Abstract
Tn5397 is a conjugative transposon that was originally isolated from Clostridium difficile. Previous analysis had shown that the central region of Tn5397 was closely related to the conjugative transposon Tn916. However, in this work we obtained the DNA sequence of the ends of Tn5397 and showed that they are completely different to those of Tn916. Tn5397 did not contain the int and xis genes, which are required for the excision and integration of Tn916. Instead, the right end of Tn5397 contained a gene, tndX, that appears to encode a member of the large resolvase family of site-specific recombinases. TndX is closely related to the TnpX resolvase from the mobilizable but nonconjugative chloramphenicol resistance transposons, Tn4451 from Clostridium perfringens and Tn4453 from C. difficile. Like the latter elements, inserted copies of Tn5397 were flanked by a direct repeat of a GA dinucleotide. The Tn5397 target sites were also shown to contain a central GA dinucleotide. Excision of the element in C. difficile completely regenerated the original target sequence. A circular form of the transposon, in which the left and right ends of the element were separated by a GA dinucleotide, was detected by PCR in both Bacillus subtilis and C. difficile. A Tn5397 mutant in which part of tndX was deleted was constructed in B. subtilis. This mutant was nonconjugative and did not produce the circular form of Tn5397, indicating that the TndX resolvase has an essential role in the excision and transposition of Tn5397 and is thus the first example of a member of the large resolvase family of recombinases being involved in conjugative transposon mobility. Finally, we showed that introduction of Tn916 into a strain containing Tn5397 induced the loss of the latter element in 95.6% of recipients.
Conjugative transposons are genetic elements that encode their own transfer from the genome of a donor cell to the genome of a recipient cell. These elements are remarkably promiscuous and are capable of being transferred across large phylogenetic distances. They are important clinically because they are one of the major vectors involved in the spread of antibiotic resistance among bacterial pathogens. The most intensively studied conjugative transposon is the 18.3-kb element Tn916. This transposon was originally isolated from the chromosome of Enterococcus faecalis DS16, where it mediated tetracycline resistance via the tet(M) gene (12). There are several reviews describing the properties of conjugative transposons (9, 26, 32, 35).
The first step in the conjugative transposition of Tn916 and its closely related elements is excision from the donor DNA. This process is followed by circularization of the element and its subsequent transfer to a new host, where the transposon inserts into a new DNA target site. The products of the transposon-encoded genes int and xis are required for excision (24). Int is a site-specific recombinase of the integrase family and is essential for the excision of Tn916 (24, 31, 38, 39). Xis has also been shown to be required for excision in gram-positive hosts. Recent work has shown that both Xis and Int are required for excision but that only Int is required for integration. In fact, the presence of Xis may inhibit integration of the transposon (19). The excision of Tn916 from the donor has been compared with the excision of λ phage as both proceed by a mechanism that involves staggered cuts at both ends of the element, followed by circularization and transfer to a new host (25). However, the recombination sites of λ are homologous, while those of Tn916 are not. Tn916 excision involves the Int-mediated production of 5′-protruding staggered endonucleolytic cuts. One strand is cut six bases from the end of the transposon, and the other is cut immediately adjacent to the other end (17, 30, 40). The resulting single strand overhangs that flank the transposon are then ligated to form a covalently closed circle (4). As Tn916 does not duplicate its integration target (8), the two overhangs are not usually complementary and the resulting joint between the two ends of the conjugative transposon may be a heteroduplex. However, in E. faecalis only a homoduplex joint has been found in the circular intermediate (18).
In addition to these insertion and excision functions, members of the Tn916 family encode their own conjugative transfer. The complete DNA sequence of Tn916 has been obtained (11). Open reading frames (ORFs) that have the potential to encode polypeptides with sequence similarity to proteins known to be involved in conjugation (e.g., the anti-restriction protein Ard of plasmid Collb-P9 and the MbeA mobilization protein of plasmid ColE1) have been identified. A functional oriT site has also been located (15), and there is evidence that this site is involved in single-stranded DNA transfer to the recipient (36).
We have identified a conjugative transposon, Tn5397, from the gram-positive anaerobic pathogen Clostridium difficile (21, 22). Tn5397 was shown to be transferred by a conjugation-like process from C. difficile strain 630 to Bacillus subtilis strain CU2189 and back to C. difficile, as well as between C. difficile strains (21). Furthermore, Tn5397 has also been shown to be capable of transfer in a model oral biofilm community, indicating that the element is likely to be able to transfer in natural environments (28). Physical and genetic analysis indicates that Tn5397 is related to Tn916 (14, 21, 22). However, there are some differences between the two elements, as members of our group have recently shown that Tn5397 contains a group II intron inserted into a gene almost identical to orf14 from Tn916 (22).
In this paper we show that the ends of Tn5397 are not related to those of Tn916 and that the int and xis regions of Tn916 have been replaced by a gene that has the potential to encode a site-specific recombinase, tndX, that is related to the large resolvase family of recombinases. We demonstrate that this gene is related to the resolvase genes (tnpX) from the nonconjugative, mobilizable chloramphenicol resistance transposons Tn4451 and Tn4453 (2, 16). This is the first time that such a gene has been found on a conjugative transposon. The target sites of Tn5397 in C. difficile and B. subtilis have also been determined, which has allowed us to develop a model for the integration and excision of Tn5397.
MATERIALS AND METHODS
Bacterial strains, growth media, and plasmids.
