Abstract
Background and Aims
Over 80 monogenic causes of very early onset inflammatory bowel disease [VEOIBD] have been identified. Prior reports of the natural history of VEOIBD have not considered monogenic disease status. The objective of this study is to describe clinical phenotypes and outcomes in a large single-centre cohort of patients with VEOIBD and universal access to whole exome sequencing [WES].
Methods
Patients receiving IBD care at a single centre were prospectively enrolled in a longitudinal data repository starting in 2012. WES was offered with enrollment. Enrolled patients were filtered by age of diagnosis <6 years to comprise a VEOIBD cohort. Monogenic disease was identified by filtering proband variants for rare, loss-of-function, or missense variants in known VEOIBD genes inherited according to standard Mendelian inheritance patterns.
Results
This analysis included 216 VEOIBD patients, followed for a median of 5.8 years. Seventeen patients [7.9%] had monogenic disease. Patients with monogenic IBD were younger at diagnosis and were more likely to have Crohn’s disease phenotype with higher rates of stricturing and penetrating disease and extraintestinal manifestations. Patients with monogenic disease were also more likely to experience outcomes of intensive care unit [ICU] hospitalisation, gastrostomy tube, total parenteral nutrition use, stunting at 3-year follow-up, haematopoietic stem cell transplant, and death. A total of 41 patients [19.0%] had infantile-onset disease. After controlling for monogenic disease, patients with infantile-onset IBD did not have increased risk for most severity outcomes.
Conclusions
Monogenic disease is an important driver of disease severity in VEOIBD. WES is a valuable tool in prognostication and management of VEOIBD.
Keywords: Very early onset inflammatory bowel disease, whole exome sequencing, disease course
1. Introduction
Inflammatory bowel disease [IBD], comprising Crohn’s disease [CD], ulcerative colitis [UC], and inflammatory bowel disease-undefined [IBD-U], is characterised by chronic intestinal inflammation with a relapsing-remitting course. The aetiology of IBD is typically thought to be multifactorial, triggered by environmental factors in a genetically susceptible host. It is suspected that genetic risk plays a greater role in paediatric-onset IBD,1 with some studies showing that higher polygenic risk scores2 as well as specific IBD risk variants3–5 are associated with earlier disease onset. However, in very early onset IBD [VEOIBD], defined as disease onset at age less than 6 years, genetics are postulated to play an even greater role in disease development, with over 80 monogenic causes of disease identified to date.6–9 These monogenic causes most often represent primary immune deficiencies or epithelial barrier defects10 and are critical to identify, when present, as they often have major implications in clinical management.
The natural history of VEOIBD is not yet well established. Earlier studies suggest that VEOIBD may be characterised by a more aggressive and treatment-refractory disease course.9,11–13 However, more recent data around this have been mixed, with one large Canadian study showing decreased health care use in VEOIBD compared with later-onset disease.14 Paediatric gastroenterologists are increasingly tasked with prognosticating VEOIBD, which now represents the age group with the fastest growing incidence of disease.15 Prognostication is advanced by the field’s rapidly expanding knowledge of monogenic causes of IBD,8 and this knowledge is at the forefront of understanding disease course and precision medicine approaches. For example, interleukin-10 [IL10] signaling defects are one of the most commonly described monogenic causes of VEOIBD and are notoriously refractory to conventional IBD therapies.16 IL10 signaling defects have been characterised in human and mouse models by an enhanced interleukin-1 [IL1] signature; it has been shown that patients with IL10 signaling defects can be cured with haematopoietic stem cell transplant [HSCT] and bridged to transplant with anti-IL1 therapy.17–19 This is a shining example of how our growing knowledge of monogenic mechanisms of disease is changing the nuance with which we can understand, treat, and prognosticate disease that was once treatment-refractory.
Literature to date describing the natural history of VEOIBD has not been in patient cohorts with widely available whole exome sequencing [WES]. In this study, we report on the natural history of VEOIBD in a large, single-centre cohort with universal access to WES. We hypothesise that VEOIBD represents a heterogeneous patient population on a spectrum of mild to severe disease, and that monogenic disease is an important driver of disease severity. In our VEOIBD cohort, we describe prevalence of monogenic disease and compare IBD phenotypes and outcomes in monogenic versus non-monogenic groups. Given prior reports of infantile-onset disease having more severe course,20,21 we additionally analysed outcomes in infantile versus non-infantile VEOIBD after adjusting for monogenic disease status.
2. Materials and Methods
2.1. Study design
We conducted a retrospective chart review of patients with VEOIBD enrolled in the Boston Children’s Hospital [BCH] IBD Longitudinal Data Repository. BCH is a large, quaternary children’s hospital and IBD referral centre. Patients with a diagnosis of IBD receiving clinical care at BCH have been prospectively enrolled in the repository since 2012. The repository houses longitudinal clinical data on and biospecimens from enrolled patients.
2.2. Study cohort
The data repository was filtered by age of diagnostic endoscopy <6 years, comprising the cohort for retrospective chart review. Inclusion criteria were: [1] confirmed diagnosis of IBD based on standard endoscopic, histological, or radiographic evaluation22; [2] less than 6 years of age on date of diagnostic endoscopy; and [3] at least one clinical encounter with a gastroenterologist at BCH. Exclusion criteria included: [1] presence of an alternative diagnosis accounting for IBD-like presentation, such as allergic, lymphocytic, or collagenous colitis or a non-inflammatory congenital diarrhoea syndrome; and [2] incomplete medical record, such as missing initial diagnostic data.
2.3. Patient enrolment and whole exome sequencing
IBD patients were approached by trained research assistants at BCH clinic visits, endoscopy encounters, and inpatient hospitalisations and offered enrolment in the IBD Longitudinal Data Repository. WES was offered to all enrolled patients and their first-degree relatives. Patients who had telemedicine encounters during the COVID-19 pandemic had the option to enroll by phone, and buccal swab kits for WES were completed by mail. WES performed prior to October 2018 was done on a research basis in collaboration with pharmaceutical partners. Subsequently, WES was performed clinically through the Children’s Rare Disease Cohorts [CRDC], an internally funded BCH initiative that funded clinical laboratory improvement amendments [CLIA]-compliant WES.23 Protocol for WES performed by pharmaceutical collaborators can be found in the Supplementary Methods. Patients enrolled through the CRDC initiative had DNA collected by buccal swab and sent to GeneDx [Gaithersburg, MD] for DNA isolation and sequencing. The complete protocol for WES performed by the CRDC is as described by Rockowitz et al.23
WES data were analysed by a biostatistician in conjunction with trained gastroenterologists. Data were analysed on GRCh38 background. Proband variants were filtered by rare, loss-of-function [LOF], or missense variants in established VEOIBD genes [Supplementary Table 1]6,8–10,24–27 inherited according to standard Mendelian inheritance patterns. Rare variants were those with minor allele frequency [MAF] <1% for homozygous, hemizygous, and de novo mutations and <5% for compound heterozygous mutations according to gnomAD.28 Subjects who met these filter criteria were classified as having monogenic disease, with the caveat that compound heterozygotes were classified as monogenic only if parental WES data were available confirming inheritance of the abnormal variants in a trans position. American College of Medical Genetics and Genomics [ACMG] classifications were reported for all patients classified as having monogenic disease.29 Patients whose rare variants were classified as ‘benign’ or ‘likely benign’ were excluded from the monogenic disease group. One patient who was ultimately diagnosed with a monogenic disease had inconclusive WES; diagnosis was ultimately made by absent gene product protein levels and targeted comparative genomic hybridisation testing.
A subset of this cohort had targeted genetic testing. This testing was ordered by a primary clinician based on clinical judgement. If monogenic disease was diagnosed by targeted genetic testing prior to WES sample collection, WES was in some cases not pursued.
For purposes of this analysis, patients without genetic testing were assumed to have non-monogenic disease. To confirm the validity of this approach, all analyses presented in this work were additionally performed on the subset of this cohort who had completed WES or targeted genetic testing with identical results.
2.4. Data collection
Patient records were reviewed for demographic data, clinical phenotype, extraintestinal manifestations, medical comorbidities, anthropometric parameters, hospitalisations, surgeries, haematopoietic stem cell transplant [HSCT], and death. A single paediatric gastroenterologist [LC] with expertise in IBD performed all medical record reviews and data extraction. Medical record review occurred from May to August 2020. Study data were collected and managed using REDCap [Research Electronic Data Capture] electronic data capture tools hosted at BCH. REDCap is a secure, web-based software platform designed to support data capture for research studies.30,31 For each patient, data from initial presentation to most recent clinical encounter were included. Patients must have been followed for a minimum of 1 year after diagnosis, for inclusion in this analysis.
2.5. Definitions
Infantile-onset IBD was defined as age at diagnostic endoscopy <2 years. IBD was classified as CD, UC, or IBD-U and Paris classifications were assigned according to standard criteria.32,33 Penetrating disease was determined by the occurrence of bowel perforation, intraabdominal fistula, or abscess; isolated perianal or rectovaginal fistulae were excluded from the penetrating disease category. Perianal disease included perianal or rectovaginal fistula, anal canal ulcer, or abscess.33 Extraintestinal manifestations [EIMs] assessed included arthropathy, erythema nodosum, enthesitis, episcleritis, fever, folliculitis, hepatic granulomas, oral aphthae, orofacial Crohn’s, primary sclerosing cholangitis, pyoderma gangrenosum, thrombotic events, uveitis, and vulvar Crohn’s. All comorbid diagnoses documented in the medical record were recorded. Comorbidities with autoimmune or autoinflammatory mechanism or with known genetic basis were denoted for additional analyses. Disease severity outcome measures included hospitalisation, intensive care unit [ICU] hospitalisation, gastrostomy tube [G-tube] placement, total parenteral nutrition [TPN] use, surgery, 5-aminosalicylate [5-ASA] failure, anti-tumour necrosis factor [TNF] failure, weight-for-age, height-for-age, and body mass index [BMI] z score <-2 at 3-year follow-up, HSCT, and death. Medication failure was defined by change in or escalation of therapy. Surgeries recorded included intestinal resections, colectomy, diverting ostomy, ostomy revision, ostomy closure, stricturoplasty, lysis of adhesions, diagnostic laparotomy, and perineal resection. Perianal surgeries such as anal fistulotomy, fistulectomy, incision and drainage of perianal abscess, or seton placement were not included in this analysis. Weight-for-age, length or height-for-age, and weight-for-length or BMI z-scores were based on World Health Organization [WHO] criteria for patients aged <2 years and centres for Disease Control and Prevention [CDC] criteria for patients aged ≥2 years. Anthropometric data at diagnosis were included only if measured within 4 weeks of diagnostic endoscopy. Follow-up anthropometric data were analysed only if measured at least 3 years after diagnosis.
2.6. Statistical considerations
Standard descriptive statistics were used to characterise overall study population and specific subgroups of interest. Summary statistics including means, standard deviations, medians, and interquartile ranges were compiled for all measured variables. Welch two-sample t tests were used to compare means between two groups. Univariate logistic regression or Fisher’s exact test were used to compare dichotomous outcomes in monogenic versus non-monogenic disease groups. Multivariate logistic regression was used to compare outcomes in infantile versus non-infantile disease with monogenic disease status as a covariate; p-values less than 0.05 were considered statistically significant. All statistical analyses were conducted in R [v 4.0.5].34
2.7. Ethical statement
Subjects were enrolled and data were collected under protocol 00000529, approved by the Institutional Review Board at BCH. All patients included in this study and their parents/guardians provided informed consent and assent where applicable.
