Abstract
The Schuurs–Hoeijmakers syndrome (SHMS) or PACS1 Neurodevelopment Disorder (PACS1-NDD) is a rare autosomal dominant disease caused by mutations in the PACS1 gene. To date, only 87 patients have been reported and, surprisingly, most of them carry the same variant (c.607C>T; p.R203W). The most relevant clinical features of the syndrome include neurodevelopment delay, seizures or a recognizable facial phenotype. Moreover, some of these characteristics overlap with other syndromes, such as the PACS2 or Wdr37 syndromes. The encoded protein phosphofurin acid cluster sorting 1 (PACS-1) is able to bind to different client proteins and direct them to their subcellular final locations. Therefore, although its main function is protein trafficking, it could perform other roles related to its client proteins. In patients with PACS1-NDD, a gain-of-function or a dominant negative mechanism for the mutated protein has been suggested. This, together with the fact that most of the patients carry the same genetic variant, makes it a good candidate for novel therapeutic approaches directed to decreasing the toxic effect of the mutated protein. Some of these strategies include the use of antisense oligonucleotides (ASOs) or targeting of its client proteins.
Keywords: Schuurs–Hoeijmakers syndrome, SHMS, PACS1-NDD, trafficking protein, targeted therapy
1. Introduction
The Schuurs–Hoeijmakers syndrome (SHMS), or PACS1 Neurodevelopmental Disorder (PACS1-NDD) (MIM# 615009), is a rare autosomal dominant disease [1], which has recently been included in a group of genetic disorders of cellular trafficking [2]. It was firstly described in 2012 in two patients with intellectual disability (ID) and a striking facial resemblance; they both carried the same mutation in the PACS1 gene [1]. To date, less than a hundred patients with PACS1-NDD have been genetically diagnosed, and, surprisingly, only three pathogenic variants have been reported [1,3,4,5,6]. Furthermore, most of the patients have been described in the last few years, when the PACS1 gene was included in the gene panel analysis for neurodevelopmental disorders; therefore, it is possible that the prevalence of this syndrome could be underestimated.
Although the clinical characteristics of PACS1-NDD patients are well described, there is little evidence on the pathomolecular mechanisms of this disease. The affected protein, phosphofurin acid cluster sorting 1 (PACS-1), belongs to a family of two members of membrane and protein trafficking regulators [7]. It was initially discovered in 1998 [8], and it has been reported in metazoans, invertebrates [9] and vertebrates [10]. Lower species possess only a single gene of the PACS family; however, the PACS gene was duplicated with the appearance of vertebrates, resulting in the PACS1 and PACS2 genes [10].
PACS-1 is a multifunctional membrane traffic regulator that plays an important role in cellular homeostasis [8,10]. An initial function of PACS-1 was the transport of several proteins between endosomes and the trans-Golgi network (TGN) [7]. However, in the last few years, novel roles have been proposed, such as a Ca2+ flux regulator or its probable implication in genomic stability [10,11,12]. The knowledge of the molecular mechanisms implied in the development of the PACS1-NDD is essential to make proper phenotype–genotype correlations and to propose therapies that could help and improve the quality of life of these patients. In this review, we will carry out a detailed study of the functions already known of the PACS-1 protein, of its role in the progression of the disorder and the latest improvements in the development of successful therapeutic strategies.
2. Clinical Characteristics of PACS1-NDD
The PACS1-NDD was first described in two unrelated male patients in 2012 [1]. Since then, about 100 patients have been reported in the literature [1,3,4,5,6]. All patients are described have a neurodevelopmental delay with an intellectual disability and psychomotor retardation. The range varies from mild to severe, although most of them have a moderate delay [13]. Language skills are universally affected, more severely than motor skills. Hypotonia is reported in approximately one third of the subjects, and it improves over time [13]. However, one patient has been described as having an impairment in their walking abilities over time [3]. Individuals with the PACS1 mutation interact with others and appreciate receiving personal affection, but also show behavioral difficulties. Seizures are a common clinical feature, affecting 60 percent of patients; most of them are generalized as tonic-clonic, and are well controlled with antiepileptic drugs. Brain abnormalities have been found, mostly cerebellar. Other findings include ventriculomegaly, hydrocephalus or atrophy of the corpus callosum [3,5,14,15,16,17,18].
In addition to the neurological disorders, patients show a characteristic facial phenotype, which is easily recognizable by a clinician. It is characterized by full and arched eyebrows, hypertelorims with downslanting palpebral fissures, long eyelashes, a bulbous nose, a flat philtrum and large low-set ears [13].
Patients also suffer from other congenital anomalies, from cardiac anomalies to ocular alterations, where septal defects and coloboma stand out. They may also have skeletal abnormalities (abnormal skull shape), cryptorchidism or feeding problems, among others [3,19]. These clinic characteristics are summarized in Table 1.
Table 1.
