Abstract
In Escherichia coli, the expression of sodB, which encodes iron superoxide dismutase, has been suggested to be activated by Fur, the iron-responsive global regulator initially characterized as a transcriptional repressor. We investigated sodB regulation by functional analysis of the sodB promoter using sodB-lac fusions with various truncated and mutated promoters. Several cis- and trans-acting elements involved in sodB regulation have been identified. The β-galactosidase activity of sodB-lacZ reporter fusions and RNA analysis showed sevenfold iron-dependent, Fur-mediated activation of expression. A region just downstream from −10, including a large palindromic sequence encompassing the +1 position followed by a 14-bp AT-rich motif, is the site of Fur positive regulation, and the integrity of both sequences was required for full Fur-mediated activation. The life span of sodB mRNA was three times longer in a fur+ strain, indicating that Fur-mediated activation proceeds, at least in part, at the posttranscriptional level. The H-NS and IHF histone-like factors also affected sodB expression. IHF slightly repressed sodB expression independently of Fur regulation. In contrast, H-NS negative regulation operated only in the absence of Fur. Remarkably, psodB behaved like a “pure extended -10” promoter. Deletion of the −35 region did not affect expression, whereas expression was totally abolished by a TG-to-CC mutation in the extended −10 sequence TGcTACCCT.
Aerobic metabolism generates reactive oxygen species (ROS), such as superoxide, hydrogen peroxide, and hydroxyl radicals, which may cause oxidative damage in living cells (14). Efficient protective mechanisms have been developed by all organisms exposed to oxygen, including the specific elimination of ROS, repair of damage, and induction of global responses enabling cells to survive in periods of oxidative stress (4, 17). The toxic effects of ROS are potentiated by excess iron because iron catalyzes the Fenton reaction, leading to the formation of the most reactive species, the hydroxyl radical (OH⋅), which can attack all biological macromolecules (18, 22). Thus, strict control of iron homeostasis is required to maintain concentrations of this element, which is essential for virtually all organisms, at levels that are high enough to meet the organism's needs but prevent potential toxicity. Consistent with this, there is increasing evidence of coordination between the regulation of iron homeostasis and defense against oxidative stress (41).
In Escherichia coli, iron metabolism is regulated by the Fur (ferric uptake regulation) protein (11, 19). Fur usually functions as a transcriptional regulator, repressing the expression of target genes. It binds as a homodimer, in an Fe2+-dependent manner, to specific DNA sequences, the iron boxes, blocking access of RNA polymerase to the promoter, thereby inhibiting the initiation of transcription (3, 9, 12). Under iron shortage conditions, Fur is inactivated by the release of the iron cofactor and the genes under Fur control are induced. All genes involved in iron acquisition are Fur regulated (5), along with many other genes, for which the reasons for iron regulation are more or less obvious, including regulators of general metabolism, pathogenicity genes, and genes for defense against oxidative and acid stresses (13). In some cases, a positive effect of Fur has been observed. It activates the expression of ftnA and bfr (iron storage), acnA and fumA (tricarboxylic acid cycle enzymes), and sodB (iron superoxide dismutase [FeSOD]) (1, 16, 29). However, no putative iron boxes have been found in the promoter regions of these positively regulated genes. It is unclear whether a similar mechanism is responsible for activation of the expression of all genes positively regulated by Fur and whether it is caused by a direct interaction of Fur with the promoter or results from regulatory cascades.
SODs are metalloproteins that play a major role in protection against oxidative stress by catalyzing dismutation of the first ROS produced, the superoxide radical (O2⋅−) (15). By eliminating O2⋅−, SODs not only protect against direct damage caused by O2⋅−, but, more importantly, protect against indirect O2⋅− toxicity by preventing an O2⋅−-dependent increase in the pool of intracellular free iron, leading to the production of OH⋅via the Fenton reaction (7, 22, 26). Two cytoplasmic SODs have been identified in E. coli, a manganese SOD (MnSOD) and an FeSOD, encoded by sodA and sodB, respectively. Both are regulated by iron, in antagonistic negative and positive Fur-mediated regulations. MnSOD production is oxygen dependent and has been shown to be regulated by up to five global regulators, depending on the environment (8). Fur represses sodA expression in a classical Fe2+-dependent manner (38, 39). In contrast, FeSOD is produced in both anaerobiosis and aerobiosis and was long thought to be unregulated. In 1990, it was suggested that FeSOD synthesis is positively controlled by Fur (29). However, as for the few other later reports of Fur-mediated positive regulation, nothing is known about the way in which the positive regulation is achieved.
To gain further insight into the regulation of sodB, we carried out a functional analysis of the sodB promoter in an attempt to determine the Fur-mediated activation target(s). This analysis revealed that sodB regulation is more complex than expected, with multiple cis- and trans-acting elements. In addition to Fur, the trans-acting regulatory factors H-NS and IHF are involved. The sodB promoter functions as a pure extended −10 promoter, independently of Fur-mediated regulation. A region encompassing a large palindromic sequence overlapping the start site of transcription and followed by a 14-bp AT-rich region preceding the ribosome binding site is required for complete Fur-mediated activation, suggesting that Fur regulation itself occurs at two levels.
MATERIALS AND METHODS
Bacterial strains, phages, and plasmids.
The bacterial strains, phages, and plasmids used in this study are listed in Table 1. All of the bacterial strains used are E. coli K-12 derivatives. Basic genetic manipulations were carried out using standard procedures (27). Δfur::kan, Δfur::cat, himA::cat, hns-1001::Tn5seq1, and proC::Tn5 mutations were introduced by P1 transduction as previously described (8).
TABLE 1.
