtle eggs, removing riparian vegetation, sand mining, and drowning in fishing nets are some of the turtle’s most significant problems [129,130,131,132,133].
Figure 5.
These threat factors were compiled from IUCN data, the DOF Report, DWNP Report, TRAFFIC South-east Asia Report, species-recovery plans, federal-agency re-sponses, and miscellaneous publica-tions on species’ life history. A complete list of documents used to assign biological attributes to endangered species is available from the authors.
According to References [134,135], the most critically endangered turtle species may become the most sought after due to their scarcity, which makes them especially valuable in the pet trade, hunting, and habitat degradation. Reference [136] reports that they are eaten, collected, butchered, and traded in large numbers; they are used for pets, food, and traditional medicine—eggs, juveniles, adults, and body parts are all exploited indiscriminately, with no regard for sustainability [137]. Their habitats are being destroyed, developed, fragmented, and polluted at an alarming rate [138,139]. Species all over the world are threatened or vulnerable, with many critically endangered. Others are on the verge of extinction, and a few have already perished [140]. Humans are threatening the extinction of countless eons and turtles [141].
Aside from overt and highly impactful conservation threats such as overexploitation and habitat destruction, the global turtle fauna is also increasingly facing another insidious threat: genetic pollution caused by human-facilitated hybridization and introgression from introduced and invasive species [142,143,144,145,146]. Although it is not entirely new, the current scale is unprecedented. Some taxa have already been impacted in the past. This is most likely true for Pelodiscus Asian softshell turtles. These turtles have been farmed and traded for hundreds of years. As a result, different species and local genetic lineages have been moved, leading to other taxa and lineages in captivity and the wild [147,148].
Similarly, the historical introduction of Mauremys reevesii to Japan resulted in massive hybridization with the native [149]. Another historical case of human-mediated admixture of genetic lineages is known from European pond turtles (Emys orbicularis). The non-native populations on the Balearic Islands, which were most likely introduced during Roman times [150], are of admixed origin [151]. Another population with genetic signatures of an old or ancient introduction of Emys orbicularis hellenica was discovered near Rome [151,152] within the range of another subspecies (Emys orbicularis galloitalica). However, unlike in the past, when only a few turtle species were affected, genetic pollution has become a big problem in protecting wildlife in recent years. This is because of the huge pet and food trade and increased human mobility.
Today, genetic pollution is also caused by well-meaning augmentation of endangered local turtle populations with genetically mismatched individuals (typically, but not exclusively, from non-coordinated actions by turtle enthusiasts), the release of surplus or abandoned genetically divergent pet turtles, and also by large-scale releases of confiscated turtle shipments, especially in Southeast Asia. Some endangered Emys orbicularis populations are on the northern edge of their range [153,154], and there is genetic evidence for restocking with multiple subspecies; in southern France [152,155], there is evidence of restocking with non-native Emys orbicularis hellenica rather than native Emys orbicularis. Examples of genetic pollution caused by abandoned pet turtles include Chrysemys picta bellii from British Columbia, introgressed by non-native subspecies [49], and Antillean (Trachemys), introgressed by Red-Eared Sliders (Trachemys scripta elegans) [156]. As previously stated, some cases involving European pond turtles are related to genetic contamination caused by abandoned pet turtles. In Taiwan, hybridization between Mauremys reevesii and Mauremys sinensis has been observed in the wild in released trade animals [157]. According to Reference [158], preserving well-defined genetic lineages, subspecies, and species that are mostly pure and not hybridized is critical. Therefore, in Malaysia, the two introduced species potentially cause genetic pollution.
6. Future Research and Recommendations
Several commendable research lines would also aid in understanding the growing issues and threats driving turtles to extinction. Table 4 discusses and summarizes these points.
Table 4.
A summary of recommended future research priorities.
| Topic | Method |
|---|---|
| Ecosystem effects | Monitor key turtle habitats to generate baseline data. Mesocosm experiments team up with other research disciplines and industries. Create strategies to identify and measure the trophic exchange of plastic, related poisons, and bioaccumulation. Explore the effect of plastics on the cycle of benthopelagic coupling [159]. |
| Impacts on nesting beaches | Record perceptions of experiences with seashore garbage for females and hatchlings. Use oceanographic demonstrating to conjecture how and when key waterfront regions are prone to being affected by plastic contamination [159]. |
| New sites | The purpose of the examination is to recognise new nesting zones, especially if current nesting locales become unsatisfactory because of improvement or environmental-driven change [160,161]. |
| Embryology | Developing and assessing a reliable indicator of hatchling health, comprehending endocrine influences on embryology, and further research into the role of home site selection in hatchling development [162]. |
| Molecular | There are numerous ways to deal with understanding the spatial biology of turtles, |
| counting hereditary qualities [163], natural biomarkers like stable isotopes [164]. | |
| Conservation Management | Designing management strategies with SMART (specific, measurable, achievable, realistic and time-based) objectives that permit assessment, variation, and the advancement of proof-based preservation will be critical to deciding the board achievement of current and future ventures [162]. |
| Climate Change | Understanding cumulative impacts or developing conservation responses to climate change [165]. |
| Threats | Thought of future dangers and their management in decision processes like horizontal planning [166]. GIS should provide new insights into patterns and can greatly aid in understanding the effects of hazards and the sufficiency of relief [167,168]. |
| Habitat Restoration | The carrying capacity of territory is a significant consideration in living space reclamation [169]. A few researchers have attempted to explain the general or current-carrying capacities of specific biological systems [170,171], though much more work is needed here. |
| eDNA | Streamlining field techniques for turtle eDNA assortment, further testing primer explicitness through trials of tests containing numerous species’ DNA, and creating primers focusing on other turtle networks could extraordinarily improve the recognition rates of uncommon species [172]. |
| nDNA | Nnuclear DNA markers (e.g., microsatellites, SNPs) are expected to confirm and further assess the hereditary portrayal of turtles in the EP as the information from mtDNA markers just reflects variety among female genealogies [47,70]. |
| Microbiology | Harmful microorganisms such as viruses, bacteria, parasites, and fungus that have not yet been investigated on turtles through metabarcoding, which has the potential to spread among or between hosts. Aside from that, future research could look into the impact of the dominant phylum (Proteobacteria) and genus (Cetobacterium) [122]. |
To distinguish explicit from common-sense preservation activities, it is necessary to examine each species’ information and protection status [173]. Protection status is frequently based on IUCN-proposed measures, which consider factors such as the number of the population remaining, the size of the current geographic conveyance, and data on idle dangers [19]. The restricted assets are given for turtle preservation in many countries, including Malaysia. Reference [174] says that it is occasionally necessary to have a target method for focusing on species. We believe that our work will help to collect more reliable data on turtles and address the threats to Malaysia’s most vulnerable species. Our research can help determine which species require management plans and investment from government agencies. Moreover, the research priorities can help researchers and students figure out which topics have not been studied sufficiently but are more important for making conservation recommendations [175]. The author of Reference [176] says that it is essential to direct human and economic efforts toward those issues that are high priorities. They also say that governments and regulatory authorities should work together to ensure that laws and rules are enforced more effectively [177,178].
It is critical to encourage short-to-medium-term studies in ecology and molecular systematics, as well as medium-to-long-term demographic studies that are biologically representative and can guide conservation and management efforts. Malaysian continental turtle species face significant research challenges. They entail leaving our geographical and thematic comfort zones to face the difficulty of gaining access to lesser-known species. Developing studies necessitate significant effort over long periods of time. We should bring together a wide range of expertise, such as taxonomists, ecologists, and molecular biologists. Those research outputs may directly raise public awareness of turtles’ plight toward sustainability.
