Abstract
We isolated the catC gene, encoding catalase-peroxidase in Streptomyces coelicolor, using sequence homology with the katG gene from Escherichia coli. Upstream of the catC gene, an open reading frame (furA) encoding a homologue of ferric uptake regulator (Fur) was identified. S1 mapping analysis indicated that the furA gene was cotranscribed with the catC gene. The transcriptional start site of the furA-catC mRNA was mapped to the translation start codon ATG of the furA gene. The putative promoter contains consensus −10 and −35 elements similar to those recognized by ςHrdB, the major sigma factor of S. coelicolor. The transcripts were produced maximally at late-exponential phase and decreased at the stationary phase in liquid culture. The change in the amount of mRNA was consistent with that of CatC protein and enzyme activity. When the furA gene was introduced into S. lividans on a multicopy plasmid, the increased production of catC transcripts and protein product at late growth phase was inhibited, implying a role for FurA as the negative regulator of the furA-catC operon. FurA protein bound to its own promoter region between −59 and −39 nucleotides from the transcription start site. The binding affinity of FurA increased under reducing conditions and in the presence of metals such as Ni2+, Mn2+, Zn2+, or Fe2+. Addition of these metals to the growth medium decreased the production of CatC protein, consistent with the role of FurA as a metal-dependent repressor.
Catalase plays a crucial role in removing hydrogen peroxide generated as a byproduct of aerobic respiration in a cell. Bacterial catalases are classified into two groups depending on their enzymatic properties and amino acid sequence homology: monofunctional catalases and catalase-peroxidases. Catalase-peroxidase exhibits both catalase (decomposing H2O2 to O2 and H2O) and peroxidase (reducing H2O2 to H2O using intracellular reductants) activities. Unlike ubiquitous distribution of monofunctional catalases from prokaryotes to eukaryotes, catalase-peroxidases have been found only in bacteria and some fungi (31).
A number of bacteria possess multiple catalases whose expression pattern and biological functions are distinctly different. Escherichia coli produces two catalases: HPI, a catalase-peroxidase encoded by the katG gene, and HPII, a monofunctional catalase encoded by the katE gene. Expression of HPI is regulated by OxyR in response to H2O2 (11) and by RpoS in response to nutrient limitation (22). HPII exhibits RpoS-dependent expression in the stationary phase (27). In Bacillus subtilis, all three catalases identified so far are monofunctional catalases. KatA is induced by H2O2 or metal limitation (6, 8). Its expression is mediated by a repressor (PerR) which is one of the three Fur homologues in B. subtilis (7). KatE, an E. coli HPII homologue is induced at the stationary phase and by heat, salt, ethanol stress, or glucose starvation in a ςB-dependent manner (16). The recently identified KatX, the major catalase in dormant spores, is a member of the forespore-specific ςF regulon (4). Mycobacteria display varied distribution of catalases among different species. Only HPI-type catalase-peroxidase is detected in Mycobacterium tuberculosis (KatG) (20) and Mycobacterium fortuitum (KatGI and KatGII) (29), whereas some species produce only HPII-type catalase and others produce both types (30, 37). Research on mycobacterial catalases has been focused mainly on the role of KatG in conferring susceptibility to isoniazid (INH), an antituberculosis drug. KatG is considered to transform the drug into a toxic derivative, which inhibits the fatty acid biosynthetic enzyme encoded by inhA (15, 40). In most Mycobacterium species the katG gene, encoding catalase-peroxidase, is preceded by the furA gene, encoding a homologue of ferric uptake regulator (Fur) (33). However, the role of FurA has not been elucidated yet.
Streptomyces is a genus of gram-positive soil bacteria that undergo a complex cycle of morphological and physiological differentiation. S. coelicolor produces two monofunctional catalases: CatA, an H2O2-inducible major vegetative catalase, and CatB, a stationary phase-specific catalase inducible by osmotic stress (9, 10). In addition, two isoforms of catalase-peroxidase have been detected when cells formed aerial mycelium (26). Transient production of catalase-peroxidase also has been observed in other Streptomyces species. In Streptomyces seoulensis, two isoforms of catalase-peroxidase (StCP1 and StCP2) have been detected in substrate mycelium, and a third one (StCP3) was observed in aerial and sporulated mycelium (38). Spectroscopic analysis of catalase-peroxidase in S. seoulensis (IMSNU-1) demonstrated that it is a dimeric heme protein with a histidine as the fifth ligand (39). Recently a mycelium-associated catalase-peroxidase (CpeB), expressed at an early stage of growth, was identified in Streptomyces reticuli (42). In this study, we isolated and analyzed the furA and catC gene from S. coelicolor, encoding a Fur homologue and catalase-peroxidase, respectively. We present evidence that they constitute an operon and are negatively regulated by FurA. Metal-dependent autoregulation of the furA-catC operon by FurA was proposed on the basis of transcription inhibition by FurA in vivo and metal-dependent binding of FurA to its own promoter in vitro.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
S. coelicolor A3(2) M145 and Streptomyces lividans TK24 cells were grown as described previously (21). E. coli DH5α and BL21(DE3)pLysS were used for DNA cloning and overexpression, respectively. XL1-Blue MRA was used as a host for the λEMBL3 genomic library of M145. E. coli ET12567 was used to prepare unmethylated DNA to transform S. coelicolor (28).
Cloning and sequencing of the furA and catC genes.
To generate a genomic library, DNA was prepared from S. coelicolor M145 cells, partially digested with Sau3AI and cloned into BamHI-digested λEMBL3 (Stratagene). A 399-bp internal fragment of the E. coli katG gene was generated by PCR from E. coli genomic DNA and used as a hybridization probe to screen the S. coelicolor genomic library. A common 3.0-kb BamHI/SmaI fragment from two positive phage clones was subcloned into pUC18 to generate pJH203. A total sequence of 3,027 nucleotides (nt) was determined and deposited in the GenBank, EMBL, and DDBJ databases under accession number AF126956.
Disruption of the catC gene.
A 0.8-kb PvuII/EcoRI internal fragment of the catC gene was cloned into pKC1139 (5) to generate pJH403. pJH403 plasmid DNA was prepared from E. coli ET12567 and then introduced into S. coelicolor M145 protoplasts. Transformants were selected on an R2YE (21) plate containing apramycin (25 μg/ml) at 30°C. Spores of the transformants were plated on NA medium (9) containing apramycin and incubated at 37°C for 2 days to isolate single-crossover recombinants. Disruption of the catC gene was confirmed by genomic Southern hybridization and immunoblot analysis with anti-CatC antiserum.