All the bacterial strains and plasmids used in this study together with their relevant properties are shown in Table 1. The C. difficile and B. subtilis strains were grown on brain heart infusion (BHI) agar or in BHI broth (Oxoid, Basingstoke, United Kingdom), and Escherichia coli strains were grown in Luria-Bertani agar or broth (33). Where appropriate, the medium was supplemented with ampicillin (100 μg ml−1), chloramphenicol (30 μg ml−1), erythromycin (100 μg ml−1), tetracycline (10 μg ml−1), or rifampin (15 μg ml−1). All C. difficile strains were grown at 37°C in an anaerobic chamber (Don Whitley Scientific Ltd.) with an atmosphere of 80% N2, 10% H2, 10% CO2. B. subtilis and E. coli strains were grown at 37°C aerobically.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| C. difficile | ||
| CD37 | RifrC. difficile recipient strain Tcs Ems | 14 |
| 630 | Tcr EmrC. difficile strain containing Tn5397 and Tn5398 | 14 |
| FM30 | Tcr transconjugant from the mating 630xCD37 (CD37::Tn5397) | 21 |
| FM1A | Tcr transconjugant from the mating 630xCD37 (CD37::Tn5397) | This work |
| FM168 | Tcr Emr transconjugant from the mating BS2xFM123 (CD37::Tn5397; Tn916ΔE) | P. Mullany, unpublished |
| FM48 | Tcr transconjugant from the mating BS5xCD37 (CD37::Tn916) | P. Mullany, unpublished |
| FM128 | Tcs Emr transconjugant from the mating BS19xFM30 (CD37::Tn916ΔE deletion of Tn5397) | This work |
| FM136 | Tcs Emr transconjugant from the mating BS19xFM30 (CD37::Tn916ΔE deletion of Tn5397) | This work |
| FM123 | Emr transconjugant from the mating BS19xCD37 (CD37::Tn916ΔE) | P. Mullany, unpublished |
| B. subtilis | ||
| CU2189 | Tcs recipient strain | 7 |
| BS5 | CU2189 transformed with pAM120 (contains Tn916) | 23 |
| BS2 | Tcr transconjugant from the mating 630xCU2189 (CU2189::Tn5397) | 23 |
| BS3 | Tcr transconjugant from the mating 630xCU2189 (CU2189::Tn5397) | 21 |
| BS4 | Tcr transconjugant from the mating 630xCU2189 (CU2189::Tn5397) | 21 |
| BS5A | Tcr transconjugant from the mating 630xCU2189 (CU2189::Tn5397) | This work |
| BS19 | Emr transformed with pCER110 (CU2189::Tn916ΔE) | 23 |
| BS6A | Tcr transconjugant from the mating 630xCU2189 (CU2189::Tn5397) | This work |
| BS11A | Derivative of BS6A in which tndX has been replaced with the tndXΔcat allele | This work |
| Plasmids | ||
| T-easy vector | Vector for cloning PCR products | Promega |
| pPPM122 | T-easy vector containing the tndXΔcat allele | This work |
| pJIR62 | Contains the catP gene | 37 |
| pAM120 | Cloned copy of Tn916 | 13 |
| pCER110 | Cloned copy of Tn916ΔE | 29 |
| pJIR1508 | tndX cloned into pUC18 | This work |
| pSU39 | Low-copy-number cloning vector | 3 |
| pJIR1537 | tndX cloned into pSU39 | This work |
| pJIR683 | Target plasmid containing Tn4451ΔtnpX | 2 |
| pJIR639 | Contains a wild-type tnpX gene | 2 |
Abbreviations used: Tcr, tetracycline resistant; Tcs, tetracycline sensitive; Emr, erythromycin resistant; Ems, erythromycin sensitive; Rifr, rifampicin resistant; Rifs, rifampicin sensitive
Strategy for obtaining the DNA sequence of the ends of Tn5397.
Previous subcloning and hybridization analysis has allowed the identification of the right end of Tn5397 (21, 22, 23). The complete DNA sequence of the right end of Tn5397 was determined.
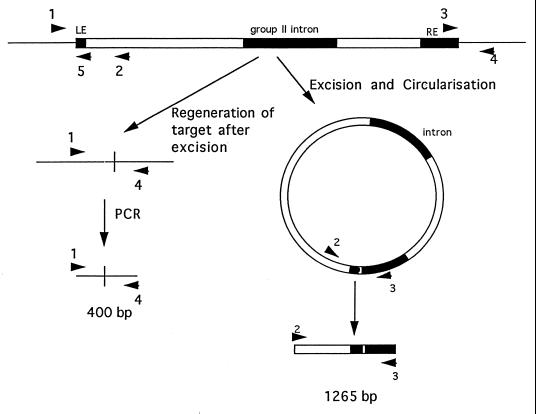
A PCR-based strategy was used to amplify the ligated ends of the circular form of the transposon (see Fig. 2). Primer 2 (5′ ACAACCAGCAGGAAAACAGG 3′) was designed from the left end of Tn916. Primer 3 (5′ ACGTGTATCAAGCAGAGGGAATCGGTAAA 3′) was designed from the sequence of the right end of Tn5397. This primer bound to Tn5397, 90 bp from the right end of the transposon. The 1,265-bp PCR product produced with primers 2 and 3 was sequenced on both strands to gain an unambiguous sequence. Based on the sequence of the left end of Tn5397, a primer reading out of the transposon (primer 5 [5′ CCACTTGATATGAAAAATCAAATGGCTC 3′]) was designed (see Fig. 2). Primers 3 and 5 were used to prime genomic DNA sequencing of C. difficile 630 DNA. The resulting genomic DNA sequence was used to design primers reading into the transposon from the flanking genomic region, specifically, primers 1 (5′ GAAAACTGCTTGGATTCAGAAG 3′) and 4 (5′ GATATTGAAAACTCCTTGAAAGTATCATATCC 3′). These primers were used to prime genomic DNA sequencing reactions in strains that contained Tn5397 and isogenic strains that did not carry the element. Primer pairs (primers 1 and 2 and primers 3 and 4) were used to amplify the junction regions, and another primer pair (primers 1 and 4) was used to amplify the target sequence in strains that did not contain Tn5397.
FIG. 2.
Strategy for obtaining the DNA sequence of the ends of Tn5397. At the top of the figure the box represents Tn5397, the white area represents the part of Tn5397 that is homologous to Tn916, and the shaded area represents the regions unique to Tn5397. The single lines represent the DNA flanking Tn5397 and (at the bottom of the figure) the genomic target of Tn5397. The arrows show the binding sites and direction of priming of oligonucleotides (the sequence of these oligonucleotides is shown in the text) that were used in PCR and genomic sequencing reactions. The circular form of the transposon is also shown. The region of the circular form that is amplified by the PCR primers is shown (again, regions unique to Tn5397 are shaded). The vertical line in the regenerated target represents the joint between sequences that were previously on each side of the transposon.
Construction of the tndX mutant tndXΔcat.