Bacterial strains, phages, and plasmids used in this study
| Strain, phage, or plasmid | Genotype or relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| MG1655 | F− λ−rph-1 | B. Bachmann |
| DPB267 | MG 1655 recD1901::Tn10 | 37 |
| TC3264 | FB8 ΔMluI lac(IZ) | 33 |
| LBK130 | proC::Tn5 | C. Berg |
| HN1491 | N99 himA::cat | H. Nash |
| BE1410 | FB8 hns-1001::Tn5seq-1 | 25 |
| QC1732 | F− ΔlacU169 rpsL fur::kan | 8 |
| QC2461 | MG1655 Δlac(IZ) | This work |
| QC2517 | DPB267 Δfur::cat | This work |
| QC2597 | QC2461 Φ(sodB-lacZ)2 | This work |
| QC2599 | QC2461 Φ(sodB′-′lacZ)2-0 | This work |
| QC2625 | QC2461 Φ(sodB-lacZ)4 | This work |
| QC2627 | QC2461 Φ(sodB-lacZ)5 | This work |
| QC2682 | QC2461 Φ(sodB-lacZ)9 | This work |
| QC2700 | QC2461 Φ(sodB-lacZ)3 | This work |
| QC2737 | QC2761 Φ(sodB-lacZ)2himA::cat | This work |
| QC2745 | QC2461 Φ(sodB-lacZ)2hns-1001::Tn5seq-1 | This work |
| QC2755 | QC2461 Φ(sodB-lacZ)4himA::cat | This work |
| QC2756 | QC2461 Φ(sodB-lacZ)4hns-1001::Tn5seq-1 | This work |
| QC2759 | QC2461 Φ(sodB-lacZ)9hns-1001::Tn5seq-1 | This work |
| QC2764 | QC2461 Φ(sodB-lacZ)11 | This work |
| QC2805 | QC2461 Φ(sodB-lacZ)12 | This work |
| QC2854 | QC2461 Φ(sodB-lacZ)5himA::cat | This work |
| QC2875 | QC2461 Φ(sodB-lacZ)3hns-1001::Tn5seq-1 | This work |
| QC2920 | QC2461 Φ(sodB-lacZ)16 | This work |
| Phages | ||
| λRS45 | 36 | |
| λSD2 to λSD16 | λRS45 recombinants carrying Φ(sodB-lacZ)2 to 16 | This work |
| Plasmids | ||
| pBT2-1 | pBR322 derivative carrying fur locus with internal fur deletion | 40 |
| pDT9 | pBR322 derivative with cat cassette in fur gene | This work |
| pHS1-8 | pBR322 derivative carrying sodB region | 34 |
| pRS414 | 36 | |
| pRS415 | 36 | |
| pSD2-1 | pRS414 with fragment containing sodB2b promoter region | This work |
| pSD2-2 to pSD2-16 | pRS415 with fragments containing sodB2 to sodB16b promoter regions | This work |
Δfur::kan derivatives of QC2461, QC2597, QC2599, QC2625, QC2682, QC2700, QC2737, QC2755, QC2764, and QC2920 are QC2558, QC2598, QC2600, QC2754, QC2683, QC2704, QC2738, QC2760, QC2765, and QC2921, respectively. Δfur::cat derivatives of QC2625, QC2627, QC2682, QC2597, QC2745, QC2756, QC2875, and QC2759 are QC2626, QC2628, QC2757, QC2735, QC2746, QC2758, QC2873, and QC2761, respectively.
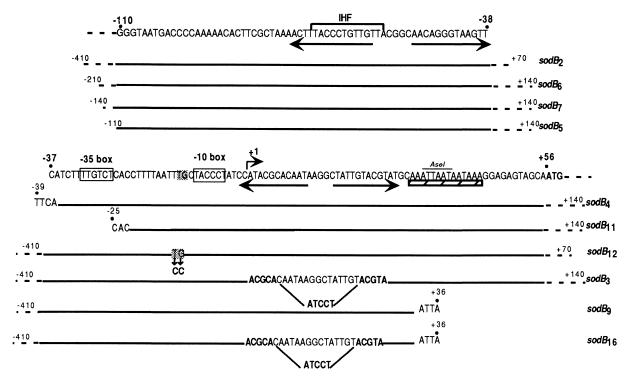
sodB2 to sodB16 refers to fragments of the sodB promoter as indicated in Fig. 1.
Specific strain and plasmid constructions.
Δfur::cat was constructed like Δfur::kan (40), except that a PstI-PstI cat cassette from Tn9 was inserted into the PstI sites of Δfur (into pBT2-1) instead of a Kanr cassette, generating pDT9. For QC2461 construction, a Δlac(IZ) deletion in MG1655 was created by successive P1 transductions. proC::Tn5 (from LBK130) was transduced into TC3264, and colonies with kanamycin resistance were selected. P1 lysate was made from a Lac− kanamycin-resistant transductant and used to transduce MG1655 with selection for kanamycin resistance and screening for the Lac− phenotype. MG1655 Δlac(IZ) proC::Tn5 was further transduced to Pro+(Kans) using a P1 lysate made from MG1655.
Media, growth conditions, and β-galactosidase assays.
Cells were grown in Luria-Bertani (LB) medium at 37°C with shaking at 200 rpm. The antibiotics added as required were ampicillin (50 μg/ml), kanamycin (40 μg/ml), and chloramphenicol (20 μg/ml). The iron chelators used were 0.25 mM 2,2′-dipyridyl and 1 mM ferrozin. The anaerobic growth conditions used and the β-galactosidase assays done were as previously described (8). β-Galactosidase activity in permeabilized cells from cultures grown under aerobiosis or anaerobiosis was assayed as described by Miller (27).
Construction of Φ(sodB-lacZ) fusions.
