Abstract
A spontaneous Xanthomonas campestris pv. phaseoli H2O2-resistant mutant emerged upon selection with 1 mM H2O2. In this report, we show that growth of this mutant under noninducing conditions gave high levels of catalase, alkyl hydroperoxide reductase (AhpC and AhpF), and OxyR. The H2O2 resistance phenotype was abolished in oxyR-minus derivatives of the mutant, suggesting that elevated levels and mutations in oxyR were responsible for the phenotype. Nucleotide sequence analysis of the oxyR mutant showed three nucleotide changes. These changes resulted in one silent mutation and two amino acid changes, one at a highly conserved location (G197 to D197) and the other at a nonconserved location (L301 to R301) in OxyR. Furthermore, these mutations in oxyR affected expression of genes in the oxyR regulon. Expression of an oxyR-regulated gene, ahpC, was used to monitor the redox state of OxyR. In the parental strain, a high level of wild-type OxyR repressed ahpC expression. By contrast, expression of oxyR5 from the X. campestris pv. phaseoli H2O2-resistant mutant and its derivative oxyR5G197D with a single-amino-acid change on expression vectors activated ahpC expression in the absence of inducer. The other single-amino-acid mutant derivative of oxyR5L301R had effects on ahpC expression similar to those of the wild-type oxyR. However, when the two single mutations were combined, as in oxyR5, these mutations had an additive effect on activation of ahpC expression.
Xanthomonas belongs to an important group of bacterial phytopathogens. In response to microbial infection, plants increase production and accumulation of reactive oxygen species (ROS), including H2O2, organic peroxide, and superoxide anions, as a component of active plant defense responses (2, 14). Moreover ROS are generated by normal aerobic metabolism (9). Exposure to high levels of ROS leads to inhibition of cell proliferation. Thus, the ability to increase ROS removal could be advantageous to bacteria (7).
OxyR is a peroxide sensor and transcription activator that regulates both catalase and alkyl hydroperoxide reductase (4, 5, 20). OxyR can be converted from the reduced to the oxidized form after exposure to oxidants by formation of a disulfide bond between the highly conserved cysteine residues C199 and C208 (1, 21). This oxidized OxyR then activates transcription of genes in the OxyR regulon (6, 7, 20). In Xanthomonas, oxyR not only regulates oxidant induction of both catalase and ahpC but also mediates the oxidant's inducible H2O2 resistance phenotype (17, 18). Xanthomonas ahpC and oxyR have atypical gene arrangements and transcription organizations. ahpC is transcribed as a monocistronic mRNA, while ahpF-oxyR and orfX are in an operon (15, 17).
We have isolated and partially characterized a spontaneous Xanthomonas campestris pv. phaseoli peroxide-resistant mutant, designated XpHR (8). The mutant is highly resistant to killing by peroxide and has over a 50-fold increase in the peroxide-scavenging enzymes catalase and alkyl hydroperoxide reductase subunit C (AhpC) (8). In this paper, we characterize the role of OxyR in the mutant XpHR. The results show not only that the level of OxyR is elevated but also that there are several mutations in the protein. These factors contribute to constitutive activation of genes in the oxyR regulon and to the H2O2 resistance phenotype.
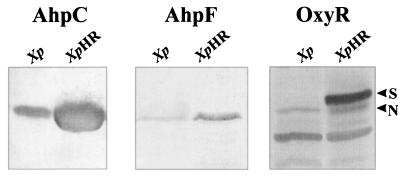
Increased levels of AhpC, AhpF, and OxyR in XpHR.
The levels of AhpC, AhpF, and OxyR in uninduced XpHR and its parental strain were compared by Western analysis (Fig. 1). AhpC, AhpF, and OxyR levels in XpHR were over 20-fold higher than in the parental strain. In Xanthomonas, exposure to oxidants leads to a severalfold increase in OxyR levels (17). The OxyR level in XpHR was threefold higher than the OxyR level in an oxidant-induced culture of the parental strain (data not shown). In addition, two forms of OxyR were detected in the mutant. One form (designated N for normal) comigrated with OxyR from the parental strain, while the other form (designated S for slow) had slower migration. In X. campestris pv. phaseoli, concentrations of catalase, AhpC, AhpF, and OxyR are increased only in response to oxidant treatments. Elevated levels of these proteins in uninduced cultures of XpHR were highly unusual and suggested deregulation of the peroxide stress response.
FIG. 1.
