Abstract
Simple Summary
Blood-based cancer biomarkers are a minimally invasive means to achieve improved assessment of risk and/or earlier detection of lung cancer. Using serum samples from individuals that went on to develop lung cancer within one year following blood draw and from matched controls, we investigated 30 microRNAs for individual and combined utility to discriminate lung cancer cases from controls. We additionally assessed the contributions of top-performing microRNA candidates for improving on the performance of a previously validated four-protein marker panel for lung cancer detection. The results of this study indicate complementary performance of combining circulating microRNA and the four-protein-marker panel in assays for early detection of lung cancer.
Abstract
There is unmet need to develop circulating biomarkers that would enable earlier interception of lung cancer when more effective treatment options are available. Here, a set of 30 miRNAs, selected from a review of the published literature were assessed for their predictive performance in identifying lung cancer cases in the pre-diagnostic setting. The 30 miRNAs were assayed using sera collected from 102 individuals diagnosed with lung cancer within one year following blood draw and 212 controls matched for age, sex, and smoking status. The additive performance of top-performing miRNA candidates in combination with a previously validated four-protein marker panel (4MP) consisting of the precursor form of surfactant protein B (Pro-SFTPB), cancer antigen 125 (CA125), carcinoembryonic antigen (CEA) and cytokeratin-19 fragment (CYFRA21-1) was additionally assessed. Of the 30 miRNAs evaluated, five (miR-320a-3p, miR-210-3p, miR-92a-3p, miR-21-5p, and miR-140-3p) were statistically significantly (Wilcoxon rank sum test p < 0.05) elevated in case sera compared to controls, with individual AUCs ranging from 0.57–0.62. Compared to the 4MP alone, the combination of 3-miRNAs + 4MP improved sensitivity at 95% specificity by 19.1% ((95% CI of difference 0.0–28.6); two-sided p: 0.006). Our findings demonstrate utility for miRNAs for early detection of lung cancer in combination with a four-protein marker panel.
Keywords: lung cancer, protein biomarkers, microRNA, early detection
1. Introduction
Lung cancer is the leading (18%) cause of cancer mortality worldwide, accounting for an estimated 1.8 million deaths [1]. Five-year relative survival rates among lung cancer subtypes are closely linked with tumor stage at time of diagnosis, with an estimated overall survival rate of 59.8% when diagnosis is made at the localized stage, 32.9% at the regional stage, and 6.3% at the distant stage, according to recent United States National Cancer Institute (NCI) Surveillance, Epidemiology, and End Results (SEER) Program data [2]. NCI National Lung Cancer Screening Trial (NLST) data indicate that a 20% reduction in lung cancer mortality is achievable in high-risk populations through individual screening with low-dose computed tomography (LDCT) [3]. Similar findings have since been reported based on results from the Dutch–Belgian Nederlands–Leuvens Longkanker Screenings Onderzoek (NELSON) lung cancer screening trial [4]. Nevertheless, translation of LDCT to the general population has proven challenging, and the implementation of imaging-based lung cancer screening to reduce lung cancer disease burden is likely to remain low in the near term. Liquid biopsies offer an ideal minimally invasive means of individualized risk assessment to address the need to improve lung cancer screening, either by identifying subjects at increased risk of lung cancer that would benefit from screening or for complementing LDCT-based screening, notably for assessment of indeterminate imaging findings [5].
MicroRNAs (miRNAs) are ~20–22-nucleotide non-coding RNAs that participate in regulation of gene expression, typically by binding to target miRNA transcripts and thus repressing miRNA translation. Dysregulation of the miRNA landscape was observed in multiple disease states, including cancer [6]. In lung cancers, altered miRNA expression has been found and characterized in tumor tissues as well as body fluids, suggesting utility for miRNAs as potential biomarkers in minimally invasive, liquid biopsy assays for assessing risk or detecting and monitoring disease status [7].
Here, using a cohort of sera (Table 1) collected up to one year prior to diagnosis of lung cancer (N = 102) and controls matched for age, sex, and smoking status (N = 212), we validated a panel of 30 miRNAs (Table 2) for predictive performance in distinguishing lung cancer cases from controls. The panel was developed based on review of previous reports of differential expression in lung cancer clinical specimens [8,9,10,11,12,13]. We further assessed the contributions of miRNAs for improving upon the predictive performance of a previously validated four-protein marker panel (4MP) consisting of cancer antigen 125 (CA125), carcinoembryonic antigen (CEA), cytokeratin-19 fragment (CYFRA 21-1), and the precursor form of surfactant protein B (Pro-SFTPB) for distinguishing lung cancer cases from controls compared to the 4MP alone [14,15].
Table 1.
Serum cohort patient and tumor characteristics. StDev, Standard Deviation.
| Variable | Cases | Controls |
|---|---|---|
| N | 102 | 212 |
| Age, mean ± StDev | 65 ± 6 | 65 ± 6 |
| Sex, N (%) | ||
| Female | 32 (31%) | 63 (30%) |
| Male | 70 (69%) | 149 (70%) |
| Smoking Status, N (%) | ||
| Current | 70 (69%) | 143 (67%) |
| Former | 32 (31%) | 69 (33%) |
| Smoking Pack Years (PYs), mean ± StDev | 53.5 ± 21.4 | 48.7 ± 20.5 |
| Histology, N (%) | ||
| Adenocarcinoma | 37 (36%) | - |
| Squamous Cell Carcinoma | 27 (26%) | - |
| Other Non-Small-Cell Lung Carcinoma | 38 (37%) | - |
Table 2.
miRNA candidates for serum-based early detection of lung cancer.
| Hsa-miRNA | Alternate ID(s) | Validated Reports of Dysregulation in Lung Cancer Clinical Specimens | TaqMan Assay ID | References | |
|---|---|---|---|---|---|
| Tissue | Biofluid | ||||
| miR-15b-5p | miR-15b | 3 | 390 | [9,10,16] | |
| miR-17-5p | miR-17 | 2 | 11 | 2308 | [9,10,11,16,17,18,19,20,21,22,23,24] |
| miR-19b-3p | miR-19b; miR-19b-1 | 1 | 9 | 396 | [9,10,22,25,26,27,28,29,30,31] |
| miR-21-5p | miR-21 | 7 | 45 | 397 | [9,10,18,23,25,28,29,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76] |
| miR-28-3p | 2 | 2446 | [9,10] | ||
| miR-30b-5p | miR-30b | 5 | 602 | [9,10,11,12,77] | |
| miR-30c-5p | 5 | 419 | [9,10,11,12,78] | ||
| miR-31-5p | miR-31 | 5 | 6 | 2279 | [25,32,34,58,68,70,79,80,81,82,83] |
| miR-92a-3p | miR-92a-2 | 6 | 431 | [9,10,11,12,16,84] | |
| miR-101-3p | miR-101 | 1 | 1 | 2253 | [10,85] |
| miR-106a-5p | miR-106a | 4 | 2169 | [9,10,53,86] | |
| miR-126-3p | miR-126 | 6 | 21 | 2228 | [9,10,11,25,28,32,34,41,51,55,68,70,71,74,87,88,89,90,91,92,93,94,95,96,97,98,99] |
| miR-133a-3p | miR-133a | 3 | 1 | 2246 | [10,100,101,102] |
| miR-140-3p | 2 | 2 | 2234 | [9,10,79,103] | |
| miR-140-5p | 4 | 1187 | [9,10,11,12] | ||
| miR-142-3p | 3 | 464 | [9,10,11] | ||
| miR-145-5p | miR-145 | 2 | 13 | 2278 | [10,13,23,32,39,49,51,57,60,71,95,96,98,104,105] |
| miR-148a-3p | miR-148; miR-148a | 2 | 5 | 470 | [10,11,12,44,63,106,107] |
| miR-155-5p | miR-155 | 2 | 9 | 2623 | [38,40,49,57,61,67,70,108,109,110,111] |
| miR-182-5p | miR-182 | 6 | 10 | 2334 | [17,23,25,34,38,55,61,68,70,71,79,93,112,113,114,115] |
| miR-197-3p | miR-197 | 5 | 497 | [9,10,31,38,61] | |
| miR-203a-3p | miR-203 | 2 | 1 | 507 | [17,116,117] |
| miR-205-5p | miR-205 | 8 | 15 | 509 | [23,28,32,34,35,51,62,68,70,72,77,80,88,95,96,99,114,116,118,119,120,121,122] |
| miR-210-3p | miR-210 | 7 | 20 | 512 | [9,17,23,34,41,51,54,55,66,67,68,70,79,83,88,89,93,95,96,98,99,107,121,123,124,125,126] |
| miR-221-3p | miR-221 | 2 | 7 | 524 | [9,10,13,23,29,94,120,127,128] |
| miR-320a-3p | miR-320 | 3 | 2277 | [9,10,13] | |
| miR-375 | 2 | 5 | 564 | [22,68,70,71,129,130,131] | |
| miR-451a | miR-451 | 5 | 7 | 1141 | [9,10,25,31,34,35,74,97,132,133,134] |
| miR-486-5p | miR-486 | 7 | 16 | 1278 | [9,10,11,34,35,41,50,52,54,55,68,70,71,75,79,88,108,122,126,134,135,136,137] |
| miR-660-5p | miR-660 | 3 | 1515 | [9,10,138] | |
2. Materials and Methods
2.1. Study Population
Serum samples for this nested case–control validation study were selected from the Carotene and Retinol Efficacy Trial (CARET) [139]. CARET was a randomized, double-blind, placebo-controlled trial conducted to assess the safety and cancer prevention efficacy of daily supplementation with β-carotene plus retinyl palmitate in 18,314 persons at high risk for lung cancer. Participants were enrolled at six United States study centers from 1985 to 1994 and were followed for cancer and mortality outcomes through 2005. Cancer status was confirmed based on review of clinical records and pathology reports. All CARET participants provided informed consent at recruitment and throughout follow-up, and the study was conducted with oversight from the respective institutional review boards at each of the study centers. CARET pre-diagnostic serum samples collected up to one year prior to diagnosis from 102 ever-smoker patients with lung cancer and 212 smoking-status-, age-, and sex-matched controls (Table 1) were used to test the individual performance of a panel of 30 candidate miRNAs (Table 2). Samples used for miRNA testing were aliquots of CARET specimens previously used for developing a four protein biomarker panel (4MP) for assessing lung cancer risk based on cancer antigen 125 (CA125), carcinoembryonic antigen (CEA), cytokeratin-19 fragment (CYFRA 21-1), and the precursor form of surfactant protein B (Pro-SFTPB) [14,15].
