Abstract
The success of checkpoint blockade therapy against cancer has unequivocally shown that cancer cells can be effectively recognized by the immune system and eliminated. However, the identity of the cancer antigens that elicit protective immunity remains to be fully explored. Over the last decade, most of the focus has been on somatic mutations derived from non-synonymous single-nucleotide variants (SNVs) and small insertion/deletion mutations (indels) that accumulate during cancer progression. Mutated peptides can be presented on MHC molecules and give rise to novel antigens or neoantigens, which have been shown to induce potent anti-tumor immune responses. A limitation with SNV-neoantigens is that they are patient-specific and their accurate prediction is critical for the development of effective immunotherapies. In addition, cancer types with low mutation burden may not display sufficient high-quality [SNV/small indels] neoantigens to alone stimulate effective T cell responses. Accumulating evidence suggests the existence of alternative sources of cancer neoantigens, such as gene fusions, alternative splicing variants, post-translational modifications, and transposable elements, which may be attractive novel targets for immunotherapy. In this review, we describe the recent technological advances in the identification of these novel sources of neoantigens, the experimental evidence for their presentation on MHC molecules and their immunogenicity, as well as the current clinical development stage of immunotherapy targeting these neoantigens.
Keywords: alternative source of neoantigen, cancer, gene fusion, RNA splicing, frameshift, dark matter
1. Introduction
The clinical success of immune checkpoint blockade (ICB) therapy has revealed that T cells are the primary mediators of anti-tumor immunity. The tumor antigens that drive protective T cell responses have been elusive until about 10 years ago when it was discovered that somatic mutations that accumulate in cancers can stimulate both CD8 and CD4 T cell responses [1,2] and are associated with clinical response to ICB therapy [3].
Somatic mutations can translate into mutated proteins, and thus generate novel antigens or neoantigens that are recognized as foreign/non-self by the immune system and elicit potent T cell responses against cancer cells. Peptides derived from these proteins can enter the major histocompatibility complex (MHC) pathways for presentation to T cells. Endogenous proteins are processed by the proteasome into peptides that enter the endoplasmic reticulum and are loaded onto MHC class I (MHCI) molecules before being transported to the cell surface for presentation to CD8 T cells. Alternatively, peptides can also be presented on MHCII to CD4 T cells. They are generally derived from exogenous proteins and are generated by lysosomal proteases and loaded on MHCII in the endosomal/lysosomal compartments [4].
Only a minority of peptide-derived proteins are presented on MHC, and antigen prediction is critical to the development of successful antigen-targeted immunotherapies. The earliest antigen prediction tools relied on peptide binding prediction on MHC molecules, and provided enrichment for immunogenic peptides [5]. However, these prediction methods also had high false positive rates [6]. More recently, improved mass spectrometry technologies associated with comprehensive genomic analyses facilitated the generation of large datasets of naturally processed antigens presented on a broad range of MHC alleles, also called human leukocyte antigen (HLA) in humans [7,8,9]. These new datasets allowed for incorporation of peptide processing and presentation and led to the significant improvement of prediction algorithms [10,11,12,13,14,15,16]. In addition, the generation of immunopeptidomics data from monoallelic cell lines allowed clear identification of eluted peptides for specific HLA types compared to multiallelic datasets that require deconvolution [17]. This strategy boosted the detection of peptide/HLA-I complexes for low-frequency HLA-Is, improving the patient’s coverage of neoantigen-prediction HLA-I presentation tools [18]. For HLA-II molecules, however, progress has been slower, mostly due to the higher polymorphism of HLA-II molecules, the more variable length of the bound peptides, and the complexity of peptide/HLA-II interaction. The field is currently amplifying the efforts to improve the limited accuracy of HLA-II neoantigen prediction algorithms, notably by increasing training data from mass spectrometry [19,20].
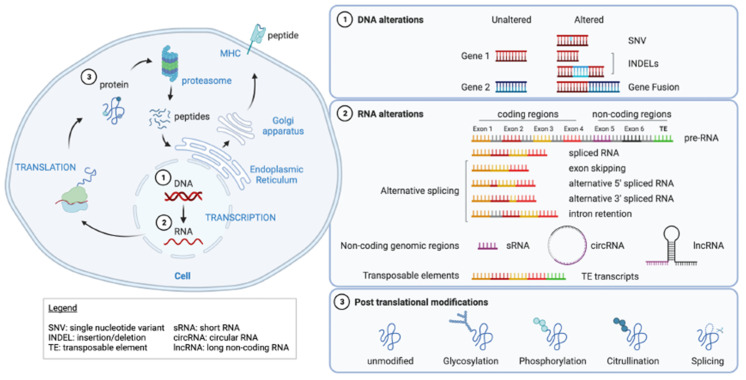
Single-nucleotide variants (SNV) and small insertion/deletion mutations (indels) are currently favored targets for neoantigen-specific immunotherapy approaches. However, the tumor mutational burden may not be sufficient to mount an efficacious anti-tumor response in many cancers. In addition, the number of SNV-predicted neoantigens reported to be immunogenic is low (~3%), as shown in the meta-analysis of 13 different publications [21]. While the newest advances in computational predictions may reflect a better ratio, it is clear that key determinants of immunogenicity are not fully understood and/or yet incorporated in neoantigen prediction models. As TCR recognition is critical to T cell activation, more disruptive aberrations than [SNVs/small indels] may generate qualitatively distinct T cell responses. Cancer cells accumulate various other types of alterations at the DNA, RNA, and protein levels that are not observed in normal cells, and may be an alternative source of cancer neoantigens with a more favorable profile (Figure 1). At the DNA level, gene fusions have been largely ignored as a source of neoantigens. At the RNA level, variants include alternative splicing events (ASE) and dysregulation of transposable element (TE) expression. Post-translational modifications include spliced peptides, glycopeptides, phosphopeptides, or citrullinated peptides.
Figure 1.
Sources of neoantigens (created with Biorender.com (accessed on 22 August 2022)).
In this review, we describe the recent updates on identification and prediction methods, as well as preclinical and clinical studies showing promising results for most of the DNA/RNA aberrations as novel sources of neoantigens.
2. DNA Alterations
2.1. SNV/Indels
SNVs and small indels are abundant in many cancers [22] and are used as the reference in neoantigen-targeted therapy.
Identification of SNV and small indels through NGS techniques and softwares is highly accurate [23], and a plethora of [SNV/small indel]-derived neoantigen predicting tools have been published in the last decade [23,24,25] with significant increased performance. However, the immunogenic fraction of predicted neoantigen candidates remains small even with improved presentation predictions, suggesting that other critical determinants are required for immunogenicity (Table 1 and Table 2).
Recent advances have focused on further identifying critical physicochemical and structural properties of immunogenic mutated peptides that led to TCR binding and T cell activation. Studies found that hydrophobicity, amino acid charge, and size of the MHCI peptide impacts TCR binding. In addition, it is important to consider the recognition of the presented peptide as foreign by the immune system. Features such as divergence between mutated (MUT) and wild-type (WT) epitopes, and similarity to microbial antigens have been suggested to contribute to peptide immunogenicity [26,27,28,29,30,31,32,33,34]. Our work showed that mutations in direct contact with the TCR were more likely to be immunogenic [32,35]. In addition, mutations at anchor positions that increase binding affinity to MHCI in comparison to the corresponding WT peptide may potentially result in novel and more immunogenic epitopes. Indeed, the difference in binding affinity between the MUT peptide and the corresponding WT (relative binding affinity) is an important predictor of immunogenicity [35,36]. Further development of artificial intelligence methods will certainly help integrate all these features into one neoantigen prediction pipeline workflow, and likely further increase immunogenicity prediction accuracy [36]. However, it is important to note that SNVs differ from the WT counterpart by only one amino acid, which may not always be sufficient to differentiate from the self and elicit potent immune responses. Indeed, we found a high frequency of T cell cross-reactivity between MUT peptides and their WT counterpart [35]. More disruptive aberrations may have a higher potential to generate a qualitative T cell response. While superior quality per se of the T cell response for non-SNV neoantigens remains to be proven, frameshift indels that generate long novel AA sequences can lead to a higher number of HLA epitopes presented on a more diverse set of HLA alleles than SNVs [37,38,39]. In addition to a higher rate of neo-epitope candidates per mutation, [long] indels-based immunotherapy may also be less subject to HLA loss-induced cancer immune evasion.
2.2. Gene Fusion
Chromosomal translocations or deletions induce DNA rearrangement that can lead to the fusion of genes, thus possibly generating fusion proteins (Figure 1). Some of these newly formed fusion products have been identified as oncogenes, such as BCR-ABL in leukemias [40,41], and used as diagnostic or predictive biomarkers as well as drug targets [42]. These oncogenic fusion proteins are also attractive neoantigen targets due to high dissimilarity to self, clonality, as well as being shared between patients (Table 1).
While more than 260 gene fusions have been reported in hematological disorders, only 70 have been identified in solid tumors [43]. Karyotypes from solid tumors are more complex, resulting in poor quality and/or less accurate cytogenetic analysis to identify chromosomal abnormalities [43]. With the development of Fluorescence In Situ Hybridization (FISH), fusion breakpoints could be detected at the molecular level and this led to new structural rearrangement discoveries. However, FISH assay is not suitable for high-throughput screening due to live material requirement, cost, and probe specificity. It is only with the advances of targeted sequencing techniques that increased reliable identification of gene fusion events could be achieved. Following the progress of high-throughput DNA and RNA sequencing, novel tools are being developed to identify gene fusions [44,45,46,47,48,49]. While some of these methods showed high accuracy and speed, the sensitivity remains to be improved especially for low expressed transcripts [45]. In order to provide more insights in identification of tumor-specific neoantigens derived from gene fusions, Rathe et al. compared the fusion sequences provided by the deFuse algorithm [50] to the transcriptome generated by the Trinity method [51], and used NetMHCpan 4.0 [10] to predict neoepitope binders [52] from osteosarcoma patient samples. The authors were able to identify candidate neoantigens associated with fusions and found the frequency of fusion events to correlate with patient outcome. Although promising, additional studies are now required to experimentally validate the approach.
Early exploratory clinical trials uncovered the actionable potential fusion-neo-antigens [53,54,55]. In one trial, three patients with Philadelphia chromosome positive acute lymphoblastic leukemia were treated with BCR/ABL-specific CD8 T cells expanded from the patients’ peripheral blood mononuclear cells or from donors’ hematopoietic stem cells. Interestingly, BCR/ABL-specific T cells were increased in the bone marrow of all three patients post infusion, and all patients achieved a molecular or hematologic complete remission [56]. In solid tumors, targeting of the EWS/FLI-1 and PAX3/FKHR breakpoint regions in Ewing sarcoma and alveolar rhabdomyosarcoma patients through vaccination showed mixed results. In a first pilot study, the vaccinated patients did not show clinical benefit, nor did they develop a T cell response [57]. However, in a subsequent study, vaccinated patients had an increased overall survival and 25% developed vaccine-induced T cell responses [58]. The different outcomes between the two studies could be due to the different vaccine platforms used or the patient population. Indeed, the patients in the second study were in remission compared to an advanced cancer stage for the first study (Table 2). This may suggest that treating healthier patients with low/no tumor burden with vaccines may be more effective.
Although these early results are encouraging, some challenges remain to be overcome in order to include gene fusion-derived neoantigens in clinical neoantigen-based immunotherapy pipelines. The sensitivity of fusion products identification would benefit from wide access to deep sequencing technologies. In addition, expanding validation sets from presented gene fusion-derived peptides to train neoantigen prediction algorithms would help prioritizing neoantigens from alternative sources (Table 1 and Table 2).
