Abstract
Streptococcus pneumoniae (the pneumococcus) is a leading cause of childhood mortality globally and in Cambodia. It is commensal in the human nasopharynx, occasionally resulting in invasive disease. Monitoring population genetic shifts, characterized by lineage and serotype expansions, as well as antimicrobial-resistance (AMR) patterns is crucial for assessing and predicting the impact of vaccination campaigns. We sought to elucidate the genetic background (global pneumococcal sequence clusters; GPSCs) of pneumococci carried by Cambodian children following perturbation by pneumococcal conjugate vaccine (PCV) 13. We sequenced pre-PCV13 (01/2013–12/2015, N=258) and post-PCV13 carriage isolates (01/2016–02/2017, N=428) and used PopPUNK and SeroBA to determine lineage prevalence and serotype composition. Following PCV13 implementation in Cambodia, we saw expansions of non-vaccine type (NVT) serotypes 23A (GPSC626), 34 (GPSC45) and 6D (GPSC16). We predicted antimicrobial susceptibility using the CDC-AMR pipeline and determined concordance with phenotypic data. The CDC-AMR pipeline had >90 % concordance with the phenotypic antimicrobial-susceptibility testing. We detected a high prevalence of AMR in both expanding non-vaccine serotypes and residual vaccine serotype 6B. Persistently high levels of AMR, specifically persisting multidrug-resistant lineages, warrant concern. The implementation of PCV13 in Cambodia has resulted in NVT serotype expansion reflected in the carriage population and driven by specific genetic backgrounds. Continued monitoring of these GPSCs during the ongoing collection of additional carriage isolates in this population is necessary.
Keywords: AMR, Cambodia, carriage, genomics, PCV, Streptococcus pneumoniae
Data Summary
The sequencing reads for the genomes analysed have been deposited in the European Nucleotide Archive and the accession numbers for each isolate are listed in Table S1 (available with the online version of this article). A phylogenetic tree and associated metadata are available for download and visualization on the Microreact website: https://microreact.org/project/mvgn3EvNgmxAcPjPBnyFMj/a32e0dc6.
Impact Statement.
Streptococcus pneumoniae was a leading cause of morbidity and mortality in Cambodian children in 2015; however, in the same year, pneumococcal conjugate vaccine (PCV) 13 was included in the routine immunization schedule. Pneumococcal disease is necessarily preceded by carriage. In this study, we analyse pre- and post-PCV13 pneumococcal carriage populations from Cambodian children. Our study is concordant with our previous research [16], which identified a decrease in vaccine serotypes after vaccination. We contextualize these serotypes by their genetic backgrounds, including antimicrobial-resistance (AMR) analysis. We identify several lineages containing non-vaccine serotypes that expanded in the year after vaccination. Furthermore, we characterize AMR, identifying persistence of a multidrug-resistant lineage despite its vaccine type serotype. We also identify >90 % concordance between the phenotypic and genotypic AMR confirming the use of in silico AMR predictions in Cambodia.
Introduction
Streptococcus pneumoniae (the pneumococcus) is an opportunistic human pathogen; colonization is a prerequisite for disease. Its occupation of the nasopharynx has a range of outcomes from asymptomatic carriage to life-threatening invasive pneumococcal disease (IPD) [1]. Estimates of pneumococcal-related deaths in 2015 were 318 000, with 3.7 million cases of severe disease worldwide [2]. IPD is under-detected in low-income countries with high child mortality rates due to limited access to healthcare, scarce diagnostic microbiology capacity and antibiotic use prior to testing, making this a likely underestimate [3, 4].
Pneumococcal conjugate vaccines (PCVs) are given to infants globally; PCVs currently target up to 13 capsular serotypes (1, 3, 4, 5, 6A, 6B, 7F, 9V, 14, 18C, 19A, 19F, 23F) that account for most invasive disease in children aged 2 years and under. PCVs were broadly used in 145 countries by 2020 and have substantially reduced vaccine-serotype-associated pneumococcal disease among children [5, 6]. Furthermore, there was an estimated 51 % decline in pneumococcal-related deaths globally from 2000 to 2015, which can likely be explained by global deployment of PCV [2].
Currently, over 100 capsular polysaccharide serotypes have been identified on the genetic backbones of over 900 global lineages, called global pneumococcal sequence clusters (GPSCs) [7]. It is not uncommon for a single GPSC to undergo capsular switching and express different serotypes. Some GPSCs have a propensity for high diversity of capsular types, while others are restricted to just a few [8]. GPSCs represent the genetic background of pneumococci and, henceforth, will be referred to as GPSCs and lineages interchangeably. Understanding the genetic background upon which vaccine-related serotype switching occurs can further elucidate lineages with the potential to perpetuate troublesome phenotypes, such as antibiotic resistance and invasiveness. Understanding the lineages driving post-vaccine serotype expansion will be useful to guide future pneumococcal disease prevention strategies [8].
PCVs are designed to decrease nasopharyngeal infection with serotypes associated with systemic illness to protect children from IPD [9]. Children are the primary reservoirs and transmission vectors for pneumococci [6, 10, 11]. As such, pneumococcal population ecology within the nasopharynges of children is an indicator of IPD prevalence and vaccine impact within a population [12].
In Cambodia in 2015, pneumococcal pneumonia was estimated to be the leading cause of lower respiratory infection associated with death in children under 5 years, and the second leading cause of morbidity and mortality [13]. In the same year, PCV13 was added to the national routine childhood immunization schedule. Several studies elucidating serotype distribution have been conducted in Cambodia; however, contextualizing these serotypes by their genetic background and continued evaluation of the population ecology is necessary to both determine vaccine impact and elucidate lineages that are driving any post-PCV upswing in disease or antimicrobial resistance (AMR) [14–18]. Disease and carriage populations do not exist in isolation from one another. The prevalence in one reflects the eventual prevalence of the other,although highly invasive serotypes such as serotype 1 are an exception to this due to their ability to rapidly cross the respiratory mucosa remaining undetectable in carriage [19, 20]. Understanding the genetic background of carriage populations regionally can indicate which serotypes may emerge following PCV13 and may elucidate future drivers of IPD. This study aims to characterize the impact of PCV13 on the GPSC and serotype prevalence pre- and post-PCV13 implementation in a Cambodian paediatric carriage population.
