Abstract
Background and Aims
Invariant natural killer T [iNKT] cells perform pleiotropic functions in different tissues by secreting a vast array of pro-inflammatory and cytotoxic molecules. However, the presence and function of human intestinal iNKT cells capable of secreting immunomodulatory molecules such as IL-10 has never been reported so far. Here we describe for the first time the presence of IL10-producing iNKT cells [NKT10 cells] in the intestinal lamina propria of healthy individuals and of Crohn’s disease [CD] patients.
Methods
Frequency and phenotype of NKT10 cells were analysed ex vivo from intestinal specimens of Crohn’s disease [n = 17] and controls [n = 7]. Stable CD-derived intestinal NKT10 cell lines were used to perform in vitro suppression assays and co-cultures with patient-derived mucosa-associated microbiota. Experimental colitis models were performed by adoptive cell transfer of splenic naïve CD4+ T cells in the presence or absence of IL10-sufficient or -deficient iNKT cells. In vivo induction of NKT10 cells was performed by administration of short chain fatty acids [SCFA] by oral gavage.
Results
Patient-derived intestinal NKT10 cells demonstrated suppressive capabilities towards pathogenic CD4+ T cells. The presence of increased proportions of mucosal NKT10 cells associated with better clinical outcomes in CD patients. Moreover, an intestinal microbial community enriched in SCFA-producing bacteria sustained the production of IL10 by iNKT cells. Finally, IL10-deficient iNKT cells failed to control the pathogenic activity of adoptively transferred CD4+ T cells in an experimental colitis model.
Conclusions
These results describe an unprecedentd IL10-mediated immunoregulatory role of intestinal iNKT cells in controlling the pathogenic functions of mucosal T helper subsets and in maintaining the intestinal immune homeostasis.
Keywords: Crohn’s disease, IL10, iNKT cells, SCFA, microbiota
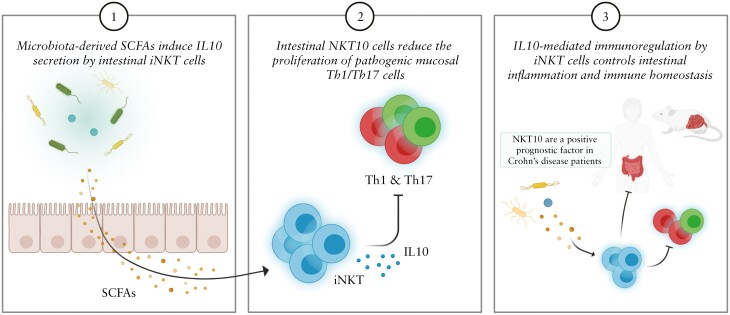
Graphical Abstract
Graphical Abstract.
1. Introduction
Invariant natural killer T [iNKT] cells are a unique T cell subset bearing a semi-invariantly rearranged αβ T Cell Receptor [TCR] and recognising lipid antigens presented in the context of the non-polymorphic, Major Histocompatibility Complex [MHC]-I-like CD1d molecules.1 In mice and men the cytokine profile and transcription factor signature subdivide iNKT cells into distinct functional populations, i.e., NKT1, NKT2, and NKT17, closely resembling CD4+ conventional T helper cells.2–4 These iNKT cell populations have been shown to participate in immune responses against infectious microorganisms [bacteria, viruses, parasites5], in autoimmune pathologies with either protective or detrimental roles,6 and in anti-tumour responses.7,8
Recently, IL10-producing iNKT cells [NKT10] have been reported in visceral adipose tissues and peripheral blood,9 but their presence has never been reported in the human intestinal compartment, either during homeostatic or in pathological conditions. In murine models of experimental autoimmune encephalomyelitis [EAE],2,10 obesity, and diabetes,9,11,12 NKT10 cells have been associated with regulatory and protective functions, to suppress pro-inflammatory macrophages9 and to promote tolerogenic dendritic cell differentiation.13 Murine iNKT cells can also directly suppress T cell pathogenicity in EAE-affected mice by inhibiting IL17A and/or IFN gamma [IFNγ] production of T helper cells in an IL4/IL10-dependent manner.10
Crohn’s disease [CD], a form of inflammatory bowel disease [IBD], is characterised by a pathological inflammation of the gastrointestinal tract which is driven by abnormal T cell responses14 in genetically susceptible individuals. Genome-wide association studies [GWAS] identified more than 200 genetic risk loci, including genes involved in IL10 signalling,15 and patients bearing mutations in the IL10 pathway [IL10, IL10R, STAT3] manifest early and more severe disease due to the expansion of pro-inflammatory Th17 cells.16 Finally, abnormalities in the IL10 signalling pathway17,18 are invariantly associated with intestinal inflammation in animal models.
The involvement of iNKT cells in the inflammatory processes occurring in IBD patients is still not completely understood.19–22 We previously observed that murine iNKT cells release pro-inflammatory cytokines, such as IFNγ, IL17 and IL13 during the acute23,24 and chronic25 phases of intestinal inflammation in mice. Moreover, iNKT cells are present in IBD patients’ lamina propria where, concomitantly with an altered gut microbiota, they secrete pro-inflammatory cytokines.22 Yet, murine iNKT cells have been shown to contribute to gut homeostasis maintenance through the CD1d-mediated induction of IL10 expression in intestinal epithelial cells.26 Recently, we showed the presence of NKT10 cells upon restoration of normobiosis by faecal microbiota transplantation [FMT] in the colons of mice suffering from intestinal inflammation.23 However, there is no evidence so far concerning the existence of human NKT10 cell in the intestinal compartment and what might be their functions during homeostatic or pathological conditions.
Building up from our previous work in mice,23 we here describe for the first time the presence of human IL10-producing iNKT cells in the lamina propria of healthy individuals and CD patients. Mechanistically, we show that murine iNKT cells are capable of exerting regulatory functions by inhibiting the proliferation of pathogenic CD4+T cells in an IL10-dependent manner. In Crohn’s disease patients, the abundance of NKT10 cells inversely correlated with pro-inflammatory Th1 and Th17 cells, and those showing higher frequencies of colonic NKT10 reported better clinical outcomes. Similarly to what observed in mice, iNKT cells isolated from NKT10-high CD patients were also suppressive towards CD4+ T cell proliferation in vitro. Moreover, the abundance of NKT10 cells associated with the presence of an intestinal mucosal microbiota enriched in health-promoting short chain fatty acid [SCFA]-producing bacteria, both in mice and in CD patients.
2. Results
2.1. iNKT cells are not colitogenic when adoptively transferred into lymphopenic and control CD4+ T cell pathogenicity in vivo
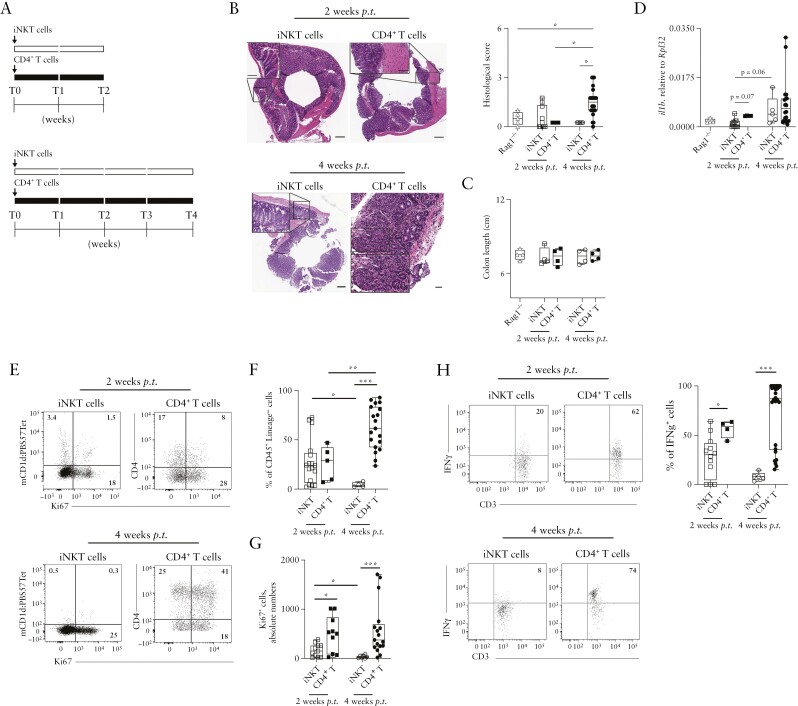
iNKT cells represent the predominant subset of CD3+ lymphocytes in murine liver and they are present in other peripheral organs including the colon [Supplementary Figure S1A–D]. Interestingly, as compared with cells isolated from liver, spleen, and blood, iNKT cells from colon lamina propria [LP], visceral adipose tissue [VAT], and mesenteric lymphnodes [mLNs] were specifically characterised by a high expression of IL1023,27 [Supplementary Figure S1E], thus suggesting that also the intestinal microenvironment might induce IL10 in resident or recently egressed intestinal iNKT cells. Consistently, IL-10 expression was driven by the E4BP4 transcription factor, as previously observed in VAT-associated iNKT cells11 [Supplementary Figure S1F and G]. Furthermore, intestinal iNKT cells expressed TIM-3, PD-1, and LAG3, surface receptors previously associated with IL10-expressing immune-suppressive mucosal cells such as Tr128,29 [Supplementary Figure S1H]. To note, the expression of IL4, an immunomodulatory cytokine produced by iNKT cells,30 has been detected only in the mLN. To test whether iNKT cells might have a suppressive phenotype in vivo, we first excluded that iNKT were colitogenic per se by adoptive transfer of splenic iNKT cells into lymphopenic Rag1-/- mice [Figure 1A; and Supplementary Figure S2A]. Whereas conventional naïve CD4+ T cells transferred into Rag1-/- mice induced a transmural colitis,31 the same number of mCD1d:PBS57 Tetramer+-sorted iNKT cells failed to do so [Figure 1B], although we did not observe significant changes in colon length [Figure 1C] and body weight [Supplementary Figure S2B]. Colonic expression of Il1β, upregulated during colitis in mice and humans,23,32 showed higher levels of induction 4 weeks after conventional CD4+ T cell transfer, compared with iNKT cell transfer [Figure 1D].