3. Results
3.1. Cohort demographics
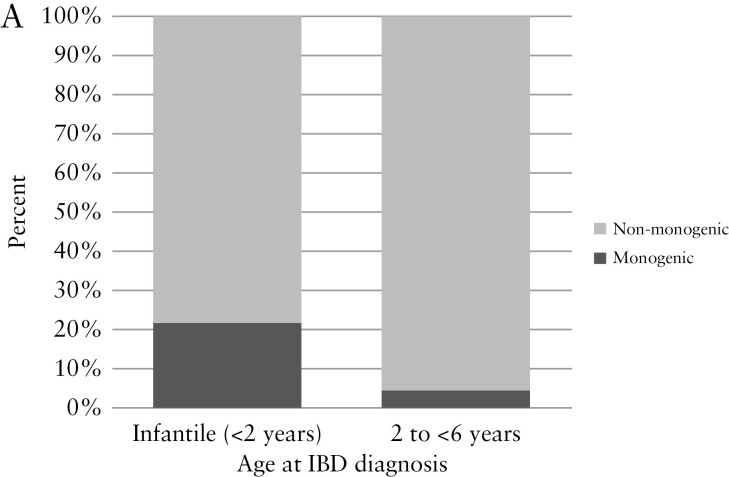
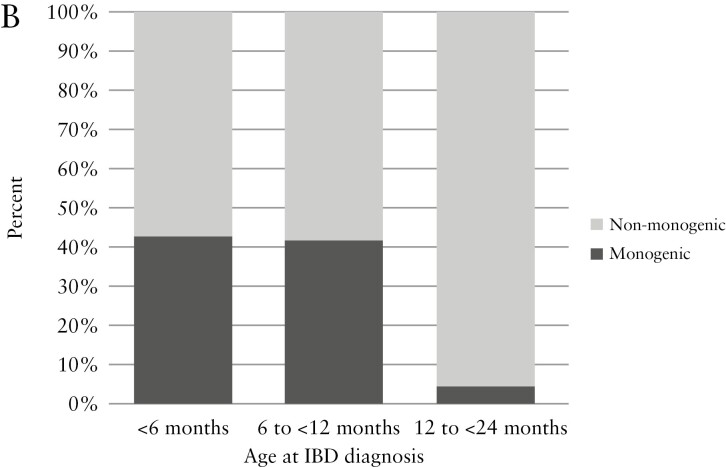
The study population included a total of 216 patients with VEOIBD [Table 1]. Patients in the cohort were diagnosed with IBD at a median age of 3.8 years (interquartile range [IQR] 2.4, 4.9) and were followed for a median of 5.8 years [IQR 2.6, 11.3] after diagnosis. Males represented 53.7% of the cohort. Transfers of care or second opinions comprised 55.1% of the cohort. WES was completed on 82.4% of the cohort. Sequencing of trios [patient and both parents] was completed in 45.8%. Targeted genetic testing was performed in 6.0%. Monogenic diagnoses were made in a total of 17 patients, comprising 7.9% of the total cohort. Of patients with infantile-onset disease, 22.0% had monogenic diagnoses, compared with 4.6% diagnosed at age 2 to 6 years [Figure 1A]. Upon further stratification of the infantile-onset disease group, monogenic disease was found in 42.9% of those diagnosed <6 months, 41.7% diagnosed 6 months to <12 months, and 4.5% diagnosed 12 to <24 months [Figure 1B].
Table 1.
Cohort demographics [n = 216].
| Age at diagnosis, years | |
| Mean ± SD | 3.53 ± 1.58 |
| Median [range] | 3.78[0.17, 5.98] |
| 25th, 75th percentile | 2.37, 4.87 |
| VEOIBD classification by age at diagnosis | |
| Infantile [<2 years] | 41 [19.0%] |
| 2 to 6 years | 175 [81.0%] |
| Male sex | 116 [53.7%] |
| Race | |
| White | 142 [65.7%] |
| Unknown/not reported | 40 [18.5%] |
| Other | 22 [10.2%] |
| Black or African American | 6 [2.8%] |
| Asian | 5[2.3%] |
| American Indian or Alaska Native | 1[0.5%] |
| Transfer of care or second opinion | 119 [55.1%] |
| Duration of follow-up, years | |
| Mean +/- SD | 7.24 ± 5.24 |
| Median [range] | 5.83 [1.02, 21.72] |
| 25th, 75th percentile | 2.59, 11.31 |
| Year of diagnosis | |
| Prior to Jan 1, 2012 | 101 [46.8%] |
| Jan 1, 2012 or later | 115 [53.2%] |
| Genetic testing | |
| Whole exome sequencing | 178 [82.4%] |
| Whole exome sequencing, trio | 99 [45.8%] |
| Targeted genetic testing | 13 [6.0%] |
SD, standard deviation; VEOIBD, very early onset inflammatory bowel disease.
Figure 1.
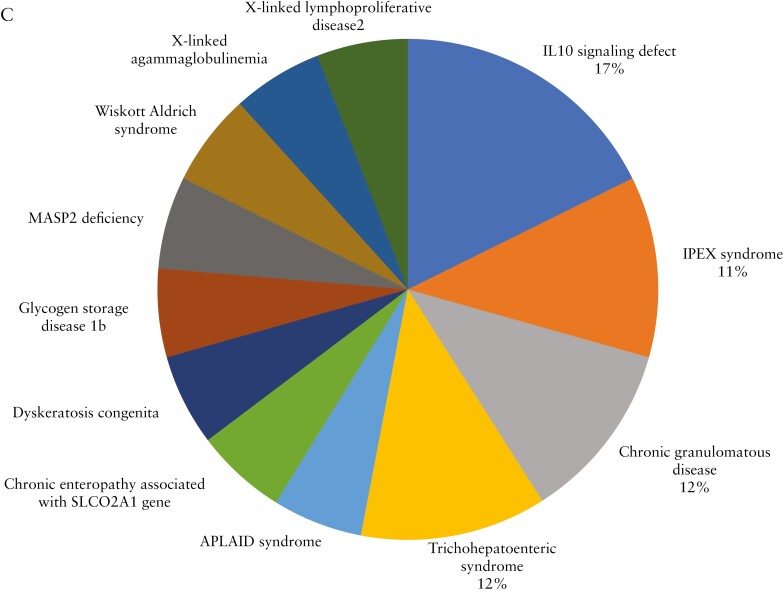
Distribution of monogenic VEOIBD by age of diagnosis and monogenic aetiology identified. [A] Proportion of patients with monogenic VEOIBD by age at diagnosis. Monogenic VEOIBD was identified in 22.0% of patients with infantile-onset IBD compared with 4.6% of patients with IBD diagnosed at age 2 to <6 years. [B] Further stratification of infantile-onset disease group identified monogenic VEOIBD in 42.9% of patients diagnosed at <6 months, 41.7% diagnosed from 6 months to <12 months, and 4.5% diagnosed from 12 to <24 months. [C] Distribution of the 17 cases of monogenic VEOIBD identified in this cohort. IL10 signaling defects were most common, followed by chronic granulomatous disease, IPEX syndrome, and trichohepatoenteric syndrome. VEOIBD, very early onset inflammatory bowel disease.
To address concern around generalisability of these prevalence data from a large referral centre, we compared prevalence of monogenic disease in the subset of patients who received their initial gastroenterology care at BCH with those who transferred care or presented for second opinion. We identified no difference in monogenic disease prevalence between these groups [8.2% in those receiving initial care at our centre, 7.6% in those who transferred care, p = 0.85], suggesting that referral bias may not have played a significant role in our prevalence findings.
3.2. Molecular characterization and clinical phenotype of patients with monogenic diagnoses
Seventeen patients had monogenic diagnoses identified [Figure 1C]. These included: IL10 signaling defects [n = 3]; chronic granulomatous disease [n = 2]; immune dysregulation, polyendocrinopathy, enteropathy, X-linked [IPEX] syndrome [n = 2]; trichohepatoenteric syndrome [n = 2]; auto-inflammation and phospholipase Cγ2-associated antibody deficiency and immune dysregulation [APLAID] syndrome [n = 1]; chronic enteropathy associated with SLCO2A1 gene [n = 1]; dyskeratosis congenita [n = 1]; glycogen storage disease type 1b [n = 1]; MASP2 deficiency [n = 1]; Wiskott–Aldrich syndrome [n = 1]; X-linked lymphoproliferative disease 2 [XLP2] [n = 1]; and X-linked agammaglobulinemia [n = 1]. Exome and clinical data for patients with monogenic diagnoses are summarised in Tables 2 and 3. An additional four patients [1.9%] inherited two heterozygous rare variants in established VEOIBD genes but were not classified as having monogenic disease, based on lack of parental sequencing to confirm compound heterozygous inheritance. For 15 of the 17 patients with monogenic disease [88.2%], IBD was a presenting feature. The remaining two had an established monogenic diagnosis at time of IBD diagnosis.
Table 2.
Rare variants in monogenic VEOIBD genes identified in cohort.
