Abstract
The bacterial two-component signal transduction systems regulate adaptation processes and are likely to play a role in Mycobacterium tuberculosis physiology and pathogenesis. The previous initial characterization of an M. tuberculosis response regulator from one of these systems, mtrA-mtrB, suggested its transcriptional activation during infection of phagocytic cells. In this work, we further characterized the mtrA response regulator from M. tuberculosis H37Rv. Inactivation of mtrA on the chromosome of M. tuberculosis H37Rv was possible only in the presence of plasmid-borne functional mtrA, suggesting that this response regulator is essential for M. tuberculosis viability. In keeping with these findings, expression of mtrA in M. tuberculosis H37Rv was detectable during in vitro growth, as determined by S1 nuclease protection and primer extension analyses of mRNA levels and mapping of transcript 5′ ends. The mtrA gene was expressed differently in virulent M. tuberculosis and the vaccine strain M. tuberculosis var. bovis BCG during infection of macrophages, as determined by monitoring of mtrA-gfp fusion activity. In M. bovis BCG, mtrA was induced upon entry into macrophages. In M. tuberculosis H37Rv, its expression was constitutive and unchanged upon infection of murine or human monocyte-derived macrophages. In conclusion, these results identify mtrA as an essential response regulator gene in M. tuberculosis which is differentially expressed in virulent and avirulent strains during growth in macrophages.
Tuberculosis remains the leading cause of death in the world from a single infectious agent (2). The capacity of Mycobacterium tuberculosis to establish infection within an individual and efficiently disseminate within the human population is mediated in part by its ability to survive within professional phagocytic cells, remain dormant over long periods of latent infection, and resume growth upon disease reactivation (27). The physiological and environmental signals during periods of active disease, dormancy, or disease reactivation are likely to contribute to M. tuberculosis adaptation during various stages of infection. One well-recognized class of ubiquitous bacterial regulatory elements associated with signal recognition and adaptive responses is that of the two-component signal transduction systems. Bacterial two-component systems regulate various functions, including transient adaptations, developmental phenomena, and production of secondary metabolites (reviewed in reference 16). In pathogenic organisms, two-component systems can also regulate expression of virulence determinants or factors that contribute to disease pathogenesis (reviewed in references 16 and 30). In addition to the majority of two-component systems, which modulate nonvital albeit important cellular functions, a limited number of essential two-component systems have also been described. Such systems, although rare, have been shown to regulate genes involved in cell cycle control (29) and membrane permeability (23).
M. tuberculosis encodes a number of two-component signal transduction systems. The MtrA-MtrB system was the first such system to be characterized in the tubercle bacillus (5, 7, 35). Since then, an additional 11 complete and 8 unlinked sensor kinase and response regulator homologs have been identified in the M. tuberculosis H37Rv genome (4, 12, 14, 21, 22, 33). Some of these two-component systems appear to be differentially regulated during growth within cultured macrophages in vitro. For example, expression of the mtrA response regulator (Rv3246c), which has been studied in M. bovis BCG, is induced in infected murine macrophages (7, 35). In addition, cDNAs corresponding to transcripts encoding the prrA response regulator (Rv0903c) and the sensor kinase prrB (Rv0902c) have been recovered from M. tuberculosis grown in human peripheral blood monocyte-derived macrophages but not from bacteria grown in standard laboratory medium (12). These limited examples reflect the preliminary nature of the initial analyses of M. tuberculosis two-component systems. In continuation of our characterization of the mtrA-mtrB system, we attempted to disrupt the mtrA gene in M. tuberculosis. Here we present data suggesting that mtrA is an essential gene in M. tuberculosis H37Rv. We also report the mapping of the 5′ end of the mtrA mRNA and its in vivo expression profiles in M. tuberculosis H37Rv and M. bovis BCG.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and electrotransformation.
M. tuberculosis H37Rv (ATCC 27294) and M. bovis BCG Pasteur (ATCC 27291) were used. All transformations done with Escherichia coli were performed with strain DH5α. Mycobacteria were grown under standard conditions in Middlebrook 7H9 broth or on Middlebrook 7H10 agar (Difco Laboratories) supplemented with 0.5% glycerol, 10% ADC or OADC (oleic acid-albumin-dextrose-catalase) (Difco) and 0.05% Tween 80 (Sigma) at 37°C in the presence of 5% CO2. E. coli was grown in LB medium (Difco) and incubated at 37°C. When required, Middlebrook or LB medium was supplemented with 25 or 50 μg of kanamycin sulfate (Sigma) per ml, 50 or 200 μg of hygromycin B (Boehringer Mannheim) per ml, 25 or 100 μg of streptomycin sulfate (Sigma) per ml, and 2 or 10% sucrose, respectively. Preparation of electrocompetent cells and transformation of M. tuberculosis were performed as previously described (18).
Construction of plasmid vectors.
Plasmid pmtrA-gfp has been described previously (7). Plasmid pTZ113 was used for the disruption of mtrA and was constructed as follows. A 2.7-kb SalI fragment containing the entire mtrA gene and the 5′ end of mtrB was filled in by treatment with Klenow enzyme and ligated into the SmaI site of pSM243, a mycobacterial suicide vector carrying the sacB gene. Next, a 1.2-kb NheI-SpeI fragment carrying the Kmr gene from pMV206 (31) was filled in and cloned into a blunt-ended BglII site in mtrA. Finally, a 2.0-kb NheI-XbaI fragment encoding xylE from pHSX-1 (5) was ligated into the XbaI site of the pSM243-derived polylinker. Temperature-sensitive (ts), mtrA+-complementing plasmid pTZ178 was constructed by first ligating the SalI fragment carrying mtrA into the SalI site of pUC12 to create pTZ100. An SpeI-NotI fragment encoding Hygr from pOLYG, a derivative of p16R1 (11), was filled in and ligated into the SmaI site of pTZ100 to create pTZ175. Finally, a filled-in EcoRV-KpnI fragment encoding the ts origin of replication from pCG63 (13) was ligated into the ScaI site of pTZ175 to create pTZ178. pTZ195 is a derivative of pTZ178 that lacks the SalI fragment encoding mtrA. Plasmid pTZ199 was constructed by ligating a 3.4-kb SpeI fragment encoding Strr from pSM240 into an SpeI-NheI fragment carrying xylE+ from pHSX-1 (5).