To generate the deletion mutant of tndX (tndXΔcat) the region from bp 285 to bp 1281 (bp 1 is the first base in the ORF [see Fig. 1D]) of the tndX gene was replaced with the catP gene from pJIR62 (37). Primers 5′ TTGTTAAAACAGCAAGC 3′ and 5′ CCCACTTCGACTGCACTCCCCCACATAGTACATGAATAGTGC 3′ were used to amplify bp 1 to 285, primers 5′ GCACTATTCATGTACTATGTGGGGGAGTGCAGTCGAAGTGGG 3′ and 5′ GCATTTTGCTCTATAAGTTTGGGGTCTTTGTACTAACCTGTGG 3′ were used to amplify the catP gene, and primers 5′ CCACAGGTTAGTACAAAGACCCCAAACTTATAGAGCAAAATGC 3′ and 5′ TATCAATGAGACACTGC 3′ were used to amplify bp 1281 to 1599 from tndX. These PCR products were joined together by subsequent PCR reactions. The resulting recombinant was cloned into the pGEM-T Easy vector (Promega) to generate pPPM122. To replace the tndX allele with tndXΔcat, pPPM122 was used to transform the B. subtilis strain BS6A (CU2189::Tn5397), and selection for chloramphenicol-resistant transformants was done. Transformation of B. subtilis was carried out using the method of Anagnostopoulos and Spizizen (1). One transformant, in which tndXΔcat had replaced tndX and no vector sequences were present, as verified by PCR and DNA sequencing, was designated BS11A and chosen for further study.
FIG. 1.
Comparison of the ends of Tn916 and Tn5397. (A) Schematic diagram of the structure of the left and right ends of Tn916 and Tn5397. The genes labeled xis and int in Tn916 are the excisionase and the integrase, respectively. The functions of the genes labeled orf23 and orf24 in Tn916 are not yet known, and they are labeled as Flannagan et al. labeled them (11). In Tn5397, the gene labeled tndX encodes a putative site-specific recombinase related to the large resolvase family and orf23* is almost identical to orf23 from Tn916. Intergenic regions that are unique to Tn5397 are shown (thick line), as are the intergenic regions common to both transposons (thin line). The regions that are shown in more detail in panels B, C, and D are indicated. The sizes of the transposons are also shown. Bar, 1 kb. (B) DNA sequence of the left end of Tn5397. The first 328 bp is shown. At the beginning of the transposon, the GA dinucleotide that flanks Tn5397 in direct repeat is underlined. The region that is homologous to Tn916 is shown in boldface. Where the Tn916 sequence differs by a single base, the Tn916 sequence is written above the Tn5397 sequence in lowercase. Δ10, a region where there has been a deletion of 10 bp compared to Tn916; ΔA, a site where there has been a deletion of an adenine residue compared to Tn916. The region written in italics is not homologous to Tn916, and here 25 bp of Tn5397 have replaced 19 bp of Tn916 sequence. The ATG start codon of orf23* is underlined. The direction of transcription of orf23* is indicated by an arrow under the sequence. An imperfect inverted repeat that occurs at the left and right ends of Tn5397 is shown (boxed). The inverted repeat sequence AAATAG is also boxed. (C) DNA sequence of the right end of Tn5397, where it diverges from Tn916. The region written in bold is homologous to Tn916. Where Tn916 differs from Tn5397 by a single base, the Tn916 sequence is written above the Tn5397 sequence in lowercase. The region in italics labeled 1 is not homologous to Tn916; at this site 21 bp of Tn5397 have replaced 125 bp from Tn916. The region of Tn916 that has been replaced contains the promoter for xisTn and the palindromes at the end of orf8 (6). The region labeled 2 is not homologous to Tn916, and 18 bp of Tn5397 have replaced 22 bp of Tn916. ΔT, a site where there has been a deletion of a thymine derivative compared to Tn916. The last part of the sequence shown in plain text is not homologous to Tn916; the underlined TTG is the putative start codon of tndX. The direction of transcription of tndX is indicated by the arrow. The inverted repeat TAAAAC is boxed. (D) DNA sequence of the last 40 bases of the right end of Tn5397, showing the end of tndX. The tndX stop codon is shown, and the GA dinucleotide that is present in direct repeat at the end of the transposon is underlined. The direction of transcription of tndX is shown by the arrow. An imperfect inverted repeat that occurs at the left and right ends of Tn5397 is shown (boxed).
DNA manipulations.
Plasmid constructions (with the exception of those described above) were all carried out essentially as previously described (33). Genomic DNA from C. difficile and B. subtilis was prepared using a gram-positive DNA isolation kit (Puregene). Plasmid DNA was prepared using a plasmid Miniprep kit (Qiagen, Crawley, United Kingdom).
Sequencing methods.
Genomic sequencing was carried out with a reaction mixture containing 8 μl of Big Dye Mix (ABI), 15 to 30 pmol of primer, 3 to 6 μg of purified genomic DNA, and 1 μl of ThermoFidelase (Fidelity Systems, Inc.), with water added to obtain a final volume of 20 μl. The thermocycling conditions were as follows: 95°C for 5 min, followed by 99 cycles of a rapid thermal ramp to 95°C, 95°C for 30 sec, a rapid thermal ramp to 55°C, 55°C for 20 sec, and a rapid thermal ramp to 65°C, 65°C for 4 min. This was followed by a rapid thermal ramp to 4°C, and the mixture was held at that temperature. These samples were ethanol precipitated and analyzed on an ABI PRISM 310 genetic analyzer. For all PCR-based sequencing, the DNA sequences of the leading and lagging strands were determined with products from independent PCR experiments.
PCR.
The PCR reaction consisted of 30 cycles of denaturation (94°C, 1 min), annealing (50°C, 1 min), and extension (72°C, 1 to 3 min). The products were stored at 4°C until ready for analysis. The Taq polymerase was obtained from Promega, and reactions were carried out in buffer provided by the manufacturer.
Filter matings.