Figure 1.
iNKT cells are not pro-inflammatory when adoptively transferred in Rag1-/- hosts. [A] Schematic representation of the experiment. [B] H&E staining [left, scalebar 100 µm] and cumulative histological score [right plots] of colon specimens obtained at sacrifice 2 weeks [square symbols] or 4 weeks [round symbols] post iNKT [white symbols] or naïve CD4+ T [black symbols] transfer; Rag1-/- mice controls are also shown. [C] Colon length. [D] Il1β colonic expression at sacrifice 2- or 4-weeks post transfer. [E-G] Frequency of colonic iNKT cells and CD4+ T cells at sacrifice. Representative dot plots [E] for total [F] and Ki67+ [G] colonic iNKT cells [white symbols] and CD4+ T cells [black symbols] 2 or 4 weeks after transfer gated on CD45+ Lin- cells. [H] Representative dot plots and cumulative percentages of IFNγ-producing colonic iNKT cells and CD4+ T cells 2 or 4 weeks after adoptive transfer. Statistical significance was calculated using the Mann-Whitney U test corrected for multiple comparisons by controlling the false-discovery rate [FDR]; *p ≤0.05, **p ≤0.01, ***p ≤0.001. H&E, haematoxylin and eosin.
Moreover, colonic CD4+ T cell frequencies steadily increased over time, whereas iNKT cells were detectable for the 2 weeks after transfer but their presence decreased thereafter [Figure 1F]. The observed lack of expansion of iNKT cells was in line with their reduced proliferative capacity [Figure 1E and G] and activation status [Supplementary Figure S2C] as compared with conventional CD4+ T cells. The vast majority of transplanted iNKT cells homed to the intestine and expressed IL-10 [Supplementary Figure S2D-G]. With respect to colonic CD4+ T cells, iNKT cells expressed also significantly lower levels of the pro-inflammatory cytokine IFNγ [Figure 1H].
These results collectively indicate that iNKT cells isolated from healthy mice are not pro-inflammatory per se when adoptively transferred into a lymphopenic host.
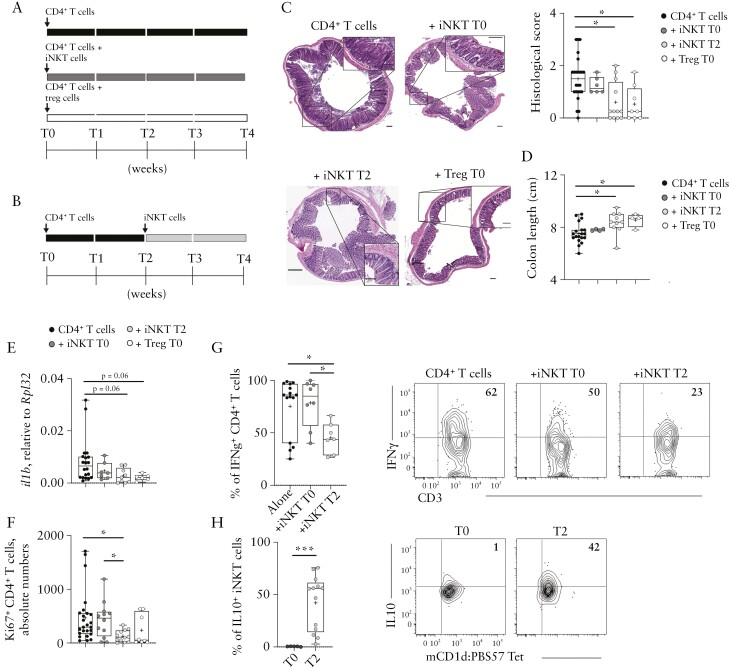
We next wondered whether iNKT cells might be directly endowed with regulatory functions, similar to Treg or Tr1 cells.29,31 According to the protocol described originally for Tregs,31 we co-transferred iNKT cells with naïve CD4+ T cells [T0] [Figure 2A]. Because of the low expansion capacity that iNKT cells showed in vivo [Figure 1E and F], we tested the suppressive capacity of iNKT cells also by injecting them 2 weeks after [T2] CD4+ T cells colitis induction [T0] [Figure 2B], i.e., when CD4+ T cell expansion was already detectable.
Figure 2.
iNKT cells suppress colitogenic CD4+T cells in vivo. [A, B] Schematic representation of the experiment.[C] H&E staining [C, left, scalebar 100 µm] and cumulative histological score [C, right], [D] colon length, [E] Il1β colonic expression, and [F] Ki67+CD4+ T cells absolute numbers in colon specimens of the indicated experimental groups. [G] Representative dot plots and cumulative frequency of IFNγ-producing colonic CD4+ T cells and [H] IL10-producing colonic iNKT cells. Statistical significance was calculated using the Kruskal-Wallis test corrected for multiple comparisons by controlling the false-discovery rate [FDR]; *p ≤0.05, **p ≤0.01, ***p ≤0.001. H&E, haematoxylin and eosin.
Histological examination and colon length variation revealed that inflammation and tissue damage induced by pathogenic CD4+ T cells were reduced upon a delayed iNKT cell transfer, but not in the original setting, i.e., injected at T0 [Figure 2C] as described by Powrie and colleagues.31 As expected, the co-transfer of Treg cells prevented the pathogenic CD4+ T cell-induced colitis [Figure 2C and D]. iNKT cells injected at T2 showed also a comparable ability to modulate the colonic expression of Il1β with respect to Treg cells [Figure 2E]. Furthermore, upon transfer of iNKT cells at T2, colonic CD4+ T cells showed a reduced proliferative capacity, not observed when iNKT cells were simultaneously co-transferred with naïve CD4+ T cells [T0] [Figure 2F]. Consistently, CD4+ T cells expressed less IFNγ when iNKT cells were injected at T2 but not when they were co-injected at T0 [Figure 2G], a condition associated with the highest levels of IL10 secretion by iNKT cells [Figure 2H]. iNKT cells injected at T0 were mostly absent 4 weeks after transfer in the colon, mLN, and spleen.
2.2. iNKT cells control CD4+ T cell pathogenicity through IL10 secretion
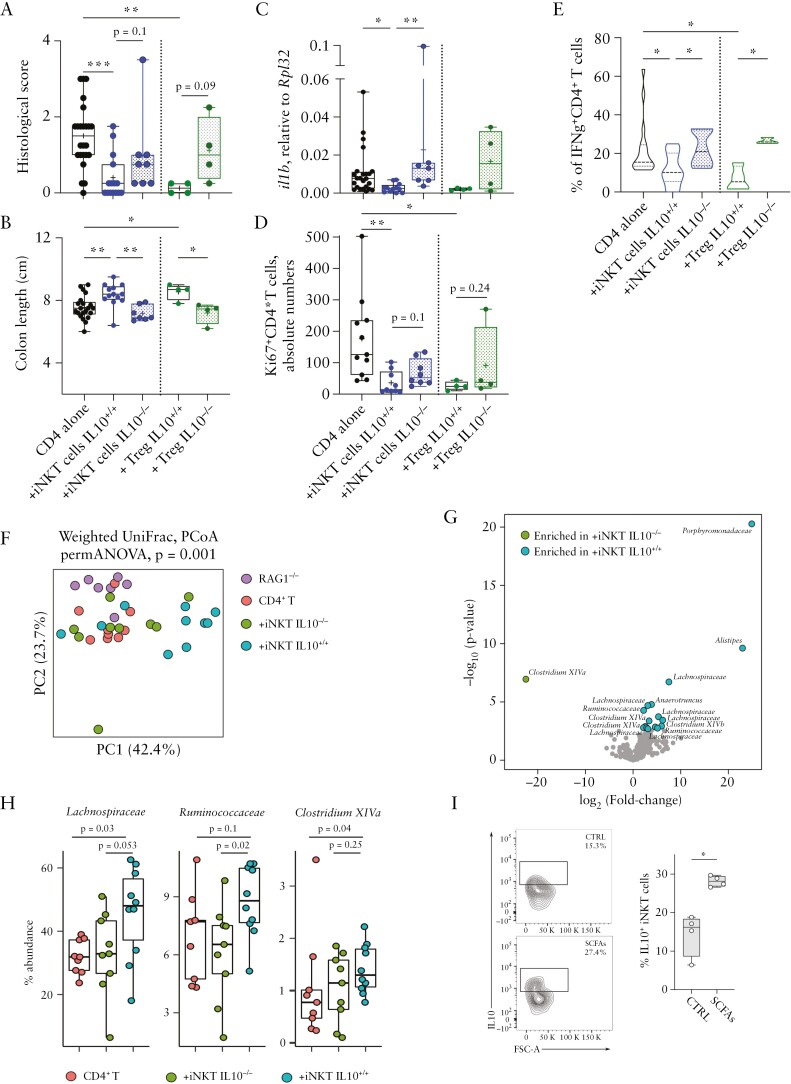
To evaluate if IL10 secretion by iNKT cells was required for their immunoregulatory function, we adoptively transferred IL10-deficient or -sufficient iNKT cells into colitic mice. The characterisation of IL10-\- mice showed no differences in the frequency and phenotype of iNKT cells compared with wild-type [WT] animals [Supplementary Figure S2H–K]. The IL10-proficient iNKT cells were able to control the CD4+ T cell-driven inflammation as measured by histoscores, colon length, and colonic Il1β expression analysis [Figure 3A–C], and reduced proliferation status of pathogenic CD4+ T cells [Figure 3D], similarly to Tregs. The production of IFNγ by colonic CD4+ T cells was also significantly reduced in presence of iNKT cell-derived IL10 [Figure 3E].