| Pt | Clinical syndrome | Gene | Mode of inheritance | Genomic location [Chr. Pos.] | Variant cDNA change | Amino acid change | Effect | ACMG classification |
|---|---|---|---|---|---|---|---|---|
| 1 | APLAID syndrome | PLCG2 | Autosomal dominant | 16:81919551 | c.2122G > C | p.Ala708Pro | Missense | Pathogenic |
| 2 | Chronic enteropathy associated with SLCO2A1 gene | SLCO2A1 | Compound heterozygous | 3:133935844 | c.1744C > T | p.Arg582Trp | Missense | Likely pathogenic |
| 3:133951215 | c.854G > A | p.Gly285Glu | Missense | VUS | ||||
| 3 | CGD | CYBB | X-linked | X:37805092 | c.1237dup | p.Val413Glyfs*18 | Frameshift | Pathogenic |
| 4 | CGD | NCF2 | Autosomal recessive | Exon 1 | Unavailable | Partial deletion of NCF2 gene that includes exon 1 | Unavailable | Pathogenic |
| 5 | Dyskeratosis congenita | RTEL1 | Compound heterozygous | 20:63661344 | c.149T > C | p.Leu50Pro | Missense | Likely pathogenic |
| 20:63688168 | c.1628G > T | p.Gly543Val | Missense | Likely pathogenic | ||||
| 6 | GSD1b | SLC37A4 | Autosomal recessive | 11:119026655 | c.818del | p.Gly273AlafsStop3 | Frameshift | Pathogenic |
| 7 | IL10 signaling defect | IL10RA | Autosomal recessive | 11:117986469 | c.2T > G | p.Met1Arg | Missense | Likely pathogenic |
| 8 | IL10 signaling defect | IL10RA | Autosomal recessive | 11:117989554 | c.301C > T | p.Arg101Trp | Missense | Pathogenic |
| 9 | IL10 signaling defect | IL10RB | Autosomal recessive | 21:33268392 | c.50-2A > T | Unavailable | Splice acceptor | Pathogenic |
| 10 | IPEX syndrome | FOXP3 | X-linked | X:49251403 | c.1226A > C | p.D409A | Missense | Likely pathogenic |
| 11 | IPEX syndrome | FOXP3 | X-linked | X:49251467 | c.1163A > G | p.N3885S | Missense | Likely pathogenic |
| 12 | MASP2 deficiency | MASP2 | Compound heterozygous | 1:11030763 | c.1207A > C | p.Met403Leu | Missense | VUS |
| 1:11046609 | c.359A > G | p.Asp120Gly | Missense | VUS | ||||
| 13 | Trichohepatoenteric syndrome | TTC37 | Compound heterozygous | 5:95517309 | c.2128C > T | p.Arg710Stop | Nonsense | Pathogenic |
| 5:95494727 | c.3757_3758delAAinsG | p.Asn1253ValfxX4 | Frameshift | Pathogenic | ||||
| 14 | Trichohepatoenteric syndrome | TTC37 | Autosomal dominant | 5:95547097 | c.53_54del | p.Arg18Lysfs*12 | Frameshift | Pathogenic |
| 15 | WAS | WASP | X-linked | Estimated deletion of 18-220Kb | Unavailable | Unavailable | Large gene deletion | Pathogenic |
| 16 | XLA | BTK | X-linked | X:101354688 | c.1573C > T | p.Arg525Ter | Stop gained | Pathogenic |
| 17 | XLP2 | BIRC4 | X-linked | X:123860449 | c. 951_961 del | p.W317fsX318 | Frameshift | Pathogenic |
VEOIBD, very early onset inflammatory bowel disease; ACMG, American College of Medical Genetics and Genomics; APLAID, auto-inflammation and phospholipase Cγ2-associated antibody deficiency and immune dysregulation; cDNA, complementary DNA; CGD, chronic granulomatous disease; Chr.Pos., chromosome position; GSD1b, glycogen storage disease 1b; IPEX, immune dysregulation, polyendocrinopathy, enteropathy, X-linked; Pt, patient; VUS, variant of unknown significance; WAS, Wiskott–Aldrich syndrome; XLA, X-linked agammaglobulinaemia; XLP2, X-linked lymphoproliferative disease 2.
Table 3.
Clinical phenotypes and outcomes of monogenic VEOIBD patients.
| Pt | Syndrome | Sex | Age at diagnosis [y] | Duration of follow-up [y] | IBD diagnosis | Paris classification | Perianal | IBD diagnosis preceded monogenic disease diagnosis | Diagnosis weight z-score | EIM | Atypical/opportunistic infections | Comorbidities | Surgery | Prior medication exposure | HSCT | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | APLAID syndrome | M | 0.6 | 1.1 | CD | L2B1 | Y | Y | -2.80 | Erythema nodosum, pyoderma gangrenosum | Multiple respiratory infections requiring intubation | Asthma, haemangioma, recurrent C.diff, hypogamma-globulinaemia, tracheal stenosis, subglottic granuloma, subglottic stenosis | N | Sulphasalazine [partial response] | N | Partial response to anakinra and immuno-globulin; pursuing match for HSCT |
| 2 | Chronic enteropathy associated with SLCO2A1 gene | M | 3.3 | 1.5 | UC | E3S0 | N | Y | 0.08 | None | None | None | N | None | N | Clinical and endoscopic remission on sulphasalazine |
| 3 | CGD | M | 2.3 | 1.5 | CD | L2B1 | N | Y | -1.05 | Fever | Fungal pulmonary nodules, multiloculated neck abscess | None | N | 5-ASA [PNR], methotrexate [PNR], sulphasalazine [AE—rash], vedolizumab [partial response, did not complete induction prior to HSCT] | Y, age 2.9 y | Curative HSCT |
| 4 | CGD | F | 4.7 | 8.7 | CD | L2B3 | Y | Y | -1.47 | None | Recurrent abscesses, pulmonary aspergillosis, osteomyelitis | Vertebral compression fracture, attention-deficit disorder, oppositional defiant disorder | N | Sulphasalazine [PNR], balsalazide [PNR], azathioprine [LOR], anakinra [PNR], vedolizumab [responder—discontinued at HSCT] | Y, age 10.0 y | Curative HSCT |
| 5 | Dyskeratosis congenita | M | 3.8 | 1.7 | CD | L2L4aB2 | Y | Y | Unknown | Oral aphthous ulcers | Recurrent pneumonias | Recurrent oesophageal stricture with multiple balloon dilations, large tongue lesions requiring excision, pancytopenia | N | n/a | N | Clinical and endoscopic remission on budesonide monotherapy; lost to follow-up |
| 6 | GSD1b | F | 2.2 | 9.0 | CD | L3L4aB2 | N | N | -0.50 | Oral ulcers, pyoderma gangrenosum | Candidal oesophagitis | Chronic neutropenia, osteoporosis, vertebral compression fractures | N | 5-ASA [LOR] | N | Partial response to infliximab |
| 7 | IL10 signaling defect | F | 0.5 | 3.6 | CD | L2B2B3 | Y | Y | Unknown | Thrombotic complication | Candidal sepsis, Gram-negative septicaemia | Eczema | Y [colonic perforation requiring segmental colonic resection and colostomy, subsequent colostomy takedown] | Azathioprine [PNR], infliximab [PNR], mesalamine [PNR], tacrolimus [AE—opportunistic infection], anakinra [clinical and endoscopic remission—bridge to HSCT] | Y, age 2.7 y | Curative HSCT |
| 8 | IL10 signaling defect | M | 3.7 | 1.8 | CD | L2L4aB1 | Y | Y | Unknown | Arthropathy, thrombotic complication, fever | None | Developmental delay, cortical visual blindness, diabetes insipidus, significant cavities at young age requiring dental extractions, left femur fracture | N | n/a | N | Remission on steroids and ciprofloxacin/metronidazole; matched HSCT donor being cautiously pursued due to concern about neurological sequelae of conditioning |
| 9 | IL10 signaling defect | F | 0.2 | 16.5 | CD | L2L4aB3 | Y | Y | Unknown | Arthropathy, folliculitis, fever | None | Severe pulmonary disease characterised by recurrent pneumonias, bronchiectasis, lobectomy, and lingulectomy; selective IgG2 deficiency, mannose-binding lectin deficiency | Y [intestinal perforation prompting emergent colectomy with permanent ileostomy] | Infliximab [failure] | N | Clinical remission on 6MP and immunoglobulin |
| 10 | IPEX syndrome | M | 0.7 | 8.3 | CD | L3L4aB1 | N | Y | 0.21 | None | None | Eczema, food allergies [anaphylaxis], medication allergies requiring desensitisation, hypogamma-globulinaemia, obsessive compulsive disorder | N | Tacrolimus [AE—HTN] | Y, age 1.2 y | HSCT complicated by graft failure, PTLD. Partial response to sirolimus, prednisone, and immunoglobulin |
| 11 | IPEX syndrome | M | 0.9 | 1.6 | CD | L2L4aB1 | N | Y | -2.87 | Thrombotic complication | Septic shock in setting of MRSA bacteraemia, EBV viraemia | Pulmonary hypertension, pulmonary hypoplasia, subpleural microcysts with tracheostomy/ventilator dependence and prior ECMO requirement | N | Tacrolimus [responder] | Y, age 2.4 y | Death at age 2.6 years. Severe gut GVHD, renal failure requiring CRRT, coagulopathy, progressive pulmonary HTN with ventilator and ECMO requirement |
| 12 | MASP2 deficiency | M | 2.6 | 1.89 | CD | L2L4aB3 | N | Y | -0.27 | None | Recurrent bacteraemia [C. tertium, ESBL E coli, Enterococcus faecalis], fungal peritonitis, recurrent thrombophlebitis and cellulitis] | Developmental delay, intestinal failure secondary to internal hernia associated with Meckel’s diverticulum complicated by abdominal compartment syndrome, DIC, sepsis | Y [small intestinal resection, colectomy, fistulectomy, enterocutaneous fistula take-down] | None | N | Clinical and endoscopic remission on vedolizumab |
| 13 | Trichohepato-enteric syndrome | F | 0.8 | 7.5 | CD | L2B1 | N | Y | -4.65 | None | None | Hepatic fibrosis, brittle hair | N | None | N | Clinical remission on subcutaneous immunoglobulin |
| 14 | Trichohepato-enteric syndrome | M | 0.3 | 8.8 | CD | L3L4aB1 | N | Y | Unknown | None | Klebsiella sepsis | Eczema, food allergies, hypogamma-globulinaemia, osteoporosis, several fractures of bilateral wrists and ankle, nephrolithiasis, nephrocalcinosis, elevated liver enzymes, trichothio-dystrophy, IUGR | N | None | N | Remission on balsalazide |
| 15 | WAS | M | 0.2 | 3.9 | CD | L2B1 | N | Y | -1.15 | None | None | Eczema, thrombocytopenia, infantile scoliosis | N | Sulphasalazine [remission—discontinued at HSCT] | Y, age 1.1 y | Curative HSCT |
| 16 | XLA | M | 5.2 | 11.2 | CD | L3L4aL4bB2 | N | N | -0.78 | Arthropathy, thrombotic complication, fever, episcleritis | Gram-negative sepsis | Psoriasis [TNF-induced], growth hormone deficiency | Y [Ileocaecectomy, proximal ileal resection, ileocolaectomy with diverting ileostomy, ileostomy revision, lysis of adhesions with right hemicolectomy and ileocolic anastomosis, ileostomy take-down] | 6MP [AE—abnormal liver enzymes], methotrexate [PNR], thalidomide [AE—peripheral neuropathy] | N | Clinical and endoscopic remission on infliximab and immunoglobulin |
| 17 | XLP2 | M | 1.5 | 1.7 | CD | L3L4aB1 | N | Y | -9.25 | None | Pulmonary aspergillosis, Stenotrophomas bacteraemia | Haemophagocytic lymphohistio-cytosis [HLH], autoimmune hepatitis, macrocystic lung disease,tracheostomy, cytopenias | N | Adalimumab [AE—clinical improvement initially but with opportunistic pneumonia requiring ventilator], immunoglobulin, cyclosporine | Y, age 2.9 y | Death at 3.1 y [cardiac arrest secondary to respiratory failure] |
VEOIBD, very early onset inflammatory bowel disease; 6MP, 6-mercaptopurine; AE, adverse event; CRRT, continuous renal replacement therapy; EBV, Epstein-Barr virus; ECMO, extracorporeal membrane oxygenation; GVHD, graft versus host disease; HTN, hypertension; IUGR, intrauterine growth restriction; LOR, loss of response; MRSA, methicillin-resistant Staphlococcus aureus; PNR, primary non-response; PTLD, post-transplant lymphoproliferative disorder; Y, years; CD, Crohn’s disease; UC, ulcerative colitis; F, female; M, male; Y, yes; N, no; HSCT, hematopoietic stem cell transplant; EIM, extraintestinal manifestation.