Western analysis of AhpC, AhpF, and OxyR in XpHR and the parental strain. X. campestris pv. phaseoli (Xp) and XpHR were grown aerobically to mid-log phase in Silva Buddenhagen (SB) medium at 28°C. Cell lysate preparation, gel electrophoresis, blotting to nitrocellulose membranes, and antibody reactions were performed as previously described (17). Antibody reactions were subsequently detected with an anti-rabbit antibody conjugated to alkaline phosphate. Total protein (50 μg) was loaded into each lane. Western blots were treated with an anti-AhpC (AhpC), an anti-AhpF (AhpF), and an anti-OxyR (OxyR) antibody, respectively.
Construction of an XpHR oxyR mutant.
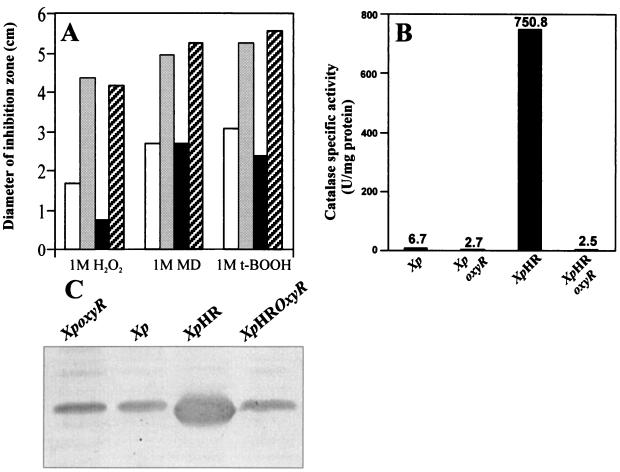
To determine whether the high level of OxyR in the uninduced growth of the mutant was responsible for the H2O2 resistance phenotype, a marker-exchanged oxyR mutant of XpHR was constructed as previously described (18). XpHR oxyR had resistance levels to H2O2, organic peroxide, and menadione killing similar to those of X. campestris pv. phaseoli oxyR (Fig. 2A). We extended these observations by determining the levels of the peroxide-scavenging enzymes catalase and AhpC in these bacteria (Fig. 2B and C). The increases in catalase activities and the amount of AhpC in XpHR were abolished in the XpHR oxyR mutant (Fig. 2B and C).
FIG. 2.
Resistance to oxidant killing and levels of peroxide-scavenging enzymes in XpHR and its parental strain. (A) Qualitative determination of levels of resistance to killing concentrations of H2O2, menadione (MD), and tert-butyl hydroperoxide (t-BOOH) in X. campestris pv. phaseoli (Xp; □), X. campestris pv. phaseoli oxyR (Xp oxyR; ▒), XpHR (■), and XpHR oxyR ( ). Essentially, log-phase cells were mixed with Silva Buddenhagen (SB) top agar and poured onto SB plates. Six microliters of the indicated concentrations of oxidants were spotted on paper disks and placed on top of cell lawns. The zone of growth inhibition was measured after 30 h of incubation (16). Experiments were repeated at least three times, and representative data are shown. (B) Catalase levels of various Xanthomonas strains. (C) Western analysis of AhpC levels in various Xanthomonas strains. Forty micrograms of total protein was loaded into each lane. Western analysis and catalase assays were performed as previously described (17).
Detection of mutations in XpHR oxyR5.
PCR of oxyR from the XpHR mutant (oxyR5) was performed, using primers located at the 5′ end (5′ACGCGCCAGTCGTTCCCCG 3′) and at the 3′ end (5′ ACCACAGCCAAAGCGATCGCA 3′) of the oxyR coding region, with Pfu polymerase for 25 cycles. The 960-bp PCR products were cloned into pGEM-T easy (Promega), and their nucleotide sequences were determined with ABI Prism kits on an ABI 310 automated DNA sequencer. oxyR from XpHR, designated oxyR5, showed three nucleotide changes from the parental gene. The first change, at nucleotide position T213C of the oxyR sequence, resulted in a silent mutation. The second and third single-base changes, at positions G590A and T902G, resulted in two amino acid residue changes at the highly conserved position G197 (to D197) and the nonconserved L301 (to R301). No other mutations were detected. To ascertain the effects of these mutations on gene expression, two additional oxyR5 variants, each with a single-amino-acid difference from the parental gene, were constructed. oxyR5G197D, with a single-amino-acid change, was constructed by partial digestion of poxyR5 (oxyR5 in pBluescript KS) with XhoI and XbaI. A 150-bp fragment from the internal portion of oxyR was removed and replaced by a 150-bp XhoI-XbaI fragment from poxyR (18). This replaced the mutation at L301R in oxyR5 with a wild-type sequence. oxyR5R301L, with a single-amino-acid change, was constructed by partial digestion of poxyR5 with EcoRI and XhoI. The 380-bp fragment containing mutated G197D was replaced with a 380-bp EcoRI-XhoI fragment from a wild-type oxyR. All constructs were sequenced to confirm the mutations.