Methods
Bacterial isolate collection
Nasopharyngeal S. pneumoniae were isolated from healthy (without pneumonia or IPD) children under 5 years old visiting the Angkor Hospital for Children, in Siem Reap, Cambodia, between 2013 and 2017. Children were visiting the outpatient department with minor illnesses. Children who were sick enough to see a doctor or be admitted to hospital were excluded. These isolates were collected as part of our larger carriage studies in 2015 and 2020 [16, 18].
Microbiology and sequencing
As part of the Global Pneumococcal Sequencing (GPS) Project [7, 8], a random subset (61.8 %) of pneumococcal carriage isolates from the previous studies [16, 18] were selectively re-cultured on BD trypticase soy agar II with 5 % sheep blood (Beckton Dickinson) and incubated overnight at 37 °C in 5 % CO2. Genomic DNA was then extracted manually using a modified QIAamp DNA mini kit (Qiagen) protocol. Whole-genome sequencing (WGS) was performed on the Illumina HiSeq platform to produce paired-end reads with a mean of 125–151 bases in length. Data were deposited in the European Nucleotide Archive database. WGS data were processed as previously described [7]. Phenotypic antimicrobial-susceptibility testing was conducted against penicillin (PEN), erythromycin (ERY), ceftriaxone (CRO), co-trimoxazole (COT), chloramphenicol (CAT) and tetracycline (TET), using disc diffusion and/or Etests (penicillin/ceftriaxone; bioMérieux) on 5 % citrated sheep blood Mueller–Hinton agar (Oxoid) [21, 22]. All the isolates were classified as susceptible, intermediate or resistant based on Clinical and Laboratory Standards Institute (CLSI) guidelines (M100-ED28 : 2018). The meningitis cut-off was used to interpret the penicillin susceptibility on all isolates; MIC (minimum inhibitory concentration) ≤0.06 mg l−1 was categorized as sensitive, while ≥0.12 mg l−1 was categorized as resistant. Multidrug resistance was defined as isolates’ resistance to ≥3 classes of antibiotics.
Classification and antimicrobial-susceptibility testing
In brief, in silico serotypes and pneumococcal lineages or GPSCs were determined using SeroBA [23] and PopPUNK [24], respectively. We performed predictive resistance profiling using the CDC-AMR pipeline for six antimicrobials, including penicillin (encoded by the genes pbp1A, pbp2B, pbp2X) [25, 26], chloramphenicol (cat), co-trimoxazole (folA and folP), erythromycin (ermB and mefA), fluoroquinolones (gyrA and parC), tetracycline [tet(M) and tet(O)] and vancomycin (vanA, vanB, vanC, vanD, vanE and vanG) [27–29]. Furthermore, we interrogated the Pathogenwatch pipeline for tet(32) and tet(S/M) (https://pathogen.watch/). Additionally, we determined concordance between the in silico and phenotypic data for AMR.
Statistical analysis
All statistical analyses were run with R (v.3.0.6). Fisher’s exact test was used to determine prevalence changes, Simpson’s diversity index was utilized for diversity analysis [30], and Welch’s t-test was used to determine changes in diversity. Wilcoxon’s rank sum test was used to determine MIC differences between the dominant and non-dominant lineages. Confidence intervals were calculated by bootstrapping. The prevalence of phenotypic resistance to individual antimicrobials was compared amongst lineages and serotypes using Fisher’s exact test and a significance cut-off value of 0.01.
Results and Discussion
Overview of Cambodian carriage dataset
A total of 686 carriage isolates of S. pneumoniae were included in the GPS project, collected from children ranging in age from 1 month to 5 years (mean age 18 months, sd ±13.8). The isolates from prior to the PCV introduction (pre-PCV) were collected between January 2013 until August 2015 (N=232), and post-PCV13 introduction isolates were collected between January 2016 and February 2017 (N=428). Vaccination status relied on parental recall in many cases, due to unavailable vaccine cards and lack of access to the national registry. Classifying the children as PCV13 eligible considered any child born on or after 1st December 2014 as PCV eligible (in concordance with our previous work in 2020 [16], and in discussion with the head of the national immunization programme). Specifically interrogating the 26 isolates collected in 2015, the year of vaccine rollout, 24 (92.3 %) of the children were vaccine non-eligible, while only 2 (7.7 %) were eligible. Thus, the isolates collected in 2015, during the early stages of introduction (peri-PCV13), were included in the pre-PCV13 population (N=26). Excluding the peri-PCV isolates from the study elicits the same trends at the serotype and GPSC level as including them in the pre-PCV population (Table S2). The national PCV uptake was 68 % in 2015, and increased to 87 % in 2016 and 82 % in 2017 [31]. The pre-PCV population comprised 141 (54.7 %) isolates from male children and 117 (45.3 %) from female children, while the post-PCV13 population comprised 227 (53.0 %) males and 201 (47.0 %) females. The mean age of children in the pre-PCV and post-PCV13 populations was 17.8 months (sd 13.9) and 18.1 months (sd 13.7), respectively; the maximum age was 5 years old (Table 1).
Table 1.
Description of sample collection period, gender, age and vaccine status for 686 healthy Cambodian children stratified by pre-PCV (N=258) and post-PCV13 (N=428) pre-PCV, isolates recovered prior and during PCV roll-out (before January 2016); VT, 13 serotypes that are included in PCV13; NVT, serotypes not included in PCV13 (non-vaccine type); NT, non-typable serotypes
|
Pre-PCV (N=258) |
Post-PCV13 (N=428) |
Total (N=686) |
|
|---|---|---|---|
|
Collection period |
2013–2015 |
2016–2017 |
– |
|
Female gender [N (%)] |
117 (45.3 %) |
201 (47.0 %) |
318 (46.4 %) |
|
Age (months) |
|||
|
Mean (sd) |
17.779 (13.941) |
18.079 (13.736) |
17.966 (13.804) |
|
Range |
2–48 |
1–60 |
1–60 |
|
Serotype vaccine status |
|||
|
NVT |
107 (41.5 %) |
230 (53.7 %) |
337 (49.1 %) |
|
VT |
143 (55.4 %) |
185 (43.2 %) |
328 (47.8 %) |
|
NT |
8 (3.1 %) |
13 (3.0 %) |
21 (3.0 %) |
Ongoing serotype replacement
There is evidence that following a perturbation such as vaccination, it can take as long as 10 years for the perturbed genetic composition of the pneumococcal population to settle into a stable equilibrium frequency [32]. This results in increasing diversity following vaccination at both lineage and serotype resolution [33, 34]. The Simpsons diversity (D) index increased for both serotypes (P value=0.007) [ =0.901 (95 % confidence interval 0.90–0.93)] to 0.93 (0.92–0.94) and lineages (P=0.024) [ =0.92 (0.9–0.93) to 0.94 (0.93–0.95)] from the pre-PCV to the post-PCV13 populations in Cambodia.
