Abstract
Strain PA14, a human clinical isolate of Pseudomonas aeruginosa, is pathogenic in mice and insects (Galleria mellonella). Analysis of 32 different PA14 mutants in these two hosts showed a novel positive correlation in the virulence patterns. Thus, G. mellonella is a good model system for identifying mammalian virulence factors of P. aeruginosa.
The common soil bacterium Pseudomonas aeruginosa is an opportunistic pathogen in diverse hosts, including mammals (21), insects (4), nematodes (15, 22, 23), and plants (17). In humans, P. aeruginosa is a leading cause of nosocomial infections in cystic fibrosis patients, burn victims, and other immunocompromised individuals (2). Recent work with strain UCBPP-PA14 (referred to herein as PA14), a clinical isolate of P. aeruginosa, showed that some bacterial mutants that are less virulent in plants or nematodes are also less virulent in mice (15, 17, 22, 23). However, the severities of the effects of particular mutations do not correlate well in these three host systems. Here we present the novel finding of a significant (P < 0.001) positive correlation between virulence of PA14 mutants in mice and insects. This correlation suggests that the underlying mechanisms of infection are similar in the two hosts and that insects are suitable as a model to identify and characterize bacterial genes involved in mammalian infections.
Larvae of Galleria mellonella (greater wax moth) are quite sensitive to P. aeruginosa injected into the hemolymph. Several studies have shown a 50% lethal dose (LD50) of fewer than 10 bacteria (5, 10, 12, 14). Both induced defenses in the insect (20) and bacterial inhibition of this insect's response (11) have been reported. In a study of P. aeruginosa mutants, lipopolysaccharide-deficient isolates had greatly reduced virulence in G. mellonella, whereas mutants with defects in pili, flagellae, or proteases did not (12).
Compared to previous nonvertebrate model hosts (plants and nematodes) developed in our laboratory to study P. aeruginosa pathogenicity (15, 17, 22, 23), insects have a relatively advanced system of antimicrobial defenses and are thus more likely to produce information relevant to the mammalian infection process. Like mammals, insects possess a circulatory system and a complex innate immune response (3, 8, 9, 13). Cells in the hemolymph are capable of phagocytosing or encapsulating microbial invaders (16), and humoral responses include the inducible production of lysozyme and small antibacterial proteins called cecropins (7). Several basic components of the bacterial infection process, such as toxin production, cell adhesion, and cell invasion, are also likely to be important in both insects and mammals. Other attributes which make G. mellonella larvae an attractive model system include their relatively large size (250 mg), which enables the rapid injection of defined doses of bacteria, examination of the pathology of the infection, and the calculation of LD50s; extensive literature on microbial pathogenesis, including that of P. aeruginosa (5, 19, 24); and their commercial availability at a low price.
In initial experiments, we studied the course of infection at 25°C of wild-type PA14 injected into hemolymph of fifth-instar G. mellonella larvae (Van der Horst Wholesale, St. Marys, Ohio). Overnight PA14 cultures grown in Luria-Bertani (LB) broth were diluted 1:100 in the same medium and grown to an optical density at 600 nm of 0.3 to 0.4. Cultures were pelleted and resuspended in 10 mM MgSO4. After dilution to an optical density at 600 nm of 0.1 with 10 mM MgSO4, serial 10-fold dilutions were made in 10 mM MgSO4–2 mg of rifampin/ml. A 10-μl Hamilton syringe was used to inject 5-μl aliquots into G. mellonella larvae via the hindmost left proleg. The final concentration of approximately 40 μg of rifampin per g of larva prevented infection by bacteria naturally present on the surface of the larvae. PA14 is resistant to rifampin, and only 1% of control larvae injected with 5 μl of 10 mM MgSO4–2 mg of rifampin/ml died. When plated on LB agar or Pseudomonas isolation agar (Difco), samples taken from the interior of infected larvae produced only PA14 colonies. Whereas the larvae are impervious to infection when dipped in a saturated culture of PA14 (data not shown), even one wild-type PA14 bacterium injected through the cuticle can result in death (Table 1). After an injection of relatively small doses of PA14 (up to 10,000 CFU), G. mellonella larvae behaved and moved normally until roughly 48 h after infection, when there was a rapid melanization (Fig. 1) followed by death, often in as little as 2 h. Larvae that were not dead 60 h postinfection had no sign of PA14 in hemolymph samples and showed no sign of infection, even if they were left for an additional week at 25°C (data not shown).
TABLE 1.