Figure 3.
IL10 secretion by iNKT is required to suppress colitogenic CD4+T cells. [A] Cumulative histological scores, [B] colon length, [C] Il1β colonic expression, [D] Ki67+CD4+ T cells absolute numbers, and [E] frequency of IFNγ-producing colonic CD4+ T cells in colon specimens of the indicated experimental groups. Statistical significance was calculated using the Kruskal-Wallis test corrected for multiple comparisons by controlling the false-discovery rate [FDR]; *p ≤0.05, **p ≤0.01, ***p ≤0.001. [F] PCoA of microbiota beta-diversity as measured by Weighted UniFrac distance. [G] Volcano plot representing the significantly enriched bacterial taxa [FDR p <0.05] in transplanted IL10+/+ vs IL10-/- iNKT animals by DEseq2 analysis. The names of the significantly enriched amplicon sequence variants [ASVs] classified to the genus and family levels are reported. [H] Relative abundance of the significantly enriched SCFA-producing bacteria identified by DEseq2 analysis in mice injected with naïve CD4+T, IL10-deficient iNKT [+iNKT IL10-/-] and IL10-proficient iNKT [+iNKT IL10+/+] cells. [I] Representative dot plots and cumulative graphs of IL10 production by murine colonic iNKT cells upon in vivo exposure to SCFAs. Statistical significance was calculated using the Kruskal-Wallis test corrected for multiple comparisons by controlling the false-discovery rate [FDR]; *p ≤0.05. SCFA, short chain fatty acid.
The gut microbiota is implicated in the functional plasticity of intestinal T cells,33 including iNKT cells.23 Analysis of the microbial community structure in iNKT and Treg-transplanted animals [Figure 3F; and Supplementary Figure S3] revealed a distinct clustering of the gut microbiota isolated from mice receiving IL10+/+ or IL10-/- iNKT cells, the latter being more similar to CD4+ T cell-induced colitic mice [Figure 3F].
In particular, mice transferred with IL10-producing iNKT cells were enriched in SCFA-producing bacteria, such as Clostridium XIVa, Lachnospiraceae, and Ruminococcaceae [Figure 3G and H]. Importantly, in vivo administration of SCFAs induced sustained secretion of IL10 by colonic iNKT cells [Figure 3I].
Altogether, these results suggest that iNKT cells can control T cell-mediated intestinal inflammation by suppressing CD4+ T cell pathogenicity through the secretion of IL10, favouring the structure of a microbial community enriched in SCFA-producing gut microbes.
2.3. Crohn’s disease patient-derived intestinal NKT10 cells suppress CD4+ T cells
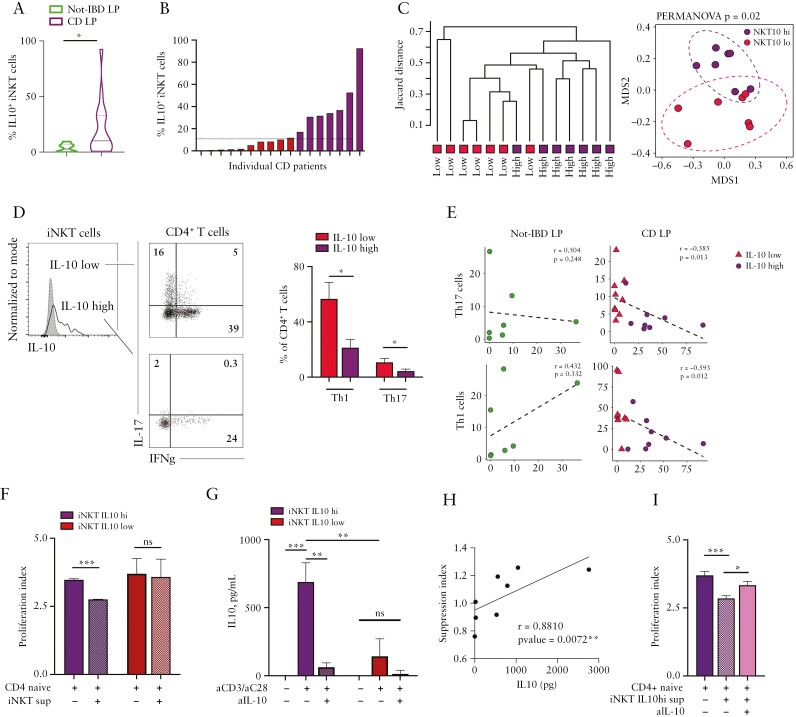
Next, we tested if IL10+iNKT [NKT10] cells were also present in the human gut and if they were capable of modulating gut inflammation. To do so, lamina propria [LP] mononuclear cells from patients suffering from Crohn’s Disease [CD] were isolated and analysed by multiparametric flow cytometry [Supplementary FigureS4A and B]; uninflamed specimens from patients undergoing surgical resection for intestinal tumours [non-IBD] were analysed as controls [Table 1]. We observed a significantly higher abundance of NKT10 cells in the LP of CD patients [Figure 4A] which was unrelated to the age of the enrolled subjects [r = -0.27, p = 0.14]. Since CD patients showed also a wider frequency distribution of NKT10 than non-IBD subjects [Figure 4A], we subdivided CD patients according to their median NKT10 abundance as threshold, i.e., in NKT10-high [NKT10hi] and NKT10-low [NKT10lo] patients [Figure 4B]. NKT10hi and NKT10lo patients differed not only because of their IL10+iNKT cell abundance, but clustered apart also according to their intestinal T cell immunophenotype [Figure 4C]. NKT10hi patients showed reduced infiltration of pro-inflammatory Th1 and Th17 cells in the intestinal LP [Figure 4D] as well as a reduction of infiltrating antigen-presenting cells [APC cells] including dendritic cells [DCs], macrophages, and monocytes [Supplementary Figure S4C]. Furthermore, the frequency of NKT10 correlated negatively with the abundance of Th1 and Th17 in the LP of CD patients, whereas no significant correlations were observed in the LP of non-IBD patients [Figure 4E]. To note, the frequency of NKT10 cells mirrored the expression of IL10 and of TGFB in the colonic tissue of CD patients, although to a less significant extent [Supplementary Figure S4D]; there was no overlap between IL10-producing iNKT cells and IL13-, IFNγ- or IL17-producing ones [Supplementary Figure S5].
Table 1.
Clinical parameters
| Clinical parameter | Crohn’s disease | Healthy donors |
|---|---|---|
| n = 17 | n = 7 | |
| Male [%] | 14 [82.35] | 4 [57.1] |
| Age at enrolment [mean ± SD, yr] | 41.9 [±11.8] | 64.9 [±8.1] |
| Disease duration [mean ± SD, yr] | 15.4 [±10] | - |
| Smoking status [yes/no/ex] | 5/2/10 | - |
| Crohn’s disease location [] | ||
| L1 [ileal] | 2 [11.76] | - |
| L2 [colonic] | 1 [5.88] | - |
| L3 [ileocolonic] | 14 [82.35] | - |
| L4 [upper disease] | 0 [0] | - |
| Crohn’s disease behaviour [%] | ||
| B1 [non-stricturing, non-penetrating] | 3 [17.64] | - |
| B2 [stricturing] | 2 [11.76] | - |
| B3 [penetrating] | 12 [70.58] | - |
| Concomitant therapy at enrolment [%] | 3 [17.64] | - |
| Antibiotics [%] | ||
| Mesalazine [%] | 2 [11.76] | - |
| Thiopurines[%] | 3 [17.64] | - |
| Corticosteroids [%] | 2 [11.76] | - |
| Anti-TNF | 6 [35.29] | - |
| Vedolizumab | 1 [5.88] | - |
| Anti-IL23 | 1 [5.88] | - |
| None | 4 [23.52] | - |
| HBI score [median; IQR] | 6 [3.5] | - |
| SES-CD [median; IQR] | 8.5 [4.25] | - |
SD, standard deviation; yr, year; TNF, tumour necrosis factor; HBI, Harvey-Bradshaw Index; SES-CD, Simple endoscopic score for Crohn’s disease.
Figure 4.