Both B. subtilis and C. difficile were grown on BHI agar plates for 18 h. The cells were scraped off the plates and resuspended in 20 ml of BHI broth. The cultures were grown at 37°C until mid-exponential phase (optical density at 600 nm of 0.45). Cultures of donor and recipient were mixed and centrifuged in an anaerobic environment to form a cell pellet. The pellet was resuspended in 1 ml of BHI broth, and 100 μl was spread on nitrocellulose 0.45-μm-pore-size filters on BHI agar plates which were incubated for 18 h at 37°C in an anaerobic environment. The filters were removed from the agar plates and placed in 20-ml bottles containing (each) 1 ml of sterile BHI broth (at 37°C) and vortexed for 10 to 20 s. When C. difficile was the recipient, 100-μl aliquots were spread on BHI agar supplemented with the appropriate antibiotics and incubated anaerobically for 48 h. When B. subtilis was the recipient, 100-μl aliquots were spread on BHI agar supplemented with the appropriate antibiotics and incubated aerobically for 48 h, with checking for growth at 24 and 48 h.
Excision assays.
The ability of TndX to promote the excision of the Tn4451 derivative Tn4451ΔtnpX (2) was determined as follows. The tndX gene was amplified by PCR from C. difficile 630 using the primers 5′ CGTGATAATGATACTCC 3′ and 5′ ATATGTCCTTCTGTTGCTGA 3′. The resulting PCR fragment was filled using T4 DNA polymerase (Boehringer Mannheim) and ligated into the pUC18 SmaI site to generate pJIR1508. The region containing tndX was subcloned into the EcoRI-XbaI site of pSU39 to generate pJIR1537. The excision assays were performed essentially as previously described (2, 10), with some modifications. The target plasmid carrying Tn4451ΔtnpX in these assays was pJIR683 (Apr Cmr), which is a pBluescript derivative (2). Therefore, the strains constructed for analysis were DH12S sublines carrying pJIR683 together with either pSU39 (the negative control) or pJIR1537 (the tndX+ test plasmid). The DH12S sublines were exposed to 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h prior to the extraction of plasmid DNA in order to induce expression of tndX. Miniprep DNA was prepared and subsequently used to transform DH5α to ampicillin resistance. Loss of the transposon was then monitored by plating onto media containing chloramphenicol.
Nucleotide sequence accession numbers.
In this study, the complete DNA sequence of the right end (GenBank accession number AF193610) and the sequence of the left end (GenBank accession number AF193609) of Tn5397 were determined, as was the DNA sequence surrounding the C. difficile insertion site in the transconjugants FM168 and FM30 (559 bp) (GenBank accession number AF249883).
RESULTS
Genetic organization of the ends of Tn5397.
Previous hybridization analysis of Tn5397 had shown that the central region of the element was very closely related to Tn916 but that the ends of the two elements were different (22, 23). To determine the nature of these differences, the ends of Tn5397 were completely sequenced on both strands. The regions where the two transposons diverge are shown schematically in Fig. 1A. Analysis of the first 328 bp of the left end of Tn5397 (Fig. 1B) showed that the first 201 bp of Tn916 was absent and was replaced by 180 bp of unrelated DNA sequence in Tn5397. Thereafter, the two transposons are closely related, although there are some insertions and deletions (Fig. 1B). In addition, there is an inverted repeat, 5′ AAATAG 3′, (Fig. 1B) on either side of the regions where the two transposons diverge.
The DNA sequence of the right end of Tn5397 was also determined (Fig. 1C and D). The last 1,724 bp of Tn916 is not present and there is 1,925 bp of Tn5397-specific DNA. As a result, the int and xis genes are not present in Tn5397. They have been replaced with an ORF of 1,599 bp, encoding a putative 533-amino-acid protein of 61.5 kDa, which we have designated tndX. The tndX ORF ends 10 bp from the end of Tn5397 (Fig. 1D).
In the right end region of Tn5397 that is related to Tn916 (Fig. 1C), there are numerous insertions and deletions. The largest is the replacement of 125 bp from Tn916 with 21 bp of Tn5397-specific sequence. This has resulted in the loss of the xis promoter and the palindromes at the end of orf8 (6) (Fig. 1C). In addition, there is an TAAAAC inverted repeat on either side of the regions where the two transposons diverge. There is also an imperfect inverted repeat (13 of 20 nucleotides) at the left and right ends of Tn5397 (Fig. 1B and D).
Determination of site specificity of Tn5397 in C. difficile and B. subtilis.
To determine the target site specificity of Tn5397 we determined the sequence of the transposon-host genome junctions from C. difficile strain 630, the original donor strain (Fig. 2 shows the strategy used), and two independently isolated C. difficile CD37 transconjugants, FM168 and FM30 (Table 1). These sequences were identical (Fig. 3A). The DNA sequence surrounding the C. difficile insertion site in these strains was also determined. It was found that Tn5397 inserted into an ORF that was predicted to encode a polypeptide that has limited homology to the pilin gene inverting protein PivNM-2 from Neisseria meningitidis (accession number AE0025251). The insertion sites in four independently isolated B. subtilis strains were also sequenced, in this host Tn5397 had inserted into different sites. At each site the transposon was flanked by a directly repeated GA dinucleotide (Fig. 3A). The sites of insertion in the B. subtilis genome are also shown in Fig. 3. It is of interest to note that two insertions in different regions of the padC gene were isolated.
FIG. 3.
(A) The DNA sequence of the transposon genome junction regions in C. difficile and B. subtilis, the junction regions were the same in the donor strain C. difficile 630, and in the C. difficile transconjugants. The DNA sequence of the transposon is written in bold text and the GA dinucleotides flanking the element are written in a larger font. (B) Comparison of the target sites in C. difficile and B. subtilis with the ends of Tn5397 and the circular form of the transposon. The insertion sites of Tn5397 in the B. subtilis genome are also shown. ∗, This target is also present after excision of Tn5397 in strains FM128 and FM136 (see text for more details). Superscripts indicate the following: 1, Tn5397 has inserted into the padC gene which encodes phenolic acid decarboxylase; 2, Tn5397 has inserted in an intergenic region between the genes yxjB and yxjC; 3, Tn5397 has inserted into the gene encoding threonine dehydratase; 4, Tn5397 has inserted into padC (a different part of padC to the insertion in BS2).