From the pre- to the post-PCV13 populations, we found a significant decrease in PCV13 serotypes (vaccine type; VT) [P<0.01, odds ratio (OR) 0.57 (95 % confidence interval 0.41–0.79)] and a significant increase in non-PCV13 serotypes (non-vaccine type; NVT) [P<0.01, OR 1.79 (1.28–2.52)] by Fisher's exact test. Together with the increasing genetic diversity, these findings suggest ongoing serotype replacement in Cambodia 3 years following PCV13 roll out. This observation is consistent with what is expected in a vaccinated population [8, 18, 34, 35] (Table 1).
Emerging serotypes and their associated pneumococcal lineages
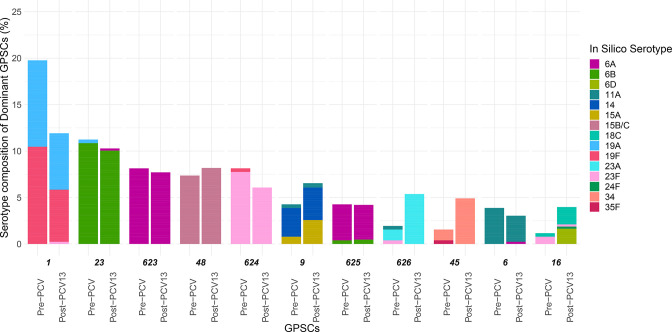
Similar to our previous studies, increases from pre- to post-PCV13 were observed in NVTs 23A [pre-PCV N=5; post-PCV N=23; OR 2.86 (1.05–9.79)] and 34 [pre-PCV N=3; post-PCV N=21; OR 4.38 (1.29–23.15)]. In addition, this study also identified a small number of isolates with NVT 6D (pre-PCV N=0; post-PCV N=9; OR Inf, [1.2-Inf]), which was only detected in this population after PCV13 introduction; however, this serotype decreased in a 2020 study of invasive disease isolates [15, 16] (Table S3). The same study also reported an increase in NVT 15B/C in 2018, which was beyond the period of collection in this study. The major pneumococcal sequence clusters expressing emerging NVTs 23A, 34, 6D and 15B/C were GPSC626 (accounting for 96.3 % serotype 23A), GPSC45 (100 %), GPSC16 (87.5 %) and GPSC48 (96.4 %), respectively. The odds of these lineages inclusion post-PCV as compared to pre-PCV were: GPSC626 OR=2.84 (1.04–9.69), GPSC45 OR=3.24 (1.08–13.13), GPSC16 OR=3.48 (0.99–18.7), GPSC48 OR=1.11 (0.6–2.1) (Fig. 2) (Table S4). This implies that GPSC626 and GPSC45 are the major drivers for the increase of their respective serotypes (Table 2).
Table 2.
Odds of change from the pre- to the post-PCV populations for dominant lineages (GPSCs with N>20)
Significance calculated using Fisher's exact test and confidence intervals via bootstrapping. GPSC1 decreased, while GPSC626 and GPSC45 expanded.
|
GPSC |
Pre-PCV (N=258) |
Post-PCV13 (N=428) |
OR [95 % confidence intervals] |
|---|---|---|---|
|
1 |
51 (19.8 %) |
51 (11.9 %) |
0.54 [0.35–0.85] |
|
626 |
5 (1.9 %) |
23 (5.4 %) |
2.84 [1.04–9.69] |
|
45 |
4 (1.6 %) |
21 (4.9 %) |
3.24 [1.08–13.13] |
|
16 |
3 (1.2 %) |
17 (4.0 %) |
3.48 [0.99–18.7] |
|
9 |
11 (4.3 %) |
28 (6.5 %) |
1.56 [0.73–3.53] |
|
624 |
21 (8.1 %) |
26 (6.1 %) |
0.72 [0.38–1.38] |
|
6 |
10 (3.9 %) |
13 (3.0 %) |
0.77 [0.31–1.99] |
|
23 |
29 (11.2 %) |
44 (10.3 %) |
0.9 [0.53–1.53] |
|
48 |
19 (7.4 %) |
35 (8.2 %) |
1.11 [0.6–2.1] |
|
623 |
21 (8.1 %) |
33 (7.7 %) |
0.93 [0.51–1.74] |
|
625 |
11 (4.3 %) |
18 (4.2 %) |
0.98 [0.43–2.33] |
Pneumococcal lineages in Cambodia
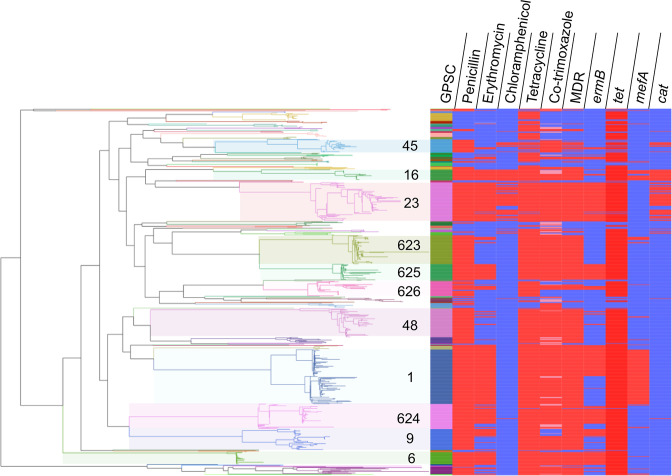
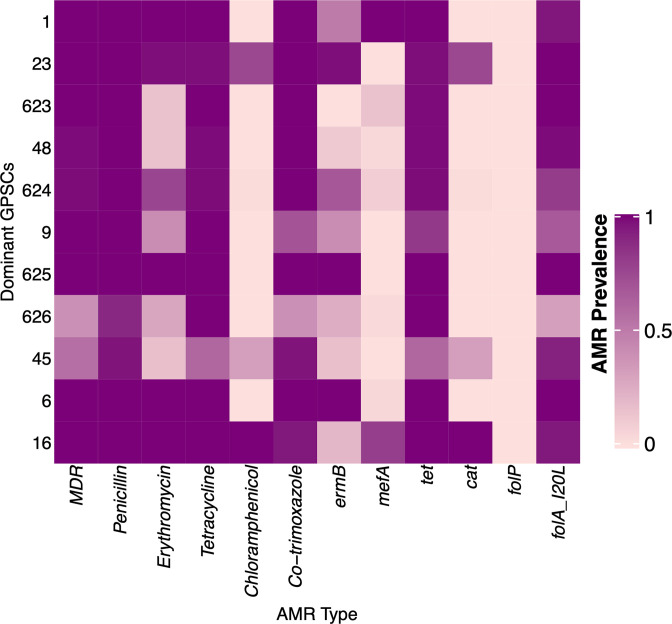
The pneumococcal collection in this study comprised 70 distinct GPSCs (10 unique to pre-PCV, 25 unique to post-PCV13 and 35 in both) and 40 distinct serotypes. A single serotype 18F was unique to 2015, the year of PCV administration, 12 serotypes emerged following PCV13, and 27 were present both before and after PCV (Tables S3 and S4). There were 11 GPSCs with N 20 overall in Cambodia comprising 72.01 % of the carriage population (N=494), which are, henceforth, referred to as dominant lineages (highlighted in Figs 1 and 2, Table 2). All GPSCs are included in (Fig. S1). All dominant lineages were predicted to be penicillin non-susceptible by the cut-off for meningitis at 0.06 mg l−1 MIC. Overall in the population, the MIC50 and MIC90 were 0.25 and 2 mg l−1, respectively. Dominant lineages accounted for 70 % of the total penicillin resistance in the Cambodian carriage population and had higher levels of resistance (P value <2.2×10−16). For the dominant lineages, the penicillin MIC50 was 1 mg l−1 and MIC90 was 4 mg l−1. Whereas, for the non-dominant lineages, the MIC50 and MIC90 were 0.06 and 1 mg l−1, respectively.