Overall, patients with monogenic IBD were younger at diagnosis compared with those with non-monogenic IBD [mean age 2.0 years versus 3.7 years, p = 0.001; Table 4]. Males comprised 70.6% of the monogenic cohort compared with 52.3% of the non-monogenic cohort, but this difference did not reach significance. There was no difference in duration of follow-up for monogenic versus non-monogenic sub-groups.
Table 4.
IBD phenotype in monogenic versus non-monogenic disease [n = 216].
| Monogenic [n = 17] | Non-monogenic [n = 199] | p-value | |
|---|---|---|---|
| Age at diagnosis in years, mean | 1.97 | 3.66 | 0.001 |
| Duration of follow-up in years, mean | 5.31 | 7.40 | 0.09 |
| Male sex | 12 [70.6%] | 104 [52.3%] | 0.15 |
| Infantile onset | 9 [52.9%] | 32 [16.1%] | 0.001 |
| IBD phenotype | <0.001 | ||
| Crohn’s disease [CD] | 16 [94.1%] | 92 [46.2%] | |
| Ulcerative colitis [UC] | 1 [5.9%] | 96 [48.2%] | |
| IBD-unclassified [IBD-U] | 0 [0.0%] | 11 [5.5%] | |
| IBD re-classified during study period | 1 [5.9%] | 29 [14.6%] | 0.48 |
| CD, location | |||
| L1: terminal ileal ± limited cecal disease | 0 [0.0%] | 3 [3.3%] | 1.00 |
| L2: colonic | 11 [68.8%] | 35 [38.0%] | <0.001 |
| L3: ileocolonic | 5 [31.3%] | 47 [51.1%] | 0.56 |
| L4a: upper disease proximal to ligament of Treitz | 10 [62.5%] | 35 [38.0%] | <0.001 |
| L4b: upper disease distal to ligament of Treitz and proximal to distal 1/3 ileum | 1 [6.3%] | 7 [7.6%] | 0.49 |
| Isolated upper tract disease [L4a/b only] | 0 [0.0%] | 3 [3.3%] | 1.00 |
| Isolated perianal disease | 0 [0.0%] | 3 [3.3%] | 1.00 |
| CD, behaviour | |||
| B1: non-stricturing, non-penetrating | 9 [56.3%] | 73 [79.3%] | 0.18 |
| B2: stricturing | 3 [18.8%] | 11 [12.0%] | 0.09 |
| B3: penetrating | 3 [18.8%] | 4 [4.3%] | 0.01 |
| B2B3: stricturing and penetrating | 1 [6.3%] | 3 [3.3%] | 0.28 |
| Perianal disease modifier | 5 [31.3%] | 25 [27.2%] | 0.07 |
| Stricturing, any | 4 [25.0%] | 14 [15.2%] | 0.04 |
| Penetrating, any | 4 [25.0%] | 7 [7.6%] | 0.01 |
| UC/IBD-U, location | 0.24 | ||
| E1: ulcerative proctitis | 0 [0.0%] | 1 [0.9%] | |
| E2: left-sided UC | 0 [0.0%] | 9 [8.4%] | |
| E3: extensive UC | 1 [100%] | 11 [10.3%] | |
| E4: pancolitis | 0 [0.0%] | 82 [76.6%] | |
| Unknown | 0 [0.0%] | 4 [3.7%] | |
| UC/IBD-U, severity | 1.00 | ||
| S0: never-severe | 1 [100%] | 49 [45.8%] | |
| S1: ever-severe | 0 [0.0%] | 43 [40.2%] | |
| Unknown | 0 [0.0%] | 15 [14.0%] | |
| Extraintestinal manifestations | |||
| ≥1 EIM | 9 [52.9%] | 55 [27.6%] | 0.04 |
| ≥2 EIM | 5 [29.4%] | 13 [6.5%] | 0.01 |
| Arthropathy | 3 [17.6%] | 26 [13.1%] | 0.71 |
| Enthesitis | 0 [0.0%] | 1 [0.5%] | 1.00 |
| Episcleritis | 1 [5.9%] | 0 [0.0%] | 0.08 |
| Erythema nodosum | 1 [5.9%] | 4 [2.0%] | 0.34 |
| Fever | 4 [23.5%] | 5 [2.5%] | 0.003 |
| Folliculitis | 1 [5.9%] | 1 [0.5%] | 0.15 |
| Hepatic granuloma | 0 [0.0%] | 1 [0.5%] | 1.00 |
| Oral aphthous ulcers | 2 [11.8%] | 11 [5.5%] | 0.27 |
| Orofacial Crohn’s | 0 [0.0%] | 2 [1.0%] | 1.00 |
| Primary sclerosing cholangitis | 0 [0.0%] | 7 [3.5%] | 1.00 |
| Pyoderma gangrenosum | 2 [11.8%] | 1 [0.5%] | 0.02 |
| Thrombotic complication | 4 [23.5%] | 5 [2.5%] | 0.003 |
| Uveitis | 0 [0.0%] | 1 [0.5%] | 1.00 |
| Vulvar/vaginal Crohn’s | 0 [0.0%] | 4 [2.0%] | 1.00 |
| Comorbidities | |||
| Non-IBD autoimmune/autoinflammatory disease | 2 [11.8%] | 29 [14.6%] | 1.00 |
| ≥2 non-IBD autoimmune/autoinflammatory disease | 0 [0.0%] | 7 [3.5%] | 1.00 |
| Non-IBD genetic disorder | 0 [0.0%] | 10 [5.0%] | 1.00 |
| Family history in 1st deg relative | 4 [23.5%] | 50 [25.1%] | 1.00 |
| IBD | 1 [5.9%] | 31 [15.6%] | 0.48 |
| Other autoimmune disease | 3 [17.6%] | 20 [10.1%] | 0.40 |
| Immunodeficiency | 0 [0.0%] | 1 [0.5%] | 1.00 |
3.3. IBD phenotype at follow-up
Table 4 summarises the comparison of IBD phenotypes in monogenic versus non-monogenic disease. Patients with monogenic disease overwhelmingly had a CD phenotype at last follow-up [94.1%], whereas in patients with non-monogenic disease the distribution of phenotype between CD and UC was more evenly split. Thirty patients [13.9%] had their IBD phenotype re-classified during the study period. Only 11 patients [5.1%], had a diagnosis of IBD-U at follow-up. Reasons for IBD-U diagnosis included atypical upper tract findings [n = 2], atypical ileal findings [n = 3], rectal sparing [n = 1], discontinuous colitis [n = 2], non-specific small bowel abnormality on imaging [n = 2], and colitis with growth failure [n = 1].
In terms of CD location, patients with monogenic disease were more likely to have primarily colonic [L2] disease [p <0.001], whereas ileocolonic [L3] disease was most common in the non-monogenic group. Upper tract involvement [L4a or L4b] was common in both groups, but occurred with greater frequency in patients with monogenic disease [p = 0.001]. Four patients with non-monogenic IBD did not fit standard Paris classification criteria35; three patients [1.4%] had isolated perianal CD and one patient [0.5%] had histologically confirmed orofacial CD without intestinal involvement. Stricturing disease and penetrating disease were both significantly more common in the monogenic disease group [p = 0.04 and p = 0.01, respectively]. There was a trend toward perianal disease being more common in the monogenic disease group [p = 0.07]. The majority of patients with non-monogenic CD had non-stricturing, non-penetrating disease [79.3%].
The majority of UC/IBD-U patients in the cohort had pancolitis [E4], consistent with prior literature.36,37 A subset [9.3%] had left-sided disease only [E1 or E2]. For four patients [3.7%], Paris disease location could not be assigned due to incomplete ileocolonoscopy at diagnosis. Nearly half of the UC/IBD-U sub-group had never-severe [S0] disease, whereas 40.2% had ever-severe [S1] disease, defined as Paediatric Ulcerative Colitis Activity Index [PUCAI] ≥ 65. There was no significant difference in duration of follow-up in patients with S0 compared with S1 disease [mean follow-up 6.8 years versus 7.3 years, p = 0.65]. Notably, there was only one patient with monogenic disease in the UC group and he had extensive [E3], never-severe disease.
3.4. Extraintestinal manifestations
Over half of patients with monogenic disease had at least one extraintestinal manifestation [EIM] compared with 27.6% of patients with non-monogenic disease [p = 0.04; Table 4]. A subset of patients in both monogenic and non-monogenic groups had two or more EIMs, and this feature was again more prevalent in the monogenic disease group [p = 0.01; Table 4]. Of EIMs observed in this cohort, fever, pyoderma gangrenosum, and thrombotic complications were more common in the monogenic disease group, though after controlling for parenteral nutrition use, the difference in thrombotic complications resolved. Primary sclerosing cholangitis [PSC], which affected a total of seven subjects, was found exclusively in the non-monogenic group. Orofacial and vulvar Crohn’s disease were also found exclusively in the non-monogenic group.
3.5. Comorbidities
At least one non-IBD autoimmune/autoinflammatory diagnosis was observed in 11.8% of the monogenic disease group and 14.6% of the non-monogenic disease group. Two or more non-IBD autoimmune/autoinflammatory diagnoses were observed in 3.5% of the non-monogenic disease group [Table 4]. Comorbid autoimmune/autoinflammatory diagnoses observed included psoriasis [n = 14], primary sclerosing cholangitis [n = 7], autoimmune hepatitis [n = 3], coeliac disease [n = 3], chronic regional multifocal osteomyelitis [n = 2], juvenile rheumatoid arthritis or spondyloarthropathy [n = 2], autoimmune dysautonomia [n = 1], drug-induced lupus [n = 1], psoriatic arthritis [n = 1], hidradenitis suppurativa [n = 1], idiopathic thrombocytopenic purpura [n = 1], IgA vasculitis [n = 1], periodic fever syndrome [n = 1], and pericarditis [n = 1]; see Supplementary Table 2.
A genetic disorder not causally linked to IBD was observed in 10 patients with non-monogenic IBD and zero patients with monogenic IBD [Table 4]. These disorders included choroideremia, retinitis pigmentosa, Ep-GRIN1-associated neurodevelopmental syndrome, epidermolysis bullosa, glucose-6-phosphate dehydrogenase [G6PD] deficiency, hereditary spherocytosis, thalassaemia, trisomy 9, Turner syndrome, Williams syndrome, and velocardiofacial [DiGeorge] syndrome [see Supplementary Table 2]. Epidermolysis bullosa has previously been described in association with monogenic VEOIBD6,9,38; however, the epidermolysis bullosa patient in this cohort did not harbour rare or established causative variants in any gene with known associated VEOIBD and epidermolysis bullosa phenotype, including FERMT1 and COL7A1. To our knowledge, the other genetic disorders have not been described in association with monogenic VEOIBD.
Allergic/atopic, dermatological, neurological, neuropsychiatric, and renal comorbidities were observed at similar rates in monogenic and non-monogenic disease groups. Endocrine, haematological, infectious, immunological, and pulmonary comorbidities were more common in the monogenic group. In terms of endocrine comorbidities, osteopenia/osteoporosis were diagnosed at similar rates in the monogenic group compared with the non-monogenic group [p = 0.13]. However, vertebral compression fractures were significantly more common in the monogenic IBD group [11.8% compared with 0.5%, p = 0.01]; see Supplementary Table 2.