3. RNA Aberrations
3.1. Alternative Splicing
Splicing is the essential step that creates mature mRNAs by removing introns from pre-mRNAs made of both introns and exons, and allows protein synthesis. Alternative splicing is a multiplexing process enabling the generation of multiple proteins from a single gene (different combination of exons), allowing protein diversity (Figure 1). Splicing events are involved in all major cell functions and, therefore, the splicing machinery or spliceosome is highly regulated.
Alternative splicing events (ASE) are detected at the transcriptome level and include skipped exons, alternative 5′ splice sites (donor), alternative 3′ splice sites (acceptor), retained intron, and mutually exclusive exon usage [59,60,61] (Figure 1). Computational methods based on RNA sequencing are used to detect and quantify ASE (reviewed elsewhere [62,63,64,65,66,67,68]). However, precise quantification of transcript isoforms remains challenging due to limited read length. The depth and quality of sequencing is also at play to improve the detection of novel transcripts and limiting false negative events.
Dysregulation of splicing events is involved in oncogenesis [61] and associated with drug resistance. Furthermore, tumor-specific ASEs have been used as predictive bio-markers for therapeutic response and/or clinical outcome in various cancers [69,70,71,72,73,74,75]. Mutations occurring in the spliceosome machinery, in addition to directly providing a source of SNV-neoantigens, also produce potential abnormal splicing events that may generate novel antigens. Kahles et al. identified MHCI epitopes derived from ASEs in breast and ovarian cancers from the TGCA immunopeptidomic database [76]. In these two cancer indications with relatively low SNV numbers, epitopes derived from ASEs were more abundant than from SNVs. In addition, ASE-derived peptides have been shown to be immunogenic in vitro [77], and in vivo with specific T cells found in the blood of cancer patients [78]. Some tumors also have an increased number of splicing events compared to normal tissue [76], suggesting that ASEs may generate neoantigens that are tumor-specific. Indeed, mutation of the splicing factor SF3B1 has been shown to induce tumor-specific ASE-derived neoepitopes that are recognized by CD8 T cells from uveal melanoma patients [79]. Altogether, these studies highlight that ASE-derived peptides can be immunogenic and tumor-specific, thus being suitable for neoantigen-based immunotherapy (Table 1 and Table 2).
Currently, most of the therapeutic approaches rely on pharmacological compounds targeting the spliceosome machinery. While targeting ASE-derived neoantigens for immunotherapy is attractive, several questions need to be addressed before their use in the clinic: (1) Can these novel epitopes generate strong and lasting anti-tumor responses? ASEs seem less expressed than SNVs [76], thus possibly limiting ASE-derived neoepitope presentation. In addition, some splicing events induce only minor changes to the protein sequence and may not bypass immune tolerance. Similarly to SNVs, evaluating the dissimilarity to self-peptides may improve prediction and selection of immunogenic ASE-neoantigens for targeted immunotherapies [35]. (2) How truly tumor-specific are the predicted ASE and are they not occurring in normal cells in a tissue-specific manner? (3) How many of ASEs are shared between tumor cells and/or patients? ASEs seem to mostly derive from passenger- rather than driver- mutations [76], suggesting that they are likely private (Table 1).
3.2. Non-Coding Genomic Regions
Most of the human genome is considered as non-protein coding genes. With the development of sequencing technologies (WGS, RNA-seq, ChIP-seq, ATAC-seq, Hi-C, …), annotations for the non-coding genome have widely increased in the last decades. This led to discovery and characterization of RNA transcripts such as short and long non-coding RNAs (lncRNAs) or circular RNAs (Figure 1), and their role in regulating transcription, splicing, and translation [80]. Surprisingly, peptides derived from annotated non-coding sequences have been identified. In some instances, mislabeled annotations as “non-coding” seem to explain this observation [81,82]. Some of these regions have also been suggested to produce non-stable and thus non-functional proteins, leading to a quick degradation [83]. Other regions, known as pseudogenes, have recovered a lost protein-coding function in cancer cells [84], suggesting tumor-specificity to these “dark matter” antigens.
The recent advances in peptidomics and proteogenomics have increased the sensitivity of the techniques, thus detecting lower amounts of presented peptides [85]. However, most of these techniques refer to annotated proteins. It is only recently that new tools have emerged to identify unconventional antigens [86]. Interestingly, ribosome profiling has identified long non-coding RNA with both 5′ cap and polyA tails bound to ribosomes. Although the resulting translated proteins may not be functional, epitopes have been shown to be presented on the tumor cell surface [87]. Ribo-seq tool has also allowed identification of HLA-I presented peptides derived from small or novel unannotated open reading frames (smORF or nuORF) [83,88,89]. Importantly, these dark matter antigens have shown tumor-specificity to some extent [89,90] (Table 1). Using a new workflow combining immunopeptidomics, RNA-seq and Ribo-seq, Chong et al. suggested that 23% of the identified non-canonical HLA-Ip can be considered as tumor-specific [87]. These results, with other studies demonstrating tumor-specificity of HLA epitopes derived from non-coding regions [91] are encouraging for cancer patient immunotherapy. Another recent study, in a preclinical colorectal tumor model, showed delayed CT26 tumor growth in mice prophylactically vaccinated with peptides derived from the cryptic or “non-coding” transcriptome identified by mass spectrometry [92]. Although this study lacks definitive proof of specific T cell response, it suggests that these cryptic peptides can generate an anti-tumor response. Importantly, only a very limited number of these cryptic antigens showed anti-tumor effects, and the combination of three cryptic peptides were required for reducing tumor growth post vaccination, suggesting either limited presentation and/or weak T cell responses for each of the antigens. Similarly, only one of the identified non-canonical peptides from the Chong et al. study was found to be immunogenic [87]. Thus, sufficient clonality and quantity of these antigens from non-coding regions to generate efficacious anti-tumor T cell responses remains to be demonstrated before being clinically tested for immunotherapy in cancer patients (Table 2). Another concern is that non-functional proteins may be more easily subject to immunoediting, thus leading to tumor escape.
3.3. Transposable Elements
Transposable elements (TEs) or jumping genes are DNA-repetitive sequences integrated into the human genome that perform essential functions in driving genome evolution [93]. There are two different classes of TEs: (1) DNA transposons that mobilize through a DNA intermediate in a “cut and paste” mechanism, and (2) retrotransposons that undergo reverse transcription with a “copy and paste” mechanism. Although DNA transposons are part of the human genome, they are no longer active [94]. Retrotransposons are subdivided into two types: long terminal repeat (LTR) or non-LTR retrotransposons. LTR retrotransposons include human endogenous retroviruses (hERVs) derived in part from ancient retroviruses that infected germ cell progenitors, and the mammalian apparent LTRs retrotransposons (MaLRs). Non-LTR transposons include long interspersed nuclear elements (LINEs) that are autonomously active retrotransposons, or short interspersed nuclear elements (SINEs) and SINE-VNTR-Alu (SVAs), with both needing a specific protein for activation [95,96]. While most retrotransposons are silenced through DNA methylation, histone modifications, or RNA-mediated silencing, dysregulation can occur and lead to cancer initiation or progression through various mechanisms (reviewed elsewhere [96,97]).
Genomic TE insertions are identified through whole-genome sequencing, RNA-seq or Ribo-seq, and specific bioinformatic tools showing variable sensitivity and specificity [98]. Most methods are designed for short-read sequencing, which limits the detection of TE’s repetitive sequences. The combination of both short-read and long-read sequencing data seem to increase the detection of both germline and somatic TEs [99]. However, the development of comprehensive pipelines combining different tools is recommended for higher performance [98,100] (Table 1).
Several studies have shown that the amount of TE transcripts and their derived peptides presented by HLA-I is abnormally increased in human cancer cells [101,102], and associated with tumor-infiltrating T cells as well as enhanced responses to ICB in cancer patients [102,103]. Immunogenicity of TE-derived epitopes was reported a long time ago in preclinical models and showed to protect against tumor challenge [104,105]. In humans, recognition of HERV antigens by CD8 T cells from healthy donor PBMCs has been demonstrated [106,107], as well as from cancer patients [108,109]. In addition, selective expression of HERV-E antigens has been associated with tumor regression in metastatic renal cell carcinoma [110]. These results suggest that TE-derived peptides are suitable for immunotherapy. Moreover, this targeted strategy may be used across patients since presented peptides can be found from conserved TE families (HERV, LINE, SINE, and SINE-VNTR-Alu) [101], and specific T cells against the same HERV antigens found in tumors from multiple patients [102] (Table 2). However, since TE can be expressed in normal cells [103] and many share sequence homologies, their immunogenicity may be limited. In addition, the expression pattern of TEs varies between cancer types [103] and since most computational tools seem to identify TE subfamilies with specificity [98], further development of computational workflows is required for high throughput TE identification and selection pipelines (Table 1). Interestingly, increased expression of TE-transcripts and TE-derived epitopes have been shown in cancer cells after in vitro treatment with DNA methylation inhibitors [101,111,112,113], suggesting that DNA-demethylating therapy (hypomethylating drugs such as azacytidine and decitabine) could be used in combination with immunotherapies. Further studies have shown synergistic anti-tumor effects with ICB therapies in preclinical models [114]. Epigenetic repression of TE-transcripts has also recently been associated with resistance to anti-PD1 therapy [115], and blocking the KDM5B–SETDB1 interaction could lead to effective anti-tumor responses mediated by TE-specific T cells [115,116]. Many epigenetic therapies are currently tested in clinical trials with some already approved for hematological malignancies [117], and it will be interesting to determine whether the objective clinical responses are associated with modulation of the immune response against TEs.
4. Post-Translational Modifications
Tumor antigens can derive from post-translational modifications (PTM) that induce peptides that differ from the parental protein sequence. Different PTM have been involved as potential sources of candidate neoantigens such as glycosylation [118], phosphorylation [119,120,121,122], citrullination [123], or peptide splicing (Figure 1).
Glycosylation is the covalent attachment of a carbohydrate or glycan to a protein by a glycosyltransferase, and the most common PTM occurring in cells. Tumor cells present altered glycosylation patterns [124,125], and N- (Asn-linked) or O- (Ser/Thr-linked) glycosylations have been shown to generate neo- or overexpressed glyco-antigens that can be presented on the cell surface in many cancer types (reviewed in [126,127]) (Table 1). Altered glycosylation (and expression) of mucin 1 (MUC1) is one of the most studied PTM events and has been associated with several cancers [128]. Many therapeutic strategies targeting MUC1-derived truncated O-glycans such as Tn, sialyl-Tn, or Thomsen-Friedenreich (TF) antigens have been tested in clinical trials (phases 1 to 3), including dendritic cell, peptide-, or virus-based vaccines [128]. In contrast to early trials showing encouraging results, the most advanced trials showed mitigated results. No significant difference in overall survival was observed with non-small cell lung cancer (NSCLC) patients receiving Tecemotide (L-BLP25) peptide vaccine in a phase 3 trial [129]. In contrast, progression-free survival was significantly improved for patients receiving a modified vaccinia Ankara (MVA) expressing MUC1 and interleukin-2 (TG4010) plus chemotherapy compared to the control arm in a phase 2b/3 trial for advanced NSCLC. However, the survival benefit was only 0.8 months [130] (Table 2).