Clinical characteristics of the PACS1-NDD, PACS2, Wdr37, Kabuki and Cornelia de Lange Syndromes.
| Clinical Feature | HPO ID * | PACS1-NDD | PACS2 Syndrome | Wdr37 Syndrome | Kabuki Syndrome | CdLS |
|---|---|---|---|---|---|---|
| Neurodevelopmental features | ||||||
| Intellectual disability | 0001249 | Obligate | Very frequent | Obligate | Obligate | Very frequent |
| Autism spectrum disorder | 000729 | Occasional | Occasional | Occasional | Occasional | Frequent |
| Development delay | 0012758 | Obligate | Very frequent | Obligate | Obligate | Occasional |
| Speech delay | 0000750 | Very frequent | Very frequent | Frequent | Occasional | Frequent |
| Hypotonia | 0001252 | Frequent | Frequent | Frequent | Frequent | Occasional |
| Seizures | 0001250 | Frequent | Very frequent | Very frequent | Occasional | Occasional |
| Congenital malformations | ||||||
| Dysmorphic facial features | ||||||
| Full and arched eyebrows | 0002553 | Frequent | Frequent | Frequent | Very frequent | Very frequent |
| Hypertelorism | 0000316 | Frequent | Frequent | Frequent | Occasional | Very rare |
| Downslanting palpebral fissures | 0000494 | Frequent | Frequent | Frequent | Very frequent | Very rare |
| Bulbous nasal tip | 0000414 | Frequent | Very frequent | Obligate | Frequent | Very frequent |
| Downturned mouth | 0002714 | Frequent | Frequent | Frequent | Very rare | Very frequent |
| Thin upper lip | 0000219 | Frequent | Very frequent | Very frequent | Occasional | Very frequent |
| Brain abnormalities | ||||||
| Hypoplasia or partial agenesis of the cerebellar dermis | 0006817 | Frequent | Frequent | Obligate | Occasional | Occasional |
| Ophthalmologic | ||||||
| Coloboma | 0000589 | Occasional | Occasional | Very frequent | Occasional | Very rare |
| Congenital heart anomalies | ||||||
| Atrial or ventricular septal defects | 0001671 | Frequent | Occasional | Frequent | Frequent | Frequent |
| Others | ||||||
| Feeding/GI issues | 0011968 | Occasional | Occasional | Very frequent | Frequent | Frequent |
| Skeletal anomalies | 0000924 | Occasional | Frequent | Frequent | Frequent/ very frequent | Frequent |
| Cryptorchidism | 0000028 | Frequent | Frequent | Very frequent | Occasional | frequent |
* HPO ID, Human Phenotype Ontology Identifier. Obligate 100%; Very frequent 80–99%; Frequent 30–79%; Occasional 5–29%; Very rare 1–4%; Excluded 0%. Light grey: same disease spectrum.
The strong similarity between PACS1-NDD, PACS2 syndrome and the recently diagnosed Wdr37 syndrome is striking. The three disorders share a very similar facial gestalt, intellectual disability, neurodevelopmental delay and seizures, suggesting that they might be included in the same disease spectrum [20,21,22,23,24,25]. Moreover, a new group of diseases, caused by cellular trafficking defects and characterized by neurodevelopmental disorders and skeletal abnormalities, has been recently proposed [2,20,21,22,23,24,25,26]. On the other hand, there are other closely related clinical syndromes, including Cornelia de Lange, Kabuki or Coffin-Siris, although they do not share the facial gestalt (Table 1).
3. Molecular Basis of the Disease
3.1. Genetic Update
To date, 87 patients with a PACS1 deficiency have been genetically diagnosed, and, surprisingly, most of them carry the same pathogenic variant, the missense c.607C>T (p.R203W). This variant has been clearly demonstrated to be pathogenic because none of the parents tested were carriers [3], which means that all patients had a de novo mutation. There is one other pathogenic variant, c.608G>A, reported in only one patient, which results in a change in the same position, but the amino acid change is different, p.R203Q [5]. These patients have most of the typical features of the PACS1-NDD (Table 1). Lately, two novel missense variants have been reported, one in the ClinVar database, the c.1574G>A (p.R525K), whose relationship with the PACS1-NDD has to be studied [27]. The other is in a broad study about the autism spectrum disorder (p.R245W) without a specific phenotype of the disease [28].
More recently, a multi-exon deletion of PACS1 has been reported [6]. Liu and Cols found in four members of a three-generation pedigree the deletion of the exons 12 to 24 in the PACS1 gene. However, the phenotype of this variant was milder, with slight speech and cognitive delay, only affecting two members of the last studied generation [6]. Furthermore, several databases have reported chromosomal reorganizations where the PACS1 gene is involved (ClinVar, DECIPHER) [27,29]. Nevertheless, the huge number of affected genes does not allow us to directly relate the patients’ phenotype with the PACS1 gene.
3.2. PACS1 Gene Regulation
PACS1 is a gene that is broadly expressed in human tissues (GTEx database) [30]. According to the BrainSpan and EvoDevo databases, its mRNA expression is upregulated during fetal brain and cerebellum development, and it decreases after birth to slightly increase in puberty [31,32]. Its expression level is also important in pubertal testis tissues [31,32]. This specific tissue distribution could be the reason for some of the clinical characteristics of PACS1-NDD patients.
The PACS1 gene is located on 11q13.1 and contains 24 exons and at least 16 transcripts (ensembl) [33]. Its expression regulation has been poorly studied, but some proteins have been proposed as regulators of PACS-1 expression, such as the P300/CBP-associated factor (PCAF) or the transcriptional adaptor protein 3 (ADA3). Chromatin inmuno precipitation (ChIP) experiments showed an enrichment in the promoter of PACS1 of the proteins PCAF and ADA3. The downregulation of both factors decreased the relative PACS1 expression level in HeLa and HCT116 cell lines. These facts point to a plausible role of PCAF and ADA3 as gene regulators of PACS1 [34].
In addition to that, the study of its 3′-UTR sequence showed two putative binding sites for the miRNAs, 34a and 449a. In tumor tissues, an overexpression of these miRNAs and a downexpression of the PACS-1 protein has been reported [35]. Furthermore, the overexpression of another miRNA, miR-485-5p, induces a decrease of PACS-1 in pericytes that has been related to Alzheimer’s disease progression [36].