Mutations in oxyR affect gene expression.
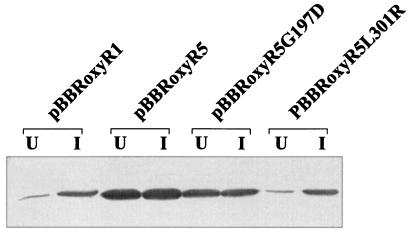
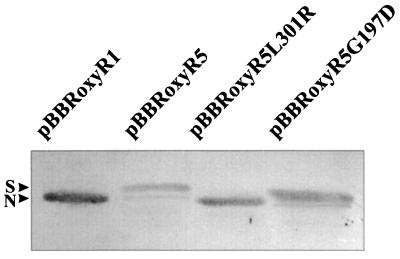
The effects of different oxyR mutations on the expression of an oxyR-regulated gene, ahpC, were determined. In Xanthomonas, ahpC has a unique pattern of regulation. Its expression can be increased 50-fold in response to oxidants in an oxyR-dependent fashion (17, 18). Moreover, expression of the gene is affected by both oxidized and reduced forms of OxyR (18; S. Mougkolsuk, unpublished data). High levels of reduced OxyR lead to repression of ahpC (Mougkolsuk, unpublished), while oxidized OxyR activates expression of ahpC (18). Thus, expression analysis of the gene would also give an indication of the redox status of the cells and OxyR. In X. campestris pv. phaseoli under noninducing growth conditions, ahpC is expressed at low levels. By contrast, ahpC is expressed at high levels in XpHR without any inducing signals (Fig. 1). We tested whether mutations in oxyR were responsible for the altered ahpC expression. An X. campestris pv. phaseoli oxyR mutant was transformed with expression plasmids containing pBBRoxyR5, pBBRoxyR1, pBBRoxyR5G197D, and pBBRoxyR5L301R, and the AhpC levels were monitored (Fig. 3). The oxyR mutant harboring pBBRoxyR5 showed a greater-than-50-fold increase in AhpC levels in the uninduced state. On the other hand, cells harboring pBBRoxyR1 showed fivefold repression of AhpC levels. The OxyR mutant harboring pBBRoxyR5L301R repressed AhpC levels in a fashion similar to that of cells harboring pBBRoxyR1, while the mutant harboring pBBRoxyR5G197D produced AhpC at levels 20 times higher than those of a control strain in the absence of inducing signals. Nonetheless, AhpC levels in strains harboring pBBRoxyR5G197D were still about twofold less than the level attained in cells harboring pBBRoxyR5. Next, we examined the effects of an oxidant on mutant OxyR proteins. The levels of AhpC were monitored in X. campestris pv. phaseoli oxyR cells harboring various oxyR-containing plasmids grown under noninducing and inducing conditions (100 μM menadione) (Fig. 3). AhpC levels in cells harboring pBBRoxyR1 or pBBRoxyR5L301R showed strong induction after menadione treatment. By contrast, cells harboring pBBRoxyR5 or pBBRoxyR5G197D expressed ahpC at constitutive high levels, and menadione treatment did not result in further increases in the amount of AhpC (Fig. 3).
FIG. 3.
Effects of various mutations in oxyR on levels of AhpC during uninduced and menadione-induced growth. Western analysis of AhpC levels in X. campestris pv. phaseoli oxyR harboring various oxyR genes on an expression vector from a parental strain (pBBRoxyR1), the XpHR mutant (pBBRoxyR5), and the gene with one amino acid changed (pBBRoxyR5G197D and pBBR oxyR5L301R). These cells were grown uninduced (U) in Silva Buddenhagen (SB) medium or induced with 100 μM menadione (I) for 30 min. Total protein (20 μg) was loaded into each lane. Lysate preparation, gel electrophoresis, and antibody reactions were performed as described previously (17) and in the legend to Fig. 1.