7. Conclusions
In a small number of species, we face a turtle endurance crisis that is both severe and urgent. Without intervention, valuable species will become extinct over the next few decades. Some of the future research requirements listed in this review may assist researchers in collaborating to save these turtles for which we are so passionate. This review’s benefits may help readers understand the risks and threats to turtles and set goals for how long we should be able to last and how safe we should be, as well as making new standards for practical conservation techniques.
Acknowledgments
We want to acknowledge the Universiti Putra Malaysia for funding this study through a Graduate Research Fellowship awarded to MHMS. Furthermore, we thank the Turtle Conservation Society of Malaysia and the Department of Wildlife and National Parks, Peninsular Malaysia, for their collaboration. Last but not least, we thank the anonymous reviewers for their insights and suggestions to improve this paper.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ani12172184/s1. Table S1: The list of turtle types in Malaysia.
Author Contributions
Conceptualization, Y.E. and S.A.M.S.; methodology, M.H.M.S.; validation, S.M.S. and Y.E.; formal analysis, M.H.M.S.; investigation, M.H.M.S. and S.M.S.; resources, Y.E. and S.A.M.S.; data curation, S.M.S. and Y.E.; writing—original draft preparation, M.H.M.S.; writing—review and editing, Y.E. and S.M.S.; visualization, Y.E. and S.A.M.S.; supervision, Y.E.; project administration, Y.E. and S.A.M.S.; funding acquisition, Y.E. and S.A.M.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was performed in accordance with the rules and with permission from Department of Wildlife and National Parks, Peninsular Malaysia (B-00335-16-20).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding, and the APC was funded by the Publication Fund, Research Management Centre, Universiti Putra Malaysia.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Forero-Medina G., Páez V.P., Garcés-Restrepo M.F., Carr J.L., Giraldo A., Vargas-Ramírez M. Research and conservation priorities for tortoises and freshwater turtles of Colombia. Trop. Conserv. Sci. 2016;9:3708. doi: 10.1177/1940082916673708. [DOI] [Google Scholar]
- 2.Lau M.M., Ruqaiyah S., Devadasan A., Duraisingham G.S., Zulkifli R. Report on the Third Technical Consultation on Research for Stock Enhancement of Sea Turtles (Japanese Trust Fund IV Program) World Wildlife Fund; Petaling Jaya, Selangor, Malaysia: 2009. Satellite tracking of Green turtles and Hawksbill turtles in Peninsular Malaysia by WWF-Malaysia; pp. 101–114. [Google Scholar]
- 3.Peters M.E. Master’s Thesis. Utrecht University; Utrecht, The Netherlands: 2018. Identification and taphonomy of a Miocene Leatherback Turtle (Testudines: Dermochelyidae) from the Westerschelde. [Google Scholar]
- 4.Chan E.H. Marine turtles in Malaysia: On the verge of extinction? Aquat. Ecosyst. Health Manag. 2006;9:175–184. doi: 10.1080/14634980600701559. [DOI] [Google Scholar]
- 5.Chan E.H., Liew H.C. The Leatherback Turtle-A Malaysian Heritage. Tropical Press Sdn. Bhd.; Kuala Lumpur, Malaysia: 1989. [Google Scholar]
- 6.Ernst C.H., Lovich J.E. Turtles of the United States and Canada. Johns Hopkins University Press; Baltimore, MD, USA: 2009. [Google Scholar]
- 7.Zug G.R. Turtle. Encyclopædia Britannica. [(accessed on 2 March 2022)]. Available online: https://www.britannica.com/animal/turtle-reptile.
- 8.Gollmann G. Turtles: The Animal Answer Guide. Amphib. Reptil. 2011;32:293. doi: 10.1163/017353710X541904. [DOI] [Google Scholar]
- 9.Wyneken J., Lutz P.L., Musick J.A. The Biology of Sea Turtles. Taylor & Francis Group (CRC Press); London, UK: 1997. Sea turtle locomotion: Mechanisms, behavior, and energetics; pp. 165–198. [Google Scholar]
- 10.Branch B. Tortoises, Terrapins & Turtles of Africa. Penguin Random House South Africa; Cape Town, South Africa: 2012. [Google Scholar]
- 11.Buhlmann K., Tuberville T., Gibbons J.W. Turtles of the Southeast. University of Georgia Press; Athens, GA, USA: 2008. [Google Scholar]
- 12.Jualaong S., Songnui A., Thongprajukaew K., Ninwat S., Khwanmaung S., Hahor W., Khunsaeng P., Kanghae H. Optimal salinity for head-starting Northern river terrapins (Batagur baska Gray, 1831) Animals. 2019;9:855. doi: 10.3390/ani9110855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speck F.G. Reptile lore of the Northern Indians. J. Am. Folk. 1923;36:273–280. doi: 10.2307/534993. [DOI] [Google Scholar]
- 14.Ibrahim N.S., Sham B.H.B., Shafie N.J., Ahmad A. Species Diversity of Freshwater Turtles and Tortoises in Terengganu, Malaysia. J. Sustain. Sci. Manag. 2018;1:1–27. [Google Scholar]
- 15.Aun P.K. Malayan testudines. J. Wildl. Parks. 1990;9:20–31. [Google Scholar]
- 16.International Union for Conservation of Nature . IUCN Red List Categories and Criteria. IUCN; Cambridge, UK: 2001. p. 30. Species Survival Commission. [Google Scholar]
- 17.Chan E.H. A report on the first 16 years of a long-term marine turtle conservation project in Malaysia. Asian J. Conserv. Biol. 2013;2:129–135. [Google Scholar]
- 18.Pollock C., Mace G.M., Hilton-Taylor C., de Iongh H.H., Bánki O.S., Bergmans W., van der Werff ten Bosch M.J. The Harmonisation of Red Lists for Threatened Species in Europe. Bakhuijs Publishers; Leiden, The Netherlands: 2003. pp. 33–48. [Google Scholar]
- 19.IUCN RED LIST CATEGORIES AND CRITERIA Version 3.1 Second edition” (PDF). International Union for Conservation of Nature and Natural Resources. 2012. [(accessed on 28 February 2022)]. Available online: http://www.iucnredlist.org/documents/redlist_cats_crit_en.Pdf.
- 20.Knight A.T., Bode M., Fuller R.A., Grantham H.S., Possingham H.P., Watson J.E., Wilson K.A. Barometer of life: More action, not more data. Science. 2010;329:141. doi: 10.1126/science.329.5988.141-a. [DOI] [PubMed] [Google Scholar]
- 21.IUCN The IUCN Red List of Threatened Species. Version 2020-3. 2020. [(accessed on 16 December 2021)]. Available online: https://www.iucnredlist.org.