LD50s of wild-type mutant PA14 in G. mellonella
| Mutant bacterial strain and/or genea | LD50 in G. mellonellab | Reference or source |
|---|---|---|
| PA14 | 1 | 17 |
| 41C1 (aefA) | 0.8 | 23 |
| 49H2 | 0.9 | M.-W. Tan, unpublished |
| 35A9 (mtrR) | 2 | 23 |
| toxA | 2 | 17 |
| 34H4 | 2 | 18 |
| 41A5 | 3 | 23 |
| 16G12 | 3 | 18 |
| 34B12 | 3 | 18 |
| phnA, phnB | 4 | 15 |
| 12A1 (lasR) | 5 | 23 |
| mucD | 8 | Yorgey, unpublished |
| 25A12 | 9 | 18 |
| 3E8 (phzB) | 10 | 15 |
| algD | 10 | Yorgey, unpublished |
| 35H7 (gacA) | 10 | 23 |
| 36A4 (hrpM) | 10 | 15 |
| 1D7 | 20 | 18 |
| plcS | 20 | 17 |
| 1G2 | 20 | 15 |
| 23A2 (mexA) | 20 | 15 |
| 33A9 | 40 | 18 |
| 34B12, gacA | 50 | S. Wong, unpublished |
| pho15 (dsbA) | 60 | 18 |
| 6A6 (phzB) | 60 | 15 |
| mucD, algD | 80 | Yorgey, unpublished |
| 8C12 | 100 | 15 |
| gacA | 100 | 17 |
| 44B1 | 500 | 23 |
| 50E12 (ptsP) | 600 | 23 |
| 25F1 | 2,000 | 18 |
| 33C7 | 2,000 | 18 |
| pho23 | 70,000 | M.-W. Tan |
Name of mutant in earlier work and the affected gene, if previously identified.
Number of bacteria, rounded to one significant digit.
FIG. 1.
G. mellonella 60 h after infection with a mean of one PA14 bacterium. Dead larvae show increased melanization.
To monitor the growth of PA14 in infected larvae, we measured the bacterial concentration in the larvae at 8-h intervals following injection of a lethal dose of 25 PA14 cells per larva. At each time point, 100 larvae were harvested and groups of 10 larvae were homogenized in 200 ml of 10 mM MgSO4 in a Waring blender for 1 min at maximum speed. Serial dilutions of the homogenate were plated on Pseudomonas isolation agar to determine bacterial concentrations. The infection followed a classic pattern of exponential growth, reaching a final density of more than 1010 bacteria/g (Fig. 2). Larval death occurred at a bacterial density of approximately 109 bacteria/g.
FIG. 2.
Time course of PA14 infection in G. mellonella. Twenty-five bacteria per larva were injected at time zero. Each data point represents the mean and standard deviation of bacterial counts from 10 groups of 10 larvae. Larvae died approximately 48 h after injection, when bacterial concentrations reached 109/g.
Previous work in our laboratory resulted in a collection of more than 30 mutants of PA14 with reduced virulence in plant or nematode model systems (15, 17, 18, 22, 23; P. S. Yorgey, L. Rahme, M. W. Tan, and F. M. Ausubel, unpublished data). Whereas some of these mutations affect known bacterial virulence factors, most are in previously uncharacterized genes that are likely to encode novel virulence factors. We determined the LD50s of these PA14-derived mutants in G. mellonella. A series of 10-fold serial dilutions containing from 106 to 0 bacteria in 10 mM MgSO4–2 mg of rifampin/ml were injected into G. mellonella larvae. Ten larvae were injected at each dilution and larvae were scored as live or dead after 60 h at 25°C. For each mutant, data from three independent experiments were combined. The Systat computer program was used to fit a curve to the infection data of the following form: Y = A + (1 − A)/(1 + exp[B − G · ln(X)]), where X is the number of bacteria injected, Y is the fraction of larvae killed by the infection, A is the fraction of larvae killed by control injections, and B and G are parameters which are varied for optimal fit of the curve to the data points. LD50s were calculated from the curve and are presented in Table 1. Due to circumstances beyond our control, in particular the condition of the larvae when we received them, there was as much as a 10-fold variation in the assays from one day to the next and the data in Table 1 could be presented only with one significant digit. Isolates with different mutations in the same gene (e.g., gacA) showed similar variation in their LD50s.
Whereas wild-type PA14 has an LD50 of 1 bacterium, the mutant isolates have LD50s of up to 70,000 bacteria. In all cases when mutant isolates were able to kill G. mellonella larvae, the final titers were similar to those shown for wild-type PA14 in Fig. 1, an indication that the mutants had managed to escape the immune system. Most larvae that had not died 60 h after being injected showed no sign of PA14 in blood samples. The unusually low LD50 of the pho23 mutant can be explained at least in part by the fact that this isolate grows more slowly than wild-type P. aeruginosa in culture. All other mutants grew normally in both LB broth and minimal medium.