Genetic scheme for mtrA::Kmr gene replacement.
Plasmid pTZ113 (mtrA::Kmr sacB+ xylE+) was used for allelic exchange of mtrA+ with mtrA::Kmr in M. tuberculosis H37Rv via a two-step recombination process (Fig. 1). In the first step (integration), pTZ113 was transformed into M. tuberculosis H37Rv and recombinants were selected on 7H10 agar containing kanamycin. Merodiploid transformants were distinguished from spontaneous Kmr mutants by spraying colonies with 100 mM catechol (Fisher Scientific) (5). Transformants expressing xylE (detected as yellow colonies upon spraying with catechol) were screened for legitimate single-crossover homologous recombination by PCR using primers mtrAupstm2 (5′-CTGACCAAGCTGACCAAGGA-3′), which is a primer upstream of mtrA (and not carried on pTZ113), and KmUP2 (5′-GTAAGCAGACAGTTTTATTGTTCATGA-3′), which is a primer specific to and amplifying out of the Kmr cassette. mtrA::Kmr-mtrA+ merodiploids resulting from single-crossover homologous recombination were subsequently resolved of pTZ113 to leave mtrA::Kmr or mtrA+ in the chromosome by growth on 7H10 agar (with or without kanamycin) and sucrose, respectively (28). Colonies resistant to sucrose and white upon spraying with catechol (loss of sacB and xylE markers) were further screened by PCR using primers that flank the Kmr disruption site in mtrA (RC4 [5′-ACGTACCGGCGCGCACAAGGT-3′] and RC13 [5′-TCACGGAGGTCCGGCC-3′]) and primers that amplify an internal portion of xylE (xylEstart [5′-ATGAACAAAGGTGTAATGCG-3′] and xylEend [5′-GCGGTCGTGGTAAAAGATCG-3′]).
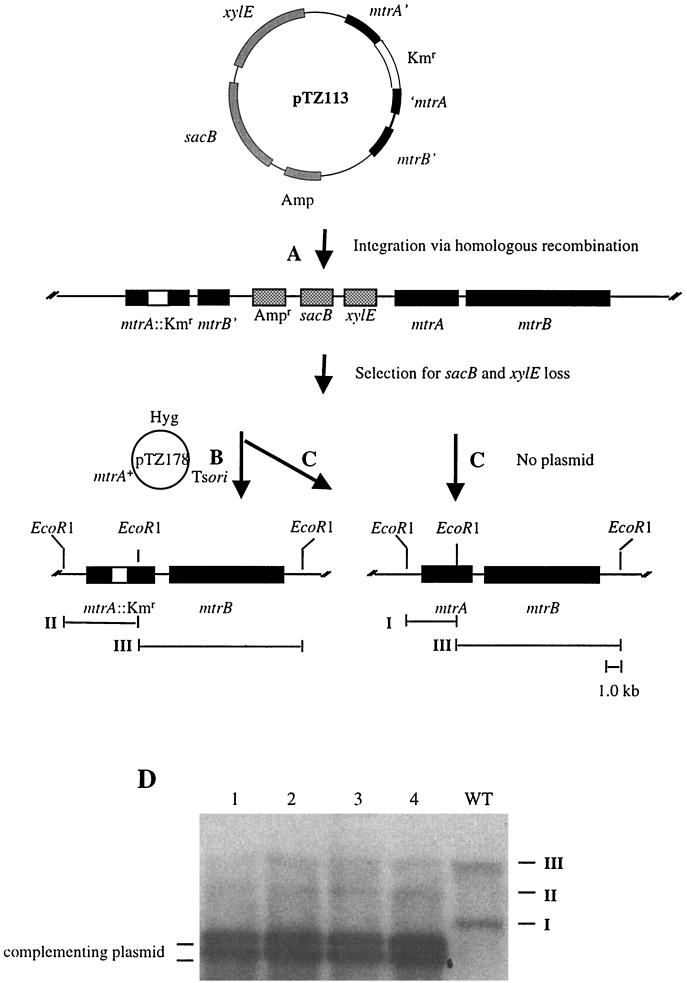
FIG. 1.
Genetic scheme for mtrA gene replacements in M. tuberculosis. (A) Mycobacterial suicide plasmid pTZ113 was integrated into the chromosome of M. tuberculosis H37Rv via legitimate single-crossover homologous recombination. (B and C) In the presence of plasmid pTZ178 (mtrA+), the merodiploid strain can be resolved upon selection on sucrose to either side of the Kmr cassette to leave the mtrA::Kmr disruption in the chromosome (B) or leave wild-type mtrA+ in the chromosome (C). In the absence of pTZ178 (no plasmid), only strains retaining wild-type mtrA+ are recovered (C). (D) Southern blot analysis of four independent strains with mtrA::Kmr allelic replacements on the chromosome obtained in the presence of pTZ178 (scheme B). EcoRI fragments: I, 3.4 kb; II, 4.6 kb; III, 7.1 kb. WT, wild type.