Intestinal NKT10 cells derived from CD patients suppress naïve CD4+ T cell proliferation through IL10 production. [A] Frequency of IL10+ iNKT cells among intestinal LPMC of non-IBD patients [green, n = 7] and CD patients [purple, n = 17]; *p ≤0.05, Mann-Whitney U test. [B] Individual IL10+iNKT cells abundance in LPMC of CD patients [red bars, NKT10-low patients; purple bars, NKT10-high patients]; the horizontal line indicates the median IL10+iNKT frequency used to stratify patients according to NKT10, i.e., NKT10-high and -low. [C] Hierarchical clustering and multidimensional scaling analysis of immunophenotyped NKT10-high and -low CD patients were calculated by samples’ distance similarities [Jaccard index] from LPMC FACS analysis on TNF+, IFNγ+, IL17+, IL10+iNKT, and CD4+T cells. [D] Representative dot plots [left] and cumulative histograms [right, n = 17 CD patients] of Th1 and Th17 CD4+ T cell frequency in NKT10-high and -low CD patients. [E] Spearman correlation analyses between IL10-producing iNKT cells and Th17 [upper panels] or Th1 [lower panels] cell frequencies in non-IBD LPMC [left plots] and CD LPMC [right plots]. [F] Proliferation indexes of naïve CD4+ T cells exposed to supernatants of aCD3/aCD28-stimulated iNKT cell lines isolated from the LP of iNKT10-high [purple bars] and -low [red bars] CD patients. [G] IL10 protein concentration in the supernatants of unstimulated or aCD3/28-stimulated iNKT cell lines isolated from the LP of NKT10-high [purple bars] and -low [red bars] CD patients in the presence or absence of IL10 blocking antibodies. [H] Two-tailed Spearman’s correlation test between IL10 protein concentration and suppression indexes. [I] Proliferation indexes of naïve CD4+ T cells exposed to supernatants of aCD3/aCD28-stimulated iNKT cell lines isolated from the LP of NKT10-high CD patients in the presence or absence of IL10 blocking antibodies. Statistical significance was calculated using the one-way ANOVA test corrected for multiple comparisons by controlling the false-discovery rate [FDR]; *p ≤0.05, **p ≤0.01, ***p ≤0.001. LPMC, lamina propria mononuclear cells; IBD, inflammatory bowel disease.
To evaluate if the IL10 produced by human iNKT cells might directly regulate T cell proliferation,34 we performed an in vitro suppression assay. The supernatants of stimulated mucosal iNKT cell lines derived from the intestinal LP of NKT10hi and NKT10lo patients were used for in vitro suppression assays with naïve CD4+CD62L+CD25-T cells [Supplementary Figure S6A]. We observed that only the exposure of naïve T cells to the secretome of iNKT cell lines from NKT10hi patients resulted in a significant suppression of naïve CD4+ T cell proliferation [Figure 4F]. Indeed, the supernatants of iNKT cells derived from iNKT10hi patients had higher levels of soluble IL10 compared with iNKT cells derived from iNKT10lo patients [Figure 4G]. The suppression effect was mediated by the secreted IL10 [Figure 4H], since reduction of available IL10 by using an anti-IL10 blocking antibody [Figure 4G] impaired the suppressive capacity of the iNKT-derived supernatant from NKT10hi patients [Figure 4I].
TCR stimulation of iNKT cell lines induced the upregulation of FOXP3 and E4BP4 transcription factors [Supplementary Figure S6B and D], classically associated with IL10 production and tolerogenic activity of T and iNKT cells35 and of surface receptors associated with a regulatory phenotype, including PD1, CTLA4, CD200, and 41BB, but not T-bet and RORγ [Supplementary Figure S6C and E].
Altogether, these data indicate that a subset of human LP iNKT cells characterised by the expression of IL10 are present in the mucosa of CD patients and are able to suppress naïve CD4+ T cell proliferation in vitro.
2.4. NKT10hi-derived microbiota promotes IL10 secretion in iNKT cells
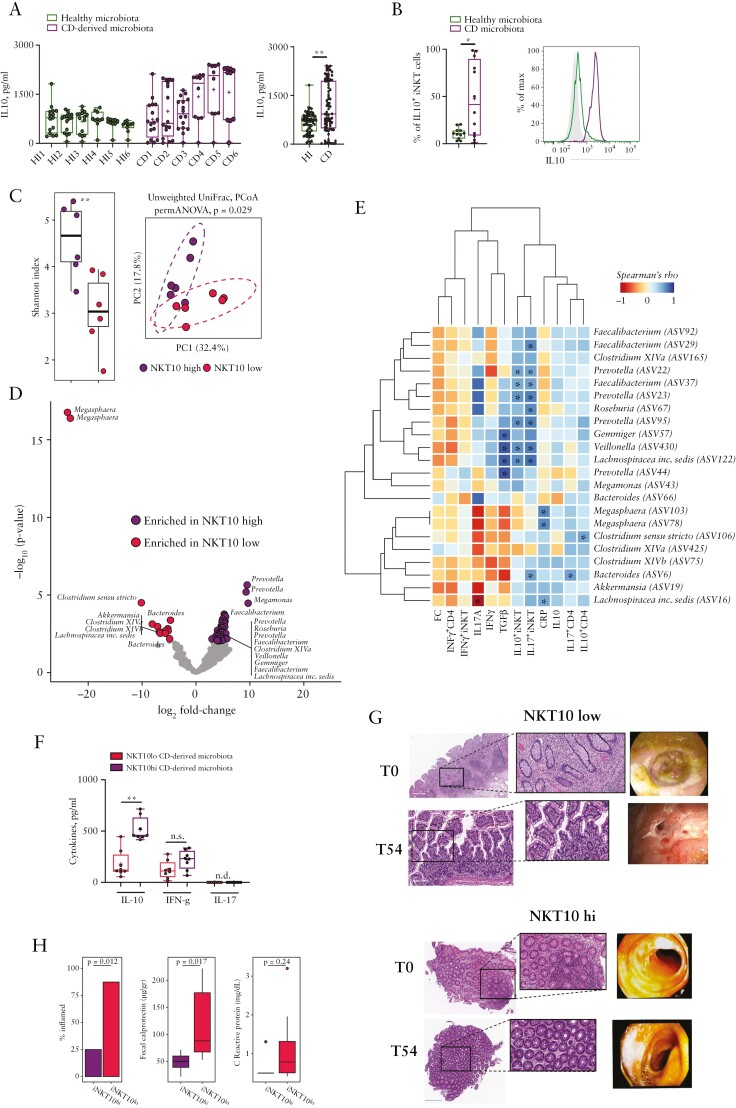
We next asked whether the gut microbiota might be linked to IL10 production by mucosal iNKT cells in CD patients. CD mucosa-associated microbiota induced a more sustained IL10 production by intestinal human iNKT cells as compared with microbiota derived from healthy individuals [Figure 5A and B] and monocyte-derived dendritic cells [moDC] contributed marginally to the production of IL10 compared with iNKT cells [Figure 5B; Supplementary Figure S6F]. An in-depth analysis of the gut microbiota from NKT10hi and NKT10lo patients showed a different microbial community structures between these patients [Figure 5C]. Furthermore, the mucosa-associated microbiota from NKT10hi patients was highly enriched in SCFA-producing bacteria36,37 [Figure 5D]. We observed the significant positive correlation of these SCFA-producing bacteria [including Faecalibacterium, Lachnospiraceae, Veillonella Gemmiger, and Prevotella,] with IL10+iNKT cells abundance and the mucosal expression of TGFβ [Figure 5E]. Remarkably, the mucosa-associated microbiota from NKT10hi patients was able to promote IL10 secretion upon stimulation of healthy donor-derived intestinal iNKT cells, whereas no differences were observed in the expression levels of IFNγ and IL17 [Figure 5F].
Figure 5.
A microbiota enriched in SCFA-producing bacteria sustain IL-10 production by iNKT cells. [A] IL10 protein concentration in the supernatants of mucosal iNKT cells co-cultured with moDCs and mucosa-associated microbiota isolated from healthy donors [n = 6] or CD patients [n = 6]. Left panel, single donors plot representation. Right panel, cumulative plot representation. [B] Frequency of IL10-producing iNKT cells upon co-culture with moDC exposed to healthy- or CD-derived mucosa-associated microbiota. [C] PCoA of beta-diversity of mucosa-associated microbiota as measured by Unweighted UniFrac distance [right] and alpha-diversity as measured by Shannon entropy index [left]; **p <0.01. [D] Volcano plot representing the significantly enriched bacterial taxa [FDR p <0.05] in NKT10-high vs -low patients by the DEseq2 analysis. The names of the significantly enriched ASVs described as SCFA-producing bacteria and classified to the genus are reported. [E] Heatmap of Spearman’s rho correlations between the significantly enriched SCFA-producing bacteria in iNKT10-high and -low patients with the indicated cell populations, clinical and gene expression data. The significant correlations with p-value <0.05 are indicated with an asterisk [*]. FC, faecal calprotectin; CRP, C-reactive protein. [F] Cytokines production by iNKT cells upon co-culture with moDC and mucosa-associated microbiota derived from iNKT10-high or -low patients; *p ≤0.05, **p ≤0.01, ***p ≤0.001, Mann-Whitney U test. G] Histological and endoscopic analyses at the beginning of the enrolment [T0] and 54 weeks post-enrolment [T54] in iNKT10-high and -low CD patients. [H] % of CD patients classified as inflamed [with a cumulative score ≥2 calculated summing 1 point with CRP >0.5 mg/dl, 1 point with FC >50 ug/g and 1 point for evident signs of inflammation/tissue damage], levels of faecal calprotectin and serum C-reactive protein 54 weeks post-enrolment; exact p-values for Fisher’s exact test [% inflamed] and Mann-Whitney test [FC and CRP] are shown. ASV, amplicon sequence variant; SCFA, short chain fatty acid; FDR, false-discovery rate; PCoA, principal coordinate analysis; moDC, monocyte-derived dendritic cells.