For both B. subtilis and C. difficile, the DNA sequence of the target site before insertion of the transposon was also obtained (Fig. 3B). Each of these targets had a single GA dinucleotide at the site of insertion. In C. difficile there are also imperfect inverted repeats present on either side of the central GA motif. Of the four B. subtilis target sites analyzed, two of four had imperfect inverted repeats flanking the central GA dinucleotide (Fig. 3). Tn5397 inserted in the same site in all C. difficile transconjugants examined; the site may represent an insertional hot spot. A database search of the B. subtilis genome sequence (http://bioweb.pasteur.fr/GenoList/SubtiList/) with the 22-bp target site (Fig. 3) from C. difficile was performed using the Blastn program (data not shown). This analysis showed that there were no sequences in the B. subtilis genome that completely matched this sequence. Furthermore, the sequences that most closely matched the C. difficile target were not those that were identified by the genetic studies reported here.
Tn5397 is found in a circular form in C. difficile.
Given that the conjugative transposon Tn916 and the mobilizable transposons from Clostridium perfringens and C. difficile, Tn4451 and Tn4453, respectively, have been shown to produce circular forms (2, 16, 36), we decided to see if Tn5397 also has a circular form. PCR with outward firing primers 2 and 3 (Fig. 2) would yield a PCR product only if the left and right ends of the transposon were ligated together. Products of the appropriate size (1265 bp) were produced when DNA from the C. difficile strains 630 and FM168 (each contains a single copy of Tn5397) were used as templates. No products were observed when DNA from the conjugation recipients CD37 or FM48 (CD37::Tn916) were used as templates (results not shown).
Five independently derived PCR products were cloned and sequenced. The sequence obtained from each recombinant was identical. The termini of Tn5397 could be seen in an orientation consistent with the formation of a circular molecule. The two ends of the transposon were separated by a GA dinucleotide at the circular form joint (Fig. 3B).
Introduction of Tn5397 into a C. difficile strain containing Tn916ΔE induces the loss of Tn916ΔE and vice versa.
When the B. subtilis strain BS2 (CU2189::Tn5397) was used as the donor and C. difficile strain FM123 (CD37::Tn916ΔE) was used as the recipient in filter mating experiments, tetracycline-resistant strains arose at a frequency of 2 × 10−7 transconjugants per donor cell. Only 65% (91 of 140) of the transconjugants were found to be resistant to erythromycin. When B. subtilis BS19 (CU2189::Tn916ΔE) was used as the donor and C. difficile FM30 (CD37::Tn5397) was used as the recipient, erythromycin-resistant transconjugants arose at a frequency of 3 × 10−7 per donor cell but only 4.3% (6 of 141) of the transconjugants were found to be tetracycline resistant. Both Tn5397 and Tn916ΔE were stable, i.e., after daily subculture for 14 days all of 100 colonies from the final subculture were tetracycline resistant and/or erythromycin resistant, respectively. This result agrees with previous work that shows that Tn5397 is stably inherited in C. difficile (21).
To see if the introduction of a second copy of Tn916 into C. difficile resulted in the loss of the resident Tn916, B. subtilis BS5 (CU2189::Tn916) was mated with C. difficile FM123 (CD37::Tn916ΔE). Tetracycline-resistant transconjugants were obtained at a frequency of 2 × 10−7 per donor cell. All of the 365 transconjugants tested from this mating were resistant to erythromycin, the nonselected marker. Therefore, introducing a second copy of Tn916 into C. difficile did not induce the loss of the resident Tn916ΔE element.
To investigate if doubly resistant transconjugants contained two functional elements, FM149 (CD37::Tn916ΔE Tn5397) was tested for its ability to transfer erythromycin and tetracycline resistance to B. subtilis. All of the transconjugants selected on erythromycin were tetracycline sensitive (142 of 142 transconjugants), and those selected on tetracycline were erythromycin sensitive (192 of 192 transconjugants), showing that Tn5397 and Tn916ΔE transferred separately and independently. The transconjugants from these matings could also transfer either Tn916ΔE or Tn5397 in a further round of mating, indicating that both transposons are still functionally intact.
Tn5397 excises precisely from its genomic target, regenerating the original target sequence.
The experiments described above provided us with the opportunity of investigating the target site after the excision of Tn5397 in C. difficile. Genomic DNA was prepared from two of the strains which had lost Tn5397, FM128 and FM136, and used in a PCR reaction with primers 1 and 4 (Fig. 2). A product of 400 bp, i.e., the size expected if Tn5397 had excised from the genome, was obtained (results not shown). The DNA sequence of the PCR product was obtained, and the DNA sequence was identical to that of the C. difficile CD37 target site (Fig. 3). These results demonstrate that upon excision, Tn5397 precisely regenerates the original target site.
Identification of a potential site-specific recombinase TndX.
Database searches with the TndX sequence revealed that it is related to TnpX (37% identity and 61% similarity), a site-specific recombinase from the C. perfringens chloramphenicol resistance transposon Tn4451 (2). In common with TnpX, TndX contains amino acid residues in the N-terminal domain that are highly conserved within the resolvase/invertase family of site-specific recombinases. The conserved TnpX resolvase residues previously shown to be required for TnpX function (10) are also present in TndX (Fig. 4). Most members of the resolvase family of proteins are less than half the size of TndX and in their C-termini contain a helix-turn-helix motif which is not found in TndX or TnpX. TndX and TnpX are members of a new family of large resolvase/invertase proteins (Fig. 4). These proteins have sequence similarity to typical resolvase/invertases in their N-terminal domains but have greatly extended C-terminal domains (10, 41).
FIG. 4.
Multiple alignment of TndX and other large resolvase/invertase proteins. The first two lines show a comparison between TndX (Tn5397) and TnpX (Tn4451). Amino acid residues that are identical in these two proteins are underlined, and residues that are similar are in italics. ∗, conserved residues that have previously been shown to be required for TnpX function and are conserved in the recombinases shown here (10). TnpX, site-specific recombinase from Tn4451 (GenBank accession number U15027); Int, integrase from phage TP901-1 (GenBank accession number X85213); XisF, site specific recombinase from Anabaena species (GenBank accession number L23220); CisA, site-specific recombinase from B. subtilis (GenBank accession number A43656); Rv1586c, hypothetical recombinase from Mycobacterium tuberculosis (GenBank accession number Z95586); YokA, site-specific recombinase from B. subtilis phage SPBc2 (GenBank accession number AF020713).
TndX is required for excision of Tn5397.