3.3. Characteristics of the PACS-1 Protein
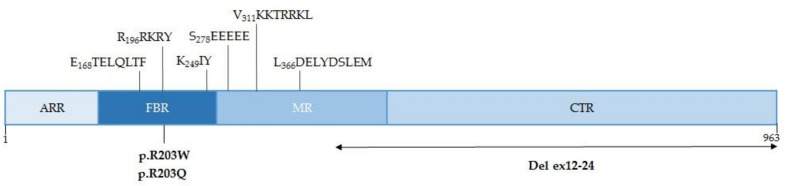
PACS-1 is a protein of 963 amino acids with several domains and key regions located in the cytosol and nucleus [12,37]. Its N-terminal region, called ARR (atrophin-1-related region), is followed by the FBR (furin-binding), the MR (middle) and the CTR (C-terminal) domains (Figure 1). The FBR binds client proteins, such as furin, as well as the cytoplasmic membrane trafficking machinery. Several specific sequences have been described, which are responsible for the binding of PACS-1 to the clathrin adaptors AP-1 and AP-3 (E168TELQLTF) or to the monomeric adaptor GGA3 (K249IY). Moreover, in the FBR is the binding sequence of the protein kinase CK2 (R196RKRY) that phosphorylates the S278 residue located in the MR autoregulatory domain (S278EEEEE). Furthermore, the MR also has a nuclear localization sequence (NLS) (V311KKTRRKL) and a nuclear export sequence (NES) (L366DELYDSLEM) [37]. The entry and exit from the nucleus of PACS-1 depends on the receptors importin alpha 5 (IPO5/KPNA1) and exportin 1 (XPO1) [37].
Figure 1.
Schematic representation of the PACS-1 protein. Key sequences and pathological variants are located in the sequence. ARR, atrophin-1-related region; FBR, furin-binding region; MR, middle region; CTR, C-terminal region.
The three-dimensional (3D) structure of PACS-1 is still unknown. Some attempts have been made using ab-initio modeling, such as the one described for residues located between V117 and D300, which are predicted as a globular domain [7]. Different unpublished analyses performed using structure prediction systems using hidden Markov models [38,39] roughly coincide in predicting a globular structure for the C-terminal subdomain (residues from R622 to L956), similar to a regulatory subunit of phosphoinositide 3-kinase. They also predict a structure similar to a C2 domain (calcium/lipid-binding domain) for the segment between amino acids P98 and E261, which would include the amino acid R203 (variant p.R203W). Unfortunately, not all of these data are conclusive. Future knowledge of the structure of PACS-1 would be key to knowing more accurately how the mutations in the protein affect its structure and to developing therapies that are more precise for these patients.
3.4. Functions of PACS-1
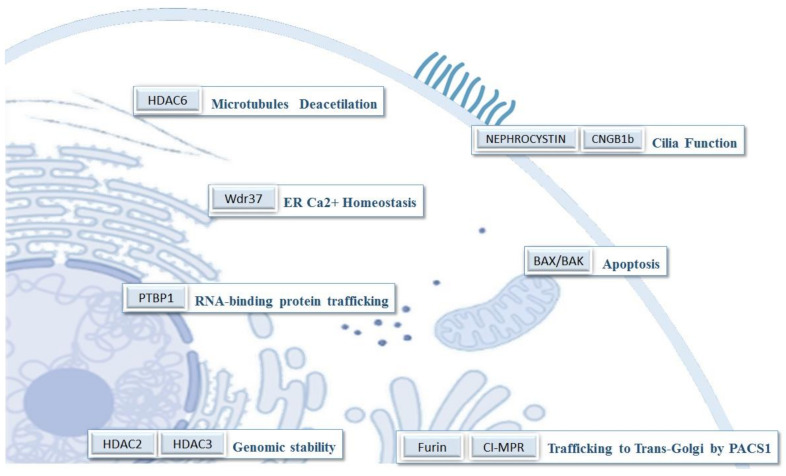
Nowadays, more than 100 proteins have been described that can putatively bind to PACS-1 [11]. This could explain, in part, the number of different processes where PACS-1 might be involved. Therefore, although the main function of PACS-1 is related to protein trafficking, sometimes it is difficult to discern between the PACS-1 function and the role of some of its client proteins (Figure 2) (Supplementary Table S1).
Figure 2.
Some of PACS-1’s client proteins and their subcellular location and function.
The first described function of the PACS-1 protein was its role in the regulation of membrane traffic proteins. This specific function is well known and conserved among vertebrates in PACS proteins. The activation mechanism involves the phosphorylation of its S278 residue by CK2. The activated PACS-1 protein is able to bind its client proteins, transferring them to their final location. It mediates the trafficking of proteins from the plasma membrane to the early endosomes (SorLA) [40] and from the late endosomes to the trans-Golgi network (TGN) (furin, CI-MPR) [8,41]. Moreover, some proteins related to the function of the primary cilium, such as nephrocystin or CNGB1b, are also trafficking [42,43]. Besides, PACS-1 has been linked to the cytosolic HDAC6, which is involved in the deacetilation of microtubules, in the Golgi integrity and cilium retraction [44,45].
Another function for the PACS family of proteins is connected to the trafficking of proteins related to the apoptosis pathway [10]. PACS2 allows the transferring of Bid to the mitochondria, where it is cleaved to tBid, facilitating the triggering of apoptosis [46]. On the other hand, the downexpression of PACS-1 increases cell survival. The mechanism implies a failure in the BAX/BAK oligomerization, avoiding the mitochondrial outer membrane permeabilization (MOMP) [34]. The increase in cell survival when PACS-1 is downregulated has been linked with a worst prognosis in gastric cancers, and it has been proposed as a biomarker [47].