Although the frequency of mucosal NKT10 cells was not significantly correlated with clinical and endoscopic scores at enrolment [Supplementary Figure S6G], evaluation of the clinical outcomes of NKT10-high and -low CD patients showed that a high baseline frequency of NKT10 cells was associated with a significantly less severe disease during follow-up, as confirmed by histological and endoscopic activity [Figure 5G] and reduced inflammatory biomarkers, including faecal calprotectin [Figure 5H].
Altogether, these data suggest that human NKT10 cells accumulate in the colonic mucosa of CD patients, promote an intestinal microbial community structure enriched in health-promoting, SCFA-producing bacteria, and contribute to controlling the pro-inflammatory activity of Th1 and Th17 cells.
3. Discussion
The multifaceted nature of iNKT cells has been widely reported. Here we provide, for the first time, evidence supporting a direct IL10-dependent suppressive role of human, and murine, intestinal iNKT cells towards pathogenic CD4+T helper cells. Accordingly, in Crohn’s disease patients, IL10 expression by colonic iNKT cells inversely correlated with the frequency of pathogenic Th17 and Th1 cells. Moreover, NKT10 cells were associated with a particular health-promoting microbiota and a better clinical outcome at a 54-week follow-up examination.
Functional iNKT cell subsets have been thoroughly described in mice2,4 but not as meticulously in humans,3 possibly because human lipid-specific T cells are more diversified than their murine counterparts.1
iNKT cell functions have been classically associated with pro-inflammatory/cytotoxic phenotypes, which differentially contribute to exacerbate or resolve pathological conditions according to each unique disease aetiopathology.38 In the gut, iNKT cells have been shown to manifest pro-inflammatory NKT2-39 or NKT1/NKT17-associated phenotypes21,23–25 at the peak of inflammation. We also recently reported that the recognition of IBD patients’ dysbiotic mucosa-associated microbiota drives the secretion of pro-inflammatory cytokines by human intestinal iNKT cells.22
However, iNKT cells do not always show pro-inflammatory phenotypes.23,31,35,40 Indeed, they have been associated in the maintenance of gut homeostasis in murine models.23,31,35,40 Accordingly, we demonstrated that maintenance of normobiosis by faecal microbiota transplantation requires also murine colonic iNKT cell-mediated production of IL10.23
NKT10 cells were previously identified among murine splenocytes and in human peripheral blood,2 and Lynch and colleagues elegantly demonstrated the immunoregulatory functions of NKT10 cells in the adipose tissue.9 Here, we report that IL10-producing iNKT cells are present in the colonic mucosa of mice and, most importantly, of healthy and CD individuals. Several lines of evidence suggested the presence of intestinal NKT10 cells. In mice, interaction of iNKT cells with the intestinal epithelium has been shown to contribute to maintenance of intestinal homeostasis, and CD1d signalling between iNKT cells and intestinal epithelial cells [IECs] induced IL10 production in the former.26 IL10 mRNA, but not protein, was also detected within iNKT cells in that context. It has also been reported that B. fragilis, a human gut commensal, is capable of suppressing murine mucosal iNKT cell proliferation,41 albeit no direct proof of IL-10 production by iNKT cells exposed to B. fragilis or to its lipid antigens has been provided. IL10-producing iNKT cells have been detected also in the mLN. We hypothesise that, as occurs for Tregs, the lower frequency of IL10+iNKT cells in the mLN compared with the colon could be related to the role of the mLN in T cells gut-homing.42,43 This could partially explain why mLN and colonic NKT10 cells are phenotypically related whereas their frequency is significantly different.
IL10 expression and resting/Ki67-low phenotypes can be considered key features of NKT10 cells. Indeed in our murine model, IL10-producing iNKT cells were mostly not proliferating and this phenotype was associated with IL10 production and suppressive capabilities.
Interestingly, we observed that human NKT10 cells were more frequent in Crohn’s disease than in healthy subjects, similarly to Treg cells where frequencies have been reported to be increased in the intestinal lamina propria of IBD patients.44,45 Different explanations might account for this finding. On one side, pre-existing, tissue-resident, naturally-occurring NKT10 cells might expand in CD patients to control inflammation, as a negative feedback mechanism already described for Tregs.46 On the other hand, NKT10 cell frequency might increase upon the conversion of pro-inflammatory NKT cells into IL10-producing cells similarly to how colitogenic Th17 cells can convert into IL10-producing cells and acquire regulatory functions.28 The presence of a tolerogenic microenvironment [high IL10 and TGFB] may favour both hypotheses. Conversely, murine NKT10 cells have been shown to greatly expand in mice upon chronic antigen stimulation, either with the agonist ɑGalCer47 or within the lipid-enriched adipose environment.9 Albeit the nature of the antigens capable of activating iNKT cells in the gut of IBD patients is still undiscovered, it is known that iNKT cells recognise antigens of microbial5,22,48 or self49 origin. Chronic antigenic stimulation has been previously demonstrated to be responsible for the conversion of IFNγ-producing LCMV-CD8+activated T cells into low-proliferating/anergic cells, a mechanism to self-regulate CD8+ pro-inflammatory potential.50
The presence of a specific commensal core ecology consisting of bacteria from different phyla, but sharing similar metabolic functions, has been identified as the mechanistic driver to support host health. Among these commensals, SCFA-producing bacteria have been associated with IL10 secretion and intestinal homeostasis promotion.33 Indeed, restoration of a microbial core ecology enriched in SCFA-producing taxa by FMT effectively controlled experimental intestinal inflammation23,25 and promoted IL10-secretion also by murine intestinal iNKT cells. Microbe-derived SCFA promoted the production of IL10 also by murine conventional Th1 cells, endowing them with protective functions against severe experimental colitis outcomes.23 Moreover, we recently demonstrated that metronidazole treatment shaped a microbial community promoting IL10 secretion by human colonic iNKT cells.51
Here, we demonstrate that NKT10 cells suppress the pathogenic functions of IFNγ-producing Th1 cells in vitro and in vivo in an IL10-dependent manner. At present, only Treg cells and Tr1 cells have shown to directly suppress intestinal inflammation in the gut.29,52,53 In IBD patients, Tregs are able to suppress proliferation of effector T cells but are less efficient in suppressing secretion of Il1b,54 similarly to what we observed for NKT10 cells. We reported the presence of NKT10 cells in the gut of CD patients inversely correlating with Th1 and Th17 responses; iNKT cells are able to control Th1 or Th17 activity in murine models of autoimmune diseases, albeit iNKT cells regulatory function was not directly ascribed to IL10 production.12,17 Conversely, in a murine model of multiple sclerosis where the pathogenic functions of Th17 cells have been widely described,55 NKT10 cells were able to modulate the disease outcome.2,10
Here, we suggest that a fine balance between pro-inflammatory and anti-inflammatory signals, including the gut microbiota and their metabolic products, might involve intestinal iNKT cells, which in turn may enhance or suppress inflammatory responses according to the cytokine milieu and to their capability to interact with other immune cell types in health and disease. The functional plasticity of mucosal iNKT cells might be relevant to maintaining intestinal homeostasis, while remaining responsive to dysbiosis and inflammatory insults also in the context of intestinal neoplasia, where the tumour microenvironment might heavily influence iNKT cell functions.56
In conclusion, our study sheds light on previously unknown functions of human iNKT cells, providing evidence for the importance of NKT10 cells in the homeostatic regulation of the intestinal compartment. Therapies involving IL10-producing iNKT cells in intestinal disorders might be envisioned, although further knowledge is needed to fully elucidate their mechanisms of activation in the periphery.
4. Materials and Methods
4.1. Human samples
Buffy coat blood from healthy donors was obtained from the IRCCS Policlinico San Matteo, Pavia, Italy. Intestinal specimens of Crohn’s disease [n = 17] and of patients undergoing intestinal surgical resection for pathologies unrelated to IBD [n = 7] were obtained from the IRCCS Policlinico Ospedale Maggiore, Milan, Italy. The clinical characteristics and concomitant therapies of patients are summarised in Table 1.
4.2. Study approval
The Institutional Review Board approved the study [permission ref. no. EA1/107/10] and informed consent was obtained from the subjects involved in the study. Animal procedures were approved by Italy’s Ministry of Health [Authorisations no. 913/16, 415/17].
4.3. Mice
C57BL/6 mice [Charles River, Italy] and Rag1-/- mice [The Jackson laboratory] of 8-10 weeks of age were housed at the IEO [Milan, Italy] animal facility in specific pathogen-free [SPF] conditions. IL10-/- mice were kindly gifted by Dr Marinos Kallikurdis [Humanitas, Milan, Italy]. All mice were housed either at the European Institute of Oncology [EIO] or at the Humanitas animal facility in SPF conditions. Littermates of same sex and age were randomly assigned to the different experimental groups. A summary of the animals used for experimentation is reported in Supplementary Table S1.
4.4. Murine adoptive transfer experiments
Naïve CD4+ T cells [CD3+CD4+CD25-CD62L+] were sorted from pre-enriched [CD4 mouse microbeads, Miltenyi] splenocytes with BD FACSAria. Approximately 2.5 x 105 naïve CD4+ T cells were injected intraperitoneally in a male Rag1-/- recipient. iNKT and Treg cells were sorted from CD19-depleted splenocytes [CD19 mouse microbeads, Miltenyi] respectively as CD11b-CD11c-CD3+mCD1dTetPBS57+CD25- and CD11b-CD11c-CD3+CD4+CD25+, and injected intraperitoneally at a 1:1 naïve CD4+T: iNKT/Treg cell ratio. The purity of sorted cells was on average over 95%.