Activity staining for catalase and peroxidase.
A cell extract was prepared and electrophoresed on a nondenaturing 7% polyacrylamide gel. Staining of catalase or peroxidase activity in the gel was carried out as described previously (12, 36).
RNA isolation and S1 nuclease protection analysis.
RNA was isolated from M145 cells grown in YEME as described (21). The probe for S1 mapping was prepared by cutting pJH2033, a pUC18 derivative containing a 0.6-kb SalI/PvuII fragment of the furA-catC junction (Fig. 1A), with NarI and labeling with [γ-32P]ATP and T4 polynucleotide kinase. Following cleavage with PvuII at the pUC18 vector body, the labeled 747-bp probe was eluted from an agarose gel. For high-resolution S1 mapping of the furA 5′ end, probe DNA was generated by PCR from pJH2032 containing a 0.6-kb BamHI/SalI fragment on pUC18, using FS1 primer (5′ AGCAGCGCGACACGGGCGGCGG 3′) and universal primer (Fig. 1). The amplified fragment was end labeled and cut with EcoRI at the multicloning site to prepare the 415-bp probe. For S1 mapping of the catC 5′ end, the probe DNA was generated by PCR from pJH2031 containing a 1.2-kb BamHI/PvuII fragment, using CS1 primer (5′ TCCTCGGTCTTCGCGTCGGTCAC 3′) and universal primer (Fig. 1). The amplified DNA was labeled at the 5′ ends and digested with EcoRI to generate the 856-bp probe. The S1 nuclease protection assay was done as described previously (34). The protected products were electrophoresed on a sequencing gel along with the sequencing ladder generated from the primers FS1 and CS1.
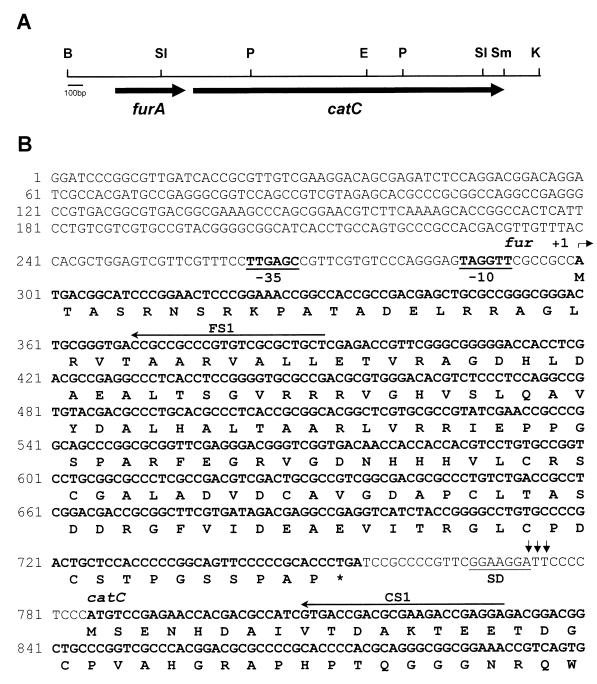
FIG. 1.
Restriction map and partial nucleotide sequence of the furA and catC genes. (A) Restriction map of the 3.3-kb BamHI/KpnI fragment containing the furA and catC genes. Thick arrows indicate the positions and directions of the furA and catC coding regions. Abbreviations for restriction enzymes: B, BamHI, K, KpnI; P, PvuII; SI, SalI; Sm, SmaI. (B) Nucleotide and predicted amino acid sequences of the furA and N-terminal portion of the catC gene. The full sequence of 3,027 nt encompassing the entire furA and catC genes was deposited in databases under accession number AF126956. The −35 and −10 hexamers of a putative promoter for the furA-catC operon are in boldface type and underlined. The transcriptional initiation site (+1) is indicated by a bent arrow. Vertical arrows in the intercistronic region indicate putative cleavage sites in the furA-catC transcript. The putative ribosome-binding site (SD) is underlined. Horizontal arrows (FS1, CS1) indicate primers used for high-resolution S1 nuclease mapping.
Overexpression of the furA and catC gene products in E. coli.
The furA coding region was amplified by PCR using mutagenic primers FON (5′ GTTCGCCCATATGACGGCATCCCG 3′ [the NdeI site is underlined]) and FOB (5′ TGGGAGGGGGATCCTTCCGAACGG 3′ [the BamHI site is underlined]) and cloned into pET3a (Novagen) to yield pJH1. An N-terminal portion of the catC gene (1.5 kb) was amplified by PCR with mutagenic primers CON (5′ TTCCCCTCATATGTCCGAGAAC 3′ [the NdeI site is underlined]) and COE (5′ AAGGCGTCCGCGAATTCCTCCG 3′ [the EcoRI site is underlined]). The PCR product was cut with NdeI and EcoRI and cloned into pET21c (Novagen) to generate pJH2. To construct the CatC overexpression plasmid pJH3, the C-terminal region of the catC gene (1.0 kb) was excised from pJH203 as an EcoRI fragment and cloned into pJH2. E. coli BL21(DE3)pLysS cells harboring recombinant plasmids were grown to an A600 of 0.5 and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h before harvest.
Western blot analysis.
Antibodies against CatA and CatC were raised in mice using CatA protein purified from a CatA-overproducing S. coelicolor strain (HR40) (Hahn et al., unpublished data) and CatC protein overproduced in E. coli, respectively. The reacting signal was detected by goat anti-mouse immunoglobulin G conjugated with horseradish peroxidase using the Western ECL detection system (Amersham Life Science).
Partial purification of S. coelicolor FurA from E. coli.
E. coli BL21(DE3)pLysS cells harboring pJH1 were grown in Luria broth and induced with IPTG. After harvest, cells were resuspended in lysis buffer (20 mM Tris-HCl [pH 7.9], 0.15 M NaCl, 5 mM EDTA, 0.1 mM dithiothreitol [DTT], 10 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, and 10% [vol/vol] glycerol) and disrupted by sonication. The lysate was centrifuged at 10,000 × g for 10 min, and the viscous transparent pellet was eluted with extraction buffer (50 mM Tris-HCl [pH 7.9], 0.1 M NaCl, 10 mM EDTA, and 0.5% Triton X-100). The eluate was enriched with FurA protein, which constitutes more than 10% of total proteins. The eluate was dialyzed twice for 8 h each against 10 volumes of TGED buffer (10 mM Tris-HCl [pH 7.9], 0.1 mM EDTA, 1 mM DTT, and 10% glycerol) and then against the storage buffer (10 mM Tris-HCl [pH 7.9], 0.1 mM EDTA, 10 mM MgCl2, 0.1 M KCl, 1 mM DTT, and 50% glycerol) at 4°C.