Cancer cells present dysregulated signaling pathways inducing an increased protein phosphorylation level and phosphopeptide presentation at the cell surface in various tumors [119,122,131,132]. Interestingly, phosphopeptides can be recognized by CD8 T cells [119,121] in a specific manner (in comparison to the non-phosphorylated counterpart) [119], and CD4 T cells [120] (Table 1). In a recent phase 1 clinical trial, 6 out of 15 melanoma patients showed evidence of CD8 T cell responses post vaccination with 2 phosphopeptides and adjuvants, including 2 pre-existing responses [133] (Table 2). While these results are encouraging, further studies are needed to demonstrate that targeting phospho-neoantigens for immunotherapy can generate anti-tumor activity in cancer patients.
Citrullination is the conversion of arginine into citrulline by peptidylarginine deiminases, which can alter the protein structure. Thus, the interest for this PTM has recently increased and its evaluation as an immunotherapy target is ongoing. Citrullinated vimentin and enolase peptides have been shown to be presented on MHCII and recognized by CD4 T cells [134,135] (Table 1). Brentville et al. demonstrated that transduced tumor cells can present a citrullinated vimentin peptide on HLA-DR4 molecules, and that differential recognition by CD4 T cells occurs compared to the WT counterpart peptide [136]. Citrullinated vimentin peptide vaccines delayed tumor growth and increased survival rates of HLA-DR4 transgenic mice implanted with B16F1 tumors expressing HLA-DR4 [136] (Table 2). Interestingly, CD4 Th1 T cell responses from PBMCs from ovarian cancer patients and healthy donors were observed against citrullinated vimentin and enolase peptides [137]. While these results suggest promising candidates for immunotherapy, further studies are required to demonstrate tumor-specificity. In addition, the presentation of citrullinated peptides presented on MHCI remains to be demonstrated.
Finally, studies have identified CD8 T cells recognizing spliced peptides in renal cell carcinoma [138], in melanoma [139], or from EBV-B cells [140], thus suggesting their immunogenic potential (Table 2). Interestingly, Liepe et al. suggested that spliced peptides represent about 25% of the HLA-I peptidome of human cancer cell lines, thus increasing the possibility of novel source of antigens [141]. However, another study only estimated it to be 2–6% of the HLA-I ligandome [142], suggesting that further work is required for accurate identification of these events. While new identification tools and workflows have emerged in the past years [141,143,144,145], biological validations remain to be demonstrated (Table 1). In addition, the mechanism behind peptide splicing remains unclear. Some studies have suggested that the proteasome can form spliced peptides by ligation of two fragments from the same source protein sequence [138,139], with deamidation thus changing amino acid residues from asparagine to aspartate [146] or by transpeptidation and trimming that led to a spliced peptide with short C-terminal fragment [147]. However, very few spliced peptides have been validated so far, leading to the hypothesis that splicing peptide events are rare, and thus possibly not relevant for immunotherapy.
In conclusion, the shared nature of PTM neoantigens makes them interesting targets but many challenges remain to be addressed to prove the relevance for immunotherapy. Increasing the precision and sensitivity in identification of most of these events is required in order to accurately evaluate the frequency of PTM-derived presented peptides on MHC molecules. In addition, most PTM are commonly used in cell metabolism, and determining their tumor-specificity is essential. Nevertheless, as many of the identified PTM-neoantigen candidates were shared between tumors and patients [118,121,128,133,136], it seems important to pursue the efforts in characterizing tumor-specific PTM-derived peptides, as well as the development of predictive methods.
5. Conclusions
Early clinical studies using [SNV/small indels]-neoantigen based-vaccines have shown that T cell responses can be induced in patients but their clinical benefit is rather disappointing [2,148,149,150,151,152]. It is noticeable that the magnitude and breadth of the T cell responses induced by these vaccines is generally low, at least in the blood, and appears insufficient for efficacy. Improved neoantigen prediction algorithms as well as the design or superior vaccine platforms may help overcome this limitation. Alternatively, the quality of the T cell response may be suboptimal. Novel and improved methods to explore the contribution of alternative sources of neoantigens to further refine our understanding of the tumor antigen landscape, as well as the characterization of tumor-specific T cell responses in cancer patients developed spontaneously or after CIB treatment, will be critical to define the determinants of effective T cell responses beyond immunogenicity, and design improved immunotherapies.
Clonal neoantigens are expressed in a higher number of tumor cells, and the fraction of clonal neoantigens correlates with ICB response [153]. As driver mutations are more clonal than passenger mutations, as well as more likely to be shared between patients, they seem to be an ideal target. However, studies have shown that shared predicted neo-epitopes between patients are rare [154,155] and less presented by MHC molecules [155,156,157]. Nevertheless, clonal neoantigens seem to generate more CD8 T cell responses compared to subclonal neoantigens [158,159]. This may be of importance when choosing the source of neoantigens for immunotherapy as many RNA or PTM aberrations are not likely to be shared within the whole tumor, unless it is induced by an oncogenic event.
The expression of neoantigens is a key immunogenic criterion. As DNA aberrations may lead to non-transcript events, most of the neoantigen prediction pipelines include filters for RNA expression levels. However, high throughput identification of translated products is challenging for all sources of neoantigens. Since presentation on MHCI by tumor cells is required for CD8-induced tumor cell killing, several clinical neoantigen selection pipelines include direct neoantigen identification through mass spectrometry. Nevertheless, identification of some neoantigens by MS such as “non-coding”, TE- or PTM-derived peptides remains to be improved before being included in clinical pipelines. In addition, lack of tumor recognition by SNV-neoantigen-specific CD8 T cells has been linked to the insufficient amount of presentation [160]. Thus, development of new quantitative approaches such as the TOMAHAQ-targeted MS [9] are needed to correlate the amount of presented neoantigens and CD8 T cell-induced anti-tumor efficacy, especially for comparing sources of neoantigens.
Another important consideration is the likelihood of neoantigen expression loss that may differ between sources. Indeed, HLA loss is observed in cancers and suggested to lead to tumor immune evasion [161]. Thus, neoantigens binding to multiple HLA may offer a therapeutic advantage. In addition, potent and dominant neoantigen-specific T cell response may favor the loss of neoantigen expression due to immune pressure [162]. Therefore, it remains to be shown that immunotherapy targeting neoantigens derived from more disruptive aberrations would generate long and lasting memory T cell responses.
An additional aspect modulating neoantigen quality is the platform used for immunotherapy. Many platforms have demonstrated potent vaccine-induced neoantigen-specific T cell responses in clinical trials, including RNA [149,152], dendritic cell [148,163,164,165], or peptide [150]. However, specific T cell responses can vary with neoantigen-based therapies [35,166], and some sources of neoantigen may have limited platform options. For example, some PTM-derived neoantigens would preferably require peptide-based vaccines or adoptive cell therapy, which have shown manufacturing challenges and production delays.
Finally, studies suggest that T helper cells are required to generate an efficacious anti-tumor-specific CD8 response. In two different preclinical tumor models (T3 MCA-induced sarcoma and SMA560 glioma), anti-tumor activity of SNV-neoantigen specific CD8 T cells could only be observed with MHCII neoantigen co-expression by tumor cells [167,168]. In addition, Swartz’s study suggests that vaccines encoding a non-tumor-specific MHCII-restricted antigen associated with MHCI neoantigen may be sufficient to generate anti-tumor responses [168], which would facilitate vaccine design and manufacturing. While additional studies need to confirm these findings and the mechanisms, the type (MHCI and/or MHCII) of T cell response generated by different sources of neoantigen may not be equal. It is important to note that combined with the vaccine platform, which also impacts immune cells differently, the balance between CD4 and CD8 T cell responses may be highly modified.
Table 1.
Summary of neoantigen reactivities.
| Alterations | Presentation | Immunogenicity | Shared between Patients | Tumor-Specificity | Tumor Alteration Burden | Main Challenges |
|---|---|---|---|---|---|---|
| SNV/indels | MHCI, MHCII | CD8, CD4 | Mostly private | Yes | Low to high depending on cancer type | Immunogenicity (similarity to self) |
| Gene fusion | MHCI, MHCII [53,54,55] |
CD8, CD4 [53,54,55,56] |
Yes | Yes | Low | Identification, prediction |
| Alternative splicing | MHCI, MHCII [76,169] |
CD8, CD4 [77,78,79,169] |
TBD | TBD | TBD | Identification, tumor-specificity |
| Non-coding genomic regions | MHCI, MHCII [83,86,87,88,89] |
CD8 [87] | Yes | Yes | TBD | Immunogenicity, tumor-specificity |
| Transposable Elements | MHCI, MHCII [104,105,106,107,108,109] |
CD8, CD4 [104,105,106,107,108,109] |
Yes | No | TBD | Identification, tumor-specificity |
| Glycosylation | MHCI [118] | CD8 [118] | Yes | TBD | Low | Identification, prediction, tumor-specificity |
| Phosphorylation | MHCI, MHCII [119,120,121,122] |
CD8, CD4 [119,120,121,122] |
Yes | TBD | Low | Identification, prediction, tumor-specificity |
| Citrullination | MHCII [134,135,136] |
CD4 [134,135,136] |
Yes | TBD | Low | Identification, prediction, tumor-specificity |
| Peptide splicing | MHCI | CD8 [137,138,139] |
TBD | TBD | TBD | Identification, prediction, tumor-specificity |
SNV: Single nucleotide variant; TBD: To be demonstrated.
Table 2.
Clinical development stage of neoantigen-targeted therapies.
| Alterations | Altered Molecule | Identification | Prediction | Most Advanced Development Stage | Example |
|---|---|---|---|---|---|
| SNV/indels | DNA | WES + RNA-seq | Available (many) | Phase 1/1b; several ongoing Phases 2/3 | Immunogenic responses observed in patients receiving peptide/DC/mRNA vaccines; or adoptive T cell therapy in different cancer types [147,148,149,170,171] |
| Gene fusion | DNA | WES + RNA-seq | Available (few) | Phase 2 | Immunogenic response but no clinical efficacy observed in patients with CML following bcr-abl peptide vaccination [172] |
| Alternative splicing | RNA | RNA-seq, Ribo-seq |
Available (few) | Preclinical | CD8 T cell recognition of the mutated splicing factor SF3B1 in patients with uveal melanoma [79] |
| Non-coding genomic regions | RNA | RNA-seq, Ribo-seq |
NA | Preclinical | Delayed tumor growth of CT26 tumors following cryptic peptide vaccination without proof of specific T cell response [92] |
| Transposable Elements | RNA | WES + RNA-seq, Ribo-seq |
Available (few) | Preclinical ongoing Phase 1 | Recognition of HERV antigens by CD8 T cells from patients [108,109] HERV-E TCR Transduced Autologous T Cells in Metastatic Kidney cancer patients (*) |
| Glycosylation | Protein | Mass spectrometry | NA | Phase 3 | No overall survival benefit with L-BLP25 peptide vaccine in NSCLC patients [129]; Improved progression free survival post TG4010 vaccine + chemotherapy in NSCLC patients [130] |
| Phosphorylation | Protein | Mass spectrometry | NA | Phase 1 | Some specific CD8 T cell responses were observed in melanoma patients who received pIRS2 and pBCAR3 peptide vaccines [133] |
| Citrullination | Protein | Mass spectrometry | NA | Preclinical | Delayed B16F1 tumor growth in HLA-DR4 transgenic mice following citrullinated peptide vaccination. Citrullinated-specific CD4 T cell responses also observed in PBMC from ovarian cancer patients [136] |
| Peptide splicing | Protein | Mass spectrometry | NA | Preclinical | Spliced peptide identified and recognized by CD8 T cells in renal cell carcinoma [137] or melanoma [138] patients, and from EBV-B cells [139] |
SNV: Single nucleotide variant; WES: Whole exome sequencing; seq: sequencing; NA: Not available. * ClinicalTrials.gov.
