Abstract
Unsaturated biofilms of Pseudomonas putida, i.e., biofilms grown in humid air, were analyzed by atomic force microscopy to determine surface morphology, roughness, and adhesion forces in the outer and basal cell layers of fresh and desiccated biofilms. Desiccated biofilms were equilibrated with a 75.5% relative humidity atmosphere, which is far below the relative humidity of 98 to 99% at which these biofilms were cultured. In sharp contrast to the effects of drying on biofilms grown in fluid, we observed that drying caused little change in morphology, roughness, or adhesion forces in these unsaturated biofilms. Surface roughness for moist and dry biofilms increased approximately linearly with increasing scan sizes. This indicated that the divides between bacteria contributed more to overall roughness than did extracellular polymeric substances (EPS) on individual bacteria. The EPS formed higher-order structures we termed mesostructures. These mesostructures are much larger than the discrete polymers of glycolipids and proteins that have been previously characterized on the outer surface of these gram-negative bacteria.
Bacteria in the environment and in vivo frequently live as biofilms where cells are embedded in a matrix of bacterially produced extracellular polymeric substances (EPS) (7, 10). Biofilms occur extensively in aquatic systems, where they are implicated in industrial concerns such as biofouling and corrosion (21), and where they are also implicated in drug resistance (17) and dental caries (37). In waste treatment systems such as trickling filters, wet biofilms are studied for their roles in catalyzing pollutant transformations (58). Generally, the morphologies of biofilms that occur in aquatic systems are mushroom-like (11) with stalked EPS-encapsulated cells growing from the substratum and channels for fluid flow (54) around and through the stalks (11). This well-characterized structure of aquatic biofilms is now understood to facilitate cell-cell communication (12).
In the absence of fluid flow, however, such as in soil systems (8, 19), in food, or on plant leaf surfaces (38), biofilms may appear as patchy films or dense microcolonies. These biofilms are unsaturated; i.e., they grow in an environment that is only transiently wet. From continuously wet to mostly dry environments, biofilms probably show a range of morphologies that are environment dependent. Correspondingly, the view of biofilms from studying aquatic systems may be only a window to the variety of morphological and functional forms that biofilms may take.
The immediate purpose of this work was to build upon earlier work regarding mass transfer characteristics (31) of unsaturated Pseudomonas putida biofilms by examining physical characteristics at the air-biofilm interface that could contribute to high overall mass transfer resistance for substances diffusing through unsaturated biofilms. In aquatic systems, channels in the biofilm matrix act as conduits for nutrient resupply and waste removal (54). In unsaturated systems, however, air-biofilm interphase mass transfer and diffusion within the biofilm matrix are the primary mechanisms for mass delivery (31). Our perspective is that just as mass transfer studies and confocal microscopy of aquatic biofilms have facilitated a relatively sophisticated understanding of biofilm structure in aquatic systems, a better understanding of unsaturated biofilm structure and function can be facilitated through combined mass transfer and morphological studies. We regard investigation of the air-biofilm interface as one step to understanding mass transfer-related characteristics of dense biofilms that occur in unsaturated systems. In response, the work reported here was to characterize the roughness, force characteristics, and morphology of the unsaturated biofilm-air interface. To perform our work, we relied on a contemporary tool in high-resolution biomaterials probing and imaging, the atomic force microscope (AFM). AFM can provide surface morphological and physical information in essentially a nondestructive manner (5, 22, 25, 52). AFM has been used to study bacterial colonization of submerged steel (53), to examine growth at oil-water interfaces (20), and to probe cell membrane elasticity (1, 59) and the adhesive properties of artificial bacterial lawns (47, 48). Here, we extend the use of bacterial AFM through our studies of the morphology and surface characteristics of native unsaturated biofilms that have been cultured and treated using conditions that commonly occur in unsaturated environments. By studying unsaturated biofilms directly and at high resolution, we strengthen our hypothesis that biofilms are morphologically different in unsaturated systems and thus merit further study and description.
MATERIALS AND METHODS
Biofilm culturing.
P. putida mt-2 (Gary Sayler, University of Tennessee) was sampled from −80°C stock cultures (in 70% Luria broth [LB]–30% glycerol) and inoculated directly onto Nuclepore 0.1-μm-pore-diameter polycarbonate filters (Corning, Acton, Mass.) overlaying LB agar. By visual observation, the amount of biomass inoculated onto the membrane was insignificantly small compared to the fully mature biofilms used in our studies. Biofilms were cultured at 27°C and harvested at various time intervals in order to study the effects of biofilm age on biofilm surface properties and morphology.
Sample preparation.
Biofilms on filters were handled under sterile conditions until just prior to imaging and were prepared according to one of four treatment combinations: fresh and unwashed, dried and unwashed, fresh and washed, or fresh and dried. These treatments were selected because they mimic the environmental conditions that biofilms in unsaturated environments experience during transient cycles of wetting and drying. Fresh biofilms on filters were imaged without any alteration to the sample. Biofilms were dried on filters by equilibrating with a 75.5% relative humidity (RH) air atmosphere. This RH is equivalent to a water potential of approximately −38 MPa (41), which is on the lower end of water stress tolerable by any bacteria (27). Washed biofilms on filters were rinsed with 100 μl of sterile MilliQ water (Millipore, Burlington, Mass.). Following preparation, filters with biofilms were transferred onto freshly cleaved mica disks (33) for imaging. Adhesion of unwashed biofilms on filters to the mica disk was facilitated by placing either a 10-μl drop of MilliQ water underneath the filter or a piece of double-stick Scotch tape (3M Corp., St. Paul, Minn.) between the mica and filter. The difference in method of adhesion did not appear to affect the data, but the double-stick tape provided a more stable substrate for AFM imaging.
AFM operation.