2.2. miRNA Isolation
Expression analysis of 30 total micro RNAs were performed using validation cohort of pre-diagnostic CARET serum samples (N = 102) collected up to one year prior to diagnosis of lung cancer and corresponding controls (N = 212) matched for age, sex, and smoking status. Total RNAs were isolated from 200 µL of serum using Norgen silicon-carbide resin Total RNA Purification Kit 17250 (Norgen Biotek, Thorold, ON, Canada) and eluted with 25 µL of elution solution. For normalization of sample-to-sample variation in the RNA isolation step, 10 fmol of Caenorhabditis elegans cel-miR-39 and cel-miR-54 (mirVana miRNA mimic, Applied Biosystems, Foster City, CA, USA); in a total volume of 5 µL were added to each denatured sample after mixing the serum sample with lysis buffer. Total RNA concentrations were quantified in each sample via a NanoDrop ND-1000 spectrophotometer.
2.3. TaqMan Assay
RNA was reverse transcribed using the TaqMan miRNA Reverse Kit (Applied Biosystems) in a 15 μL multiplex RT reaction. Relative quantification of targets was performed by qRT-PCR in reaction buffer containing SYBR Green dye (SsoAdvanced Universal SYBR SuperMix 1725270, Bio-Rad Laboratories, Hercules, CA, USA). The intercalation of SYBR Green into the PCR products was monitored in real time using a CFX384 Touch Real-time PCR Detection System (Bio-Rad Laboratories). Ct values were normalized to miR-16-5p.
2.4. Statistical Analyses
Receiver operating characteristic (ROC) curve analysis was performed to evaluate the predicative performance of biomarkers for distinguishing lung cancer cases from matched controls. A combination rule for miRNA was developed using logistic regression models. The combined model of the miRNA panel plus the protein 4MP was developed by fitting a logistic regression with the miRNA panel score and the 4MP scores as two separate predictors. The estimated AUC of the proposed 3-marker miRNA panel and miRNA panel plus 4MP were derived by using the empirical ROC estimator of the linear combination corresponding to the respective models. One thousand bootstrap iterations with replacement were implemented to estimate the confidence intervals and p values. Statistical significance was considered at p < 0.05.
3. Results
3.1. Candidate miRNA Selection
A panel of 30 candidate miRNAs was developed for testing based on review of literature reporting of differential expression in lung cancer patient biofluids or tumor tissues relative to control specimens. Priority was given to candidates identified across multiple independent studies. Table 2 summarizes the test panel candidates, with observed up- or downregulation relative to controls, and the number of studies in lung cancer that meet the criteria of: (i) inclusion of clinical samples and (ii) relevant controls, (iii) a validation component, and (iv) significance p ≤ 0.05 for the indicated miRNA. Relevant references for previous reports of dysregulated miRNA expression in clinical samples are also summarized in Table 2.
3.2. Predictive Performance of miRNAs for Early Detection of Lung Cancer
Assay of the panel of 30 miRNAs was performed using 102 case serum samples collected one-year preceding diagnosis of lung cancer and 212 control serum samples matched on the basis of age, sex, and smoking history (Table 1).
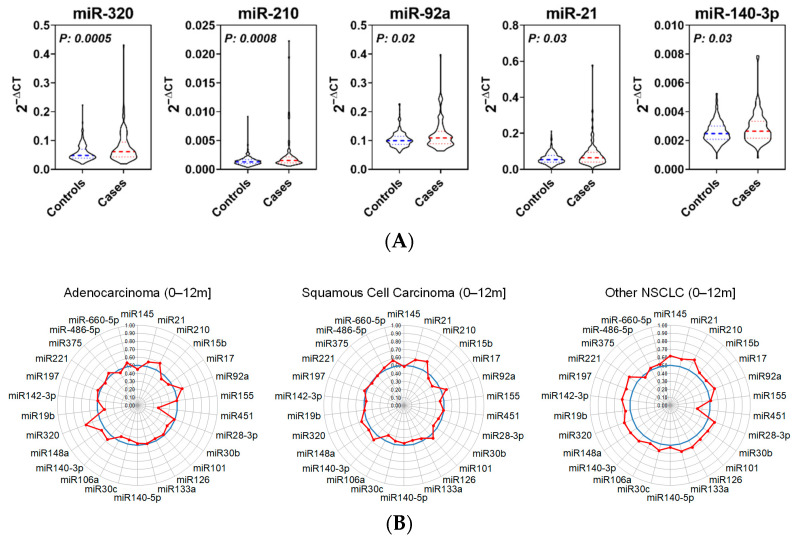
Four of the 30 miRNAs were below the limit of quantitation. Five miRNAs (miR-320a-3p, miR-210-3p, miR-92a-3p, miR-21-5p, and miR-140-3p) were statistically significantly (Wilcoxon’s rank sum p < 0.05) elevated in cases compared to matched controls with individual AUCs ranging from 0.57–0.62 (Figure 1A and Table 3; Tables S1 and S2). The performance of individual miRNA candidates was associated with stage at time of diagnosis, (Table S1). Stratification of cases according to lung cancer histological subtype revealed the predictive performance of individual miRNA candidates was conserved (Figure 1B).
Figure 1.
Expression and performance of panel miRNAs in lung cancer validation cohort. (A) Relative expression of top-performing upregulated miRNAs in prediagnostic lung cancer case sera (N = 102) and matched controls (N = 212). (B) Radar charts indicating predictive performance of panel miRNAs stratified according to lung cancer subtype. NSCLC, Non-Small-Cell Lung Carcinoma.
Table 3.
Performance of miRNAs for distinguishing lung cancer cases diagnosed within one year following blood draw from matched controls. AUC, Area Under the Receiver Operating Characteristic curve; Ctrl, Control.
| Case:Ctrl | |||
|---|---|---|---|
| Hsa-miRNA | Fold-Change | AUC | p |
| miR-451a | 0.88 | 0.37 | 0.0002 |
| miR-320a-3p | 1.36 | 0.62 | 0.0005 |
| miR-210-3p | 1.63 | 0.62 | 0.0008 |
| miR-92a-3p | 1.16 | 0.58 | 0.0217 |
| mir-21-5p | 1.33 | 0.58 | 0.0258 |
| miR-140-3p | 1.11 | 0.57 | 0.0318 |
| miR-148a-3p | 1.14 | 0.56 | 0.1135 |
| miR-660-5p | 1.24 | 0.55 | 0.1264 |
| miR-106a-5p | 0.97 | 0.46 | 0.2274 |
| miR-197-3p | 1.23 | 0.54 | 0.2274 |
| miR-17-5p | 1.01 | 0.47 | 0.3960 |
| miR-101-3p | 1.05 | 0.53 | 0.4178 |
| miR-221-3p | 1.04 | 0.52 | 0.4789 |
| miR-15b-5p | 0.98 | 0.48 | 0.4880 |
| miR-30b-5p | 1.02 | 0.48 | 0.5065 |
| miR-155-5p | 1.02 | 0.48 | 0.5686 |
| miR-142-3p | 1.06 | 0.52 | 0.6024 |
| miR-30c-5p | 1.02 | 0.48 | 0.6238 |
| miR-19b-3p | 1 | 0.49 | 0.6705 |
| miR-140-5p | 0.96 | 0.49 | 0.6988 |
| miR-145-5p | 1.07 | 0.51 | 0.7983 |
| miR-375 | 1.11 | 0.51 | 0.8771 |
| miR-28-3p | 1.07 | 0.5 | 0.8949 |
| miR-486-5p | 1.08 | 0.5 | 0.9118 |
| miR-126-3p | 1.05 | 0.5 | 0.9762 |
| miR-133a-3p | 1.1 | 0.5 | 0.9815 |
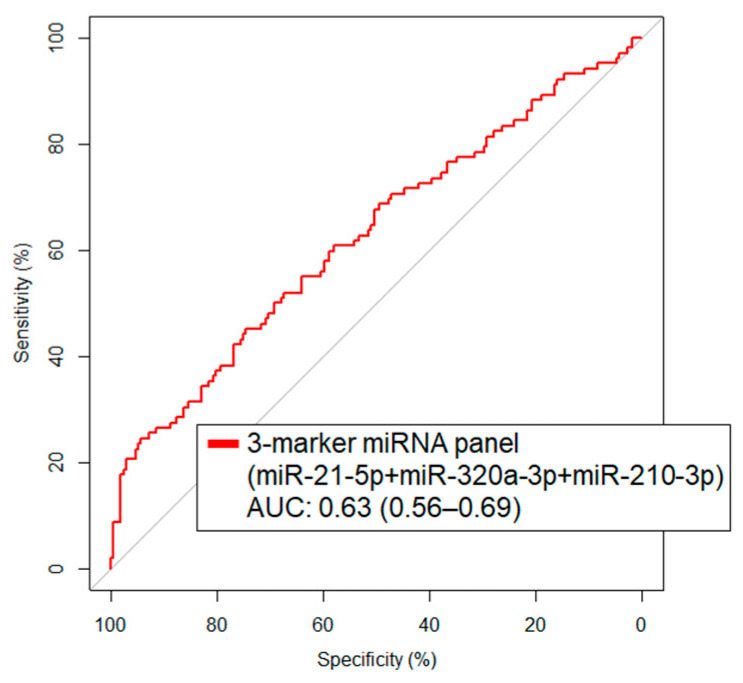
We next focused on those miRNAs that were statistically significantly elevated in case sera and, using logistic regression models, developed a combination rule. The combination of miR-21-5p + miR-320a-3p + miR-210-3p was identified as the highest performing panel with a resultant AUC of 0.63 (95% CI: 0.56–0.69) and 22.6% sensitivity at 95% specificity (Figure 2).