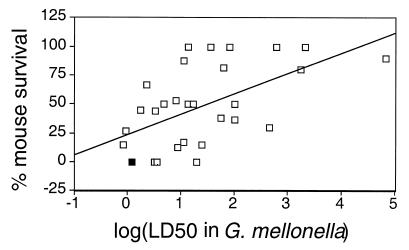
The virulence of PA14-derived mutants in a burned mouse model system has been described previously (15, 17, 18, 22, 23; Yorgey et al., unpublished). Whereas wild-type PA14 is 100% lethal at a dose of 5 × 105 cells, most of the mutant isolates are significantly less virulent. For each mutant isolate, we plotted the LD50 in G. mellonella versus percent mouse survival (Fig. 3). Analysis of variance shows that there is a significant correlation (P < 0.001) between increased LD50 in G. mellonella and reduced lethality in mice, i.e., mutant PA14 isolates that are less virulent in wax moths tend to also be less virulent in mice. When a similar analysis was performed comparing infection with PA14 mutants in plants or nematodes with infection in mice, there was no significant correlation in the severity of the defects of individual mutants, an indication that G. mellonella is a more predictive model system for studying the mammalian infection process.
FIG. 3.
Comparison of LD50 in G. mellonella and percent survival of burned mice after infection with 105 P. aeruginosa bacteria. Solid square, PA14; open squares, individual mutants derived from PA14. y = 17.7 · x + 23.7, P < 0.001, R = 0.565.
All PA14 mutants with LD50s greater than 25 bacteria in G. mellonella showed reduced virulence in mice (Fig. 3). It therefore seems likely that the use of G. mellonella as a host to screen for less virulent PA14 mutants will identify additional P. aeruginosa genes necessary for the mammalian infection process. Similar screens performed with plants (18) and nematodes (15, 23) have resulted in only partially overlapping sets of bacterial mutants. Thus, a screen for P. aeruginosa mutants with reduced virulence in G. mellonella is likely to identify genes that were not found previously.
The study of P. aeruginosa virulence in insects is not limited to G. mellonella or to direct injection of the bacteria into the host. A diet of Arabidopsis thaliana leaves infiltrated with approximately 50 μl of a suspension of 106 CFU of PA14/ml is 100% lethal to larvae of Plutella xylostella (diamondback moth). Moreover, some of the PA14 mutants described here exhibit reduced levels of diamondback moth killing (data not shown). In addition, PA14 can kill adult Drosophila melanogaster following an abdominal prick with a sharp pin that has been dipped in a PA14 culture. As in G. mellonella and P. xylostella, some PA14 mutants show reduced virulence in Drosophila (G. Lau, S. Mahajan-Miklos, F. M. Ausubel, and L. Rahme, unpublished results). Since specific Drosophila mutants with increased sensitivity to microbial infections are known (1, 13), combining such insect mutants with avirulent PA14 mutants might help to elucidate the process of infection, perhaps showing which host defensive systems are needed to counteract specific bacterial virulence factors.
While insects are generally impervious to specialist human pathogens (19, 24), several opportunistic pathogens can infect both insects and humans. G. mellonella is sensitive to the mammalian pathogens Fusarium oxysporum (G. Jander, unpublished results), Aspergillus fumigatus, Proteus vulgaris, Proteus mirabilis, and Serratia marcescens (5). It is likely that these other opportunistic pathogens, like P. aeruginosa, use common virulence factors to infect more than one host.
To the best of our knowledge, a significant correlation between bacterial virulence in insects and mammals has not been reported previously. While there are numerous cases of known mammalian pathogens infecting insects (6, 24), this is the first study in which the virulence of otherwise-isogenic bacterial mutants was compared in mammalian and insect model systems. Our results indicate that insect model systems can provide a new and powerful tool for the identification and characterization of microbial virulence factors involved in causing disease in mammals.
Acknowledgments
This work was supported by National Research Service Award GM18735 and Aventis S.A.