In terms of infectious comorbidities, atypical or opportunistic infections were significantly more common in patients with monogenic compared with non-monogenic disease [p < 0.001]; see Supplementary Table 2. Infections observed in monogenic disease are listed in Table 3. Atypical or opportunistic infections seen in non-monogenic disease included cavitary pneumonia secondary to legionnaire’s disease, toxic shock syndrome, pyomyositis, severe rotavirus infection with 6-week hospitalisation, severe or recurrent sepsis in some cases complicated by acute respiratory distress syndrome [ARDS], severe herpes simplex virus [HSV] infection, nocardiosis, and refractory warts.
Hypogammaglobulinaemia was observed more frequently in monogenic [17.6%] compared with non-monogenic [0.5%] cases, whereas isolated IgA deficiency occurred only in non-monogenic disease [1.5%]; see Supplementary Table 2. Two patients in the non-monogenic group were heart transplant recipients; one was transplanted for complex congenital heart disease prior to diagnosis of VEOIBD and the other had established VEOIBD at time of transplant. Differences in haematological and pulmonary comorbidities were driven by cytopenias and inflammatory pulmonary processes, respectively, which occurred more commonly in the monogenic group.
3.6. Family history
The majority of patients with both monogenic and non-monogenic disease had no known family history of IBD, other autoimmune disease, or immunodeficiency in a first-degree relative. Positive family history of IBD in a first-degree relative was reported in 5.9% of patients with monogenic disease compared with 15.6% in non-monogenic disease [p = 0.48].
3.7. Anthropometric parameters at diagnosis
Complete anthropometric data from time of IBD diagnosis were available for 148 patients in this cohort [68.5%]. Mean weight-for-age z-score at diagnosis was lower in monogenic disease compared with in non-monogenic disease [-2.04 compared with -0.28, p = 0.04]. Four patients with monogenic VEOIBD [23.5%] and nine patients with non-monogenic VEOIBD [4.5%] were underweight or severely underweight [weight-for-age z-score < -2.0] at diagnosis [p = 0.01]. Mean length- or height-for-age z-score was -1.83 in monogenic disease compared with -0.21 in non-monogenic [p = 0.09]. Three patients with monogenic VEOIBD [17.6%] and eight patients with non-monogenic VEOIBD [4.0%] were stunted or severely stunted [length/height-for-age z-score < -2.0] at diagnosis [p = 0.04]. Mean weight-for-length or BMI z-score was -0.87 in monogenic disease and -0.20 in non-monogenic [p = 0.20]. Two patients with monogenic VEOIBD [11.8%] and nine patients with non-monogenic VEOIBD [4.5%] were wasted or severely wasted [weight-for-length/BMI z-score < -2.0] at diagnosis [p = 0.21]. Notably, seven patients with non-monogenic VEOIBD [3.5%] were overweight or obese [weight-for-length/BMI z-score > 2.0] at diagnosis.
3.8. Outcomes stratified by monogenic disease
Table 5 summarises outcomes in monogenic versus non-monogenic disease.
Table 5.
Outcomes by monogenic versus non-monogenic disease.
| Monogenic [n = 17] | Non-monogenic [n = 199] | OR [95% CI] | p-value | |
|---|---|---|---|---|
| H=Hospitalisation | 15 [88.2%] | 139 [69.8%] | 3.24 [0.88, 20.95] | 0.12 |
| ICU hospitalisation | 9 [52.9%] | 13 [6.5%] | 14.54 [4.56, 49.98] | <0.001 |
| G-tubea | 7 [41.2%] | 11 [5.5%] | 11.96 [3.74, 37.82] | <0.001 |
| TPN in hospital | 8 [47.1%] | 23 [11.6%] | 6.80 [2.35, 19.58] | <0.001 |
| TPN at home | 2 [11.8%] | 2 [1.0%] | 13.13 [1.49, 115.98] | 0.01 |
| Surgery | 4 [23.5%] | 37 [18.6%] | 1.35 [0.36, 4.06] | 0.62 |
| 5-ASA exposure | 8 [47.1%] | 164 [82.4%] | 0.19 [0.07, 0.53] | 0.001 |
| 5-ASA failure | 5 [62.5%] | 111 [67.7%] | 0.92 [0.19, 6.62] | 0.93 |
| Anti-TNF exposure | 5 [29.4%] | 111 [55.8%] | 0.33 [0.10, 0.93] | 0.04 |
| Anti-TNF failure | 3 [60.0%] | 56 [50.5%] | 2.57 [0.32, 52.92] | 0.42 |
| Weight z-score <-2 at 3-year follow-up | 2/9 [22.2%] | 9/146 [6.2%] | 3.00 [0.43,13.13] | 0.19 |
| Height z-score <-2 at 3-year follow-up | 4/9 [44.4%] | 12/146[8.2%] | 5.17 [1.30, 17.60] | 0.01 |
| BMI z-score <-2 at 3-year follow-up | 0/9 [0.0%] | 3/146 [2.1%] | n.e. | 1.00 |
| HSCT | 7 [41.2%] | 0 [0.0%] | n.e. | <0.001 |
| Death | 2 [11.8%] | 0 [0.0%] | n.e. | 0.006 |
n.e. = not estimable.
Includes one patient with monogenic disease who had gastrojejunal [GJ] tube.
3.8.1. Hospitalisation
There were high rates of hospitalisation in both monogenic and non-monogenic groups, but intensive care unit [ICU] hospitalisation was significantly more common in monogenic disease [52.9% compared with 6.5%, p < 0.001].
3.8.2. Medication exposure and failure
Exposure to 5-ASAs was more common in the non-monogenic group, perhaps related to higher proportion of UC/IBD-U diagnoses in this group. Rates of 5-ASA failure were similar in those with monogenic and non-monogenic disease who were 5-ASA exposed. There was a higher rate of anti-TNF exposure in non-monogenic IBD, though rates of anti-TNF failure were similar between groups.
3.8.3. Surgery
Intestinal surgery was performed at similar rates in monogenic versus non-monogenic disease groups [23.5% versus 18.6%, p = 0.62].
3.8.4. Nutritional and anthropometric outcomes
G-tubes and total parenteral nutrition [TPN] were both used more commonly in patients with monogenic disease [p < 0.001]. There were nine patients with monogenic disease and 146 patients with non-monogenic disease for whom anthropometric follow-up data at least 3 years after diagnosis were available. Stunting/severe stunting, defined as height-for-age z-score less than -2.0, was more common at 3-year follow-up in monogenic disease compared with non-monogenic disease [p = 0.01]. However, underweight/severely underweight [weight-for-age z-score < -2.0] and wasting/severe wasting [BMI z-score <-2.0] at 3-year follow up occurred at similar rates in both groups.
3.8.5. Haematopoietic stem cell transplant [HSCT] and death
Both HSCT and death occurred exclusively in the monogenic disease group, at rates of 41.2% and 11.8%, respectively. HSCT was performed at median age 2.7 years [IQR 1.8, 2.9; the youngest patient was 1.1 years at time of transplant and the oldest was 10.0 years]. Median time from IBD diagnosis to HSCT was 1.4 years [IQR 0.7, 1.8]. Two patients in this cohort died at mean age 2.9 years. No patients in this cohort had a malignancy or an intestinal transplant.
3.9. Outcomes stratified by infantile-onset disease
Overall, 22.0% of patients with infantile-onset disease had a monogenic diagnosis, compared with 4.6% of non-infantile onset [p = 0.001]. After controlling for monogenic disease status, infantile-onset disease was independently associated with increased odds of hospitalisation [OR 4.14, p = 0.01] and increased odds of G-tube use [OR 3.39, p = 0.03]. After adjusting for monogenic disease, there was no difference in rates of ICU hospitalisation, TPN use, surgery, 5-ASA exposure, 5-ASA failure, anti-TNF exposure, anti-TNF failure, underweight, stunting, or wasting at 3-year follow-up, HSCT, or death in the infantile-onset group compared with the group diagnosed at age 2–6 [Table 6]. These data suggests that monogenic disease status, as opposed to age of disease onset, is the more important driver of some severe outcomes in VEOIBD.
Table 6.
Outcomes by infantile versus non-infantile onset disease, adjusted for monogenic disease.
| Infantile [n = 41] | Non-infantile [n = 175] | OR unadjusted | OR adjusteda | p-value [adjusted]b | |
|---|---|---|---|---|---|
| Monogenic | 9 [22.0%] | 8 [4.6%] | 4.60 [1.67, 12.92] | -- | 0.001* |
| Hospitalisation | 37 [90.2%] | 117 [66.9%] | 4.59 [1.73, 15.85] | 4.14 [1.54, 14.45] | 0.01 |
| ICU hospitalisation | 9 [22.0%] | 13 [7.4%] | 2.57 [0.97, 6.60] | 1.40 [0.42, 4.12] | 0.56 |
| G-tube | 9 [22.0%] | 9 [5.1%] | 5.19 [1.89, 14.30] | 3.39 [1.10, 10.07] | 0.03 |
| TPN in hospital | 11 [26.8%] | 20 [11.4%] | 2.84 [1.21, 6.47] | 2.05 [0.80, 4.93] | 0.12 |
| TPN at home | 1 [2.4%] | 3 [1.7%] | 1.43 [0.07, 11.53] | 0.56 [0.02, 5.69] | 0.65 |
| Surgery | 8 [19.5%] | 33 [18.9%] | 1.04 [0.42, 2.38] | 0.99 [0.38, 2.32] | 0.98 |
| ASA exposure | 30 [73.2%] | 142 [81.1%] | 0.63 [0.29, 1.44] | 0.89 [0.39, 2.21] | 0.78 |
| ASA failure | 18 [43.9%] | 98 [56.0%] | 0.80 [0.33, 2.11] | 0.80 [0.33, 2.13] | 0.65 |
| Anti-TNF exposure | 21 [51.2%] | 95 [54.3%] | 0.88 [0.45, 1.75] | 1.07 [0.52, 2.22] | 0.85 |
| Anti-TNF failure | 13 [31.7%] | 46 [26.3%] | 1.70 [0.63, 4.89] | 1.56 [0.56, 4.64] | 0.41 |
| Weight z-score <-2 at 3-year follow-up | 3/19 [15.8%] | 8/136 [5.9%] | 1.68 [0.36, 6.13] | 1.32 [0.25, 5.29] | 0.71 |
| Height z-score <-2 at 3-year follow-up | 4/19 [21.1%] | 12/136 [8.8%] | 1.50 [0.40, 4.59] | 0.94 [0.22, 3.25] | 0.93 |
| BMI z-score <-2 at 3-year follow-up | 0/19 [0.0%] | 3/136 [2.2%] | n.e. | n.e. | 1.00 |
| HSCT | 5 [12.2%] | 2 [1.1%] | 12.01 [2.48, 86.22] | n.e. | 0.21 |
| Death | 2 [4.9%] | 0 [0.0%] | n.e. | n.e. | 1.00 |
n.e. = not estimable.
Adjusted with monogenic disease status as covariate.
p-value corresponds to adjusted OR.