Figure 2.
Predictive performance as indicated by area under the curve (AUC) for 3-marker miRNA panel miRNAs in lung cancer prediagnostic serum validation cohort.
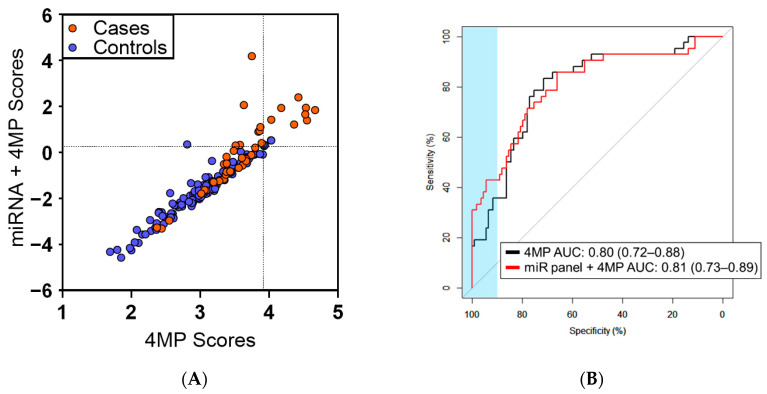
Using logistic regression models, we further determined whether the combination of miRNAs plus our previously validated 4MP would yield improved classification performance in differentiating lung cancer cases from controls when compared to the 4MP alone. A combined panel of the three-marker miRNA panel plus the 4MP yielded an AUC of 0.81 (95% CI: 0.73–0.89) (Figure 3A,B). In comparison to the 4MP alone, the combined miRNA panel plus 4MP resulted in statistically significantly improved sensitivity (19.1% (95% CI of difference 0.0–28.6); 2-sided p: 0.006) at 95% specificity (Table 4).
Figure 3.
Predictive performance of miRNA panel for distinguishing lung cancer cases diagnosed within one year of blood draw from controls. (A) Scatter plot comparing combined three-marker miRNA plus protein 4MP panel scores (y-axis) and the 4MP scores (x-axis) among cases diagnosed within one year of blood draw (orange nodes) and controls (blue nodes). Dashed lines indicate 95% Specificity thresholds. (B) ROC curves illustrating discrimination performance of the 3-marker miRNA plus protein 4MP panel and the 4MP alone.
Table 4.
Improvement in predictive performance of 4MP panel when combined with 3 miRNAs for distinguishing lung cancer cases from controls in prediagnostic serum validation cohort. CI, Confidence Interval.
| Specificity | 4MP Sensitivity | 4MP + 3 miRNA Sensitivity | Δ (95% CI) | p |
|---|---|---|---|---|
| 99% | 19.0% | 31.0% | 11.9% (2.38, 23.9) | 0.031 |
| 98% | 19.0% | 33.3% | 14.3% (4.8, 26.2) | 0.019 |
| 95% | 19.0% | 38.1% | 19.1% (0.0, 28.6) | 0.006 |
| 90% | 35.7% | 42.9% | 7.1% (-4.8, 26.2) | 0.369 |
4. Discussion
Earlier interception of lung cancer is directly linked with a more favorable outcome. Blood-based lung cancer biomarkers offer utility for assessment of individual risk, detection of asymptomatic disease and stratification of equivocal findings from PET or CT imaging. Such blood-based assays would integrate directly into existing clinical workflows and are especially ideal for implementation in a primary care setting. Here, we evaluated 30 miRNA candidates and identified a marker panel comprising three miRNAs for identifying individuals at high risk of developing or harboring disease. Importantly, the combination of miRNAs with a previously validated 4MP protein panel yielded significantly improved sensitivity at the highest specificity thresholds. When applied in a screening context, such performance would allow for identification of additional cases with a minimal false-positive burden.
We note some further considerations with respect to this study. Serum specimens were sourced from Carotene and Retinol Efficacy Trial (CARET) cohort samples for which enrolment was restricted to individuals at high-risk for developing lung cancer, based on smoking-history and/or asbestos exposure and age between 45 and 69 years. Accordingly, the predictive performance of the combined model outside of this age range or for lower-risk populations could not be determined. The performance of the combined miRNA plus 4MP among different ethnic and racial groups was likewise not assessed. Our study did not include an external cohort for further validation of the combined biomarker panels.
Indeterminate lung nodules identified by imaging present a challenging diagnostic scenario. Both the 4MP protein panel and miRNA candidates have been previously shown to contribute towards assessing indeterminate pulmonary nodules and offer improved performance compared to nodule size alone in predicting likelihood of cancer, supporting the observed association between the miRNA plus 4MP panel markers and lung cancer [10,15]. Furthermore, miRNA is known to regulate protein expression and mechanistic link between dysregulated miRNA and proteins in the context of cancer pathophysiology has been previously established [140,141,142,143,144].
5. Conclusions
A panel of miRNA plus an existing four-protein marker panel yielded notable improvements in sensitivity at high specificity compared to the protein panel alone. Our findings demonstrate the merits of including miRNAs when developing biomarker panels for lung cancer detection or risk assessment and suggest important implications for improving lung cancer screening and detection that warrant additional study.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers14174221/s1, Table S1: Candidate miRNA Panel, Cohort AUROCs; Table S2: Candidate miRNA Panel, PCR Assay Raw Data.
Author Contributions
Conceptualization, J.V., J.F.F., N.P., G.A.C. and S.M.H.; formal analysis, J.V. and J.F.F.; investigation, N.P. and M.S.; resources, G.E.G., M.D.T. and M.J.B.; data curation, N.P., M.S. and C.I.; writing—original draft preparation, J.V. and J.F.F.; writing—review and editing, N.P., M.S., E.J.O., J.B.D., G.E.G., M.D.T., M.J.B., Z.F., G.A.C. and S.M.H.; visualization, J.V., J.F.F. and N.P.; funding acquisition, G.A.C. and S.M.H. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and with approval and annual review by the respective Institutional Review Board at each of the six CARET Study Centers. An external Safety and Endpoints Monitoring Committee (SEMC) provided additional oversight and semiannual review of each CARET Study Center. Clinical Trial NCT number: NCT00712647, 1985–2005.
Informed Consent Statement
All human subjects research reported herein was conducted consistent with National Institutes of Health Informed Consent Exemption category 4 guidance and involved retrospective study of existing publicly available, deidentified banked serum specimens obtained from the CARET Biorepository.
Data Availability Statement
Relevant data supporting the findings of this study are available within the Article and Supplementary Materials or are available from the authors upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The CARET cohort was funded by NIH grants U01-CA063673, UM1-CA167462 and U01-CA167462 (NCI, NIH). This study was supported in part by grants 1U19CA203654 and UO1194733 from the National Cancer Institute and the National Cancer Institute Early Detection Research Network, and by the MD Anderson Cancer Center Lung Cancer Moon Shot Program. G.A.C. is the Felix L. Haas Endowed Professor in Basic Science and is supported by the NCI grants 1R01 CA182905-01 and 1R01CA222007-01A1, an NIGMS 1R01GM122775-01 grant, a DoD Idea Award W81XWH2110030, a Team DOD grant in Gastric Cancer, a Chronic Lymphocytic Leukemia Moonshot Flagship project, a CLL Global Research Foundation 2019 grant, a CLL Global Research Foundation 2020 grant, The G. Harold & Leila Y. Mathers Foundation, a grant from Torrey Coast Foundation, an Institutional Research Grant and Development Grant associated with the Brain SPORE 2P50CA127001.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.SEER*Explorer. [(accessed on 11 August 2021)]; Available online: https://seer.cancer.gov/explorer/index.html.