Author Contributions
Conceptualization, A.-H.C. and L.D.; writing—original draft preparation, A.-H.C., R.H. and L.D.; writing—review and editing, A.-H.C. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors are employee of Genentech and declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gros A., Robbins P.F., Yao X., Li Y.F., Turcotte S., Tran E., Wunderlich J.R., Mixon A., Farid S., Dudley M.E., et al. PD-1 Identifies the Patient-Specific CD8+ Tumor-Reactive Repertoire Infiltrating Human Tumors. J. Clin. Investig. 2014;124:2246–2259. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen C.J., Gartner J.J., Horovitz-Fried M., Shamalov K., Trebska-McGowan K., Bliskovsky V.V., Parkhurst M.R., Ankri C., Prickett T.D., Crystal J.S., et al. Isolation of Neoantigen-Specific T Cells from Tumor and Peripheral Lymphocytes. J. Clin. Investig. 2015;125:3981–3991. doi: 10.1172/JCI82416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zou X.L., Li X.B., Ke H., Zhang G.Y., Tang Q., Yuan J., Zhou C.J., Zhang J.L., Zhang R., Chen W.Y. Prognostic Value of Neoantigen Load in Immune Checkpoint Inhibitor Therapy for Cancer. Front. Immunol. 2021;12:689076. doi: 10.3389/fimmu.2021.689076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santambrogio L. Molecular Determinants Regulating the Plasticity of the MHC Class II Immunopeptidome. Front. Immunol. 2022;13:878271. doi: 10.3389/fimmu.2022.878271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen M., Lundegaard C., Blicher T., Lamberth K., Harndahl M., Justesen S., Røder G., Peters B., Sette A., Lund O., et al. NetMHCpan, a Method for Quantitative Predictions of Peptide Binding to Any HLA-A and -B Locus Protein of Known Sequence. PLoS ONE. 2007;2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters B., Nielsen M., Sette A. T Cell Epitope Predictions. Annu. Rev. Immunol. 2020;38:123–145. doi: 10.1146/annurev-immunol-082119-124838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gfeller D., Bassani-Sternberg M. Predicting Antigen Presentation-What Could We Learn from a Million Peptides? Front. Immunol. 2018;9:1716. doi: 10.3389/fimmu.2018.01716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verma A., Halder A., Marathe S., Purwar R., Srivastava S. A Proteogenomic Approach to Target Neoantigens in Solid Tumors. Expert. Rev. Proteom. 2020;17:797–812. doi: 10.1080/14789450.2020.1881889. [DOI] [PubMed] [Google Scholar]
- 9.Pollock S.B., Rose C.M., Darwish M., Bouziat R., Delamarre L., Blanchette C., Lill J.R. Sensitive and Quantitative Detection of MHC-I Displayed Neoepitopes Using a Semiautomated Workflow and TOMAHAQ Mass Spectrometry. Mol. Cell. Proteom. 2021;20:100108. doi: 10.1016/j.mcpro.2021.100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreatta M., Nielsen M. Gapped Sequence Alignment Using Artificial Neural Networks: Application to the MHC Class i System. Bioinformatics. 2016;32:511–517. doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bassani-Sternberg M., Chong C., Guillaume P., Solleder M., Pak H.S., Gannon P.O., Kandalaft L.E., Coukos G., Gfeller D. Deciphering HLA-I Motifs across HLA Peptidomes Improves Neo-Antigen Predictions and Identifies Allostery Regulating HLA Specificity. PLoS Comput. Biol. 2017;13:e1005725. doi: 10.1371/journal.pcbi.1005725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gfeller D., Guillaume P., Michaux J., Pak H.-S., Daniel R.T., Racle J., Coukos G., Bassani-Sternberg M. The Length Distribution and Multiple Specificity of Naturally Presented HLA-I Ligands. J. Immunol. 2018;201:3705–3716. doi: 10.4049/jimmunol.1800914. [DOI] [PubMed] [Google Scholar]
- 13.Freudenmann L.K., Marcu A., Stevanović S. Mapping the Tumour Human Leukocyte Antigen (HLA) Ligandome by Mass Spectrometry. Immunology. 2018;154:331–345. doi: 10.1111/imm.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulik-Sullivan B., Busby J., Palmer C.D., Davis M.J., Murphy T., Clark A., Busby M., Duke F., Yang A., Young L., et al. Deep Learning Using Tumor HLA Peptide Mass Spectrometry Datasets Improves Neoantigen Identification. Nat. Biotechnol. 2019;37:55–71. doi: 10.1038/nbt.4313. [DOI] [PubMed] [Google Scholar]
- 15.O’Donnell T.J., Rubinsteyn A., Bonsack M., Riemer A.B., Laserson U., Hammerbacher J. MHCflurry: Open-Source Class I MHC Binding Affinity Prediction. Cell Syst. 2018;7:129–132.e4. doi: 10.1016/j.cels.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 16.Boehm K.M., Bhinder B., Raja V.J., Dephoure N., Elemento O. Predicting Peptide Presentation by Major Histocompatibility Complex Class I: An Improved Machine Learning Approach to the Immunopeptidome. BMC Bioinform. 2019;20:7. doi: 10.1186/s12859-018-2561-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Mattos-Arruda L., Vazquez M., Finotello F., Lepore R., Porta E., Hundal J., Amengual-Rigo P., Ng C.K.Y., Valencia A., Carrillo J., et al. Neoantigen Prediction and Computational Perspectives towards Clinical Benefit: Recommendations from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020;31:978–990. doi: 10.1016/j.annonc.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abelin J.G., Keskin D.B., Sarkizova S., Hartigan C.R., Zhang W., Sidney J., Stevens J., Lane W., Zhang G.L., Eisenhaure T.M., et al. Mass Spectrometry Profiling of HLA-Associated Peptidomes in Mono-Allelic Cells Enables More Accurate Epitope Prediction. Immunity. 2017;46:315–326. doi: 10.1016/j.immuni.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Racle J., Michaux J., Rockinger G.A., Arnaud M., Bobisse S., Chong C., Guillaume P., Coukos G., Harari A., Jandus C., et al. Robust Prediction of HLA Class II Epitopes by Deep Motif Deconvolution of Immunopeptidomes. Nat. Biotechnol. 2019;37:1283–1286. doi: 10.1038/s41587-019-0289-6. [DOI] [PubMed] [Google Scholar]
- 20.Chen B., Khodadoust M.S., Olsson N., Wagar L.E., Fast E., Liu C.L., Muftuoglu Y., Sworder B.J., Diehn M., Levy R., et al. Predicting HLA Class II Antigen Presentation through Integrated Deep Learning. Nat. Biotechnol. 2019;37:1332–1343. doi: 10.1038/s41587-019-0280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bjerregaard A.M., Nielsen M., Jurtz V., Barra C.M., Hadrup S.R., Szallasi Z., Eklund A.C. An Analysis of Natural T Cell Responses to Predicted Tumor Neoepitopes. Front. Immunol. 2017;8:1566. doi: 10.3389/fimmu.2017.01566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence M.S., Stojanov P., Polak P., Kryukov G.V., Cibulskis K., Sivachenko A., Carter S.L., Stewart C., Mermel C.H., Roberts S.A., et al. Mutational Heterogeneity in Cancer and the Search for New Cancer-Associated Genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richters M.M., Xia H., Campbell K.M., Gillanders W.E., Griffith O.L., Griffith M. Best Practices for Bioinformatic Characterization of Neoantigens for Clinical Utility. Genome Med. 2019;11:56–66. doi: 10.1186/s13073-019-0666-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou C., Zhu C., Liu Q. Toward in Silico Identification of Tumor Neoantigens in Immunotherapy. Trends Mol. Med. 2019;25:980–992. doi: 10.1016/j.molmed.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Finotello F., Rieder D., Hackl H., Trajanoski Z. Next-Generation Computational Tools for Interrogating Cancer Immunity. Nat. Rev. Genet. 2019;20:724–746. doi: 10.1038/s41576-019-0166-7. [DOI] [PubMed] [Google Scholar]
- 26.Ebrahimi-Nik H., Moussa M., Englander R.P., Singhaviranon S., Michaux J., Pak H.S., Miyadera H., Corwin W.L., Keller G.L.J., Hagymasi A.T., et al. Reversion Analysis Reveals the in Vivo Immunogenicity of a Poorly MHC I-Binding Cancer Neoepitope. Nat. Commun. 2021;12:6423. doi: 10.1038/s41467-021-26646-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chowell D., Krishna S., Becker P.D., Cocita C., Shu J., Tan X., Greenberg P.D., Klavinskis L.S., Blattman J.N., Anderson K.S. TCR Contact Residue Hydrophobicity Is a Hallmark of Immunogenic CD8+ T Cell Epitopes. Proc. Natl. Acad. Sci. USA. 2015;112:E1754–E1762. doi: 10.1073/pnas.1500973112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calis J.J.A., Maybeno M., Greenbaum J.A., Weiskopf D., de Silva A.D., Sette A., Keşmir C., Peters B. Properties of MHC Class I Presented Peptides That Enhance Immunogenicity. PLoS Comput. Biol. 2013;9:e1003266. doi: 10.1371/journal.pcbi.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balachandran V.P., Łuksza M., Zhao J.N., Makarov V., Moral J.A., Remark R., Herbst B., Askan G., Bhanot U., Senbabaoglu Y., et al. Identification of Unique Neoantigen Qualities in Long-Term Survivors of Pancreatic Cancer. Nature. 2017;551:S12–S16. doi: 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luksza M., Riaz N., Makarov V., Balachandran V.P., Hellmann M.D., Solovyov A., Rizvi N.A., Merghoub T., Levine A.J., Chan T.A., et al. A Neoantigen Fitness Model Predicts Tumour Response to Checkpoint Blockade Immunotherapy. Nature. 2017;551:517–520. doi: 10.1038/nature24473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snyder A., Makarov V., Merghoub T., Yuan J., Zaretsky J.M., Desrichard A., Walsh L.A., Postow M.A., Wong P., Ho T.S., et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N. Engl. J. Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yadav M., Jhunjhunwala S., Phung Q.T., Lupardus P., Tanguay J., Bumbaca S., Franci C., Cheung T.K., Fritsche J., Weinschenk T., et al. Predicting Immunogenic Tumour Mutations by Combining Mass Spectrometry and Exome Sequencing. Nature. 2014;515:572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 33.Duan F., Duitama J., al Seesi S., Ayres C.M., Corcelli S.A., Pawashe A.P., Blanchard T., McMahon D., Sidney J., Sette A., et al. Genomic and Bioinformatic Profiling of Mutational Neoepitopes Reveals New Rules to Predict Anticancer Immunogenicity. J. Exp. Med. 2014;211:2231–2248. doi: 10.1084/jem.20141308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt J., Smith A.R., Magnin M., Racle J., Devlin J.R., Bobisse S., Cesbron J., Bonnet V., Carmona S.J., Huber F., et al. Prediction of Neo-Epitope Immunogenicity Reveals TCR Recognition Determinants and Provides Insight into Immunoediting. Cell Rep. Med. 2021;2:100194. doi: 10.1016/j.xcrm.2021.100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Capietto A.H., Jhunjhunwala S., Pollock S.B., Lupardus P., Wong J., Hänsch L., Cevallos J., Chestnut Y., Fernandez A., Lounsbury N., et al. Mutation Position Is an Important Determinant for Predicting Cancer Neoantigens. J. Exp. Med. 2020;217:e20190179. doi: 10.1084/jem.20190179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gartner J.J., Parkhurst M.R., Gros A., Tran E., Jafferji M.S., Copeland A., Hanada K.I., Zacharakis N., Lalani A., Krishna S., et al. A Machine Learning Model for Ranking Candidate HLA Class I Neoantigens Based on Known Neoepitopes from Multiple Human Tumor Types. Nat. Cancer. 2021;2:563–574. doi: 10.1038/s43018-021-00197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turajlic S., Litchfield K., Xu H., Rosenthal R., McGranahan N., Reading J.L., Wong Y.N.S., Rowan A., Kanu N., al Bakir M., et al. Insertion-and-Deletion-Derived Tumour-Specific Neoantigens and the Immunogenic Phenotype: A Pan-Cancer Analysis. Lancet Oncol. 2017;18:1009–1021. doi: 10.1016/S1470-2045(17)30516-8. [DOI] [PubMed] [Google Scholar]