Addition of mtrA+ to mtrA::Kmr-mtrA+ merodiploid strains.
mtrA::Kmr-mtrA+ merodiploid recombinants resulting from single-crossover homologous recombination of pTZ113 into the M. tuberculosis H37Rv chromosome were transformed with pTZ178 [mtrA+ oriM (ts) Hygr], a conditionally replicating plasmid carrying mtrA+. Resolution of pTZ113 in these strains to leave mtrA::Kmr or mtrA+ in the chromosome was achieved by plating on 7H10 agar (with or without kanamycin) containing sucrose and hygromycin and growth at 30°C (permissive temperature for pTZ178 replication). The resulting recombinants were subjected to the screens previously described. Because pTZ178 contained a ts origin of replication, loss of the mtrA+ complementing plasmid in mtrA::Kmr mutants was attempted by growing strains at the nonpermissive temperature of 39°C. In addition, loss of pTZ178 was also attempted by introduction of a second plasmid, pTZ199 (pMV261 oriM xylE+ Strr), carrying the same origin of replication as pTZ178.
DNA extraction and Southern analysis.
Mycobacterial genomic DNA was prepared as previously described (18). A 4-μg sample of genomic DNA was digested overnight with EcoRI (Gibco BRL), separated by electrophoresis on a 0.8% agarose gel, transferred onto a Duralon-UV membrane (Stratagene), and used in subsequent high-stringency hybridization and washing steps (26). An mtrA-specific probe was generated by random-primed labeling (Gibco) with [α-32P]dCTP (3,000 Ci mmol−1; NEN Dupont) using PCR products generated with oligonucleotides RC4 and RC10 (5′-CCCATCACCCGGCACC-3′).
S1 nuclease protection and primer extension.
Total RNA from M. tuberculosis H37Rv was isolated as previously described (8). To generate a uniformly labeled single-stranded DNA (ssDNA) probe for S1 nuclease protection, a 1.7-kb SalI-PstI fragment carrying the M. tuberculosis H37Rv mtrA gene and upstream sequences was directionally cloned into an M13-based phagemid vector (24). 32P-radiolabeled ssDNA probes were prepared (25) using primers mtrAS5 (5′-TCGCCGATGACCGCGGTGTC-3′) and mtrAS6 (5′-AGCGGCTACTCCGCGGTGTCGAAGCCTTCC-3′). Probes generated from primer mtrAS6 contain a 10-bp overhang (underlined nucleotides) at the 5′ end that is not homologous to mtrA mRNA. Radiolabeled ssDNA polymerization products were digested with AgeI, heat denatured in formamide, and gel purified. Hybridization reactions were performed using 75 μg of total RNA from M. tuberculosis H37Rv, and S1 nuclease protection was carried out as previously described (25). Products of S1 digestion were analyzed on sequencing gels and compared with the corresponding sequencing ladders to locate mRNA 5′ ends. For primer extension, primer mtrAS8 (5′-TCCCCCCGCAGCACGATGGTGAGCATCTCA-3′) was end labeled with [γ-32P]ATP (6,000 Ci mmol−1; NEN Dupont) and purified on a Sephadex G-25 spin column (Boehringer Mannheim). Radiolabeled primer was added to 10 μg of total M. tuberculosis H37Rv RNA in hybridization buffer (0.5 M KCl, 0.25 M Tris-HCl, pH 8.3), and aliquots were denatured, annealed, and extended by the addition of 0.1 M dithiothreitol, 2.5 mM deoxynucleoside triphosphates, reverse transcription buffer, and Superscript II reverse transcriptase (Gibco). Extension reactions were carried out at 44°C for 45 min, and samples were loaded on a sequencing gel alongside the corresponding sequence ladder.
Preparation of mycobacteria, infection of macrophage monolayers, and fluorescence microscopy.
M. tuberculosis H37Rv or M. bovis BCG Pasteur was grown in static cultures until cells reached mid-exponential phase (optical density at 600 nm of 0.5). Bacterial cells were prepared for macrophage infection by washing in phosphate-buffered saline (PBS; pH 7.2) and resuspension either in Dulbecco's modified Eagle's medium (Bio Whittaker) supplemented with 10% fetal bovine serum (Hyclone) and 4 mM l-glutamine (Bio Whittaker) or in RPMI 1640 medium (Bio Whittaker) supplemented with 5% human AB serum (Sigma) and 2 mM l-glutamine. Single-cell bacterial suspensions were obtained by vortexing bacteria with 3-mm glass beads (Fisher), low-speed centrifugation, and passage of the resulting supernatant through a 5-μm-pore-size filter (Micron Separations Inc.). The number of organisms was determined by staining with Bac-Light (Molecular Probes) and counting in a hemocytometer. Mycobacteria were used to infect murine BALB/c macrophage cell line J774A (ATCC TIB-67) or human macrophages derived from peripheral blood monocytes (3) obtained from the American Red Cross. J774 cells and human monocyte-derived macrophages were cultured before infection and maintained during infection in supplemented Dulbecco's modified Eagle's medium and RPMI medium, respectively. Both murine and human macrophage monolayers were maintained at 37°C in humidified air containing 5% CO2. For infections, macrophage monolayers were established by plating 105 cells per well in 12-well tissue culture plates (Corning) containing no. 1 thickness, 18-mm-diameter glass coverslips (Fisher). Macrophages were infected with mycobacteria at a multiplicity of infection of 10 bacilli per macrophage. Macrophages were allowed to take up bacteria for 2 h before extracellular bacteria were removed by washing in PBS. Macrophages were incubated for 2 h, 3 days, or 5 days before harvest with no apparent damage. At harvest, macrophage monolayers were washed in PBS, fixed in 3.8% paraformaldehyde, and mounted on glass slides with Permafluor (Lipshaw Immunon). Epifluorescence images were captured using a Kodak Kaf 1400-2 Olympix camera connected to an Olympus BX60 microscope. Images were captured with a shutter speed of 500 ms and analyzed using Espirit software (Life Sciences Resources). NIH Image (version 1.62; National Institutes of Health) was used to quantitate mean pixel density from individual bacilli present within monolayers. At the settings used, macrophage autofluorescence was not observed. Statistical analysis (analysis of variance [ANOVA] and Fisher's protected least significant difference) was performed with ANOVA (version 1.11; Abicus Software).