- 3.National Lung Screening Trial Research Team. Aberle D.R., Adams A.M., Berg C.D., Black W.C., Clapp J.D., Fagerstrom R.M., Gareen I.F., Gatsonis C., Marcus P.M., et al. Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. N. Engl. J. Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Koning H.J., van der Aalst C.M., de Jong P.A., Scholten E.T., Nackaerts K., Heuvelmans M.A., Lammers J.-W.J., Weenink C., Yousaf-Khan U., Horeweg N., et al. Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N. Engl. J. Med. 2020;382:503–513. doi: 10.1056/NEJMoa1911793. [DOI] [PubMed] [Google Scholar]
- 5.Dama E., Colangelo T., Fina E., Cremonesi M., Kallikourdis M., Veronesi G., Bianchi F. Biomarkers and Lung Cancer Early Detection: State of the Art. Cancers. 2021;13:3919. doi: 10.3390/cancers13153919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vannini I., Fanini F., Fabbri M. Emerging Roles of MicroRNAs in Cancer. Curr. Opin. Genet. Dev. 2018;48:128–133. doi: 10.1016/j.gde.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anfossi S., Babayan A., Pantel K., Calin G.A. Clinical Utility of Circulating Non-Coding RNAs—An Update. Nat. Rev. Clin. Oncol. 2018;15:541–563. doi: 10.1038/s41571-018-0035-x. [DOI] [PubMed] [Google Scholar]
- 8.Zhong S., Golpon H., Zardo P., Borlak J. MiRNAs in Lung Cancer. A Systematic Review Identifies Predictive and Prognostic MiRNA Candidates for Precision Medicine in Lung Cancer. Transl. Res. 2021;230:164–196. doi: 10.1016/j.trsl.2020.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Boeri M., Verri C., Conte D., Roz L., Modena P., Facchinetti F., Calabrò E., Croce C.M., Pastorino U., Sozzi G. MicroRNA Signatures in Tissues and Plasma Predict Development and Prognosis of Computed Tomography Detected Lung Cancer. Proc. Natl. Acad. Sci. USA. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sozzi G., Boeri M., Rossi M., Verri C., Suatoni P., Bravi F., Roz L., Conte D., Grassi M., Sverzellati N., et al. Clinical Utility of a Plasma-Based MiRNA Signature Classifier within Computed Tomography Lung Cancer Screening: A Correlative MILD Trial Study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2014;32:768–773. doi: 10.1200/JCO.2013.50.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bianchi F., Nicassio F., Marzi M., Belloni E., Dall’Olio V., Bernard L., Pelosi G., Maisonneuve P., Veronesi G., Di Fiore P.P. A Serum Circulating MiRNA Diagnostic Test to Identify Asymptomatic High-Risk Individuals with Early Stage Lung Cancer. EMBO Mol. Med. 2011;3:495–503. doi: 10.1002/emmm.201100154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montani F., Marzi M.J., Dezi F., Dama E., Carletti R.M., Bonizzi G., Bertolotti R., Bellomi M., Rampinelli C., Maisonneuve P., et al. MiR-Test: A Blood Test for Lung Cancer Early Detection. J. Natl. Cancer Inst. 2015;107:djv063. doi: 10.1093/jnci/djv063. [DOI] [PubMed] [Google Scholar]
- 13.Chen X., Hu Z., Wang W., Ba Y., Ma L., Zhang C., Wang C., Ren Z., Zhao Y., Wu S., et al. Identification of Ten Serum MicroRNAs from a Genome-Wide Serum MicroRNA Expression Profile as Novel Noninvasive Biomarkers for Nonsmall Cell Lung Cancer Diagnosis. Int. J. Cancer. 2012;130:1620–1628. doi: 10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- 14.Integrative Analysis of Lung Cancer Etiology and Risk (INTEGRAL) Consortium for Early Detection of Lung Cancer. Guida F., Sun N., Bantis L.E., Muller D.C., Li P., Taguchi A., Dhillon D., Kundnani D.L., Patel N.J., et al. Assessment of Lung Cancer Risk on the Basis of a Biomarker Panel of Circulating Proteins. JAMA Oncol. 2018;4:e182078. doi: 10.1001/jamaoncol.2018.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostrin E.J., Bantis L.E., Wilson D.O., Patel N., Wang R., Kundnani D., Adams-Haduch J., Dennison J.B., Fahrmann J.F., Chiu H.T., et al. Contribution of a Blood-Based Protein Biomarker Panel to the Classification of Indeterminate Pulmonary Nodules. J. Thorac. Oncol. 2021;16:228–236. doi: 10.1016/j.jtho.2020.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan L., Qi H., Teng J., Su B., Chen H., Wang C., Xia Q. Identification of Serum MiRNAs by Nano-Quantum Dots Microarray as Diagnostic Biomarkers for Early Detection of Non-Small Cell Lung Cancer. Tumor Biol. 2016;37:7777–7784. doi: 10.1007/s13277-015-4608-3. [DOI] [PubMed] [Google Scholar]
- 17.Raponi M., Dossey L., Jatkoe T., Wu X., Chen G., Fan H., Beer D.G. MicroRNA Classifiers for Predicting Prognosis of Squamous Cell Lung Cancer. Cancer Res. 2009;69:5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 18.Qi Z., Yang D.-Y., Cao J. Increased Micro-RNA 17, 21, and 192 Gene Expressions Improve Early Diagnosis in Non-Small Cell Lung Cancer. Med. Oncol. 2014;31:195. doi: 10.1007/s12032-014-0195-1. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y., Zhang Y., Yin Y., Li S. Detection of Circulating Exosomal MiR-17-5p Serves as a Novel Non-Invasive Diagnostic Marker for Non-Small Cell Lung Cancer Patients. Pathol. Res. Pract. 2019;215:152466. doi: 10.1016/j.prp.2019.152466. [DOI] [PubMed] [Google Scholar]
- 20.Cazzoli R., Buttitta F., Di Nicola M., Malatesta S., Marchetti A., Rom W.N., Pass H.I. MicroRNAs Derived from Circulating Exosomes as Noninvasive Biomarkers for Screening and Diagnosing Lung Cancer. J. Thorac. Oncol. 2013;8:1156–1162. doi: 10.1097/JTO.0b013e318299ac32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hetta H.F., Zahran A.M., El-Mahdy R.I., Nabil E.E., Esmaeel H.M., Elkady O.A., Elkady A., Mohareb D.A., Mahmoud Mostafa M., John J. Assessment of Circulating MiRNA-17 and MiRNA-222 Expression Profiles as Non-Invasive Biomarkers in Egyptian Patients with Non-Small-Cell Lung Cancer. Asian Pac. J. Cancer Prev. 2019;20:1927–1933. doi: 10.31557/APJCP.2019.20.6.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu S., Kong H., Hou Y., Ge D., Huang W., Ou J., Yang D., Zhang L., Wu G., Song Y., et al. Two Plasma MicroRNA Panels for Diagnosis and Subtype Discrimination of Lung Cancer. Lung Cancer. 2018;123:44–51. doi: 10.1016/j.lungcan.2018.06.027. [DOI] [PubMed] [Google Scholar]
- 23.Xi K.-X., Zhang X.-W., Yu X.-Y., Wang W.-D., Xi K.-X., Chen Y.-Q., Wen Y.-S., Zhang L.-J. The Role of Plasma MiRNAs in the Diagnosis of Pulmonary Nodules. J. Thorac. Dis. 2018;10:4032–4041. doi: 10.21037/jtd.2018.06.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Q., Si Q., Xiao S., Xie Q., Lin J., Wang C., Chen L., Chen Q., Wang L. Prognostic Significance of Serum MiR-17-5p in Lung Cancer. Med. Oncol. 2012;30:353. doi: 10.1007/s12032-012-0353-2. [DOI] [PubMed] [Google Scholar]
- 25.Gallach S., Jantus-Lewintre E., Calabuig-Fariñas S., Montaner D., Alonso S., Sirera R., Blasco A., Usó M., Guijarro R., Martorell M., et al. MicroRNA Profiling Associated with Non-Small Cell Lung Cancer: Next Generation Sequencing Detection, Experimental Validation, and Prognostic Value. Oncotarget. 2017;8:56143–56157. doi: 10.18632/oncotarget.18603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu C., Cao Y., He Z., He J., Hu C., Duan H., Jiang J. Serum Levels of MiR-19b and MiR-146a as Prognostic Biomarkers for Non-Small Cell Lung Cancer. Tohoku J. Exp. Med. 2014;232:85–95. doi: 10.1620/tjem.232.85. [DOI] [PubMed] [Google Scholar]