- 38.Litchfield K., Reading J.L., Lim E.L., Xu H., Liu P., Al-Bakir M., Wong Y.N.S., Rowan A., Funt S.A., Merghoub T., et al. Escape from Nonsense-Mediated Decay Associates with Anti-Tumor Immunogenicity. Nat. Commun. 2020;11:3800. doi: 10.1038/s41467-020-17526-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roudko V., Bozkus C.C., Orfanelli T., McClain C.B., Carr C., O’Donnell T., Chakraborty L., Samstein R., Huang K.-L., Blank S.V., et al. Shared Immunogenic Poly-Epitope Frameshift Mutations in Microsatellite Unstable Tumors. Cell. 2020;183:1634–1649.e17. doi: 10.1016/j.cell.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rowley J.D. A New Consistent Chromosomal Abnormality in Chronic Myelogenous Leukaemia Identified by Quinacrine Fluorescence and Giemsa Staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 41.Rabbihs T.H. Chromosomal Translocations in Human Cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 42.Druker B.J. Translation of the Philadelphia Chromosome into Therapy for CML. Nature. 2008;372:143–149. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- 43.Mitelman F., Johansson B., Mertens F. The Impact of Translocations and Gene Fusions on Cancer Causation. Nat. Rev. Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 44.Heyer E.E., Deveson I.W., Wooi D., Selinger C.I., Lyons R.J., Hayes V.M., O’Toole S.A., Ballinger M.L., Gill D., Thomas D.M., et al. Diagnosis of Fusion Genes Using Targeted RNA Sequencing. Nat. Commun. 2019;10:1388. doi: 10.1038/s41467-019-09374-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haas B.J., Dobin A., Li B., Stransky N., Pochet N., Regev A. Accuracy Assessment of Fusion Transcript Detection via Read-Mapping and de Novo Fusion Transcript Assembly-Based Methods. Genome Biol. 2019;20:213. doi: 10.1186/s13059-019-1842-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dehghannasiri R., Freeman D.E., Jordanski M., Hsieh G.L., Damljanovic A., Lehnert E., Salzman J. Improved Detection of Gene Fusions by Applying Statistical Methods Reveals Oncogenic RNA Cancer Drivers. Proc. Natl. Acad. Sci. USA. 2019;116:15524–15533. doi: 10.1073/pnas.1900391116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim P., Jang Y.E., Lee S. Fusionscan: Accurate Prediction of Fusion Genes from RNA-Seq Data. Genom. Inform. 2019;17:e26. doi: 10.5808/GI.2019.17.3.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uhrig S., Ellermann J., Walther T., Burkhardt P., Fröhlich M., Hutter B., Toprak U.H., Neumann O., Stenzinger A., Scholl C., et al. Accurate and Efficient Detection of Gene Fusions from RNA Sequencing Data. Genome Res. 2021;31:448–460. doi: 10.1101/gr.257246.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balan J., Jenkinson G., Nair A., Saha N., Koganti T., Voss J., Zysk C., Barr Fritcher E.G., Ross C.A., Giannini C., et al. SeekFusion—A Clinically Validated Fusion Transcript Detection Pipeline for PCR-Based Next-Generation Sequencing of RNA. Front. Genet. 2021;12:739054. doi: 10.3389/fgene.2021.739054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McPherson A., Hormozdiari F., Zayed A., Giuliany R., Ha G., Sun M.G.F., Griffith M., Moussavi A., Senz J., Melnyk N., et al. Defuse: An Algorithm for Gene Fusion Discovery in Tumor Rna-Seq Data. PLoS Comput. Biol. 2011;7:e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rathe S.K., Popescu F.E., Johnson J.E., Watson A.L., Marko T.A., Moriarity B.S., Ohlfest J.R., Largaespada D.A. Identification of Candidate Neoantigens Produced by Fusion Transcripts in Human Osteosarcomas. Sci. Rep. 2019;9:358. doi: 10.1038/s41598-018-36840-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cullis J., Barrett A., Goldman J., Lechler R. Binding of BCR/ABL Junctional Peptides to Major Histocompatibility Complex (MHC) Class I Molecules: Studies in Antigen-Processing Defective Cell Lines. Leukemia. 1994;8:165–170. [PubMed] [Google Scholar]
- 54.Worley B., van den Broeke L., Goletz T., Pendleton C., Daschbach E., Thomas E., Marincola F., Helman L., Berzofsky J. Antigenicity of Fusion Proteins from Sarcoma-Associated Chromosomal Translocations. Cancer Res. 2001;61:6868–6875. [PubMed] [Google Scholar]
- 55.van den Broeke L.T., Pendleton C.D., Mackall C., Helman L.J., Berzofsky J.A. Identification and Epitope Enhancement of a PAX-FKHR Fusion Protein Breakpoint Epitope in Alveolar Rhabdomyosarcoma Cells Created by a Tumorigenic Chromosomal Translocation Inducing CTL Capable of Lysing Human Tumors. Cancer Res. 2006;66:1818–1823. doi: 10.1158/0008-5472.CAN-05-2549. [DOI] [PubMed] [Google Scholar]
- 56.Comoli P., Basso S., Riva G., Barozzi P., Guido I., Gurrado A., Quartuccio G., Rubert L., Lagreca I., Vallerini D., et al. Brief Report BCR-ABL-Specific T-Cell Therapy in Ph 1 ALL Patients on Tyrosine-Kinase Inhibitors. Blood J. Am. Soc. Hematol. 2007;129:582–586. doi: 10.1182/blood-2016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dagher R., Long L., Read E., Leitman S., Carter C., Tsokos M., Goletz T., Avila N., Berzofsky J., Helman L., et al. Pilot Trial of Tumor-Specific Peptide Vaccination and Continuous Infusion Interleukin-2 in Patients with Recurrent Ewing Sarcoma and Alveolar Rhabdomyosarcoma: An Inter-Institute NIH Study. Med. Pediatr. Oncol. 2002;38:158–164. doi: 10.1002/mpo.1303. [DOI] [PubMed] [Google Scholar]
- 58.Mackall C.L., Rhee E.H., Read E.J., Khuu H.M., Leitman S.F., Bernstein D., Tesso M., Long L.M., Grindler D., Merino M., et al. A Pilot Study of Consolidative Immunotherapy in Patients with High-Risk Pediatric Sarcomas. Clin. Cancer Res. 2008;14:4850. doi: 10.1158/1078-0432.CCR-07-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y., Liu J., Huang B., Xu Y.-M., Li J., Huang L.-F., Lin J., Zhang J., Min Q.-H., Yang W.-M., et al. Mechanism of Alternative Splicing and Its Regulation. Biomed. Rep. 2015;3:152–158. doi: 10.3892/br.2014.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Desterro J., Bak-Gordon P., Carmo-Fonseca M. Targeting MRNA Processing as an Anticancer Strategy. Nat. Rev. Drug Discov. 2020;19:112–129. doi: 10.1038/s41573-019-0042-3. [DOI] [PubMed] [Google Scholar]
- 61.Bonnal S.C., López-Oreja I., Valcárcel J. Roles and Mechanisms of Alternative Splicing in Cancer—Implications for Care. Nat. Rev. Clin. Oncol. 2020;17:457–474. doi: 10.1038/s41571-020-0350-x. [DOI] [PubMed] [Google Scholar]
- 62.Mehmood A., Laiho A., Venälaïnen M.S., McGlinchey A.J., Wang N., Elo L.L. Systematic Evaluation of Differential Splicing Tools for RNA-Seq Studies. Brief. Bioinform. 2020;21:2052–2065. doi: 10.1093/bib/bbz126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang W., Chen L. Alternative Splicing: Human Disease and Quantitative Analysis from High-Throughput Sequencing. Comput. Struct. Biotechnol. J. 2021;19:183–195. doi: 10.1016/j.csbj.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cummings B.B., Marshall J.L., Tukiainen T., Lek M., Donkervoort S., Foley A.R., Bolduc V., Waddell L.B., Sandaradura S.A., O’grady G.L., et al. Improving Genetic Diagnosis in Mendelian Disease with Transcriptome Sequencing Genotype-Tissue Expression Consortium. Sci. Transl. Med. 2017;386:eaal5209. doi: 10.1126/scitranslmed.aal5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kremer L.S., Bader D.M., Mertes C., Kopajtich R., Pichler G., Iuso A., Haack T.B., Graf E., Schwarzmayr T., Terrile C., et al. Genetic Diagnosis of Mendelian Disorders via RNA Sequencing. Nat. Commun. 2017;8:15824. doi: 10.1038/ncomms15824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frésard L., Smail C., Ferraro N.M., Teran N.A., Li X., Smith K.S., Bonner D., Kernohan K.D., Marwaha S., Zappala Z., et al. Identification of Rare-Disease Genes Using Blood Transcriptome Sequencing and Large Control Cohorts. Nat. Med. 2019;25:911–919. doi: 10.1038/s41591-019-0457-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ferraro N.M., Strober B.J., Einson J., Abell N.S., Aguet F., Barbeira A.N., Bonazzola R., Brown A., Castel S.E., Jo B., et al. Transcriptomic Signatures across Human Tissues Identify Functional Rare Genetic Variation. Science. 2020;369:eaaz5900. doi: 10.1126/science.aaz5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jenkinson G., Li Y.I., Basu S., Cousin M.A., Oliver G.R., Klee E.W. LeafCutterMD: An Algorithm for Outlier Splicing Detection in Rare Diseases. Bioinformatics. 2020;36:4609–4615. doi: 10.1093/bioinformatics/btaa259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suo C., Hrydziuszko O., Lee D., Pramana S., Saputra D., Joshi H., Calza S., Pawitan Y. Integration of Somatic Mutation, Expression and Functional Data Reveals Potential Driver Genes Predictive of Breast Cancer Survival. Bioinformatics. 2015;31:2607–2613. doi: 10.1093/bioinformatics/btv164. [DOI] [PubMed] [Google Scholar]
- 70.Shen S., Wang Y., Wang C., Wu Y.N., Xing Y. SURVIV for Survival Analysis of MRNA Isoform Variation. Nat. Commun. 2016;7:11548. doi: 10.1038/ncomms11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Y., Sahni N., Pancsa R., McGrail D.J., Xu J., Hua X., Coulombe-Huntington J., Ryan M., Tychhon B., Sudhakar D., et al. Revealing the Determinants of Widespread Alternative Splicing Perturbation in Cancer. Cell Rep. 2017;21:798–812. doi: 10.1016/j.celrep.2017.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu J., Chen Z., Yong L. Systematic Profiling of Alternative Splicing Signature Reveals Prognostic Predictor for Ovarian Cancer. Gynecol. Oncol. 2018;148:368–374. doi: 10.1016/j.ygyno.2017.11.028. [DOI] [PubMed] [Google Scholar]