Gel mobility shift assay for FurA binding.
To generate a series of furA promoter fragments of varying lengths, the following forward primers were used for PCR: D1 (5′ CCGCCACGACGCTTGTTTAC 3′; 5′ end at nt −79 relative to the furA start codon), D2 (5′ CACGCTGGAGTCGTTCGTTT 3′; 5′ end at nt −59), and D3 (5′ CCTTGAGCCGTTCGTGTCCC 3′; 5′ end at nt −39). FS1 primer (Fig. 1B) was used as a backward primer. The amplified fragments were end labeled with [γ-32P]ATP using T4 polynucleotide kinase. Unincorporated nucleotides were removed through a Sephadex G-50 spun column. The labeled probe was incubated with 1 μg of partially purified FurA in 20 μl of binding buffer (10 mM Tris-HCl [pH 7.5], 1 mM MgCl2, 40 mM KCl, 100 μg of poly(dI-dC) per ml and 5% glycerol) at 30°C for 10 min. To examine the effect of various metals on FurA binding affinity, a 100 μM concentration each of FeSO4, CuCl2, MnCl2, ZnCl2, and NiCl2 was added in the binding buffer. The binding mixture was electrophoresed on a 5% native polyacrylamide gel in 20 mM Tris-borate buffer, and this was followed by autoradiography.
RESULTS
Cloning and sequence analysis of the furA and catC genes, encoding a Fur homologue and catalase-peroxidase, respectively, in S. coelicolor A3(2) M145.
Genomic Southern analysis of the M145 chromosome with the E. coli katG gene fragment revealed a specifically hybridizing DNA band. We screened the λEMBL library of the M145 genome with the katG gene probe and selected two positive phage clones. The nucleotide sequence of the 3,027-bp (BamHI/SmaI) fragment common to both clones was determined. Sequence analysis revealed the presence of two coding regions (Fig. 1). The first one (furA) is predicted to encode a protein of 151 amino acids with a calculated molecular mass of 15,976 Da. The amino acid sequence exhibits homology to Fur proteins of other bacteria. In particular, the amino acid sequence of FurA showed high similarity with that of S. reticuli FurS and M. tuberculosis FurA, whose genes are located upstream of the genes for catalase-peroxidase, cpeB and katG, respectively (15, 42). Alignment of the amino acid sequence of FurA with other bacterial Fur sequences revealed a conserved HXHXXCXXC motif which is likely to participate in binding metals (Fig. 2). An additional cysteine pair near the C terminus is also conserved among the Fur proteins. The second coding region (catC), 30 bp downstream of the furA gene, encodes a protein of 740 amino acids with a molecular mass of 80,860 Da. The predicted amino acid sequence is highly homologous to all known bacterial catalase-peroxidases.
FIG. 2.
Comparison of predicted amino acid sequences of FurA with those of other Fur homologues. The compared Fur homologues are S. coelicolor (Sco) FurA (AF126956), S. reticuli (Sre) FurS (Y14317), M. tuberculosis (Mtu) FurA (Z97193), B. subtilis (Bsu) PerR (Z99108), M. tuberculosis FurB (Z95208), and E. coli (Eco) Fur (D90708). Genes for the first three Fur proteins (shaded) share similar locations, preceding the gene for catalase-peroxidase. Conserved residues are shaded. Asterisks and dots indicate identical and similar amino acids, respectively. The residue numbers are shown on the right. Identical residues among the three closest Fur proteins are underlined in the Mtu FurA sequence.
Production of catalase-peroxidase from the catC gene.
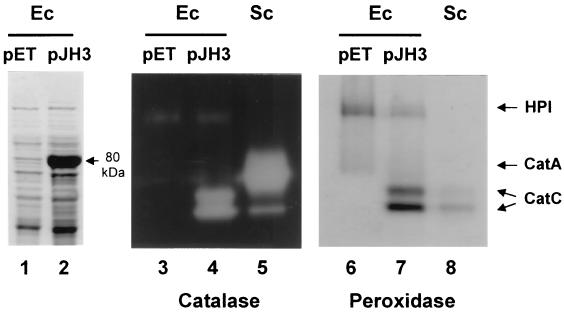
The enzymatic activity of the catC gene product was examined in E. coli and S. lividans. The CatC protein was overproduced in E. coli using the pET overexpression system as described in Materials and Methods. The overproduced protein migrated with a relative molecular mass of 80 kDa during sodium dodecyl sulfate-polyacrylamide gel electrophoresis, which is in good agreement with its calculated size (Fig. 3, lane 2). The soluble fraction of E. coli cell extracts was electrophoresed on a native polyacrylamide gel and examined for either catalase or a peroxidase activities. The control E. coli cell extract exhibited the catalase-peroxidase activity HPI (Fig. 3, lanes 3 and 6). The overproduced CatC protein was detected as two activity bands of catalase and peroxidase (lanes 4 and 7), comigrating with those from S. coelicolor below the prominent CatA band (lanes 5 and 8). These results clearly demonstrate that the catC gene encodes the two isoforms of catalase-peroxidase. Expression of the catC gene was also examined in cells of S. lividans TK24, which harbors the recombinant plasmid pJH7031 containing the furA and catC genes (BamHI/SmaI fragment in Fig. 1) on the multicopy plasmid pIJ702. Overproduction of CatC was detected by activity staining and immunoblot analysis (data not shown).
FIG. 3.