RESULTS AND DISCUSSION
Gene replacements with mtrA::Kmr in M. tuberculosis H37Rv.
To further examine the role of mtrA in M. tuberculosis, we set out to disrupt mtrA in strain H37Rv. We constructed a mycobacterial suicide plasmid, pTZ113 (see Materials and Methods), that carried a copy of mtrA disrupted by a Kmr cassette, the counterselectable marker sacB, and the xylE gene as a convenient scorable marker for subsequent recombination steps. Following electroporation of pTZ113 into M. tuberculosis H37Rv, we obtained recombinants expressing xylE (detected as yellow colonies upon spraying with catechol), of which 2.6% had undergone legitimate single-crossover homologous recombination into the chromosome, resulting in a tandem mtrA::Kmr-mtrA+ merodiploid (Fig. 1A and Table 1, row A). The low frequency of legitimate single-crossover homologous recombination was most likely a result of illegitimate integration of pTZ113 into the M. tuberculosis genome (1, 19). Subsequent attempts to produce the desired mtrA::Kmr gene replacements by resolving the mtrA::Kmr-mtrA+ merodiploids via a second crossover after selection against sacB failed (Fig. 1B and Table 1, row B). None of the colonies recovered as resistant to sucrose (loss of the sacB marker) and not expressing xylE (white upon spraying with catechol) were true double-crossover recombinants and most likely harbored other types of mutations eliminating or precluding sacB and xylE activity (data not shown). The inability to obtain an mtrA::Kmr gene replacement was not simply the result of inefficient resolution by homologous recombination, because double-crossover recombinants which had lost the plasmid moiety along with mtrA::Kmr, leaving the wild-type copy of mtrA in the chromosome, occurred efficiently (Fig. 1C and Table 1, row C).
TABLE 1.
Screening for mtrA::Kmr gene replacements in M. tuberculosis H37Rv
| Row | Stepa | Parental strain plasmid(s)b | Selection/ screen | Expected genotype | Resulting strain plasmidc | No. of colonies screened | % Legitimate eventsd |
|---|---|---|---|---|---|---|---|
| A | Integration | pTZ113 (mtrA::KmrxylE+ sacB+) | Kmr/XylE+ Sucs | mtrA::Kmr-mtrA+ | Cointegrate | 191 | 2.6 |
| B | Resolution | Cointegrate | Sucr/XylE− Kmr | mtrA::Kmr | — | 500 | 0 |
| C | Resolution | Cointegrate | Sucr/XylE− Kms | mtrA+ | — | 600 | 100 |
| D | Resolution | pTZ178 (mtrA+ Hygr) | Sucr/XylE− Kmr | mtrA::Kmr | pTZ178 (mtrA+ Hygr) | 200 | 96 |
| E | pTZ178 loss | pTZ178 (mtrA+ Hygr) | 39°C/Hygs | mtrA::Kmr | — | 100 | 0 |
| F | pTZ193 loss | pTZ193 (Hygr) | 39°C/Hygs | mtrA::Kmr-mtrA+ | — | 200 | 100 |
| G | pTZ178 loss | pTZ178 (mtrA+ Hygr), pTZ199 (Strr) | Strr/Hygs | mtrA::Kmr | pTZ199 (Strr) | 100 | 0 |
| H | pTZ193 loss | pTZ193 (Hygr), pTZ199 (Strr) | Strr/Hygs | mtrA::Kmr-mtrA+ | pTZ199 (Strr) | 100 | 100 |
Steps involved in the construction or resolution of the mtrA disruption. Parental strains were M. tuberculosis H37Rv (row A), M. tuberculosis H37Rv mtrA::Kmr-mtrA+ sacB+ xylE+ (rows B and C), M. tuberculosis H37Rv mtrA::Kmr-mtrA+(pTZ178) (rows D, E, and G), and M. tuberculosis H37Rv mtrA::Kmr-mtrA+(pTZ193) (rows F and H).
Plasmids were pTZ113 (pUC18 mtrA::Kmr sacB+ xylE+ [Fig. 1]), pTZ178 [pUC12 oriM (ts) Hygr mtrA+], pTZ193 [pUC12 oriM (ts) Hygr], and pTZ199 (pMV261 oriM xylE+ Strr). Cointegrate refers to pTZ113 integrated on the chromosome of H37Rv via single-crossover homologous recombination with the mtrA gene.
—, no plasmid.
A legitimate event is achievement of the expected genotype.
The mtrA gene is an essential response regulator in M. tuberculosis.
To test further whether the encountered difficulties in inactivating mtrA could be explained by its potentially essential function, we introduced into the mtrA::Kmr-mtrA+ merodiploids plasmid pTZ178 containing an mtrA+ copy on a conditionally replicating (ts) mycobacterial shuttle vector and repeated the selection procedure for mtrA::Kmr gene replacements. This time, in the presence of plasmid-borne mtrA+, true mtrA::Kmr gene replacements were obtained via a second crossover on the chromosome (Fig. 1B and Table 1, row D). Southern hybridization analysis performed on four randomly selected recombinants confirmed the replacement of mtrA+ with the mtrA::Kmr allele on the M. tuberculosis chromosome (Fig. 1D). Two EcoRI chromosomal fragments in wild-type M. tuberculosis H37Rv hybridized with the mtrA probe (3.4- and 7.1-kb fragments I and III, respectively). In mtrA::Kmr recombinants, fragment I was lost but, instead, a new fragment (fragment II) of 4.6 kb (corresponding to 3.4-kb EcoRI fragment I carrying the 1.2-kb Kmr insert) was detected in each of the four recombinants tested. These strains, however, also harbored a plasmid (pTZ178) borne wild-type mtrA gene.