These results raised the question of the mechanisms responsible for this deregulation. Expression of oxyR5 from XpHR in X. campestris pv. phaseoli oxyR led to activation of ahpC expression in uninduced cultures, indicating that mutations in oxyR5 were responsible for unregulated gene expression. oxyR5 had amino acid changes at two positions, G197D and L301R. G197 is a highly conserved position found in all OxyR proteins (15, 21). The observation that Xanthomonas harboring pBBRoxyR5G197D activated ahpC expression in the absence of inducing signals confirmed the importance of this mutation in producing altered gene expression. The position of this mutation is in close proximity to the redox-active cysteine C199 (21) and may be responsible for the conversion of OxyR from a reduced to an oxidized form in uninduced cells. In Escherichia coli, mutations located close to redox-active C199 (i.e., H198Y, R201C, and C208Y) (13), produce constitutively active proteins similar to G197D in Xanthomonas. Oxidation of OxyR occurs at C199 via a sulphenic intermediate and subsequent formation of a disulfide bond with C208 (1, 21). Also, the highly conserved basic residues (H198 and R291) could enhance the activity of C199 (13). Thus, an amino acid change from a neutral G to an acidic D could alter OxyR structure so that either the C199 is more easily accessible to cellular oxidants or the charged residue promotes and stabilizes the formation of sulphenic intermediates. Alternatively, the presence of a carboxylate group at D197 close to the SH group of C199 could result in proton transfer from the SH group to the carboxylate group, resulting in thiolate formation. Thiolate groups are more reactive than SH groups and can subsequently react with carboxyl groups to form relatively stable thiolester bonds. The second mutation, at L301R, introduced a basic residue that had no effect on the transcription activation activity of OxyR. The mutated protein can also be activated by exposure to oxidants (Fig. 3). However, when L301R was combined with the mutation at G197D, as in oxyR5, the double mutation enhanced the ability of OxyR to activate transcription of ahpC to levels greater than the levels attained by oxyR5G197D. The carboxy terminus regions of OxyR and a subclass of LysR transcription activators have been shown to be crucial to protein binding to DNA (12, 19) and in tetramerization or oligomerization of OxyR (12). Mutation at L301R did not seem to affect the ability of mutated oxyR to repress ahpC expression. Thus, mutation at L301R might possibly affect tetramerization and might enhance the DNA binding of OxyR. Together with G197D, it may enhance binding of OxyR5 and recruiting of RNA polymerase to the promoter. We are attempting to purify the mutated proteins and examine their abilities to bind to the promoter.
G197D mutation was responsible for altered OxyR mobility.
We next compared the proteins from several OxyR variants to determine if the mutations in oxyR were responsible for the altered protein mobility. The X. campestris pv. phaseoli oxyR-minus mutant was transformed with a broad-host-range expression vector (pBBR1MCS-4 [11]) containing various constructs of oxyR. OxyR Western analysis of lysates prepared from these cells were performed, and the results (Fig. 4) showed that wild-type oxyR produced a single OxyR form (N form) that reacted against an anti-OxyR antibody. By contrast, oxyR5 from XpHR (pBBRoxyR5) produced both S and N forms. This finding was similar to that shown in Fig. 1. Results for oxyR variants with single-amino-acid changes (Fig. 4) showed that cells harboring the plasmid containing pBBRoxyR5(G197D) produced S and N forms of OxyR with the S form accounting for greater than 90% of the total OxyR, while cells harboring plasmids containing pBBRoxyR5L301R produced OxyR with mobility similar to that of plasmids containing wild-type oxyR. We believe that the S form arises from oxidation of mutant OxyR proteins in the polyacrylamide gel.
FIG. 4.
Effects of mutations in oxyR on protein mobility. Cell growth, lysate preparation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Western analysis of OxyR were performed as described previously (17) and in Fig. 1. Proteins from lysates (20 μg) were loaded into each lane. N and S indicate two forms of OxyR.
All members of the LysR family, including oxyR, are autoregulated (19). In Xanthomonas, unlike other bacteria, OxyR increased severalfold in concentration as well as changing form in response to oxidants (17). Preliminary data suggest that oxyR expression is activated by the oxidized form of the protein (Mongkolsuk, unpublished). This autoregulation could account for the high levels of mutant OxyR detected in XpHR. We are investigating the autoactivation of Xanthomonas oxyR. Mutation and deregulation of oxyR lead to uncontrolled gene activation in XpHR that is responsible for the H2O2-resistant phenotype. In an analogous situation, a Bacillus subtilis H2O2-resistant mutant (10) has been shown to arise from deregulation of a peroxide repressor, perR (3).
Acknowledgments
We thank Tim Flegel for reviewing the manuscript and G. Storz and S. Ruchirawat for helpful comments, strains, and an anti-AhpC antibody.