Fig. 1.
Phylogenetic tree of 686 carriage isolates from healthy children in Cambodia, isolated between 2013 and 2017. GPSCs with N>20 in the population are highlighted. The tree was built from a nucleotide alignment in FastTree using a generalized time reversible model. Branches are coloured by GPSC corresponding with the first colour strip. ermB and mefA are macrolide (e.g. erythromycin) resistance genes, cat causes resistance to chloramphenicol, tet results in resistance to tetracycline and refers to presence of either tet(M) or tet(O), and folA I20L and folP cause resistance to trimethoprim and sulfamethoxazole (the components of co-trimoxazole), respectively. Antimicrobials are coloured as follows: resistant, red; susceptible, blue; and in the case of co-trimoxazole – intermediate, pink. This figure can be visualized interactively using webtool Microreact at: https://microreact.org/project/mvgn3EvNgmxAcPjPBnyFMj/a32e0dc6.
Fig. 2.
Prevalence of dominant GPSCs (N>20) in pre- and post-PCV13 periods. The bars are coloured by in silico serotype. GPSCs in descending order by count, each with pre-PCV (left) and post-PCV13 (right), are along the x-axis. The prevalence of each GPSC in each vaccine period is along the y-axis. Prevalence changes in GPSC (increasing, except for GPSC 1) notably occurred for 1, 626, 45 and 16. Notable changes in serotype prevalence between pre- and post-PCV13 were observed in serotypes 19F, 23A, 34, 6D and 18C.
Reviewing the GPS database, of the 11 dominant lineages, 3 (GPSC623, 625 and 626) are mainly observed in Cambodia, with 1 (GPSC624) also found in Thailand (GPS database last accessed on 11th May 2021). These regional lineages mainly express VTs, except for GPSC626 expressing NVT 23A, and to a lesser extent 11A. In contrast, six of eight globally spreading lineages recognized in the previous GPS study [7] were found in the current Cambodian carriage collection with a prevalence of 16.0 % (n=146, GPSC1), 11.9 % (n=109, GPSC23), 3.0 % (n=30, GPSC6), 2.3 % (n=21, GPSC16), 0.1 % (n=1, GPSC18, England14-9 PMEN clone) and 0.1 % (n=1, GPSC12).
Distinct lineages in Cambodia
Decreasing lineage – GPSC1
GPSC1 expressing VT 19F and 19A, and to a much lesser extent 23F, decreased during the post-PCV13 period (Fig. 2). Both serotypes 19A and 19F were on the GPSC1 backbone and are targeted by PCV13; however, only 19F decreased in prevalence both within GPSC1 and overall during vaccination in Cambodia. The post-PCV13 population had half the odds of being GPSC1 [OR 0.54 (0.35–0.85)] or to comprise VT 19F serotypes [OR 0.49 (0.26–0.90)] as the pre-PCV population. GPSC1 is multidrug resistant (MDR), exhibiting resistance to penicillin, tetracycline, erythromycin and co-trimoxazole. The predicted penicillin MICs were greater than 2 mg l−1 for all except for one isolate, which had a predicted MIC of 0.5 mg l−1. Tetracycline and erythromycin resistance were due to the presence of tet(M) and mefA, respectively. Additionally, all but one of 19A and 23F genomes carried an extra erythromycin-resistance gene ermB (https://microreact.org/project/mvgn3EvNgmxAcPjPBnyFMj/26d22792).