- 73.Bjørklund S.S., Panda A., Kumar S., Seiler M., Robinson D., Gheeya J., Yao M., Alnæs G.I.G., Toppmeyer D., Riis M., et al. Widespread Alternative Exon Usage in Clinically Distinct Subtypes of Invasive Ductal Carcinoma. Sci. Rep. 2017;7:798–812. doi: 10.1038/s41598-017-05537-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robertson A.G., Shih J., Yau C., Gibb E.A., Oba J., Mungall K.L., Hess J.M., Uzunangelov V., Walter V., Danilova L., et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell. 2017;32:204–220.e15. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marcelino Meliso F., Hubert C.G., Favoretto Galante P.A., Penalva L.O. RNA Processing as an Alternative Route to Attack Glioblastoma. Hum. Genet. 2017;136:1129–1141. doi: 10.1007/s00439-017-1819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kahles A., van Lehmann K., Toussaint N.C., Hüser M., Stark S.G., Sachsenberg T., Stegle O., Kohlbacher O., Sander C., Caesar-Johnson S.J., et al. Comprehensive Analysis of Alternative Splicing Across Tumors from 8705 Patients. Cancer Cell. 2018;34:211–224.e6. doi: 10.1016/j.ccell.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lupetti R., Pisarra P., Verrecchia A., Farina C., Nicolini G., Anichini A., Bordignon C., Sensi M., Parmiani G., Traversari C. Translation of a Retained Intron in Tyrosinase-Related Protein (TRP) 2 MRNA Generates a New Cytotoxic T Lymphocyte (CTL)-Defined and Shared Human Melanoma Antigen Not Expressed in Normal Cells of the Melanocytic Lineage. J. Exp. Med. 1998;188:1005–1016. doi: 10.1084/jem.188.6.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Slager E.H., van der Minne C.E., Krüse M., Krueger D.D., Griffioen M., Osanto S. Identification of Multiple HLA-DR-Restricted Epitopes of the Tumor-Associated Antigen CAMEL by CD4 + Th1/Th2 Lymphocytes. J. Immunol. 2004;172:5095–5102. doi: 10.4049/jimmunol.172.8.5095. [DOI] [PubMed] [Google Scholar]
- 79.Bigot J., Lalanne A.I., Lucibello F., Gueguen P., Houy A., Dayot S., Ganier O., Gilet J., Tosello J., Nemati F., et al. Splicing Patterns in Sf3b1mutated Uveal Melanoma Generate Shared Immunogenic Tumor-Specific Neoepitopes. Cancer Discov. 2021;11:1938–1951. doi: 10.1158/2159-8290.CD-20-0555. [DOI] [PubMed] [Google Scholar]
- 80.Qin T., Li J., Zhang K.Q. Structure, Regulation, and Function of Linear and Circular Long Non-Coding RNAs. Front. Genet. 2020;11:150. doi: 10.3389/fgene.2020.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ingolia N.T., Brar G.A., Stern-Ginossar N., Harris M.S., Talhouarne G.J.S., Jackson S.E., Wills M.R., Weissman J.S. Ribosome Profiling Reveals Pervasive Translation Outside of Annotated Protein-Coding Genes. Cell Rep. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilhelm M., Schlegl J., Hahne H., Gholami A.M., Lieberenz M., Savitski M.M., Ziegler E., Butzmann L., Gessulat S., Marx H., et al. Mass-Spectrometry-Based Draft of the Human Proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 83.Chen J., Brunner A.-D., Cogan Z., Nuñez J.K., Fields A.P., Adamson B., Itzhak D.N., Li J.Y., Mann M., Leonetti M.D., et al. Pervasive Functional Translation of Noncanonical Human Open Reading Frames. Science. 2020;367:1140–1146. doi: 10.1126/science.aay0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Poliseno L., Marranci A., Pandolfi P.P. Pseudogenes in Human Cancer. Front. Med. 2015;2:68. doi: 10.3389/fmed.2015.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lill J.R., Mathews W.R., Rose C.M., Schirle M. Proteomics in the Pharmaceutical and Biotechnology Industry: A Look to the next Decade. Expert. Rev. Proteom. 2021;18:503–526. doi: 10.1080/14789450.2021.1962300. [DOI] [PubMed] [Google Scholar]
- 86.Erhard F., Dölken L., Schilling B., Schlosser A. Identification of the Cryptic HLA-I Immunopeptidome. Cancer Immunol. Res. 2020;8:1018–1026. doi: 10.1158/2326-6066.CIR-19-0886. [DOI] [PubMed] [Google Scholar]
- 87.Chong C., Müller M., Pak H.S., Harnett D., Huber F., Grun D., Leleu M., Auger A., Arnaud M., Stevenson B.J., et al. Integrated Proteogenomic Deep Sequencing and Analytics Accurately Identify Non-Canonical Peptides in Tumor Immunopeptidomes. Nat. Commun. 2020;11:1293. doi: 10.1038/s41467-020-14968-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ruiz Cuevas M.V., Hardy M.P., Hollý J., Bonneil É., Durette C., Courcelles M., Lanoix J., Côté C., Staudt L.M., Lemieux S., et al. Most Non-Canonical Proteins Uniquely Populate the Proteome or Immunopeptidome. Cell Rep. 2021;34:108815. doi: 10.1016/j.celrep.2021.108815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ouspenskaia T., Law T., Clauser K.R., Klaeger S., Sarkizova S., Aguet F., Li B., Christian E., Knisbacher B.A., Le P.M., et al. Unannotated Proteins Expand the MHC-I-Restricted Immunopeptidome in Cancer. Nat. Biotechnol. 2022;40:209–217. doi: 10.1038/s41587-021-01021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiang R., Ma L., Yang M., Zheng Z., Chen X., Jia F., Xie F., Zhou Y., Li F., Wu K., et al. Increased Expression of Peptides from Non-Coding Genes in Cancer Proteomics Datasets Suggests Potential Tumor Neoantigens. Commun. Biol. 2021;4:3236384. doi: 10.1038/s42003-021-02007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Laumont C.M., Vincent K., Hesnard L., Audemard É., Bonneil É., Laverdure J.-P., Gendron P., Courcelles M., Hardy M.-P., Côté C., et al. Noncoding Regions Are the Main Source of Targetable Tumor-Specific Antigens. Sci. Transl. Med. 2018;10:eaau5516. doi: 10.1126/scitranslmed.aau5516. [DOI] [PubMed] [Google Scholar]
- 92.Tokita S., Kanaseki T., Torigoe T. Therapeutic Potential of Cancer Vaccine Based on MHC Class I Cryptic Peptides Derived from Non-Coding Regions. Immuno. 2021;1:424–431. doi: 10.3390/immuno1040030. [DOI] [Google Scholar]
- 93.Senft A.D., Macfarlan T.S. Transposable Elements Shape the Evolution of Mammalian Development. Nat. Rev. Genet. 2021;22:691–711. doi: 10.1038/s41576-021-00385-1. [DOI] [PubMed] [Google Scholar]
- 94.Lander S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial Sequencing and Analysis of the Human Genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 95.Vargiu L., Rodriguez-Tomé P., Sperber G.O., Cadeddu M., Grandi N., Blikstad V., Tramontano E., Blomberg J. Classification and Characterization of Human Endogenous Retroviruses Mosaic Forms Are Common. Retrovirology. 2016;13:7. doi: 10.1186/s12977-015-0232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pradhan R.K., Ramakrishna W. Transposons: Unexpected Players in Cancer. Gene. 2022;808:145975. doi: 10.1016/j.gene.2021.145975. [DOI] [PubMed] [Google Scholar]
- 97.Burns K.H. Our Conflict with Transposable Elements and Its Implications for Human Disease. Annu. Rev. Pathol. Mech. Dis. 2020;15:51–70. doi: 10.1146/annurev-pathmechdis-012419-032633. [DOI] [PubMed] [Google Scholar]
- 98.Vendrell-Mir P., Barteri F., Merenciano M., González J., Casacuberta J.M., Castanera R. A Benchmark of Transposon Insertion Detection Tools Using Real Data. Mob. DNA. 2019;10:53. doi: 10.1186/s13100-019-0197-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chu C., Borges-Monroy R., Viswanadham V.V., Lee S., Li H., Lee E.A., Park P.J. Comprehensive Identification of Transposable Element Insertions Using Multiple Sequencing Technologies. Nat. Commun. 2021;12:3836. doi: 10.1038/s41467-021-24041-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ou S., Su W., Liao Y., Chougule K., Agda J.R.A., Hellinga A.J., Lugo C.S.B., Elliott T.A., Ware D., Peterson T., et al. Benchmarking Transposable Element Annotation Methods for Creation of a Streamlined, Comprehensive Pipeline. Genome Biol. 2019;20:275. doi: 10.1186/s13059-019-1905-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kong Y., Rose C.M., Cass A.A., Williams A.G., Darwish M., Lianoglou S., Haverty P.M., Tong A.J., Blanchette C., Albert M.L., et al. Transposable Element Expression in Tumors Is Associated with Immune Infiltration and Increased Antigenicity. Nat. Commun. 2019;10:5228. doi: 10.1038/s41467-019-13035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Smith C.C., Beckermann K.E., Bortone D.S., Cubas A.A., Bixby L.M., Lee S.J., Panda A., Ganesan S., Bhanot G., Wallen E.M., et al. Endogenous Retroviral Signatures Predict Immunotherapy Response in Clear Cell Renal Cell Carcinoma. J. Clin. Investig. 2018;128:4804–4820. doi: 10.1172/JCI121476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rooney M.S., Shukla S.A., Wu C.J., Getz G., Hacohen N. Molecular and Genetic Properties of Tumors Associated with Local Immune Cytolytic Activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zeh H.J., III, Perry-Lalley D., Dudley M.E., Rosenberg S.A., Yang J.C. High Avidity CTLs for Two Self-Antigens Demonstrate Superior In Vitro and In Vivo Antitumor Efficacy. J. Immunol. 1999;162:989–994. [PubMed] [Google Scholar]
- 105.Kershaw M.H., Hsu C., Mondesire W., Parker L.L., Wang G., Overwijk W.W., Lapointe R., Yang J.C., Wang R.-F., Restifo N.P., et al. Immunization against Endogenous Retroviral Tumor-Associated Antigens 1. Cancer Res. 2001;61:7920–7924. [PMC free article] [PubMed] [Google Scholar]
- 106.Mullins C.S., Linnebacher M. Endogenous Retrovirus Sequences as a Novel Class of Tumor-Specific Antigens: An Example of HERV-H Env Encoding Strong CTL Epitopes. Cancer Immunol. Immunother. 2012;61:1093–1100. doi: 10.1007/s00262-011-1183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cherkasova E., Scrivani C., Doh S., Weisman Q., Takahashi Y., Harashima N., Yokoyama H., Srinivasan R., Linehan W.M., Lerman M.I., et al. Detection of an Immunogenic HERV-E Envelope with Selective Expression in Clear Cell Kidney Cancer. Cancer Res. 2016;76:2177–2185. doi: 10.1158/0008-5472.CAN-15-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schiavetti F., Thonnard J., Colau D., Boon T., Coulie P.G. A Human Endogenous Retroviral Sequence Encoding an Antigen Recognized on Melanoma by Cytolytic T Lymphocytes 1. Cancer Res. 2002;62:5510–5516. [PubMed] [Google Scholar]