The biofilms were imaged in either contact mode or tapping mode, using a Nanoscope III MultiMode AFM (Digital Instruments, Santa Barbara, Calif.). The contact mode is the imaging mode used to provide morphological data in conjunction with force mapping (33). Contact mode images were obtained using V-shaped silicon nitride (Si3N4) Nanoprobe cantilevers (Digital Instruments). In tapping mode, the tip oscillates, touching the sample surface at the bottom of each oscillation (22, 26), and both height and phase images are captured. Tapping mode images were obtained using silicon cantilevers with resonance frequencies of ca. 200 to 350 kHz (Digital Instruments). New cantilevers were used for each experiment to prevent sample cross-contamination.
Force mapping.
The AFM was operated in force mode to analyze the adhesion forces across the biofilm surface. The Nanoscope force mapping was performed as previously described (33). Force mapping was conducted in contact mode, using short, wide, V-shaped silicon nitride cantilevers with spring constants of ca. 0.6 N/m. Force maps were captured in the relative trigger mode, using trigger thresholds, or maximum cantilever deflections, of 10 and 50 nm. The force maps included 64 × 64 force plots containing 64 data points per force plot. A Z-scan speed of 13.0 Hz was used, allowing a force map to be completed every 10 min. Before force plots were acquired, each sample was imaged in contact mode to ensure initial stability. Force maps were captured using X-Y scan sizes of 4,000 nm or smaller. This provides spacing between force plots of 62 nm or less. Force maps were analyzed graphically to determine the median adhesion force for each force map, i.e., the adhesion force such that half the points on the force map had a lower adhesion force and half the points had a higher adhesion force. This analysis of force maps represents a new approach to systematize the data and to use the data to comparatively analyze biofilm treatment effects.
Roughness analysis.
We used Nanoscope software (Digital Instruments) to calculate the surface roughness parameters for the height images. The images were flattened and plane fitted prior to analysis. The surface roughness parameters calculated included the Z range (the difference between the highest and lowest points within a given area), the mean (the average of all the Z values), the root mean square (RMS; the standard deviation of the Z values), and the mean roughness (Ra; mean value of the surface relative to the center plane) (50). In addition to the roughness analysis of the entire biofilm image, the surface roughness parameters were also calculated for a 200- to 300-nm × 200- to 300-nm box on all 2,000-nm or lower scan sizes to provide a local roughness analysis of the extracellular polymers.
Estimating molecular masses of extracellular polymers.
For molecules of known molecular mass, molecular volumes measured by AFM correlate well with calculated molecular volumes (18, 45, 51). We measured diameters of extracellular structures but not volumes, because the height of these crowded structures is unknown. Molecular masses were estimated from these diameters by assuming densities for the globular extracellular structures of 1 to 1.3 g/ml.
Statistical analysis.
Statistics were performed using the Wilcoxon signed-rank test (34) to find any differences in the surface roughness based on the effect of preparation.
RESULTS
Over 40 separate P. putida biofilms were analyzed by AFM, with approximately equivalent representation of the various treatments. The method of biofilm preparation affected the stability of AFM imaging. Stabler, higher-quality AFM images were obtained with dried biofilms than with fresh biofilms. Also, stabler AFM images were obtained with washed biofilms than with unwashed biofilms. When biofilms grown on filters were washed with water, the upper layers of the biofilm were usually removed, leaving a single layer of bacterial cells on the filter. These washed biofilms offered an opportunity to study bacterial cells at the base of the biofilm, rather than the top layer of the biofilm with its many layers underneath. Age also affected the image quality, causing the older specimens to become thick with clumps of bacteria and, therefore, more unstable during imaging.
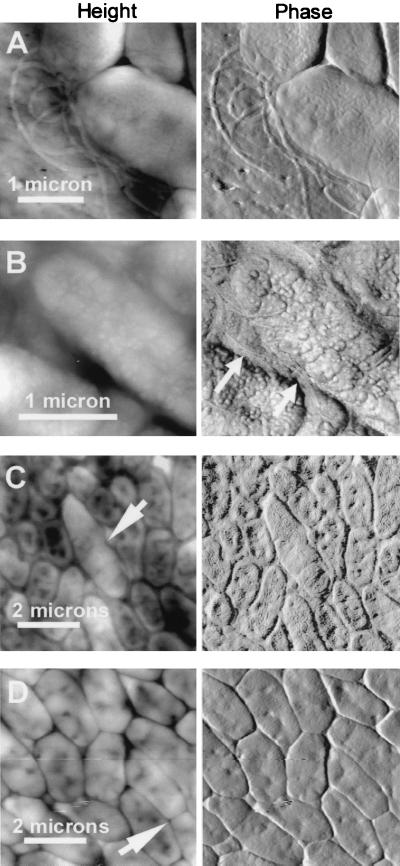
Individual bacteria varied around the expected dimensions of 1 by 2 μm (Fig. 1). Height images in Fig. 1 show the topographic profile of the biofilm surface, while phase images show the substructural features in more detail. Bacterial flagella were readily seen in some images (Fig. 1A and B), especially in areas devoid of cells. The flagella were most easily seen on the filter where biofilms had flaked off the filter after drying. Flagella were also seen in single-cell layer images at the edges of washed biofilms, as in Fig. 1A, which shows a single cell thickness of the biofilm on the edge. The flagella in Fig. 1B are nestled between two bacteria in the biofilm; other images show elongated bacteria that appear to be in the process of dividing (Fig. 1C and D, arrows). These dividing cells were seen in both the top and bottom cell layers in biofilms that had been growing for 2 to 4 days.
FIG. 1.
Bacterial flagella (A and B) and bacteria during cell division (C and D, arrows). Flagella were seen both on the filter adjacent to biofilm bacteria (A) and nestled between bacteria (B, arrows). Shown are height (left) and phase (right) AFM images of unsaturated P. putida mt-2 biofilms. Before AFM imaging, biofilms were 1 day old, unwashed and undesiccated (A), 3 days old, unwashed and desiccated (B), 2 days old, washed and desiccated (C), and 4 days old, unwashed and desiccated (D).