4. Discussion
We present the first comparison of phenotypes and outcomes in monogenic versus non-monogenic VEOIBD, using a large single-centre VEOIBD cohort with universal access to WES. Monogenic VEOIBD was found in 7.9% of our cohort, with rates of monogenic VEOIBD in those diagnosed at less than 12 months of age exceeding 40%. Prior literature has reported prevalence of monogenic disease in VEOIBD ranging from <1% to over 30%.13,21,24,36,39–47 It is challenging to extrapolate from these reports, which used different sequencing methodologies, assessed different genes, and were affected by selection bias, in some cases selecting only patients with infantile-onset disease or more severe features for genetic testing. Geographical differences can also have significant impact on monogenic VEOIBD distribution. For example, IL10 signaling defects are significantly more common in East Asia and therefore are more highly represented in these study populations.18,47 Consanguinity can also affect monogenic IBD prevalence, with consanguinity reported in 20–30% of patients, comprising two cohorts with prevalence of monogenic IBD over 30%.21,41 Our finding of monogenic diagnoses in 7.9% of our VEOIBD cohort is quite consistent with a similar Canadian cohort study by Crowley et al., which performed WES on a large unselected paediatric IBD cohort and made monogenic diagnoses in 7.8% of the ~140 children diagnosed at age less than 6 years.39
Caution should be used in applying the prevalence of monogenic IBD in our non-population based cohort to the greater VEOIBD population. With half of our cohort comprising transfers of care or second opinions, potential for referral bias skewing toward overestimation of monogenic disease prevalence exists, though we have shown similar rates of monogenic disease diagnosis in the transfer of care group compared with those who received their initial care at BCH. We additionally classified two patients whose rare variants in VEOIBD genes were variants of unknown significance [VUS] as per ACMG criteria as having monogenic disease [Table 2], which also creates potential for overestimation of monogenic disease prevalence. However, although ACMG standards provide valuable guidance for interpretation of sequence variants, they do have important limitations relevant to how we considered these VUS in our analysis. ACMG standards tend to yield conservative estimates of pathogenicity, with stringent application of these criteria in some cases leading to false-negative results.48,49 This is particularly true when it comes to interpretation of missense variants and variants inherited in compound heterozygous fashion. By ACMG criteria, missense variants are weighted less heavily than nonsense variants, and the burden of proof with additional functional, segregation, population, or computational data is significantly greater—and in some cases not available.29 ACMG standards additionally use MAF threshold of <1% to define rare variants for all modes of inheritance, whereas we applied a threshold of <5% for compound heterozygous inheritance; our approach is supported by literature which highlights that traditional MAF filtering criteria of <1% may be overly stringent for application to compound heterozygous variants.50,51 Moreover, there is a great deal of discretion in the application and interpretation of ACMG criteria to sequence variants, with interlaboratory variability reported.49 Finally, discrepancy exists among ACMG and other systems of variant classification—for example, some variants classified as VUS in our cohort [Table 2] are classified as ‘likely pathogenic’ by ClinVar.52
Whereas the above provides rationale around how our work may overestimate monogenic disease, an alternative hypothesis is that our finding of monogenic diagnoses in 7.9% underestimates the true prevalence of monogenic disease in VEOIBD. First, despite universal availability of WES to patients in our cohort, only 82.4% completed sequencing and 6.0% completed targeted genetic testing. Second, novel genetic causes of VEOIBD are rapidly being discovered, resulting in an ever-changing landscape of monogenic IBD. This is highlighted by two high-profile reviews of monogenic VEOIBD spanning 7 years: in 2014, 50 VEOIBD genes were reported,6 whereas by 2021 there was consensus on 75 VEOIBD genes.8 For purposes of this study, disease was classified as monogenic only if rare variants were identified in established VEOIBD genes. We anticipate that as the list of functionally validated VEOIBD genes continues to grow, so too will the prevalence of monogenic IBD. Third, we did not classify four patients who were compound heterozygous for rare variants in established VEOIBD genes as having monogenic disease, given lack of parental sequencing data necessary for complete interpretation. Availability of sequencing data on trios in 45.8% of our cohort was a strength overall, but improvement in diagnostic rates of up to 16% has been reported with sequencing the proband concurrently with both biological parents, and we expect that expanding trio testing would have increased diagnostic yield.53 Finally, one patient in our cohort who was ultimately diagnosed with monogenic IBD secondary to Wiskott–Aldrich syndrome was not identified by initial sequencing. Due to high clinical suspicion, inconclusive sequencing was followed up with Wiskott–Aldrich syndrome protein [WASP protein] levels and targeted comparative genomic hybridisation testing of the gene of interest, ultimately revealing absent WASP protein levels and a large deletion in the gene of interest. Inadequate coverage for detection of large deletions or large copy number variations is a well described shortcoming of WES, and in these cases targeted gene panels can be higher yield.40–42,53 Our experience highlights the expanding role of monogenic disease in VEOIBD as well as the nuance of WES interpretation and the value of a multidisciplinary team, including clinicians and bioinformaticians, in analysing and interpreting WES data.
The majority of monogenic VEOIBD patients in our cohort exhibited a complicated Crohn’s-like phenotype, characterised by higher rates of stricturing and penetrating disease and EIMs, compared with non-monogenic VEOIBD. Prior literature around monogenic VEOIBD phenotype has had mixed findings. Two smaller cohorts of monogenic VEOIBD found relatively high rates of CD, ranging from 67% to 72%, though still lower than 94% in our cohort.43,45 In contrast, Crowley, et al. reported that nine out of 11 patients with monogenic VEOIBD had UC/IBD-U phenotype.39 This is similar to a report on an infantile-onset cohort in which monogenic disease was overwhelmingly characterised as IBD-U [14/19 cases].21 It is possible that these differences in monogenic IBD phenotype among cohorts may be related to differences in genetic defects identified or duration of follow-up. In terms of disease location, whereas it has previously been reported that VEO-CD is more likely to present with isolated colonic disease,11,37,54 this work offers the additional distinction that colonic [L2] CD is more common in monogenic CD specifically. Finally, to our knowledge, this is the first report of higher rates of stricturing and penetrating disease in monogenic VEOIBD compared with non-monogenic VEOIBD.
Notably, our cohort only had one patient with monogenic VEO-UC, characterised by extensive [E3], never-severe [S0] disease in remission on sulphasalazine. This example highlights that, although complicated CD was the most common phenotype observed in monogenic VEOIBD, even monogenic VEOIBD is clinically heterogeneous and can present with mild disease.
To our knowledge, this is the first report of outcomes in monogenic versus non-monogenic VEOIBD, with several severe outcomes, including ICU hospitalisation, gastrostomy tube use, TPN use, stunting at follow-up, HSCT, and death all significantly more common in monogenic disease. Our analysis did not detect significant differences in 5-ASA or anti-TNF failure or surgery in monogenic versus non-monogenic disease. Previous work has highlighted infantile-onset disease as a risk factor for severe disease.20,21 It is well known that infantile-onset disease is a risk factor for monogenic disease, and interestingly we found that after controlling for monogenic disease, patients with infantile-onset disease had comparable risk of most severity outcomes compared with those diagnosed at age 2 to 6 years. This supports the hypothesis that monogenic disease, as opposed to age at diagnosis, is the more important driver of disease severity and severe outcomes in VEOIBD. Furthermore, although the narrative has traditionally been that VEOIBD patients have more severe disease, it is important to recognise that there are meaningful subsets of VEOIBD patients with mild disease course. For example within non-monogenic VEO-UC, remarkably, nearly half of our cohort had never-severe [S0] disease over a mean follow-up period of 6.8 years. Other features suggestive of mild disease course highlighted in this work include subsets of patients with no hospitalisations, no surgical intervention, normal growth parameters, and 5-ASA non-failure. This adds nuance to prior literature which has focused on severe phenotypes and outcomes in VEOIBD.
It is critical to highlight that, whereas this work identifies several phenotypic features and outcomes which were more common in the monogenic disease group, monogenic VEOIBD itself is quite heterogeneous. Once monogenic disease is identified, management and prognostication should be based on current knowledge of the unique genetic defect. The value of grouping monogenic patients together in this study was 2-fold. First, it offers additional data around features potentially suggestive of monogenic disease, which are valuable as guidelines around how genetic testing should be prioritised in continuing to evolve. A recent position paper from the European Society of Paediatric Gastroenterology, Hepatology, and Nutrition [ESPGHAN] has recommended genetic screening for all patients with infantile-onset IBD and consideration of genetic screening in VEOIBD patients, especially if relevant comorbidity, EIMs, or positive family history are present.8 Our results strongly support the recommendation for genetic testing in patients with infantile-onset IBD, with particularly strong evidence for testing in those diagnosed at less than 12 months of age, where monogenic disease prevalence was over 40%. Our results also support the recommendation for genetic testing in those with EIMs and relevant comorbidities, including but not limited to cytopenias, hypogammaglobulinaemia, atypical or unusual infections, and inflammatory pulmonary conditions. Based on our findings, we would add that patients with VEOIBD and complicated Crohn’s disease phenotype [e.g. stricturing, penetrating, or perianal disease], or with anthropometric parameters consistent with moderate-to-severe underweight or stunting, should also be considered for genetic testing. Finally, it is worth noting that for 15/17 patients in our monogenic disease group, IBD was an initial presenting feature of their monogenic disease. It is therefore important to recognise that features suggestive of a monogenic disorder may evolve over time, and consideration of genetic testing should be ongoing, extending beyond the initial visit following IBD diagnosis.
The second advantage of grouping patients into heterogeneous groups of monogenic and non-monogenic VEOIBD is to add nuance to existing VEOIBD natural history literature which highlights severe phenotypes and outcomes. These reports group VEOIBD—itself a heterogeneous population—as a single entity in their analyses.13,36 Other reports that stratify VEOIBD tend to do so by infantile-onset disease.21,37 These approaches may serve to reinforce the notion that earlier onset of disease correlates to more severe disease or worse outcomes. Here we offer an alternative approach to stratifying VEOIBD, which we hope will aid in defining a wide spectrum of outcomes which may not be as dependent on age of diagnosis as initially presumed. Additional studies, including a paediatric-onset IBD [6 to 18 years] comparison group, are needed to more clearly delineate the effect of age at diagnosis on outcomes after controlling for monogenic disease status.
As access to genetic testing expands and patients with VEOIBD are increasingly able to be categorised as having monogenic versus non-monogenic disease, the value of classifying patients as having ‘VEOIBD’ is also evolving. The classification of VEOIBD offers benefit in that it may heighten provider awareness to risk of monogenic disease and offers a concrete parameter for use in guidelines to support genetic testing. However, it is critical for the clinician to recognise that not all patients with VEOIBD have monogenic disease—our current understanding is that the vast majority do not—and many patients with VEOIBD actually behave like ‘typical’ IBD. It is additionally essential for the clinician to recognise that monogenic IBD can present after age 6,6 so consideration of monogenic disease should not be limited to VEOIBD.