Expression of S. coelicolor catC gene in E. coli. E. coli BL21(DE3)pLysS cells carrying catC-overproducing plasmid pJH3 (lanes 2, 4, and 7) or the parental vector pET21c (lanes 1, 3, and 6) were grown and induced with 1 mM IPTG for 3 h. Cell extracts were electrophoresed on sodium dodecyl sulfate–10% polyacrylamide gels followed by Coomassie brilliant blue staining (lanes 1 and 2), or on nondenaturing 7% polyacrylamide gel followed by catalase activity staining (lanes 3 to 5) or peroxidase activity staining (lanes 6 to 8). Lanes 5 and 8 contained cell extracts of S. coelicolor grown in YEME medium for 40 h as a control. The overproduced CatC protein is indicated by an arrow with the predicted molecular mass (lane 2). Activity bands for catalase-peroxidase of E. coli (HPI) and S. coelicolor (CatC) as well as monofunctional catalase of S. coelicolor (CatA) are indicated.
Analysis of furA and catC transcription.
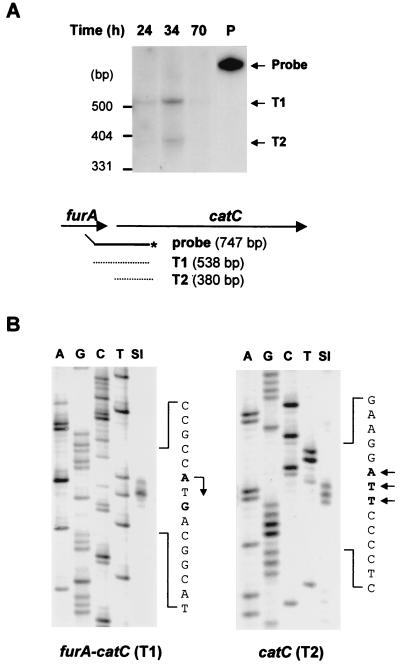
We examined the transcripts from the furA and catC genes by S1 nuclease mapping. For initial assessment, we used a 747-bp DNA probe encompassing the intercistronic region (Fig. 4A). It consists of a 538-bp furA-catC gene and a 209-bp vector DNA. A fully protected 538-bp band was generated from a transcript (T1) spanning the furA and catC coding regions. Its presence suggests that the catC gene is cotranscribed with furA, together forming an operon. Another protected (380-bp) band was generated from a transcript (T2) whose 5′ end lies immediately upstream of the catC gene. Both T1 and T2 transcripts were expressed transiently during growth in liquid culture, reaching maximum levels at the late exponential phase and decreasing at the stationary phase.
FIG. 4.
Analysis of furA and catC mRNAs by S1 nuclease mapping. RNA was prepared from S. coelicolor A3(2) M145 cells grown in YEME medium for various lengths of time as indicated. (A) S1 nuclease mapping analysis was done with the 747-bp PvuII/NarI probe uniquely labeled at the NarI site. Two protected bands (T1 and T2) are indicated by arrows. Schematic representations of the probe and protected fragments are also indicated. (B) High-resolution S1 mapping of 5′ ends of the furA-catC transcripts. For mapping the 5′ end of the T1 transcript, the 415-bp probe uniquely labeled at the 5′ end of FS1 (Fig. 1 [at position +92 relative to the furA start codon]) was used. A DNA sequencing ladder was generated with primer FS1. For mapping the 5′ end of the T2 transcript, the 856-bp probe uniquely labeled at the 5′ end of CS1 (Fig. 1 [at position +47 relative to the catC start codon]) was used. A DNA sequencing ladder was generated with primer CS1. The positions of transcript 5′ ends are designated in boldface type and by arrows on the sense sequence.
The 5′ ends of both transcripts were determined by high-resolution S1 mapping (Fig. 4B). The end site for the T1 transcript was mapped to A and G residues of the ATG translation start codon of the furA coding region. Upstream of the start site, we identified putative −10 (TAGGTT) and −35 (TTGAGC) elements of consensus promoters recognized by ςHrdB, the major sigma factor of S. coelicolor (Fig. 1B). The nucleotide sequence alignment of the furA promoter region with the furS gene sequence from S. reticuli revealed that they share nearly identical putative promoter elements located at the same position (see Fig. 7B). We believe that the transcription initiated from residue A of the ATG start codon, considering the proper distance from the putative −10 box. The G-ended RNAs could have been generated by degradation of 2 nt from the 5′ end. The 5′ end of the T2 transcript was mapped to A and T residues located 11 to 9 nt upstream of the ATG start codon of the catC coding region. No consensus promoter elements were found in the adjacent upstream region, nor was any promoter activity detected by monitoring the promoter-driven catechol dioxygenase activity from recombinant promoter-probing vector pXE4 (data not shown). A consensus ribosome binding site was located just upstream of the 5′ end of the T2 transcript. From these observations, we postulate that the T2 transcript might have been generated not from genuine transcriptional initiation, but from cleavage of the multicistronic T1 transcript.
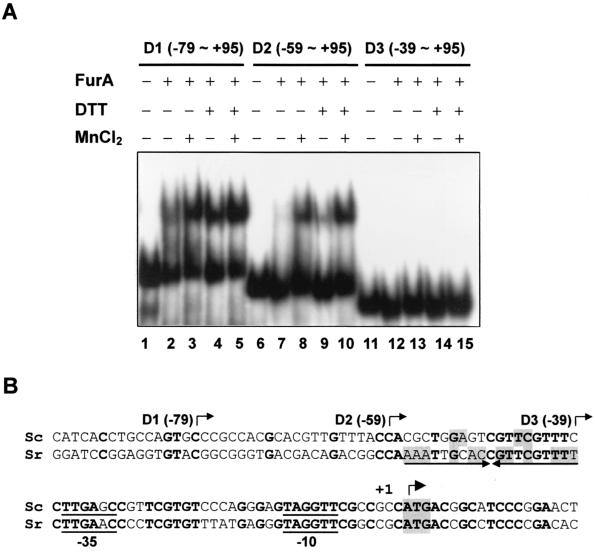
FIG. 7.
Mapping of the FurA binding site within the furA-catC promoter. (A) Gel shift assay for FurA binding. End-labeled DNA fragments (D1 to D3) containing the indicated region of the furA-catC promoter were analyzed for binding with partially purified FurA, which had been preincubated with (+) or without (−) 100 mM DTT. DNA and the FurA mixture were incubated at 30°C for 10 min in the presence or absence of 100 μM MnCl2 as indicated. The final concentration of DTT in the binding mixture was 10 mM. (B) Putative binding site of FurA. The nucleotide sequence alignment of the S. coelicolor (Sc) furA promoter region with the corresponding region of S. reticuli (Sr) furS is presented. The 5′ boundaries of the furA promoter fragments (D1, D2, and D3) used for gel shift assay are indicated by bent arrows. The inverted repeat sequence prominent in the furS gene is marked by arrows. The nucleotides identical between the two species are in boldface type. The inverted repeat nucleotides within the binding site are shaded. The translational start codon, ATG, is also shaded, and the transcription start site (+1) is indicated with another bent arrow.