In most biofilms there were valleys between the individual bacteria (Fig. 1 and 2A). The valleys between bacteria were typically 40 to 120 nm deep, measured from the highest point on the adjacent bacteria. The depth of these valleys may be even greater because the width of the AFM tip makes it impossible to measure the depth of valleys that are too deep and narrow for the tip to reach the bottom. Bacteria such as those in Fig. 1 and 2A typically also showed indentations on their surfaces, with depths of ∼10 to 60 nm. These indentations are broad enough that their depth can be measured accurately by AFM.
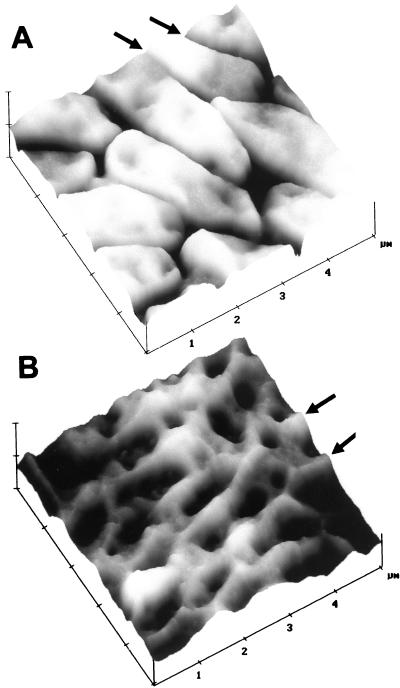
FIG. 2.
Biofilm bacteria with crevasses (A) and ridges (B) between cells. Surface plots of AFM height images emphasized surface corrugations. Z (height) scales are 250 (A) and 150 (B) nm/division. Unsaturated P. putida mt-2 biofilms were unwashed, desiccated, and 4 (A) or 3 (B) days old before AFM imaging.
Instead of valleys, some biofilms exhibited ridges surrounding the individual bacteria (Fig. 2B); these ridges were typically 20 to 70 nm above the lowest point in the adjacent bacteria. Ridges were seen only on the top layer of biofilm bacteria. Valleys, not ridges, were observed in basal cell layers following washing. The ridges may be due to variable production of EPS, such that EPS is sometimes produced in such great quantity that it protrudes around the edges of the bacteria and is more pronounced after the bacterial cytoplasm has shrunk following biofilm drying (Fig. 2B). In other biofilms (Fig. 2A), there may be much less EPS, so that the bacteria shrink and separate from one another upon drying.
Extracellular mesostructures.
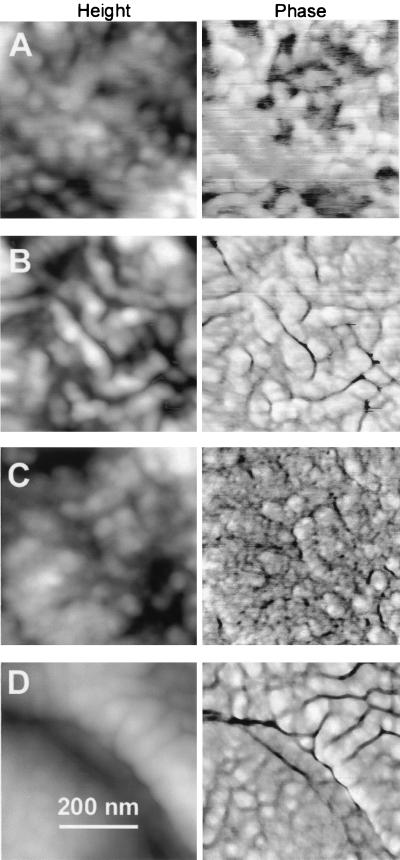
The EPS exhibited mesostructures on the cells. Mesostructures were typically 40 to 70 nm in diameter (Fig. 3), although some as small as 10 to 20 nm were occasionally observed (Fig. 3B). EPS mesostructures were arranged in arrays that showed differences in appearance, varying from rounded, spheroid structures (Fig. 3A and B) to worm-like structures (Fig. 3C). Sometimes adjacent cells or adjacent regions on the same cell showed both worm-like and spheroid structures (Fig. 3D). Some of the mesostructures appeared in a hexagonal array (data not shown). In general, the morphologies of mesostructures appeared to be independent of the conditions used to prepare the biofilm for AFM imaging.
FIG. 3.
EPS by AFM on unsaturated P. putida mt-2 biofilms according to sample preparation treatment and age. Before AFM imaging, biofilms were 1 day old, unwashed and desiccated (A), 3 days old, unwashed and desiccated (B), 2 days old, washed and desiccated (C), and 2 days old, washed and undesiccated (D).
The extracellular mesostructures that we observed were not artifacts from the scanning of the AFM tip, as have been seen on the surface of polymer films (35). Such tip artifacts are readily detected because, upon zooming out, one sees a characteristic square pattern in the center of the larger scan that clearly shows the tip-induced changes during scanning of the smaller square area. In contrast, the mesostructures on the surface of these biofilms are extremely stable and resistant to damage or distortion by repeated scanning.
Biofilm roughness.
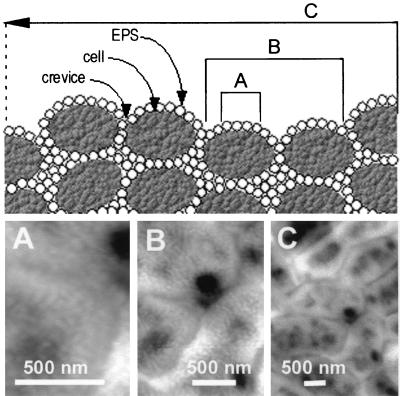
The surface roughness of the biofilms was analyzed from over 450 AFM images on length scales ranging from 0.5 to 10 μm. These different length scales are visually depicted in the diagram at the top of Fig. 4 and the AFM images of Fig. 4A to C. Figure 4, top, depicts a conceptual model of three scales of surface roughness: across the extracellular polymers on a single bacterium (A), across one or a few bacteria (B), and across many bacteria on the biofilm surface (C). Images of small scan size (Fig. 4A) reveal the organization of the mesostructures on the surface of an individual bacterium in detail. Medium scan sizes (Fig. 4B) depict the individual bacteria, the crevices between the bacteria, and pores or pits on the surface of the bacteria. Large scan sizes (Fig. 4C) reveal the packing of bacteria at the biofilm surface, with bacteria grouped next to each other in random organization. Unwashed, multilayer biofilms also reveal bacteria emerging out from underneath other bacteria, giving an impression of considerable crowding and variation in cell size (e.g., Fig. 1C).