Various fragments of the sodB promoter (as shown Fig. 1) were generated by PCR using primers (see Table 2) carrying restriction sites at their extremities. The PCR products were digested and inserted between the corresponding sites in pRS415 for transcriptional fusions and in pRS414 for the translational fusion. The fusions were transferred to lambda phage (λRS45) and integrated into the chromosome of strain QC2461 at the lambda attachment sites as previously described (36). Monolysogens were selected with a PCR test described elsewhere (32), with slight modification for DNA amplification from single colonies. Briefly, single colonies were picked separately into 500 μl of LB medium, suspended by vortexing, and centrifuged for 10 min at 10,000 × g. The supernatant was removed, and the washing procedure was repeated twice. The final cell pellets were resuspended in a volume of 100 μl of sterile water. PCR was performed with 10 μl of cell suspension as the template and an equimolar mixture of primers at a final concentration of 4.5 μM. Taq polymerase, its buffer, and deoxynucleoside triphosphates were added at the recommended concentrations. The following thermal cycling program was used: 95°C for 1 min, followed by 25 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min and then 72°C for 10 min. Primers sodB2 5′ and sodB2 3′, sodB4 and sodB-BamHI, sodB5 and sodB-BamHI, and sodB11 and sodB-BamHI were used to construct Φ(sodB-lacZ)2, Φ(sodB-lacZ)4, Φ(sodB-lacZ)5, and Φ(sodB-lacZ)11, respectively. To generate the sodB3 fragment, in which the CAATAAGGCTATTGT region (+8 to +22) is replaced with ATCCT, destabilizing the palindromic sequence (Fig. 1), two PCR fragments were synthesized, one with sodB2 5′ and sodB-BglII1 and the other with sodB-BglII2 and sodB-BamHI. Both fragments were digested with BglII and ligated into the BglII site. The resulting fragment was further digested with EcoRI and BamHI, integrated into the corresponding sites of pRS415, and transferred to the chromosome as described above. To generate the Φ(sodB-lacZ)9 fusion, pSD2-2 was digested with EcoRI and AseI (the AseI site is shown in Fig. 1) and the resulting fragment was inserted between the EcoRI and SmaI sites of pRS415. The Φ(sodB-lacZ)16 fusion was generated by digesting the sodB3-containing plasmid with EcoRI and AseI and inserting it between the EcoRI and SmaI sites of pRS415. The Quick Change Site-Directed Mutagenesis Kit (Stratagene) was used to construct the Φ(sodB-lacZ)12 fusion, in which the TG dinucleotide 1 bp upstream from the −10 box (position −13 to −12) was replaced with CC in the sodB2 fragment. The primers carrying the mutated sequence were mut12 and mut12 inv (Table 2). All chromosomal fusions were checked by DNA sequencing after amplification by PCR of the chromosomal DNA region from a single colony as described above.
FIG. 1.
Nucleotide sequence of the sodB promoter region. Position +1 is the start site for transcription. The −35 and −10 regions are boxed. The ATG start site of translation is in bold. Inverted repeats are shown by arrows. The AT-rich region is underlined by a hatched bar. The putative IHF box is indicated by a thick line. Fragments of the sodB promoter indicated below the sequence were fused to lacZ to create the corresponding fusions.
TABLE 2.
Sequences of the oligonucleotides used in this study
| Oligonucleotide | Sequence | Location |
|---|---|---|
| sodB2 5′ | 5′-ATTTGGTGAAAATTCGAATTCCGCGTACGG-3′ | −345 to −315 |
| sodB2 3′ | 5′-GGTAGTGCAGGGAATTCGAAATGACATTGC-3′ | +52 to +80a |
| sodB4 | 5′-GGAATTCATCTTTTGTCTCACC-3′ | −45 to −22 |
| sodB5 | 5′-CGGGTAATGACCCCAAAAAGAATTCGC-3′ | −102 to −77 |
| sodB11 | 5′-GAATTCACCTTTTAATTTGCTACCCTATCC-3′ | −25 to −2 |
| sodB-BglII1 | 5′-GAAGATCTGCGTATGGATAGGG-3′ | +8 to −8 |
| sodB-BglII2b | 5′-GAAGATCTTACGTATGCAAATTAA-3′ | +19 to +35 |
| sodB-BamHIb | 5′-CGGGATCCGTAGTGATACTCGATGG-3′ | +148 to +122 |
| mut12 | 5′-GTCTCACCTTTTAATTCCCTACCCTATCCATACGC-3′ | −29 to +8 |
| mut12 inv | 5′-GCGTATGGATAGGGTAGGGAATTAAAAGGTGAGAC-3′ | +8 to −29 |
| 4218 | 5′-GGTCAGCGGAGTACCCGCG-3′ | +477 to +495 |
| 4219 | 5′-CGCCGGTGGCGAACCGACTGG-3′ | +306 to +326 |
Two modifications were introduced into the sodB2 3′ oligonucleotide with respect to the original +52 to +80 sodB sequence: an additional base was added (between +62 and +63) to keep the same reading frame in the translation fusion, and the base in position +70 was modified to create an EcoRI restriction site. Sequence modifications and the start site for translation are in bold type.
Restriction sites (EcoRI, BglII, and BamHI) introduced at the 5′ end of oligonucleotides for further digestions are underlined. Mutated bases in mut12 and mut12 inv are in bold type.
RNA manipulations. (i) RNA purification.
Cells were grown in morpholinepropanesulfonic acid (MOPS)-Tricine medium (28) at 37°C. The medium contained 0.4% glucose, 2 mM potassium phosphate, 1% Casamino Acids, 0.1 mM FeSO4, and 0.0001% thiamine. When cultures reached an optical density at 600 nm (OD600) of 1, we added 2 ml of the bacterial suspension to 2 ml of boiling lysis buffer containing 2% (wt/vol) sodium dodecyl sulfate, 4 mM EDTA, and 3 M sodium acetate (pH 5) (42). The samples were heated at 100°C for 5 min and subjected to hot phenol RNA extraction, followed by RNA precipitation with ethanol.
(ii) Primer extension reactions.
Reverse transcription was carried out at 42°C with avian myeloblastosis virus reverse transcriptase as previously described (42), using a 5′-end-labeled 28-mer oligonucleotide (sodB-BamHI [Table 2]) complementary to nucleotides +122 to +148 of the sodB gene. The DNA was sequenced with the same primer using the dideoxy-chain termination method and the Sequenase kit, version 2.0 (U.S. Biochemical Corp.), with [α-35S]dATP (ICN).
(iii) Determination of mRNA half-life.