Increasing lineages – GPSC45, GPSC626, GPSC16
(i) GPSC45 – 34, 35F
After the introduction of PCV13, the increase in GPSC45 (pre-PCV13, 1.9 %; post-PCV13, 4.9 %) had 3.24 (1.08–13.13) increased odds of being in the post-PCV13 compared with the pre-PCV population and was the major driver for the increase in NVT 34. Prior to PCV13 use, GPSC45 comprised NVTs 34 (N=3, 75 %) and 15B/C (N=1, 25 %), whereas following PCV13 it only comprised NVT 34 (N=21, 100 %) (Fig. 2). GPSC45 lineages exhibit resistance to penicillin (predominant pbp profile 0-238-383) and co-trimoxazole (I100L in folA and 178 insertion in folP), except for one isolate. Chloramphenicol and tetracycline resistance were sporadically observed across the phylogeny, indicating multiple gains and losses of cat and tet(M) gene, respectively. A potential single acquisition of erythromycin-resistance gene ermB was detected in a cluster of four isolates, recovered in 2016 (n=1) and 2017 (n=3) (https://microreact.org/project/mvgn3EvNgmxAcPjPBnyFMj/5a20d817).
(ii) GPSC626 – 23A, 23F, 11A
Despite its increase, GPSC626 underwent a loss of serotype diversity following PCV13 [N=28, P=0.03, OR 2.84 (1.04–9.69)] from comprising VT 23F (1, 20 %), NVT 11A (N=1, 20 %) and NVT 23A (3, 60 %) to only NVT 23A (N=23, 100 %). This may indicate an epistatic interaction between the cps locus and the rest of the GPSC626 genetic background favouring NVT 23A. A majority (87 %) of GPSC626 isolates were penicillin resistant, and 42 % were MDR. There were two sub-clades of isolates expressing NVT 23A (n=12) that were predicted to be non-susceptible to cefotaxime and ceftriaxone; one of them (n=3) was MDR. All but two of the isolates belonging to these two clades were detected after PCV13 (https://microreact.org/project/mvgn3EvNgmxAcPjPBnyFMj/f4375564).
(iii) GPSC16 – 18C, 6D, 24, 23F
Although the odds of GPSC16 inclusion in the post-PCV population have confidence intervals intersecting OR=1, its composition of four different serotypes including two which expanded post-PCV is notable. GPSC16 increased in prevalence from 1.2 % in the pre-PCV13 population to 4.0 % in the post-PCV population [N=20, P=0.04, OR 3.48 (0.99–18.7)], likely due to the expansions of serotype 6D [OR 1.3 (0.97–1.75)] and serotype 18C [OR 1.08 (0.76–1.56)] (Fig. 2). The GPSC16 expansion of NVT 6D indicates that the 6A and 6B inclusion in PCV13 may not be cross-protective amongst these carriage isolates. All GPSC16 isolates, except for one expressing NVT 24, were resistant to penicillin (a uniform pbp profile of 15-12-18), chloramphenicol (cat), co-trimoxazole (I100L in folA and 178 insertion in folP), erythromycin (ermB in VT 23F isolates, while mefA in 6D and 18C isolates) and tetracycline [tet(M)] (https://microreact.org/project/mvgn3EvNgmxAcPjPBnyFMj/bb468401). Dominant lineages that persist at a stable prevalence from the pre- to post-PCV populations are described in the Supplementary Appendix 1.
AMR
Concordance between whole-genome sequence and phenotypic AMR for all assessed antibiotics was greater than 90 % (Table 3). The AMR prevalence and particularly high occurrence of multidrug resistance (77.1%) in Cambodia are higher than described overall globally (20.1 %) [7]; however, they are similar to previously described in South-East Asia [36, 37].
Table 3.
Concordance rates between genotypic and phenotypic AMR data
False positive is referred to as ‘major discrepancy' by the US-FDA, while false negative is referred to as ‘very major discrepancy’ by the US-FDA. Overall, the prevalence of predicted AMR in Cambodia is summarized in Table 4) . There was no significant difference in AMR or multidrug resistance from the pre-PCV13 (79 % MDR) to post-PCV13 (76 % MDR) populations (Table 4); however, there was a higher prevalence of mutlidrug resistance in the VT serotypes (91 % MDR) compared with the NVT serotypes (62 % MDR) (Table 5). Assuming the previously demonstrated vaccine effectiveness for pneumococcal colonization of 39.2 % (95 % confidence interval 26.7–46.1) for VT serotypes in Cambodia maintains, the prevalence of AMR should decrease [16]. Alternatively, despite high vaccine efficacy, the genetic backgrounds containing VT and AMR may persist via serotype switching and expansion of previously low prevalence NVT lineages, resulting in vaccine escape (Fig. 1).
|
Antibiotic |
Concordance (%) |
Discordance (%) |
|
|---|---|---|---|
|
False positive by WGS |
False negative by WGS |
||
|
Multidrug resistance |
651 (94.9) |
16 (2.3) |
6 (0.9) |
|
Penicillin (meningitis threshold) |
685 (99.9) |
0 |
1 (0.1) |
|
Erythromycin |
682 (99.4) |
3 (0.4) |
1 (0.1) |
|
Chloramphenicol |
684 (99.7) |
2 (0.3) |
0 |
|
Tetracycline |
646 (94.2) |
22 (3.2) |
18 (2.6) |
|
Co-trimoxazole |
661 (96.4) |
25 (3.6) |
– |
Table 4.
Prevalence of antimicrobial non-susceptible isolates pre- and post-PCV13 in Cambodia for multidrug resistance (non-susceptible for three or more antimicrobials), penicillin (using the resistance threshold for meningitis), erythromycin, chloramphenicol, tetracycline, co-trimoxazole and specific genes including cat (chloramphenicol) and tet (tetracycline), as well as macrolide resistance genes mef and ermB
|
Antimicrobial/gene |
pre-PCV (N=258) |
post-PCV13 (N=428) |
Total (N=686) |
|---|---|---|---|
|
Multidrug resistance |
204 (79.1 %) |
325 (75.9 %) |
529 (77.1 %) |
|
Penicillin (meningitis threshold) |
210 (81.4 %) |
355 (82.9 %) |
565 (82.4 %) |
|
Erythromycin |
142 (55.0 %) |
211 (49.3 %) |
353 (51.5 %) |
|
Chloramphenicol |
30 (11.6 %) |
61 (14.3 %) |
91 (13.3 %) |
|
Tetracycline |
231 (89.5 %) |
392 (91.6 %) |
623 (90.8 %) |
|
Co-trimoxazole |
228 (88.4 %) |
346 (80.8 %) |
574 (83.7 %) |
|
mef presence |
61 (23.6 %) |
82 (19.2 %) |
143 (20.8 %) |
|
ermB presence |
107 (41.5 %) |
155 (36.2 %) |
262 (38.2 %) |
|
cat presence |
30 (11.6 %) |
61 (14.3 %) |
91 (13.3 %) |
|
tet presence |
226 (87.6 %) |
383 (89.5 %) |
609 (88.8 %) |
Table 5.