FIG. 4.
Three scales of roughness on biofilms. (Top) Conceptual diagram. (Bottom) AFM height images of unsaturated P. putida mt-2 biofilms. Each of the three length scales provides different values for roughness, which probably vary due to the predominating surface feature at that scale: (A) within cells (0.5-μm scale), mesoscale structures of EPS determine roughness; (B) across 1 or 2 cells (2-μm scale), roughness is from EPS and boundaries between bacteria; (C) roughness averaged over many cells (5-μm and greater scale) is likely dominated by fluctuations in biofilm thickness and boundaries between bacteria.
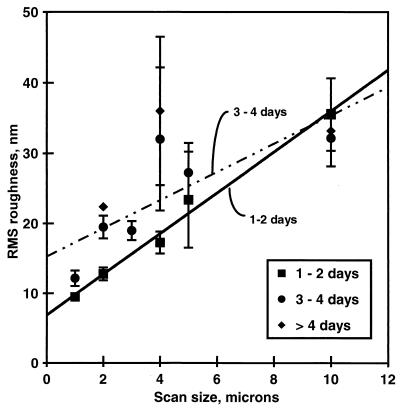
Roughness showed a direct correlation with scan size, as measured by both RMS roughness (Fig. 5) and Ra. There were no clear effects of biofilm preparation on biofilm roughness (data not shown), but older biofilms were slightly rougher than younger biofilms at scan sizes of 5 μm and smaller (Fig. 5).
FIG. 5.
Surface roughness (RMS) of unwashed, unsaturated P. putida mt-2 biofilms as a function of AFM scan size. The RMS roughness for similar biofilm age groups is displayed. Within each age group, n > 3; standard errors of the means are shown. Within each age group, roughness did not vary with sample preparation treatment.
Biofilm force maps.
Force maps of biofilms generally showed median lift-off forces of ∼30 to 60 nN. There was no consistent trend for differences in this median adhesion force between washed versus unwashed biofilms, dry versus fresh biofilms, or new versus older biofilms.
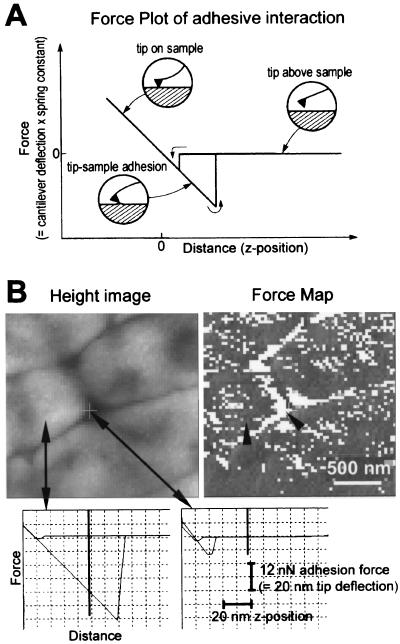
In the force map of Fig. 6, each pixel contains the information for a pair of force-versus-distance curves (a force plot), such as those in Fig. 6A and at the bottom of Fig. 6B. Force plots display the tip-sample interaction forces as the AFM tip moves toward and away from the sample surface. Each force plot contains one force-versus-distance curve for the approach of the cantilever toward and into the sample and a second force-versus-distance curve for the retraction of the cantilever from the sample. Force plots show whether the tip-sample interactions are primarily attractive, as in Fig. 6, or repulsive. Attractive force plots typically show some hysteresis between the approach and retract curves. This hysteresis is typically of the type shown in Fig. 6A, where the tip is slightly attracted to the surface upon approaching and then adheres more strongly to the surface as it retracts from the surface. Force plots also show whether the sample surface is hard or elastic (33, 46). The biofilm surface in Fig. 6 shows no elasticity that can be detected with the cantilever of 0.6-N/m spring constant that was used.
FIG. 6.
Biofilm force map. (A) Diagram of the tip-sample interactions occurring as the AFM tip approaches and retracts from an adhesive surface. The force plot consists of two curves: one for the tip approaching the sample and one for the tip retracting from the sample. During the approach, the cantilever moves from right to left in the diagram, and the cantilever deflects as it presses into the biofilm. In the retract curve, the cantilever deflects sharply before separating from the biofilm as it moves from left to right in the diagram. The horizontal region of the curve represents the tip not in contact with the biofilm, and the sloped region of the curve represents the tip in contact with the biofilm. The slope for a hard surface is approximately 1 nm of deflection/nm of z distance, while softer surfaces show a more gradual deflection (33). (B) Force map analysis of unsaturated P. putida mt-2 biofilm. Biofilm was 2 days old, unwashed, and desiccated at 75.5% RH. Height image arrows show where example force plots were acquired. Lighter regions are higher than darker regions. Force map (force-volume image) shows patterns of adhesion on surface of biofilm. Darker regions are more adhesive than lighter regions. Arrowheads indicate pixels on force map where force plots were acquired. Scale bar indicates the x-y scales of the force map and height image. Force-versus-distance curves on cell surface (left) and between cells (right) show the large adhesion on the bacterial surface and small adhesion between bacteria. Vertical bars on force plots show the z position represented in the force map (= 50 nm above the position of maximum cantilever deflection into surface). Scale bars give x and y dimensions of force plots.