Cells were grown as described above. When cultures reached an OD600 of 1, rifampin was added to a final concentration of 150 μg/ml and samples were taken at various times following incubation at 37°C. RNAs were extracted from samples as described above. RNAs (5 to 10 μg) were separated on a 1% agarose gel. Analysis of the RNA was done by Northern blot assay and hybridization (at 42°C in 50% [vol/vol] formamide) performed essentially as described by Sambrook et al. (35). Radioactivity in bands was quantitated with a Molecular Dynamics PhosphorImager. An internal sodB fragment, reaching from +310 to +500, amplified by PCR with oligonucleotides 4218 and 4219 (see Table 4) was used as a probe. The oligonucleotide 5′-ACTACCATCGGCGCTACGGC-3′ was used as a probe for the 5S rRNA to normalize the quantity of RNA in each lane.
TABLE 4.
Effects of IHF and H-NS on expression of various sodB-lacZ fusions
| Strains | Relevant genotype | β-Galactosidase activity (U)a
|
|
|---|---|---|---|
| fur+ strains | Δfur strains | ||
| QC2597, QC2598 | (sodB-lacZ)2 | 950 | 130 |
| QC2737, QC2738 | (sodB-lacZ)2himA::cat | 1,300 | 350 |
| QC2745, QC2746 | (sodB-lacZ)2hns-1001 | 980 | 300 |
| QC2627, QC2628 | (sodB-lacZ)5 | 900 | 150 |
| QC2854, QC2855 | (sodB-lacZ)5himA::cat | 1,300 | 320 |
| QC2625, QC2626 | (sodB-lacZ)4 | 1,350 | 400 |
| QC2755, QC2760 | (sodB-lacZ)4himA::cat | 1,400 | 450 |
| QC2756, QC2758 | (sodB-lacZ)4hns-1001 | 1,350 | 630 |
| QC2700, QC2704 | (sodB-lacZ)3 | 2,000 | 770 |
| QC2875, QC2873 | (sodB-lacZ)3hns-1001 | 2,000 | 770 |
| QC2682, QC2683 | (sodB-lacZ)9 | 110 | 110 |
| QC2759, QC2761 | (sodB-lacZ)9hns-1001 | 110 | 110 |
Values (Miller units) were calculated as described in Table 3, footnote b.
RESULTS
Fur activates sodB expression.
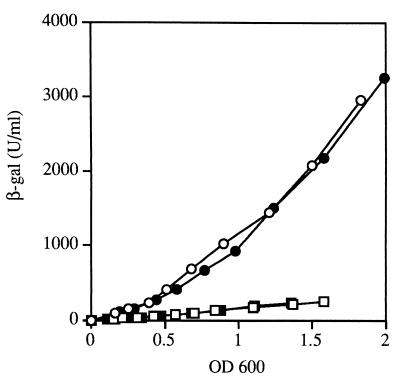
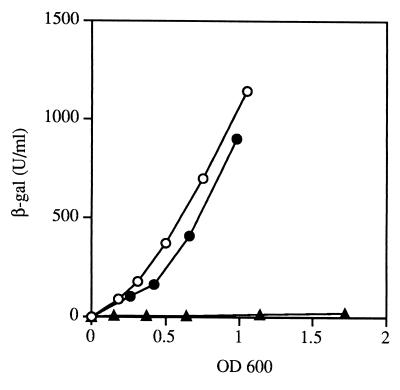
To determine the level at which Fur-mediated activation of sodB occurs, we constructed transcriptional and translational fusions to promoterless lacZ genes, (sodB-lacZ)2 and (sodB′-′lacZ)2-0, respectively. Comparison of the expression of the two fusions in the fur mutant and the wild-type strain showed that the increases in β-galactosidase activity (six- to sevenfold) were similar for both fusion types in the presence of Fur (Fig. 2), ruling out a translational effect. The fusions were constructed using a lambda phage, and therefore the lysogen strains retained their wild-type sodB allele. Introduction of the sodBΔ2 mutation into the fusion strains did not change fusion expression, showing that sodB expression is not autoregulated (data not shown).
FIG. 2.
Effect of a Δfur mutation on expression of Φ(sodB-lacZ) transcriptional and translational fusions. Strains QC2597, QC2598 [Φ(sodB-lacZ)2], QC2599, and QC2600 [Φ(sodB′-′lacZ)2-0] were grown in LB medium and assayed for β-galactosidase (β-gal) activity as described in Materials and Methods. β-Galactosidase activity, expressed in units per milliliter, is plotted against units of OD600. The values shown are means of three experiments, and individual values did not differ by more than 15% from the means. Symbols: ○, QC2597 (wild type); □, QC2598 (Δfur mutant); ●, QC2599 (wild type); ■, QC2600 (Δfur mutant).
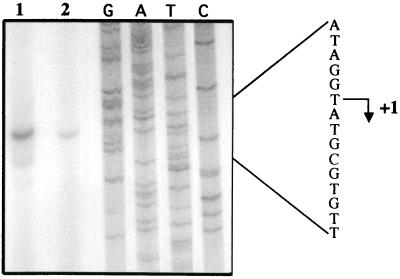
The effect of Fur was confirmed by primer extension analysis. Quantification of primer extension products for RNA from the wild-type (QC2461) and Δfur (QC2558) strains showed that there was eight times more product in the wild-type strain than in the Δfur strain (Fig. 3). A unique transcription start site was identified. Its location did not depend on the presence of Fur in the cell.
FIG. 3.
Analysis by primer extension of the sodB transcript in wild-type and Δfur strains. RNAs isolated from E. coli strains QC2461 and QC2558 were hybridized with the 32P-labeled 28-mer sodB-BamHI oligonucleotide and used as a template for avian myeloblastosis virus reverse transcriptase. Lanes: 1, primer extension product of RNA from QC2461; 2, primer extension product of RNA from QC2558. GATC is the sequence obtained with the same primer. Position +1 is the start site for transcription.
Fur-mediated activation of sodB expression is iron dependent.