Moreover, it has been reported that the complex PACS1–Wdr37 facilitates the regulation of calcium flux between the endoplasmic reticulum and the cytosol [11]. The endoplasmic reticulum’s Ca2+ release, mediated by the inositol 1,4,5-trisphosphate receptor (IP3R), is regulated by this complex. The deletion of PACS-1 provoked a decrease in the Wdr37 protein, and the deficiency of both caused a reduction in the IP3R expression level. This could explain the overlapping phenotypic characteristics found between the PACS1 and Wdr37 deficiency patients (Table 1).
In the nucleus, PACS-1 binds to PTBP1, a protein involved in the binding and trafficking of RNA [36]. Moreover, it interacts with and could stabilize HDAC2 and HDAC3, contributing to genomic stability [12,35].
4. Relationship between PACS-1 Function and the PACS1-NDD Patient’s Phenotype
Nowadays, there are a few studies regarding the relationship between PACS-1 function and the PACS1-NDD phenotype. It is proposed that the most frequent PACS-1 variant, p.R203W, causes a gain-of-function (GOF) or a dominant negative mechanism [1,48]. However, the truncation of the protein (del ex12-24) with a loss-of-function has been described in two patients with a mild and not characteristic PACS1-NDD clinic [6].
The mutated amino acid (R203W) is in the furin cargo-binding domain and in close proximity to the CK2 binding motif (Figure 1), so it is possible that the correct binding and phosphorylation function of CK2 could be compromised [14]. Several experimental approaches have been carried out with a mutant PACS-1 to explore the differences in the binding of PACS-1 to its client proteins. In this sense, some protein–protein interactions are not affected (AP3D1, CLCN7, HDAC2/3) [1,12], whilst others decrease their binding level (TRPV4v2) [1] and others increase it (HDAC6) [45]. On the other hand, experiments in zebrafish embryos where the mutant mRNA of PACS1 was injected showed a decrease in cranial cartilaginous structures compared to the control. This could be due to the fact that the migration of cranial-neural-crest cells (CNCCs) is related to PACS1 [1].
An experimental model of the PACS1-NDD of forebrain organoids has been developed (PACS1(+/R203W)), besides the knock-out of the PACS1 gene (PACS1(-/-)). Gene expression pattern experiments showed differences between the PACS1(+/R203W) and the control model, but not between the PACS1(-/-) and the control, supporting the idea that this specific mutation confers a GOF and a toxic effect on the protein. Genes directly related to the autism spectrum disorder (ASD) and to the development of GABAergic synapses are upregulated in the disease model [48]. These results support the previous findings that about 40% of the PACS1-NDD patients have been formally diagnosed with autism [49].
These results, in zebrafish and forebrain organoids, could explain, in part, the neurologic impairment development of PACS1-NDD patients. Moreover, other proteins related to intracellular trafficking have also been associated with craniofacial diseases [26] and neurodevelopmental disorders [2].
5. Therapy for PACS1 Deficiency Patients
To date, the PACS1-NDD syndrome has been symptomatically treated by a multidisciplinary team. The neurodevelopmental alterations recommend an early interventional program, which includes occupational, physical and speech therapy. Anxiety and behavioral problems have been managed with psychotropic drugs. Seizures respond well to the classical epilepticus treatment. It is suggested that early physical therapy treatment for motor dysfunction can improve mobility and decrease the risk of later orthopedic complications. Feeding problems might need nutritional intervention therapy and, in severe cases, a gastrostomy tube [13].
In recent years, more targeted therapy approaches are being developed for some rare diseases. In this sense, the PACS1-NDD is a good candidate [50] because most of the patients carry the same mutation, and a gain-of-function or a dominant negative mechanism has been proposed [1,3,48]. However, the highest expression level of PACS-1 during fetal brain development could limit the effectiveness of the treatment [31,32]. Nowadays, there are four approaches which can be performed; two of them are in more advanced steps due to the use of antisense oligonucleotides (ASOs) (Figure 3) or inhibitors against HDAC6 [45,51]. The other two are subject to advances in the knowledge of the 3D PACS-1 structure [7], as the proteolysis-targeting chimeras (PROTACs) or molecules, which specifically target the mutated protein.
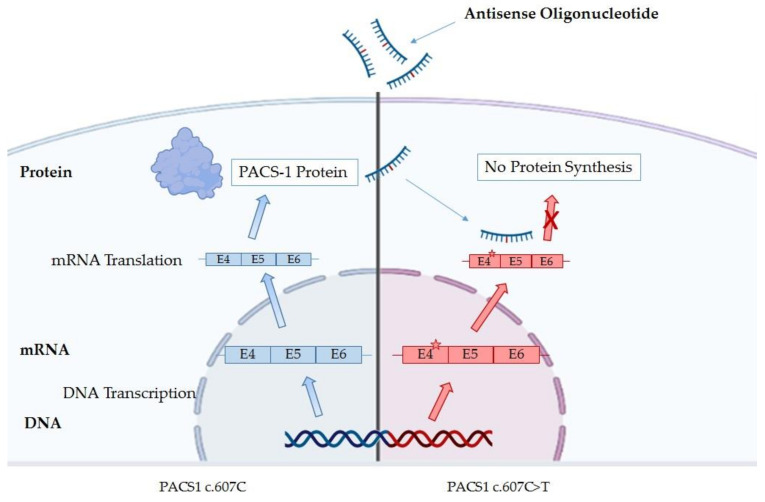
Figure 3.
Schematic mechanism of antisense oligonucleotides (ASOs) therapy in order to avoid the translation of the mutant PACS-1 protein.