Mice were sacrificed 2 or 4 weeks following the adoptive transfer. At sacrifice colons were collected, their length measured and divided in portions to be fixed in 10% formalin for histological analyses as described in Cribiu et al.57 and in Supplementary Table S2, snap-frozen for RNA extraction, and used for lamina propria mononuclear cell [LPMC] immunophenotyping.
4.5. In vivo administration of SCFA
SPF C57BL/6 mice 4-5 weeks old were pre-treated with an antibiotic mixture containing vancomycin [1.25 mg], ampicillin [2.5 mg], and metronidazole [1.25 mg] [VAM] in 100 µl water per mouse by oral gavage for 14 days. Starting from the second week of VAM treatment, mice were treated with a short chain fatty acids [SCFAs] mix [67.5 mM acetate, 40 mM butyrate, 25.9 mM propionate] in the drinking water for 21 days, and water solutions [pH- and sodium-matched] were prepared and changed every 5 days. At the end of the experiment, cells from colonic lamina propria were isolated and analysed as described below.
4.6. Human and murine cell isolation
Human LPMC [lamina propria mononuclear cells] were isolated as previously described.58 Murine LPMC were isolated as in Burrello et al.23 Briefly, the dissected human intestinal mucosa was freed of mucus and epithelial cells in sequential steps with DTT [0.1 mmol/L] and EDTA [1 mmol/L] [both from Sigma] and then digested with collagenase D [400 U/ml] [Worthington Biochemical Corporation, Lakewood, NJ] for 5 h at 37°. LPMCs were then separated with a Percoll gradient and cultured in complete RPMI 1640 medium containing 5% human serum [Sigma] and 100 UI/ml IL-2 [Proleukin].
For murine LPMC isolation, colonic lamina propria mononuclear cells were isolated via incubation with 5 mM EDTA at 37°C for 30 min, followed by mechanical disruption with GentleMACS [Miltenyi Biotec]. After filtration with 100-µm and 70-µm nylon strainers [BD], the LPMC were counted and stained for immunophenotyping.
4.7. Flow cytometry
Human and murine cells were stained with combinations of directly conjugated antibodies as specified in Supplementary Table S3. The gating strategy using the antibodies mCD1d/PBS-57 and hCD1d/PBS-57 to identify murine and human iNKT cells is described in Supplementary Figures S1A and S4A, respectively. The PBS-57 ligand is an analogue of the a-galactosylceramide complexed to CD1d tetramers, provided by the NIH Tetramer Facility.
Intracellular cytokines were detected after stimulation of human cells for 3 h with 50 ng/ml PMA [Sigma] and 1 µg/ml ionomycin [Sigma] in the presence of 10 µg/ml Brefeldin A [Sigma]. Cells were fixed and permeabilised with Cytofix/Cytoperm [BD] before the addition of the antibodies detecting the cytokine released. For the staining of transcription factors, cells were fixed and permeabilised with FOXP3 Transcription Factor Staining Buffer Set [eBioscience].
Samples were analysed with a FACSCelesta flow cytometer [BD], gated to exclude singlets on the basis of light scatter and non-viable cells by Zombie Dye labelling. Data were analysed using FlowJo v10 software [BD].
4.8. Human and murine histological analyses
For histological analyses, samples were placed in neutral-buffered 4% formalin solution for 24-36 h. All tissue samples were processed with a LEICA PELORIS processor before paraffin embedding. Human samples were included either manually or with an automated system [SAKURA], based on the size of the specimen. After haematoxylin and eosin staining, scans of the histological slides were obtained using an Aperio AT2 scanner with a resolution of 0.5 µm/pixel with 20x objective and 0.25 µm/pixel at 40x, using a doubler.
Histological scoring of disease activity included evaluation of the severity and extent of the inflammatory cell infiltrate, epithelial hyperplasia, goblet cell loss, and alterations of the mucosal architecture [complete scoring criteria, definitions, and values are available in Supplementary Table S2].
4.9. Supernatant and tissue ELISA for protein detection
Human IL10 concentration in the supernatant of stimulated and unstimulated iNKT cells was evaluated by ELISA assay [Biolegend, see Supplementary Table S3] performed following the manufacturer’s instructions. Colonic tissues were homogenised in 300 µl RIPA buffer [Cell Signaling Technology] supplemented with phosphatase inhibitors [Sigma] and protease inhibitors [Complete Ultra tablets, Roche]. The samples were then incubated at 4°C for 30 min under slow rotation and then centrifuged at 13 000 g for 15 min at 4°C. The supernatant was quantified at the NanoDrop with Bradford Assay [BioRad]. Cytokines were measured by ELISA [Biolegend, see Supplementary Table S4] performed following the manufacturer’s instructions. Correlation has been performed upon normalisation of the samples to 1 mg/ml on the total protein extract for each sample.
4.10. RNA isolation, cDNA synthesis, quantitative PCR, and gene expression
Total RNA from intestinal tissues was isolated using TRIzol [Invitrogen] and Quick-RNA MiniPrep [ZymoResearch] according to manufacturer’s specifications. cDNAs were generated from 1 µg of total RNA with EasyScript Plus cDNA Synthesis kit [abm]. Gene expression levels were evaluated by qPCR and normalised to GAPDH [human] or Rpl32 [mouse] gene expression. Murine and human primers are listed in SupplementaryTable S5.
4.11. Intestinal iNKT cell lines generation
Human iNKT cell lines were generated as previously described in Burrello et al.22 Briefly, sorted CD45+CD3+CD1d:PBS57Tet+ cells from total LPMCs of CD patients or PBMCs of healthy donors were expanded in vitro for 2 weeks in the presence of irradiated peripheral blood feeders, hIL2 [100 IU/ml, Proleukin] and PHA [1 µg/ml, Sigma].
4.12. In vitro suppression assay
In vitro suppression was performed as previously described59 with some modifications. Naïve CD4+ T cells were sorted as CD3+CD4+CD25-CD62L+ cells on a BD FACSAria starting from pre-enriched CD3+ PBMCs of healthy donors [CD3 human microbeads, Miltenyi Biotec] [Supplementary Figure 3]. Cells were labelled with 1 nM Far Red CellTrace [ThermoFisher], re-suspended in medium containing hIL2 [Proleukin] and anti-CD28 antibody [2 µg/ml, Tonbo] and plated in 96-well plates [NUNC Maxisorp] pre-coated with anti-CD3 antibody [2 µg/ml, Tonbo] at a concentration of 2.5 x 104 cells/well; 2 x 105 cells/well were plated as unstimulated negative control. iNKT cell lines were stimulated with anti-CD3 and anti-CD28 antibodies [2 µg/ml each, Tonbo] on NUNC Maxisorp plates for 5 days. Their supernatant was collected, applied to proliferating naïve CD4+ T cells in the presence or absence of anti-IL10 neutralising antibody [10 µg/ml], and in part used to evaluate cytokine concentration, as described in Supplementary Methods. After 5 days, proliferating naïve CD4+ T cells were labelled with Zombie vital dye [Biolegend] and analysed with [BD] FACS Celesta. Proliferation analyses were performed with a FlowJo v10 [BD] proliferation tool. The suppression index was calculated by dividing the proliferation index of maximum proliferation controls by that of the wells under investigation.
4.13. Human ileocolonoscopies analyses
All ileocolonoscopies were performed at the Endoscopy Unit of the IRCCS Policlinico Ospedale Maggiore, Milan, Italy, by expert endoscopists with particular expertise in endoscopy of IBD patients.
4.14. Patients outcomes
CD patients outcomes were evaluated at 54 weeks after enrolment. We collected data of objective measures of disease activity, i.e., ileocolonscopy and/or faecal calprotectin and/or C-reactive protein, assessed according to usual clinical practice. Endoscopic remission was defined by complete mucosal healing in the absence of any ulcers. Biochemical remission was defined by a faecal calprotectin <50 µg/g and/or C-reactive protein <0.5 mg/dL. We considered patients without any sign of significant inflammatory activity if they reached both endoscopic and biochemical remission when all data were available. When endoscopy at 54 weeks was not performed, we evaluated only biochemical remission.
4.15. 16S rRNA gene sequencing and data analysis
Intestinal mucus scraped from the human colon and faecal pellets from mice were stored at -80°C until the DNA was extracted. DNA extraction, 16S rRNA gene amplification, purification, library preparation, and pair-end sequencing on the Illumina MiSeq platform were performed as described in Burrello et al.25 Reads were pre-processed using the MICCA pipeline [v.1.7.0] [http://www.micca.org].60 Forward and reverse primers trimming and quality filtering were performed using micca trim and micca filter, respectively. Filtered sequences were de-noised using the UNOISE61 algorithm implemented in micca otu to determine true biological sequences at the single nucleotide resolution by generating amplicon sequence variants [ASVs]. Bacterial ASVs were taxonomically classified using micca classify and the Ribosomal Database Project [RDP] Classifier v2.11.62 Multiple sequence alignment [MSA] of 16S sequences was performed using the Nearest Alignment Space Termination [NAST] algorithm63 implemented in micca msa with the template alignment clustered at 97% similarity of the Greengenes database64 [release 13_08]. Phylogenetic trees were inferred using micca tree.65 Sampling heterogeneity was reduced rarefying samples at the depth of the less abundant sample, using micca tablerare. Alpha- [within-sample richness] and beta-diversity [between-sample dissimilarity] estimates were computed using the phyloseq R package.66 A permutational multivariate analysis of variance [PERMANOVA] test was performed using the adonis function in the R package vegan with 999 permutations. Differential abundance testing was carried out using the R package DESeq267 using the non-rarefied data,68 and p-values were false-discovery rate-corrected using the Benjamini-Hochberg procedure implemented in DESeq2.