As Fur has been shown to require ferrous iron as a cofactor for repressor activity (3), we investigated whether the effect of Fur on sodB expression is iron dependent. Culture in the presence of 2,2′-dipyridyl, a cell-permeating iron chelator, decreased the level of β-galactosidase activity of the sodB-lacZ fusion in wild-type strain QC2597 to a level similar to that in the corresponding Δfur strain (QC2598) (Table 3). This result indicates that the activation of sodB expression by Fur is iron dependent. The addition of an extracellular iron chelator, ferrozin, was insufficient to completely abolish Fur-mediated activation of sodB expression (Table 3). Presumably, intracellular iron depletion with ferrozin is less severe than that with 2,2′-dipyridyl and only high levels of iron deprivation completely prevent Fur activation.
TABLE 3.
Effect of Δfur mutation on expression of (sodB-lacZ)2 under various growth conditions
| Growth conditiona | β-Galactosidase activity (U)b in strain:
|
|
|---|---|---|
| QC2597 (fur+) | QC2598 (Δfur) | |
| Aerobiosis | 900 | 130 |
| Aerobiosis + 0.25 mM 2,2′-dipyridyl | 112 | 112 |
| Aerobiosis + 1 mM ferrozin | 570 | 115 |
| Anaerobiosis | 2,750 | 300 |
Cells were grown and assayed for β-galactosidase as described in Materials and Methods and the legend to Fig. 2.
The values shown (in Miller units) were deduced from the slopes of the curves and represent the averages of at least three experiments with variations of no more than 15% from the mean.
Fur activation is equally efficient under aerobic and anaerobic conditions.
In studies of the expression of several Fur-repressed genes, we have previously observed that repression is much stronger in anaerobiosis than in aerobiosis (39). We investigated whether the iron-dependent Fur activation of sodB was stronger under anaerobiosis. We measured β-galactosidase activity from a Φ(sodB-lacZ)2 fusion in anaerobiosis and found Fur activation by a factor of about seven, as in aerobiosis.
A Fur-independent increase of sodB expression was observed. Attempts to identify a regulatory factor responsible for this anaerobic induction failed, excluding possible ArcAB- and Fnr-related effects (data not shown).
Location of cis-acting regulatory elements involved in Fur-mediated activation.
Specific palindromic sequences, the iron boxes, have been found in all of the characterized promoter regions of genes negatively regulated by Fur. Analysis of the sodB promoter (psodB) sequence revealed two palindromic DNA sequences but no putative iron box. One of the palindromic sequences was located upstream from the −35 motif, and the other encompassed the −1 to +29 region (Fig. 1). A set of Φ(sodB-lacZ) chromosomal fusions with various deletions in the sodB promoter were generated (Fig. 1). The Fur-mediated activation of sodB expression was not affected by promoter deletions upstream from position −39, as shown by the β-galactosidase activity resulting from the Φ(sodB-lacZ)7, Φ(sodB-lacZ)6, Φ(sodB-lacZ)5, and Φ(sodB-lacZ)4 fusions (data not shown). This suggested that the mechanism of Fur-mediated activation is not of a classical type, with an upstream −35 bound regulatory protein. Interestingly, Fur activation was also not affected by the Φ(sodB-lacZ)11 fusion, in which the −35 box was also deleted (see below), locating cis-acting activation elements downstream from position −25.
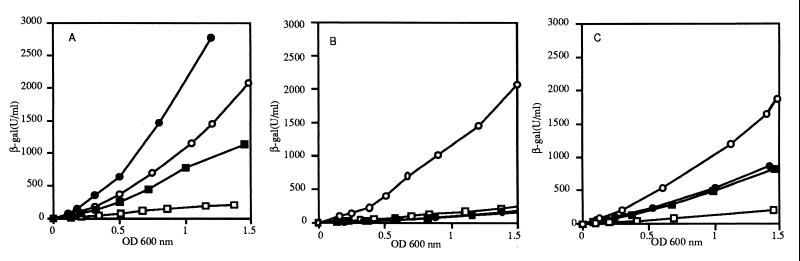
The Φ(sodB-lacZ)3 fusion, from which the palindromic DNA sequence overlapping the start site of transcription was partially deleted, retained some Fur regulation. Fur-mediated activation by a factor of only 2.5 was seen. However Fur-independent activation of expression by a factor of about four was observed (Fig. 4A), indicating that the palindromic sequence interferes with both basic expression and Fur regulation.
FIG. 4.
Effects of deletions in the sodB promoter on sensitivity to Fur regulation. Conditions were as described in the legend to Fig. 2. Symbols: circles, wild type; squares, Δfur strains. Panels: A, Φ(sodB-lacZ)3 (QC2700 and QC2704); B, Φ(sodB-lacZ)9 (QC2682 and QC2683); C, Φ(sodB-lacZ)16 (QC2920 and QC2921). Open symbols correspond to expression from the wild-type promoter [Φ(sodB-lacZ)2 strains QC2597 and QC2598].
In contrast, the Φ(sodB-lacZ)9 fusion, which retained only the part of the promoter upstream from +36 (Fig. 1), resulting in partial deletion of a 14-bp AT-rich region preceding the ATG start codon, exhibited a slightly lower level of Fur-independent expression and was not Fur regulated (Fig. 4B). Similar results were obtained with a fusion in which the AT-rich region was completely deleted (data not shown).
However, partial deletion of both the AT-rich and palindromic sequences in Φ(sodB-lacZ)16 resulted in a higher level of Fur-independent expression and loss of Fur-mediated activation of sodB expression (Fig. 4C). Thus, the AT-rich region is essential for positive regulation by Fur and the palindromic sequence controls basal, Fur-independent expression. However, the integrity of both regions seems to be necessary for full Fur-mediated activation.
Effect of Fur on sodB mRNA stability.