The antisense oligonucleotides (ASOs) can specifically target the mutated mRNA of PACS1, avoiding the translation of the pathologic protein [52] (Figure 3). The company IONIS is collaborating with the PACS1 foundation in order to develop an ASO targeting the p.R203W variant [51], and, nowadays, it is an active research field. However, ASOs are incapable of crossing the blood brain barrier (BBB) and require direct central nervous system (CNS) administration, so intrathecal delivery is recommended [53].
Other approaches propose the targeting of some of the client proteins of PACS-1, whose function is altered in these patients. In this sense, the research carried out by Dr. Thomas′s group is interesting [45]. They suggest the specific targeting of HDAC6 through its inhibition. They have delivered a patent application in which, in fibroblasts derived from patients, the stronger binding between the mutated PACS-1 and HDAC6 could be associated with a fragmented Golgi and a different microtubule network compared to the controls. They confirm that the use of general (TSA) or selective (tubacin, ACY-1215 or SW-100) HDAC6 inhibitors rescues the normal cellular phenotype [45].
Finally, two more approaches can be developed that focus on the specific targeting of the mutated PACS-1 protein. The proteolysis-targeting chimeras (PROTACs) bind simultaneously to the target protein and to an E3 ligase, forming a ternary complex, which promotes the ubiquitination of the protein of interest, thereby, inducing proteasomal degradation [54]. On the other hand, the use of molecules that specifically bind to the mutant PACS-1 protein is another way to produce protein degradation. However, the proper development of both technologies needs knowledge of the 3D structure and of the differences between the PACS-1 wild-type and mutated proteins [7].
6. Conclusions
In recent years, an extraordinary advance in the study of cellular protein trafficking has been made. PACS-1 is a connector multifunctional protein with a role that trespasses the trafficking between endosomes and the TGN and could be key in cellular homeostasis, interacting with proteins related to apoptosis, genomic stability or calcium flux in the endoplasmic reticulum. Since 2012, when PACS1-NDD was first characterized, deep phenotyping analysis has allowed us to establish a potential relationship with other neurodevelopmental disorders, such as the PACS2 or Wdr37 syndromes. However, it is necessary to find a more profound approach that allows us to connect the physiological mechanisms of cellular trafficking and clinical features in order to reclassify and properly understand these genetic disorders.
Exceptionally, most of the PACS1-NDD patients share the same mutation, and a gain-of-function or a dominant negative mechanism has been proposed. This makes it a great model for the therapy of rare diseases caused by this mechanism. In this sense, strategies focused on decreasing the toxic effect of the mutated protein by inhibiting its expression with ASOs or its client proteins seem to be promising.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23179649/s1. References [55,56,57,58,59,60,61,62] are cited in the supplementary materials.
Author Contributions
M.A. and Á.A., manuscript writing, collection and assembly of data; A.L.-P., C.L.-C., M.G.-S., A.A.-C., M.J.P., G.B.-L. and F.J.R., Literature review, collection and assembly of data; P.G.-P., structural studies; J.P. and B.P., conception and design, manuscript editing and final approval of the manuscript. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Schuurs-Hoeijmakers J.H., Oh E.C., Vissers L.E., Swinkels M.E., Gilissen C., Willemsen M.A., Holvoet M., Steehouwer M., Veltman J.A., de Vries B.B., et al. Recurrent De Novo Mutations in PACS1 Cause Defective Cranial-Neural-Crest Migration and Define a Recognizable Intellectual-Disability Syndrome. Am. J. Hum. Genet. 2012;91:1122–1127. doi: 10.1016/j.ajhg.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.García-Cazorla A., Oyarzábal A., Saudubray J.-M., Martinelli D., Dionisi-Vici C. Genetic disorders of cellular trafficking. Trends Genet. 2022;38:724–751. doi: 10.1016/j.tig.2022.02.012. [DOI] [PubMed] [Google Scholar]
- 3.Schuurs-Hoeijmakers J.H.M., Landsverk M.L., Foulds N., Kukolich M.K., Gavrilova R.H., Greville-Heygate S., Hanson-Kahn A., Bernstein J.A., Glass J., Chitayat D., et al. Clinical delineation of thePACS1-related syndrome-Report on 19 patients. Am. J. Med. Genet. Part A. 2016;170:670–675. doi: 10.1002/ajmg.a.37476. [DOI] [PubMed] [Google Scholar]
- 4.Tenorio-Castaño J., Morte B., Nevado J., Martinez-Glez V., Santos-Simarro F., García-Miñaúr S., Palomares-Bralo M., Pacio-Míguez M., Gómez B., Arias P., et al. Schuurs–Hoeijmakers Syndrome (PACS1 Neurodevelopmental Disorder): Seven Novel Patients and a Review. Genes. 2021;12:738. doi: 10.3390/genes12050738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyake N., Ozasa S., Mabe H., Kimura S., Shiina M., Imagawa E., Miyatake S., Nakashima M., Mizuguchi T., Takata A., et al. A novel missense mutation affecting the same amino acid as the recurrent PACS1 mutation in Schuurs-Hoeijmakers syndrome. Clin. Genet. 2017;93:929–930. doi: 10.1111/cge.13105. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y., Ding H., Yan T., Liu L., Yu L., Huang Y., Li F., Zeng Y., Huang W., Zhang Y., et al. A Novel Multi-Exon Deletion of PACS1 in a Three-Generation Pedigree: Supplements to PACS1 Neurodevelopmental Disorder Spectrum. Front. Genet. 