4.16. Microbiota in vitro assays
Mucosa-associated microbiota was scraped from surgical resections of CD patients and stored at -80. Three cycles of heat inactivation and freezing/thawing were performed before plating with moDC and iNKT cell lines [1:1 ratio]. Microbial load was normalised by qPCR. After 36 h, T cell activation was estimated by measuring cytokine released in culture supernatants by ELISA assays or intracellular staining.
4.17. Statistics
Wilcoxon rank- sum tests and Spearman’s correlations were performed by using the R software [version 3.6.2] through the stats R package and the psych R package, respectively. Immunological data from iNKT10-high and -low patients have been used to calculate samples’ distance similarity according to the Jaccard index by mean of the ‘vegdist’ function within the vegan R package. A permutational MANOVA [PERMANOVA] test was performed using the adonis function of the vegan R package with 999 permutations. Samples have been clustered hierarchically according to the UPGMA method by using the ‘hclust’ function within the stats R package.
Statistical analyses were performed with GraphPad Prism 7 [GraphPad Software] and R. Statistical significance was calculated by using the Mann-Whitney U test and the unpaired Student’s t test for comparison within two groups, or the Kruskal-Wallis and ANOVA tests for comparison within more than two groups. Multiple comparisons were corrected by controlling for false-discovery rate [FDR]; p ≤0.05 [*], p ≤0.01 [**], and p ≤0.001 [***] were regarded as statistically significant.
Supplementary Material
Acknowledgements
We thank the EIO Animal Facility for the excellent animal husbandry, the NIH Tetramer Facility for providing h/mCD1d:PBS57 Tetramers, and Prof. Pier Paolo Di Fiore for critical discussions. We thank Chiara Amoroso for the help during in vivo experiments.
Contributor Information
Claudia Burrello, Department of Experimental Oncology, European Institute of Oncology IRCCS, Milan, Italy.
Francesco Strati, Department of Experimental Oncology, European Institute of Oncology IRCCS, Milan, Italy.
Georgia Lattanzi, Department of Experimental Oncology, European Institute of Oncology IRCCS, Milan, Italy; Department of Oncology and Hemato-oncology, Università degli Studi di Milano, Milan, Italy.
Angelica Diaz-Basabe, Department of Experimental Oncology, European Institute of Oncology IRCCS, Milan, Italy; Department of Oncology and Hemato-oncology, Università degli Studi di Milano, Milan, Italy.
Erika Mileti, Department of Experimental Oncology, European Institute of Oncology IRCCS, Milan, Italy.
Maria Rita Giuffrè, Department of Experimental Oncology, European Institute of Oncology IRCCS, Milan, Italy.
Gianluca Lopez, Pathology Unit, Fondazione IRCCS Cà Granda, Ospedale Maggiore Policlinico, Milan, Italy.
Fulvia Milena Cribiù, Pathology Unit, Fondazione IRCCS Cà Granda, Ospedale Maggiore Policlinico, Milan, Italy.
Elena Trombetta, Clinical Chemistry and Microbiology Laboratory Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy.
Marinos Kallikourdis, Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy; Laboratory of Adaptive Immunity, IRCCS Humanitas Research Hospital, Milan, Italy.
Marco Cremonesi, Laboratory of Adaptive Immunity, IRCCS Humanitas Research Hospital, Milan, Italy.
Francesco Conforti, Gastroenterology and Endoscopy Unit, Fondazione IRCCS Cà Granda, Ospedale Maggiore Policlinico, Milan, Italy.
Fiorenzo Botti, Gastroenterology and Endoscopy Unit, Fondazione IRCCS Cà Granda, Ospedale Maggiore Policlinico, Milan, Italy; General and Emergency Surgery Unit, Fondazione IRCCS Ca’ Granda, Ospedale Maggiore Policlinico, Milan, Italy.
Laura Porretti, Clinical Chemistry and Microbiology Laboratory Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy.
Maria Rescigno, Department of Biomedical Sciences, Humanitas University, Pieve Emanuele, Milan, Italy.
Maurizio Vecchi, Gastroenterology and Endoscopy Unit, Fondazione IRCCS Cà Granda, Ospedale Maggiore Policlinico, Milan, Italy; Department of Pathophysiology and Transplantation, Università degli Studi di Milano, Milan, Italy.
Massimo C Fantini, Department of Medical Science and Public Health, University of Cagliari, Cagliari, Italy.
Flavio Caprioli, Gastroenterology and Endoscopy Unit, Fondazione IRCCS Cà Granda, Ospedale Maggiore Policlinico, Milan, Italy; Department of Pathophysiology and Transplantation, Università degli Studi di Milano, Milan, Italy.
Federica Facciotti, Department of Experimental Oncology, European Institute of Oncology IRCCS, Milan, Italy; Department of Biotechnology and Biosciences, University of Milan-Bicocca, Milan, Italy.
Funding
This work was made possible thanks to the financial support of Associazione Italiana per la Ricerca sul Cancro [Start-Up 2013 14378, Investigator Grant - IG 2019 22923 to FF] and of Italy’s Ministry of Health [GR-2016-0236174 to FF and FC]. This work has been and partially supported by the Italian Ministry of Health with Ricerca Corrente and 5 X1000 funds.
Conflict of Interest
The authors have declared that no conflict of interest exists.
Author Contributions
Concept: FF, CB; methodology: CB, FS, EM, ET, LP, FMC, GL; investigation: CB, GL, ADB, FS, EM, MRG, MC; funding acquisition: FF, FC; patient recruitment: FC, FB, FSC; writing original draft: FF, FS, CB, GL, ADB, MRG; review and editing: FF, FMC, FC, MK, MR, MV, FS.
Data and materials availability
All data are available in the main text or the Supplementary materials. 16S rRNA gene sequencing raw data are available in the European Nucleotide Archive with accession numbers PRJEB42828 and PRJEB42829.
References
- 1. Bendelac A, Savage PB, Teyton L.. The biology of NKT cells. Annu Rev Immunol 2007;25:297–336. [DOI] [PubMed] [Google Scholar]
- 2. Sag D, Krause P, Hedrick CC, Kronenberg M, Wingender G.. IL-10-producing NKT10 cells are a distinct regulatory invariant NKT cell subset. J Clin Invest 2014;124:3725–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Crosby CM, Kronenberg M.. Tissue-specific functions of invariant natural killer T cells. Nat Rev Immunol 2018;18:559–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee YJ, Wang H, Starrett GJ, Phuong V, Jameson SC, Hogquist KA.. Tissue-specific distribution of iNKT cells impacts their cytokine response. Immunity 2015;43:566–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tupin E, Kinjo Y, Kronenberg M.. The unique role of natural killer T cells in the response to microorganisms. Nat Rev Microbiol 2007;5:405–17. [DOI] [PubMed] [Google Scholar]
- 6. Van Kaer LWL. Therapeutic potential of invariant natural killer T cells in autoimmunity. Front Immunol. 2018;9:519. Doi: 10.3389/fimmu.2018.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Terabe M, Berzofsky JA.. Tissue-specific roles of NKT Cells in tumor immunity. Front Immunol 2018;9:1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cortesi F, Delfanti G, Grilli A, et al. Bimodal CD40/Fas-dependent crosstalk between iNKT cells and tumor-associated macrophages impairs prostate cancer progression. Cell Rep 2018;22:3006–20. [DOI] [PubMed] [Google Scholar]
- 9. Lynch L, Nowak M, Varghese B, et al. Adipose tissue invariant NKT cells protect against diet-induced obesity and metabolic disorder through regulatory cytokine production. Immunity 2012;37:574–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oh SJ, Chung DH.. Invariant NKT cells producing IL-4 or IL-10, but not IFN-gamma, inhibit the Th1 response in experimental autoimmune encephalomyelitis, whereas none of these cells inhibits the Th17 response. J Immunol 2011;186:6815–21. [DOI] [PubMed] [Google Scholar]
- 11. Lynch L, Michelet X, Zhang S, et al. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of T[reg] cells and macrophages in adipose tissue. Nat Immunol 2015;16:85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baev DV, Caielli S, Ronchi F, et al. Impaired SLAM-SLAM homotypic interaction between invariant NKT cells and dendritic cells affects differentiation of IL-4/IL-10-secreting NKT2 cells in nonobese diabetic mice. J Immunol 2008;181:869–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hegde S, Jankowska-Gan E, Roenneburg DA, Torrealba J, Burlingham WJ, Gumperz JE.. Human NKT cells promote monocyte differentiation into suppressive myeloid antigen-presenting cells. J Leukoc Biol 2009;86:757–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaser A, Zeissig S, Blumberg RS.. Inflammatory bowel disease. Annu Rev Immunol 2010;28:573–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wawrzyniak M, Scharl M.. Genetics and epigenetics of inflammatory bowel disease. Swiss Med Wkly 2018;148:w14671. [DOI] [PubMed] [Google Scholar]
- 16. Shouval DS, Konnikova L, Griffith AE, et al. Enhanced TH17 responses in patients with IL10 receptor deficiency and infantile-onset IBD. Inflamm Bowel Dis 2017;23:1950–61. [DOI] [PubMed] [Google Scholar]
- 17. Shouval DS, Ouahed J, Biswas A, et al. Interleukin 10 receptor signaling: master regulator of intestinal mucosal homeostasis in mice and humans. Adv Immunol 2014;122:177–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shouval DS, Biswas A, Kang YH, et al. Interleukin 1β mediates intestinal inflammation in mice and patients with interleukin 10 receptor deficiency. Gastroenterology 2016;151:1100–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marrero I, Maricic I, Feldstein AE, et al. Complex network of NKT cell subsets controls immune homeostasis in liver and gut. Front Immunol 2018;9:2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Middendorp S, Nieuwenhuis EE.. NKT cells in mucosal immunity. Mucosal Immunol 2009;2:393–402. [DOI] [PubMed] [Google Scholar]