Relationship between AMR and VT serotypes for five different antimicrobials and macrolide-resistance genes
Adjusting for multiple testing (threshold 0.005), all except tet are significantly associated with VT serotypes. Antimicrobials and genes: penicillin (using the resistance threshold for meningitis), erythromycin, tetracycline, chloramphenicol, co-trimoxazole and specific genes including cat (chloramphenicol) and tet (tetracycline), as well as the macrolide-resistance genes mef and ermB.
|
Antibiotic/gene |
Odds ratio [95 % confidence intervals] |
P value |
|---|---|---|
|
Multidrug resistance |
6.46 [4.09–10.5] |
<0.01 |
|
Penicillin |
6.39 [3.79–11.24] |
<0.01 |
|
Erythromycin |
6.76 [4.76–9.68] |
<0.01 |
|
Tetracycline |
2.65 [1.46–4.98] |
<0.01 |
|
Chloramphenicol |
3.56 [2.12–6.17] |
<0.01 |
|
Co-trimoxazole |
6.55 [3.8–11.86] |
<0.01 |
|
ermB |
3.55 [2.52–5.02] |
<0.01 |
|
mefA |
9.11 [5.44–15.96] |
<0.01 |
|
cat |
3.56 [2.12–6.17] |
<0.01 |
|
tet |
2.07 [1.23–3.56] |
0.0050 |
A total of 93.9 % of the dominant lineages (comprising 72 % total population) are MDR; however, among these only 18 % carry resistance to chloramphenicol. GPSC23 and GPSC16 were the predominant chloramphenicol resistant-MDR isolates. GPSC16 expanded post-PCV, concurrently carrying chloramphenicol resistance in all isolates, while GPSC23 maintained a stable prevalence from the pre-PCV (10.3 %) to post-PCV (11.2 %) population, respectively. GPSC23 comprises VT 6B (97.2 %), with one VT 19A and one VT 6A isolate. All GPSC23 isolates had high prevalence resistance to all described antimicrobials (Figs 1 and 3).
Fig. 3.
The prevalence of AMR among the 11 dominant pneumococcal lineages or GPSCs in Cambodia. GPSCs are ordered by their overall prevalence in the population (rows) and colours indicate the prevalence of AMR and resistance determinants (columns) for each dominant GPSC in Cambodia. Coloured by AMR prevalence from 0 (light purple) to 100 % (dark purple). ermB and mefA are macrolide (e.g. erythromycin) resistance genes, cat causes resistance to chloramphenicol, tet results in resistance to tetracycline and refers to presence of either tet(M) or tet(O), and folA I20L and folP cause resistance to trimethoprim and sulfamethoxazole (the components of co-trimoxazole), respectively.
The variability in GPSC stability can be attributed to the invasive potential of the serotypes they carry and their AMR profile [7, 38, 39]. For example, serotype 14 persists post-PCV13 on a GPSC9 genetic backbone despite its inclusion in the vaccine. This serotype on this backbone has been previously reported to have lower invasiveness than on other backbones [7]. Lower invasiveness may contribute to its success on the basis that lower invasiveness can translate to longer carriage duration, which in turn may counteract the impact of the vaccine [40]. The rise and fall of GPSCs following perturbation by vaccination may also be explained by negative frequency dependent selection. Pneumococcal populations have genes that persist at a stable, intermediate frequency. After mass-vaccination, the prevalence of these genes is often perturbed. Over time selection acts on the lineages that carry these genes to return them to their initial, intermediate frequency [32, 41].
Additionally, the gene tet(32) was identified in GPSC158 (100 % NVT 16F) – a low prevalence lineage (N=4). This gene was first described in S. pneumoniae in 2020 in genomes isolated in Liverpool on a GPSC12 backbone with serotype 3 [42].
Conclusions
This study demonstrates that the Cambodian pneumococcal carriage population was perturbed within 2 years following the PCV13 vaccination campaign. This presents with a decrease in some vaccine serotypes, and emergence and expansion of NVTs. The expanding NVTs are among both regional and international lineages. There is a high prevalence of AMR and MDR isolates in both the newly expanding lineages and those with persistent high prevalence. The relationship between the carriage and disease populations is unclear; however, as IPD is an outcome of pneumococcal carriage, we would expect that mitigating disease-causing lineages in carriage will have an impact on pneumococcal disease. Pneumococcal surveillance is ongoing in Cambodia and may further elucidate this question. Additionally, further genomic surveillance will likely reveal which NVTs emerge, and whether such emergence is driven by a specific genetic background [43].
Supplementary Data
Funding information
This study was co-funded by the Bill and Melinda Gates Foundation (grant code OPP1034556), the Wellcome Sanger Institute (core Wellcome grants 098051 and 206194), and the US Centres for Disease Control and Prevention. P.T. and Cambodia Oxford Medical Research Unit (COMRU) are supported by core funding from Wellcome (grant 220211). The funding sources had no role in isolate selection, analysis nor data interpretation. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centres for Disease Control and Prevention. The corresponding author had full access to the data and is responsible for the final decision to submit for publication.
Acknowledgements
We would like to thank all members of the GPS consortium for their collaborative spirit and determination during the monumental task of sampling, extracting and sequencing this dataset, and all contributions to experimental design and input into this manuscript. We also would like to thank all the clinicians for submitting specimens and the Pathogen Informatics Team at the Wellcome Sanger Institute for technical support of bioinformatic analyses.