- 22.Fukushima C., Mendoza J.I., West R.C., Longhorn S.J., Rivera E., Cooper E.W., Hénaut Y., Henriques S., Cardoso P. Species conservation profiles of tarantula spiders (Araneae, Theraphosidae) listed on CITES. Biodivers. Data J. 2019;7:e39342. doi: 10.3897/BDJ.7.e39342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith M.J., Benítez-Díaz H., Clemente-Muñoz M.Á., Donaldson J., Hutton J.M., McGough H.N., Medellin R.A., Morgan D.H., O’Criodain C., Oldfield T.E., et al. Assessing the impacts of international trade on CITES-listed species: Current practices and opportunities for scientific research. Biol. Conserv. 2011;144:82–91. doi: 10.1016/j.biocon.2010.10.018. [DOI] [Google Scholar]
- 24.Nijman V., Shepherd C.R. Trade in non-native, CITES-listed, wildlife in Asia, as exemplified by the trade in freshwater turtles and tortoises (Chelonidae) in Thailand. Contrib. Zool. 2007;76:207–211. doi: 10.1163/18759866-07603007. [DOI] [Google Scholar]
- 25.Wyatt T. Is CITES Protecting Wildlife?: Assessing Implementation and Compliance. Taylor and Francis Group, Routledge; London, UK: 2021. p. 192. [Google Scholar]
- 26.Sharma D.S. Tortoise and Freshwater Turtle Trade and Utilisation in Peninsular Malaysia. TRAFFIC Southeast Asia; Petaling Jaya, Malaysia: 1999. p. 39. [Google Scholar]
- 27.Challender D.W., MacMillan D.C. Investigating the influence of non-state actors on amendments to the CITES appendices. J. Int. Wildl. Law Policy. 2019;22:90–114. doi: 10.1080/13880292.2019.1638549. [DOI] [Google Scholar]
- 28.Bowen B., Avise J.C., Richardson J.I., Meylan A.B., Margaritoulis D., Hopkins-Murphy S.R. Population structure of loggerhead turtles (Caretta caretta) in the northwestern Atlantic Ocean and Mediterranean Sea. Conserv. Biol. 1993;7:834–844. doi: 10.1046/j.1523-1739.1993.740834.x. [DOI] [Google Scholar]
- 29.Duchene S., Frey A., Alfaro-Núñez A., Dutton P.H., Gilbert M.T.P., Morin P.A. Marine turtle mitogenome phylogenetics and evolution. Mol. Phylogenet. Evol. 2012;65:241–250. doi: 10.1016/j.ympev.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 30.Robinson N.J., Paladino F.V. Sea Turtles, Reference Module in Earth Systems and Environmental Sciences. Elsevier; Amsterdam, The Netherlands: 2013. pp. 1–13. [DOI] [Google Scholar]
- 31.Nick B. Turtles of Southeast Asia. [(accessed on 20 March 2021)]. Available online: https://www.ecologyasia.com/verts/turtles.htm.
- 32.Böhme M., Vasilyan D. Ectothermic vertebrates from the late Middle Miocene of Gratkorn (Austria, Styria) Palaeobiodivers. Palaeoenviron. 2014;94:21–40. doi: 10.1007/s12549-013-0143-7. [DOI] [Google Scholar]
- 33.Stanford C.B., Iverson J.B., Rhodin A.G., van Dijk P.P., Mittermeier R.A., Kuchling G., Berry K.H., Bertolero A., Bjorndal K.A., Blanck T.E., et al. Turtles and tortoises are in trouble. Curr. Biol. 2020;320:R721–R735. doi: 10.1016/j.cub.2020.04.088. [DOI] [PubMed] [Google Scholar]
- 34.Ferronato B.O., Morales V.M. Biology and conservation of the freshwater turtles and tortoises of Peru. IRCF Reptiles Amphib. Conserv. Nat. Hist. 2012;19:103–116. doi: 10.17161/randa.v19i2.13889. [DOI] [Google Scholar]
- 35.Le M., Duong H.T., Dinh L.D., Nguyen T.Q., Pritchard P.C., McCormack T. A phylogeny of softshell turtles (Testudines: Trionychidae) with reference to the taxonomic status of the critically endangered, giant softshell turtle, Rafetus swinhoei. Org. Divers. Evol. 2014;14:279–293. doi: 10.1007/s13127-014-0169-3. [DOI] [Google Scholar]
- 36.Pritchard P.C.H. Carapacial pankinesis in the Malayan softshell turtle, Dogania subplana. Chelonian Conserv. Biol. 1993;1:31–36. [Google Scholar]
- 37.Thomson S., Georges A. A new species of freshwater turtle of the genus Elseya (Testudinata: Pleurodira: Chelidae) from the Northern Territory of Australia. Zootaxa. 2016;4061:18–28. doi: 10.11646/zootaxa.4061.1.2. [DOI] [PubMed] [Google Scholar]
- 38.Devi N.A., Eprilurahman R., Yudha D.S., Raharjo S., As-Singkily M., Gunalen D., Arida E. IOP Conference Series: Earth and Environmental Science. Volume 948. IOP Publishing; Bristol, UK: 2021. Genetic diversity and species identity of the critically endangered Rote Island snake-necked turtle, Chelodina mccordi Rhodin; p. 012001. [Google Scholar]
- 39.Rivera A.R., Rivera G., Blob R.W. Forelimb kinematics during swimming in the pig-nosed turtle, Carettochelys insculpta, compared with other turtle taxa: Rowing versus flapping, convergence versus intermediacy. J. Exp. Biol. 2013;216:668–680. doi: 10.1242/jeb.079715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shepherd C.R., Gomez L., Nijman V. Illegal wildlife trade, seizures and prosecutions: A 7.5-year analysis of trade in pig-nosed turtles in and from Indonesia. Glob. Ecol. Conserv. 2020;24:e01249. doi: 10.1016/j.gecco.2020.e01249. [DOI] [Google Scholar]
- 41.Tuxbury K.A., Clayton L.A., Snakard E.P., Fishman E.K. Multiple skull fractures in a captive fly river turtle (Carretochelys insculpta): Diagnosis, surgical repair, and medical management. J. Herpetol. Med. Surg. 2010;20:11–19. doi: 10.5818/1529-9651-20.1.11. [DOI] [Google Scholar]
- 42.Meyer L., Du Preez L., Verneau O., Bonneau E., Héritier L. Parasite host-switching from the invasive American red-eared slider, Trachemys scripta elegans, to the native Mediterranean pond turtle, Mauremys leprosa, in natural environments. Aquat. Invasions. 2015;10:79–91. doi: 10.3391/ai.2015.10.1.08. [DOI] [Google Scholar]
- 43.Plymale H.H., Jackson C.G.G., Jr., Collier G. Kyphosis in Chrysemys scripta yaquia (Testudines: Emydidae) and other turtles. Southwest. Nat. 1978:457–461. doi: 10.2307/3670252. [DOI] [Google Scholar]
- 44.Fritz U. Introduction to zoogeography and subspecific differentiation in Emys orbicularis (Linnaeus, 1758); Proceedings of the EMYS Symposium Dresden; Dresden, Germany. 4–6 October 1998; pp. 1–27. [Google Scholar]
- 45.Seidel M.E., Ernst C.H. A systematic review of the turtle family Emydidae. Vertebr. Zool. 2017;67:1–122. [Google Scholar]
- 46.Pearse D.E., Avise J.C. Turtle mating systems: Behavior, sperm storage, and genetic paternity. J. Hered. 2001;92:206–211. doi: 10.1093/jhered/92.2.206. [DOI] [PubMed] [Google Scholar]
- 47.Bowen B.W., Karl S.A. Population genetics and phylogeography of sea turtles. Mol. Ecol. 2007;16:4886–4907. doi: 10.1111/j.1365-294X.2007.03542.x. [DOI] [PubMed] [Google Scholar]
- 48.Lee P.L. Molecular ecology of marine turtles: New approaches and future directions. J. Exp. Mar. Biol. Ecol. 2008;356:25–42. doi: 10.1016/j.jembe.2007.12.021. [DOI] [Google Scholar]