- 27.Bulgakova O., Zhabayeva D., Kussainova A., Pulliero A., Izzotti A., Bersimbaev R. MiR-19 in Blood Plasma Reflects Lung Cancer Occurrence but Is Not Specifically Associated with Radon Exposure. Oncol. Lett. 2018;15:8816–8824. doi: 10.3892/ol.2018.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaporozhchenko I.A., Morozkin E.S., Skvortsova T.E., Ponomaryova A.A., Rykova E.Y., Cherdyntseva N.V., Polovnikov E.S., Pashkovskaya O.A., Pokushalov E.A., Vlassov V.V., et al. Plasma MiR-19b and MiR-183 as Potential Biomarkers of Lung Cancer. PLoS ONE. 2016;11:e0165261. doi: 10.1371/journal.pone.0165261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou X., Wen W., Shan X., Zhu W., Xu J., Guo R., Cheng W., Wang F., Qi L.-W., Chen Y., et al. A Six-MicroRNA Panel in Plasma Was Identified as a Potential Biomarker for Lung Adenocarcinoma Diagnosis. Oncotarget. 2016;8:6513–6525. doi: 10.18632/oncotarget.14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma J., Lin Y., Zhan M., Mann D.L., Stass S.A., Jiang F. Differential MiRNA Expressions in Peripheral Blood Mononuclear Cells for Diagnosis of Lung Cancer. Lab. Investig. 2015;95:1197–1206. doi: 10.1038/labinvest.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pu Q., Huang Y., Lu Y., Peng Y., Zhang J., Feng G., Wang C., Liu L., Dai Y. Tissue-Specific and Plasma MicroRNA Profiles Could Be Promising Biomarkers of Histological Classification and TNM Stage in Non-Small Cell Lung Cancer. Thorac. Cancer. 2016;7:348–354. doi: 10.1111/1759-7714.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Begum S., Hayashi M., Ogawa T., Jabboure F.J., Brait M., Izumchenko E., Tabak S., Ahrendt S.A., Westra W.H., Koch W., et al. An Integrated Genome-Wide Approach to Discover Deregulated MicroRNAs in Non-Small Cell Lung Cancer: Clinical Significance of MIR-23b-3p Deregulation. Sci. Rep. 2015;5:13236. doi: 10.1038/srep13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao W., Yu Y., Cao H., Shen H., Li X., Pan S., Shu Y. Deregulated Expression of MiR-21, MiR-143 and MiR-181a in Non Small Cell Lung Cancer Is Related to Clinicopathologic Characteristics or Patient Prognosis. Biomed. Pharmacother. 2010;64:399–408. doi: 10.1016/j.biopha.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 34.Li C., Yin Y., Liu X., Xi X., Xue W., Qu Y. Non-Small Cell Lung Cancer Associated MicroRNA Expression Signature: Integrated Bioinformatics Analysis, Validation and Clinical Significance. Oncotarget. 2017;8:24564–24578. doi: 10.18632/oncotarget.15596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solomides C.C., Evans B.J., Navenot J.-M., Vadigepalli R., Peiper S.C., Wang Z. MicroRNA Profiling in Lung Cancer Reveals New Molecular Markers for Diagnosis. Acta Cytol. 2012;56:645–654. doi: 10.1159/000343473. [DOI] [PubMed] [Google Scholar]
- 36.Tian F., Li R., Chen Z., Shen Y., Lu J., Xie X., Ge Q. Differentially Expressed MiRNAs in Tumor, Adjacent, and Normal Tissues of Lung Adenocarcinoma. BioMed Res. Int. 2016;2016:1428271. doi: 10.1155/2016/1428271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian L., Shan W., Zhang Y., Lv X., Li X., Wei C. Up-Regulation of MiR-21 Expression Predicate Advanced Clinicopathological Features and Poor Prognosis in Patients with Non-Small Cell Lung Cancer. Pathol. Oncol. Res. 2016;22:161–167. doi: 10.1007/s12253-015-9979-7. [DOI] [PubMed] [Google Scholar]
- 38.Abd-El-Fattah A.A., Sadik N.A.H., Shaker O.G., Aboulftouh M.L. Differential MicroRNAs Expression in Serum of Patients with Lung Cancer, Pulmonary Tuberculosis, and Pneumonia. Cell Biochem. Biophys. 2013;67:875–884. doi: 10.1007/s12013-013-9575-y. [DOI] [PubMed] [Google Scholar]
- 39.Aiso T., Ohtsuka K., Ueda M., Karita S., Yokoyama T., Takata S., Matsuki N., Kondo H., Takizawa H., Okada A.A., et al. Serum Levels of Candidate MicroRNA Diagnostic Markers Differ among the Stages of Non-Small-Cell Lung Cancer. Oncol. Lett. 2018;16:6643–6651. doi: 10.3892/ol.2018.9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao F., Chang J., Wang H., Zhang G. Potential Diagnostic Value of MiR-155 in Serum from Lung Adenocarcinoma Patients. Oncol. Rep. 2014;31:351–357. doi: 10.3892/or.2013.2830. [DOI] [PubMed] [Google Scholar]
- 41.Świtlik W.Z., Karbownik M.S., Suwalski M., Kozak J., Szemraj J. Serum MiR-210-3p as a Potential Noninvasive Biomarker of Lung Adenocarcinoma: A Preliminary Study. Genet. Test. Mol. Biomark. 2019;23:353–358. doi: 10.1089/gtmb.2018.0275. [DOI] [PubMed] [Google Scholar]
- 42.Wang X., Jia Z., Shi H., Pan C. Identification and Evaluation of 2 Circulating MicroRNAs for Non-Small Cell Lung Cancer Diagnosis. Clin. Exp. Pharmacol. Physiol. 2018;45:1083–1086. doi: 10.1111/1440-1681.12977. [DOI] [PubMed] [Google Scholar]
- 43.Wang Z.-X., Bian H.-B., Wang J.-R., Cheng Z.-X., Wang K.-M., De W. Prognostic Significance of Serum MiRNA-21 Expression in Human Non-Small Cell Lung Cancer. J. Surg. Oncol. 2011;104:847–851. doi: 10.1002/jso.22008. [DOI] [PubMed] [Google Scholar]
- 44.Yang J., Li B., Lu H., Chen Y., Lu C., Zhu R., Liu S., Yi Q., Li J., Song C. Serum MiR-152, MiR-148a, MiR-148b, and MiR-21 as Novel Biomarkers in Non-Small Cell Lung Cancer Screening. Tumor Biol. 2015;36:3035–3042. doi: 10.1007/s13277-014-2938-1. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y., Chen K., Zhou Y., Hu Z., Chen S., Huang Y. Application of Serum MicroRNA-9-5p, 21–25p, and 223-3p Combined with Tumor Markers in the Diagnosis of Non-Small-Cell Lung Cancer in Yunnan in Southwestern China. OncoTargets Ther. 2018;11:587–597. doi: 10.2147/OTT.S152957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye M., Yang W., Wang W., Hu W., Zhu C., Yang H. Serum MicroRNA-21 Is a Potential Diagnostic Marker for Earlier Lung Squamous Cell Carcinoma Detection. Int. J. Clin. Exp. Med. 2017;10:3352–3358. [Google Scholar]
- 47.Zhao W., Zhao J.-J., Zhang L., Xu Q.-F., Zhao Y.-M., Shi X.-Y., Xu A.-G. Serum MiR-21 Level: A Potential Diagnostic and Prognostic Biomarker for Non-Small Cell Lung Cancer. Int. J. Clin. Exp. Med. 2015;8:14759–14763. [PMC free article] [PubMed] [Google Scholar]
- 48.Abu-Duhier F.M., Javid J., Sughayer M.A., Mir R., Albalawi T., Alauddin M.S. Clinical Significance of Circulatory MiRNA-21 as an Efficient Non-Invasive Biomarker for the Screening of Lung Cancer Patients. Asian Pac. J. Cancer Prev. 2018;19:2607–2611. doi: 10.22034/APJCP.2018.19.9.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geng Q., Fan T., Zhang B., Wang W., Xu Y., Hu H. Five MicroRNAs in Plasma as Novel Biomarkers for Screening of Early-Stage Non-Small Cell Lung Cancer. Respir. Res. 2014;15:149. doi: 10.1186/s12931-014-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin X., Chen Y., Chen H., Fei S., Chen D., Cai X., Liu L., Lin B., Su H., Zhao L., et al. Evaluation of Tumor-Derived Exosomal MiRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using next-Generation Sequencing. Clin. Cancer Res. 2017;23:5311–5319. doi: 10.1158/1078-0432.CCR-17-0577. [DOI] [PubMed] [Google Scholar]
- 51.Lin Y., Leng Q., Jiang Z., Guarnera M.A., Zhou Y., Chen X., Wang H., Zhou W., Cai L., Fang H., et al. A Classifier Integrating Plasma Biomarkers and Radiological Characteristics for Distinguishing Malignant from Benign Pulmonary Nodules. Int. J. Cancer. 2017;141:1240–1248. doi: 10.1002/ijc.30822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mozzoni P., Banda I., Goldoni M., Corradi M., Tiseo M., Acampa O., Balestra V., Ampollini L., Casalini A., Carbognani P., et al. Plasma and EBC MicroRNAs as Early Biomarkers of Non-Small-Cell Lung Cancer. Biomarkers. 2013;18:679–686. doi: 10.3109/1354750X.2013.845610. [DOI] [PubMed] [Google Scholar]
- 53.Shan X., Zhang H., Zhang L., Zhou X., Wang T., Zhang J., Shu Y., Zhu W., Wen W., Liu P. Identification of Four Plasma MicroRNAs as Potential Biomarkers in the Diagnosis of Male Lung Squamous Cell Carcinoma Patients in China. Cancer Med. 2018;7:2370–2381. doi: 10.1002/cam4.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen J., Liu Z., Todd N.W., Zhang H., Liao J., Yu L., Guarnera M.A., Li R., Cai L., Zhan M., et al. Diagnosis of Lung Cancer in Individuals with Solitary Pulmonary Nodules by Plasma MicroRNA Biomarkers. BMC Cancer. 2011;11:374. doi: 10.1186/1471-2407-11-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen J., Todd N.W., Zhang H., Yu L., Lingxiao X., Mei Y., Guarnera M., Liao J., Chou A., Lu C.L., et al. Plasma MicroRNAs as Potential Biomarkers for Non-Small-Cell Lung Cancer. Lab. Investig. 2011;91:579–587. doi: 10.1038/labinvest.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun Y., Mei H., Xu C., Tang H., Wei W. Circulating MicroRNA-339-5p and -21 in Plasma as an Early Detection Predictors of Lung Adenocarcinoma. Pathol. Res. Pract. 2018;214:119–125. doi: 10.1016/j.prp.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 57.Tang D., Shen Y., Wang M., Yang R., Wang Z., Sui A., Jiao W., Wang Y. Identification of Plasma MicroRNAs as Novel Noninvasive Biomarkers for Early Detection of Lung Cancer. Eur. J. Cancer Prev. 2013;22:540–548. doi: 10.1097/CEJ.0b013e32835f3be9. [DOI] [PubMed] [Google Scholar]