2021;12:690216. doi: 10.3389/fgene.2021.690216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Youker R.T., Shinde U., Day R., Thomas G. At the crossroads of homoeostasis and disease: Roles of the PACS proteins in membrane traffic and apoptosis. Biochem. J. 2009;421:1–15. doi: 10.1042/BJ20081016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wan L., Molloy S.S., Thomas L., Liu G., Xiang Y., Rybak S.L., Thomas G. PACS-1 Defines a Novel Gene Family of Cytosolic Sorting Proteins Required for trans-Golgi Network Localization. Cell. 1998;94:205–216. doi: 10.1016/S0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- 9.Sieburth D., Ch’Ng Q., Dybbs M., Tavazoie M., Kennedy S., Wang D., Dupuy D., Rual J.-F., Hill D.E., Vidal M., et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- 10.Thomas G., Aslan J., Thomas L., Shinde P., Shinde U., Simmen T. Caught in the act—Protein adaptation and the expanding roles of the PACS proteins in tissue homeostasis and disease. J. Cell Sci. 2017;130:1865–1876. doi: 10.1242/jcs.199463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nair-Gill E., Bonora M., Zhong X., Liu A., Miranda A., Stewart N., Ludwig S., Russell J., Gallagher T., Pinton P., et al. Calcium flux control by PACS1-Wdr37 promotes lymphocyte quiescence and lymphoproliferative diseases. EMBO J. 2021;40:e104888. doi: 10.15252/embj.2020104888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mani C., Tripathi K., Luan S., Clark D.W., Andrews J.F., Vindigni A., Thomas G., Palle K. The multifunctional protein PACS-1 is required for HDAC2- and HDAC3-dependent chromatin maturation and genomic stability. Oncogene. 2020;39:2583–2596. doi: 10.1038/s41388-020-1167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lusk L., Smith S., Martin C., Taylor C., Chung W. PACS1 Neurodevelopmental Disorder. In: Adam M.P., Mirzaa G.M., Pagon R.A., Wallace S.E., Bean L.J., Gripp K.W., Amemiya A., editors. GeneReviews®. University of Washington; Seattle, WA, USA: 1993. [(accessed on 23 June 2022)]. Available online: http://www.ncbi.nlm.nih.gov/books/NBK559434/ [PubMed] [Google Scholar]
- 14.Stern D., Cho M., Chikarmane R., Willaert R., Retterer K., Kendall F., Deardorff M., Hopkins S., Bedoukian E., Slavotinek A., et al. Association of the missense variant p.Arg203Trp in PACS1 as a cause of intellectual disability and seizures. Clin. Genet. 2017;92:221–223. doi: 10.1111/cge.12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Monseny A., Bolasell M., Arjona C., Martorell L., Yubero D., Arsmtrong J., Maynou J., Fernandez G., Salgado M.D.C., Palau F., et al. Mutation of PACS1: The milder end of the spectrum. Clin. Dysmorphol. 2018;27:148–150. doi: 10.1097/MCD.0000000000000237. [DOI] [PubMed] [Google Scholar]
- 16.Pefkianaki M., Schneider A., Capasso J.E., Wasserman B.N., Bardakjian T., Levin A.V. Ocular manifestations of PACS1 mutation. J. Am. Assoc. Pediatr. Ophthalmol. Strabismus. 2018;22:323–325. doi: 10.1016/j.jaapos.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Dutta A.K. Schuurs-Hoeijmakers syndrome in a patient from India. Am. J. Med. Genet. Part A. 2019;179:522–524. doi: 10.1002/ajmg.a.61058. [DOI] [PubMed] [Google Scholar]
- 18.Hoshino Y., Enokizono T., Imagawa K., Tanaka R., Suzuki H., Fukushima H., Arai J., Sumazaki R., Uehara T., Takenouchi T., et al. Schuurs-Hoeijmakers syndrome in two patients from Japan. Am. J. Med. Genet. Part A. 2018;179:341–343. doi: 10.1002/ajmg.a.9. [DOI] [PubMed] [Google Scholar]
- 19.Seto M.T., Bertoli-Avella A.M., Cheung K.W., Chan K.Y., Yeung K.S., Fung J.L., Beetz C., Bauer P., Luk H.M., Lo I.F., et al. Prenatal and postnatal diagnosis of Schuurs-Hoeijmakers syndrome: Case series and review of the literature. Am. J. Med. Genet. Part A. 2020;185:384–389. doi: 10.1002/ajmg.a.61964. [DOI] [PubMed] [Google Scholar]
- 20.Hay E., Henderson R.H., Mansour S., Deshpande C., Jones R., Nutan S., Mankad K., Young R.M., Moosajee M., Genomics England Research Consortium et al. Expanding the phenotypic spectrum consequent upon de novo WDR37 missense variants. Clin. Genet. 2020;98:191–197. doi: 10.1111/cge.13795. [DOI] [PubMed] [Google Scholar]
- 21.Kanca O., Andrews J.C., Lee P.-T., Patel C., Braddock S.R., Slavotinek A.M., Cohen J.S., Gubbels C.S., Aldinger K., Williams J., et al. De Novo Variants in WDR37 Are Associated with Epilepsy, Colobomas, Dysmorphism, Developmental Delay, Intellectual Disability, and Cerebellar Hypoplasia. Am. J. Hum. Genet. 2019;105:413–424. doi: 10.1016/j.ajhg.2019.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reis L.M., Sorokina E.A., Thompson S., Muheisen S., Velinov M., Zamora C., Aylsworth A.S., Semina E.V. De Novo Missense Variants in WDR37 Cause a Severe Multisystemic Syndrome. Am. J. Hum. Genet. 2019;105:425–433. doi: 10.1016/j.ajhg.2019.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dentici M.L., Barresi S., Niceta M., Ciolfi A., Trivisano M., Bartuli A., Digilio M.C., Specchio N., Dallapiccola B., Tartaglia M. Expanding the clinical spectrum associated with PACS2 mutations. Clin. Genet. 2019;95:525–531. doi: 10.1111/cge.13516. [DOI] [PubMed] [Google Scholar]
- 24.Olson H.E., Jean-Marçais N., Yang E., Heron D., Tatton-Brown K., van der Zwaag P.A., Bijlsma E.K., Krock B.L., Backer E., Kamsteeg E.-J., et al. A Recurrent De Novo PACS2 Heterozygous Missense Variant Causes Neonatal-Onset Developmental Epileptic Encephalopathy, Facial Dysmorphism, and Cerebellar Dysgenesis. Am. J. Hum. Genet. 2018;102:995–1007. doi: 10.1016/j.ajhg.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terrone G., Marchese F., Vari M.S., Severino M., Madia F., Amadori E., Del Giudice E., Romano A., Gennaro E., Zara F., et al. A further contribution to the delineation of epileptic phenotype in PACS2-related syndrome. Seizure. 2020;79:53–55. doi: 10.1016/j.seizure.2020.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Lu C.-L., Kim J. Craniofacial Diseases Caused by Defects in Intracellular Trafficking. Genes. 2021;12:726. doi: 10.3390/genes12050726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.ClinVar. [(accessed on 22 August 2022)]; Available online: https://www.ncbi.nlm.nih.gov/clinvar/?term=PACS1%5Bgene%5D&redir=gene.