- 49.Jensen E.L., Govindarajulu P., Madsen J., Russello M.A. Extirpation by introgression? Genetic evidence reveals hybridisation between introduced Chrysemys picta and endangered Western Painted turtles (C. p. bellii) in British Columbia. Herpetol. Conserv. Biol. 2014;9:342–353. [Google Scholar]
- 50.Meylan A.B., Donnelly M. Status justification for listing the hawksbill turtle (Eretmochelys imbricata) as critically endangered on the 1996 IUCN Red List of Threatened Animals. Chelonian Conserv. Biol. 1999;3:200–224. [Google Scholar]
- 51.Mortimer J.A., Donnelly M. Eretmochelys imbricata The IUCN Red List of Threatened Species 2008: e.T8005A12881238. IUCN; Gland, Switzeland: 2008. pp. 1–42. [Google Scholar]
- 52.Joseph J., Shaw P.W. Multiple paternity in egg clutches of hawksbill turtles (Eretmochelys imbricata) Conserv. Genet. 2011;12:601–605. doi: 10.1007/s10592-010-0168-7. [DOI] [Google Scholar]
- 53.Phillips K.P., Jorgensen T.H., Jolliffe K.G., Jolliffe S.M., Henwood J., Richardson D.S. Reconstructing paternal genotypes to infer patterns of sperm storage and sexual selection in the hawksbill turtle. Mol. Ecol. 2013;22:2301–2312. doi: 10.1111/mec.12235. [DOI] [PubMed] [Google Scholar]
- 54.Tedeschi J.N., Kennington W.J., Berry O., Whiting S., Meekan M., Mitchell N.J. Increased expression of Hsp70 and Hsp90 mRNA as biomarkers of thermal stress in loggerhead turtle embryos (Caretta caretta) J. Therm. Biol. 2015;47:42–50. doi: 10.1016/j.jtherbio.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 55.Peare T., Parker P.G. Local genetic structure within two rookeries of Chelonia mydas (the green turtle) J. Hered. 1996;77:619–628. doi: 10.1038/hdy.1996.189. [DOI] [PubMed] [Google Scholar]
- 56.Schroth W., Streit B., Schierwater B. Evolutionary handicap for turtles. Nature. 1996;384:521–522. doi: 10.1038/384521a0. [DOI] [Google Scholar]
- 57.Smith M.H., Hillstad H.O., Manlove M.N., Straney D.O., Dean J.M. Management implications of genetic variability in loggerhead and green sea turtles; Proceedings of the 13th International Congress of Game Biologists; Sofia, Bulgaria. 4–8 September 1977. [Google Scholar]
- 58.Wood J.R., Wood F.E.E., Critchley K. Hybridisation of Chelonia mydas and Eretmochelys imbricata. Copeia. 1983:839–842. doi: 10.2307/1444361. [DOI] [Google Scholar]
- 59.Harry J.L., Briscoe D.A. Multiple paternity in the loggerhead turtle (Caretta caretta) J. Hered. 1988;79:96–99. doi: 10.1093/oxfordjournals.jhered.a110480. [DOI] [PubMed] [Google Scholar]
- 60.Conceicao M.B., Levy J.A., Marins L.F.F., Marcovaldi M. A. Electro- phoretic characterisation of a hybrid between Eretmochelys imbricata and Caretta caretta (Cheloniidae) Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1990;97:275–278. doi: 10.1016/0305-0491(90)90280-7. [DOI] [Google Scholar]
- 61.Karl S.A., Bowen B.W., Avise J.C.C. Global population genetic structure and male mediated gene flow in the green turtle (Chelonia mydas)—RFLP analyses of anonymous nuclear loci. Genetics. 1992;131:163–173. doi: 10.1093/genetics/131.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karl S.A., Bowen B.W., Avise J.C. Hybridisation among the ancient mariners: Characterisation of marine turtle hybrids with molecular genetic assays. J. Hered. 1995;86:262–268. doi: 10.1093/oxfordjournals.jhered.a111579. [DOI] [PubMed] [Google Scholar]
- 63.Karl S.A., Avise J.C. PCR based assays of mendelian polymorphisms from anonymous single-copy nuclear DNA—techniques and applications for population genetics. Mol. Biol. Evol. 1993;10:342–361. doi: 10.1093/oxfordjournals.molbev.a040002. [DOI] [PubMed] [Google Scholar]
- 64.FitzSimmons N.N., Moritz C., Limpus C.J., Pope L., Prince R. Geographic structure of mitochondrial and nuclear gene polymorphisms in Australian Green turtle populations and male-biased gene flow. Genetics. 1997;147:1843–1854. doi: 10.1093/genetics/147.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karl S.A., Bowen B.W. Evolutionary significant units versus geopolitical taxonomy: Molecular systematics of an endangered sea turtle (genus Chelonia) Conserv. Biol. 1999;13:990–999. doi: 10.1046/j.1523-1739.1999.97352.x. [DOI] [Google Scholar]
- 66.Chassin-Noria O., Abreu-Grobois A., Dutton P.H., Oyama K. Conservation genetics of the East Pacific green turtle (Chelonia mydas) in Michoacan, Mexico. Genetica. 2004;121:195–206. doi: 10.1023/B:GENE.0000040394.47843.e4. [DOI] [PubMed] [Google Scholar]
- 67.Roberts M.A., Schwartz T.S., Karl S.A. Global population genetic structure and male-mediated gene flow in the green sea turtle (Chelonia mydas): Analysis of microsatellite loci. Genetics. 2004;166:1857–1870. doi: 10.1093/genetics/166.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bowen B.W., Bass A.L., Soares L., Toonen R.J. Conservation implications of complex population structure: Lessons from the loggerhead turtle (Caretta caretta) Mol. Ecol. 2005;14:2389–2402. doi: 10.1111/j.1365-294X.2005.02598.x. [DOI] [PubMed] [Google Scholar]
- 69.Rivalan P., Pradel R., Choquet R., Girondot M., Prévot-Julliard A.C. Estimating clutch frequency in the sea turtle Dermochelys coriacea using stopover duration. Mar. Ecol. Prog. Ser. 2006;317:285–295. doi: 10.3354/meps317285. [DOI] [Google Scholar]
- 70.Carreras C., Pascual M., Cardona L., Aguilar A., Margaritoulis D., Rees A., Turkozan O., Levy Y., Gasith A., Aureggi M., et al. The genetic structure of the loggerhead sea turtle (Caretta caretta) in the Mediterranean as revealed by nuclear and mitochondrial DNA and its conservation implications. Conserv. Genet. 2007;8:761–775. doi: 10.1007/s10592-006-9224-8. [DOI] [Google Scholar]
- 71.Naro-Maciel E., Reid B., Fitzsimmons N.N., Le M., Desalle R.O.B., Amato G. DNA barcodes for globally threatened marine turtles: A registry approach to documenting biodiversity. Mol. Ecol. Resour. 2010;10:252–263. doi: 10.1111/j.1755-0998.2009.02747.x. [DOI] [PubMed] [Google Scholar]
- 72.Revelles M., Carreras C., Cardona L., Marco A., Bentivegna F., Castillo J.J., De Martino G., Mons J.L., Smith M.B., Rico C., et al. Evidence for an asymmetrical size exchange of loggerhead sea turtles between the Mediterranean and the Atlantic through the Straits of Gibraltar. J. Exp. Mar. Biol. Ecol. 2007;349:261–271. doi: 10.1016/j.jembe.2007.05.018. [DOI] [Google Scholar]
- 73.Dutton P.H. Ph.D. Thesis. Texas A&M University; College Station, TX, USA: Dec, 1995. Molecular Evolution of Sea Turtles with Special Reference to the leatherback, Dermochelys coriacea. [Google Scholar]
- 74.FitzSimmons N.N., Moritz C., Moore S.S. Conservation and dynamics of microsatellite loci over 300 million years of marine turtle evolution. Mol. Biol. Evol. 1995;12:432–440. doi: 10.1093/oxfordjournals.molbev.a040218. [DOI] [PubMed] [Google Scholar]