Author contributions
S.B.: data curation, formal analysis, writing – original draft. S.S.: writing – review and editing, methodology. C.S.: writing – review and editing, methodology. R.G.: conceptualization. P.A.H.: data curation, validation. R.F.B.: conceptualization, writing – review and editing. L.M.: conceptualization, data curation, funding acquisition, writing – review and editing. S.D.B.: conceptualization, data curation, funding acquisition, writing – review and editing. S.W.L.: conceptualization, data curation, writing – review and editing. P.T.: conceptualization, data curation, funding acquisition, writing – review and editing.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Protocols for the nasopharyngeal colonization studies were approved by the adventist health care (AHC) Institutional Review Board (422/13, 371/14, 0348/15), Cambodia National Ethics Committee for Health Research (210NECHR, 289NECHR, 150NECHR, 137NECHR), WHO Western Pacific Regional Office IRB (2015.6.CAM.1.EPI), and the University of Oxford Tropical Research Ethics Committee (1009-13, 559-15).
Footnotes
Abbreviations: AMR, antimicrobial resistance; GPS, global pneumococcal sequencing; GPSC, global pneumococcal sequence cluster; IPD, invasive pneumococcal disease; MDR, multidrug resistant; MIC, minimum inhibitory concentration; NVT, non-vaccine type; OR, odds ratio; PCV, pneumococcal conjugate vaccine; VT, vaccine type; WGS, whole-genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Supplementary material is available with the online version of this article.
References
- 1.Weiser JN, Ferreira DM, Paton JC. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol. 2018;16:355–367. doi: 10.1038/s41579-018-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wahl B, O’Brien KL, Greenbaum A, Majumder A, Liu L, et al. Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000–15. Lancet Glob Health. 2018;6:e744–e757. doi: 10.1016/S2214-109X(18)30247-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009;374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 4.GBD 2015 LRI Collaborators Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory tract infections in 195 countries: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis. 2017;17:1133–1161. doi: 10.1016/S1473-3099(17)30396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pilishvili T, Lexau C, Farley MM, Hadler J, Harrison LH, et al. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J Infect Dis. 2010;201:32–41. doi: 10.1086/648593. [DOI] [PubMed] [Google Scholar]
- 6.von Gottberg A, de Gouveia L, Tempia S, Quan V, Meiring S, et al. Effects of vaccination on invasive pneumococcal disease in South Africa. N Engl J Med. 2014;371:1889–1899. doi: 10.1056/NEJMoa1401914. [DOI] [PubMed] [Google Scholar]
- 7.Gladstone RA, Lo SW, Lees JA, Croucher NJ, van Tonder AJ, et al. International genomic definition of pneumococcal lineages, to contextualise disease, antibiotic resistance and vaccine impact. EBioMedicine. 2019;43:338–346. doi: 10.1016/j.ebiom.2019.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lo SW, Gladstone RA, van Tonder AJ, Lees JA, du Plessis M, et al. Pneumococcal lineages associated with serotype replacement and antibiotic resistance in childhood invasive pneumococcal disease in the post-PCV13 era: an international whole-genome sequencing study. Lancet Infect Dis. 2019;19:759–769. doi: 10.1016/S1473-3099(19)30297-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lakshman R, Murdoch C, Race G, Burkinshaw R, Shaw L, et al. Pneumococcal nasopharyngeal carriage in children following heptavalent pneumococcal conjugate vaccination in infancy. Arch Dis Child. 2003;88:211–214. doi: 10.1136/adc.88.3.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wyllie AL, Rümke LW, Arp K, Bosch AATM, Bruin JP, et al. Molecular surveillance on Streptococcus pneumoniae carriage in non-elderly adults; little evidence for pneumococcal circulation independent from the reservoir in children. Sci Rep. 2016;6:34888. doi: 10.1038/srep34888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camargos P, Drumond E, Nascimento-Carvalho CM. Effect of pneumococcal conjugate vaccines on invasive pneumococcal disease. Lancet Infect Dis. 2021;21:453. doi: 10.1016/S1473-3099(21)00051-7. [DOI] [PubMed] [Google Scholar]
- 12.Hamaluba M, Kandasamy R, Ndimah S, Morton R, Caccamo M, et al. A cross-sectional observational study of pneumococcal carriage in children, their parents, and older adults following the introduction of the 7-valent pneumococcal conjugate vaccine. Medicine. 2015;94:e335. doi: 10.1097/MD.0000000000000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.GBD 2015 Mortality and Causes of Death Collaborators Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1459–1544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gladstone RA, Jefferies JM, Tocheva AS, Beard KR, Garley D, et al. Five winters of pneumococcal serotype replacement in UK carriage following PCV introduction. Vaccine. 2015;33:2015–2021. doi: 10.1016/j.vaccine.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inghammar M, By Y, Farris C, Phe T, Borand L, et al. Serotype distribution of clinical Streptococcus pneumoniae isolates before the introduction of the 13-valent pneumococcal conjugate vaccine in Cambodia. Am J Trop Med Hyg. 2018;98:791–796. doi: 10.4269/ajtmh.17-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turner P, Leab P, Ly S, Sao S, Miliya T, et al. Impact of 13-valent pneumococcal conjugate vaccine on colonization and invasive disease in cambodian children. Clin Infect Dis. 2020;70:1580–1588. doi: 10.1093/cid/ciz481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore CE, Giess A, Soeng S, Sar P, Kumar V, et al. Characterisation of invasive Streptococcus pneumoniae isolated from Cambodian children between 2007 – 2012. PLoS One. 2016;11:e0159358. doi: 10.1371/journal.pone.0159358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner P, Turner C, Suy K, Soeng S, Ly S, et al. Pneumococcal infection among children before introduction of 13-valent pneumococcal conjugate vaccine, Cambodia. Emerg Infect Dis. 2015;21:2080–2083. doi: 10.3201/eid2111.150914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacques LC, Panagiotou S, Baltazar M, Senghore M, Khandaker S, et al. Increased pathogenicity of pneumococcal serotype 1 is driven by rapid autolysis and release of pneumolysin. Nat Commun. 