Analysis of the DNA sequence of several Tn5397 target sites suggested that the element integrated and excised by using a method typical of resolvases, i.e., strand exchange occurs by a concerted four-strand break-and-rejoining mechanism involving 2-bp-staggered ends. To demonstrate that TndX was required for the conjugative transposition of Tn5397, a portion (bp 285 to 1281) of the tndX gene was replaced with a chloramphenicol resistance gene (catP) by allelic replacement in B. subtilis; the resulting mutant allele was designated tndXΔcat. That the expected homologous recombination reaction had occurred was verified by PCR on DNA extracted from BS6A (parent strain) and BS11A (mutant containing tndXΔcat) (results not shown), demonstrating unambiguously that tndXΔcat had replaced the tndX gene in BS11A. Subsequent conjugation experiments showed that strain BS11A could not act as a donor for the transfer of Tn5397 to C. difficile strain CD37 in filter mating experiments. By contrast, the parent strain BS6A yielded 2 × 10−7 tetracycline-resistant transconjugants per donor cell. Moreover, the circular form of the transposon could not be amplified from BS11A but could be amplified from the parent strain BS6A (results not shown). These results indicate that TndX is required for formation of the circular form of Tn5397, presumably by catalyzing transposon excision.
TndX cannot replace TnpX in a trans complementation assay.
TndX and TnpX are related at the amino acid sequence level, and both appear to excise their respective transposons by a similar resolvase-mediated mechanism. Therefore, TndX was used in a trans complementation assay in E. coli to see if it could excise a derivative of Tn4451, Tn4451tnpXΔ1, in which an internal fragment of tnpX had been deleted (2). Although transposon excision was detected when this derivative was complemented with a plasmid (pJIR639) carrying a wild-type tnpX gene, no excision was detected when we used a similar plasmid (pJIR1537) carrying a wild-type tndX gene (data not shown). Evidence that the cloning procedure had not introduced a mutation in tndX was confirmed by completely sequencing the tndX gene in pJIR1537. Expression of TndX in cells containing pJIR1537 was confirmed by Western blotting (results not shown).
DISCUSSION
In this paper, we have examined the ends of Tn5397 in detail. At the right end of Tn5397, the xis and int genes that are present in the equivalent positions in Tn916 have been replaced by tndX, which appears to encode a protein related to the large resolvase family of site-specific recombinases. The members of this family include enzymes involved in excision of DNA during spore formation (34), heterocyst development (5), excision of transposons (2, 10, 16), and the integration and excision of bacteriophage genomes (41). Each of these proteins has the resolvase/invertase catalytic domain at its N terminus, but they are much larger than typical resolvase/invertase proteins, being greatly extended in the C-terminal region. TndX is most closely related the TnpX resolvase, which promotes the excision of the C. perfringens transposon Tn4451 and the closely related C. difficile transposon Tn4453 (2, 10, 16). All of the residues that Crellin and Rood (10) showed were essential for TnpX function were also present in TndX. However, we demonstrated in this work that TndX could not promote the excision of Tn4451ΔtnpX. Previous work demonstrated that TnpX efficiently excises Tn4451ΔtnpX under the same conditions (2).
The difference in activity between two proteins probably reflects sequence differences and differences in the target site specificity of the TndX and TnpX enzymes, specifically the sequences of the transposon ends and insertion sites. With the exception of the GA dinucleotide at the ends of the elements and at the center of the target sites, there is no obvious sequence similarity between the ends of Tn5397 and Tn4451 or between the target sites of the two transposons. However, the target sites of both transposons, at least in their clostridial hosts, resemble the ends of their cognate transposon (see reference 10 for the Tn4451 sites in C. perfringens). For Tn5397 in C. difficile, 8 out of 10 bases of the target to the right of the GA are identical to the right end of the transposon and 5 out of 10 bases to the left of the GA are identical to the left end of the transposon. However, in B. subtilis Tn5397 can insert at sites that do not resemble the ends of the transposon and the only obvious requirement is the GA dinucleotide at the centre of the target site. Interestingly, a search of the B. subtilis genome revealed that the target site found in C. difficile is not present in B. subtilis, which may account for the ability of Tn5397 to insert into different sites in this host.
We have shown that Tn5397, in addition to existing in an integrated form, produces a circular molecule. Production of the circular form was dependent on TndX since a mutant of Tn5397 in which part of tndX had been deleted could not produce this circular molecule. Furthermore, the tndX mutant was not capable of conjugative transfer. In the related conjugative transposon, Tn916, a circular form was shown to be the transposition intermediate (36). Tn4451 and Tn4453 also produce circular forms (2, 16) which have now been shown to be the transposition intermediates (D. Lyras and J. I. Rood, unpublished results). Other large resolvase-like proteins have been shown to catalyze the formation of circular DNA molecules (41). Taken together, these results strongly suggest that the circular form of Tn5397 is the transposition intermediate.
On the basis of the above discussion we propose that TndX is responsible for the insertion and excision of Tn5397. The available information indicates that TndX mediates the excision of Tn5397 by introducing 2-bp-staggered cuts at the 3′ ends of the directly repeated GA dinucleotides at the ends of the transposon. Strand exchange occurs, resulting in excision of the transposon as a circular molecule and regeneration of the original target site. The circular form can then be transferred to a new host cell by conjugation and can transpose into the new genome. Integration occurs by TndX recognizing a suitable target site that contains a central GA dinucleotide and promoting site-specific recombination with the joint of the circular form. Strand exchange and ligation result in the insertion of Tn5397 flanked by directly repeated GA dinucleotides.
Tn5397 can induce the loss of Tn916 when introduced into a C. difficile cell by conjugation and vice versa. The mechanism of this interaction is not known. However, recent work has allowed a model to be proposed for the regulation of Tn916 transcription by tetracycline (6). These authors identified at least three possible trans-acting regulatory proteins, the positive regulators encoded by orf7 and orf8 and the putative negative regulator encoded by orf9. When Tn916 is transferred to C. difficile, these proteins are presumably synthesized and have an effect on the stability of the resident copy of Tn5397. The fact that the introduction of a marked copy of Tn916 does not induce the loss of a resident Tn916 element in C. difficile must reflect differences in the regulation of the int and xis genes in Tn916 and the tndX gene in Tn5397.