Growth phase-dependent expression of the CatC protein.
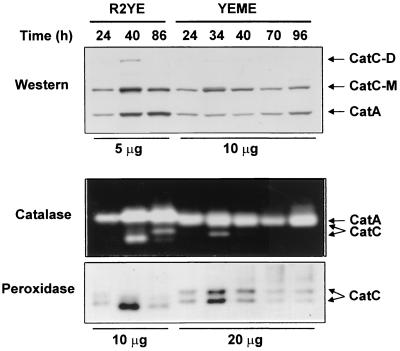
We examined the change in the expression of the CatC protein during growth. Cells were grown either on an R2YE plate or in liquid YEME medium. The level of CatC expression was determined by immunoblotting and activity staining (Fig. 5). In YEME medium, CatC protein increased as cell growth proceeded from early exponential (24 h) to late exponential (34 h) phase and then decreased slowly until late stationary phase (Fig. 5), consistent with the change in the furA-catC mRNA level presented above (Fig. 4). On surface culture, cells were harvested when they formed substrate mycelium (24 h), aerial mycelium (40 h), and spores (86 h). The production of CatC protein and catalase-peroxidase activity increased when cells formed aerial mycelium and decreased when cells sporulated. The presence of dimer-sized CatC suggests the dimeric nature of CatC as observed in S. seoulensis (IMSNU-1) (39). In comparison with CatC, the expression of the major catalase CatA was relatively constitutive in liquid culture, as previously observed (10). On surface culture, CatA increased as cells formed aerial mycelium and the increased level was maintained during differentiation. The amount of both CatA and CatC in surface-grown cells was more than twofold higher than in liquid-grown cells, suggesting that both catalases are induced by the same aerobic cues.
FIG. 5.
Transient expression of CatC on liquid or solid culture. M145 cells were grown on R2YE plates or in YEME liquid medium for the indicated lengths of time. Mycelial cells grown on R2YE plates were harvested when they formed substrate (24 h), aerial (40 h), or sporulated (86 h) mycelium. The amount of cell extracts analyzed was either 5 to 10 μg for plate cultures or 10 to 20 μg for liquid cultures. The amounts of CatA and CatC protein were determined by Western blot analysis. Either catalase or peroxidase activities were detected on 7% native polyacrylamide gels. Positions of monomeric (CatC-M) and dimeric (CatC-D) CatC protein are indicated.
However, transient (1-h) treatment with oxidants such as 200 μM H2O2, cumene hydroperoxide, or superoxide generators, including paraquat, plumbagin, and menadione, did not induce CatC expression. Neither heat (42°C), nor osmotic shock (0.5 M NaCl) induced its expression (data not shown). These results indicate that CatC is not an immediately responding catalase against stress, unlike CatA and CatB.
The catC gene was disrupted in S. coelicolor M145 by integration of a 0.8-kb PvuII/EcoRI internal fragment of the catC gene (Fig. 1A). The catC mutant did not produce any catalase-peroxidase activity bands on native gel or CatC protein on immunoblot analysis (data not shown). The catC mutant cells grew slowly but differentiated normally, suggesting that CatC catalase-peroxidase is not critically required for differentiation of S. coelicolor but is necessary for efficient growth. The sensitivity of the catC mutant to H2O2 was compared with that of the wild type by spotting spores in serial dilution on NA plates containing 0.1 to 0.5 mM H2O2 or cumene hydroperoxide. No significant change in sensitivity was observed. Since CatC contributes less than 10% to the total catalase activity as judged from the intensity of activity bands (Fig. 3, lane 5), the lack of sensitivity change is understandable. In contrast to catalase-peroxidase (CpeB) from S. reticuli, which is associated with mycelium (42), CatC was found to be a cytosolic protein, unextractable with 0.1% Triton X-100 (data not shown).
Repression of furA-catC transcription by multicopy expression of the furA gene.
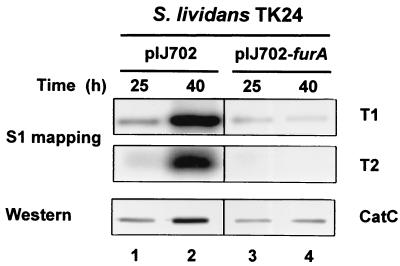
The regulatory role of FurA for its own operon was postulated in analogy with other known Fur proteins. To examine whether FurA acts as a regulator, the furA gene (1.2-kb BamHI/PvuII fragment) cloned on multicopy plasmid pIJ702 was introduced into S. lividans TK24. S. lividans cells containing the parental vector produced T1 and T2 transcripts as well as CatC protein in a growth phase-dependent manner as observed in S. coelicolor (Fig. 6, lanes 1 and 2). When the furA gene was introduced, the growth phase-dependent increase in T1 and T2 transcripts was inhibited (Fig. 6, lanes 3 and 4), implying that the furA gene product negatively regulates its own operon in vivo.
FIG. 6.
Repression of CatC expression by multicopy furA gene. S. lividans cells harboring pIJ702 or pIJ702-furA (pJH7032) carrying the furA gene were grown in YEME medium containing thiostrepton (50 μg/ml) for 25 or 40 h. S1 nuclease mapping analysis was carried out with a 851-bp probe uniquely labeled at the 5′ end of primer CS1. The level of CatC protein was determined by Western blot analysis.
Binding of FurA protein to its own promoter region.