- 58.Wang S., Wang Z., Wang Q., Cui Y., Luo S. Clinical Significance of the Expression of MiRNA-21, MiRNA-31 and MiRNA-Let7 in Patients with Lung Cancer. Saudi J. Biol. Sci. 2019;26:777–781. doi: 10.1016/j.sjbs.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei J., Gao W., Zhu C.-J., Liu Y.-Q., Mei Z., Cheng T., Shu Y.-Q. Identification of Plasma MicroRNA-21 as a Biomarker for Early Detection and Chemosensitivity of Non-Small Cell Lung Cancer. Chin. J. Cancer. 2011;30:407–414. doi: 10.5732/cjc.010.10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang H., Mao F., Shen T., Luo Q., Ding Z., Qian L., Huang J. Plasma MiR-145, MiR-20a, MiR-21 and MiR-223 as Novel Biomarkers for Screening Early-stage Non-small Cell Lung Cancer. Oncol. Lett. 2017;13:669–676. doi: 10.3892/ol.2016.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng D., Haddadin S., Wang Y., Gu L.-Q., Perry M.C., Freter C.E., Wang M.X. Plasma Micrornas as Novel Biomarkers for Early Detection of Lung Cancer. Int. J. Clin. Exp. Pathol. 2011;4:575–586. [PMC free article] [PubMed] [Google Scholar]
- 62.Azimi S.A., Sadegh Nia H.R., Mosavi Jarrahi A., Jamaati H.R., Kazempour Dizaji M., Dargahi H., Bahrami N., Pasdar A., Khosravi A., Bahrami N., et al. Ectopic Expression of MiRNA-21 and MiRNA-205 in Non-Small Cell Lung Cancer. Int. J. Cancer Manag. 2019;12:e85456. doi: 10.5812/ijcm.85456. [DOI] [Google Scholar]
- 63.Abdollahi A., Rahmati S., Ghaderi B., Sigari N., Nikkhoo B., Sharifi K., Abdi M. A Combined Panel of Circulating MicroRNA as a Diagnostic Tool for Detection of the Non-Small Cell Lung Cancer. QJM. 2019;112:779–785. doi: 10.1093/qjmed/hcz158. [DOI] [PubMed] [Google Scholar]
- 64.Li Y., Li W., Ouyang Q., Hu S., Tang J. Detection of Lung Cancer with Blood MicroRNA-21 Expression Levels in Chinese Population. Oncol. Lett. 2011;2:991–994. doi: 10.3892/ol.2011.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qiu F., Gu W.-G., Li C., Nie S.-L., Yu F. Analysis on Expression Level and Diagnostic Value of MiR-19 and MiR-21 in Peripheral Blood of Patients with Undifferentiated Lung Cancer. Eur. Rev. Med. Pharmacol. Sci. 2018;22:8367–8373. doi: 10.26355/eurrev_201801_16534. [DOI] [PubMed] [Google Scholar]
- 66.Razzak R., Bédard E.L.R., Kim J.O., Gazala S., Guo L., Ghosh S., Joy A., Nijjar T., Wong E., Roa W.H. MicroRNA Expression Profiling of Sputum for the Detection of Early and Locally Advanced Non-Small-Cell Lung Cancer: A Prospective Case–Control Study. Curr. Oncol. 2016;23:86–94. doi: 10.3747/co.23.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roa W.H., Kim J.O., Razzak R., Du H., Guo L., Singh R., Gazala S., Ghosh S., Wong E., Joy A.A., et al. Sputum MicroRNA Profiling: A Novel Approach for the Early Detection of Non-Small Cell Lung Cancer. Clin. Investig. Med. 2012;35:E271–E281. doi: 10.25011/cim.v35i5.18700. [DOI] [PubMed] [Google Scholar]
- 68.Shen J., Liao J., Guarnera M.A., Fang H., Cai L., Stass S.A., Jiang F. Analysis of MicroRNAs in Sputum to Improve Computed Tomography for Lung Cancer Diagnosis. J. Thorac. Oncol. 2014;9:33–40. doi: 10.1097/JTO.0000000000000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xie Y., Todd N.W., Liu Z., Zhan M., Fang H., Peng H., Alattar M., Deepak J., Stass S.A., Jiang F. Altered MiRNA Expression in Sputum for Diagnosis of Non-Small Cell Lung Cancer. Lung Cancer. 2010;67:170–176. doi: 10.1016/j.lungcan.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xing L., Su J., Guarnera M.A., Zhang H., Cai L., Zhou R., Stass S.A., Jiang F. Sputum MicroRNA Biomarkers for Identifying Lung Cancer in Indeterminate Solitary Pulmonary Nodules. Clin. Cancer Res. 2015;21:484–489. doi: 10.1158/1078-0432.CCR-14-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu L., Todd N.W., Xing L., Xie Y., Zhang H., Liu Z., Fang H., Zhang J., Katz R.L., Jiang F. Early Detection of Lung Adenocarcinoma in Sputum by a Panel of MicroRNA Markers. Int. J. Cancer. 2010;127:2870–2878. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang M., Zhang P., Hu G., Xiao Z., Xu F., Zhong T., Huang F., Kuang H., Zhang W. Relative Expressions of MiR-205-5p, MiR-205-3p, and MiR-21 in Tissues and Serum of Non-Small Cell Lung Cancer Patients. Mol. Cell. Biochem. 2013;383:67–75. doi: 10.1007/s11010-013-1755-y. [DOI] [PubMed] [Google Scholar]
- 73.Liu X.-G., Zhu W.-Y., Huang Y.-Y., Ma L.-N., Zhou S.-Q., Wang Y.-K., Zeng F., Zhou J.-H., Zhang Y.-K. High Expression of Serum MiR-21 and Tumor MiR-200c Associated with Poor Prognosis in Patients with Lung Cancer. Med. Oncol. 2012;29:618–626. doi: 10.1007/s12032-011-9923-y. [DOI] [PubMed] [Google Scholar]
- 74.Markou A., Sourvinou I., Vorkas P.A., Yousef G.M., Lianidou E. Clinical Evaluation of MicroRNA Expression Profiling in Non Small Cell Lung Cancer. Lung Cancer. 2013;81:388–396. doi: 10.1016/j.lungcan.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 75.Tian F., Shen Y., Chen Z., Li R., Lu J., Ge Q. Aberrant MiR-181b-5p and MiR-486-5p Expression in Serum and Tissue of Non-Small Cell Lung Cancer. Gene. 2016;591:338–343. doi: 10.1016/j.gene.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 76.Zhu H., Chen W., Xu J., Yin L., Zhu H., Liu J., Wang T., He X. MiR-21 Expression Significance in Non-Small Cell Lung Cancer Tissue and Plasma. Int. J. Clin. Exp. Med. 2017;10:2918–2924. [Google Scholar]
- 77.Yang X., Su W., Chen X., Geng Q., Zhai J., Shan H., Guo C., Wang Z., Fu H., Jiang H., et al. Validation of a Serum 4-MicroRNA Signature for the Detection of Lung Cancer. Transl. Lung Cancer Res. 2019;8:636–648. doi: 10.21037/tlcr.2019.09.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rani S., Gately K., Crown J., O’Byrne K., O’Driscoll L. Global Analysis of Serum MicroRNAs as Potential Biomarkers for Lung Adenocarcinoma. Cancer Biol. Ther. 2013;14:1104–1112. doi: 10.4161/cbt.26370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tan X., Qin W., Zhang L., Hang J., Li B., Zhang C., Wan J., Zhou F., Shao K., Sun Y., et al. A 5-MicroRNA Signature for Lung Squamous Cell Carcinoma Diagnosis and Hsa-MiR-31 for Prognosis. Clin. Cancer Res. 2011;17:6802–6811. doi: 10.1158/1078-0432.CCR-11-0419. [DOI] [PubMed] [Google Scholar]
- 80.Wang J., Li Z., Ge Q., Wu W., Zhu Q., Luo J., Chen L. Characterization of MicroRNA Transcriptome in Tumor, Adjacent, and Normal Tissues of Lung Squamous Cell Carcinoma. J. Thorac. Cardiovasc. Surg. 2015;149:1404–1414.e4. doi: 10.1016/j.jtcvs.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 81.Yang M., Xiao L., Zhang Y., Li G., Zhou J. Clinical Significance of Serum MiR-31 as a Predictive Biomarker for Lung Adenocarcinoma. Int. J. Clin. Exp. Pathol. 2017;10:4668–4674. [Google Scholar]
- 82.Yan H.-J., Ma J.-Y., Wang L., Gu W. Expression and Significance of Circulating MicroRNA-31 in Lung Cancer Patients. Med. Sci. Monit. 2015;21:722–726. doi: 10.12659/MSM.893213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li N., Ma J., Guarnera M.A., Fang H., Cai L., Jiang F. Digital PCR Quantification of MiRNAs in Sputum for Diagnosis of Lung Cancer. J. Cancer Res. Clin. Oncol. 2014;140:145–150. doi: 10.1007/s00432-013-1555-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu Y., Zuo J., Tan Q., Thin K.Z., Li P., Zhu M., Yu M., Fu Z., Liang C., Tu J. Plasma MiR-92a-2 as a Biomarker for Small Cell Lung Cancer. Cancer Biomark. 2017;18:319–327. doi: 10.3233/CBM-160254. [DOI] [PubMed] [Google Scholar]
- 85.Luo L., Zhang T., Liu H., Lv T., Yuan D., Yao Y., Lv Y., Song Y. MiR-101 and Mcl-1 in Non-Small-Cell Lung Cancer: Expression Profile and Clinical Significance. Med. Oncol. 2012;29:1681–1686. doi: 10.1007/s12032-011-0085-8. [DOI] [PubMed] [Google Scholar]
- 86.Zhang L., Shan X., Wang J., Zhu J., Huang Z., Zhang H., Zhou X., Cheng W., Shu Y., Zhu W., et al. A Three-MicroRNA Signature for Lung Squamous Cell Carcinoma Diagnosis in Chinese Male Patients. Oncotarget. 2017;8:86897–86907. doi: 10.18632/oncotarget.19666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen S.-W., Wang T.-B., Tian Y.-H., Zheng Y.-G. Down-Regulation of MicroRNA-126 and MicroRNA-133b Acts as Novel Predictor Biomarkers in Progression and Metastasis of Non Small Cell Lung Cancer. Int. J. Clin. Exp. Pathol. 2015;8:14983–14988. [PMC free article] [PubMed] [Google Scholar]