- 75.Aggarwal R.K., Velavan T.P., Udaykumar D., Hendre P.S., Shanker K., Choudhury B.C., Singh L. Development and characterisation of novel microsatellite markers from the Olive Ridley sea turtle (Lepidochelys olivacea) Mol. Ecol. Notes. 2004;4:77–79. doi: 10.1046/j.1471-8286.2003.00574.x. [DOI] [Google Scholar]
- 76.Shamblin B.M., Faircloth B.C., Dodd M., Wood-Jones A., Castleberry S.B., Carroll J.P., Nairn C.J. Tetranucleotide microsatellites from the loggerhead sea turtle (Caretta caretta) Mol. Ecol. Notes. 2007;7:784–787. doi: 10.1111/j.1471-8286.2007.01701.x. [DOI] [Google Scholar]
- 77.Uysal S., Petridis G., Ozcan S., Faikoglu R., Barcak D., Yukseloglu H., Abaci-Kalfoglou E., Atasoy S. The use of DNA microsatellite loci for “Caretta caretta” identification. J. Environ. Sci. Health Part A. 2006;41:1981–1987. doi: 10.1080/10934520600779273. [DOI] [PubMed] [Google Scholar]
- 78.Ennen J.R., Lovich J.E., Kreiser B.R., Selman W., Qualls C.P. Genetic and morphological variation between populations of the Pascagoula map turtle (Graptemys gibbonsi) in the Pearl and Pascagoula rivers with description of a new species. Chelonian Conserv. Biol. 2010;9:98–113. doi: 10.2744/CCB-0835.1. [DOI] [Google Scholar]
- 79.Santos R.G., Martins A.S., da Nobrega Farias J., Horta P.A., Pinheiro H.T., Torezani E., Baptistotte C., Seminoff J.A., Balazs G.H. Work, TMM Coastal habitat degradation and green sea turtle diets in Southeastern Brazil. Mar. Pollut. Bull. 2011;62:1297–1302. doi: 10.1016/j.marpolbul.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 80.Gillespie J.H. The molecular clock may be an episodic clock. Proc. Natl. Acad. Sci. USA. 1984;81:8009–8013. doi: 10.1073/pnas.81.24.8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gaos A.R., Lewison R.L., Liles M.J., Gadea V., Altamirano E., Henríquez A.V., Torres P., Urteaga J., Vallejo F., Baquero A., et al. Hawksbill turtle terra incognita: Conservation genetics of eastern Pacific rookeries. Ecol. Evol. 2016;6:1251–1264. doi: 10.1002/ece3.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pritchard P.C., Lutz P.L., Musick J.A. Evolution, phylogeny, and current status. Biol. Sea Turt. 1997;1:1–28. [Google Scholar]
- 83.Witherington B., Hirama S., Mosier A. Sea turtle responses to barriers on their nesting beach. J. Exp. Mar. Biol. Ecol. 2011;401:1–6. doi: 10.1016/j.jembe.2011.03.012. [DOI] [Google Scholar]
- 84.Spielman D., Brook B.W., Frankham R. Most species are not driven to extinction before genetic factors impact them. Proc. Natl. Acad. Sci. USA. 2004;101:15261–15264. doi: 10.1073/pnas.0403809101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frankham R., Ballou S.E.J.D., Briscoe D.A., Ballou J.D. Introduction to Conservation Genetics. Cambridge University Press; Cambridge, UK: 2002. [Google Scholar]
- 86.Allendorf F.W., Berry O., Ryman N. So long to genetic diversity, and thanks for all the fish. Mol. Ecol. 2014;23:23–25. doi: 10.1111/mec.12574. [DOI] [PubMed] [Google Scholar]
- 87.Nunney L. The influence of mating system and overlapping generations on effective population size. Evolution. 1993;47:1329–1341. doi: 10.1111/j.1558-5646.1993.tb02158.x. [DOI] [PubMed] [Google Scholar]
- 88.Hillis D.M., Mable B.K., Moritz C. Applications of molecular systematics: The state of the field and a look to the future. In: Hillis D.M., Moritz C., Mable B.K., editors. Molecular Systematics. 2nd ed. Sinauer Associates, Oxford University Press; Oxford, UK: 1996. pp. 515–543. [Google Scholar]
- 89.Zhang D.X., Hewitt G.M. Nuclear DNA analyses in genetic studies of populations: Practice, problems and prospects. Mol. Ecol. 2003;12:563–584. doi: 10.1046/j.1365-294X.2003.01773.x. [DOI] [PubMed] [Google Scholar]
- 90.Salleh M.H.M., Esa Y. Microsatellite Loci Reveal Heterozygosis and Population Structure in the Critically Endangered Southern River Terrapin (Batagur affinis ssp.) of Peninsular Malaysia. Chem. Proc. 2022;10:11. doi: 10.3390/IOCAG2022-12230. [DOI] [Google Scholar]
- 91.Zane L., Bargelloni L., Patarnello T. Strategies for microsatellite isolation: A review. Mol. Ecol. 2002;11:1–16. doi: 10.1046/j.0962-1083.2001.01418.x. [DOI] [PubMed] [Google Scholar]
- 92.Glenn T.C., Schable N.A. Isolating microsatellite DNA loci. Methods Enzymol. 2005;395:202–222. doi: 10.1016/S0076-687995013-1. [DOI] [PubMed] [Google Scholar]
- 93.Jeffreys A.J., Wilson V., Thein S.L. Hypervariable minisatellite regions in human DNA. Nature. 1985;314:67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- 94.Allard M.W., Miyamoto M.M., Bjorndal K.A., Bolten A.B., Bowen B.W. Support for natal homing in green turtles from mitochondrial DNA sequences. Copeia. 1994;1:34–41. doi: 10.2307/1446668. [DOI] [Google Scholar]
- 95.Norman J.A., Moritz C., Limpus C.J. Mitochondrial DNA control region polymorphisms-genetic markers for ecological studies of marine turtles. Mol. Ecol. 1994;3:363–373. doi: 10.1111/j.1365-294X.1994.tb00076.x. [DOI] [PubMed] [Google Scholar]
- 96.Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Williams J.G., Kubelik A.R., Livak K.J., Rafalski J.A., Tingey S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Encalada S.E., Lahanas P.N., Bjorndal K.A., Bolten A.B., Miyamoto M.M., Bowen B.W. Phylogeography and population structure of the Atlantic and Mediterranean green turtle Chelonia mydas: A mitochondrial DNA control region sequence assessment. Mol. Ecol. 1996;5:473–483. doi: 10.1111/j.1365-294X.1996.tb00340.x. [DOI] [PubMed] [Google Scholar]
- 99.Encalada S.E., Bjorndal K.A., Bolten A.B., Zurita J.C., Schroeder B., Possardt E., Sears C.J., Bowen B.W. Population structure of loggerhead turtle (Caretta caretta) nesting colonies in the Atlantic and Mediterranean as inferred from mitochondrial DNA control region sequences. Mar. Biol. 1998;130:567–575. doi: 10.1007/s002270050278. [DOI] [Google Scholar]
- 100.Bland L.M., Keith D.A., Miller R.M., Murray N.J., Rodríguez J.P. Guidelines for the Application of IUCN Red List of Ecosystems Categories and Criteria, Version 1.1. International Union for the Conservation of Nature; Gland, Switzerland: 2017. pp. 1–100. [Google Scholar]
- 101.Lim B.L., Indraneil D. Turtles of Borneo and Peninsular Malaysia. Natural History Publications (Borneo); Kota Kinabalu, Sabah, Malaysia: 1999. p. 151. [Google Scholar]
- 102.Hasnan L. Turtles Crying Foul in Malaysia. The Asean Post. [(accessed on 2 June 2019)]. Available online: https://theaseanpost.com/article/turtles-crying-foul-malaysia.