Many of the force maps showed an unexpected feature (Fig. 6): the spaces between cells were less adhesive than the surfaces of the cells. This can be seen from the two force plots in Fig. 6, which show the large adhesion on the top of the cell (left) and the small adhesion in the crack between the cells (right). Similarly, the force map or force volume image on the right shows a pattern of light pixels (less adhesive force plots) following the spaces between the cells, with most of the top of the cell surface covered by dark pixels (more adhesive force plots). There are at least two possible explanations for this observation. One possibility is that the bacterial surfaces differ from the spaces between bacteria in properties such as moisture content or quantity of EPS. An alternate possibility is that the AFM tip makes less contact with the biofilm in the spaces between the cells, so that the reduced adhesion is due to the smaller tip-sample contact area between the bacteria. Further research will be needed to distinguish between these two explanations.
Another unexpected feature of the biofilm force maps is that the adhesion forces between the tip and the biofilm surface were not strongly dependent on the maximum force that was applied to the biofilm surface during each force plot. The maximum force applied to the biofilm during each force plot is set by the trigger threshold. The trigger threshold is the maximum cantilever deflection that can occur as the cantilever advances into the biofilm surface. Trigger thresholds were set at 10 to 50 nm, which corresponds to ∼6- to 30-nN applied force for cantilevers with a spring constant of 0.6 N/m.
DISCUSSION
This study of surface characteristics of P. putida unsaturated biofilms is part of an ongoing investigation into why these biofilms have appeared to be highly resistant to mass transfer (31). Generally biofilms are studied as part of an aqueous, fluid flowing system; in contrast, biofilms grown in air have been studied less. Because biofilm is an important growth habit in unsaturated systems, the factors that limit the transfer of nutrients and waste to unsaturated biofilms should be understood. We relied on AFM, a high-resolution tool in surface imaging and probing, to make our investigations.
AFM has been a useful tool in furthering many areas of biological research, including the imaging of DNA (6, 24), proteins (39, 52), and cells (1, 14, 28, 29, 56). The biofilms that have been imaged previously by AFM were all grown in fluid (2, 4, 55) or, in the case of P. putida, at an oil-water interface (20). Many of our AFM images of unsaturated P. putida are reminiscent of the published AFM images of P. putida in fluid (20, 53), although the prior methods were quite different from ours.
Our work offers three significant new contributions that have not been addressed previously. First, we studied specifically the air-biofilm interface using a range of preparation conditions representative of those that unsaturated biofilms could be exposed to in the natural environment (e.g., wet to dry, unwashed to washed). Second, we measured the roughness of unsaturated biofilms and showed that, in contrast to roughness measures of biofilms in saturated, fluid flowing environments, unsaturated biofilms are very smooth. Third, we measured the adhesive properties of the biofilms and developed an approach for analyzing the force mapping data so that we could demonstrate how adhesive the biofilms were as a function of treatment. In this discussion, we address these three contributions to biofilm science and provide additional interpretation of our morphological observations.
Reproducing the unsaturated biofilm environment.
We observed that our biofilms had similar morphologies under all conditions used for growth and imaging—desiccated versus fresh; older versus younger; top versus bottom layer of bacterial cells. Such comparisons, particularly dry versus wet, can provide a sense of how biofilm morphologies in unsaturated environments are stable with the changing environmental conditions that commonly occur in soils subject to seasonal wetting and drying. Through washing, we also found that extracellular mesostructures, presumably clumped exopolymers, were present in both the top and basal cell layers of the biofilm.
Smooth biofilms.
Biofilm roughness has previously been reported either as RMS (3) or as Ra* (40, 44), where Ra* is calculated by dividing Ra by the biofilm thickness (40). We reported only RMS as our index of biofilm roughness because Ra was numerically similar in all cases. Although aquatic biofilms shrink when dried such that the hydrated parts of EPS condense to 1% of the original volume (55), drying did not increase the surface roughness of our biofilms. This may imply that shrinkage or condensation of the polymers was uniform.
Age may indirectly relate to roughness: older aquatic biofilms are generally thicker and rougher (40, 42). Consistently, the surface structures of biofilms growing in water become continuously more complex as they age, over a period of weeks and months (44). In our studies of unsaturated biofilm, we found a slight increase in surface roughness at smaller scan sizes (below 10 μm) for biofilms over 2 days old versus younger biofilms. Overall, however, our biofilms are less than 0.2% as rough at all scan sizes compared to biofilms cultivated in aqueous fluid flowing systems. Thus, the smoothness of our biofilms, measured here by surface roughness, generally confirms our previous observations by transmission electron microscopy that the air-biofilm interface of P. putida biofilms cultured under unsaturated conditions was uniformly flat over distances ranging from one cell to tens of cells (31).
In aquatic systems, rough biofilms facilitate external (boundary layer) mass transport to biofilms (7, 10) and improve the rate of nutrient resupply into biofilms (16). Improved mass transfer of nutrients will tend to improve biofilm growth rate (32, 42). In aquatic systems, there may be a sequence of roughness-mass transfer relationships that begins with rough biofilms whose texture is a consequence of nutrient deprivation; then, with rapid fluid flow, rough biofilms improve boundary layer mass transfer of nutrients to the end that the biofilms grow and become smooth (43). We interpreted a previously defined (43) ratio of growth rate to mass transfer, G, for the case of unsaturated biofilms. In our biofilms, G = μ(L2/D), where μ is the first-order growth rate constant for exponentially growing cells, L is a characteristic length for external mass transfer, and D is the molecular diffusivity for the diffusing substrate. We assumed the limiting mass transfer process external to unsaturated biofilms to be gas-phase substrate (e.g., oxygen) diffusion (as opposed to flowing air). Our calculations using a molecular diffusivity for oxygen (ambient temperature) of 0.175 cm2/s (57), a first-order growth rate constant for P. putida mt-2 of approximately 0.5/h (30), and a characteristic length L of 1 μm result in a value for G that is very low, i.e., on the order of 10−11. One interpretation of this calculation is that diffusive transport of oxygen to the biofilm should not limit growth; another interpretation is that the biofilm should be smooth given the high external mass transfer rate relative to cellular growth rate (43). Thus the smoothness of P. putida air-biofilm interfaces reported here and previously by transmission electron microscopy (31) is indicative of relatively low mass transfer resistance across the air-biofilm interfacial boundary layer.