Overall, we found a rate of monogenic VEOIBD of 7.9% in a large single-centre cohort of patients with universal access to WES. Identification of monogenic disease allows for important prognostication of outcomes and, in some cases, precision medicine approaches, with curative HSCT performed in 41.2% of this monogenic disease cohort. Our findings reinforce the emerging recommendation that WES be standard-of-care in infantile-onset IBD and strongly considered in VEOIBD, as it permits optimal prognostication of disease course and advancement of tailored therapeutic approaches. Non-monogenic disease in our cohort was heterogeneous in phenotype, with disease ranging from mild to severe. Our work has additionally revealed subsets of VEOIBD patients with disease currently classified as non-monogenic but with phenotypic features [e.g., infantile-onset, complicated disease, with multiple EIMs] overlapping with certain monogenic disease groups. These patients will serve as the foundation for future translational work aimed at discovery of novel genetic causes of disease.
Supplementary Material
Acknowledgements
The authors thank all of the BCH patients and their families who have consented and participated in this study and the health care professionals at BCH who care for these IBD patients. Regeneron Genetics centre and Merck Research Labs supported whole exome sequencing for this cohort. Regeneron Genetics: RGC Management and Leadership Team: Goncalo Abecasis, Aris Baras, Michael Cantor, Giovanni Coppola, Andrew Deubler, Aris Economides, Katia Karalis, Luca A. Lotta, John D. Overton, Jeffrey G. Reid, Katherine Siminovitch, Alan Shuldiner,. Sequencing and Lab Operations: Christina Beechert, Caitlin Forsythe, Erin D. Fuller, Zhenhua Gu, Michael Lattari, Alexander Lopez, John D. Overton, Maria Sotiropoulos Padilla, Manasi Pradhan, Kia Manoochehri, Thomas D. Schleicher, Louis Widom, Sarah E. Wolf, Ricardo H. Ulloa. Clinical Informatics: Amelia Averitt, Nilanjana Banerjee, Michael Cantor, Dadong Li, Sameer Malhotra, Deepika Sharma, Jeffrey Staples. Genome Informatics: Xiaodong Bai, Suganthi Balasubramanian, Suying Bao, Boris Boutkov, Siying Chen, Gisu Eom, Lukas Habegger, Alicia Hawes, Shareef Khalid, Olga Krasheninina, Rouel Lanche, Adam J. Mansfield, Evan K. Maxwell, George Mitra, Mona Nafde, Sean O’Keeffe, Max Orelus, Razvan Panea, Tommy Polanco, Ayesha Rasool, Jeffrey G. Reid, William Salerno, Jeffrey C. Staples, Kathie Sun, Jiwen Xin. Analytical Genomics and Data Science: Goncalo Abecasis, Joshua Backman, Amy Damask, Lee Dobbyn, Manuel Allen Revez Ferreira, Arkopravo Ghosh, Christopher Gillies, Lauren Gurski, Eric Jorgenson, Hyun Min Kang, Michael Kessler, Jack Kosmicki, Alexander Li, Nan Lin, Daren Liu, Adam Locke, Jonathan Marchini, Anthony Marcketta, Joelle Mbatchou, Arden Moscati, Charles Paulding, Carlo Sidore, Eli Stahl, Kyoko Watanabe, Bin Ye, Blair Zhang, Andrey Ziyatdinov. Therapeutic Area Genetics: Ariane Ayer, Aysegul Guvenek, George Hindy, Giovanni Coppola, Jan Freudenberg, Jonas Bovijn Julie Horowitz, Katherine Siminovitch, Kavita Praveen, Luca A. Lotta, Manav Kapoor, Mary Haas, Moeen Riaz, Niek Verweij, Olukayode Sosina, Parsa Akbari, Priyanka Nakka, Sahar Gelfman, Sujit Gokhale, Tanima De, Veera Rajagopal, Alan Shuldiner, Bin Ye, Gannie Tzoneva, Juan Rodriguez-Flores. RGC Biology: Shek Man Chim, Valerio Donato, Aris Economides, Daniel Fernandez, Giusy Della Gatta, Alessandro Di Gioia, Kristen Howell, Katia Karalis, Lori Khrimian, Minhee Kim, Hector Martinez, Lawrence Miloscio, Sheilyn Nunez, Elias Pavlopoulos, Trikaldarshi Persaud. Research Program Management & Strategic Initiatives: Esteban Chen, Marcus B. Jones, Michelle G. LeBlanc, Jason Mighty, Lyndon J. Mitnaul, Nirupama Nishtala, Nadia Rana.
Contributor Information
Lauren V Collen, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Boston Children’s Hospital and Harvard Medical School, Boston, MA, USA.
David Y Kim, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Boston Children’s Hospital and Harvard Medical School, Boston, MA, USA.
Michael Field, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Boston Children’s Hospital and Harvard Medical School, Boston, MA, USA.
Ibeawuchi Okoroafor, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Boston Children’s Hospital and Harvard Medical School, Boston, MA, USA.
Gwen Saccocia, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Boston Children’s Hospital and Harvard Medical School, Boston, MA, USA.
Sydney Driscoll Whitcomb, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Boston Children’s Hospital and Harvard Medical School, Boston, MA, USA.
Julia Green, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Boston Children’s Hospital and Harvard Medical School, Boston, MA, USA.
Michelle Dao Dong, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Boston Children’s Hospital and Harvard Medical School, Boston, MA, USA.
Jared Barends, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Boston Children’s Hospital and Harvard Medical School, Boston, MA, USA.
Bridget Carey, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Boston Children’s Hospital and Harvard Medical School, Boston, MA, USA.
Madison E Weatherly, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Boston Children’s Hospital and Harvard Medical School, Boston, MA, USA.
Regeneron Genetics centre, Regeneron Genetics centre, Tarrytown, NY, USA.
Shira Rockowitz, Manton centre for Orphan Disease Research, Boston Children’s Hospital, Boston, MA, USA.
Piotr Sliz, Manton centre for Orphan Disease Research, Boston Children’s Hospital, Boston, MA, USA; Division of Molecular Medicine, Boston Children’s Hospital, Boston, MA, USA.
Enju Liu, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Boston Children’s Hospital and Harvard Medical School, Boston, MA, USA; Institutional centres for Clinical and Translational Research, Boston Children’s Hospital, Boston, MA, USA.
Alal Eran, Computational Health Informatics Program, Boston Children’s Hospital, Boston, MA, USA; Harvard Medical School, Department of Biomedical Informatics, Boston, MA, USA; Department of Life Sciences and Zlotowski centre for Neuroscience, Ben Gurion University of the Negev, Beer-Sheva, Israel.
Leslie Grushkin-Lerner, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Boston Children’s Hospital and Harvard Medical School, Boston, MA, USA.
Athos Bousvaros, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Boston Children’s Hospital and Harvard Medical School, Boston, MA, USA.
Aleixo M Muise, SickKids Inflammatory Bowel Disease centre, Research Institute, Hospital for Sick Children, Toronto, ON, Canada; Division of Gastroenterology, Hepatology and Nutrition, Department of Pediatrics, University of Toronto, Toronto, ON, Canada; Institute of Medical Science, University of Toronto, Toronto, ON, Canada.
Christoph Klein, Department of Pediatrics, Dr. von Hauner Children’s Hospital, LMU Klinikum, and Gene centre, Ludwig Maximilians Universität München, München,Germany.
Vanessa Mitsialis, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Boston Children’s Hospital and Harvard Medical School, Boston, MA, USA; Division of Gastroenterology, Department of Medicine, Brigham & Women’s Hospital and Harvard Medical School, Boston, MA, USA.
Jodie Ouahed, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Boston Children’s Hospital and Harvard Medical School, Boston, MA, USA.
Scott B Snapper, Division of Gastroenterology, Hepatology, and Nutrition, Department of Pediatrics, Boston Children’s Hospital and Harvard Medical School, Boston, MA, USA; Division of Gastroenterology, Department of Medicine, Brigham & Women’s Hospital and Harvard Medical School, Boston, MA, USA.
Rare variants identified in monogenic VEOIBD patients are available in the article and in its online Supplementary material. Raw whole exome sequencing data will be shared on reasonable request to the corresponding author.
Funding
LVC is supported by National Institute of Diabetes and Digestive Kidney Diseases of the National Institutes of Health [Award Number T32 DK007477]. LVC received support from Harvard Catalyst (Harvard Clinical and Translational Science Centre, National Centre for Advancing Translational Sciences of the National Institutes of Health [Award UL1 TR002541]). JO is supported by National Institute of Diabetes and Digestive Kidney Diseases of the National Institutes of Health [Award Number K08DK122133] and a Career Development Award from the Office of Faculty Development at Boston Children’s Hospital. AMM is supported by National Institute of Diabetes and Digestive Kidney Diseases of the National Institutes of Health [Award Number RC2DK118640], a Canada Research Chair [Tier 1] in Pediatric IBD, and Canadian Institutes of Health Research Foundation Grant. CK is supported by the Care-for-Rare Foundation. AMM, CK, and SBS are supported by National Institute of Diabetes and Digestive Kidney Diseases of the National Institutes of Health [Award Number RC2DK122532] and the Leona M. and Harry B. Helmsley Charitable Trust. SBS is supported by National Institute of Diabetes and Digestive Kidney Diseases of the National Institutes of Health [Award Number P30DK03485], the Wolpow Family Chair in IBD Treatment and Research, the Translational Investigator Service at Boston Children’s Hospital, and the Children’s Rare Disease Cohort [CRDC] Study.
Conflict of Interest
JO declares the following interests: independent contractor as ‘speaker’ for Janssen and consultant for Skygenics. SBS declares the following interests: scientific advisory board participation for Pfizer, BMS, Lilly, IFM Therapeutics, Merck, and Pandion; grant support from Pfizer, Novartis, Takeda; consulting for Hoffman La Roche, Takeda, and Amgen. AB declares the following interests: grant support from Prometheus, Janssen, Abbvie, Arena, Lilly; gonsulting for Arena, Takeda, Lilly.
Author Contributions
LVC: study design, patient identification, data collection, genetic analysis, statistical analysis, manuscript and figure preparation. DYK: study design, patient identification, sample acquisition, data collection. MF: patient identification, genetic analysis, manuscript and figure preparation. IO: patient identification, sample acquisition. GS: patient identification, sample acquisition. SDW: patient identification, sample acquisition. JG: patient identification, sample acquisition. MDD: patient identification, sample acquisition. JB: patient identification, sample acquisition. BC: patient identification, sample acquisition. MW: patient identification, sample acquisition. RGC: genetic analysis. SR: genetic analysis. PS: genetic analysis. EL: statistical analysis. AE: genetic analysis, manuscript and figure preparation. LGL: patient identification, manuscript review. AB: study design, patient identification, manuscript review. AMM: study design, manuscript review. CK: study design, manuscript review. VM: statistical analysis, manuscript and figure preparation. JO: study design, patient identification, genetic analysis, manuscript and figure preparation. SBS: study design, patient identification, genetic analysis, manuscript and figure preparation.