- 88.Świtlik W., Karbownik M.S., Suwalski M., Kozak J., Szemraj J. MiR-30a-5p Together with MiR-210-3p as a Promising Biomarker for Non-Small Cell Lung Cancer: A Preliminary Study. Cancer Biomark. 2018;21:479–488. doi: 10.3233/CBM-170767. [DOI] [PubMed] [Google Scholar]
- 89.Tafsiri E., Darbouy M., Shadmehr M.B., Zagryazhskaya A., Alizadeh J., Karimipoor M. Expression of MiRNAs in Non-Small-Cell Lung Carcinomas and Their Association with Clinicopathological Features. Tumor Biol. 2015;36:1603–1612. doi: 10.1007/s13277-014-2755-6. [DOI] [PubMed] [Google Scholar]
- 90.Grimolizzi F., Monaco F., Leoni F., Bracci M., Staffolani S., Bersaglieri C., Gaetani S., Valentino M., Amati M., Rubini C., et al. Exosomal MiR-126 as a Circulating Biomarker in Non-Small-Cell Lung Cancer Regulating Cancer Progression. Sci. Rep. 2017;7:15277. doi: 10.1038/s41598-017-15475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin Q., Mao W., Shu Y., Lin F., Liu S., Shen H., Gao W., Li S., Shen D. A Cluster of Specified MicroRNAs in Peripheral Blood as Biomarkers for Metastatic Non-Small-Cell Lung Cancer by Stem-Loop RT-PCR. J. Cancer Res. Clin. Oncol. 2012;138:85–93. doi: 10.1007/s00432-011-1068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang P., Yang D., Zhang H., Wei X., Ma T., Cheng Z., Hong Q., Hu J., Zhuo H., Song Y., et al. Early Detection of Lung Cancer in Serum by a Panel of MicroRNA Biomarkers. Clin. Lung Cancer. 2015;16 doi: 10.1016/j.cllc.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 93.Zhu W., Zhou K., Zha Y., Chen D., He J., Ma H., Liu X., Le H., Zhang Y. Diagnostic Value of Serum MiR-182, MiR-183, MiR-210, and MiR-126 Levels in Patients with Early-Stage Non-Small Cell Lung Cancer. PLoS ONE. 2016;11:e0153046. doi: 10.1371/journal.pone.0153046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu Y., Li T., Chen G., Yan G., Zhang X., Wan Y., Li Q., Zhu B., Zhuo W. Identification of a Serum MicroRNA Expression Signature for Detection of Lung Cancer, Involving MiR-23b, MiR-221, MiR-148b and MiR-423-3p. Lung Cancer. 2017;114:6–11. doi: 10.1016/j.lungcan.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 95.Leng Q., Lin Y., Jiang F., Lee C.-J., Zhan M., Fang H., Wang Y., Jiang F. A Plasma MiRNA Signature for Lung Cancer Early Detection. Oncotarget. 2017;8:111902–111911. doi: 10.18632/oncotarget.22950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li J., Fang H., Jiang F., Ning Y. External Validation of a Panel of Plasma MicroRNA Biomarkers for Lung Cancer. Biomark. Med. 2019;13:1557–1564. doi: 10.2217/bmm-2019-0213. [DOI] [PubMed] [Google Scholar]
- 97.Niu Y., Su M., Wu Y., Fu L., Kang K., Li Q., Li L., Hui G., Li F., Gou D. Circulating Plasma MiRNAs as Potential Biomarkers of Non–Small Cell Lung Cancer Obtained by High-Throughput Real-Time PCR Profiling. Cancer Epidemiol. Prev. Biomark. 2019;28:327–336. doi: 10.1158/1055-9965.EPI-18-0723. [DOI] [PubMed] [Google Scholar]
- 98.Bagheri A., Khorshid H.R.K., Tavallaie M., Mowla S.J., Sherafatian M., Rashidi M., Zargari M., Boroujeni M.E., Hosseini S.M. A Panel of Noncoding RNAs in Non–Small-Cell Lung Cancer. J. Cell. Biochem. 2019;120:8280–8290. doi: 10.1002/jcb.28111. [DOI] [PubMed] [Google Scholar]
- 99.Xing L., Todd N.W., Yu L., Fang H., Jiang F. Early Detection of Squamous Cell Lung Cancer in Sputum by a Panel of MicroRNA Markers. Mod. Pathol. 2010;23:1157–1164. doi: 10.1038/modpathol.2010.111. [DOI] [PubMed] [Google Scholar]
- 100.Bjaanæs M.M., Halvorsen A.R., Solberg S., Jørgensen L., Dragani T.A., Galvan A., Colombo F., Anderlini M., Pastorino U., Kure E., et al. Unique MicroRNA-Profiles in EGFR-Mutated Lung Adenocarcinomas. Int. J. Cancer. 2014;135:1812–1821. doi: 10.1002/ijc.28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lan D., Zhang X., He R., Tang R., Li P., He Q., Chen G. MiR-133a Is Downregulated in Non-Small Cell Lung Cancer: A Study of Clinical Significance. Eur. J. Med. Res. 2015;20:50. doi: 10.1186/s40001-015-0139-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Y., Li J., Chen H., Mo Y., Ye H., Luo Y., Guo K., Mai Z., Zhang Y., Chen B., et al. Down-Regulation of MiR-133a as a Poor Prognosticator in Non-Small Cell Lung Cancer. Gene. 2016;591:333–337. doi: 10.1016/j.gene.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 103.Qu Y., Zhou L., Li L., Liu P., Jiao S. MicroRNA-140-3p Expression Level Is an Independent Prognostic Factor in NSCLC. Int. J. Clin. Exp. Pathol. 2016;9:8565–8569. [Google Scholar]
- 104.Shen H., Shen J., Wang L., Shi Z., Wang M., Jiang B., Shu Y. Low MiR-145 Expression Level Is Associated with Poor Pathological Differentiation and Poor Prognosis in Non-Small Cell Lung Cancer. Biomed. Pharmacother. 2015;69:301–305. doi: 10.1016/j.biopha.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 105.Wang R.-J., Zheng Y.-H., Wang P., Zhang J.-Z. Serum MiR-125a-5p, MiR-145 and MiR-146a as Diagnostic Biomarkers in Non-Small Cell Lung Cancer. Int. J. Clin. Exp. Pathol. 2015;8:765–771. [PMC free article] [PubMed] [Google Scholar]
- 106.Chen Y., Min L., Ren C., Xu X., Yang J., Sun X., Wang T., Wang F., Sun C., Zhang X. MiRNA-148a Serves as a Prognostic Factor and Suppresses Migration and Invasion through Wnt1 in Non-Small Cell Lung Cancer. PLoS ONE. 2017;12:e0171751. doi: 10.1371/journal.pone.0171751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.He Y., Yang Y., Kuang P., Ren S., Rozeboom L., Rivard C.J., Li X., Zhou C., Rhirsch F. Seven-MicroRNA Panel for Lung Adenocarcinoma Early Diagnosis in Patients Presenting with Ground-Glass Nodules. OncoTargets Ther. 2017;10:5915–5926. doi: 10.2147/OTT.S151432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mohamed M.A., Mohamed E.I., El-Kaream S.A.A., Badawi M.I., Darwish S.H. Underexpression of MiR-486-5p but Not Overexpression of MiR-155 Is Associated with Lung Cancer Stages. MicroRNA. 2018;7:120–127. doi: 10.2174/2211536607666180212124532. [DOI] [PubMed] [Google Scholar]
- 109.Arife Z., Necdet Ö., Serdar K., Tuba E., Mehmet T.K., Kürşad T., Mustafa T., Leyla T., Mehmet E.E. Diagnostic Value of MiR-125b as a Potential Biomarker for Stage I Lung Adenocarcinoma. Curr. Mol. Med. 2019;19:216–227. doi: 10.2174/1566524019666190314113800. [DOI] [PubMed] [Google Scholar]
- 110.Cui E., Li H., Hua F., Wang B., Mao W., Feng X., Li J., Wang X. Serum MicroRNA 125b as a Diagnostic or Prognostic Biomarker for Advanced NSCLC Patients Receiving Cisplatin-Based Chemotherapy. Acta Pharmacol. Sin. 2013;34:309–313. doi: 10.1038/aps.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Roth C., Kasimir-Bauer S., Pantel K., Schwarzenbach H. Screening for Circulating Nucleic Acids and Caspase Activity in the Peripheral Blood as Potential Diagnostic Tools in Lung Cancer. Mol. Oncol. 2011;5:281–291. doi: 10.1016/j.molonc.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Luo J., Shi K., Yin S., Tang R., Chen W., Huang L., Gan T., Cai Z., Chen G. Clinical Value of MiR-182-5p in Lung Squamous Cell Carcinoma: A Study Combining Data from TCGA, GEO, and RT-QPCR Validation. World J. Surg. Oncol. 2018;16:76. doi: 10.1186/s12957-018-1378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shi Y., Liu X., Liu J., Liu H., Du X. Up-Regulation of MiR-182-5p Predicts Poor Prognosis in Patients with Lung Cancer and Associates with Tumor Cell Growth and Migration. Int. J. Clin. Exp. Pathol. 2017;10:3061–3068. [Google Scholar]
- 114.Zou J.-G., Ma L.-F., Li X., Xu F.-L., Fei X.-Z., Liu Q., Bai Q.-L., Dong Y.-L. Circulating MicroRNA Array (MiR-182, 200b and 205) for the Early Diagnosis and Poor Prognosis Predictor of Non-Small Cell Lung Cancer. Eur. Rev. Med. Pharmacol. Sci. 2019;23:1108–1115. doi: 10.26355/eurrev_201902_17001. [DOI] [PubMed] [Google Scholar]