2020;11:1892. doi: 10.1038/s41467-020-15751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner P, Turner C, Jankhot A, Helen N, Lee SJ, et al. A longitudinal study of Streptococcus pneumoniae carriage in a cohort of infants and their mothers on the Thailand-Myanmar border. PLoS One. 2012;7:e38271. doi: 10.1371/journal.pone.0038271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russell FM, Biribo SSN, Selvaraj G, Oppedisano F, Warren S, et al. As a bacterial culture medium, citrated sheep blood agar is a practical alternative to citrated human blood agar in laboratories of developing countries. J Clin Microbiol. 2006;44:3346–3351. doi: 10.1128/JCM.02631-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeh E, Pinsky BA, Banaei N, Baron E. Hair sheep blood, citrated or defibrinated, fulfills all requirements of blood agar for diagnostic microbiology laboratory tests. PLoS One. 2009;4:e6141. doi: 10.1371/journal.pone.0006141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Epping L, van Tonder AJ, Gladstone RA, Bentley SD, Page AJ, et al. SeroBA: rapid high-throughput serotyping of Streptococcus pneumoniae from whole genome sequence data. Microb Genom. 2018;4:e000186. doi: 10.1099/mgen.0.000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lees JA, Harris SR, Tonkin-Hill G, Gladstone RA, Lo SW, et al. Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Res. 2019;29:304–316. doi: 10.1101/gr.241455.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Metcalf BJ, Chochua S, Li Z, Gertz RE, Jr, et al. Penicillin-binding protein transpeptidase signatures for tracking and predicting β-lactam resistance levels in Streptococcus pneumoniae . mBio. 2016;7:e00756-16. doi: 10.1128/mBio.00756-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Metcalf BJ, Chochua S, Li Z, Gertz RE, Jr, et al. Validation of β-lactam minimum inhibitory concentration predictions for pneumococcal isolates with newly encountered penicillin binding protein (PBP) sequences. BMC Genomics. 2017;18:621. doi: 10.1186/s12864-017-4017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metcalf BJ. Sanger_SPN_Scripts-Ref. 2016. https://github.com/BenJamesMetcalf/Sanger_SPN_Scripts-Ref
- 28.Metcalf BJ, Gertz RE, Gladstone RA, Walker H, Sherwood LK, et al. Strain features and distributions in pneumococci from children with invasive disease before and after 13-valent conjugate vaccine implementation in the USA. Clin Microbiol Infect. 2016;22:P60.E9–P60.E29. doi: 10.1016/j.cmi.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom. 2017;3:e000131. doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simpson EH. Measurement of diversity. Nature. 1949;163:688. doi: 10.1038/163688a0. [DOI] [Google Scholar]
- 31.WHO UNICEF WHO Pneumococcal Vaccination Coverage. WHO World Health Organization: immunization, vaccines and biologicals. Vaccine preventable diseases. Vaccines monitoring system 2020. Global summary reference time series: PCV3. 2020. https://immunizationdata.who.int/pages/coverage/PCV.html?CODE=KHM&ANTIGEN=PCV3&YEAR=
- 32.Corander J, Fraser C, Gutmann MU, Arnold B, Hanage WP, et al. Frequency-dependent selection in vaccine-associated pneumococcal population dynamics. Nat Ecol Evol. 2017;1:1950–1960. doi: 10.1038/s41559-017-0337-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanage WP, Finkelstein JA, Huang SS, Pelton SI, Stevenson AE, et al. Evidence that pneumococcal serotype replacement in Massachusetts following conjugate vaccination is now complete. Epidemics. 2010;2:80–84. doi: 10.1016/j.epidem.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Polain De Waroux O, Edmunds WJ, Takahashi K, Ariyoshi K, Mulholland EK, et al. Predicting the impact of pneumococcal conjugate vaccine programme options in Vietnam. Hum Vaccin Immunother. 2018;14:1939–1947. doi: 10.1080/21645515.2018.1467201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodrigo C, Bewick T, Sheppard C, Greenwood S, Mckeever TM, et al. Impact of infant 13-valent pneumococcal conjugate vaccine on serotypes in adult pneumonia. Eur Respir J. 2015;45:1632–1641. doi: 10.1183/09031936.00183614. [DOI] [PubMed] [Google Scholar]
- 36.Zellweger RM, Carrique-Mas J, Limmathurotsakul D, Day NPJ, Thwaites GE, et al. A current perspective on antimicrobial resistance in Southeast Asia. J Antimicrob Chemother. 2017;72:2963–2972. doi: 10.1093/jac/dkx260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim SH, Song J-H, Chung DR, Thamlikitkul V, Yang Y, et al. Changing trends in antimicrobial resistance and serotypes of Streptococcus pneumoniae isolates in Asian countries: an Asian Network for Surveillance of Resistant Pathogens (ANSORP) study. Antimicrob Agents Chemother. 2012;56:1418–1426. doi: 10.1128/AAC.05658-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davies NG, Flasche S, Jit M, Atkins KE. Modeling the effect of vaccination on selection for antibiotic resistance in Streptococcus pneumoniae . Sci Transl Med. 2021;13:aaz8690. doi: 10.1126/scitranslmed.aaz8690. [DOI] [PubMed] [Google Scholar]
- 39.Lo SW, Mellor K, Cohen R, Alonso AR, Belman S, et al. Emergence of a multidrug resistant and virulent Streptococcus pneumoniae lineage mediates serotype replacement after PCV13. medRxiv. 2021:21266813. doi: 10.1101/2021.11.24.21266813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bricio-Moreno L, Chaguza C, Yahya R, Shears RK, Cornick JE, et al. Lower density and shorter duration of nasopharyngeal carriage by pneumococcal serotype 1 (ST217) may explain its increased invasiveness over other serotypes. mBio. 2020;11:e00814-20. doi: 10.1128/mBio.00814-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colijn C, Corander J, Croucher NJ. Designing ecologically optimized pneumococcal vaccines using population genomics. Nat Microbiol. 2020;5:473–485. doi: 10.1038/s41564-019-0651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nikolaou E, Hubbard ATM, Botelho J, Marschall TAM, Ferreira DM, et al. Antibiotic resistance is associated with integrative and conjugative elements and genomic islands in naturally circulating Streptococcus pneumoniae isolates from adults in Liverpool, UK. Genes. 2020;11:E625. doi: 10.3390/genes11060625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bentley SD, Lo SW. Global genomic pathogen surveillance to inform vaccine strategies: a decade-long expedition in pneumococcal genomics. Genome Med. 2021;13:84. doi: 10.1186/s13073-021-00901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.