- 109.Wang-Johanning F., Radvanyi L., Rycaj K., Plummer J.B., Yan P., Sastry K.J., Piyathilake C.J., Hunt K.K., Johanning G.L. Human Endogenous Retrovirus K Triggers an Antigen-Specific Immune Response in Breast Cancer Patients. Cancer Res. 2008;68:5869–5877. doi: 10.1158/0008-5472.CAN-07-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Takahashi Y., Harashima N., Kajigaya S., Yokoyama H., Cherkasova E., McCoy J.P., Hanada K.I., Mena O., Kurlander R., Abdul T., et al. Regression of Human Kidney Cancer Following Allogeneic Stem Cell Transplantation Is Associated with Recognition of an HERV-E Antigen by T Cells. J. Clin. Investig. 2008;118:1099–1109. doi: 10.1172/JCI34409C1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chiappinelli K.B., Strissel P.L., Desrichard A., Li H., Henke C., Akman B., Hein A., Rote N.S., Cope L.M., Snyder A., et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via DsRNA Including Endogenous Retroviruses. Cell. 2015;162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Roulois D., Loo Yau H., Singhania R., Wang Y., Danesh A., Shen S.Y., Han H., Liang G., Jones P.A., Pugh T.J., et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shraibman B., Kadosh D.M., Barnea E., Admon A. Human Leukocyte Antigen (HLA) Peptides Derived from Tumor Antigens Induced by Inhibition of DNA Methylation for Development of Drug-Facilitated Immunotherapy. Mol. Cell. Proteom. 2016;15:3058–3070. doi: 10.1074/mcp.M116.060350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang K., Leung S.Y. Abstract PR06: Genomic Characterization of Immune Escape Pathways in Gastric Cancer. Cancer Immunol. Res. 2015;3:PR06. doi: 10.1158/2326-6074.TUMIMM14-PR06. [DOI] [Google Scholar]
- 115.Griffin G.K., Wu J., Iracheta-Vellve A., Patti J.C., Hsu J., Davis T., Dele-Oni D., Du P.P., Halawi A.G., Ishizuka J.J., et al. Epigenetic Silencing by SETDB1 Suppresses Tumour Intrinsic Immunogenicity. Nature. 2021;595:309–314. doi: 10.1038/s41586-021-03520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang S.M., Cai W.L., Liu X., Thakral D., Luo J., Chan L.H., McGeary M.K., Song E., Blenman K.R.M., Micevic G., et al. KDM5B Promotes Immune Evasion by Recruiting SETDB1 to Silence Retroelements. Nature. 2021;598:682–687. doi: 10.1038/s41586-021-03994-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nepali K., Liou J.P. Recent Developments in Epigenetic Cancer Therapeutics: Clinical Advancement and Emerging Trends. J. Biomed. Sci. 2021;28:27. doi: 10.1186/s12929-021-00721-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Malaker S.A., Penny S.A., Steadman L.G., Myers P.T., Loke J.C., Raghavan M., Bai D.L., Shabanowitz J., Hunt D.F., Cobbold M. Identification of Glycopeptides as Posttranslationally Modified Neoantigens in Leukemia. Cancer Immunol. Res. 2017;5:376–384. doi: 10.1158/2326-6066.CIR-16-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zarling A.L., Polefrone J.M., Evans A.M., Mikesh L.M., Shabanowitz J., Lewis S.T., Engelhard V.H., Hunt D.F. Identification of Class I MHC-Associated Phosphopeptides as Targets for Cancer Immunotherapy. Proc. Natl. Acad. Sci. USA. 2006;103:14889–14894. doi: 10.1073/pnas.0604045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Depontieu F.R., Qian J., Zarling A.L., Mcmiller T.L., Salay T.M., Norris A., English A.M., Shabanowitz J., Engelhard V.H., Hunt D.F., et al. Identification of Tumor-Associated, MHC Class II-Restricted Phosphopeptides as Targets for Immunotherapy. Proc. Natl. Acad. Sci. USA. 2009;106:12073–12078. doi: 10.1073/pnas.0903852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Penny S.A., Abelin J.G., Malaker S.A., Myers P.T., Saeed A.Z., Steadman L.G., Bai D.L., Ward S.T., Shabanowitz J., Hunt D.F., et al. Tumor Infiltrating Lymphocytes Target HLA-I Phosphopeptides Derived From Cancer Signaling in Colorectal Cancer. Front. Immunol. 2021;12:723566. doi: 10.3389/fimmu.2021.723566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cobbold M., De H., Peña L., Norris A., Polefrone J.M., Qian J., English A.M., Cummings K.L., Penny S., Turner J.E., et al. MHC Class I-Associated Phosphopeptides Are the Targets of Memory-like Immunity in Leukemia. Sci. Transl. Med. 2013;5:203ra125. doi: 10.1126/scitranslmed.3006061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brentville V.A., Atabani S., Cook K., Durrant L.G. Novel Tumour Antigens and the Development of Optimal Vaccine Design. Ther. Adv. Vaccines Immunother. 2018;6:31–47. doi: 10.1177/2515135518768769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kudelka M.R., Ju T., Heimburg-Molinaro J., Cummings R.D. Advances in Cancer Research. Volume 126. Academic Press Inc.; Cambridge, MA, USA: 2015. Simple Sugars to Complex Disease-Mucin-Type O-Glycans in Cancer; pp. 53–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Peixoto A., Relvas-Santos M., Azevedo R., Lara Santos L., Ferreira J.A. Protein Glycosylation and Tumor Microenvironment Alterations Driving Cancer Hallmarks. Front. Oncol. 2019;9:380. doi: 10.3389/fonc.2019.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Reily C., Stewart T.J., Renfrow M.B., Novak J. Glycosylation in Health and Disease. Nat. Rev. Nephrol. 2019;15:346–366. doi: 10.1038/s41581-019-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mereiter S., Balmaña M., Campos D., Gomes J., Reis C.A. Glycosylation in the Era of Cancer-Targeted Therapy: Where Are We Heading? Cancer Cell. 2019;36:6–16. doi: 10.1016/j.ccell.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 128.Gao T., Cen Q., Lei H. A Review on Development of MUC1-Based Cancer Vaccine. Biomed. Pharmacother. 2020;132:110888. doi: 10.1016/j.biopha.2020.110888. [DOI] [PubMed] [Google Scholar]
- 129.Butts C., Socinski M.A., Mitchell P.L., Thatcher N., Havel L., Krzakowski M., Nawrocki S., Ciuleanu T.E., Bosquée L., Trigo J.M., et al. Tecemotide (L-BLP25) versus Placebo after Chemoradiotherapy for Stage III Non-Small-Cell Lung Cancer (START): A Randomised, Double-Blind, Phase 3 Trial. Lancet Oncol. 2014;15:59–68. doi: 10.1016/S1470-2045(13)70510-2. [DOI] [PubMed] [Google Scholar]
- 130.Quoix E., Lena H., Losonczy G., Forget F., Chouaid C., Papai Z., Gervais R., Ottensmeier C., Szczesna A., Kazarnowicz A., et al. TG4010 Immunotherapy and First-Line Chemotherapy for Advanced Non-Small-Cell Lung Cancer (TIME): Results from the Phase 2b Part of a Randomised, Double-Blind, Placebo-Controlled, Phase 2b/3 Trial. Lancet Oncol. 2016;17:212–223. doi: 10.1016/S1470-2045(15)00483-0. [DOI] [PubMed] [Google Scholar]
- 131.Mohammed F., Cobbold M., Zarling A.L., Salim M., Barrett-Wilt G.A., Shabanowitz J., Hunt D.F., Engelhard V.H., Willcox B.E. Phosphorylation-Dependent Interaction between Antigenic Peptides and MHC Class I: A Molecular Basis for the Presentation of Transformed Self. Nat. Immunol. 2008;9:1236–1243. doi: 10.1038/ni.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bassani-Sternberg M., Bräunlein E., Klar R., Engleitner T., Sinitcyn P., Audehm S., Straub M., Weber J., Slotta-Huspenina J., Specht K., et al. Direct Identification of Clinically Relevant Neoepitopes Presented on Native Human Melanoma Tissue by Mass Spectrometry. Nat. Commun. 2016;7:13404. doi: 10.1038/ncomms13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Engelhard V.H., Obeng R.C., Cummings K.L., Petroni G.R., Ambakhutwala A.L., Chianese-Bullock K.A., Smith K.T., Lulu A., Varhegyi N., Smolkin M.E., et al. MHC-Restricted Phosphopeptide Antigens: Preclinical Validation and First-in-Humans Clinical Trial in Participants with High-Risk Melanoma. J. Immunother. Cancer. 2020;8:e000262. doi: 10.1136/jitc-2019-000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Feitsma A.L., van der Voort E.I.H., Franken K.L.M.C., el Bannoudi H.E., Elferink B.G., Drijfhout J.W., Huizinga T.W.J., de Vries R.R.P., Toes R.E.M., Ioan-Facsinay A. Identification of Citrullinated Vimentin Peptides as T Cell Epitopes in HLA-DR4-Positive Patients with Rheumatoid Arthritis. Arthritis Rheum. 2010;62:117–125. doi: 10.1002/art.25059. [DOI] [PubMed] [Google Scholar]
- 135.Gerstner C., Dubnovitsky A., Sandin C., Kozhukh G., Uchtenhagen H., James E.A., Rönnelid J., Ytterberg A.J., Pieper J., Reed E., et al. Functional and Structural Characterization of a Novel HLA-DRB1*04: 01-Restricted α-Enolase T Cell Epitope in Rheumatoid Arthritis. Front. Immunol. 2016;7:494. doi: 10.3389/fimmu.2016.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Brentville V.A., Metheringham R.L., Gunn B., Symonds P., Daniels I., Gijon M., Cook K., Xue W., Durrant L.G. Citrullinated Vimentin Presented on MHC-II in Tumor Cells Is a Target for CD4+ T-Cell-Mediated Antitumor Immunity. Cancer Res. 2016;76:548–560. doi: 10.1158/0008-5472.CAN-15-1085. [DOI] [PubMed] [Google Scholar]
- 137.Brentville V.A., Metheringham R.L., Daniels I., Atabani S., Symonds P., Cook K.W., Vankemmelbeke M., Choudhury R., Vaghela P., Gijon M., et al. Combination Vaccine Based on Citrullinated Vimentin and Enolase Peptides Induces Potent CD4-Mediated Anti-Tumor Responses. J. Immunother. Cancer. 2020;8:e000560. doi: 10.1136/jitc-2020-000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hanada K.-I., Yewdell J.W., Yang J.C. Immune Recognition of a Human Renal Cancer Antigen through Post-Translational Protein Splicing. Nature. 2004;427:252–256. doi: 10.1038/nature02240. [DOI] [PubMed] [Google Scholar]
- 139.Vigneron N., Stroobant V., Chapiro J., Ooms A., Degiovanni G., Morel S., van der Bruggen P., Boon T., van den Eynde B.J. An Antigenic Peptide Produced by Peptide Splicing in the Proteasome. Science. 2004;304:587–590. doi: 10.1126/science.1095522. [DOI] [PubMed] [Google Scholar]
- 140.Warren E.H., Vigneron N.J., Gavin M.A., Coulie P.G., Stroobant V., Dalet A., Tykodi S.S., Xuereb S.M., Mito J.K., Riddell S.R., et al. An Antigen Produced by Splicing of Noncontiguous Peptides in the Reverse Order. Science. 2006;313:1444–1447. doi: 10.1126/science.1130660. [DOI] [PubMed] [Google Scholar]