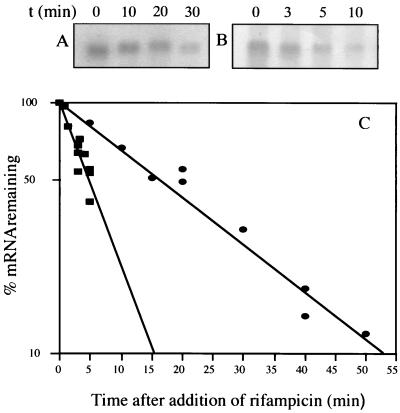
The location of the Fur effect in a target region downstream of the transcription start site questioned whether Fur-mediated activation is due to sodB mRNA stabilization. The half-life of the sodB transcript in fur+ strain QC2461 was found to be 14 min. In fur mutant strain QC2558, the transcript level, seven times lower, was too low for exact half-life measurement (data not shown). To amplify the signal, we used as a template the complete sodB region carried by a plasmid derived from pBR322 (pHS1-8). In the fur+ strain, the half-life of sodB mRNA was 14 min and it was almost threefold lower in the Δfur strain, at 4.75 min (Fig. 5). This difference in decay rate is consistent with the higher expression of the Φ(sodB-lacZ) fusion in the fur+ than in the Δfur strain. However, it is not clear whether the threefold higher stability of sodB mRNA in the fur+ strain could account for sevenfold higher sodB expression and an additive transcriptional regulatory mechanism cannot be excluded. Unspecific nucleotide oxidative damage generated in fur mutants (40) was not responsible for a lower sodB mRNA life span, since the same difference in mRNA stability between the wild-type and Δfur strains was seen in anaerobiosis as well (data not shown).
FIG. 5.
Effect of Fur on sodB mRNA stability. Strains containing plasmid pHS1-8 were grown at 37°C to an OD600 of about 1. Rifampin was added to a final concentration of 150 μg/ml, and samples were taken following incubation at 37°C for Northern blot analysis. Panels: A and B, Northern blots with RNAs from fur+ and Δfur strains, respectively; C, quantitative Northern blot analysis of fur+ (circles) and Δfur (squares) strains. Half-lives were calculated from the slope of each plot, and half-life errors were estimated from the standard deviation of the slopes. The measured half-lives were 14 ± 2 min in the fur+ strain and 4.75 ± 1 min in the Δfur strain.
The sodB promoter belongs to the extended −10 class.
The sodB promoter presents a 5′-TGN-3′ motif immediately upstream of the −10 hexamer. This is characteristic of the so-called extended −10 promoters, which do not need a −35 consensus sequence to be transcribed (24). Two sodB-lacZ reporter fusions were generated in an attempt to determine whether the TG dinucleotide, in association with the −10 box, is both necessary and sufficient for sodB transcription. No significant difference in sodB expression was observed when the Φ(sodB-lacZ)11 fusion, in which the promoter region upstream from position −25 has been deleted, was used as the reporter (Fig. 6). Thus, psodB retains almost optimal activity with no −35 region. To confirm that the TGN motif is essential for psodB activity, TGN was mutated to CCN by site-directed mutagenesis (sodB12 in Fig. 1). This mutation abolished psodB activity (Fig. 6). Similar results were obtained with a Δfur strain (data not shown). Thus, psodB is a member of the extended −10 class of promoters.
FIG. 6.
sodB expression from the extended −10 promoter. sodB expression was measured, as described in the legend to Fig. 2, from the sodB promoter truncated at position −25 and the sodB promoter mutated from TGN to CCN (as shown in Fig. 1). Symbols: ○, Φ(sodB-lacZ)2; ●, Φ(sodB-lacZ)11; ▴, Φ(sodB-lacZ)12. β-gal, β-galactosidase.
Regulation of sodB expression by the histone-like proteins IHF and H-NS.
Histone-like proteins, such as IHF, H-NS, HU, and Fis, play a major role in modifying DNA conformation, changing the access of certain regulators and/or RNA polymerase to promoters (2, 10). We investigated whether these proteins affect the Fur-mediated activation of sodB. Inactivation of the hupA-hupB loci and of the fis gene affected neither the Fur-mediated activation nor the Fur-independent expression of the Φ(sodB-lacZ)2 fusion (data not shown).
A mutation in himA, which encodes one of the two subunits of IHF, caused a slight increase in expression of the Φ(sodB-lacZ)2 fusion in both the fur+ and Δfur strains (Table 4). To identify the target sequence of IHF-mediated repression, we used several reporter fusions between sodB-truncated promoters and lacZ. The fusion carrying the sodB5 fragment, the 5′ extremity of which was located at position −110, was repressed by IHF, whereas the fragment carrying the sodB4 fragment, the 5′ extremity of which was located at position −39, was not. Thus, the target sequence for IHF-mediated repression in the sodB promoter is located between positions −39 and −110. A putative IHF box is present in this region, suggesting that IHF directly interacts with the sodB promoter (Fig. 1).
A mutation in hns resulted in 2.5 times more expression of the Φ(sodB-lacZ)2 fusion in the Δfur mutant but had no effect on the expression of this fusion in the wild-type strain (Table 4). The effect of H-NS was retained by Φ(sodB-lacZ)4 but not by Φ(sodB-lacZ)3 and Φ(sodB-lacZ)9. This indicates that the DNA sequence required for complete Fur-mediated activation of sodB is also required for an H-NS-mediated effect.
DISCUSSION
FeSOD synthesis, which was initially thought to be unregulated and then to be possibly activated by Fur (29), now appears to be under the control of multiple trans- and cis-regulatory elements. Functional analysis of psodB, which was primarily undertaken to identify the DNA target of Fur-mediated activation, revealed several rather unusual features of psodB.
Nucleotide sequence analysis showed two large palindromic sequences, one just upstream from the −35 region and a second (28 bp) just downstream from the −10 box and directly followed by a 14-bp AT-rich sequence. The upstream palindromic sequence is not involved in Fur-mediated sodB regulation and may be the transcription termination site of an upstream gene. However, its deletion slightly modifies sodB expression, alleviating a minor repressive effect of IHF.