We propose that Tn5397 was produced by a recombination event between different mobile genetic elements. In this process there was an exchange of recombination modules, resulting in Tn5397 retaining a Tn916-like conjugation system but acquiring a completely different resolvase-mediated integration and excision system. The recent finding in C. difficile of a nonconjugative transposon closely related to Tn4451 (i.e., Tn4453 [16]) indicates that there is a clear possibility for interaction between Tn916-like elements and transposons containing TnpX genes in C. difficile. However, the resolvase gene from Tn5397 has diverged significantly from those of Tn4451 and Tn4453, suggesting that the formation of Tn5397 was not a relatively recent genetic event.
As well as swapping of modules, conjugative transposons have been shown to associate with each other and other mobile elements to produce composite transposons (20, 27). This type of cooperation between different mobile elements is a powerful means of generating rapid evolutionary change. In Tn5397 we have seen the generation of a novel conjugative transposon with different mechanisms of integration/excision and target site preferences to the ancestral Tn916-like element. Tn5397 is also a composite element as it has acquired a group II intron which is very likely to be mobile (22).
In conclusion, we have shown that Tn5397 is a modular mobile element, with the central portion of the element, the region involved in conjugation, being very closely related to the promiscuous enterococcal element Tn916 and the ends of the element, which are involved in integration and excision, being unique to Tn5397 but closely related to the recombination region of the nonconjugative mobilizable clostridial transposons Tn4451 and Tn4453. Tn5397 inserts and excises from the host replicon by the action of an enzyme of the large resolvase/invertase family. Although Tn4451 and Tn4453 can be mobilized, they are not conjugative. Tn5397 is the only conjugative transposon discovered so far whose translocation appears to be dependent on resolvase-mediated site-specific recombination events.
ACKNOWLEDGMENTS
H.W. and A.P.R. contributed equally to the work.
We thank the Wellcome Trust for supporting the work in one of our laboratories (P.M.) and facilitating the collaboration between P.M. and J.I.R. A.P.R. was the recipient of a BBSRC research studentship. Work in one of our laboratories (J.I.R.) was supported by grants from the Australian National Health and Medical Research Council.
We thank Don Clewell for pAM120 and Craig Rubens for pCER110. We thank Diane Massie for her excellent assistance with some of the DNA sequencing.
REFERENCES
- 1.Anagnostopoulos G, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannam T L, Crellin P K, Rood J I. Molecular genetics of the chloramphenicol-resistance transposon Tn4451 from Clostridium perfringens: the TnpX site-specific recombinase excises a circular transposon molecule. Mol Microbiol. 1995;16:535–551. doi: 10.1111/j.1365-2958.1995.tb02417.x. [DOI] [PubMed] [Google Scholar]
- 3.Bartolome B, Jubete Y, Martinez E, de la Cruz F. Construction and properties of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 4.Caparon M G, Scott J R. Excision and insertion of the conjugative transposon Tn916 involves a novel recombination mechanism. Cell. 1989;59:1027–1034. doi: 10.1016/0092-8674(89)90759-9. [DOI] [PubMed] [Google Scholar]
- 5.Carrasco C D, Ramaswamy K S, Ramasubramanian T S, Golden J W. Anabaena xisF gene encodes a developmentally regulated site-specific recombinase. Genes Dev. 1993;8:74–83. doi: 10.1101/gad.8.1.74. [DOI] [PubMed] [Google Scholar]
- 6.Celli J, Trieu-Cuot P. Circularisation of Tn916 is required for expression of the transposon-encoded transfer functions: characterisation of long tetracycline-inducible transcripts reading through the attachment site. Mol Microbiol. 1998;28:103–117. doi: 10.1046/j.1365-2958.1998.00778.x. [DOI] [PubMed] [Google Scholar]
- 7.Christie P J, Korman R Z, Zahler S A, Adsit J C, Dunny G M. Two conjugation systems associated with Streptococcus faecalis plasmid pCF10: identification of a conjugative transposon that transfers between S. faecalis and Bacillus subtilis. J Bacteriol. 1987;169:2529–2536. doi: 10.1128/jb.169.6.2529-2536.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clewell D B, Flannagan S E, Ike Y, Jones J M, Gawron-Burke C. Sequence analysis of the termini of conjugative transposon Tn916. J Bacteriol. 1988;170:3046–3052. doi: 10.1128/jb.170.7.3046-3052.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clewell D, Flannagan S A, Jaworski D D. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 1995;229:229–236. doi: 10.1016/s0966-842x(00)88930-1. [DOI] [PubMed] [Google Scholar]
- 10.Crellin P K, Rood J I. The resolvase/invertase domain of the site-specific recombinase TnpX is functional and recognises a target sequence that resembles the junction of the circular form of the Clostridium perfringens transposon Tn4451. J Bacteriol. 1997;179:5148–5156. doi: 10.1128/jb.179.16.5148-5156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flannagan S E, Zitzow L A, Su Y A, Clewell D B. Nucleotide sequence of the 18 kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid. 1994;32:350–354. doi: 10.1006/plas.1994.1077. [DOI] [PubMed] [Google Scholar]