Biofilm adhesiveness.
AFM force mapping reveals properties of a sample that are not revealed in the AFM height images (46). For example, force mapping of cholinergic synaptic vesicles has revealed that the centers of these vesicles are harder or stiffer than the peripheries of the vesicles (33). Force maps combine force plot data with the visual images and thus enable one to correlate specific tip-sample interactions with specific features on the sample surface.
Our biofilm force maps showed that the spaces between biofilm bacteria were less adhesive than the bacterial surfaces and that the median adhesion forces were 30 to 60 nN. These median adhesion forces can be compared to what one would expect for the surface tension of an aqueous solution, such as the thin water layer on the surface of the biofilm. Surface tensions of aqueous Triton X detergent solutions range from 70 (dilute) to 30 (concentrated) mN/m, as measured with a liquid tensiometer. A biofilm adhesion force of 30 nN corresponds to an air-liquid surface tension of 30 mN/m if the circumference of contact between the tip and the biofilm is 1 μm.
AFM images, depending on the mode of acquisition, lend themselves to physical interpretations not available through other high-resolution imaging approaches such as electron microscopy. For example, phase images in tapping AFM, such as those in Fig. 1 and 3, show the phase difference between the oscillation driving the cantilever and the oscillation of the cantilever as it interacts with the sample surface (36) (http://www.di.com/appnotes/Phase/PhaseMain.html). Phase images are a map of the energy dissipated by the tip-sample interaction at each point on the sample surface (9). Changes in energy dissipation over the sample surface are related to changes in such surface properties as adhesiveness and stiffness. Phase images of lysed synpatic vesicles (15), wood pulp fiber, and DNA (23) show distinctively varying patterns of energy dissipation. In contrast to these biomaterials, the biofilm phase images in Fig. 1 and 3 are relatively uniform.
Extracellular mesostructures.
Although the overall morphology of the biofilms is smooth, the surface morphology of individual biofilm bacteria indicated the presence of extracellular structures larger than the glycolipids and glycoproteins reported previously, which generally range in size from ∼1 kDa for glycolipids to 100 kDa for glycoproteins (13, 49) (http://www.cmdr.ubc.ca/bobh/genomics.htm). The extracellular mesostructures in our images resembled the “orange peel” appearance previously reported for P. putida grown at an oil-water interface (20). The smallest mesostructures in Fig. 3 have diameters of 10 nm, which gives them a molecular mass of >100 kDa if they are globular. Typical mesostructures of 40 nm or larger may have masses of at least 10,000 kDa. Therefore, either there are larger macromolecules on the surface of these biofilm bacteria than have been identified previously or the observed mesostructures are actually aggregates of much smaller molecules.
ACKNOWLEDGMENTS
We thank the reviewers for their valuable suggestions.
This work was supported by NSF grant MCB9604566 (H.G.H., C.S., and I.D.A.), the UCSB Donald Bren School of Environmental Science and Management (I.D.A.), and U.S. EPA grant R827133-01.
REFERENCES
- 1.Arnoldi M, Kacher C M, Baeuerlein E, Radmacher M, Fritz M. Elastic properties of the cell wall of Magnetospirillum gryphiswaldense investigated by atomic force microscopy. Appl Phys A. 1998;66:S613–S617. [Google Scholar]
- 2.Beech I B. The potential use of atomic force microscopy for studying corrosion of metals in the presence of bacterial biofilms—an overview. Int Biodeterior Biodegrad. 1996;37:141–149. [Google Scholar]
- 3.Bishop P L, Gibbs J T, Cunningham B E. Relationship between concentration and hydrodynamic boundary layers over biofilms. Environ Technol. 1997;18:375–386. [Google Scholar]
- 4.Bremer P J, Geesey G G, Drake B. Atomic force microscopy examination of the topography of a hydrated bacterial biofilm on a copper surface. Curr Microbiol. 1992;24:223–230. [Google Scholar]
- 5.Bustamante C, Keller D. Scanning force microscopy in biology. Phys Today. 1995;48:32–38. [Google Scholar]
- 6.Bustamante C, Rivetti C. Visualizing protein-nucleic acid interactions on a large scale with the scanning force microscope. Annu Rev Biophys Biomol Struct. 1996;25:395–429. doi: 10.1146/annurev.bb.25.060196.002143. [DOI] [PubMed] [Google Scholar]
- 7.Characklis W G, Marshall K C. Biofilms: a basis for an interdisciplinary approach. In: Characklis W G, Marshall K C, editors. Biofilms. New York, N.Y: John Wiley & Sons, Inc.; 1990. pp. 3–15. [Google Scholar]
- 8.Chenu C. Extracellular polysaccharides: an interface between microorganisms and soil constituents. In: Huang P M, Berthelin J, Bollag J-M, McGill W B, Page A L, editors. Environmental impact of soil component interactions. 1. Natural and anthropogenic organics. Boca Raton, Fla: CRC Press, Inc.; 1995. pp. 217–233. [Google Scholar]
- 9.Cleveland J P, Anczykowski B, Schmid A E, Elings V B. Energy dissipation with a tapping-mode atomic force microscope. Appl Phys Lett. 1998;72:2613–2615. [Google Scholar]
- 10.Costerton J W, Lappin-Scott H M. Introduction to microbial biofilms. In: Lappin-Scott H M, Costerton J W, editors. Microbial biofilms. Cambridge, England: Cambridge University Press; 1995. pp. 1–11. [Google Scholar]