Because pTZ178 carried a mycobacterial ts origin of replication, we next tried to eliminate the plasmid by growing the mtrA::Kmr (pTZ178) strains at the nonpermissive temperature of 39°C. However, none of the colonies arising following growth at the nonpermissive temperature lost the complementing plasmid (Table 1, row E). In contrast, loss of pTZ193 (a derivative of pTZ178 lacking the mtrA gene) from the mtrA::Kmr-mtrA+ merodiploid parental strain was observed at high frequency (100%) upon growth at 39°C (Table 1, row F). Because two plasmids containing the same origin of replication cannot be maintained simultaneously in the same cell, we also tried to eliminate pTZ178 from mtrA::Kmr mutants by introduction of plasmid pTZ199. This approach also failed to cure the mtrA+ plasmid from mtrA::Kmr mutants despite the fact that the merodiploid parental strain carrying pTZ193 could be cured of this plasmid (Table 1, rows G and H). Based on these experiments, we conclude that mtrA encodes a response regulator that is essential for M. tuberculosis viability in vitro. The viability-associated function resided within the mtrA gene and was not due to polar effects on mtrB, as it was possible to knockout mtrA on the chromosome in the presence of the mtrA+ complementing plasmid, which did not contain a complete mtrB gene (Fig. 1D).
Expression of mtrA in M. tuberculosis grown outside the host.
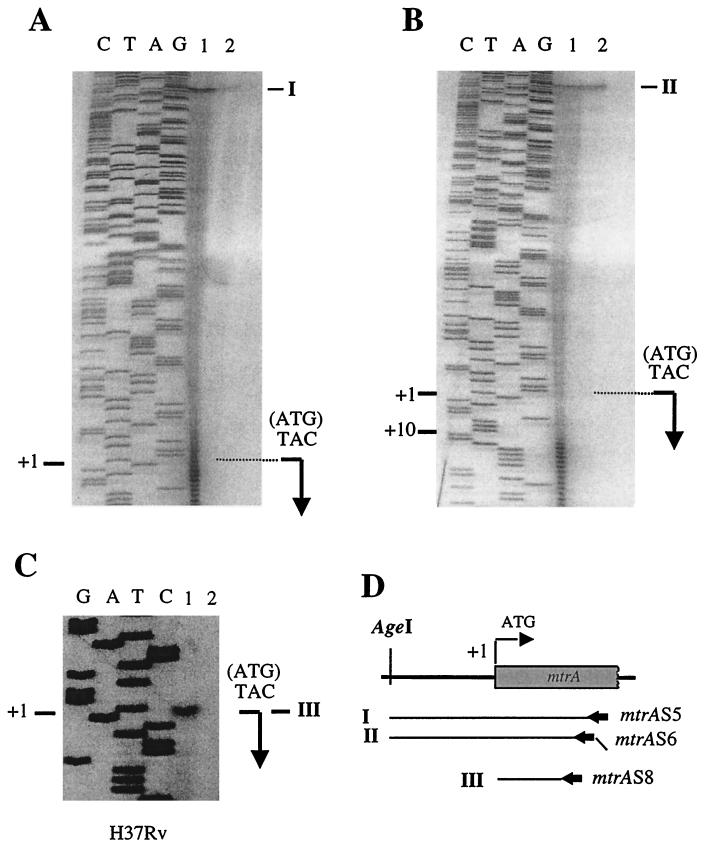
The inability to recover viable mtrA mutants would require that mtrA be expressed in vitro. Expression of the mtrA gene is inducible in M. bovis BCG during growth in cultured J774 macrophages (35). However, this does not preclude the possibility that mtrA is expressed at baseline levels in vitro outside the host. To test this possibility, we analyzed mtrA transcription and mapped mtrA mRNA 5′ ends in M. tuberculosis H37Rv by S1 nuclease protection and primer extension analyses. Total cellular RNA was isolated from bacteria grown in 7H9 medium and hybridized with a uniformly labeled ssDNA probe, and the products of the hybridization reaction were digested with S1 nuclease. In keeping with our prediction that mtrA was expressed during in vitro growth, a band of protection corresponding to mtrA transcripts was observed (Fig. 2A). The 5′ end of the protected fragments corresponded to the mtrA initiation codon. Similar results were obtained using a probe (Fig. 2D) generated with a primer that contained a 5′ (10-bp) overhang that did not correspond to the mtrA sequence. The addition of the heterologous 10-bp overhang sequence resulted in the corresponding reduction in the size of the protected fragment obtained upon S1 nuclease treatment (Fig. 2B), confirming that the assigned transcript was mtrA specific and indicating that its 5′ end coincided with the translational initiation site. Due to the intrinsic heterogeneity of the products of uniformly labeled S1 nuclease probes, which introduced some uncertainty regarding the exact position of the mRNA 5′ end, it was important to test whether the mtrA mRNA 5′ end included the mtrA translational start. With this aim, we performed primer extension analysis. The results of these experiments indicated that the translation and transcriptional initiation start sites of mtrA overlap (Fig. 2C). Thus, in M. tuberculosis H37Rv, mtrA is expressed in vitro from a transcript with the 5′ mRNA end overlapping the translational start site. Overlapping transcriptional and translational start sites have been identified for several other mycobacterial genes, including the major oxyR transcript of M. leprae (6) and the furA promoter from M. tuberculosis (unpublished data). In addition, a large number of genes from actinomycetes, a phylogenetic group closely related to mycobacteria, also contain overlapping transcriptional and translational sites (32). In conclusion, the in vitro expression of mtrA in M. tuberculosis H37Rv was in keeping with our finding that mtrA is essential for the viability of this organism.