- 28.Lim E.T., Uddin M., De Rubeis S., Chan Y., Kamumbu A.S., Zhang X., D’Gama A.M., Kim S.N., Hill R.S., Goldberg A.P., et al. Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat. Neurosci. 2017;20:1217–1224. doi: 10.1038/nn.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DECIPHER. [(accessed on 22 August 2022)]. Available online: https://www.deciphergenomics.org/gene/PACS1/overview/clinical-info.
- 30.GTEx. [(accessed on 22 August 2022)]. Available online: https://gtexportal.org/home/gene/PACS1.
- 31.Cardoso-Moreira M., Halbert J., Valloton D., Velten B., Chen C., Shao Y., Liechti A., Ascenção K., Rummel C., Ovchinnikova S., et al. Gene expression across mammalian organ development. Nature. 2019;571:505–509. doi: 10.1038/s41586-019-1338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.EvoDevo. [(accessed on 22 August 2022)]. Available online: https://apps.kaessmannlab.org/evodevoapp/
- 33.Ensembl. [(accessed on 22 August 2022)]. Available online: https://www.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000175115;r=11:66070272-66244744.
- 34.Brasacchio D., Alsop A.E., Noori T., Lufti M., Iyer S., Simpson K.J., Bird P.I., Kluck R.M., Johnstone R.W., Trapani J.A. Epigenetic control of mitochondrial cell death through PACS1-mediated regulation of BAX/BAK oligomerization. Cell Death Differ. 2017;24:961–970. doi: 10.1038/cdd.2016.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veena M.S., Raychaudhuri S., Basak S.K., Venkatesan N., Kumar P., Biswas R., Chakrabarti R., Lu J., Su T., Gallagher-Jones M., et al. Dysregulation of hsa-miR-34a and hsa-miR-449a leads to overexpression of PACS-1 and loss of DNA damage response (DDR) in cervical cancer. J. Biol. Chem. 2020;295:17169–17186. doi: 10.1074/jbc.RA120.014048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He C., Su C., Zhang W., Wan Q. miR-485-5p alleviates Alzheimer’s disease progression by targeting PACS1. Transl. Neurosci. 2021;12:335–345. doi: 10.1515/tnsci-2020-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trothen S.M., Zang R.X., Lurie A., Dikeakos J.D. PACS-1 contains distinct motifs for nuclear-cytoplasmic transport and interacts with the RNA-binding protein PTBP1 in the nucleus and cytosol. FEBS Lett. 2021;596:232–248. doi: 10.1002/1873-3468.14243. [DOI] [PubMed] [Google Scholar]
- 38.Zheng W., Wuyun Q., Zhou X., Li Y., Freddolino P.L., Zhang Y. LOMETS3: Integrating deep learning and profile alignment for advanced protein template recognition and function annotation. Nucleic Acids Res. 2022;50:W454–W464. doi: 10.1093/nar/gkac248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt V., Sporbert A., Rohe M., Reimer T., Rehm A., Andersen O.M., Willnow T.E. SorLA/LR11 Regulates Processing of Amyloid Precursor Protein via Interaction with Adaptors GGA and PACS-1. J. Biol. Chem. 2007;282:32956–32964. doi: 10.1074/jbc.M705073200. [DOI] [PubMed] [Google Scholar]
- 41.Scott G.K., Fei H., Thomas L., Medigeshi G.R., Thomas G. A PACS-1, GGA3 and CK2 complex regulates CI-MPR trafficking. EMBO J. 2006;25:4423–4435. doi: 10.1038/sj.emboj.7601336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schermer B., Höpker K., Omran H., Ghenoiu C., Fliegauf M., Fekete A., Horvath J., Köttgen M., Hackl M., Zschiedrich S., et al. Phosphorylation by casein kinase 2 induces PACS-1 binding of nephrocystin and targeting to cilia. EMBO J. 2005;24:4415–4424. doi: 10.1038/sj.emboj.7600885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jenkins P.M., Zhang L., Thomas G., Martens J.R. PACS-1 Mediates Phosphorylation-Dependent Ciliary Trafficking of the Cyclic-Nucleotide-Gated Channel in Olfactory Sensory Neurons. J. Neurosci. 2009;29:10541–10551. doi: 10.1523/JNEUROSCI.1590-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seidel C., Schnekenburger M., Dicato M., Diederich M. Histone deacetylase 6 in health and disease. Epigenomics. 2015;7:103–118. doi: 10.2217/epi.14.69. [DOI] [PubMed] [Google Scholar]
- 45.Thomas G., Thomas L., Villar-Pazos S. Methods of Treating PACS1 and Pacs2 Syndromes. WO 2020/018647 A1. [(accessed on 19 August 2022)]. Available online: https://patents.google.com/patent/WO2020018647A1/en.