- 21. Biancheri P, Di Sabatino A, Ammoscato F, et al. Absence of a role for interleukin-13 in inflammatory bowel disease. Eur J Immunol 2014;44:370–85. [DOI] [PubMed] [Google Scholar]
- 22. Burrello C, Pellegrino G, Giuffrè MR, et al. Mucosa-associated microbiota drives pathogenic functions in IBD-derived intestinal iNKT cells. Life Sci Alliance 2019;2:e201800229. Doi: 10.26508/lsa.201800229. PMID: 30760554; PMCID: PMC6374994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burrello C, Garavaglia F, Cribiù FM, et al. Therapeutic faecal microbiota transplantation controls intestinal inflammation through IL10 secretion by immune cells. Nat Commun 2018;9:5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burrello C, Garavaglia F, Cribiù FM, et al. Short-term oral antibiotics treatment promotes inflammatory activation of colonic invariant natural KILLER T and conventional CD4+ T cells. Front Med 2018;5:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burrello C, GiuffrèMR, MacandogAD, et al. Fecal microbiota transplantation controls murine chronic intestinal inflammation by modulating immune cell functions and gut microbiota composition. Cells 2019;8. Doi: 10.3390/cells8060517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Olszak T, Neves JF, Dowds CM, et al. Protective mucosal immunity mediated by epithelial CD1d and IL-10. Nature 2014;509:497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Geginat J, Larghi P, Paroni M, et al. The light and the dark sides of Interleukin-10 in immune-mediated diseases and cancer. Cytokine Growth Factor Rev 2016;30:87–93. [DOI] [PubMed] [Google Scholar]
- 28. Gagliani N, Magnani CF, Huber S, et al. Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nat Med 2013;19:739–46. [DOI] [PubMed] [Google Scholar]
- 29. Alfen JS, Larghi P, Facciotti F, et al. Intestinal IFN-γ-producing type 1 regulatory T cells coexpress CCR5 and programmed cell death protein 1 and downregulate IL-10 in the inflamed guts of patients with inflammatory bowel disease. J Allergy Clin Immunol 2018;142:1537–47.e8. [DOI] [PubMed] [Google Scholar]
- 30. Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA.. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol 2013;14:1146–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Powrie F, Leach MW, Mauze S, Caddle LB, Coffman RL.. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int Immunol 1993;5:1461–71. [DOI] [PubMed] [Google Scholar]
- 32. Coletta M, Paroni M, Alvisi MF, et al. Immunological variables associated with clinical and endoscopic response to vedolizumab in patients with inflammatory bowel diseases. J Crohns Colitis 2020;14:1190–201. [DOI] [PubMed] [Google Scholar]
- 33. Amoroso C, PerilloF, Strati F, et al. The role of gut microbiota biomodulators on mucosal immunity and intestinal inflammation. Cells 2020;9. Doi: 10.3390/cells9051234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thornton AM, Shevach EM.. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J Exp Med 1998;188:287–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Motomura Y, Kitamura H, Hijikata A, et al. The transcription factor E4BP4 regulates the production of IL-10 and IL-13 in CD4+ T cells. Nat Immunol 2011;12:450–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van der Hee B, Wells JM.. Microbial regulation of host physiology by short-chain fatty acids. Trends Microbiol 2021;29:700–12. [DOI] [PubMed] [Google Scholar]
- 37. Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F.. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 2016;165:1332–45. [DOI] [PubMed] [Google Scholar]
- 38. Mori L, Lepore M, De Libero G.. The immunology of CD1- and MR1-restricted T cells. Annu Rev Immunol 2016;34:479–510. [DOI] [PubMed] [Google Scholar]
- 39. Heller F, Fuss IJ, Nieuwenhuis EE, Blumberg RS, Strober W.. Oxazolone colitis, a Th2 colitis model resembling ulcerative colitis, is mediated by IL-13-producing NK-T cells. Immunity 2002;17:629–38. [DOI] [PubMed] [Google Scholar]
- 40. Fuss IJ, Heller F, Boirivant M, et al. Nonclassical CD1d-restricted NK T cells that produce IL-13 characterize an atypical Th2 response in ulcerative colitis. J Clin Invest 2004;113:1490–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. An D, Oh SF, Olszak T, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 2014;156:123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hammerschmidt SI, Ahrendt M, Bode U, et al. Stromal mesenteric lymph node cells are essential for the generation of gut-homing T cells in vivo. J Exp Med 2008;205:2483–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Geem D, Ngo V, Harusato A, et al. Contribution of mesenteric lymph nodes and GALT to the intestinal Foxp3+ regulatory T-cell compartment. Cell Mol Gastroenterol Hepatol 2016;2:274–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maul J, Loddenkemper C, Mundt P, et al. Peripheral and intestinal regulatory CD4+ CD25[high] T cells in inflammatory bowel disease. Gastroenterology 2005;128:1868–78. [DOI] [PubMed] [Google Scholar]
- 45. Holmen N, Lundgren A, Lundin S, et al. Functional CD4+CD25high regulatory T cells are enriched in the colonic mucosa of patients with active ulcerative colitis and increase with disease activity. Inflamm Bowel Dis 2006;12:447–56. [DOI] [PubMed] [Google Scholar]
- 46. Garetto S, Trovato AE, Lleo A, et al. Peak inflammation in atherosclerosis, primary biliary cirrhosis and autoimmune arthritis is counter-intuitively associated with regulatory T cell enrichment. Immunobiology 2015;220:1025–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wingender G, Stepniak D, Krebs P, et al. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology 2012;143:418–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zajonc DM, Girardi E.. Recognition of microbial glycolipids by natural killer T cells. Front Immunol 2015;6:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Facciotti F, Ramanjaneyulu GS, Lepore M, et al. Peroxisome-derived lipids are self antigens that stimulate invariant natural killer T cells in the thymus. Nat Immunol 2012;13:474–80. [DOI] [PubMed] [Google Scholar]
- 50. Zajac AJ, Blattman JN, Murali-Krishna K, et al. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med 1998;188:2205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Strati F, Pujolassos M, Burrello C, et al. Antibiotic-associated dysbiosis affects the ability of the gut microbiota to control intestinal inflammation upon fecal microbiota transplantation in experimental colitis models. Microbiome 2021;9:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huber S, Gagliani N, Esplugues E, et al. Th17 cells express interleukin-10 receptor and are controlled by Foxp3- and Foxp3+ regulatory CD4+ T cells in an interleukin-10-dependent manner. Immunity 2011;34:554–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rubtsov YP, Rasmussen JP, Chi EY, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 2008;28:546–58. [DOI] [PubMed] [Google Scholar]
- 54. Cook L, Stahl M, Han X, et al. Suppressive and gut-reparative functions of human Type 1 T regulatory cells. Gastroenterology 2019;157:1584–98. [DOI] [PubMed] [Google Scholar]
- 55. Frohman EM, Racke MK, Raine CS.. Multiple sclerosis – the plaque and its pathogenesis. N Engl J Med 2006;354:942–55. [DOI] [PubMed] [Google Scholar]
- 56. Wang Y, Cardell SL.. The yin and yang of invariant natural killer T cells in tumor immunity-suppression of tumor immunity in the intestine. Front Immunol 2017;8:1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cribiu FM, et al. Using robotic systems to process and embed colonic murine samples for histological analyses. J Vis Exp 2019. Doi: 10.3791/58654. [DOI] [PubMed] [Google Scholar]
- 58. Caprioli F, Sarra M, Caruso R, et al. Autocrine regulation of IL-21 production in human T lymphocytes. J Immunol 2008;180:1800–7. [DOI] [PubMed] [Google Scholar]
- 59. Facciotti F, Gagliani N, Häringer B, et al. IL-10-producing forkhead box protein 3-negative regulatory T cells inhibit B-cell responses and are involved in systemic lupus erythematosus. J Allergy Clin Immunol 2016;137:318–21.e5. [DOI] [PubMed] [Google Scholar]
- 60. Albanese D, Fontana P, De Filippo C, Cavalieri D, Donati C.. MICCA: a complete and accurate software for taxonomic profiling of metagenomic data. Sci Rep 2015;5:9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Edgar RC. UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv 2016. Doi: 10.1101/081257. [DOI] [Google Scholar]
- 62. Wang Q, Garrity GM, Tiedje JM, Cole JR.. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. DeSantis TZ Jr, Hugenholtz P, Keller K, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res 2006;34:W394–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006;72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Price MN, Dehal PS, Arkin AP.. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 2010;5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. McMurdie PJ, Holmes S.. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. McMurdie PJ, Holmes S.. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 2014;10:e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.