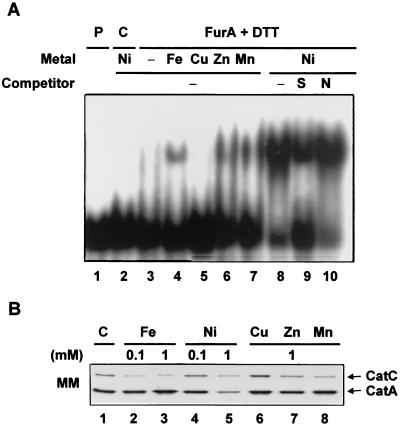
To examine whether FurA acts directly as a negative regulator for its own operon, a gel mobility shift assay was carried out. The FurA protein was overproduced in E. coli, where it accumulated as a 21.5-kDa protein. The protein was partially purified and assayed for DNA binding activity with the furA promoter fragments of various lengths as described in Materials and Methods (Fig. 7). The longer DNA fragments, D1 and D2, containing nucleotides up to −79 and −59 relative to the furA start codon, formed specific complexes with FurA (Fig. 7A, lanes 2 to 5 and 7 to 10). Both MnCl2 (0.1 mM) and DTT (10 mM) enhanced the complex formation, with their effects being additive. With the D3 fragment, in which nucleotides from −59 to −40 were further deleted, FurA binding was not detected, suggesting that this region contains the binding site for FurA (Fig. 7A, lanes 11 to 15).
Comparison of the furA promoter region with the furS gene sequence from S. reticuli revealed some conservation in several regions: −10, −35, and the spacer regions of the putative promoter as well as from nt −60 to −40 upstream from the translational start codon (Fig. 7B). Whereas the furS gene exhibits salient dyad symmetry from residues −57 to −39, the furA gene contains only the half-site. The sequence similarity within the putative FurA binding region predicts that FurS protein may also bind to this region and regulate the furS gene in a way similar to that of furA of S. coelicolor.
Metal-dependent activity of FurA.
We examined whether metals other than Mn2+ enhance the binding activity of FurA to the D2 fragment. We observed that 0.1 mM Fe2+, Mn2+, Zn2+, and Ni2+ all enhanced FurA binding in the following order: Ni2+ > Mn2+ ≈ Zn2+ > Fe2+ (Fig. 8A). Cu2+, however, did not enhance FurA binding. Proteins purified in the same way from control cells containing the parental vector did not produce any complex, confirming that the band retardation is caused by the FurA protein (lane 2). The specificity of FurA binding was demonstrated by its sensitivity to a 350-fold molar excess of specific competitors (unlabeled probe DNA; lane 9) and resistance to nonspecific competitors [pGEM-3Zf(+) DNA digested with HpaII; lane 10] in the presence of Ni2+.
FIG. 8.
Effects of various metals on FurA binding in vitro and CatC production in vivo. (A) A gel mobility shift assay was carried out with the D2 fragment of the furA promoter and FurA protein, which was preincubated with 100 mM DTT as described in the legend to Fig. 7. A 100 μM concentration each of NiCl2, FeSO4, CuSO4, ZnCl2, and MnCl2 was added in the binding reaction (lanes 4 to 10). In order to demonstrate the specificity of binding, a 350-fold molar excess of unlabeled D2 fragment (S, lane 9) or HpaII-digested pGEM-3Zf(+) DNA (N, lane 10) was added as a specific or nonspecific competitor, respectively. As a control, a cell extract was prepared from E. coli containing the parental pET3a vector by the same method used to prepare the FurA protein, and the gel mobility shift assay was performed in the presence of DTT and Ni2+ (lane 3). Lane 1 contains only the labeled probe. (B) Effects of various metals on the production of CatC on surface culture. M145 cells were grown on minimal medium plates (MM) containing the above metals at the indicated concentrations. Cells were grown at 30°C for 40 h, and the amounts of CatA and CatC proteins were detected by Western blotting.
When M145 cells were grown on minimal medium in the presence of a 0.1 or 1 mM concentration of each metal, we observed that the CatC production was reduced about twofold by all the metals except Cu2+ (Fig. 8B), consistent with the observation in vitro. When EDTA (1 mM) was added to the plate in an effort to deplete metals and thereby inhibit the action of FurA, the cell growth was retarded and masked the effect of metal depletion on CatC production, if any. However, addition of 1 mM EDTA for 1 h to liquid medium enhanced the production of catC transcripts by two- to threefold in FurA-overproducing cells, as judged by S1 mapping analysis under the same experimental conditions as in Fig. 6 (lanes 3 to 4; data not shown). These results support our proposal that FurA inhibits catC transcription in vivo in a metal-dependent manner.
DISCUSSION
In this study, we demonstrated that the catC gene encodes a catalase-peroxidase in S. coelicolor and constitutes an operon with the upstream furA gene. Similar gene organization has been reported in S. reticuli (furS-cpeB) (42) and Mycobacterium species (furA-katG) (15, 33). However, the role of Fur homologues has not been well understood in these genes. In spite of high similarity in amino acid sequences between CpeB of S. reticuli (42) and CatC of S. coelicolor, they are distinguished in both cellular localization and expression profile during growth. CpeB is expressed at the early stage of growth as a mycelium-associated protein released by detergent treatment, whereas CatC is maximally expressed at the transition period from the late exponential phase to the stationary phase as a cytosolic protein.
Fur was initially discovered as a transcriptional repressor of a large number of genes for iron uptake systems in response to iron sufficiency in E. coli (3). In the presence of divalent metal ions, such as ferrous iron, Fur protein binds to the ∼19-bp dyad symmetric operator region (Fur box) and inhibits transcription (3, 14). In addition to iron uptake systems, the Fur regulon includes genes for oxidative defense enzymes such as the sodA (encoding Mn superoxide dismutase [SOD]) gene in E. coli (32, 35) and fagA (Fur-associated gene)-fumC (encoding fumarase)-orfX-sodA (encoding MnSOD) operon in Pseudomonas aeruginosa (19). Recently it has been shown that the expression of the fur gene in E. coli was activated by both OxyR and SoxRS, the global regulators for H2O2 and O2− stress responses, respectively (41). Although metals are necessary for cell growth, surplus Fe2+ or Cu2+ ions are deleterious because they promote production of hydroxyl radicals from H2O2 by the Harber-Weiss-Fenton reaction (18). Therefore, a regulatory system coordinating metal metabolism and oxidative stress response might be required, and Fur-like proteins are suggested to be responsible for this regulatory role.
We proposed in this study that the furA-catC operon is negatively regulated by FurA on the grounds that (i) introduction of the furA gene into S. lividans on a multicopy plasmid inhibits the increased expression of the furA-catC operon at a later growth phase, (ii) FurA protein binds to the promoter region between nt −59 and −39 upstream from the transcription start site in a metal-dependent manner, (iii) addition of FurA-activating metals (Ni, Mn, Zn, and Fe) to the growth medium represses the production of CatC protein, and (iv) addition of EDTA to FurA-overproducing cells enhances production of catC transcripts.