- 103.Walker T. Plant Conservation: Why It Matters and How It Works. Timber Press; Portland, OR, USA: London, UK: 2013. p. 303. [Google Scholar]
- 104.CITES, UNEP-WCMC. The Checklist of CITES Species Website. Appendices I, II and III valid from 04 April 2017. CITES Secretariat, Geneva, Switzerland. Compiled by UNEP-WCMC, Cambridge, UK. 2017. [(accessed on 1 August 2020)]. Available online: https://www.cites.org/eng/app/appendices.php.
- 105.Reid B.N., LE M., McCord W.P., Iverson J.B., Georges A., Bergmann T., Amato G., Desalle R., Naro-Maciel E. Comparing and combining distance-based and character-based approaches for barcoding turtles. Mol. Ecol. Resour. 2011;11:956–967. doi: 10.1111/j.1755-0998.2011.03032.x. [DOI] [PubMed] [Google Scholar]
- 106.Çilingir F.G., Seah A., Horne B.D., Som S., Bickford D.P., Rheindt F.E. Last exit before the brink: Conservation genomics of the Cambodian population of the critically endangered southern river terrapin. Ecol. Evol. 2019;9:9500–9510. doi: 10.1002/ece3.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kundu S., Kumar V., Laskar B.A., Tyagi K., Chandra K. Pet and turtle: DNA barcoding identified twelve Geoemydid species in northeast India. Mitochondrial DNA Part B. 2018;3:513–518. doi: 10.1080/23802359.2018.1467215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kundu S., Das K.C., Ghosh S.K. Taxonomic rank of Indian tortoise: Revisit with DNA barcoding perspective. DNA Barcodes. 2013;1:39–45. doi: 10.2478/dna-2013-0003. [DOI] [Google Scholar]
- 109.Ouso D.O., Otiende M.Y., Jeneby M.M., Oundo J.W., Bargul J.L., Miller S.E., Wambua L., Villinger J. Three-gene PCR and high-resolution melting analysis for differentiating vertebrate species mitochondrial DNA for biodiversity research and complementing forensic surveillance. Sci. Rep. 2020;10:4741. doi: 10.1038/s41598-020-61600-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vella N., Vella A. The first genetic analyses of the leatherback turtle, Dermochelys coriacea from a stranding in central Mediterranean. Rapp. Comm. int. Mer Médit. 2016;41 [Google Scholar]
- 111.Kundu S., Das K.C., Ghosh S.K. Amino acid analysis of cytochrome c oxidase subunit 1(COI) of Indian testudines’. J. Environ. Sociobiol. 2013;10:43–48. [Google Scholar]
- 112.Xin C.N. Species Identification of Turtles Using Mitochondrial DNA Markers. 2009. in press .
- 113.Xin C.N., Peng J.J., Wang Y., Wang L. Application of Cyt b gene as a molecular marker in species identification. Chin. J. Wildlife. 2009;30:217–221. [Google Scholar]
- 114.Jeong T.J., Jun J., Han S., Kim H.T., Oh K., Kwak M. DNA barcode reference data for the Korean herpetofauna and their applications. Mol. Ecol. Resour. 2013;13:1019–1032. doi: 10.1111/1755-0998.12055. [DOI] [PubMed] [Google Scholar]
- 115.Farajallah A., Suryobroto B., Setyadji R., Perwitasari-Farajallah D., Osamu T. The Complete Nucleotide Sequence of Malayan Soft-Shelled Turtle (Dogania subplana) Mitochondrial Genome. [(accessed on 14 May 2021)]; Available online: https://www.ncbi.nlm.nih.gov/nuccore/AF366350.
- 116.Zhang H., Yao H., Cui L., DU H., Lin Z., Gao X., Lang X., Song J., Luo K., Shi L., et al. Application of COI-based DNA barcoding for identifying animal medical materials in the Chinese pharmacopoeia. World Sci. Technol. Mod. Tradit. Chin. Med. 2013;12:371–380. [Google Scholar]
- 117.Yudha S.D., Eprillurahman R., Irwanjasmoro. Supramono Y. Survei Awal Analisa Habitat Ditemukannya Labi-labi Bintang (Chitra chitra) di Sungai Sempor, Sleman, DIY. War. Herpetof. 2019;6:1–34. [Google Scholar]
- 118.Goldberg S.R., Mahrdt C.R. Bogertophis rosaliae: Reproduction. Herpetol. Rev. 2012;43:655. [Google Scholar]
- 119.Friedlander A.M. Biodiversity and Ecosystem health of the Aldabra Group, Southern Seychelles: Scientific Report to the Government of Seychelles. National Geographic Pristine Seas; Washington, DC, USA: 2015. [Google Scholar]
- 120.Guntoro J. The Body Size and Some Field Notes of Painted Terrapin (Batagur borneoensis) in District of Aceh Tamiang, Indonesia. Asian J. Conserv. Biol. 2012;1:74–77. [Google Scholar]
- 121.Ihlow F., Dawson J.E., Hartmann T., Som S. Conservation Biology of Freshwater Turtles and Tortoises. Indotestudo elongata (Blyth 1854)–Elongated Tortoise, Yellow-headed Tortoise, Yellow Tortoise. Chelonian Res. Monogr. 2017;5:1–13. [Google Scholar]
- 122.Mo M. Only in captivity?: An interaction between two threatened chelonians, an Asian Giant Tortoise (Manouria emys) and a Malaysian Giant Turtle (Orlitia borneensis) Reptiles Amphib. 2020;27:89–90. doi: 10.17161/randa.v27i1.14470. [DOI] [Google Scholar]
- 123.Salleh M.H.M., Esa Y., Ngalimat M.S., Chen P.N. Faecal DNA metabarcoding reveals novel bacterial community patterns of critically endangered Southern River Terrapin, Batagur affinis. PeerJ. 2022;10:e12970. doi: 10.7717/peerj.12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Inskipp T., Gillett H. Checklist of CITES Species and Annotated CITES Appendices and Reservations: A Reference to the Appendices to the Convention on International Trade in Endangered Species of Wild Fauna and Flora. IUCN—International Union for Conservation of Nature; Gland, Switzerland: 2005. [(accessed on 22 April 2022)]. Available online: https://policycommons.net/artifacts/1376877/checklist-of-cites-species-and-annotated-cites-appendices-and-reservations/1991140/ [Google Scholar]
- 125.Sajeva M., Augugliaro C., Smith M.J., Oddo E. Regulating internet trade in CITES species. Conserv. Biol. 2013;27:429. doi: 10.1111/cobi.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Challender D.W., Harrop S.R., MacMillan D.C. Understanding markets to conserve trade-threatened species in CITES. Biol. Conserv. 2015;187:249–259. doi: 10.1016/j.biocon.2015.04.015. [DOI] [Google Scholar]
- 127.Basintal P., Lakim M. Population status and management of sea turtles at the Sabah Turtle Island Park; Proceedings of the 1st ASEAN Symposium-Workshop on Marine Turtle Conservation; Manila, Philippines. 6–10 December 1993. [Google Scholar]
- 128.Sharma D.S., Tisen O.B. Freshwater turtle and tortoise utilisation and conservation status in Malaysia. Chelonian Res. Monogr. 2000;2:120–128. [Google Scholar]
- 129.Hendrickson J.R. The green sea turtle, Chelonia mydas (Linn.) in Malaya and Sarawak. Proc. Zool. Soc. Lond. 1958;130:455–535. doi: 10.1111/j.1096-3642.1958.tb00583.x. [DOI] [Google Scholar]