- 11.Costerton J W, Lewandowski Z, De Beer D, Caldwell D, Korber D, James G. Biofilms, the customized microniche. J Bacteriol. 1994;176:2137–2142. doi: 10.1128/jb.176.8.2137-2142.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 13.Desai J D, Banat I M. Microbial production of surfactants and their commercial potential. Microbiol Mol Biol Rev. 1997;61:47–64. doi: 10.1128/mmbr.61.1.47-64.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fritz M, Radmacher M, Gaub H E. Granula motion and membrane spreading during activation of human platelets imaged by atomic force microscopy. Biophys J. 1994;66:1328–1334. doi: 10.1016/S0006-3495(94)80963-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia R A, Laney D E, Parsons S M, Hansma H G. Substructure and responses of cholinergic synaptic vesicles in the atomic force microscope. J Neurosci Res. 1998;52:350–355. doi: 10.1002/(SICI)1097-4547(19980501)52:3<350::AID-JNR11>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 16.Gibbs J T, Bishop P L. A method for describing biofilm surface roughness using geostatistical techniques. Water Sci Technol. 1995;32:91–98. [Google Scholar]
- 17.Gilbert P, Brown M R W. Mechanisms of the protection of bacterial biofilms from antimicrobial agents. In: Lappin-Scott H M, Costerton J W, editors. Microbial biofilms. Cambridge, England: Cambridge University Press; 1995. pp. 118–130. [Google Scholar]
- 18.Golan R, Pietrasanta L I, Hsieh W, Hansma H G. DNA toroids: stages in condensation. Biochemistry. 1999;38:14069–14076. doi: 10.1021/bi990901o. [DOI] [PubMed] [Google Scholar]
- 19.Gray T R G, Baxby P, Hill I R, Goodfellow M. Direct observation of bacteria in soil. In: Gray T R G, Parkinson D, editors. The ecology of soil bacteria. Toronto, Canada: University of Toronto Press; 1968. pp. 171–192. [Google Scholar]
- 20.Gunning P A, Kirby A R, Parker M L, Gunning A P, Morris V J. Comparative imaging of Pseudomonas putida bacterial biofilms by scanning electron microscopy and both DC contact and AC non-contact atomic force microscopy. J Appl Bacteriol. 1996;81:276–282. [Google Scholar]
- 21.Hamilton W A. Biofilms and microbially influenced corrosion. In: Lappin-Scott H M, Costerton J W, editors. Microbial biofilms. Cambridge, England: Cambridge University Press; 1995. pp. 171–182. [Google Scholar]
- 22.Hansma H G, Hoh J. Biomolecular imaging with the atomic force microscope. Annu Rev Biophys Biomol Struct. 1994;23:115–139. doi: 10.1146/annurev.bb.23.060194.000555. [DOI] [PubMed] [Google Scholar]
- 23.Hansma H G, Kim K J, Laney D E, Garcia R A, Argaman M, Parsons S M. Properties of biomolecules measured from atomic force microscope images: a review. J Struct Biol. 1997;119:99–108. doi: 10.1006/jsbi.1997.3855. [DOI] [PubMed] [Google Scholar]
- 24.Hansma H G, Pietrasanta L. Atomic force microscopy and other scanning probe microscopies. Curr Opin Chem Biol. 1998;2:579–584. doi: 10.1016/s1367-5931(98)80086-0. [DOI] [PubMed] [Google Scholar]
- 25.Hansma, H. G., L. I. Pietransanta, I. D. Auerbach, C. Sorenson, R. Golan, and P. A. Holden. Probing biopolymers with the atomic force microscope: a review. J. Biomater. Sci. Polym. Ed., in press. [DOI] [PubMed]
- 26.Hansma P K, Cleveland J P, Radmacher M, Walters D A, Hillner P, Bezanilla M, Fritz M, Vie D, Hansma H G, Prater C B, Massie J, Fukunaga L, Gurley J, Elings V. Tapping mode atomic force microscopy in liquids. Appl Phys Lett. 1994;64:1738–1740. [Google Scholar]
- 27.Harris R F. Effect of water potential on microbial growth and activity. In: Parr J F, Gardner W R, Elliott L F, editors. Water potential relations in soil microbiology. SSSA Special Publication no. 9. Madison, Wis: Soil Science Society of America; 1981. pp. 23–95. [Google Scholar]
- 28.Henderson E, Haydon P G, Sakaguchi D S. Actin filament dynamics in living glial cells imaged by atomic force microscopy. Science. 1992;257:1944–1946. doi: 10.1126/science.1411511. [DOI] [PubMed] [Google Scholar]
- 29.Hoh J H, Schoenenberger C-A. Surface morphology and mechanical properties of MDCK monolayers by atomic force microscopy. J Cell Sci. 1994;107:1105–1114. doi: 10.1242/jcs.107.5.1105. [DOI] [PubMed] [Google Scholar]
- 30.Holden P A, Halverson L J, Firestone M K. Water stress effects on toluene biodegradation by Pseudomonas putida. Biodegradation. 1997;8:143–151. doi: 10.1023/a:1008237819089. [DOI] [PubMed] [Google Scholar]
- 31.Holden P A, Hunt J R, Firestone M K. Toluene diffusion and reaction in unsaturated Pseudomonas putida biofilms. Biotechnol Bioeng. 1997;56:656–670. doi: 10.1002/(SICI)1097-0290(19971220)56:6<656::AID-BIT9>3.0.CO;2-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korber D R, Choi A, Wolfaardt G M, Ingham S C, Caldwell D E. Substratum topography influences susceptibility of Salmonella enteritidis biofilms to trisodium phosphate. Appl Environ Microbiol. 1997;63:3352–3358. doi: 10.1128/aem.63.9.3352-3358.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laney D E, Garcia R A, Parsons S M, Hansma H G. Changes in the elastic properties of cholinergic synaptic vesicles as measured by atomic force microscopy. Biophys J. 1997;72:806–813. doi: 10.1016/s0006-3495(97)78714-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lapin L L. Statistics: meaning and method. San Francisco, Calif: Harcourt Brace Jovanovich, Inc.; 1975. [Google Scholar]