The gel mobility shift assay indicates that both the thiol-reducing condition and the presence of metals enhance the binding activity of FurA. Among the metals tested, Ni2+ was found to be most effective in activating FurA. In S. coelicolor, nickel has been identified as an active metal that ensures the enzyme activity of Ni-containing SOD and regulates expression of the sodN and sodF genes, encoding Ni-containing and Fe-containing SODs, respectively (24, 25). Moreover, a high concentration of Ni (1 mM) also inhibited the major catalase (CatA) expression (Fig. 8B). Therefore nickel seems to play a pleiotropic role in S. coelicolor in regulating oxidative defense enzymes.
Recently, E. coli Fur has been identified as a zinc metalloprotein containing either a single zinc ion (Zn1Fur) or two (Zn2Fur) with similar DNA binding activity (2). The tightly associated zinc ion, which seems to have a structural role, is coordinated with two sulfur ligands and two N or O ligands (23). The two sulfur ligands are identified as Cys92 and Cys95 (17), known to be essential for Fur activity by mutagenesis (13). The two cysteine residues are also conserved in most Fur-homologous proteins, including S. coelicolor FurA (Cys99 and Cys102) (Fig. 2). The second zinc-binding site with lower affinity could be a regulatory site which senses the divalent cations. Studies with Co(II)-incorporated Fur from E. coli revealed that the cobalt is in an octahedral environment with at least two histidines, one aspartate or glutamate, and no cysteine ligands (1). Although there could be some difference in metal binding sites between E. coli Fur and S. coelicolor FurA, the reduced sulfhydryl group of cysteine residues of FurA might be necessary to bind metals and/or to maintain the proper tertiary structure of FurA. Thus, it is likely that the metal binding to FurA is regulated by the redox state of the protein. The fact that higher levels of CatC expression were obtained on surface culture than from liquid culture supports this idea that the redox state might affect FurA activity. However, the FurA protein does not seem to respond sensitively to H2O2, since we observed no significant induction of catC transcription by H2O2. This is consistent with the observation that expression of the katG gene in Mycobacterium species is insensitive to H2O2 (28). Such a characteristic differentiates the FurA-type regulators from those of the B. subtilis PerR type, which regulates genes in a manner highly sensitive to H2O2 (7).
The regulation of catalase-peroxidase gene expression by FurA in S. coelicolor in a metal- and redox-dependent manner fortifies the suggested role of FurA as an effector for both oxidation- and metal-dependent responses. Further studies are anticipated to reveal the mechanism of FurA activation and the interplay between metal- and redox-dependent activation.
ACKNOWLEDGMENTS
We thank You-Hee Cho, Jae-Bum Bae, and Yeonsoo Cho for helpful discussions, assistance in protein purification, and antibody preparation.
This work was supported by a Basic Research Grant for interdisciplinary researches (1999-2-202-002-5) from KOSEF. S.-O. Oh was supported by BK21 Research Fellowship from the Korean Ministry of Education.
REFERENCES
- 1.Adrait A, Jacquamet L, Le Pape L, Gonzalez de Peredo A, Aberdam D, Hazemann J L, Latour J M, Michaud-Soret I. Spectroscopic and saturation magnetization properties of the manganese- and cobalt-substituted Fur (ferric uptake regulation) protein from Escherichia coli. Biochemistry. 1999;38:6248–6260. doi: 10.1021/bi9823232. [DOI] [PubMed] [Google Scholar]
- 2.Althaus E W, Outten C E, Olson K E, Cao H, O'Halloran T V. The ferric uptake regulation (Fur) repressor is a zinc metalloprotein. Biochemistry. 1999;38:6559–6569. doi: 10.1021/bi982788s. [DOI] [PubMed] [Google Scholar]
- 3.Bagg A, Neilands J B. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol Rev. 1987;51:509–518. doi: 10.1128/mr.51.4.509-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagyan I, Casillas-Martinez L, Setlow P. The katX gene, which codes for the catalase in spores of Bacillus subtilis, is a forespore-specific gene controlled by ςF, and KatX is essential for hydrogen peroxide resistance of the germinating spore. J Bacteriol. 1998;180:2057–2062. doi: 10.1128/jb.180.8.2057-2062.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bierman M, Logan R, O'Brien K, Seno E T, Rao R N, Schoner B E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 6.Bol D K, Yasbin R E. The isolation, cloning and identification of a vegetative catalase gene from Bacillus subtilis. Gene. 1991;109:31–37. doi: 10.1016/0378-1119(91)90585-y. [DOI] [PubMed] [Google Scholar]
- 7.Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann J D. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Keramati L, Helmann J D. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc Natl Acad Sci USA. 1995;92:8190–8194. doi: 10.1073/pnas.92.18.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho Y-H, Lee E-J, Roe J-H. A developmentally regulated catalase required for proper differentiation and osmoprotection of Streptomyces coelicolor. Mol Microbiol. 2000;35:150–160. doi: 10.1046/j.1365-2958.2000.01685.x. [DOI] [PubMed] [Google Scholar]
- 10.Cho Y-H, Roe J-H. Isolation and expression of the catA gene encoding the major vegetative catalase in Streptomyces coelicolor Müller. J Bacteriol. 1997;179:4049–4052. doi: 10.1128/jb.179.12.4049-4052.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christman M F, Morgan R W, Jacobson F S, Ames B N. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell. 1985;41:753–762. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- 12.Claiborne A, Fridovich I. Purification of the o-dianisidine peroxidase from Escherichia coli B. Physicochemical characterization and analysis of its dual catalatic and peroxidatic activities. J Biol Chem. 1979;254:4245–4252. [PubMed] [Google Scholar]
- 13.Coy M, Doyle C, Besser J, Neilands J B. Site-directed mutagenesis of the ferric uptake regulation gene of Escherichia coli. Biometals. 1994;7:292–298. doi: 10.1007/BF00144124. [DOI] [PubMed] [Google Scholar]