FIG. 2.
S1 nuclease mapping and primer extension analysis of the mtrA promoter. (A and B) S1 nuclease protection was performed with total cellular RNA from M. tuberculosis H37Rv and uniformly radiolabeled, single-stranded mtrA probes synthesized using primer mtrAS5 (I) or mtrAS6 (II) ending at the AgeI site. mtrAS6 contained a 10-bp overhang of an irrelevant sequence at the 5′ end which did not correspond to mtrA. Lanes: 1, S1 digestion; 2, probe loaded in amounts diluted 100× relative to lane 1. The bent arrow indicates the location of the translational start site (+1). Note that S1 nuclease products in panel A coincide with +1 while in B they are at +10 due to the 5′ overhang of probe II that is not complementary to mtrA mRNA. (C) Primer extension analysis. Reverse transcription was performed using 10 μg of total cellular RNA from M. tuberculosis H37Rv and end-labeled primer mtrAS8 (extension product III). Lanes: 1, reaction mixture containing RNA; 2, control reaction mixture without RNA. (D) Schematic representation of S1 nuclease probes (I and II), reverse transcription products (III), and the primers (mtrAS5, -S6, and -S8) used to generate them.
The mtrA promoter is induced in M. bovis BCG but is constitutively active in M. tuberculosis H37Rv within macrophages.
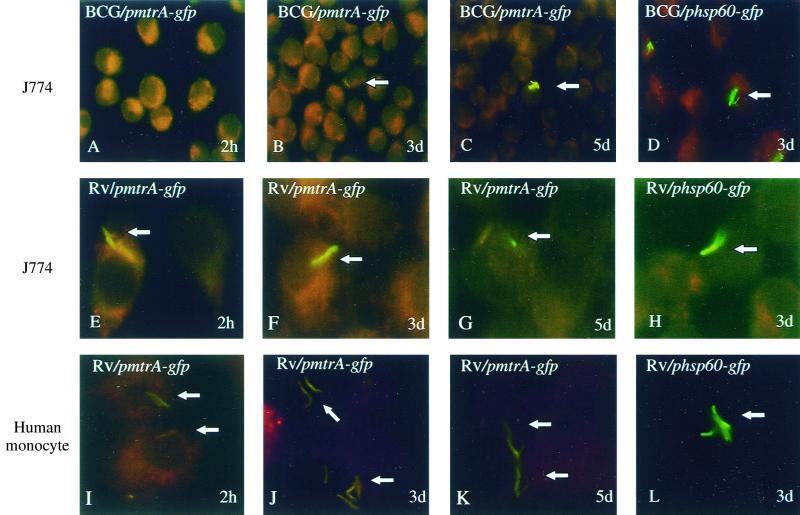
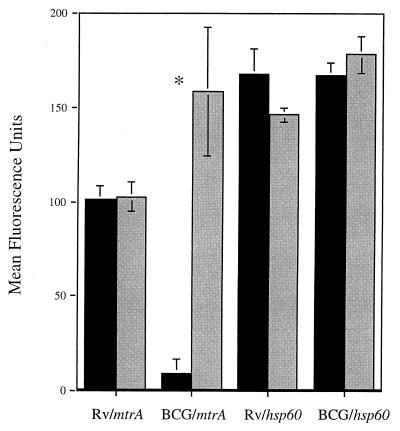
To examine mtrA expression in M. tuberculosis during intracellular growth, we tested whether the mtrA induction previously detected in M. bovis BCG could also be observed in M. tuberculosis H37Rv during infection of J774 murine macrophages. As reported previously, green fluorescent protein fluorescence in M. bovis BCG carrying pmtrA-gfp became detectable after 3 days of incubation in J774 cells (Fig. 3A to C) (35). However, green fluorescent protein fluorescence from pmtrA-gfp was bright in M. tuberculosis H37Rv even prior to infection and no further induction was observed during growth in J774 murine macrophages over a 5-day period (Fig. 3E to G). As a control, the previously characterized hsp60-gfp fusion (7) was fluorescent in both M. bovis BCG and M. tuberculosis H37Rv during macrophage infection (Fig. 3D and H and data not shown). Next we tested the expression of the mtrA-gfp fusion in M. tuberculosis H37Rv cells in human macrophages derived from peripheral blood monocytes. These experiments showed similar results (Fig. 3I to L). A quantitative analysis of fluorescence in J774 cells is shown in Fig. 4. Different expression in M. bovis BCG and M. tuberculosis H37Rv has also been observed with other two-component response regulators from M. tuberculosis (T.C.Z. and V.D., unpublished results). In conclusion, mtrA is expressed during in vitro growth in virulent M. tuberculosis and its expression differs significantly between M. tuberculosis H37Rv and the vaccine strain M. bovis BCG during growth in macrophages.
FIG. 3.
Expression of mtrA in infected macrophages monitored by mtrA-gfp fusion and epifluorescence microscopy. M. tuberculosis H37Rv or M. bovis BCG carrying the pmtrA-gfp or phsp60-gfp reporter plasmid was used to infect macrophages, which were incubated for 2 h, 3 days, or 5 days before the cells were fixed and prepared for fluorescence microscopy analysis. Panels: A to H, J774 murine macrophage-like cell line; I to L, human peripheral blood monocyte-derived macrophages. Arrows point at the bacteria within the macrophages.