- 115.Zhu W., Liu X., He J., Chen D., Hunag Y., Zhang Y.K. Overexpression of Members of the MicroRNA-183 Family Is a Risk Factor for Lung Cancer: A Case Control Study. BMC Cancer. 2011;11:393. doi: 10.1186/1471-2407-11-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Halvorsen A.R., Bjaanæs M., LeBlanc M., Holm A.M., Bolstad N., Rubio L., Peñalver J.C., Cervera J., Mojarrieta J.C., López-Guerrero J.A., et al. A Unique Set of 6 Circulating MicroRNAs for Early Detection of Non-Small Cell Lung Cancer. Oncotarget. 2016;7:37250–37259. doi: 10.18632/oncotarget.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tang R., Zhong T., Dang Y., Zhang X., Li P., Chen G. Association between Downexpression of MiR-203 and Poor Prognosis in Non-Small Cell Lung Cancer Patients. Clin. Transl. Oncol. 2016;18:360–368. doi: 10.1007/s12094-015-1377-9. [DOI] [PubMed] [Google Scholar]
- 118.Võsa U., Vooder T., Kolde R., Fischer K., Välk K., Tõnisson N., Roosipuu R., Vilo J., Metspalu A., Annilo T. Identification of MiR-374a as a Prognostic Marker for Survival in Patients with Early-Stage Nonsmall Cell Lung Cancer. Genes Chromosomes Cancer. 2011;50:812–822. doi: 10.1002/gcc.20902. [DOI] [PubMed] [Google Scholar]
- 119.Zhang Y., Sui J., Shen X., Li C., Yao W., Hong W., Peng H., Pu Y., Yin L., Liang G. Differential Expression Profiles of MicroRNAs as Potential Biomarkers for the Early Diagnosis of Lung Cancer. Oncol. Rep. 2017;37:3543–3553. doi: 10.3892/or.2017.5612. [DOI] [PubMed] [Google Scholar]
- 120.Zhang Y.-K., Zhu W.-Y., He J.-Y., Chen D.-D., Huang Y.-Y., Le H.-B., Liu X.-G. MiRNAs Expression Profiling to Distinguish Lung Squamous-Cell Carcinoma from Adenocarcinoma Subtypes. J. Cancer Res. Clin. Oncol. 2012;138:1641–1650. doi: 10.1007/s00432-012-1240-0. [DOI] [PubMed] [Google Scholar]
- 121.Leng Q., Wang Y., Jiang F. A Direct Plasma MiRNA Assay for Early Detection and Histological Classification of Lung Cancer. Transl. Oncol. 2018;11:883–889. doi: 10.1016/j.tranon.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sromek M., Glogowski M., Chechlinska M., Kulinczak M., Szafron L., Zakrzewska K., Owczarek J., Wisniewski P., Wlodarczyk R., Talarek L., et al. Changes in Plasma MiR-9, MiR-16, MiR-205 and MiR-486 Levels after Non-Small Cell Lung Cancer Resection. Cell. Oncol. 2017;40:529–536. doi: 10.1007/s13402-017-0334-8. [DOI] [PubMed] [Google Scholar]
- 123.He R., Cen W., Cen J., Cen W., Li J., Li M., Gan T., Hu X., Chen G. Clinical Significance of MiR-210 and Its Prospective Signaling Pathways in Non-Small Cell Lung Cancer: Evidence from Gene Expression Omnibus and the Cancer Genome Atlas Data Mining with 2763 Samples and Validation via Real-Time Quantitative PCR. Cell. Physiol. Biochem. 2018;46:925–952. doi: 10.1159/000488823. [DOI] [PubMed] [Google Scholar]
- 124.Li Z.-H., Zhang H., Yang Z.-G., Wen G.-Q., Cui Y.-B., Shao G.-G. Prognostic Significance of Serum MicroRNA-210 Levels in Nonsmall-Cell Lung Cancer. J. Int. Med. Res. 2013;41:1437–1444. doi: 10.1177/0300060513497560. [DOI] [PubMed] [Google Scholar]
- 125.Li X.-X., Liu Y., Meng H.-H., Wang X.-W. Expression of MiR-210 in Senile COPD Complicating Primary Lung Cancer. Eur. Rev. Med. Pharmacol. Sci. 2017;21:38–42. [PubMed] [Google Scholar]
- 126.Wang X., Zhi X., Zhang Y., An G., Feng G. Role of Plasma MicroRNAs in the Early Diagnosis of Non-Small-Cell Lung Cancers: A Case-Control Study. J. Thorac. Dis. 2016;8:1645–1652. doi: 10.21037/jtd.2016.06.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ge X., Liu S., Liu H., Li D., Wang C. Increased Levels of Tissue MicroRNA-221 in Human Non-Small Cell Lung Cancer and Its Clinical Significance. Int. J. Clin. Exp. Pathol. 2016;9:2003–2008. [Google Scholar]
- 128.Lv S., Xue J., Wu C., Wang L., Wu J., Xu S., Liang X., Lou J. Identification of A Panel of Serum MicroRNAs as Biomarkers for Early Detection of Lung Adenocarcinoma. J. Cancer. 2017;8:48–56. doi: 10.7150/jca.16644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen L., Li X., Zhao Y., Liu W., Wu H., Liu J., Mu X., Wu H. Down-Regulated MicroRNA-375 Expression as a Predictive Biomarker in Non-Small Cell Lung Cancer Brain Metastasis and Its Prognostic Significance. Pathol. Res. Pract. 2017;213:882–888. doi: 10.1016/j.prp.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 130.Jin Y., Liu Y., Zhang J., Huang W., Jiang H., Hou Y., Xu C., Zhai C., Gao X., Wang S., et al. The Expression of MiR-375 Is Associated with Carcinogenesis in Three Subtypes of Lung Cancer. PLoS ONE. 2015;10:e0144187. doi: 10.1371/journal.pone.0144187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yu H., Jiang L., Sun C., Guo L., Lin M., Huang J., Zhu L. Decreased Circulating MiR-375: A Potential Biomarker for Patients with Non-Small-Cell Lung Cancer. Gene. 2014;534:60–65. doi: 10.1016/j.gene.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 132.Goto A., Tanaka M., Yoshida M., Umakoshi M., Nanjo H., Shiraishi K., Saito M., Kohno T., Kuriyama S., Konno H., et al. The Low Expression of MiR-451 Predicts a Worse Prognosis in Non-Small Cell Lung Cancer Cases. PLoS ONE. 2017;12:e0181270. doi: 10.1371/journal.pone.0181270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang X., Tian L.-L., Jiang X.-Y., Wang Y.-Y., Li D.-G., She Y., Chang J.-H., Meng A.-M. The Expression and Function of MiRNA-451 in Non-Small Cell Lung Cancer. Cancer Lett. 2011;311:203–209. doi: 10.1016/j.canlet.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 134.Yao B., Qu S., Hu R., Gao W., Jin S., Liu M., Zhao Q. A Panel of MiRNAs Derived from Plasma Extracellular Vesicles as Novel Diagnostic Biomarkers of Lung Adenocarcinoma. FEBS Open Bio. 2019;9:2149–2158. doi: 10.1002/2211-5463.12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hu Z., Wang X., Yang Y., Zhao Y., Shen Z., Huang Y. MicroRNA Expression Profiling of Lung Adenocarcinoma in Xuanwei, China: A Preliminary Study. Medicine. 2019;98:e15717. doi: 10.1097/MD.0000000000015717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhu J., Zeng Y., Xu C., Qin H., Lei Z., Shen D., Liu Z., Huang J.-A. Expression Profile Analysis of MicroRNAs and Downregulated MiR-486-5p and MiR-30a-5p in Non-Small Cell Lung Cancer. Oncol. Rep. 2015;34:1779–1786. doi: 10.3892/or.2015.4141. [DOI] [PubMed] [Google Scholar]
- 137.Li W., Wang Y., Zhang Q., Tang L., Liu X., Dai Y., Xiao L., Huang S., Chen L., Guo Z., et al. MicroRNA-486 as a Biomarker for Early Diagnosis and Recurrence of Non-Small Cell Lung Cancer. PLoS ONE. 2015;10:e0134220. doi: 10.1371/journal.pone.0134220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhou C., Chen Z., Dong J., Li J., Shi X., Sun N., Luo M., Zhou F., Tan F., He J. Combination of Serum MiRNAs with Cyfra21-1 for the Diagnosis of Non-Small Cell Lung Cancer. Cancer Lett. 2015;367:138–146. doi: 10.1016/j.canlet.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 139.Goodman G.E., Thornquist M.D., Balmes J., Cullen M.R., Meyskens F.L., Jr., Omenn G.S., Valanis B., Williams J.H., Jr. The Beta-Carotene and Retinol Efficacy Trial: Incidence of Lung Cancer and Cardiovascular Disease Mortality During 6-Year Follow-up After Stopping β-Carotene and Retinol Supplements. J. Natl. Cancer Inst. 2004;96:1743–1750. doi: 10.1093/jnci/djh320. [DOI] [PubMed] [Google Scholar]
- 140.Peng Y., Croce C.M. The Role of MicroRNAs in Human Cancer. Signal Transduct. Target. Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Inoue J., Inazawa J. Cancer-Associated MiRNAs and Their Therapeutic Potential. J. Hum. Genet. 2021;66:937–945. doi: 10.1038/s10038-021-00938-6. [DOI] [PubMed] [Google Scholar]
- 142.Van Roosbroeck K., Calin G.A. Chapter Four—Cancer Hallmarks and MicroRNAs: The Therapeutic Connection. In: Croce C.M., Fisher P.B., editors. Advances in Cancer Research. Volume 135. Academic Press; Cambridge, MA, USA: 2017. pp. 119–149. miRNA and Cancer; [DOI] [PubMed] [Google Scholar]
- 143.Hanna J., Hossain G.S., Kocerha J. The Potential for MicroRNA Therapeutics and Clinical Research. Front. Genet. 2019;10:478. doi: 10.3389/fgene.2019.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shah M.Y., Calin G.A. MicroRNAs as Therapeutic Targets in Human Cancers. WIREs RNA. 2014;5:537–548. doi: 10.1002/wrna.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Relevant data supporting the findings of this study are available within the Article and Supplementary Materials or are available from the authors upon reasonable request.