- 14.de Lorenzo V, Wee S, Herrero M, Neilands J B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol. 1987;169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deretic V, Song J, Pagán-Ramos E. Loss of oxyR in Mycobacterium tuberculosis. Trends Microbiol. 1997;5:367–372. doi: 10.1016/S0966-842X(97)01112-8. [DOI] [PubMed] [Google Scholar]
- 16.Engelmann S, Lindner C, Hecker M. Cloning, nucleotide sequence, and regulation of katE encoding a ςB-dependent catalase in Bacillus subtilis. J Bacteriol. 1995;177:5598–5605. doi: 10.1128/jb.177.19.5598-5605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez de Peredo A, Saint-Pierre C, Adrait A, Jacquamet L, Latour J M, Michaud-Soret I, Forest E. Identification of the two zinc-bound cysteines in the ferric uptake regulation protein from Escherichia coli: chemical modification and mass spectrometry analysis. Biochemistry. 1999;38:8582–8589. doi: 10.1021/bi9902283. [DOI] [PubMed] [Google Scholar]
- 18.Halliwell B, Gutteridge J M. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassett D J, Howell M L, Ochsner U A, Vasil M L, Johnson Z, Dean G E. An operon containing fumC and sodA encoding fumarase C and manganese superoxide dismutase is controlled by the ferric uptake regulator in Pseudomonas aeruginosa: fur mutants produce elevated alginate levels. J Bacteriol. 1997;179:1452–1459. doi: 10.1128/jb.179.5.1452-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heym B, Zhang Y, Poulet S, Young D, Cole S T. Characterization of the katG gene encoding a catalase-peroxidase required for the isoniazid susceptibility of Mycobacterium tuberculosis. J Bacteriol. 1993;175:4255–4259. doi: 10.1128/jb.175.13.4255-4259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: John Innes Foundation; 1985. [Google Scholar]
- 22.Ivanova A, Miller C, Glinsky G, Eisenstark A. Role of rpoS (katF) in oxyR-independent regulation of hydroperoxidase I in Escherichia coli. Mol Microbiol. 1994;12:571–578. doi: 10.1111/j.1365-2958.1994.tb01043.x. [DOI] [PubMed] [Google Scholar]
- 23.Jacquamet L, Aberdam D, Adrait A, Hazemann J L, Latour J M, Michaud-Soret I. X-ray absorption spectroscopy of a new zinc site in the Fur protein from Escherichia coli. Biochemistry. 1998;37:2564–2571. doi: 10.1021/bi9721344. [DOI] [PubMed] [Google Scholar]
- 24.Kim E-J, Chung H-J, Suh B, Hah Y C, Roe J-H. Expression and regulation of the sodF gene encoding iron- and zinc-containing superoxide dismutase in Streptomyces coelicolor Müller. J Bacteriol. 1998;180:2014–2020. doi: 10.1128/jb.180.8.2014-2020.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim E-J, Chung H-J, Suh B, Hah Y C, Roe J-H. Transcriptional and post-transcriptional regulation by nickel of sodN gene encoding nickel-containing superoxide dismutase from Streptomyces coelicolor Müller. Mol Microbiol. 1998;27:187–195. doi: 10.1046/j.1365-2958.1998.00674.x. [DOI] [PubMed] [Google Scholar]
- 26.Lee J-H. M.S. thesis. Seoul, Korea: Seoul National University; 1995. [Google Scholar]
- 27.Loewen P C, Triggs B L. Genetic mapping of katF, a locus that with katE affects the synthesis of a second catalase species in Escherichia coli. J Bacteriol. 1984;160:668–675. doi: 10.1128/jb.160.2.668-675.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacNeil D J, Gewain K M, Ruby C L, Dezeny G, Gibbons P H, MacNeil T. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene. 1992;111:61–68. doi: 10.1016/0378-1119(92)90603-m. [DOI] [PubMed] [Google Scholar]
- 29.Menéndez M C, Ainsa J A, Martín C, García M J. katGI and katGII encode two different catalases-peroxidases in Mycobacterium fortuitum. J Bacteriol. 1997;179:6880–6886. doi: 10.1128/jb.179.22.6880-6886.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milano A, de Ross E, Gusberti L, Heym B, Marone P, Riccardi G. The katE gene, which encodes the catalase HPII of Mycobacterium avium. Mol Microbiol. 1996;19:113–123. doi: 10.1046/j.1365-2958.1996.352876.x. [DOI] [PubMed] [Google Scholar]
- 31.Nadler V, Goldberg I, Hochman A. Comparative study of bacterial catalases. Biochim Biophys Acta. 1986;882:234–241. [Google Scholar]
- 32.Niederhoffer E C, Naranjo C M, Bradley K L, Fee J A. Control of Escherichia coli superoxide dismutase (sodA and sodB) genes by the ferric uptake regulation (fur) locus. J Bacteriol. 1990;172:1930–1938. doi: 10.1128/jb.172.4.1930-1938.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pagán-Ramos E, Song J, McFalone M, Mudd M H, Deretic V. Oxidative stress response and characterization of the oxyR-ahpC and furA-katG loci in Mycobacterium marinum. J Bacteriol. 1998;180:4856–4864. doi: 10.1128/jb.180.18.4856-4864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith C P. Methods for mapping transcribed DNA sequences. In: Brown T A, editor. Essential molecular biology, a practical approach. New York, N.Y: Oxford University Press; 1991. pp. 237–252. [Google Scholar]
- 35.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wayne L G, Diaz G A. A double staining method for differentiating between two classes of mycobacterial catalase in polyacrylamide electrophoresis gels. Anal Biochem. 1986;157:89–92. doi: 10.1016/0003-2697(86)90200-9. [DOI] [PubMed] [Google Scholar]
- 37.Wayne L G, Diaz G A. Serological, taxonomic, and kinetic studies of the T and M classes of mycobacterial catalase. Int J Syst Bacteriol. 1982;32:296–304. [Google Scholar]
- 38.Youn H-D. Ph.D. thesis. Seoul, Korea: Seoul National University; 1995. [Google Scholar]
- 39.Youn H-D, Yim Y-I, Kim K, Hah Y C, Kang S-O. Spectral characterization and chemical modification of catalase-peroxidase from Streptomyces sp. J Biol Chem. 1995;270:13740–13747. doi: 10.1074/jbc.270.23.13740. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 41.Zheng M, Doan B, Schneider T D, Storz G. OxyR and SoxRS regulation of fur. J Bacteriol. 1999;181:4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zou P, Borovok I, Ortiz de Orué Lucana D, Müller D, Schrempf H. The mycelium-associated Streptomyces reticuli catalase-peroxidase, its gene and regulation by FurS. Microbiology. 1999;145:549–559. doi: 10.1099/13500872-145-3-549. [DOI] [PubMed] [Google Scholar]