FIG. 4.
Quantitation of fluorescence intensities of mtrA-gfp and hsp60-gfp fusions in M. tuberculosis H37Rv and M. bovis BCG infecting J774 cells. Mean fluorescence intensities (± the standard errors) were determined as described in Materials and Methods. Macrophages were infected for 2 h (black bars) or 3 days (grey bars) with M. tuberculosis H37Rv or M. bovis BCG Pasteur carrying the pmtrA-gfp or phsp60-gfp reporter plasmid. ∗, significant difference in fluorescence intensity (P < 0.05 by ANOVA).
Essential two-component systems in bacteria.
To our knowledge, this is the first report of an essential two-component signal transduction system in M. tuberculosis. Although rare, essential two-component systems have been reported in other bacterial species. For example, Caulobacter crescentus encodes two essential two-component systems, CtrA-CckA (17, 29) and DivK-DivJ (15, 36), that are required for cell cycle regulation in this organism. The CtrA-CckA system has been shown to regulate genes involved in at least five distinct cell cycle events, including flagellar biogenesis, DNA methylation, and DNA replication (29). The CtrA response regulator also controls the differentiation of the swarmer cell type into the stalked cell type by directly binding to sites present within the chromosomal origin of replication, thus blocking an essential DnaA box and promoter necessary for replication initiation (29). Interestingly, the other essential two-component system, DivK-DivJ, appears to mediate cell cycle regulation through the CtrA-CckA system, adding further complexity to this already multicomponent regulatory system (17, 36). In Bacillus subtilis, an essential two-component signal transduction system, yycF-yycG, has also been described (9, 10); however, the process(es) regulated by this system remains unknown. In addition, essential two-component systems have also been identified in pathogenic organisms, as an essential two-component system from Staphylococcus aureus showing high similarity to yycF-yycG from B. subtilis has recently been reported (23). Initial characterization of this system in S. aureus suggests that its role includes the proper regulation of bacterial cell wall or membrane composition (23). In addition, among the 13 two-component signal transduction systems present in Streptococcus pneumoniae, one two-component response regulator could not be inactivated (20, 34), suggesting that this system is also required for an essential cellular function.
Although mtrB is located immediately downstream of mtrA and probably encodes its cognate sensor histidine kinase (35), our results suggest that mtrB is not essential for the growth of M. tuberculosis in vitro. The identification of a nonessential histidine kinase closely linked to an essential response regulator has also been observed in S. pneumoniae (34); however, other essential two-component systems appear to require both a response regulator and its cognate histidine kinase for growth in vitro (9, 10, 17, 23, 29). It is not known whether mtrA is also essential in M. bovis BCG. As low-level expression of mtrA may be sufficient to exert its function, we anticipate that mtrA could be essential for the growth of M. bovis BCG in vitro. Regardless, the presence of an essential two-component signal transduction system in M. tuberculosis underscores the need for further characterization of the functions controlled by this and other regulators of this type in the tubercle bacillus.
ACKNOWLEDGMENTS
We thank V. Vishwanath for the preparation of human peripheral blood monocyte-derived macrophages. Plasmid pCG63 was kindly provided by Brigitte Gicquel.
This work was supported by a National Research Service award (AI10278) to T.C.Z. and by grants (AI35217 and AI42999) from the National Institute of Allergy and Infectious Diseases to V.D.
REFERENCES
- 1.Aldovini A, Husson R N, Young R A. The uraA locus and homologous recombination in Mycobacterium bovis BCG. J Bacteriol. 1993;175:7282–7289. doi: 10.1128/jb.175.22.7282-7289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloom B R, Murray C J. Tuberculosis: commentary on a reemergent killer. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 3.Boyum A. Separation of leukocytes from blood and bone marrow. Introduction. Scand J Clin Lab Investig Suppl. 1968;97:7. [PubMed] [Google Scholar]
- 4.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 5.Curcic R, Dhandayuthapani S, Deretic V. Gene expression in mycobacteria: transcriptional fusions based on xylE and analysis of the promoter region of the response regulator mtrA from Mycobacterium tuberculosis. Mol Microbiol. 1994;13:1057–1064. doi: 10.1111/j.1365-2958.1994.tb00496.x. [DOI] [PubMed] [Google Scholar]
- 6.Dhandayuthapani S, Mudd M, Deretic V. Interactions of OxyR with the promoter region of the oxyR and ahpC genes from Mycobacterium leprae and Mycobacterium tuberculosis. J Bacteriol. 1997;179:2401–2409. doi: 10.1128/jb.179.7.2401-2409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhandayuthapani S, Via L E, Thomas C A, Horowitz P M, Deretic D, Deretic V. Green fluorescent protein as a marker for gene expression and cell biology of mycobacterial interactions with macrophages. Mol Microbiol. 1995;17:901–912. doi: 10.1111/j.1365-2958.1995.mmi_17050901.x. [DOI] [PubMed] [Google Scholar]