- 130.Kalyar K., Thorbjarnarson J., Thirakhupt K. An overview of the current population and conservation status of the Critically Endangered River Terrapin, Batagur baska (Gray, 1831) in Myanmar, Thailand and Malaysia. Trop. Nat. Hist. 2007;7:51–65. [Google Scholar]
- 131.Moll E.O., Platt S.G., Chan E.H., Horne B.D., Platt K., Praschag P., Chen P.N., van Dijk P.P. Batagur affinis (Cantor 1847)–Southern River Terrapin, Tuntong. In: Rhodin A.G.J., Pritchard P.C.H., van Dijk P.P., Saumure R.A., Buhlmann K.A., Iverson J.B., Mittermeier R.A., editors. Conservation Biology of Freshwater Turtles and Tortoises: A Compilation Project of the IUCN/SSC Tortoise and Freshwater Turtle Specialist Group. Volume 5. Chelonian Res. Monogr; Luneburg, MA, USA: 2015. pp. 1–17. [DOI] [Google Scholar]
- 132.Salleh M.H.B.M., Sah S.A.B.M. Morphometric variations in distinguishing sex differences of River Terrapin–Batagur affinis. Malay. Nat. J. 2019;71:413–422. [Google Scholar]
- 133.Salleh M.H.B.M., Sah S.A.B.M. Twenty-First Years Breeding Performance of Southern River Terrapin (Batagur affinis) at Perak River, Peninsular Malaysia. Int. J. Nat. Life Sci. 2019;3:52–58. [Google Scholar]
- 134.Lozano M., Baro J., García T., Frías A., Rey J., Báez J.C. Loggerhead sea turtle bycatch data in artisanal fisheries within a marine protected area: Fishermen surveys versus scientific observations. Anim. Biodivers. Conserv. 2011;34:31–34. doi: 10.32800/abc.2011.34.0031. [DOI] [Google Scholar]
- 135.Murray J.P. Bachelor’s Thesis. Universiti Malaysia Terengganu; Kuala Terengganu, Terengganu, Malaysia: 2006. Testudinidae and Trionychidae (Animalia: Reptilia) in the Pet Trade with Special Emphasis on Indian Star Tortoises (Geochelone elegans) in Peninsular Malaysia. [Google Scholar]
- 136.Chen P.N. Conservation of the Southern River Terrapin Batagur affinis (Reptilia: Testudines: Geoemydidae) in Malaysia: A case study involving local community participation. J. Threat. Taxa. 2017;9:10035–10046. doi: 10.11609/jott.3267.9.4.10035-10046. [DOI] [Google Scholar]
- 137.Turtle Conservation Fund . A Global Action Plan for Conservation of Tortoises and Freshwater Turtles. Strategy and Funding Prospectus 2002–2007. Conservation International and Chelonian Research Foundation; Washington, DC, USA: 2022. p. 30. [Google Scholar]
- 138.Lambert F.R., Howes J.R. Ranging, breeding behaviour and food of the Asian brown tortoise Manouria emys in Borneo. Malay. Nat. J. 1994;48:125–131. [Google Scholar]
- 139.Jasmi A., Vidyadaran M.K. Wildlife Conservation in Peninsular Malaysia. UPM Press; Helsinki Finland: 1993. pp. 193–201. The Animal Industry in Malaysia. [Google Scholar]
- 140.Rahman M.F.A., Manaf L.A. Conservation of river terrapin (Batagur affinis) in Malaysia: Status and challenges; Proceedings of the Malaysia International Biology Symposium; Putrajaya, Malaysia. 26–27 October 2016. [Google Scholar]
- 141.Jolis G., Min L.M., Mustafa S.R.S., Sumamporuw M., Rajan S.G., Jumin R., Sharma D.S. Sea Turtle Conservation in Malaysia: Issues, Challenges and Recommendations; Proceedings of the Seminar and Workshop on Sea Turtle Conservation in Malaysia; Kuala Terengganu, Terengganu, Malaysia. 1 September 2015. [Google Scholar]
- 142.Levell J.P. Commercial exploitation of Blanding’s Turtle, Emydoidea blandingii, and the Wood Turtle, Clemmys insculpta, for the live animal trade. Chelonian Conserv. Biol. 2000;3:665–674. [Google Scholar]
- 143.Rhymer J.M., Simberloff D. Extinction by hybridisation and introgression. Annu. Rev. Ecol. Syst. 1996;27:83–109. doi: 10.1146/annurev.ecolsys.27.1.83. [DOI] [Google Scholar]
- 144.Simison W.B., Sellas A.B., Feldheim K.A., Parham J.F. Isolation and characterisation of microsatellite markers for identifying hybridisation and genetic pollution associated with red-eared slider turtles (Trachemys scripta elegans) Conserv. Genet. Resour. 2013;5:1139–1140. doi: 10.1007/s12686-013-9978-5. [DOI] [Google Scholar]
- 145.Spencer R.J., Georges A., Lim D., Welsh M., Reid A.M. The risk of inter-specific competition in Australian short-necked turtles. Ecol. Res. 2014;29:767–777. doi: 10.1007/s11284-014-1169-7. [DOI] [Google Scholar]
- 146.García-Díaz P., Ross J.V., Ayres C., Cassey P. Understanding the biological invasion risk posed by the global wildlife trade: Propagule pressure drives the introduction and establishment of Nearctic turtles. Glob. Chang. Biol. 2015;21:1078–1091. doi: 10.1111/gcb.12790. [DOI] [PubMed] [Google Scholar]
- 147.Nori J., Tessarolo G., Ficetola G.F., Loyola R., Di Cola V., Leynaud G. Buying environmental problems: The invasive potential of imported freshwater turtles in Argentina. Aquat. Conserv. Mar. Freshw. Ecosyst. 2017;27:685–691. doi: 10.1002/aqc.2715. [DOI] [Google Scholar]
- 148.Fritz U., Daniels S.R., Hofmeyr M.D.D., González J., Barrio-Amorós C.L., Široký P., Hundsdörfer A.K., Stuckas H. Mitochondrial phylogeography and subspecies of the wide-ranging sub-Saharan leopard tortoise Stigmochelys pardalis (Testudines: Testudinidae)–a case study for the pitfalls of pseudogenes and GenBank sequences. J. Zool. Syst. Evol. Res. 2010;48:348–359. doi: 10.1111/j.1439-0469.2010.00565.x. [DOI] [Google Scholar]
- 149.Suzuki D., Hikida T. Taxonomic status of the softshell turtles populations in Japan: A molecular approach. Curr. Herpetol. 2014;33:171–179. doi: 10.5358/hsj.33.171. [DOI] [Google Scholar]
- 150.Suzuki D., Yabe T., Hikida T. Hybridisation between Mauremys japonica and Mauremys reevesii inferred by nuclear and mitochondrial DNA analyses. J. Herpetol. 2014;48:445–454. doi: 10.1670/11-320. [DOI] [Google Scholar]
- 151.Valenzuela A., Cau M.A., Alcover J.A. Archaeological evidence for the introduction of Emys orbicularis (Testudines: Emydidae) in the Balearic Islands. Amphib. Reptil. 2016;37:229–236. doi: 10.1163/15685381-00003049. [DOI] [Google Scholar]
- 152.Lenk P., Fritz U., Joger U., Wink M. Mitochondrial phylogeography of the European pond turtle, Emys orbicularis (Linnaeus 1758) Mol. Ecol. 1999;8:1911–1922. doi: 10.1046/j.1365-294x.1999.00791.x. [DOI] [PubMed] [Google Scholar]
- 153.Vamberger M., Stuckas H., Sacco F., D’Angelo S., Arculeo M., Cheylan M., Corti C., Lo Valvo M., Marrone F., Wink M., et al. Differences in gene flow in twofold secondary contact zone of pond turtles in southern Italy (Testudines: Emydidae: Emys orbicularis galloitalica, E. o. hellenica, E. trinacris) Zool. Scr. 2015;44:233–249. doi: 10.1111/zsc.12102. [DOI] [Google Scholar]