- 46.Brasacchio D., Noori T., House C., Brennan A.J., Simpson K., Susanto O., Bird P., Johnstone R.W., Trapani J.A. A functional genomics screen identifies PCAF and ADA3 as regulators of human granzyme B-mediated apoptosis and Bid cleavage. Cell Death Differ. 2014;21:748–760. doi: 10.1038/cdd.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brasacchio D., Busuttil R.A., Noori T., Johnstone R., Boussioutas A., Trapani J.A. Down-regulation of a pro-apoptotic pathway regulated by PCAF/ADA3 in early stage gastric cancer. Cell Death Dis. 2018;9:442. doi: 10.1038/s41419-018-0470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rylaarsdam L., Guemez-Gamboa A. A gain-of-function recurrent missense variant leads to a GABAergic/glutamatergic imbalance in a forebrain organoid model of PACS1 syndrome. bioRxiv. 2022 doi: 10.1101/2022.05.13.491892. [DOI] [Google Scholar]
- 49.Van Nuland A., Reddy T., Quassem F., Vassalli J.-D., Berg A.T. PACS1-Neurodevelopmental disorder: Clinical features and trial readiness. Orphanet J. Rare Dis. 2021;16:386. doi: 10.1186/s13023-021-02001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rylaarsdam L., Reddy T., Guemez-Gamboa A. In search of a cure: PACS1 Research Foundation as a model of rare disease therapy development. Trends Genet. 2021;38:109–112. doi: 10.1016/j.tig.2021.10.010. [DOI] [PubMed] [Google Scholar]
- 51.PACS1 Foundation. [(accessed on 18 July 2022)]. Available online: https://www.PACS1foundation.org/research.
- 52.Rinaldi C., Wood M.J.A. Antisense oligonucleotides: The next frontier for treatment of neurological disorders. Nat. Rev. Neurol. 2017;14:9–21. doi: 10.1038/nrneurol.2017.148. [DOI] [PubMed] [Google Scholar]
- 53.Mendonça M.C.P., Kont A., Aburto M.R., Cryan J.F., O’Driscoll C.M. Advances in the Design of (Nano)Formulations for Delivery of Antisense Oligonucleotides and Small Interfering RNA: Focus on the Central Nervous System. Mol. Pharm. 2021;18:1491–1506. doi: 10.1021/acs.molpharmaceut.0c01238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deshaies R. Prime time for PROTACs. Nat. Chem. Biol. 2015;11:634–635. doi: 10.1038/nchembio.1887. [DOI] [PubMed] [Google Scholar]
- 55.Sun M., Zhang H. Par3 and aPKC regulate BACE1 endosome-to-TGN trafficking through PACS1. Neurobiol. Aging. 2017;60:129–140. doi: 10.1016/j.neurobiolaging.2017.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hinners I., Wendler F., Fei H., Thomas L., Thomas G., Tooze S.A. AP-1 recruitment to VAMP4 is modulated by phosphorylation-dependent binding of PACS-1. EMBO Rep. 2003;4:1182–1189. doi: 10.1038/sj.embor.7400018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piguet V., Wan L., Borel C., Mangasarian A., Demaurex N., Thomas G., Trono D. HIV-1 Nef protein binds to the cellular protein PACS-1 to downregulate class I major histocompatibility complexes. Nat. Cell Biol. 2000;2:163–167. doi: 10.1038/35004038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu H., Hu P.-W., Budhiraja S., Misra A., Couturier J., Lloyd R.E., Lewis D.E., Kimata J.T., Rice A.P. PACS1 is an HIV-1 cofactor that functions in Rev-mediated nuclear export of viral RNA. Virology. 2019;540:88–96. doi: 10.1016/j.virol.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chiu Y.-F., Sugden B., Chang P.-J., Chen L.-W., Lin Y.-J., Lan Y.-C., Lai C.-H., Liou J.-Y., Liu S.-T., Hung C.-H. Characterization and Intracellular Trafficking of Epstein-Barr Virus BBLF1, a Protein Involved in Virion Maturation. J. Virol. 2012;86:9647–9655. doi: 10.1128/JVI.01126-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Köttgen M., Benzing T., Simmen T., Tauber R., Buchholz B., Feliciangeli S., Huber T.B., Schermer B., Kramer-Zucker A., Höpker K., et al. Trafficking of TRPP2 by PACS proteins represents a novel mechanism of ion channel regulation. EMBO J. 2005;24:705–716. doi: 10.1038/sj.emboj.7600566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen J., Wang J., Meyers K.R., Enns C.A. Transferrin-Directed Internalization and Cycling of Transferrin Receptor 2. Traffic. 2009;10:1488–1501. doi: 10.1111/j.1600-0854.2009.00961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiang Y., Molloy S.S., Thomas L., Thomas G. The PC6B Cytoplasmic Domain Contains Two Acidic Clusters That Direct Sorting to distinct trans-Golgi Network/Endosomal Compartments. Mol. Biol. Cell. 2000;11:1257–1273. doi: 10.1091/mbc.11.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.