Conference presentation
Digestive Disease Week, virtual, May 2021, Pediatric IBD: Clinical and Translational Studies
References
- 1. Okou DT, Kugathasan S.. Role of genetics in pediatric inflammatory bowel disease. Inflamm Bowel Dis 2014;20[10]:1878–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen GB, Lee SH, Montgomery GW, et al. Performance of risk prediction for inflammatory bowel disease based on genotyping platform and genomic risk score method. BMC Med Genet 2017;18[1]:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cleynen I, Boucher G, Jostins L, et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet 2016;387[10014]:156–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brant SR, Panhuysen CI, Bailey-Wilson JE, et al. Linkage heterogeneity for the IBD1 locus in Crohn’s disease pedigrees by disease onset and severity. Gastroenterology 2000;119[6]:1483–90. [DOI] [PubMed] [Google Scholar]
- 5. de Ridder L, Weersma RK, Dijkstra G, et al. Genetic susceptibility has a more important role in pediatric-onset Crohn’s disease than in adult-onset Crohn’s disease. Inflamm Bowel Dis 2007;13[9]:1083–92. [DOI] [PubMed] [Google Scholar]
- 6. Uhlig HH, Schwerd T, Koletzko S, et al. The diagnostic approach to monogenic very early onset inflammatory bowel disease. Gastroenterology 2014;147[5]:990–1007 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Muise AM, Snapper SB, Kugathasan S.. The age of gene discovery in very early onset inflammatory bowel disease. Gastroenterology 2012;143[2]:285–8. [DOI] [PubMed] [Google Scholar]
- 8. Uhlig HH, Charbit-Henrion F, Kotlarz D, et al. Clinical Genomics for the Diagnosis of Monogenic Forms of Inflammatory Bowel Disease: A Position Paper From the Paediatric IBD Porto Group of European Society of Paediatric Gastroenterology, Hepatology and Nutrition. J Pediatr Gastroenterol Nutr 2021;72[3]:456–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ouahed J, Spencer E, Kotlarz D, et al. Very early onset inflammatory bowel disease: a clinical approach with a focus on the role of genetics and underlying immune deficiencies. Inflamm Bowel Dis 2020;26[6]:820–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pazmandi J, Kalinichenko A, Ardy RC, Boztug K.. Early-onset inflammatory bowel disease as a model disease to identify key regulators of immune homeostasis mechanisms. Immunol Rev 2019;287[1]:162–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oliva-Hemker M, Hutfless S, Al Kazzi ES, et al. Clinical presentation and five-year therapeutic management of very early-onset inflammatory bowel disease in a large North American cohort. J Pediatr 2015;167[3]:527–32 e1-3. [DOI] [PubMed] [Google Scholar]
- 12. Kelsen JR, Sullivan KE, Rabizadeh S, et al. North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition Position Paper on the Evaluation and Management for Patients With Very Early-onset Inflammatory Bowel Disease. J Pediatr Gastroenterol Nutr 2020;70[3]:389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kelsen JR, Conrad MA, Dawany N, et al. The unique disease course of children with very early onset-inflammatory bowel disease. Inflamm Bowel Dis 2020;26[6]:909–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Benchimol EI, Mack DR, Nguyen GC, et al. Incidence, outcomes, and health services burden of very early onset inflammatory bowel disease. Gastroenterology 2014;147[4]:803–13 e7; quiz e14-5. [DOI] [PubMed] [Google Scholar]
- 15. Benchimol EI, Bernstein CN, Bitton A, et al. Trends in Epidemiology of Pediatric Inflammatory Bowel Disease in Canada: Distributed Network Analysis of Multiple Population-Based Provincial Health Administrative Databases. Am J Gastroenterol 2017;112[7]:1120–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glocker EO, Kotlarz D, Boztug K, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med 2009;361[21]:2033–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shouval DS, Biswas A, Kang YH, et al. Interleukin 1beta mediates intestinal inflammation in mice and patients with interleukin 10 receptor deficiency. Gastroenterology 2016;151[6]:1100–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zheng C, Huang Y, Hu W, et al. Phenotypic characterization of very early-onset inflammatory bowel disease with interleukin-10 signaling deficiency: based on a large cohort study. Inflammatory Bowel Diseases 2019;25[4]:756–66. [DOI] [PubMed] [Google Scholar]
- 19. Kotlarz D, Beier R, Murugan D, et al. Loss of interleukin-10 signaling and infantile inflammatory bowel disease: implications for diagnosis and therapy. Gastroenterology 2012;143[2]:347–55. [DOI] [PubMed] [Google Scholar]
- 20. Ruemmele FM, Khoury MGE, Talbotec C, et al. Characteristics of inflammatory bowel disease with onset during the first year of life. J Pediatr Gastroenterol Nutr 2006;43[5]:603–9. [DOI] [PubMed] [Google Scholar]
- 21. Kammermeier J, Dziubak R, Pescarin M, et al. Phenotypic and genotypic characterisation of inflammatory bowel disease presenting before the age of 2 years. J Crohns Colitis 2017;11[1]:60–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levine A, Koletzko S, Turner D, et al. ESPGHAN revised Porto criteria for the diagnosis of inflammatory bowel disease in children and adolescents. J Pediatr Gastroenterol Nutr 2014;58[6]:795–806. [DOI] [PubMed] [Google Scholar]
- 23. Rockowitz S, LeCompte N, Carmack M, et al. Children’s rare disease cohorts: an integrative research and clinical genomics initiative. NPJ Genom Med 2020;5:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kammermeier J, Drury S, James CT, et al. Targeted gene panel sequencing in children with very early onset inflammatory bowel disease: evaluation and prospective analysis. J Med Genet 2014;51[11]:748–55. [DOI] [PubMed] [Google Scholar]
- 25. Dobbs K, Dominguez Conde C, Zhang SY, et al. Inherited DOCK2 deficiency in patients with early-onset invasive infections. N Engl J Med 2015;372[25]:2409–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kelsen J, Boztug K.. Autoinflammatory diseases predominantly affecting the gastrointestinal tract. In: Sullivan KE, Stiehm ER, editors. Stiehm's Immune Deficiencies. 2nd edn. Academic Press; 2020: 721–735. ISBN: 9780128167687. [Google Scholar]
- 27. Ouahed J, Kelsen JR, Spessott WA, et al. Variants in STXBP3 are associated with very early onset inflammatory bowel disease, bilateral sensorineural hearing loss, and immune dysregulation. J Crohns Colitis. 2021;15[11]:1908–19. doi: 10.1093/ecco-jcc/jjab077. PMID: 33891011; PMCID: PMC8575043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581[7809]:434–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17[5]:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG.. Research electronic data capture [REDCap]: a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42[2]:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harris PA, Taylor R, Minor BL, et al. The REDCap consortium: Building an international community of software platform partners. J Biomed Inform 2019;95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bousvaros A, Antonioli DA, Colletti RB, et al. ; North American Society for Pediatric Gastroenterology Hepatology, and Nutrition; Crohn’s and Colitis Foundation of America. Differentiating ulcerative colitis from Crohn disease in children and young adults: report of a working group of the North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition and the Crohn’s and Colitis Foundation of America. J Pediatr Gastroenterol Nutr 2007;44[5]:653–74. [DOI] [PubMed] [Google Scholar]
- 33. Levine A, Griffiths A, Markowitz J, et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis 2011;17[6]:1314–21. [DOI] [PubMed] [Google Scholar]
- 34. R Foundation for Statistical Computing. R : A Language and Environment for Statistical Computing. 2021. https://www.R-project.org/. Accessed January 31, 2022. [Google Scholar]
- 35. Van Limbergen J, Russell RK, Drummond HE, et al. Definition of phenotypic characteristics of childhood-onset inflammatory bowel disease. Gastroenterology 2008;135[4]:1114–22. [DOI] [PubMed] [Google Scholar]
- 36. Kerur B, Benchimol EI, Fiedler K, et al. Natural history of very early onset inflammatory bowel disease in North America: a retrospective cohort study. Inflamm Bowel Dis 2021;27[3]:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aloi M, Lionetti P, Barabino A, et al. Phenotype and disease course of early-onset pediatric inflammatory bowel disease. Inflamm Bowel Dis 2014;20[4]:597–605. [DOI] [PubMed] [Google Scholar]
- 38. Takeichi T, Liu L, Fong K, et al. Whole-exome sequencing improves mutation detection in a diagnostic epidermolysis bullosa laboratory. Br J Dermatol 2015;172[1]:94–100. [DOI] [PubMed] [Google Scholar]
- 39. Crowley E, Warner N, Pan J, et al. Prevalence and clinical features of inflammatory bowel diseases associated with monogenic variants, identified by whole-exome sequencing in 1000 children at a single centre. Gastroenterology 2020;158[8]:2208–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Petersen BS, August D, Abt R, et al. Targeted gene panel sequencing for early-onset inflammatory bowel disease and chronic diarrhea. Inflamm Bowel Dis 2017;23[12]:2109–20. [DOI] [PubMed] [Google Scholar]
- 41. Charbit-Henrion F, Parlato M, Hanein S, et al. Diagnostic yield of next-generation sequencing in very early onset inflammatory bowel diseases: a multicentre study. J Crohns Colitis 2018;12[9]:1104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kelsen JR, Dawany N, Moran CJ, et al. Exome sequencing analysis reveals variants in primary immunodeficiency genes in patients with very early onset inflammatory bowel disease. Gastroenterology 2015;149[6]:1415–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lega S, Pin A, Arrigo S, et al. Diagnostic approach to monogenic inflammatory bowel disease in clinical practice: a ten-year multicentric experience. Inflamm Bowel Dis 2020;26[5]:720–7. [DOI] [PubMed] [Google Scholar]
- 44. Ashton JJ, Andreoletti G, Coelho T, et al. Identification of variants in genes associated with single-gene inflammatory bowel disease by whole-exome sequencing. Inflamm Bowel Dis 2016;22[10]:2317–27. [DOI] [PubMed] [Google Scholar]
- 45. Kim KY, Lee EJ, Kim JW, et al. Higher morbidity of monogenic inflammatory bowel disease compared with the adolescent onset inflammatory bowel disease. Pediatr Gastroenterol Hepatol Nutr 2018;21[1]:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cannioto Z, Berti I, Martelossi S, et al. IBD and IBD mimicking enterocolitis in children younger than 2 years of age. Eur J Pediatr 2009;168[2]:149–55. [DOI] [PubMed] [Google Scholar]
- 47. Fang YH, Luo YY, Yu JD, Lou JG, Chen J.. Phenotypic and genotypic characterization of inflammatory bowel disease in children under six years of age in China. World J Gastroenterol 2018;24[9]:1035–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim YE, Ki CS, Jang MA.. Challenges and considerations in sequence variant interpretation for mendelian disorders. Ann Lab Med 2019;39[5]:421–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Amendola LM, Jarvik GP, Leo MC, et al. Performance of ACMG-AMP variant interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet 2016;98[6]:1067–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miller DB, Piccolo SR.. Compound heterozygous variants in pediatric cancers: a systematic review. Front Genet 2020;11:493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Miller DB, Piccolo SR.. A survey of compound heterozygous variants in pediatric cancers and structural birth defects. Front Genet 2021;12:640242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Landrum MJ, Lee JM, Benson M, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018;46[D1]:D1062–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Adams DR, Eng CM.. Next-generation sequencing to diagnose suspected genetic disorders. N Engl J Med 2018;379[14]:1353–62. [DOI] [PubMed] [Google Scholar]
- 54. Heyman MB, Kirschner BS, Gold BD, et al. Children with early-onset inflammatory bowel disease [IBD]: analysis of a pediatric IBD consortium registry. J Pediatr 2005;146[1]:35–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.