- 154.Fritz U., Guicking D., Lenk P., Joger U., Wink M. When turtle distribution tells European history: mtDNA haplotypes of Emys orbicularis reflect in Germany former division by the Iron Curtain. Biologia. 2004;59:19–25. [Google Scholar]
- 155.Velo-Antón G., Wink M., Schneeweiss N., Fritz U. Native or not? Tracing the origin of wild-caught and captive freshwater turtles in a threatened and widely distributed species (Emys orbicularis) Conserv. Genet. 2011;12:583–588. doi: 10.1007/s10592-010-0141-5. [DOI] [Google Scholar]
- 156.Raemy M., Fritz U., Cheylan M., Ursenbacher S. Hybridisation between turtle subspecies: A case study with the European pond turtle (Emys orbicularis) Conserv. Genet. 2017;18:287–296. doi: 10.1007/s10592-016-0901-y. [DOI] [Google Scholar]
- 157.Parham J.F., Papenfuss T.J., van Dijk P.P., Wilson B.S., Marte C., Schettino L.R., Simison W.B. Genetic introgression and hybridisation in Antillean freshwater turtles (Trachemys) revealed by coalescent analyses of mitochondrial and cloned nuclear markers. Mol. Phylogenet. Evol. 2013;67:176–187. doi: 10.1016/j.ympev.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 158.Fong J.J., Chen T.H. DNA evidence for the hybridisation of wild turtles in Taiwan: Possible genetic pollution from trade animals. Conserv. Genet. 2010;11:2061–2066. doi: 10.1007/s10592-010-0066-z. [DOI] [Google Scholar]
- 159.Rhodin A.G.J., Iverson J.B., Bour R., Fritz U., Georges A., Shaffer H.B. Turtles of the World: Annotated Checklist and Atlas of Taxonomy. Synon. Distrib. Conserv. Status. 2017;8:9–14. [Google Scholar]
- 160.Nelms S.E., Duncan E.M., Broderick A.C., Galloway T.S., Godfrey M.H., Hamann M., Lindeque P.K., Godley B.J.J. Plastic and marine turtles: A review and call for research. ICES J. Mar. Sci. 2016;73:165–181. doi: 10.1093/icesjms/fsv165. [DOI] [Google Scholar]
- 161.Hamann M., Limpus C., Read M. Vulnerability of Marine Reptiles to Climate Change in the Great Barrier Reef. In: Johnson J., Marshal P., editors. Climate change and the Great Barrier Reef. Great Barrier Reef Marine Park Authority and The Australian Greenhouse Office; Canberra, Australia: 2007. pp. 235–288. [Google Scholar]
- 162.Poloczanska E., Limpus C.J., Hays G. Vulnerability of marine turtles to climate change. Adv. Mar. Biol. 2009;56:1–61. doi: 10.1016/S0065-288156002-6. [DOI] [PubMed] [Google Scholar]
- 163.Hamann M., Godfrey M.H., Seminoff J.A., Arthur K., Barata P.C.R., Bjorndal K.A., Bolten A.B., Broderick A.C., Campbell L.M., Carreras C., et al. Global research priorities for sea turtles: Informing management and conservation in the 21st century. Endanger. Species Res. 2010;11:245–269. doi: 10.3354/esr00279. [DOI] [Google Scholar]
- 164.Bowen B.W. Tracking marine turtles with genetic-markers—voyages of the ancient mariners. BioScience. 1995;45:528–534. doi: 10.2307/1312697. [DOI] [Google Scholar]
- 165.Caut S., Fossette S., Guirlet E., Angulo E., Das K., Girondot M., Georges J.Y. Isotope analysis reveals foraging area dichotomy for Atlantic leatherback turtlese. PLoS ONE. 2008;3:e1845. doi: 10.1371/journal.pone.0001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Fuentes M.M., Pike D.A., Dimatteo A., Wallace B.P. Resilience of marine turtle regional management units to climate change. Glob. Chang. Biol. 2013;19:1399–1406. doi: 10.1111/gcb.12138. [DOI] [PubMed] [Google Scholar]
- 167.Sutherland W.J., Woodroof H.J. The need for environmental horizon scanning. Trends Ecol. Evol. 2009;24:523–527. doi: 10.1016/j.tree.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 168.Grech A., Marsh H. Rapid assessment of risks to a mobile marine mammal in an ecosystem-scale marine protected area. Conserv. Biol. 2008;22:711–720. doi: 10.1111/j.1523-1739.2008.00923.x. [DOI] [PubMed] [Google Scholar]
- 169.Grech A., Marsh H., Coles R. A spatial assessment of the risk to a mobile marine mammal from bycatch. Aquat. Conserv. Mar. Freshw. Ecosyst. 2008;18:1127–1139. doi: 10.1002/aqc.943. [DOI] [Google Scholar]
- 170.Elliott M., Burdon D., Hemingway K.L., Apitz S.E. Estuarine, coastal and marine ecosystem restoration: Confusing management and science—A revision of concepts. Estuar Coast Shelf Sci. 2007;74:349–366. doi: 10.1016/j.ecss.2007.05.034. [DOI] [Google Scholar]
- 171.Jackson J.B., Kirby M.X., Berger W.H., Bjorndal K.A., Botsford L.W., Bourque B.J., Bradbury R.H., Cooke R., Erlandson J., Estes J.A., et al. Historical overfishing and the recent collapse of coastal ecosystems. Science. 2001;293:629–637. doi: 10.1126/science.1059199. [DOI] [PubMed] [Google Scholar]
- 172.Girondot M., Tucker A.D., Rivalan P., Godfrey M.H., Chevalier J. Density-dependent nest destruction and population fluctuations of Guianan leatherback turtles. Anim. Conserv. 2002;5:75–84. doi: 10.1017/S1367943002001099. [DOI] [Google Scholar]
- 173.Davy C.M., Kidd A.G., Wilson C.C. Development and Validation of Environmental DNA (eDNA) Markers for Detection of Freshwater Turtles. PLoS ONE. 2015;10:e0130965. doi: 10.1371/journal.pone.0130965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Muimi E. Head, Department of Wildlife and National Parks, Maun Office. Personal Interview Conducted in the Department of Wildlife and National Parks. Maun Office; Maun, Botswana: 2002. p. 19. [Google Scholar]
- 175.Limpus C.J. Recommendations for Conservation of Marine Turtles in Peninsula Malaysia. Report to Department of Fisheries, Ministry of Agriculture, Malaysia. Queensland Department of Environment and Heritage; Brisbane, Australia: 1993. p. 60. [Google Scholar]
- 176.Hamann M., Ibrahim K., Limpus C. Status of Leatherback Turtles in Malaysia. In: Hamann M., Limpus C., Hughes G., Mortimer J., Pilcher N., editors. Indian Ocean – South-East Asian Marine Turtle Memorandum of Understanding. Assessment of the Conservation Status of the Leatherback Turtle in the Indian Ocean and South East Asia. IOSEA Species Assessment: Volume I. IOSEA Marine Turtle MoU Secretariat; Bangkok, Thailand: 2006. pp. 78–82. [Google Scholar]
- 177.Alam M.F., Omar I.H., Squires D. Sustainable fisheries development in the tropics: Trawlers and licence limitation in Malaysia. Appl. Econ. 2002;34:325–337. doi: 10.1080/00036840110036305. [DOI] [Google Scholar]
- 178.Islam G.M.N., Tai S.Y., Kusairi M.N., Ahmad S., Aswani F.M.N., Senan M.K.A.M., Ahmad A. Community perspectives of governance for effective management of marine protected areas in Malaysia. Ocean. Coast. Manag. 2017;135:34–42. doi: 10.1016/j.ocecoaman.2016.11.001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.