- 8.Dhandayuthapani S, Zhang Y, Mudd M H, Deretic V. Oxidative stress response and its role in sensitivity to isoniazid in mycobacteria: characterization and inducibility of ahpC by peroxides in Mycobacterium smegmatis and lack of expression in M. aurum and M. tuberculosis. J Bacteriol. 1996;178:3641–3649. doi: 10.1128/jb.178.12.3641-3649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabret C, Feher V A, Hoch J A. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J Bacteriol. 1999;181:1975–1983. doi: 10.1128/jb.181.7.1975-1983.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabret C, Hoch J A. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol. 1998;180:6375–6383. doi: 10.1128/jb.180.23.6375-6383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garbe T R, Barathi J, Barnini S, Zhang Y, Abou-Zeid C, Tang D, et al. Transformation of mycobacterial species using hygromycin resistance as selectable marker. Microbiology. 1994;140:133–138. doi: 10.1099/13500872-140-1-133. [DOI] [PubMed] [Google Scholar]
- 12.Graham J E, Clark-Curtiss J E. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS) Proc Natl Acad Sci USA. 1999;96:11554–11559. doi: 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guilhot C, Gicquel B, Martin C. Temperature-sensitive mutants of the Mycobacterium plasmid pAL5000. FEMS Microbiol Lett. 1992;77:181–186. doi: 10.1016/0378-1097(92)90152-e. [DOI] [PubMed] [Google Scholar]
- 14.Haydel S E, Dunlap N E, Benjamin W H., Jr In vitro evidence of two-component system phosphorylation between the Mycobacterium tuberculosis TrcR/TrcS proteins. Microb Pathog. 1999;26:195–206. doi: 10.1006/mpat.1998.0265. [DOI] [PubMed] [Google Scholar]
- 15.Hecht G B, Lane T, Ohta N, Sommer J M, Newton A. An essential single domain response regulator required for normal cell division and differentiation in Caulobacter crescentus. EMBO J. 1995;14:3915–3924. doi: 10.1002/j.1460-2075.1995.tb00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. [Google Scholar]
- 17.Jacobs C, Domian I J, Maddock J R, Shapiro L. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell. 1999;97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs W R, Jr, Kalpana G V, Cirillo J D, Pascopella L, Snapper S B, Udani R A, et al. Genetic systems for mycobacteria. Methods Enzymol. 1991;204:537–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- 19.Kalpana G V, Bloom B R, Jacobs W R., Jr Insertional mutagenesis and illegitimate recombination in mycobacteria. Proc Natl Acad Sci USA. 1991;88:5433–5437. doi: 10.1073/pnas.88.12.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lange R, Wagner C, de Saizieu A, Flint N, Molnos J, Stieger M, et al. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene. 1999;237:223–234. doi: 10.1016/s0378-1119(99)00266-8. [DOI] [PubMed] [Google Scholar]
- 21.Magdalena J, Supply P, Locht C. Specific differentiation between Mycobacterium bovis BCG and virulent strains of the Mycobacterium tuberculosis complex. J Clin Microbiol. 1998;36:2471–2476. doi: 10.1128/jcm.36.9.2471-2476.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magdalena J, Vachée A, Supply P, Locht C. Identification of a new DNA region specific for members of Mycobacterium tuberculosis complex. J Clin Microbiol. 1998;36:937–943. doi: 10.1128/jcm.36.4.937-943.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin P K, Li T, Sun D, Biek D P, Schmid M B. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J Bacteriol. 1999;181:3666–3673. doi: 10.1128/jb.181.12.3666-3673.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Misra T K. DNA sequencing: a new strategy to create ordered deletions, modified M13 vector, and improved reaction conditions for sequencing by dideoxy chain termination method. Methods Enzymol. 1987;155:119–139. doi: 10.1016/0076-6879(87)55012-1. [DOI] [PubMed] [Google Scholar]
- 25.Mohr C D, Martin D W, Konyecsni W M, Govan J R, Lory S, Deretic V. Role of the far-upstream sites of the algD promoter and the algR and rpoN genes in environmental modulation of mucoidy in Pseudomonas aeruginosa. J Bacteriol. 1990;172:6576–6580. doi: 10.1128/jb.172.11.6576-6580.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagan-Ramos E, Song J, McFalone M, Mudd M H, Deretic V. Oxidative stress response and characterization of the oxyR-ahpC and furA-katG loci in Mycobacterium marinum. J Bacteriol. 1998;180:4856–4864. doi: 10.1128/jb.180.18.4856-4864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parrish N M, Dick J D, Bishai W R. Mechanisms of latency in Mycobacterium tuberculosis. Trends Microbiol. 1998;6:107–112. doi: 10.1016/s0966-842x(98)01216-5. [DOI] [PubMed] [Google Scholar]
- 28.Pelicic V, Reyrat J M, Gicquel B. Expression of the Bacillus subtilis sacB gene confers sucrose sensitivity on mycobacteria. J Bacteriol. 1996;178:1197–1199. doi: 10.1128/jb.178.4.1197-1199.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quon K C, Marczynski G T, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 30.Rappuoli R, Scarlato V, Arico B, editors. Signal transduction and bacterial virulence. R. G. Austin, Tex: Landes Company; 1995. [Google Scholar]
- 31.Stover C K, de la Cruz V F, Fuerst T R, Burlein J E, Benson L A, Bennett L T, et al. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 32.Strohl W R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992;20:961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Supply P, Magdalena J, Himpens S, Locht C. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol Microbiol. 1997;26:991–1003. doi: 10.1046/j.1365-2958.1997.6361999.x. [DOI] [PubMed] [Google Scholar]
- 34.Throup J P, Koretke K K, Bryant A P, Ingraham K A, Chalker A F, Ge Y, et al. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol Microbiol. 2000;35:566–576. doi: 10.1046/j.1365-2958.2000.01725.x. [DOI] [PubMed] [Google Scholar]
- 35.Via L E, Curcic R, Mudd M H, Dhandayuthapani S, Ulmer R J, Deretic V. Elements of signal transduction in Mycobacterium tuberculosis: in vitro phosphorylation and in vivo expression of the response regulator MtrA. J Bacteriol. 1996;178:3314–3321. doi: 10.1128/jb.178.11.3314-3321.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu J, Ohta N, Newton A. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc Natl Acad Sci USA. 1998;95:1443–1448. doi: 10.1073/pnas.95.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]