- 141.Liepe J., Marino F., Sidney J., Jeko A., Bunting D.E., Sette A., Kloetzel P.M., Stumpf M.P.H., Heck A.J.R., Mishto M. A Large Fraction of HLA Class Iligands Are Proteasome-Generatedspliced Peptides. Science. 2016;354:354–358. doi: 10.1126/science.aaf4384. [DOI] [PubMed] [Google Scholar]
- 142.Mylonas R., Beer I., Iseli C., Chong C., Pak H.S., Gfeller D., Coukos G., Xenarios I., Müller M., Bassani-Sternberg M. Estimating the Contribution of Proteasomal Spliced Peptides to the HLA-I Ligandome. Mol. Cell. Proteom. 2018;17:2347–2357. doi: 10.1074/mcp.RA118.000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liepe J., Mishto M., Textoris-Taube K., Janek K., Keller C., Henklein P., Kloetzel P.M., Zaikin A. The 20S Proteasome Splicing Activity Discovered by SpliceMet. PLoS Comput. Biol. 2010;6:e1000830. doi: 10.1371/journal.pcbi.1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Specht G., Roetschke H.P., Mansurkhodzhaev A., Henklein P., Textoris-Taube K., Urlaub H., Mishto M., Liepe J. Large Database for the Analysis and Prediction of Spliced and Non-Spliced Peptide Generation by Proteasomes. Sci. Data. 2020;7:146. doi: 10.1038/s41597-020-0487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rolfs Z., Solntsev S.K., Shortreed M.R., Frey B.L., Smith L.M. Global Identification of Post-Translationally Spliced Peptides with Neo-Fusion. J. Proteom. Res. 2019;18:349–358. doi: 10.1021/acs.jproteome.8b00651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dalet A., Robbins P.F., Stroobant V., Vigneron N., Li Y.F., El-Gamil M., Hanada K.I., Yang J.C., Rosenberg S.A., van den Eyndea B.J. An Antigenic Peptide Produced by Reverse Splicing and Double Asparagine Deamidation. Proc. Natl. Acad. Sci. USA. 2011;108:E323–E331. doi: 10.1073/pnas.1101892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Michaux A., Larrieu P., Stroobant V., Fonteneau J.-F., Jotereau F., van den Eynde B.J., Moreau-Aubry A., Vigneron N. A Spliced Antigenic Peptide Comprising a Single Spliced Amino Acid Is Produced in the Proteasome by Reverse Splicing of a Longer Peptide Fragment Followed by Trimming. J. Immunol. 2014;192:1962–1971. doi: 10.4049/jimmunol.1302032. [DOI] [PubMed] [Google Scholar]
- 148.Carreno B.M., Magrini V., Becker-Hapak M., Kaabinejadian S., Hundal J., Petti A.A., Ly A., Lie W.R., Hildebrand W.H., Mardis E.R., et al. A Dendritic Cell Vaccine Increases the Breadth and Diversity of Melanoma Neoantigen-Specific T Cells. Science. 2015;348:803–808. doi: 10.1126/science.aaa3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sahin U., Derhovanessian E., Miller M., Kloke B.P., Simon P., Löwer M., Bukur V., Tadmor A.D., Luxemburger U., Schrörs B., et al. Personalized RNA Mutanome Vaccines Mobilize Poly-Specific Therapeutic Immunity against Cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 150.Ott P.A., Hu Z., Keskin D.B., Shukla S.A., Sun J., Bozym D.J., Zhang W., Luoma A., Giobbie-Hurder A., Peter L., et al. An Immunogenic Personal Neoantigen Vaccine for Patients with Melanoma. Nature. 2017;547:217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ott P.A., Hu-Lieskovan S., Chmielowski B., Govindan R., Naing A., Bhardwaj N., Margolin K., Awad M.M., Hellmann M.D., Lin J.J., et al. A Phase Ib Trial of Personalized Neoantigen Therapy Plus Anti-PD-1 in Patients with Advanced Melanoma, Non-Small Cell Lung Cancer, or Bladder Cancer. Cell. 2020;183:347–362.e24. doi: 10.1016/j.cell.2020.08.053. [DOI] [PubMed] [Google Scholar]
- 152.Lopez J.S., Camidge R., Iafolla M., Rottey S., Schuler M., Hellmann M., Balmanoukian A., Dirix L., Gordon M., Sullivan R., et al. Abstract CT301: A Phase Ib Study to Evaluate RO7198457, an Individualized Neoantigen Specific ImmunoTherapy (INeST), in Combination with Atezolizumab in Patients with Locally Advanced or Metastatic Solid Tumors. Cancer Res. 2020;80:CT301. doi: 10.1158/1538-7445.AM2020-CT301. [DOI] [Google Scholar]
- 153.McGranahan N., Furness A.J.S., Rosenthal R., Ramskov S., Lyngaa R., Saini S.K., Jamal-Hanjani M., Wilson G.A., Birkbak N.J., Hiley C.T., et al. Clonal Neoantigens Elicit T Cell Immunoreactivity and Sensitivity to Immune Checkpoint Blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Koboldt D.C., Fulton R.S., McLellan M.D., Schmidt H., Kalicki-Veizer J., McMichael J.F., Fulton L.L., Dooling D.J., Ding L., Mardis E.R., et al. Comprehensive Molecular Portraits of Human Breast Tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., Hackl H., Trajanoski Z. Pan-Cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 156.Marty R., Kaabinejadian S., Rossell D., Slifker M.J., van de Haar J., Engin H.B., de Prisco N., Ideker T., Hildebrand W.H., Font-Burgada J., et al. MHC-I Genotype Restricts the Oncogenic Mutational Landscape. Cell. 2017;171:1272–1283.e15. doi: 10.1016/j.cell.2017.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Claeys A., Luijts T., Marchal K., van den Eynden J. Low Immunogenicity of Common Cancer Hot Spot Mutations Resulting in False Immunogenic Selection Signals. PLoS Genet. 2021;17:e1009368. doi: 10.1371/journal.pgen.1009368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Anagnostou V., Smith K.N., Forde P.M., Niknafs N., Bhattacharya R., White J., Zhang T., Adleff V., Phallen J., Wali N., et al. Evolution of Neoantigen Landscape during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer. Cancer Discov. 2017;7:264–276. doi: 10.1158/2159-8290.CD-16-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Linette G.P., Becker-Hapak M., Skidmore Z.L., Baroja M.L., Xu C., Hundal J., Spencer D.H., Fu W., Cummins C., Robnett M., et al. Immunological Ignorance Is an Enabling Feature of the Oligo-Clonal T Cell Response to Melanoma Neoantigens. Proc. Natl. Acad. Sci. USA. 2019;116:23662–23670. doi: 10.1073/pnas.1906026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Vormehr M., Reinhard K., Blatnik R., Josef K., Beck J.D., Salomon N., Suchan M., Selmi A., Vascotto F., Zerweck J., et al. A Non-Functional Neoepitope Specific CD8 + T-Cell Response Induced by Tumor Derived Antigen Exposure in Vivo. Oncoimmunology. 2019;8:1553478. doi: 10.1080/2162402X.2018.1553478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rosenthal R., Cadieux E.L., Salgado R., Al-Bakir M., Moore D.A., Hiley C.T., Lund T., Tanić M., Reading J.L., Joshi K., et al. Neoantigen-Directed Immune Escape in Lung Cancer Evolution. Nature. 2019;567:479–485. doi: 10.1038/s41586-019-1032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lo A.A., Wallace A., Oreper D., Lounsbury N., Havnar C., Pechuan-Jorge X., Wu T.D., Bourgon R., Jones R., Krogh K., et al. Indication-Specific Tumor Evolution and Its Impact on Neoantigen Targeting and Biomarkers for Individualized Cancer Immunotherapies. J. Immunother. Cancer. 2021;9:e003001. doi: 10.1136/jitc-2021-003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sarivalasis A., Boudousquié C., Balint K., Stevenson B.J., Gannon P.O., Iancu E.M., Rossier L., Martin Lluesma S., Mathevet P., Sempoux C., et al. A Phase I/II Trial Comparing Autologous Dendritic Cell Vaccine Pulsed Either with Personalized Peptides (PEP-DC) or with Tumor Lysate (OC-DC) in Patients with Advanced High-Grade Ovarian Serous Carcinoma. J. Transl. Med. 2019;17:391. doi: 10.1186/s12967-019-02133-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bassani-Sternberg M., Digklia A., Huber F., Wagner D., Sempoux C., Stevenson B.J., Thierry A.C., Michaux J., Pak H., Racle J., et al. A Phase Ib Study of the Combination of Personalized Autologous Dendritic Cell Vaccine, Aspirin, and Standard of Care Adjuvant Chemotherapy Followed by Nivolumab for Resected Pancreatic Adenocarcinoma—A Proof of Antigen Discovery Feasibility in Three Patients. Front. Immunol. 2019;10:1832. doi: 10.3389/fimmu.2019.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chen F., Zou Z., Du J., Su S., Shao J., Meng F., Yang J., Xu Q., Ding N., Yang Y., et al. Neoantigen Identification Strategies Enable Personalized Immunotherapy in Refractory Solid Tumors. J. Clin. Investig. 2019;129:2056–2070. doi: 10.1172/JCI99538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kreiter S., Vormehr M., van de Roemer N., Diken M., Löwer M., Diekmann J., Boegel S., Schrörs B., Vascotto F., Castle J.C., et al. Mutant MHC Class II Epitopes Drive Therapeutic Immune Responses to Cancer. Nature. 2015;520:692–696. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Alspach E., Lussier D.M., Miceli A.P., Kizhvatov I., DuPage M., Luoma A.M., Meng W., Lichti C.F., Esaulova E., Vomund A.N., et al. MHC-II Neoantigens Shape Tumour Immunity and Response to Immunotherapy. Nature. 2019;574:696–701. doi: 10.1038/s41586-019-1671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Swartz A.M., Congdon K.L., Nair S.K., Li Q.J., Herndon J.E., Suryadevara C.M., Riccione K.A., Archer G.E., Norberg P.K., Sanchez-Perez L.A., et al. A Conjoined Universal Helper Epitope Can Unveil Antitumor Effects of a Neoantigen Vaccine Targeting an MHC Class I-Restricted Neoepitope. NPJ Vaccines. 2021;6:12. doi: 10.1038/s41541-020-00273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Vauchy C., Gamonet C., Ferrand C., Daguindau E., Galaine J., Beziaud L., Chauchet A., Henry Dunand C.J., Deschamps M., Rohrlich P.S., et al. CD20 Alternative Splicing Isoform Generates Immunogenic CD4 Helper T Epitopes. Int. J. Cancer. 2015;137:116–126. doi: 10.1002/ijc.29366. [DOI] [PubMed] [Google Scholar]
- 170.Keskin D.B., Anandappa A.J., Sun J., Tirosh I., Mathewson N.D., Li S., Oliveira G., Giobbie-Hurder A., Felt K., Gjini E., et al. Neoantigen Vaccine Generates Intratumoral T Cell Responses in Phase Ib Glioblastoma Trial. Nature. 2019;565:234–239. doi: 10.1038/s41586-018-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hilf N., Kuttruff-Coqui S., Frenzel K., Bukur V., Stevanović S., Gouttefangeas C., Platten M., Tabatabai G., Dutoit V., van der Burg S.H., et al. Actively Personalized Vaccination Trial for Newly Diagnosed Glioblastoma. Nature. 2019;565:240–245. doi: 10.1038/s41586-018-0810-y. [DOI] [PubMed] [Google Scholar]
- 172.Cathcart K., Pinilla-Ibarz J., Korontsvit T., Schwartz J., Zakhaleva V., Papadopoulos E.B., Scheinberg D.A. A Multivalent Bcr-Abl Fusion Peptide Vaccination Trial in Patients with Chronic Myeloid Leukemia. Blood. 2004;103:1037–1042. doi: 10.1182/blood-2003-03-0954. [DOI] [PubMed] [Google Scholar]