The sodB promoter did not present, a priori, an optimal sequence for the efficient binding of RNA polymerase. The −10 box is rather poor, with three mismatches with the consensus sequence, and the spacing (15 bp) between the −10 and −35 boxes is not optimal, but the −35 box is of moderate quality (two mismatches with the consensus sequence). A TGN motif precedes the −10 hexamer, and we found that psodB does, indeed, function as a typical −10 extended promoter. Why does the sodB gene have a −10 extended promoter? Several possible and nonexclusive functions of −10 extended promoters have been proposed (24). The extended −10 motif may be a primitive form of transcription machinery. FeSODs are thought to have appeared early in evolution, after the appearance of oxygen, and are well conserved between species, suggesting a low level of evolution. There are relatively few promoter sequences of FeSOD for which the start site of transcription has been well defined, but among these, most have a putative extended −10 box (Campylobacter jejuni, Helicobacter pylori, Pseudomonas aeruginosa, and Streptomyces coelicolor Müller) (20, 23, 30, 31). Often, in the presence of the TGN motif, the sequence of the −35 hexamer (directly or by means of regulatory proteins) still modulates the initiation of transcription. However sodB expression was unaffected by deletion of the −35 region and was completely abolished by a mutation of the TG motif to CC. Thus, for sodB, despite the presence of a moderate −35 sequence, the −10 extension motif appears to be essential and sufficient for recognition by RNA polymerase and does not simply strengthen a weak classical promoter. Another possible reason for the requirement of an extended −10 promoter is particular sequence constraints in the binding region of RNA polymerase. Further investigation is required to determine whether the palindromic sequence downstream from position −10 in psodB has forced RNA polymerase to adopt this particular binding mode. The binding of RNA polymerase to −10 extended promoters does not require the sigma factor carboxy-terminal region, which could be proteolytically cleaved under stress conditions. Therefore, it has been suggested that the TGN motif confers an advantage by facilitating transcription initiation under stress conditions. FeSOD plays a particularly important protective role, counteracting oxidative stress encountered by bacteria during the shift from anaerobiosis to aerobiosis (21). The use of a strong −10 extended promoter may confer certain advantages under such stress conditions.
Fur clearly activated sodB expression and required iron, as when acting as a repressor. Whereas the repressive mechanism has been thoroughly investigated and Fur binding to a specific DNA sequence has been demonstrated, in no case has the mechanism of positive regulation been elucidated and indirect mechanisms have not been excluded. While the sodB promoter elements involved in Fur-mediated regulation clearly emerged from our data, the mechanism of Fur activation remains obscure. The sodB mRNA is more stable in a fur+ strain, and the longevity of the sodB transcript is significantly higher than the average of E. coli messages, showing a posttranscriptional effect. However, it seems unlikely that a threefold increase in mRNA stability could account for sevenfold Fur-mediated activation of sodB expression. Thus, a possible additional Fur effect at the transcriptional level has to be considered. Both the downstream palindromic sequence and the AT-rich region are required for full positive Fur regulation. It was tempting, in view of the demonstration that the stem-loop structure at the 5′ terminus of other messages has a stabilizing effect (6), to hypothesize that the hairpin structure in the 5′ terminus of the sodB message plays a similar role. However, partial deletion of the palindromic sequence, which predicts destabilization of the putative stem-loop, in the presence or absence of the AT-rich region, increased Fur-independent expression. This finding simply did not fit the above model. Moreover, deletion of the contiguous AT-rich region had a drastic effect. Fur-mediated activation was completely abolished, and the basal levels of sodB expression fell slightly. This suggests that the AT-rich region is required for Fur-mediated activation, which may be modulated by the palindromic sequence. Further experiments are in progress to determine whether the palindromic structure is involved in the stability of sodB mRNA, whether and how Fur stabilizes it, and how the integrity of the AT-rich region interferes with it.
The two contiguous DNA regions required for full Fur-mediated activation were also found to be required, in the absence of Fur, for H-NS-mediated repression. Thus, it seems that either H-NS competes with Fur as a direct antagonist or Fur and H-NS regulate the same intermediary regulatory protein but in opposing ways.
Whereas FeSOD is positively regulated by Fur, MnSOD is negatively regulated by this protein (8). Each enzyme requires a specific metal for activity. An ad hoc explanation for their antagonistic regulation by Fur is that an effective concentration of SOD is required in the cell, whatever the iron availability. When iron is scarce, E. coli may reduce the production of iron proteins to reduce the iron demand and induce MnSOD to compensate for the FeSOD loss. Although such a system is likely, the apparently complex mechanism underlying Fur activation of sodB expression suggests that there is a more subtle relationship between the control of iron homeostasis and defense against oxygen toxicity.
ACKNOWLEDGMENTS
We thank M. Springer for stimulating discussions and critical reading of the manuscript. We thank M. Uzan for helpful comments and advice on our work.
S. Dubrac was supported by a fellowship from the Ministère de l'Enseignement et de la Recherche, France. This work was supported by a grant from the Association pour la Recherche sur le Cancer (no. 5581).