- 12.Franke A E, Clewell D B. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J Bacteriol. 1981;145:494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gawron-Burke C, Clewell D B. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn916 in E. coli: strategy for targeting and cloning of genes from gram-positive bacteria. J Bacteriol. 1984;159:214–221. doi: 10.1128/jb.159.1.214-221.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hachler H, Kayser F H, Berger-Bachi B. Homology of a transferable tetracycline resistance determinant of Clostridium difficile with Streptococcus (Enterococcus) faecalis transposon Tn916. Antimicrob Agents Chemother. 1987;31:1033–1038. doi: 10.1128/aac.31.7.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaworski D D, Clewell D B. A functional origin of transfer (oriT) on the conjugative transposon Tn916. J Bacteriol. 1995;177:6644–6651. doi: 10.1128/jb.177.22.6644-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyras D, Storie C, Huggins A S, Crellin P K, Bannam T L, Rood J I. Chloramphenicol resistance in Clostridium difficile is encoded on Tn4453 transposons that are closely related to Tn4451 from Clostridium perfringens. Antimicrob Agents Chemother. 1998;23:784–786. doi: 10.1128/aac.42.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manganelli R, Ricci S, Pozzi G. Conjugative transposon Tn916: evidence for excision with formation of 5′-protruding termini. J Bacteriol. 1996;178:5813–5816. doi: 10.1128/jb.178.19.5813-5816.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manganelli R, Ricci S, Pozzi G. The joint of Tn916 circular intermediate is a homoduplex in Enterococcus faecalis. Plasmid. 1997;38:71–78. doi: 10.1006/plas.1997.1300. [DOI] [PubMed] [Google Scholar]
- 19.Marra D, Scott J R. Regulation of excision of the conjugative transposon Tn916. Mol Microbiol. 1999;31:609–621. doi: 10.1046/j.1365-2958.1999.01201.x. [DOI] [PubMed] [Google Scholar]
- 20.McDougal L K, Tenover F C, Lee L N, Kamile J, Patterson J E, Jorgensen J H, LeBlanc D J. Detection of Tn916-like sequences within a Tn916-like conjugative transposon (Tn3872) in erythromycin-resistant isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2321–2318. doi: 10.1128/aac.42.9.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullany P, Wilks M, Lamb I, Clayton C, Wren B, Tabaqchali S. Genetic analysis of a tetracycline resistance determinant from Clostridium difficile and its conjugal transfer to and from Bacillus subtilis. J Gen Microbiol. 1990;136:1343–1349. doi: 10.1099/00221287-136-7-1343. [DOI] [PubMed] [Google Scholar]
- 22.Mullany P, Pallen M, Wilks M, Tabaqchali S. A group II intron in a conjugative transposon from the Gram-positive bacterium, Clostridium difficile. Gene. 1996;174:145–150. doi: 10.1016/0378-1119(96)00511-2. [DOI] [PubMed] [Google Scholar]
- 23.Mullany P, Wilks M, Tabaqchali S. Transfer of Tn916 and Tn916ΔE into Clostridium difficile: demonstration of a hot-spot for these elements in the C. difficile genome. FEMS Microbiol Lett. 1991;79:191–194. doi: 10.1016/0378-1097(91)90084-n. [DOI] [PubMed] [Google Scholar]
- 24.Poyart-Salmeron C, Trieu-Cuot P, Carlier C, Courvalin P. Molecular characterisation of two proteins involved in the excision of the conjugative transposon Tn1545: homologies with other site-specific recombinases. EMBO J. 1989;8:2425–2433. doi: 10.1002/j.1460-2075.1989.tb08373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poyart-Salmeron C, Trieu-Cuot P, Carlier C, Courvalin P. The integration-excision system of the conjugative transposon Tn1545 is structurally and functionally related to those of the lamboid phages. Mol Microbiol. 1990;4:1513–1521. doi: 10.1111/j.1365-2958.1990.tb02062.x. [DOI] [PubMed] [Google Scholar]
- 26.Rice L B. Tn916 family of conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob Agents Chemother. 1998;42:1871–1877. doi: 10.1128/aac.42.8.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice L B, Carias L L. Transfer of Tn5385, a composite multiresistance chromosomal element from Enterococcus faecalis. J Bacteriol. 1998;180:714–721. doi: 10.1128/jb.180.3.714-721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts A P, Pratten J, Wilson M, Mullany P. Transfer of a conjugative transposon, Tn5397 in a model oral biofilm. FEMS Microbiol Lett. 1999;177:63–66. doi: 10.1111/j.1574-6968.1999.tb13714.x. [DOI] [PubMed] [Google Scholar]
- 29.Rubens C E, Heggen L M. Tn916ΔE: a Tn916 transposon derivative expressing erythromycin resistance. Plasmid. 1988;20:137–142. doi: 10.1016/0147-619x(88)90016-9. [DOI] [PubMed] [Google Scholar]
- 30.Rudy C K, Scott J R. Length of the coupling sequence of Tn916. J Bacteriol. 1994;176:3386–3388. doi: 10.1128/jb.176.11.3386-3388.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudy C K, Scott J R, Churchward G. DNA binding by the Xis protein of the conjugative transposon Tn916. Nucleic Acids Res. 1997;25:4061–4066. doi: 10.1093/nar/25.20.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salyers A A, Shoemaker N B, Stevens A M, Lhing-Yew L. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Sato M. The cisA cistron of Bacillus subtilis spoliation gene spoIVC encodes a protein homologous to a site-specific recombinase. J Bacteriol. 1990;172:1092–1098. doi: 10.1128/jb.172.2.1092-1098.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott J R, Churchward G G. Conjugative transposition. Annu Rev Microbiol. 1995;49:367–397. doi: 10.1146/annurev.mi.49.100195.002055. [DOI] [PubMed] [Google Scholar]
- 36.Scott J R, Bringel F, Marra D, Van Alstine G, Rudy C K. Conjugative transposition of Tn916: preferred targets and evidence for conjugative transfer of a single strand and for a double-stranded circular intermediate. Mol Microbiol. 1994;11:1099–1108. doi: 10.1111/j.1365-2958.1994.tb00386.x. [DOI] [PubMed] [Google Scholar]
- 37.Sloan J, Warner T A, Scott P T, Bannam T L, Berryman D I, Rood J I. Construction of a sequenced Clostridium perfringens-Escherichia coli shuttle plasmid. Plasmid. 1992;27:207–219. doi: 10.1016/0147-619x(92)90023-4. [DOI] [PubMed] [Google Scholar]
- 38.Storrs M J, Poyart-Salmeron C, Trieu-Cuot P, Courvalin P. Conjugative transposition of Tn916 requires the excisive and integrative activities of the transposon encoded integrase. J Bacteriol. 1991;173:4347–4352. doi: 10.1128/jb.173.14.4347-4352.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su Y A, Clewell D B. Characterisation of the left 4 kb of conjugative transposon Tn916: determinants involved in excision. Plasmid. 1993;30:234–250. doi: 10.1006/plas.1993.1055. [DOI] [PubMed] [Google Scholar]
- 40.Taylor K L, Churchward G. Specific DNA cleavage mediated by the integrase of conjugative transposon Tn916. J Bacteriol. 1997;179:1117–1125. doi: 10.1128/jb.179.4.1117-1125.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thorpe H M, Smith M C M. In vitro site-specific integration of bacteriophage DNA catalysed by a recombinase of the resolvase/invertase family. Proc Natl Acad Sci USA. 1998;95:5505–5510. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]