The research was supported by grants from Chulabhorn Research Institute to the Laboratory of Biotechnology, Thailand Research Fund BRG10-40, and NSTDA career development award RCF 01-40-005 to S.M.
REFERENCES
- 1.Aslund F, Zheng M, Beckwith J, Storz G. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc Natl Acad Sci USA. 1999;96:6161–6165. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker C J, Orlandi E W. Active oxygen in plant pathogenesis. Annu Rev Phytopathol. 1995;33:299–321. doi: 10.1146/annurev.py.33.090195.001503. [DOI] [PubMed] [Google Scholar]
- 3.Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann J D. Bacillus subtiliscontains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- 4.Christman M F, Storz G, Ames B N. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc Natl Acad Sci USA. 1989;86:3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demple B. Regulation of bacterial oxidative stress genes. Annu Rev Genet. 1991;25:315–337. doi: 10.1146/annurev.ge.25.120191.001531. [DOI] [PubMed] [Google Scholar]
- 6.Dhandayuthapani S, Mudd M, Deretic V. Interactions of OxyR with the promoter region of the oxyR and ahpC genes from Mycobacterium leprae and Mycobacterium tuberculosis. J Bacteriol. 1997;179:2401–2409. doi: 10.1128/jb.179.7.2401-2409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farr S B, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuangthong M, Mongkolsuk S. Isolation and characterization of a multiple peroxide resistant mutant from Xanthomonas campestris pv. phaseoli. FEMS Microbiol Lett. 1997;152:189–194. doi: 10.1016/s0378-1097(97)00208-5. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez-Flecha B, Demple B. Homeostatic regulation of intracellular hydrogen peroxide concentration in aerobically growing Escherichia coli. J Bacteriol. 1997;179:382–388. doi: 10.1128/jb.179.2.382-388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartford O M, Dowds B C. Isolation and characterization of a hydrogen peroxide resistant mutant of Bacillus subtilis. Microbiology. 1994;140:297–304. doi: 10.1099/13500872-140-2-297. [DOI] [PubMed] [Google Scholar]
- 11.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop R M I, Peterson K M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic resistances. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 12.Kullik I, Stevens J, Toledano M B, Storz G. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for DNA binding and multimerization. J Bacteriol. 1995;177:1285–1291. doi: 10.1128/jb.177.5.1285-1291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kullik I, Toledano M B, Tartaglia L A, Storz G. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J Bacteriol. 1995;177:1275–1284. doi: 10.1128/jb.177.5.1275-1284.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine A, Tenhaken R, Dixon R, Lamb C. H2O2from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 15.Loprasert S, Atichartpongkun S, Whangsuk W, Mongkolsuk S. Isolation and analysis of the Xanthomonas alkyl hydroperoxide reductase gene and the peroxide sensor regulator genes ahpC and ahpF-oxyR-orfX. J Bacteriol. 1997;179:3944–3949. doi: 10.1128/jb.179.12.3944-3949.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mongkolsuk S, Loprasert S, Vattanaviboon P, Chanvanichayachai C, Chamnongpol S, Supsamran N. Heterologous growth phase- and temperature-dependent expression and H2O2 toxicity protection of a superoxide-inducible monofunctional catalase gene from Xanthomonas oryzaepv. oryzae. J Bacteriol. 1996;178:3578–3584. doi: 10.1128/jb.178.12.3578-3584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mongkolsuk S, Loprasert S, Whangsuk W, Fuangthong M, Atichartpongkun S. Characterization of transcription organization and analysis of unique expression patterns of an alkyl hydroperoxide reductase C gene (ahpC) and the peroxide regulator operon ahpF-oxyR-orfX from Xanthomonas campestrispv. phaseoli. J Bacteriol. 1997;179:3950–3955. doi: 10.1128/jb.179.12.3950-3955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mongkolsuk S, Sukchawalit R, Loprasert S, Praituan W, Upaichit A. Construction and physiological analysis of a Xanthomonas mutant to examine the role of the oxyRgene in oxidant-induced protection against peroxide killing. J Bacteriol. 1998;180:3988–3991. doi: 10.1128/jb.180.15.3988-3991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schell M A, Brown P H, Raju S. Use of saturation mutagenesis to localize probable functional domains in the NahR protein, a LysR-type transcription activator. J Biol Chem. 1990;265:3844–3850. [PubMed] [Google Scholar]
- 20.Toledano M B, Kullik I, Trinh F, Baird P T, Schneider T D, Storz G. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell. 1994;78:897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 21.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]