REFERENCES
- 1.Belvin M P, Anderson K V. A conserved signaling pathway: the Drosophila toll-dorsal pathway. Annu Rev Cell Dev Biol. 1996;12:393–416. doi: 10.1146/annurev.cellbio.12.1.393. [DOI] [PubMed] [Google Scholar]
- 2.Bodey G P, Bolivar R. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1983;5:279–313. doi: 10.1093/clinids/5.2.279. [DOI] [PubMed] [Google Scholar]
- 3.Boman H G. Peptide antibiotics and their role in innate immunity. Annu Rev Immunol. 1995;13:61–92. doi: 10.1146/annurev.iy.13.040195.000425. [DOI] [PubMed] [Google Scholar]
- 4.Bulla L A, Rhodes R A, St. Julian G. Bacteria as insect pathogens. Annu Rev Microbiol. 1975;29:163–190. doi: 10.1146/annurev.mi.29.100175.001115. [DOI] [PubMed] [Google Scholar]
- 5.Chadwick J S. Serological responses of insects. Fed Proc. 1967;26:1675–1679. [PubMed] [Google Scholar]
- 6.Chadwick J S, Caldwell S S, Chadwick P. Adherence patterns and virulence for Galleria mellonella larva of isolates of Serratia marcescens. J Invertebr Pathol. 1990;55:133–134. doi: 10.1016/0022-2011(90)90044-7. [DOI] [PubMed] [Google Scholar]
- 7.Hoffman D, Hultmark D, Boman H G. Insect immunity: Galleria mellonella and other Lepidoptera have cecropia-P9-like factors active against gram negative bacteria. Insect Biochem. 1981;11:537–548. [Google Scholar]
- 8.Hoffman J A. Innate immunity of insects. Curr Opin Immunol. 1995;7:4–10. doi: 10.1016/0952-7915(95)80022-0. [DOI] [PubMed] [Google Scholar]
- 9.Hultmark D. Immune reactions in Drosophila and other insects: a model for innate immunity. Trends Genet. 1993;9:178–183. doi: 10.1016/0168-9525(93)90165-e. [DOI] [PubMed] [Google Scholar]
- 10.Jarosz J. Interaction of Pseudomonas aeruginosa proteinase with the inducible non-self response system of insects. Cytobios. 1995;83:71–84. [Google Scholar]
- 11.Jarosz J. Identification of immune inhibitor from Pseudomonas aeruginosa of inducible cell-free antibacterial activity in insects. Cytobios. 1997;89:73–80. [PubMed] [Google Scholar]
- 12.Jarrell K F, Kropinski A M. The virulence of protease and cell surface mutants of Pseudomonas aeruginosa for the larvae of Galleria mellonella. J Invertebr Pathol. 1982;39:395–400. doi: 10.1016/0022-2011(82)90065-9. [DOI] [PubMed] [Google Scholar]
- 13.Lemaitre B, Nicolas E, Michaut L, Reichhart J-M, Hoffman J A. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 14.Lysenko O. The mechanisms of pathogenicity of Pseudomonas aeruginosa (Schroeter) Migula 1. The pathogenicity of strain N-06 for the larvae of the greater wax moth, Galleria mellonella (Linnaeus) J Insect Pathol. 1963;5:78–82. [Google Scholar]
- 15.Mahajan-Miklos S, Tan M-W, Rahme L G, Ausubel F M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 16.Mullett H, Ratcliffe N A, Rowley A F. Analysis of immune defences of the wax moth, Galleria mellonella, with anti-haemocytic monoclonal antibodies. J Insect Physiol. 1993;39:897–902. [Google Scholar]
- 17.Rahme L, Stevens E, Wolfort S, Shao J, Tompkins R, Ausubel F. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;269:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 18.Rahme L G, Tan M-W, Le L, Wong S M, Tompkins R G, Calderwood S B, Ausubel F M. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc Natl Acad Sci USA. 1997;94:13245–13250. doi: 10.1073/pnas.94.24.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinhaus E A. Principles of insect pathology. New York, N.Y: McGraw-Hill; 1949. [Google Scholar]
- 20.Stephens J M. Bactericidal activity of the blood of actively immunized wax moth larvae. Can J Microbiol. 1962;8:491–499. [Google Scholar]
- 21.Stevens E J, Ryan C M, Friedberg J S, Barnhill R L, Yarmush M L, Tompkins R G. A quantitative model of Pseudomonas aeruginosa infection in injury. J Burn Care Rehabil. 1994;15:232–235. doi: 10.1097/00004630-199405000-00005. [DOI] [PubMed] [Google Scholar]
- 22.Tan M, Mahajan-Miklos S, Ausubel F. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci USA. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan M, Rahme L, Sternberg J, Tompkins R, Ausubel F. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci USA. 1999;96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanada Y, Kaya H. Insect pathology. San Diego, Calif: Academic Press; 1993. [Google Scholar]