- 35.Leung O M, Goh M C. Orientational ordering of polymers by atomic force microscope tip-surface interaction. Science. 1992;255:64–66. doi: 10.1126/science.255.5040.64. [DOI] [PubMed] [Google Scholar]
- 36.Magonov S N, Elings V, Whangbo M H. Phase imaging and stiffness in tapping-mode atomic force microscopy. Surf Sci. 1997;375:L385–L391. [Google Scholar]
- 37.Marsh P D. Dental plaque. In: Lappin-Scott H M, Costerton J W, editors. Microbial biofilms. Cambridge, England: Cambridge University Press; 1995. pp. 282–300. [Google Scholar]
- 38.Morris C E, Monier J-M, Jacques M-A. Methods for observing microbial biofilms directly on leaf surfaces and recovering them for isolation of culturable organisms. Appl Environ Microbiol. 1997;63:1570–1576. doi: 10.1128/aem.63.4.1570-1576.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller D J, Amrein M, Engel A. Adsorption of biological molecules to a solid support for scanning probe microscopy. J Struct Biol. 1997;119:172–188. doi: 10.1006/jsbi.1997.3875. [DOI] [PubMed] [Google Scholar]
- 40.Murga R, Stewart P S, Daly D. Quantitative analysis of biofilm thickness variability. Biotechnol Bioeng. 1995;45:503–510. doi: 10.1002/bit.260450607. [DOI] [PubMed] [Google Scholar]
- 41.Papendick R I, Campbell G S. Theory and measurement of water potential. In: Parr J F, Gardner W R, Elliot L F, editors. Water potential relations in soil microbiology. SSSA Special Publication no. 9. Madison, Wis: Soil Science Society of America; 1981. pp. 1–22. [Google Scholar]
- 42.Peyton B M. Effects of shear stress and substrate loading rate on Pseudomonas aeruginosa biofilm thickness and density. Water Res. 1996;30:29–36. [Google Scholar]
- 43.Picioreanu C, van Loosdrecht M C M, Heijnen J J. Discrete-differential modelling of biofilm structure. Water Sci Technol. 1999;39:115–122. [Google Scholar]
- 44.Picioreanu C, van Loosdrecht M C M, Heijnen J J. Mathematical modeling of biofilm structure with a hybrid differential-discrete cellular automation approach. Biotechnol Bioeng. 1998;58:101–116. doi: 10.1002/(sici)1097-0290(19980405)58:1<101::aid-bit11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 45.Pietrasanta L I, Thrower D, Hsieh W, Rao S, Stemmann O, Lechner J, Carbon J, Hansma H G. Probing the Saccharomyces cerevisiae CBF3-CEN DNA kinetochore complex using atomic force microscopy. Proc Natl Acad Sci USA. 1999;96:3757–3762. doi: 10.1073/pnas.96.7.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radmacher M, Cleveland J P, Fritz M, Hansma H G, Hansma P K. Mapping interaction forces with the atomic force microscope. Biophys J. 1994;66:2159–2165. doi: 10.1016/S0006-3495(94)81011-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Razatos A, Ong Y-L, Sharma M M, Georgiou G. Evaluating the interaction of bacteria with biomaterials using atomic force microscopy. J Biomater Sci Polym Ed. 1998;9:1361–1373. doi: 10.1163/156856298x00442. [DOI] [PubMed] [Google Scholar]
- 48.Razatos A, Ong Y-L, Sharma M M, Georgiou G. Molecular determinants of bacterial adhesion monitored by atomic force microscopy. Proc Natl Acad Sci USA. 1998;95:11059–11064. doi: 10.1073/pnas.95.19.11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Russel M. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J Mol Biol. 1998;279:485–499. doi: 10.1006/jmbi.1998.1791. [DOI] [PubMed] [Google Scholar]
- 50.Sayles R S. The profile as a random process. In: Thomas T R, editor. Rough surfaces. London, England: Longman; 1982. pp. 91–118. [Google Scholar]
- 51.Schneider S W, Larmer J, Henderson R M, Oberleithner H. Molecular weights of individual proteins correlate with molecular volumes measured by atomic force microscopy. Pflugers Arch. 1998;435:362–367. doi: 10.1007/s004240050524. [DOI] [PubMed] [Google Scholar]
- 52.Shao Z, Mou J, Czaijkowsky D M, Yang J, Yuan J-Y. Biological atomic force microscopy: what is achieved and what is needed. Adv Phys. 1996;45:1–86. [Google Scholar]
- 53.Steele A, Goddard D T, Beech I B. An atomic force microscopy study of the biodeterioration of stainless steel in the presence of bacterial biofilms. Int Biodeterior Biodegrad. 1994;34:35–46. [Google Scholar]
- 54.Stoodley P, DeBeer D, Lewandowski Z. Liquid flow in biofilm systems. Appl Environ Microbiol. 1994;60:2711–2716. doi: 10.1128/aem.60.8.2711-2716.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Surman S B, Walker J T, Goddard D T, Morton L H G, Keevil C W, Weaver W, Skinner A, Hanson K, Caldwell D, Kurtz J. Comparison of microscope techniques for the examination of biofilms. J Microbiol Methods. 1996;25:57–70. [Google Scholar]
- 56.Ushiki T, Hitomi J, Ogura S, Umemoto T, Shigeno M. Atomic force microscopy in histology and cytology. Arch Histol Cytol. 1996;59:421–431. doi: 10.1679/aohc.59.421. [DOI] [PubMed] [Google Scholar]
- 57.Welty J R, Wicks C E, Wilson R E. Fundamentals of momentum, heat and mass transfer. 3rd ed. New York, N.Y: John Wiley & Sons; 1984. [Google Scholar]
- 58.Wyndham R C, Kennedy K J. Microbial consortia in industrial wastewater treatment. In: Lappin-Scott H M, Costerton J W, editors. Microbial biofilms. Cambridge, England: Cambridge University Press; 1995. pp. 183–195. [Google Scholar]
- 59.Xu W, Mulhern P J, Blackford B L, Jericho M H, Firtel M, Beveridge T J. Modeling and measuring the elastic properties of an archaeal surface, the sheath of Methanospirillum hungatei, and the implication for methane production. J Bacteriol. 1996;178:3106–3112. doi: 10.1128/jb.178.11.3106-3112.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]