REFERENCES
- 1.Andrews S C, Harrison P M, Guest J R. Cloning, sequencing, and mapping of the bacterioferritin gene (bfr) of Escherichia coli. J Bacteriol. 1989;171:3940–3947. doi: 10.1128/jb.171.7.3940-3947.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 3.Bagg A, Neilands J B. Ferric uptake regulation acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry. 1987;26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 4.Beyer W, Imlay J, Fridovich I. Superoxide dismutases. Prog Nucleic Acid Res Mol Biol. 1991;40:221–253. doi: 10.1016/s0079-6603(08)60843-0. [DOI] [PubMed] [Google Scholar]
- 5.Braun V, Schaffer S, Hantke K, Troger W. The molecular basis of bacterial metabolism. Heidelberg, Germany: Springer-Verlag; 1990. Regulation of gene expression by iron; pp. 164–179. [Google Scholar]
- 6.Bricker A L, Belasco J G. Importance of a 5′ stem-loop for longevity of papA mRNA in Escherichia coli. J Bacteriol. 1999;181:3587–3590. doi: 10.1128/jb.181.11.3587-3590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Compan I, Touati D. Interaction of six global transcription regulators in expression of manganese superoxide dismutase in Escherichia coli K-12. J Bacteriol. 1993;175:1687–1696. doi: 10.1128/jb.175.6.1687-1696.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lorenzo V, Wee S, Herrero M, Neilands J B. Operator sequences of the aerobactin operon of plasmid ColV-K3 binding the ferric uptake regulation (fur) repressor. J Bacteriol. 1987;169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drlica K, Rouviere-Yaniv J. Histone-like proteins of bacteria. Microbiol Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ernst J F, Bennett R L, Rothfield L I. Constitutive expression of the iron-enterochelin and ferrichrome uptake systems in a mutant strain of Salmonella typhimurium. J Bacteriol. 1978;135:928–934. doi: 10.1128/jb.135.3.928-934.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escolar L, de Lorenzo V, Perez-Martin J. Metalloregulation in vitro of the aerobactin promoter of Escherichia coli by the Fur (ferric uptake regulation) protein. Mol Microbiol. 1997;26:799–808. doi: 10.1046/j.1365-2958.1997.6211987.x. [DOI] [PubMed] [Google Scholar]
- 13.Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fridovich I. The biology of oxygen radicals. Science. 1978;201:875–880. doi: 10.1126/science.210504. [DOI] [PubMed] [Google Scholar]
- 15.Fridovich I. Superoxide anion radical(O2−), superoxide dismutases (SODs), and related matters. J Biol Chem. 1997;272:18515–18517. doi: 10.1074/jbc.272.30.18515. [DOI] [PubMed] [Google Scholar]
- 16.Gruer M J, Guest J R. Two genetically distinct and differentially-regulated aconitases (AcnA and AcnB) in Escherichia coli. Microbiology. 1994;140:2531–2541. doi: 10.1099/00221287-140-10-2531. [DOI] [PubMed] [Google Scholar]
- 17.Halliwell B. Superoxide dismutase, catalase and glutathione peroxidase: solutions to the problem of living with oxygen. New Phytol. 1974;73:1075–1086. [Google Scholar]
- 18.Halliwell B, Gutteridge M C. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hantke H. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet. 1981;182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 20.Hasset D J, Woodruff W A, Wozniak D J, Vasil M L, Cohen M S, Ohman D E. Cloning and characterization of the Pseudomonas aeruginosa sodA and sodB genes encoding manganese- and iron-cofactored superoxide dismutase: demonstration of increased manganese superoxide dismutase activity in alginate-producing bacteria. J Bacteriol. 1993;175:7658–7665. doi: 10.1128/jb.175.23.7658-7665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imlay J A. A metabolic enzyme that rapidly produces superoxide, fumarate reductase of Escherichia coli. J Biol Chem. 1995;270:19767–19777. [PubMed] [Google Scholar]
- 22.Keyer K, Imlay J A. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci USA. 1996;93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim E-J, Chung H-J, Suh B, Hah Y C, Roe J-H. Expression and regulation of the sodF gene encoding iron- and zinc-containing superoxide dismutase in Streptomyces coelicolor Müller. J Bacteriol. 1998;180:2014–2020. doi: 10.1128/jb.180.8.2014-2020.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar A, Malloch R A, Fujita N, Smillie D A, Ishihama A, Hayward R S. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J Mol Biol. 1993;232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- 25.Laurent-Winter C, Ngo S, Danchin A, Bertin P. Role of Escherichia coli histone-like nucleoid-structuring protein in bacterial metabolism and stress response. Eur J Biochem. 1997;244:767–773. doi: 10.1111/j.1432-1033.1997.00767.x. [DOI] [PubMed] [Google Scholar]
- 26.Liochev S I, Fridovich I. The role of O2°− in the production of HO°: in vitro and in vivo. Free Radic Biol Med. 1994;16:29–33. doi: 10.1016/0891-5849(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 27.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 28.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niederhoffer E C, Naranjo C M, Bradley K L, Fee J A. Control of Escherichia coli superoxide dismutase (sodA and sodB) genes by the ferric uptake regulation (fur) locus. J Bacteriol. 1990;172:1930–1938. doi: 10.1128/jb.172.4.1930-1938.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pesci E C, Cottle D L, Pickett C L. Genetic, enzymatic, and pathogenic studies of the iron superoxide dismutase of Campylobacter jejuni. Infect Immun. 1994;62:2687–2694. doi: 10.1128/iai.62.7.2687-2694.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pesci E C, Pickett C L. Genetic organization and enzymatic activity of a superoxide dismutase from the microaerophilic human pathogen, Helicobacter pylori. Gene. 1994;27:111–116. doi: 10.1016/0378-1119(94)90614-9. [DOI] [PubMed] [Google Scholar]
- 32.Powell B S, Court D L, Nakamura Y, Rivas M P, Turnbough C L. Rapid confirmation of single-copy lambda prophage integration by PCR. Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasmussen L J, Moller P L, Atlung T. Carbon metabolism regulates expression of the pfl (pyruvate formate-lyase) gene in Escherichia coli. J Bacteriol. 1991;173:6390–6397. doi: 10.1128/jb.173.20.6390-6397.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakamoto H, Touati D. Cloning of the iron superoxide dismutase gene (sodB) in Escherichia coli K-12. J Bacteriol. 1984;159:418–420. doi: 10.1128/jb.159.1.418-420.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 37.Singer M, Baker T A, Schnitzler G, Deischel S, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tardat B, Touati D. Iron and oxygen regulation of Escherichia coli MnSOD expression: competition between the global regulators Fur and ArcA for binding to DNA. Mol Microbiol. 1993;9:53–63. doi: 10.1111/j.1365-2958.1993.tb01668.x. [DOI] [PubMed] [Google Scholar]
- 39.Tardat B, Touati D. Two global regulators repress the anaerobic expression of MnSOD in E. coli: Fur (ferric uptake regulation) and Arc (aerobic respiration control) Mol Microbiol. 1991;5:455–465. doi: 10.1111/j.1365-2958.1991.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 40.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Touati D. Iron and oxidative stress in bacteria. Arch Biochem Biophys. 2000;373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 42.Uzan M, Favre R, Brody E. A nuclease that cuts specifically in the ribosome binding site of some T4 mRNAs. Proc Natl Acad Sci USA. 1988;85:8895–8899. doi: 10.1073/pnas.85.23.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]